
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol., 15 October 2021
Sec. Gastrointestinal Cancers: Gastric and Esophageal Cancers
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.734694
This article is part of the Research TopicThe Role of non-coding RNAs in Gastrointestinal CancersView all 18 articles
 Changyan Zou1†
Changyan Zou1† Jinrong Liao1†
Jinrong Liao1† Dan Hu2†
Dan Hu2† Ying Su1
Ying Su1 Huamei Lin1
Huamei Lin1 Keyu Lin1
Keyu Lin1 Xingguan Luo3
Xingguan Luo3 Xiongwei Zheng2
Xiongwei Zheng2 Lurong Zhang1*
Lurong Zhang1* Tao Huang4*
Tao Huang4* Xiandong Lin1,5*
Xiandong Lin1,5*SNHG8, a family member of small nucleolar RNA host genes (SNHG), has been reported to act as an oncogene in gastric carcinoma (GC). However, its biological function in Epstein–Barr virus (EBV)-associated gastric cancer (EBVaGC) remains unclear. This study investigated the role of SNHG8 in EBVaGC. Sixty-one cases of EBVaGC, 20 cases of non-EBV-infected gastric cancer (EBVnGC), and relative cell lines were studied for the expression of SNHG8 and BHRF1 (BCL2 homolog reading frame 1) encoded by EBV with Western blot and qRT-PCR assays. The relationship between the expression levels of SNHG8 and the clinical outcome in 61 EBVaGC cases was analyzed. Effects of overexpression or knockdown of BHRF1, SNHG8, or TRIM28 on cell proliferation, migration, invasion, and cell cycle and the related molecules were determined by several assays, including cell proliferation, colony assay, wound healing assay, transwell invasion assay, cell circle with flow cytometry, qRT-PCR, and Western blot for expression levels. The interactions among SNHG8, miR-512-5p, and TRIM28 were determined with Luciferase reporter assay, RNA immunoprecipitation (RIP), pull-down assays, and Western blot assay. The in vivo activity of SNHG8 was assessed with SNHG8 knockdown tumor xenografts in zebrafish. Results demonstrated that the following. (1) BHRF1 and SNHG8 were overexpressed in EBV-encoded RNA 1-positive EBVaGC tissues and cell lines. BHRF1 upregulated the expressions of SNHG8 and TRIM28 in AGS. (2) SNHG8 overexpression had a significant correlation with tumor size and vascular tumor thrombus. Patients with high SNHG8 expression had poorer overall survival (OS) compared to those with low SNHG8 expression. (3) SNHG8 overexpression promoted EBVaGC cell proliferation, migration, and invasion in vitro and in vivo, cell cycle arrested at the G2/M phase via the activation of BCL-2, CCND1, PCNA, PARP1, CDH1, CDH2 VIM, and Snail. (4) Results of dual-luciferase reporter assay, RNA immunoprecipitation, and pull-down assays indicated that SNHG8 sponged miR-512-5p, which targeted on TRIM28 and promoted cancer malignant behaviors of EBVaGC cells. Our data suggest that BHRF1 triggered the expression of SNHG8, which sponged miR-512-5p and upregulated TRIM28 and a set of effectors (such as BCL-2, CCND1, CDH1, CDH2 Snail, and VIM) to promote EBVaGC tumorigenesis and invasion. SNHG8 could be an independent prognostic factor for EBVaGC and sever as target for EBVaGC therapy.
Epstein–Barr virus (EBV), belonging to the herpes virus family (1, 2), is a causative virus for many malignancies including nasopharyngeal carcinoma (NPC) and Burkitt lymphoma (3–5). The existence of the EBV genome in gastric cancer (GC) was first detected using a polymerase chain reaction (PCR) in 1990 (5, 6). So far, EBV has been detected in approximately 10% of gastric cancers worldwide (6, 7).
EBVaGC is a distinct subset of GC determined by comprehensive molecular analyses, which has unique clinical pathological features, including PIK3CA mutations, DNA hypermethylation, amplification of JAK2 and PD-L1/PD-L2 (1, 7). However, the underlying molecular mechanism of the development and progression of EBVaGC is unclear.
LncRNAs, non-coding RNAs with their sizes longer than 200 nucleic acids, were initially considered as “junk” or “genomic dark matter” without function (8, 9). However, recently, lncRNAs have been found to participate widely in various physiological and pathological processes, including cancer progression (9–12). Dysregulations of lncRNAs are associated with a variety of cancer malignant behaviors, such as cell migration, invasion, metastasis, gene transcription, and tumorigenesis (13, 14). For example, SNHG8, located on 4q26 and encoding small nucleolar RNAs (snoRNAs), was detected in multiple malignant tumors, including non-small-cell lung cancer, hepatocellular carcinoma (10, 15), and pancreatic adenocarcinoma (11, 16). LncRNAs, as oncogenes or tumor suppressors, are directly involved in tumorigenesis, cell cycle arrest, apoptosis, epithelial to mesenchymal transition (EMT), cell migration, invasion metastasis, and chemoresistance via activating the JAK2/STAT3 pathway or Wnt/β-catenin signaling (17–20). Therefore, lncRNAs might be excellent candidates for individualized treatments and monitoring the prognosis of gastric cancer.
We have reported that SNHG8 was a key regulator of EBVaGC by an integrative analysis of lncRNA and mRNA expression (21). This study was to explore the molecular mechanisms of SNHG8 contributing to the progression of EBVaGC via sponging miR-512-5p and targeting TRIM28 and a set of effectors.
After GCs were surgically resected, tissues from 61 patients with EBER-1-positive EBVaGC and 20 patients with EBER-1-negative EBVnGC were identified by in situ hybridization (22) and used in this study. All GC tissue adjacent normal tissues were quickly frozen in liquid nitrogen and stored at −80°C until use. The study protocol was approved by the Ethics Committee of the Fujian University Cancer Hospital, Fujian Cancer Hospital (Fuzhou, China), and the written informed consents from all the participators were obtained. All protocols are consistent with the Helsinki declaration.
EBER1 in-situ hybridization (ISH) was carried out with the EBER1 probe ISH kit (ZsBio, Beijing, China) on FFPE (formalin-fixed and paraffin-embedded) tissue slides. The tumor cells with clear nuclear staining of EBER1 were considered EBV positive.
AGS-BX1 [EBV-infected GC cell line (23)] was obtained from Dr. HL Chen (The University of Hong Kong); MKN-28 (GC cell line) and GES-1 (normal epithelial cell line of gastric mucosa) were purchased from ATCC. Cells were cultured in DMEM/F-12 containing 10% (v/v) heat-inactivated fetal bovine serum (FBS) and 100 U/ml of streptomycin/penicillin mixture (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) in a humidified 37°C incubator with 5% of CO2.
For the overexpression or knockdown SNHG8 in cell lines, the virus vectors were constructed by Hanheng Biotechnology Co Ltd (Beijing, China). To create cell lines with SNHG8 overexpression, HBLV-SNHG8-OE (pHBLV-CMV-mcs-3flag-EF1-ZsGReen-T2A-PURO inserted with SNHG8 gene) was used. The original vector was used as vector alone control (HBLV-NC). For SNHG8 knockdown in cells, HBLV-SNHG8-shrna1, HBLV-SNHG8-shrna2, and HBLV-SNHG8-shrna3 were used and their parental vector pHBLV-U6-MCS-CMV-Zs/m cherry was used as vector alone control. AGS-BX1 cells cultured in six-well plates(5 × 105/well) were infected with 10 MOI of viral vectors in the presence of 6 μg/ml of polyamine. After 48 h, the cells were selected with 2 μg/ml of puromycin for 2 weeks to obtain stable infected cells. Newly established cells were named as AGS-BX1/NC, AGS-BX1/SNHG8-OE, or AGS-BX1/SNHG8-SH. The symbols -OE and –SH mean overexpression and knockdown, respectively.
The plasmids pCDNA3.1-flag-N-humanized-BHRF1 and pCDNA3.1-flag-NC were purchased from Hanheng Biotechnology Co. Ltd. (Beijing, China); AGS cells in six-well plates(5 × 105/well) were transfected with 2 μg plasmid. After 48 h of transfection, the cells with overexpression of BHRF1 or with empty vector were named as AGS/BHRF1-OE or AGS-NC.
Total RNAs were isolated using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA). To reversely transcribe total RNAs into complementary DNA (cDNA) of SNHG8, hsa-miR-512-5p, TRIM28, BCL-2, CCND1, PCNA, PARP1, CDH1, CDH2 VIM, and Snail in AGS-BX1 cells, the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific; Waltham, MA, USA) and miScript Reverse Transcription Kit were used. The generated cDNAs were used as templates for assessing gene expressions with miScript SYBR Green PCR Kit (Qiagen GmbH, Hilden, Germany) and LightCycler 480 SYBR Green I Master (Roche Applied Science; Indianapolis, IN, USA). The housekeeping gene glyceraldehyde phosphate dehydrogenase (GAPDH) served as the internal control of TRIM28, BCL-2, CCND1, PCNA, PARP1, CDH1, CDH2 VIM, and Snail, whereas U6 small nuclear RNA served as the internal reference for the expression of hsa-miR-512-5p. Relative gene expression was calculated with the 2−ΔΔCt method.
Relative mRNA expression levels of SNHG8 were quantified in all EBVaGC samples using the comparative 2−ΔΔCt method. GAPDH was used to normalize expression levels of SNHG8.
Twenty-four hours after infection, the cells were collected, resuspended in the medium, and inoculated into 96-well plates (5,000 cells in 100 μl/well). The cell proliferation was assessed at 24, 48, 72, and 96 h later. At every time point, 20 μl of the MTT solution (Promega; Madison, WI, USA) was added into each well, followed by incubation at 37°C for another 4 h. One hundred fifty microliters of dimethyl sulfoxide was added into each well and shaken for 10 min to fully dissolve the MTT. The absorbance of each well was detected at OD490 nm by Model 680 reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Cell cycle analysis was done using Muse™ Cell Cycle Kit™ Cell Analyzer (Millipore, USA) in accordance with the manufacturer’s instructions (24).
The transwell chambers (8 μm diameter; Corning Inc., Corning, NY, USA) were used for cell migration assessment, while the upper chambers precoated with Matrigel (BD Biosciences, USA) were used for the invasion assay. Cells were harvested, washed with PBS, and resuspended in DMEM/F-12 without FBS. One hundred microliters of 2 × 104 cells were added into the upper chambers of a 24-well plate with 500 μl of media. At 24 h after seeding, the cells in the upper chambers were gently removed, and the migrated or invaded cells in the bottom side were fixed with 100% methanol, stained with 0.5% crystal violet, washed with PBS, and observed under inverted microscope (Olympus Corporation, Tokyo, Japan).
For colony-formation assay, about 200 cells/well were seeded in six-well plates and cultured for 14 days, then the cells were fixed in methanol and stained with 0.2% crystal violet. The colonies with >50 cells were pictured with ImageScanner (GE, USA) and counted using ImageJ (NIH, MD, USA).
Cells infected with lentiviral constructors were detached with trypsin and then seeded with DMEM/F12-10% FBS in a six-well plate in triplicates. After 24 h, the middle cells of each well were scratched with a 10-μl sterile pipette tip to make an empty line via washing off the detached cell twice with PBS. After 48 h, the pictures of cells that migrated into the empty line were taken under microscope and analyzed by ImageJ.
The lncRNASNP2 database (http://bioinfo.life.hust.edu.cn/lncRNASNP#!/) was utilized for the prediction of the binding site in SNHG8 for hsa-miR-512-5p and analyzed with inmiRDB (http://mirdb.org/) and TargetScan (http://www.targetscan.org/vert_72/).
The wild-type (wt) SNHG8 and mutant (mut) SNHG8 that had the predicted hsa-miR-512-5p-binding site were chemically synthesized by Hanheng Biotechnology Co. Ltd. (Beijing, China) and inserted into pSI-check2 luciferase reporter plasmids (Promega Corporation, Madison, WI, USA) to make the pMIR-SNHG8-wt (SNHG8-wt) and pMIR-SNHG8-mut (SNHG8-mut) reporter plasmids. Similarly, the reporter plasmids, TRIM28-wt and TRIM28-mut, were also made. For the reporter assay, when cells in 24-well plates grew to 70% confluence, the reporter plasmids were co-transfected with hsa-miR-512-5p mimics or miR-NC into the cells using Lipofectamine 2000 reagent. After 48 h, the luciferase activity in the transfected cells was detected with Dual-Luciferase Reporter Assay System (Promega Corporation, Madison, WI, USA) on a Synergy H4 reader. The relative luciferase activity was normalized with Renilla luciferase activity.
For the RIP assay, the EZ-Magna RIP Kit (Sigma, St. Louis, MO, USA) was used. In brief, the lysate of 1 × 107 AGS/BX-1 cells in RIP lysis buffer was incubated with Anti-IgG or Anti-Ago2-coated magnetic beads for 6 h. RNAs of SNHG8 and miR-512-5p enriched on the beads was extracted and quantified by reverse-transcription quantitative polymerase chain reaction (qRT-PCR).
RNA pull-down assay was carried out to assess the interaction of miR-512-5p with SNHG8 using a Magnetic RNA Pull-Down Kit (Thermo Fisher Scientific). The miR-512-5p mimic (Bio-miR-512-5p) and negative control (Bio-miR-NC) were biotinylated by RiboBio Inc. (Guangzhou, China). AGS/BX-1 cells were transfected with 100-nm biotinylated probes for 48 h; then, the cell lysates were incubated with streptavidin-coated magnetic beads overnight. The RNA enriched on the beads was separated, and the content of SNHG8 was detected by qRT-PCR.
Cells were harvested and lysed with RIPA buffer (50 mM Tris–Cl, pH 8.0, 150 mM NaCl, 5 mM EDTA, 0.1% SDS, 1% NP-40) containing protease inhibitor cocktail (Abcam, Cat No. ab65621). The lysates were centrifuged at 12,000 rpm for 30 min at 4°C. The protein concentrations of supernatants were determined by BCA protein assay (Thermo Scientific, Rockford, IL, USA). The protein extract of each sample (25 μg) was electrophoresed on 10%–12% polyacrylamide gel with sodium dodecyl sulfate and then transferred onto nitrocellulose membrane (Millipore A) at 100 V for 1.5 h. After being blocked with 3% BSA in TBST (TBS-1% Tween 20) for 1 h, the membranes were incubated with 1:1,000 primary antibodies (purchased from CST, Danvers, MA, USA) of TRIM28 (Cat No. #4124), VIM (Cat No. 5741S), CCND1 (Cat No. 55506s), PCNA (Cat No. D3H8P), Bcl-2 (Cat No. 5071S), CDH1 (Cat No. 3195s), CDH2 (Cat No. 13116s), Snail (Cat No. 3879S), or PARP(Cat No. 46D11) overnight at 4°C, respectively, then washed and further incubated with secondary horseradish peroxidase-conjugated anti-rabbit IgG. Finally, protein bands were detected by developing the blots with Immobilon ECL Ultra Western HRP Substrate (Millipore, Cat No. WBULS0500) and pictured on Image Station 4000MM Pro (Carestream, Canada). ImageJ was used to quantify protein levels relative to load control β-actin.
Transgenic zebrafish TG (apo14-GFP) was provided by the Institute of Hydrobiology, Chinese Academy of Sciences. The culture condition of adult fish was 26.5°C, and the light dark ratio was 12 h:12 h. AGS-BX-1/SNHG8-SH cells and AGS-BX-1/NC cells were injected with a Pico-liter injector. Under a microscope, 1,500 cells were injected into the IVF Development Zone of apo14-EGFP zebrafish yolk sac on the second day after fertilization. After injection, the embryos were cultured in a 33°C incubator and transferred to a 35°C incubator 24 h later. The distribution and expression of red fluorescence in the abdominal cavity of zebra were observed under a laser scanning confocal microscope (LSM 710, Carl Zeiss AG, Oberkochen, Germany). The expression level of red fluorescence in the abdominal cavity was analyzed by ImageJ 1.48v. Imaging and quantification of the results were performed with an inverted SP5 STED confocal microscope (Leica, Germany). At least 40 zebrafishes in each group were analyzed, and three representative images were used. All the experiments were repeated three times.
All data were processed by SPSS 19.0 software (SPSS Company, Chicago, USA). Pearson correlation analysis was used. The two-tailed t test was used to test the difference between groups. Paired or nonparametric Kruskal–Wallis test was used to evaluate the relationship between SNHG8 level and other characteristics. The survival curve was calculated by the Kaplan–Meier method. A p value less than 0.05 (*p <0.05, **p <0.01, ***p <0.001, and ****p <0.0001) was statistically significant.
BHRF1 (EBV-encoded small ribonucleic acid 1) is an EBV oncogene confirmed recently. Figures 1A, B show that BHRF1 was expressed in EBVaGC cases, but not expressed in EBVnGC (EBV-negative GC) tissues. In addition, the BHRF1 expression was significantly higher in AGS-BX (EBV infected GC cell line) than that in non-EBV-infected GC cell lines (AGS and MKN28) (Figure 1D). Besides, SNHG8 expression was also much higher in EBVaGC and AGS-BX than in EBVnGC- and in non-EBV-infected GC cell lines (AGS, MKN28) (Figures 1C, D).
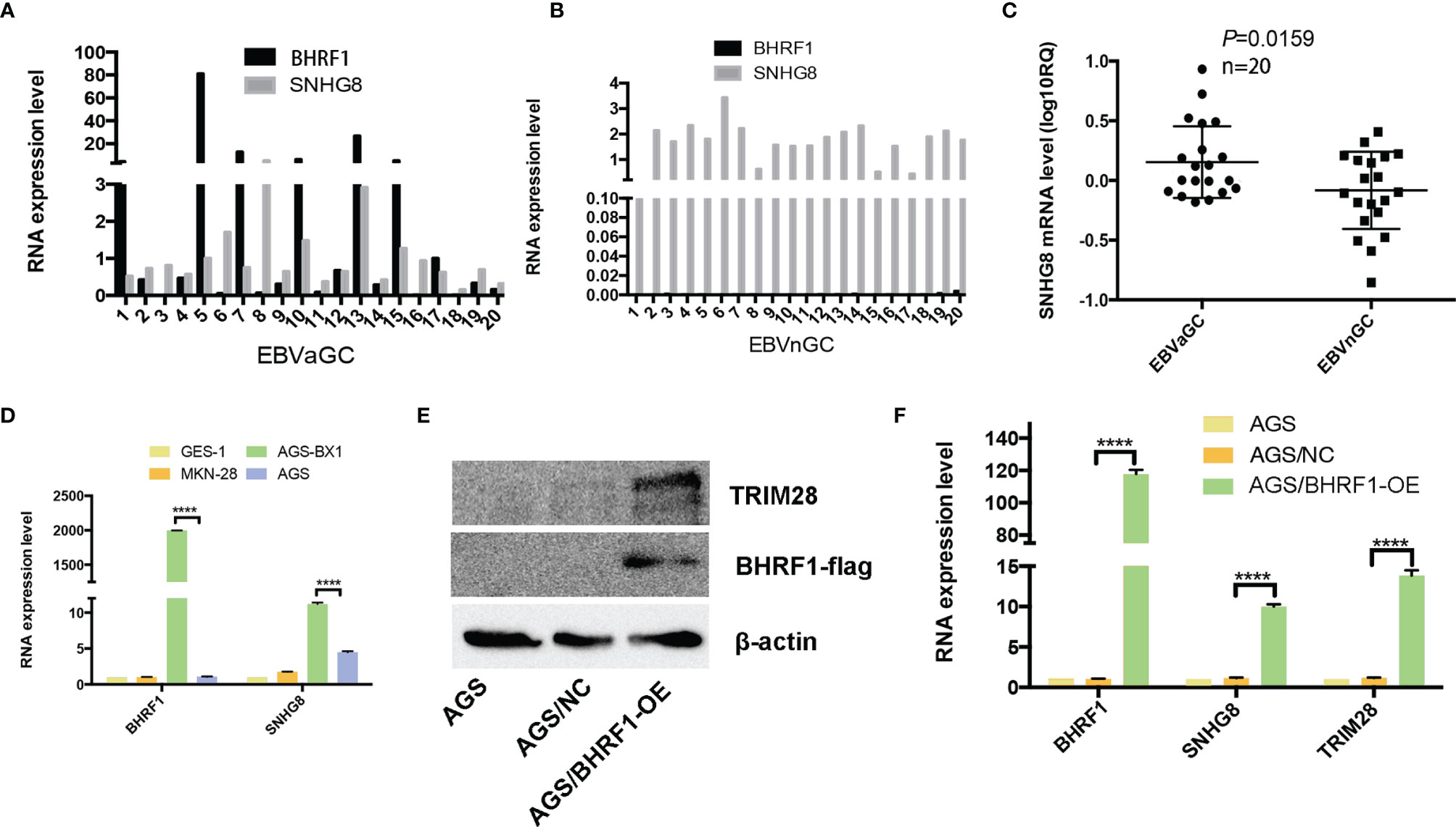
Figure 1 Expression of BFRF1 in GC and GC cell lines and its regulation with SNHG8 and TRIM28. (A, B) BHRF1 was expressed in EBVaGC tissues at different levels, while it was almost not expressed on EBVnGC. (C) The SNHG8 expression was much higher in EBVaGC tissues than that in EBVnGC tissues (p = 0.0159). RQ = 2−ΔΔCt, where Ct values were generated from qPCR. (D) The expression of BHRF1 and SNHG8 was much higher in EBV+ AGS-BX cells than in EBV- GC cell lines (AGS, MKN28). (E) Overexpression of EBV protein BFRF1 resulted in an increased TRIM28 demonstrated by Western blot analysis. (F) Overexpression of EBV protein BFRF1 enhanced the transcription of SNHG8 and TRIM28 demonstrated by qRT-PCR. ****p < 0.0001.
To determine the relationship between BHRF1 and SNHG8, we constructed a vector pcdna3.1-flag-BHRF1 to stably express human BHRF1 protein in EBV-negative AGS cells. Western blot assays (WB) show that the expression levels of BFRF1 and TRIM28 were much higher in AGS/BHRF1 cells than those in AGS/NC cells (Figure 1E). qRT-PCR assays revealed that the expression of BHRF1, SNHG8, and TRIM28 was much higher in the AGS/BHRF1 group than in the AGS/NC group (Figure 1F). These results indicated that the over-expression of EBV-related protein BFRF1 in EBV-negative AGS cells upregulated the expression of SNHG8 and TRIM28.
With in situ hybridization, EBVaGC was recognized by the expression of EBER1 in nuclei of cancer cells; EBVaGC is a group of lymphoepithelioma-like diffuse-type carcinoma with dense lymphocytic infiltration. As shown in Figure 2A, EBVaGC had EBER1-positive nuclei and surrounded with lymphoid stroma.
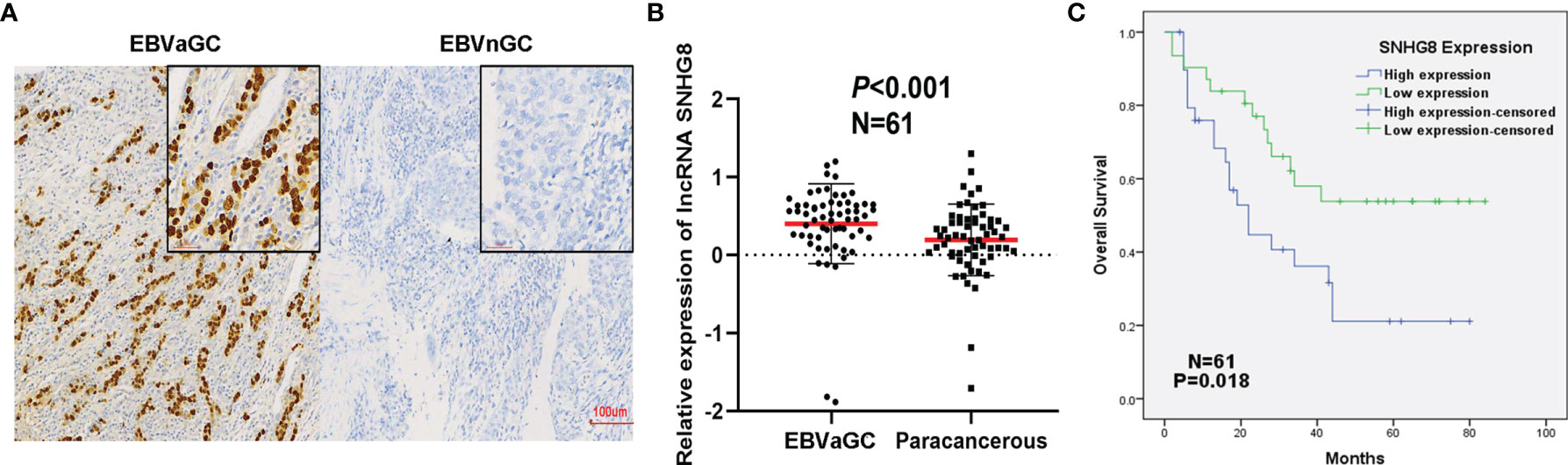
Figure 2 Poor prognosis of EBVaGC was associated with the upregulation of SNHG8. (A) EBER in gastric cancer detected by in situ hybridization. Left: EBER is positive in EBV-associated gastric cancer (EBVaGC). Right: EBER is negative in non-EBV-associated gastric cancer (EBVnGC). (B) The expression of SNHG8 was higher in 61 cases of EBVaGC than that in their paracancerous tissues (p < 0.001). (C) Kaplan–Meier curves showed that patients with a high level of SNHG8 had a poor OS (p = 0.018).
To determine the expression of SNHG8 in EBVaGC, qRT-PCR assays were performed in 61 cancer tissues and paired paracancerous tissues. SNHG8 was expressed at significantly higher levels in EBVaGC compared to the level in paracancerous tissues (Figure 2B). The average log10 relative expression levels in cancer tissues were ~0.41, a two-fold increase compared to that in paracancerous tissues (~0.20). Furthermore, EBVaGC tissue samples were divided into two groups (SNHG8 high- and low-expression groups) using the median mRNA level of SNHG8 as the cutoff value. As shown in Table 1, the overexpression of SNHG8 had a significant correlation with tumor size (p = 0.036) and vascular tumor thrombus (p = 0.024). Moreover, the overexpression of SNHG8 correlated with the poor prognosis in EBVaGC. Patients with the high levels of SNHG8 had significantly shorter overall survival (OS) than those with low SNHG8 expression (p = 0.018; Figure 2C). The results suggested that the expression of SNHG8 was positively correlated with tumor size and poor prognosis in EBVaGC.
To assess the effects of SNHG8 on the malignant behaviors of EBVaGC, EBV-positive gastric carcinoma cell lines AGS-BX1 were infected with HBLV-NC or HBLV-SNHG8-OE lentivirus. RT-qPCR analysis revealed that SNHG8 expression was efficiently increased in AGS-BX1/SNHG8-OE as compared with AGS-BX1/NC (Figure 3A). The effect of SNHG8 overexpression on the enhanced proliferation and colony formation of AGS-BX1 cells was demonstrated in MTT assay (Figure 3B) and colony assay (Figure 3C). In addition, SNHG8 overexpression markedly promoted cell cycle arrest at the G2/M phase of AGS-BX1 compared with control cell AGS-BX1/NC (Figure 3D). Furthermore, results of RT-qPCR and WB showed that the expression of proliferation-related genes CCND1, PCNA, and anti-apoptotic BCL-2, metastasis-related genes Snai1VIM and CDH2 were upregulated, while PARP1 and CDH1 were downregulated in AGS-BX1/SNHG8-OE cells (Figures 3E, F, J, K). Besides, the results of wound healing and transwell invasion assays showed that the upregulation of SNHG8 resulted in an increased abilities of wound healing (Figures 3G, H) and transwell invasiveness (Figure 3I). These findings suggested that SNHG8 might exert oncogenic effects on the aggressiveness of EBVaGC cell in vitro.
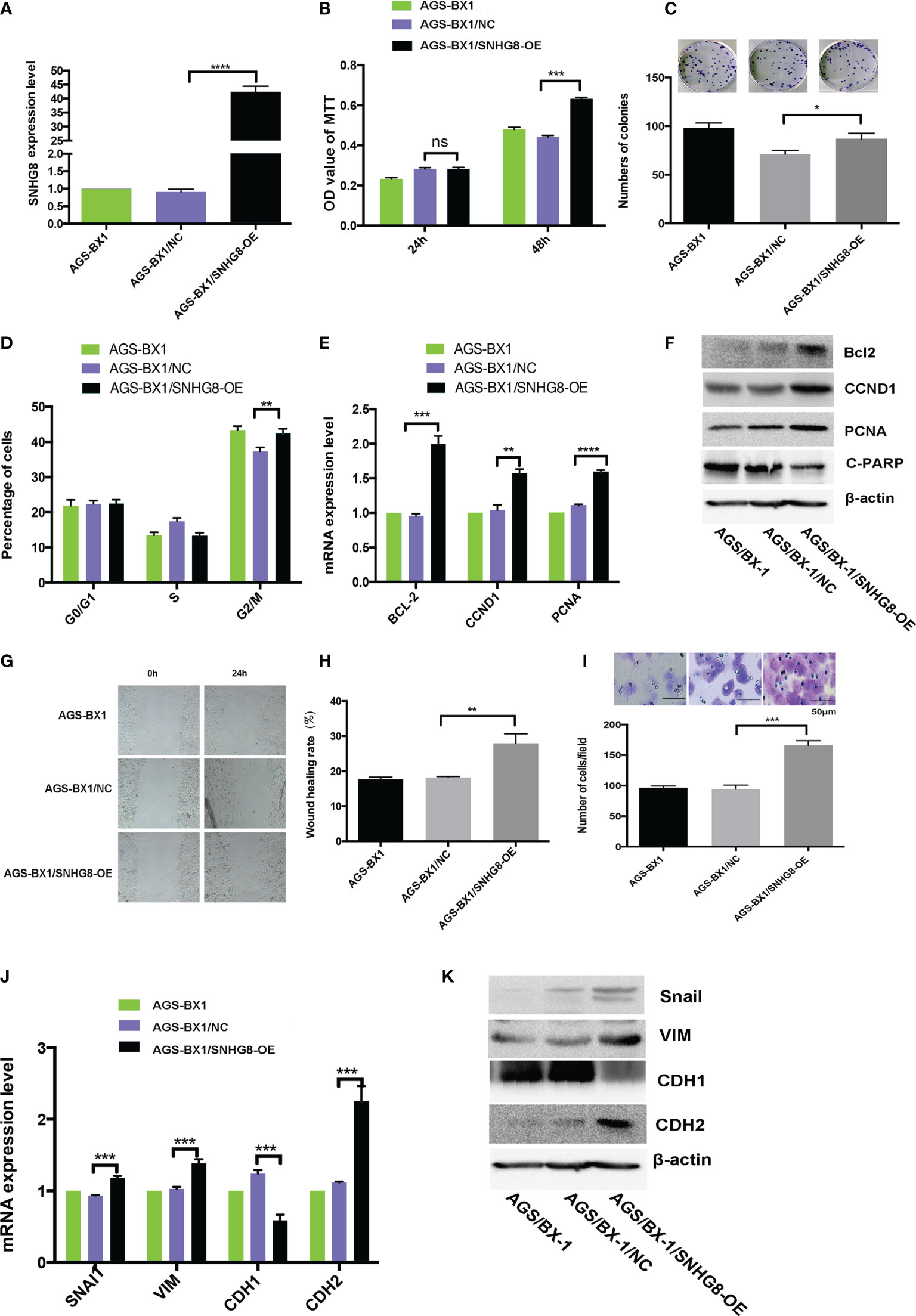
Figure 3 Overexpression of SNHG8 promoted the proliferation, migration, and invasion of EBVaGC. (A) AGS-BX1 cells with SNHG8 gene overexpression (AGS-BX1/SNHG8-OE)had a high level of the expression of SNHG8 demonstrated by qPCR as compared to vector alone control (AGS-BX1/NC) and wild-type cells (AGS-BX1). The overexpression of SNHG8 promoted the malignant behaviors of AGS-BX1/SNHG8-OE cells in the following aspects: (B) increasing cell proliferation as measured with MTS assay; (C) enhancing colony formation; (D) stopping cells at G2/M; (E, F) increasing the expression of proliferation-related genes of BCL-2, CCND1, C-PARP, and PCNA, confirmed by qRT-PCR and Western blot; (G, H) promoting the migration measured with the wound healing assay and the invasion measured with the transwell invasive assay; (I) and enhancing the expression of invasion/metastasis-related-genes, Snai1, VIM, CDH1, and CDH2 as demonstrated by qRT-PCR and Western blot (J, K). *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. ns mean not statistical significance.
To assess if downregulation of SNHG8 could reduce the malignant behaviors of EBVaGC, AGS-BX1 was infected with HBLV-NC or HBLV-SNHG8-SH lentivirus. RT-qPCR analysis showed that the SNHG8 expression was significantly reduced in AGS-BX1/SNHG8-SH as compared with AGS-BX1/NC (Figure 4A). SNHG8 silencing markedly inhibited the proliferation and colony formation of AGS-BX1 (Figures 4B, C). In addition, compared with AGS-BX1/NC, there was a significant amount of cells arrested at the GO/G1 phase in AGS-BX1/SNHG8-SH as measured by a Muse cell analyzer (Figure 4D). RT-qPCR and WB results showed that the expressions of proliferation-related genes BCL-2, CCND1, PCNA, and metastasis-related genes Snai1VIM and CDH1 were downregulated, while PARP1 and CDH1 were upregulated in AGS-BX1/SNHG8-SH cells (Figures 4E, F, J, K). Furthermore, the wound healing migratory ability and transwell invasive abilities were inhibited after the downregulation of SNHG8 (Figures 4G–I). These findings suggested that silencing of SNHG8 inhibited the proliferation, migration, and invasiveness of EBVaGC cells in vitro.
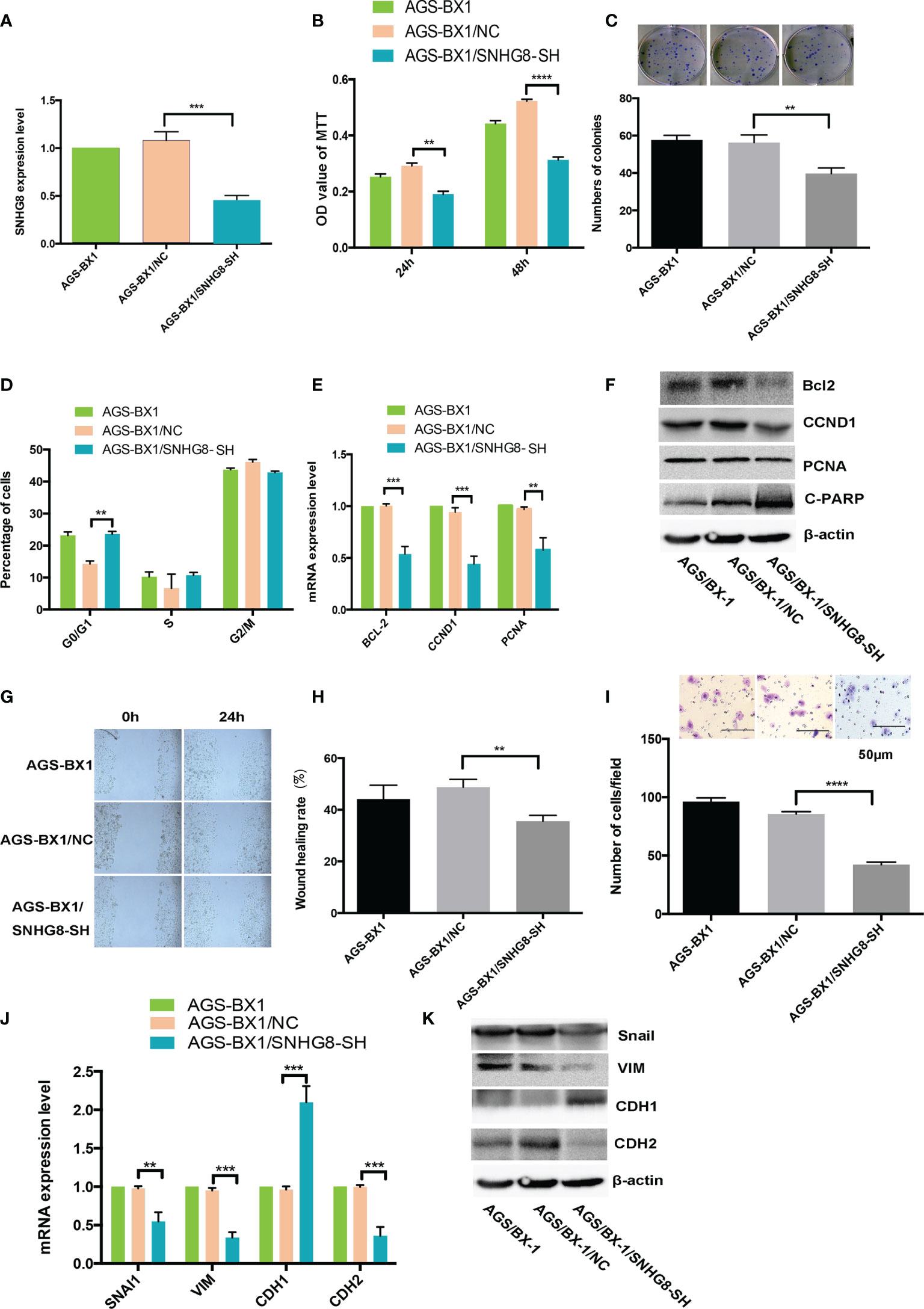
Figure 4 Silencing SNHG8 inhibited the proliferation, migration, and invasion of EBVaGC. (A) SNHG8 gene knockdown in AGS-BX1 cells (AGS-BX1/SNHG8-SH) had a low expression of SNHG8 quantified by qPCR as compared to vector alone control (AGS-BX1/NC) and wild-type cells (AGS-BX1). SNHG8 silencing reduced the malignant behaviors of AGS-BX1/SNHG8-SH cells in following aspects: the cell proliferation as measured with MTT assay (B); colony formation (C); cells in G0/1 (D); the expressions of proliferation-related-genes of BCL-2, CCND1, C=PARP, and PCNA demonstrated by qRT-PCR and Western blot (E, F); the migration measured with the wound healing assay (G, H); the invasion measured with the transwell invasive assay (I); and the expressions of invasion/metastasis-related-genes of Snail, VIM, CDH1, and CDH2 demonstrated by qRT-PCR and Western blotting (J, K). **p < 0.01; ***p < 0.001; ****p < 0.0001.
Potential miRNAs associated with SNHG8 in EBVaGC cells were examined since lncRNAs mainly functioned as miRNA spongers. Analyzed with the lncRNASNP2 database, hsa-miR-512-5p turned out to be a possible target of SNHG8. Further analyzed with the TargetScan and miRDB databases, TRIM28 turned out to be a possible target of hsa-miR-512-5p. Putative binding sites of hsa-miR-512-5p and wild-type regions of SNHG8 and TRIM28 are shown in Figures 5A, B. RT-qPCR analysis showed that the overexpression or silencing of SNHG8 significantly decreased or increased hsa-miR-512-5p expression (Figures 5C, D), respectively. In parallel, the overexpression of hsa-miR-512-5p decreased the functions of SNGH8 and TRIM28 expression, knockdown of hsa-miR-512-5p increased the functions of SNGH8 and TRIM28 expression (Figures 5E, F).
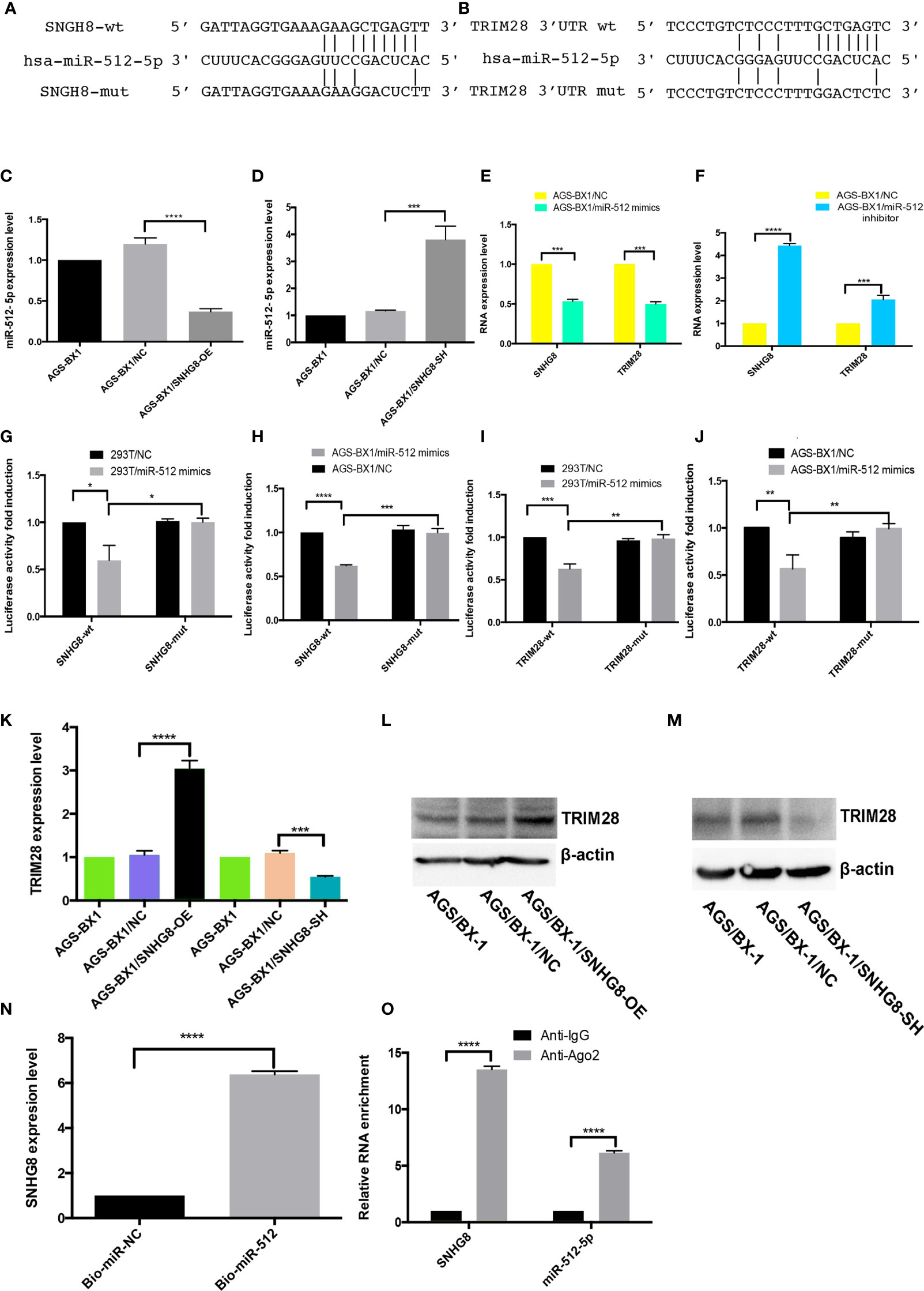
Figure 5 SNHG8 exerted its function through sponging hsa-miR-512-5p and upregulating of TRIM28. (A, B) Sequence alignment of hsa-miR-512-5p with the putative binding sites in the wild-type and mutant regions of SNHG8 and TRIM28. (C, D) Hsa-miR-512-5p expression was decreased in AGS-BX1/SNHG8-OE cells and increased in AGS-BX1/SNHG8-SH cells tested with qRT-PCR. (E, F) The expression of SNHG8 and TRIM28 was reduced in AGS-BX1/miR-512-5p mimic cells tested with qRT-PCR. (G–J) Dual-luciferase reporter assay showed that hsa-miR-512-5p mimics reduced the intensity of fluorescence in HEK293T or AGS-BX1 cells transfected with SNHG8-Wt or TRIM28-Wt, but not in the controls of SNHG8-Mut or TRIM28-Mut vector. (K–M) TRIM28 expression was increased in AGS-BX1/SNHG8-OE cells and decreased in AGS-BX1/SNHG8-SH cells tested with qRT-PCR and Western blot analysis. (N) SNHG8 expression was measured with qRT-PCR after RNA pull-down assay using Bio-miR-NC and Bio-miR-512-5p. (O) SNHG8 and miR-512-5p levels were determined by qRT-PCR after Ago 2 or IgG RIP assay. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
To explore whether SNHG8 and TRIM28 were functional targets of hsa-miR-512-5p, dual-luciferase reporter assay was performed. The luciferase activity was significantly reduced when hsa-miR-512-5p mimics were added into cultured SNHG8-Wt or TRIM28-Wt-co-transfected HEK293T and AGS-BX1 cells, while there was no effect on the same cell transfected with SNHG8-Mut or TRIM28-Mut vector (Figures 5G–J).
As expected, knocking down of SNHG8 with lentivirus significantly suppressed the expression of TRIM28 in AGS-BX1 cells (Figures 5K, M). In contrast, the overexpression of SNHG8 significantly increased the expression of TRIM28 in AGS-BX1 cells (Figures 5K, L). RNA pull-down analysis presented that SNHG8 could bind to hsa-miR-512-5p (Figure 5N). In addition, the result of the NA immunoprecipitation (RIP) assay showed that SNHG8 and hsa-miR-512-5p could be co-enriched in an Ago2-dependent manner (Figure 5O).
Taken together, these results demonstrated that SNHG8 and TRIM28’s 3 ′UTR had hsa-miR-512-5p-binding sites. Hsa-miR-512-5p was an inhibitor of SNHG8 and a blocker of TRIM28 in the EBVaGC progression.
To test our hypothesis that TRIM28 might contribute to SNHG8-mediated malignant behaviors, the effects of SNHG8 on the TRIM28 expression in AGS-BX1 cells were studied. A rescue assay by silencing TRIM28 in AGS-BX1/SNHG8-OE was performed. Results of qPCR and WB revealed that the expression level of TRIM28 in AGS-BX1/SNHG8-OE/TRIM28-SH was significantly lower than that in AGS-BX1/SNHG8-OE (Figures 6A, B). Silencing of TRIM28 reduced the proliferation and colony formation of AGS-BX1/SNHG8-OE cell as determined by MTT assay (Figure 6C) and colony assay (Figure 6D). The silencing TRIM28 markedly reduced the cell cycle arrest at the G2/M phase of AGS-BX1 compared with AGS-BX1/SNHG8-OE (Figure 6E). In addition, results of qRT-PCR and WB assay showed that the expression of a panel of proliferation or apoptosis-related genes, such as BCL-2, CCND1, and PCNA, was reduced after TRIM28 was silenced in AGS-BX1/SNHG8-OE cells (Figures 6F, G). Furthermore, the migratory and invasive abilities of AGS-BX1/SNHG8-OE cell lines greatly reduced after the silencing of TRIM28, as demonstrated in wound healing (Figures 6H, I) and transwell invasion assays (Figure 6J). The expressions of downstream molecules (Snai1, VIM, and CDH2) related with metastasis were downregulated as determined by qRT-PCR and WB, while CDH1 was upregulated silencing TRIM28 in AGS-BX1/SNHG8-OE cells (Figures 6K, L).
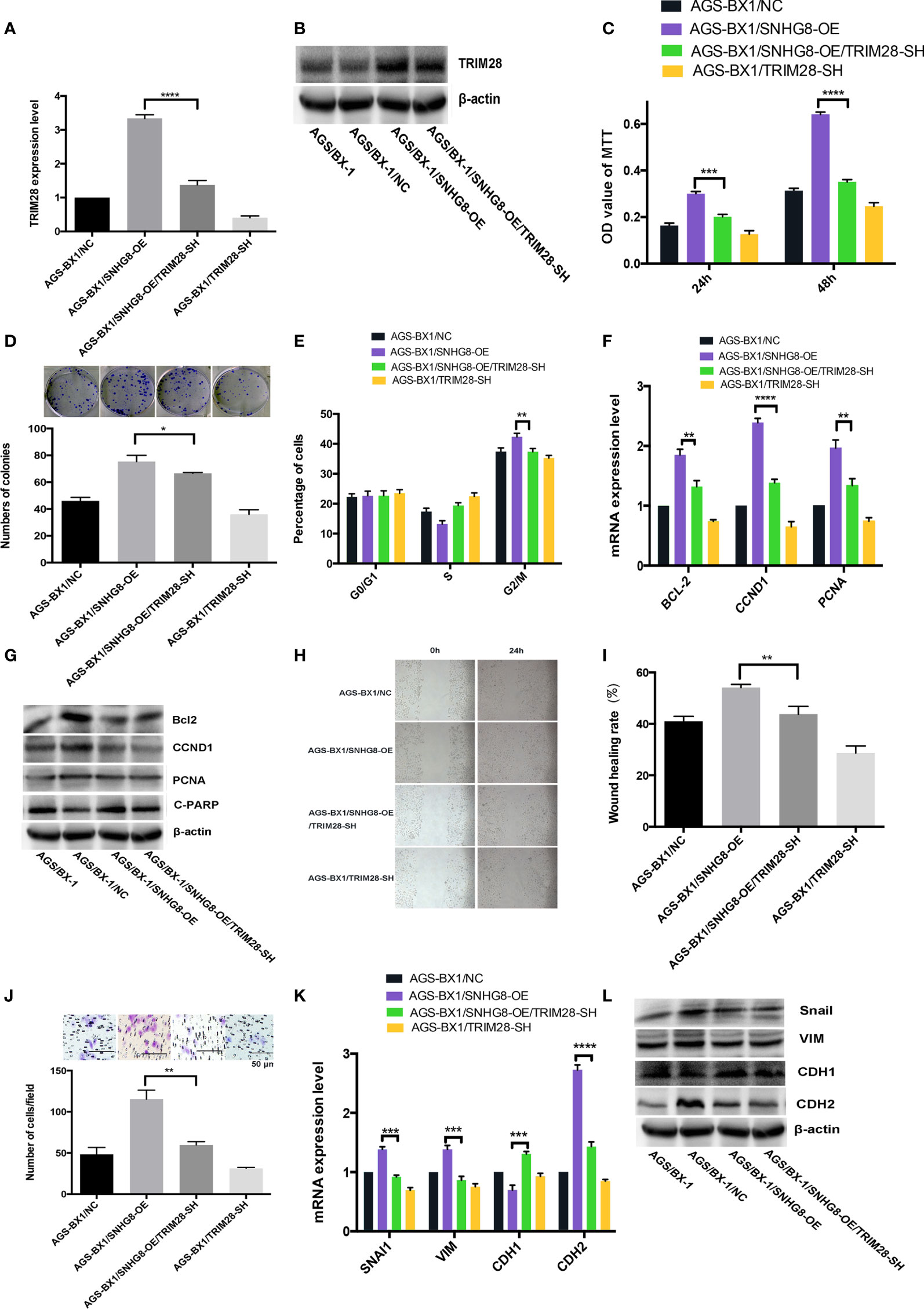
Figure 6 Enhancing of TRIM28 was critical for SNHG8-mediated malignant behaviors. (A, B) TRIM28 expression was increased in AGS-BX1/SNHG8-OE cells and reduced by silencing of TRIM28 tested by RT- PCR and Western blot. (C) SNHG8 enhanced cell proliferation could be reduced by silencing of TRIM28 tested with MTS assay. (D) SNHG8 enhanced colony formation could be reduced by silencing of TRIM28. (E) SNHG8-enhanced cell cycle stopping at G2/M could be reduced by silencing of TRIM28 as detected by flow cytometry. (F, G) SNHG8-enhanced expression of BCL-2, CCND1, C-PARP, and PCNA could be reduced by silencing of TRIM28 tested with qRT-PCR and Western Blot. (H, I) SNHG8-enhanced cell migration could be reduced by silencing of TRIM28 tested with wound healing assay. (J) SNHG8-enhanced cell invasion could be reduced by silencing of TRIM28 tested with transwell assay. (K, L) SNHG8-enhanced expressions of invasion/metastasis-related-genes of VIM and CDH2 could be reduced by silencing of TRIM28 tested with qRT-PCR and Western blot. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
These findings suggest that the enhancement of TRIM28 is critical for SNHG8-mediated malignant behaviors in EBVaGC cells in vitro.
To explore the role of SNHG8 in tumor growth of EBVaGC in vivo, the zebrafish EBVaGC tumor xenograft model was used. Injection of AGS-BX1 cells did not affect the normal development of zebrafish (Figure 7A). However, the growth of SNHG8-knocked-down mCherry-expressing AGS-BX1 cells significantly decreased compared to the vector-alone control group on 3 days after injection of the same number of cells into the circulation of zebrafish embryos, as demonstrated by the low intensity of red fluorescence (Figures 7B, C) and low proliferation rate (Figure 7D) in the AGS/BX-1/SNHG8-SH group.
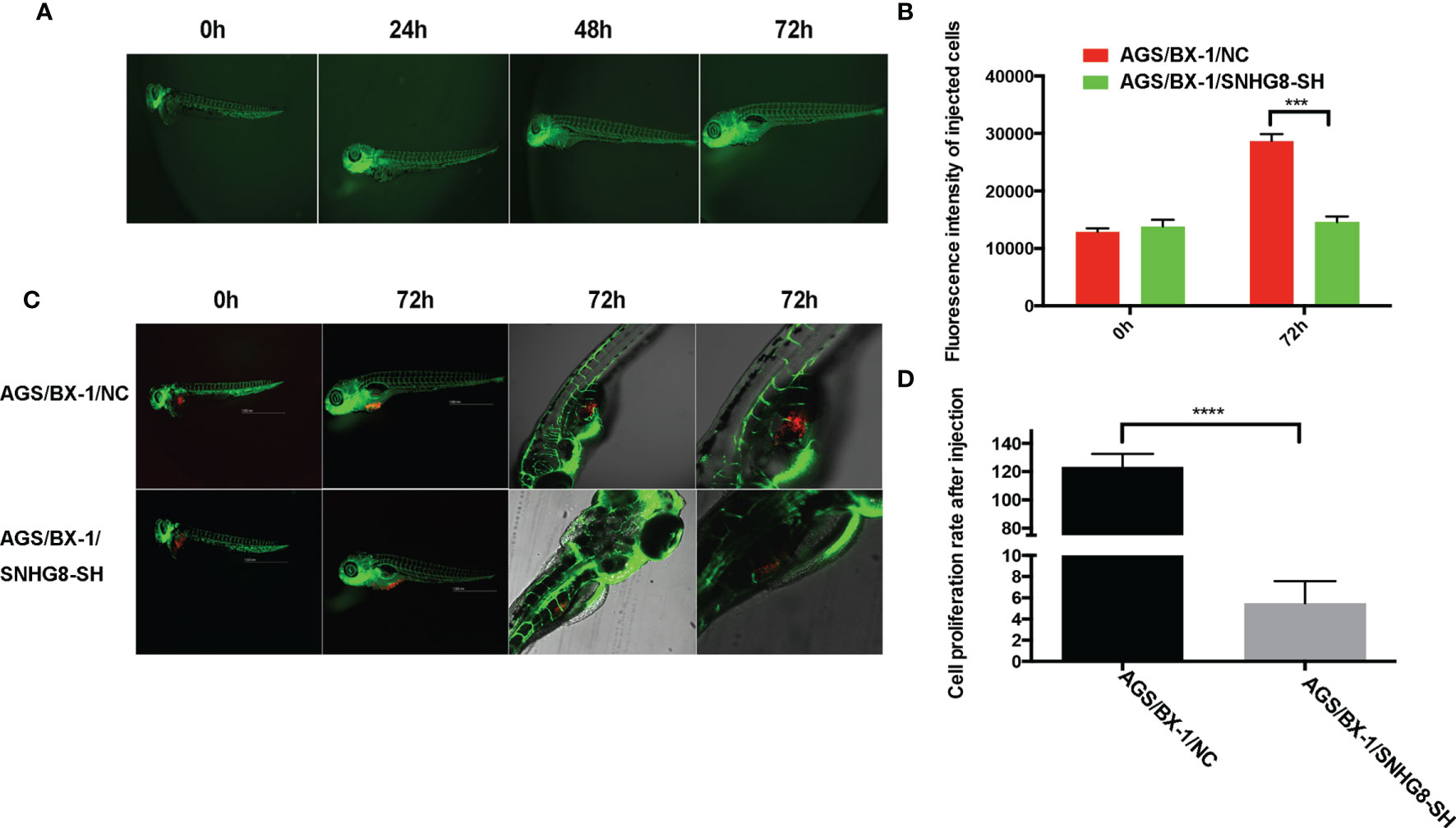
Figure 7 SNHG8 promoted the growth of EBVaGC tumor in zebrafish. (A) The growth of zebrafish was not affected by the injection of AGS-BX1 cells in 3 days. (B) Representative images of zebrafish from the control (AGS-BX1-NC) and AGS-BX1-SNHG8-SH groups with zoom-in of cancer cells. (C, D) Analysis of cancer cell numbers of each group in zebrafish assay. AGS-BX1 cells transfected with red florescence gene (AGS-BX1-NC, AGS-BX1-SNHG8-SH) could form tumor in zebrafish, which could be reduced by silencing of SNHG8 as determined by red fluorescence integral density and cancer cells in zebrafish abdominal cavity. ***p < 0.001; ****p < 0.0001.
Taken together, these data show that SNHG8 could sponge hsa-miR-512-5p and regulate the expression of TRIM28 in EBVaGC to enhance malignant behaviors.
In this study, several facts were revealed for the first time for EBVaGC, a distinct subtype of GC: (1) the existence of BHRF1 was correlated with the expression of SNHG8; (2) BHRF1+/SNHG8 high had a poor prognosis; (3) there was a coexistence of BHRF1+/SNHG8 high/miR-512-5p low/TRIM28high in EBVaGC; (4) the chain reaction was that BHRF1 triggered the high expression of SNHG8, which sponged hsa-miR-512-5p and upregulated TRIM28; (5) the malignant behaviors of BHRF1+/SNHG8 high/miR-512-5p low/TRIM28high EBVaGC were exerted by a set of proliferation-related genes, such as BCL-2, CCND1, PCNA, and PARP1 and metastasis-related genes, such as Snail, VIM, CDH1, and CDH2. This study further explored the molecular mechanism of our previous finding that BHRF1-SNHG8 was a key regulator of EBVaGC (21).
EBV is an oncogenic virus, associated with tumors from epithelia and hematopoietic cells (2, 25). Most studies have focused on genomes of EBV, such as EBER, EBNA-1, and BART (2, 26, 27), while this study further studied the role of BHRF1 in the development and progression of EBVaGC. The upregulation of BHRF1 resulted in over-expression of SNHG8, connecting the viral component to the human oncogene, possibly leading to this unique subtype of GC.
Cellular lncRNAs can be differentially induced by EBV infection. The dysregulated lncRNAs probably modulate tumorigenesis and other biological functions (28–30). Our result is consistent with others’ reports that SNHG8 is associated with the progression in multiple types of cancer in the liver, colon, lung, ovary, prostate, esophagus, and so on (15, 31–35). Our data support that the acting mechanism of SNHG8 could be multiple: (1) it activated BCL-2, preventing GC cells from apoptosis; (2) it activated CCND1, regulating the cell cycle; and (3) it activated CDH1, CDH2Snai1, and VIM, enhancing the epithelial–mesenchymal transition, thus promoting the migration, invasion, and metastasis of GC cells.
LncRNAs are involved in the regulation of gene transcription, post-transcription, and epigenetic modulation (12, 36). Since lncRNAs and mRNAs share miRNAs’ response elements, they compete for binding to these miRNAs, regulating the expression of each other. The interaction between these RNA molecules forms a network of complex posttranscriptional regulation (21, 37–40). Bioinformatics analyses revealed that miR-512-5p shared common binding sites with SNHG8 and TRIM28. Our dual-luciferase reporter assay indeed confirmed that SNHG8 functions as a ceRNA in the miR-512-5p/TRIM28 axis as SNHG8 bound to miR-512-5p, while TRIM28 bound to miR-152-5p. Furthermore, subsequent biotinylated miRNA pull-down assays confirmed the competitive binding between SNHG8 and TRIM28 to miR-512-5p. Moreover, the rescue experiment showed that SNHG8 significantly reduced the effects of miR-512-5p on TRIM28.
TRIM28, a member of a conserved family of transcription co-factors, has diverse functions for the regulation of cell proliferation, DNA repair, and differentiation (40–43). TRIM28 regulated E-cadherin and N-cadherin, resulting in EMT in lung cancer cells (44). TRIM28 prevented TRIM24 from SPOP-mediated degradation, promoting the progression of prostate cancer (42). TRIM28 also affected the mTOR signaling pathway, resulting in the growth of cervical cancer (45). Reduction of TRIM28 could reduce the malignant behaviors triggered by BHRF1-SNHG8.
Taken together, our in vitro and in vivo data showed that BHRF1 upregulated SNHG8, which sponged miR-512-5p, leading to a high level of TRIM28, carrying out the malignant behaviors. On the other hand, the silencing of SNHG8 would lead to miR-512-5p upregulation and reduction of TRIM28. This process of BHRF1+/SNHG8 high/miR-512-5p low/TRIM28high in EBVaGC directly enhances the activation of a set of tumor-promoting factors, such as proliferation-related genes (BCL-2, CCND1, PCNA, PARP1) and metastasis-related genes (Snail, VIM, CDH1, and CDH2).
Therefore, silencing of SNHG8 would greatly reduce the malignant behaviors of EBVaGC, which might pave a new road to overcome EBVaGC.
Our findings demonstrated that BHRF1 triggered the expression of SNHG8, which sponged miR-512-5p and upregulated TRIM28 and a set of effectors (such as BCL-2, CCND1, PCNA, CDH1, CDH2, Snail, and VIM) to promote the EBVaGC tumorigenesis and invasion. SNHG8 could be an independent prognostic factor for EBVaGC and sever as target for the therapy.
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
The studies involving human participants were reviewed and approved by the Ethics Committee of the Fujian University Cancer Hospital, Fujian Cancer Hospital (Fuzhou, China). The patients/participants provided their written informed consent to participate in this study.
XGL and TH designed the research. CZ, JL, DH, HL, and KL performed the experiments. YS, XGL, LZ, and TH analyzed the data. DH, XZ, LZ, and XDL conducted the histological/pathological analysis. XDL, JL, and CZ wrote the paper. LZ and TH edited the paper. All authors contributed to the article and approved the submitted version.
This study was supported by the Natural Science Foundation of Fujian Province (No. 2019J01196, 2020J011109), Fujian Provincial Clinlical Research Center for Cancer Radiotherapy and Immunotherapy (NO. 2020Y2012), the Joint Funds for the Innovation of Science and Technology, Fujian province (No. 2018Y9113), Medical Innovation Program of Fujian Province (No. 2019-CX-5), Strategic Priority Research Program of Chinese Academy of Sciences (XDB38050200, XDA26040304), National Key R&D Program of China (2018YFC0910403, 2017YFC1201200), and Shanghai Municipal Science and Technology Major Project (2017SHZDZX01).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors thank Dr. Shimin Zhang for his helpful comments.
EBV, Epstein–Barr virus; GC, gastric carcinoma; EBVaGC, EBV-positive GC; EBVnGC, non-EBV-infected gastric cancer; RIP, RNA immunoprecipitation; OS, overall survival; ISH, in situ hybridization; EMT, epithelial–mesenchymal transition; TCGA, The Cancer Genome Atlas; SNHG8, small nucleolar RNA host gene 8; BHRF1, EBV-encoded small ribonucleic acid 1; TRIM28, tripartite motif-containing 28; RIP, RNA immunoprecipitation; EBER-1, Epstein–Barr encoding region 1; FFPE, formalin-fixed and paraffin-embedded; MTS, (4,5-dimethylthiazol)-3-(4-sulfophenyl)tetrazolium, inner salt; HBLV-SNHG8, pHBLV-CMV-mcs-3flag-EF1-ZsGReen-T2A-PURO inserted with SNHG8 gene; AGS-BX1/NC, empty vector of AGS-BX1 cell infection; AGS-BX1/SNHG8-OE, SNHG8 overexpression AGS-BX1 cell; AGS-BX1/SNHG8-SH, SNHG8 knockdown AGS-BX1 cell; AGS/BHRF1-OE, BHRF1 overexpression AGS cell; SNHG8-wt, SNHG8/miR-512-5p-binding site wild-type cells; SNHG8-mut, SNHG8/miR-512-5p-binding site mutant cells; TRIM28-wt, TRIM28/miR-512-5p-binding site wild-type cells; TRIM28-mut, TRIM28/miR-512-5p-binding site mutant cells; AGS-BX1/SNHG8-OE/TRIM28-SH, SNHG8 overexpression/TRIM28 knockdown AGS-BX1 cell.
1. Farrell PJ. Epstein-Barr Virus and Cancer. Annu Rev Pathol (2019) 14:29–53. doi: 10.1146/annurev-pathmechdis-012418-013023
2. Ayee R, Ofori MEO, Wright E, Quaye O. Epstein Barr Virus Associated Lymphomas and Epithelia Cancers in Humans. J Cancer (2020) 11(7):1737–50. doi: 10.7150/jca.37282
3. Hui KF, Chiang AK. Suberoylanilide Hydroxamic Acid Induces Viral Lytic Cycle in Epstein-Barr Virus-Positive Epithelial Malignancies and Mediates Enhanced Cell Death. Int J Cancer (2010) 126(10):2479–89. doi: 10.1002/ijc.24945
4. Shannon-Lowe C, Rickinson A. The Global Landscape of EBV-Associated Tumors. Front Oncol (2019) 9:713. doi: 10.3389/fonc.2019.00713
5. Sun K, Jia K, Lv H, Wang SQ, Wu Y, Lei H, et al. EBV-Positive Gastric Cancer: Current Knowledge and Future Perspectives. Front Oncol (2020) 10:583463. doi: 10.3389/fonc.2020.583463
6. Burke AP, Yen TS, Shekitka KM, Sobin LH. Lymphoepithelial Carcinoma of the Stomach With Epstein-Barr Virus Demonstrated by Polymerase Chain Reaction. Mod Pathol (1990) 3(3):377–80.
7. Nogueira C, Mota M, Gradiz R, Cipriano MA, Caramelo F, Cruz H, et al. Prevalence and Characteristics of Epstein-Barr Virus-Associated Gastric Carcinomas in Portugal. Infect Agent Cancer (2017) 12:41. doi: 10.1186/s13027-017-0151-8
8. Wapinski O, Chang HY. Long Noncoding RNAs and Human Disease. Trends Cell Biol (2011) 21(6):354–61. doi: 10.1016/j.tcb.2011.04.001
9. Peng WX, Koirala P, Mo YY. LncRNA-Mediated Regulation of Cell Signaling in Cancer. Oncogene (2017) 36(41):5661–7. doi: 10.1038/onc.2017.184
10. Li J, Meng H, Bai Y, Kai W. Regulation of lncRNA and Its Role in Cancer Metastasis. Oncol Res (2016) 23(5):205–17. doi: 10.3727/096504016X14549667334007
11. Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell (2016) 29(4):452–63. doi: 10.1016/j.ccell.2016.03.010
12. Bhan A, Soleimani M, Mandal S. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res (2017) 77(15):3965–81. doi: 10.1016/j.eururo.2014.01.032
13. Sültmann H, Diederichs S. Long Noncoding RNA: "LNCs" to Cancer. Eur Urol (2014) 65(6):1152–3. doi: 10.1016/j.eururo.2014.01.032.
14. Liang H, Yu T, Han Y, Jiang H, Wang C, You T, et al. LncRNA PTAR Promotes EMT and Invasion-Metastasis in Serous Ovarian Cancer by Competitively Binding miR-101-3p to Regulate ZEB1 Expression. Mol Cancer (2018) 17(1):119. doi: 10.1186/s12943-018-0870-5
15. Chen C, Zhang Z, Li J, Sun Y. SNHG8 is Identified as a Key Regulator in non-Small-Cell Lung Cancer Progression Sponging to miR-542-3p by Targeting CCND1/CDK6. Onco Targets Ther (2018) 11:6081–90. doi: 10.2147/OTT.S170482
16. Song Y, Zou L, Li J, Shen Z-P, Cai Y-L, Wu X-D. LncRNA SNHG8 Promotes the Development and Chemo-Resistance of Pancreatic Adenocarcinoma. Eur Rev Med Pharmacol Sci (2018) 22(23):8161–8. doi: 10.26355/eurrev_201812_16508
17. Jin S, Yang X, Li J, Yang W, Ma H, Zhang Z, et al. P53-Targeted LincRNA-p21 Acts as a Tumor Suppressor by Inhibiting JAK2/STAT3 Signaling Pathways in Head and Neck Squamous Cell Carcinoma. Mol Cancer (2016) 18(1):38–52. doi: 10.1186/s12943-019-0993-3
18. Tang Y, Song G, Liu H, Yang S, Yu X, Shi L, et al. Silencing of Long Non-Coding RNA HOTAIR Alleviates Epithelial-Mesenchymal Transition in Pancreatic Cancer via the Wnt/β-Catenin Signaling Pathway. Cancer Manag Res (2021) 13(2):3247–57. doi: 10.2147/CMAR.S265578
19. Zhang L, Kang W, Lu X, Ma S, Dong L, Zou B. LncRNA CASC11 Promoted Gastric Cancer Cell Proliferation, Migration and Invasion In Vitro by Regulating Cell Cycle Pathway. Cell Cycle (2018) 17(15):1886–900. doi: 10.1080/15384101.2018.1502574
20. Deng W, Zhang Y, Cai J, Zhang J, Liu X, Yin J, et al. LncRNA-ANRIL Promotes Gastric Cancer Progression by Enhancing NF-kB Signaling. Exp Biol Med (Maywood) (2019) 244(12):953–9. doi: 10.1177/1535370219860207
21. Huang T, Ji Y, Hu D, Chen B, Zhang H, Li C, et al. SNHG8 is Identified as a Key Regulator of Epstein-Barr Virus(EBV)-Associated Gastric Cancer by an Integrative Analysis of lncRNA and mRNA Expression. Oncotarget (2016) 7(49):80990–1002. doi: 10.18632/oncotarget.13167
22. Shibata D, Weiss LM. Epstein-Barr Virus-Associated Gastric Adenocarcinoma. Am J Pathol (1992) 140(4):769–74.
23. Huang J, Liao G, Chen H, Wu F, Hutt-Fletcher L, Hayward G, et al. Contribution of C/EBP Proteins to Epstein-Barr Virus Lytic Gene Expression and Replication in Epithelial Cells. J Virol (2006) 80(3):1098–109. doi: 10.1128/JVI.80.3.1098-1109.2006
24. Bavelloni A, Focaccia E, Piazzi M, Errani C, Blalock W, Faenza I. Cell Cycle Arrest and Apoptosis Induced by Kinamycin F in Human Osteosarcoma Cells. Anticancer Res (2017) 37(8):4103–9. doi: 10.21873/anticanres.11797
25. Grywalska E, Rolinski J. Epstein-Barr Virus-Associated Lymphomas. Semin Oncol (2015) 42(2):291–303. doi: 10.1053/j.seminoncol.2014.12.030
26. Yau TO, Tang CM, Yu J. Epigenetic Dysregulation in Epstein-Barr Virus-Associated Gastric Carcinoma: Disease and Treatments. World J Gastroenterol (2014) 20(21):6448–56. doi: 10.3748/wjg.v20.i21.6448
27. Wang J, Zheng X, Qin Z, Wei L, Lu Y, Peng Q, et al. Epstein-Barr Virus miR-BART3-3p Promotes Tumorigenesis by Regulating the Senescence Pathway in Gastric Cancer. J Biol Chem (2019) 294(13):4854–66. doi: 10.1074/jbc.RA118.006853
28. Zhang J, Li X, Hu J, Cao P, Yan Q, Zhang S, et al. Long Noncoding RNAs Involvement in Epstein-Barr Virus Infection and Tumorigenesis. Virol J (2020) 17(1):51. doi: 10.1186/s12985-020-01308-y
29. Liu Y, Hu Z, Zhang Y, Wang C. Long Non-Coding RNAs in Epstein-Barr Virus-Related Cancer. Cancer Cell Int (2021) 21(1):278. doi: 10.1186/s12935-021-01986-w
30. Notarte KI, Senanayake S, Macaranas I, Albano PM, Mundo L, Fennell E, et al. MicroRNA and Other Non-Coding RNAs in Epstein-Barr Virus-Associated Cancers. Cancers (2021) 13(15):3909–35. doi: 10.3390/cancers13153909
31. Dong J, Teng F, Guo W, Yang J, Ding G, Fu Z. lncRNA SNHG8 Promotes the Tumorigenesis and Metastasis by Sponging miR-149-5p and Predicts Tumor Recurrence in Hepatocellular Carcinoma. Cell Physiol Biochem (2018) 51(5):2262–74. doi: 10.1159/000495871
32. Yang H, Jiang Z, Wang S, Zhao Y, Song X, Yufeng X, et al. Long Non-Coding Small Nucleolar RNA Host Genes in Digestive Cancers. Cancer Med (2019) 8(18):7693–704. doi: 10.1002/cam4.2622
33. Hen YY, Ye H, Wang Z, Xia Z, Wang B, Yi W, et al. Knockdown of SNHG8 Repressed the Growth, Migration, and Invasion of Colorectal Cancer Cells by Directly Sponging With miR-663. BioMed Pharmacother (2019) 116:109000. doi: 10.1016/j.biopha.2019.109000
34. Shi Z, Zhang H, Jie S, Yang X, Huang Q, Mao Y, et al. Long Non-Coding RNA SNHG8 Promotes Prostate Cancer Progression Through Repressing miR-384 and Up-Regulating HOXB7. J Gene Med (2021) 23(3):e3309. doi: 10.1002/jgm.3309
35. Xuan L, Sun Z, Wang J, Gao S. lncRNA SNHG8 Promotes Ovarian Cancer Progression Through Serving as Sponge for miR-1270 to Regulate S100A11 Expression. J Gene Med (2021) e3315. doi: 10.1002/jgm.3315
36. Chi Y, Wang D, Wang J, Yu W, Yang J. Long Non-Coding RNA in the Pathogenesis of Cancers. Cells (2019) 8(9):1015–59. doi: 10.3390/cells8091015
37. Wang H, Huo X, Yang XR, He J, Cheng L, Wang N, et al. STAT3-Mediated Upregulation of lncRNA HOXD-AS1 as a ceRNA Facilitates Liver Cancer Metastasis by Regulating SOX4. Mol Cancer (2017) 16(1):136. doi: 10.1186/s12943-017-0680-1
38. Le K, Guo H, Zhang Q, Huang X, Xu M, Huang Z, et al. Gene and lncRNA Co-Expression Network Analysis Reveals Novel ceRNA Network for Triple-Negative Breast Cancer. Sci Rep (2019) 9(1):15122. doi: 10.1038/s41598-019-51626-7
39. Wang L, Cho KB, Li Y, Tao G, Xie Z, Guo B. Long Noncoding RNA (lncRNA)-Mediated Competing Endogenous RNA Networks Provide Novel Potential Biomarkers and Therapeutic Targets for Colorectal Cancer. Int J Mol Sci (2019) 20(22):5758–84. doi: 10.3390/ijms20225758
40. Zhou RS, Zhang EX, Sun QF, Ye Z, Liu J, Zhou D, et al. Integrated Analysis of lncRNA-miRNA-mRNA ceRNA Network in Squamous Cell Carcinoma of Tongue. BMC Cancer (2019) 19(1):779. doi: 10.1186/s12885-019-5983-8
41. Czerwińska P, Mazurek S, Wiznerowicz M. The Complexity of TRIM28 Contribution to Cancer. J BioMed Sci (2017) 24(1):63. doi: 10.1186/s12929-017-0374-4
42. Fong KW, Zhao JC, Song B, Zheng B, Yu J. TRIM28 Protects TRIM24 From SPOP-Mediated Degradation and Promotes Prostate Cancer Progression. Nat Commun (2018) 9(1):5007. doi: 10.1038/s41467-018-07475-5
43. Jin JO, Lee GD, Nam SH, Lee TH, Kang DH, Yun JK, et al. Sequential Ubiquitination of P53 by TRIM28, RLIM, and MDM2 in Lung Tumorigenesis. Cell Death Differ (2021) 28(6):1790–803. doi: 10.1038/s41418-020-00701-y
44. Chen L, Muñoz-Antonia T, Cress WD. Trim28 Contributes to EMT via Regulation of E-Cadherin and N-Cadherin in Lung Cancer Cell Lines. PloS One (2014) 9(7):e101040. doi: 10.1371/journal.pone.0101040
Keywords: EBVaGC, SNHG8, miR-512-5p, TRIM28, cell proliferation, migration, invasion
Citation: Zou C, Liao J, Hu D, Su Y, Lin H, Lin K, Luo X, Zheng X, Zhang L, Huang T and Lin X (2021) SNHG8 Promotes the Progression of Epstein–Barr Virus-Associated Gastric Cancer via Sponging miR-512-5p and Targeting TRIM28. Front. Oncol. 11:734694. doi: 10.3389/fonc.2021.734694
Received: 01 July 2021; Accepted: 23 September 2021;
Published: 15 October 2021.
Edited by:
Kanjoormana Aryan Manu, Amala Cancer Research Centre, IndiaReviewed by:
Kin Israel Notarte, University of Santo Tomas, PhilippinesCopyright © 2021 Zou, Liao, Hu, Su, Lin, Lin, Luo, Zheng, Zhang, Huang and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiandong Lin, bGlueGRvbmcxOTcwQHllYWgubmV0; Tao Huang, dG9odWFuZ3Rhb0AxMjYuY29t; Lurong Zhang, bHo4NTA2QDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.