- 1Network of Immunity in Infection, Malignancy and Autoimmunity (NIIMA), Universal Scientific Education and Research Network (USERN), Tehran University of Medical Sciences, Tehran, Iran
- 2Cancer Research Center, Mashhad University of Medical Sciences, Mashhad, Iran
- 3Metabolic Syndrome Research Center, Mashhad University of Medical Sciences, Mashhad, Iran
- 4Vasei Clinical Research Development Unit, Sabzevar University of Medical Sciences, Sabzevar, Iran
- 5Department of Cell, Developmental and Integrative Biology, University of Alabama at Birmingham, Birmingham, AL, United States
- 6Basic Medical Sciences Institute, Mashhad University of Medical Sciences, Mashhad, Iran
Prostate cancer (P.C.) is one of the most frequent diagnosed cancers among men and the first leading cause of death with an annual incidence of 1.4 million worldwide. Prostate-specific antigen is being used for screening/diagnosis of prostate disease, although it is associated with several limitations. Thus, identification of novel biomarkers is warranted for diagnosis of patients at earlier stages. MicroRNAs (miRNAs) are recently being emerged as potential biomarkers. It has been shown that these small molecules can be circulated in body fluids and prognosticate the risk of developing P.C. Several miRNAs, including MiR-20a, MiR-21, miR-375, miR-378, and miR-141, have been proposed to be expressed in prostate cancer. This review summarizes the current knowledge about possible molecular mechanisms and potential application of tissue specific and circulating microRNAs as diagnosis, prognosis, and therapeutic targets in prostate cancer.
Introduction
Prostate cancer (P.C.) served as the second most frequent with 37.5 per 100,000 incidence in countries with a higher development index and the fifth most leading to death cancer of men with 1.4 million new cases and 375,000 deaths, worldwide in 2020 (1). The main goal in the diagnosis of P.C. is to detect cancer early and treat patients at initial stages to raise cure rates. Prostate-specific antigen (PSA) testing, a highly sensitive diagnostic test for detection of P.C., influences the diagnosis of this medical condition by early detection of prostate cancer; however, its use declined recently (1). The lack of specificity and sensitivity in the PSA index provides the demand for rectified biomarkers. In the same way, personalized medicine using genes and proteins, i.e., specific features of cells, attracts attention of clinicians and researchers in the management of cancers including P.C. (2). It is becoming manifested that microRNAs (miRNA) are associated with progression and development of prostate cancer (3–8). MiRNAs are small regulatory RNAs with an average length of 22 nucleotides that affect the expression of their target genes and their dysregulation. They play a critical role in various biological functions, maintaining hemostasis, and due to their modulatory operations, they regulate near 60% of all human genes (5).
Moreover, miRNAs are found commonly in cancer-related genomic loci or fragile sites; thus, they implicate in carcinogenesis as a result of their involvement in the expression or suppression of many oncogene genes (9). Thus, they can be used as non-invasive specific markers for estimating the prognosis of disease and classification of tumor (10, 11). MiRNAs are detectable in different body parts, including body fluids (plasma/serum/urine) and tissues. Endogenous RNase cleaves miRNAs; however, having this capability to affect single or several gene targets simultaneously results in the regulation of numerous targets in the malignant cells.
There are a lot of mechanisms that cause cancer progression through modulation of miRNAs, such as deletions, amplifications, and mutations involving miRNA loci, epigenetic silencing, the dysregulation of transcription factors that affects specific miRNAs, or the inhibition of processing. Thus, the miRNA expression profile plays a critical role as a diagnostic and prognostic tool in malignant cancers (12). Plasma and serum samples of normal participants and non-recurrent and recurrent metastatic prostate cancer were assessed for 742 microRNAs by Bryant et al. using a real-time polymerase chain reaction analysis (RT-PCR) to identify differentially quantified microRNAs confirmed considerable changes in the concentration of twelve microRNAs in these patients comparing normal ones (13).
This review summarizes the current knowledge about possible molecular mechanisms and potential application of tissue-specific and circulating microRNAs as diagnosis, prognosis, and therapeutic targets in prostate cancer.
Data Sources and Search Strategies
Searching for articles without a time limit was performed on international databases such as Science Direct, PubMed, and Google Scholar. To maximize the search consistency, we used the search terms “prostate cancer,” “miRNA,” and “circulating marker” with all possible combinations of OR and AND operators.
Study Selection
Initially, the authors provided a list of all the articles’ titles and abstracts in the above databases. After an initial review of articles’ abstracts, related topics were selected, and non-related studies were excluded. Finally, the list of references used in all the searched articles was reviewed to include other possible sources. Those matched with inclusion criteria were selected. The main inclusion criteria in this study were assessing the role of miRNAs in cancer, which were utilized in the introduction section, and the papers with a specific focus on prostate cancer for the rest of this article. Exclusion criteria included case reports, editorials, letters to editor, and vague and unclear studies or the subject of the study was not relevant.
Computational Methods for Predicting Non-Coding RNAs
The miRNAs are essential non-coding RNAs and play significant roles in lots of biological processes; thus, many methods have been developed for predicting disease-related miRNAs and their functions, such as informatic and experimental methods. However, experimental methods are not only long-delayed but also expensive. Genomic SELEX, microarray analysis, and parallel cloning of ncRNAs by specific cDNA libraries or enzymatic and chemical RNA sequencing belong to this category (14). Therefore, hundreds of computational methods were developed. Computational methods have been proposed to analyze the non-coding RNAs. We introduced, discussed, and analyzed these methods in this section. Homology-based methods (including sequence-based method, structure-based methods, and hybrid methods), de novo methods using RNA sequence and structure features (including sequence feature-based methods, structure feature-based methods, and hybrid feature-based methods), transcriptional sequencing and assembling-based methods, and RNA family-specific methods (including miRNA-specific methods and lncRNA) are among the four most available methods of computational methods in analyzing non-coding RNAs (15). Recently, some computational methods have been proposed, such as iMiRNA-PseDPC (16), iMiRNA-SSF (17), miRNA-dis (18), miRNA-deKmer (14), and 2L-piRNA (19).
In addition, all the latest biological knowledge paved the way for developing the new bioinformatics tools, algorithms, and miRNA-specific databases designed for non-coding miRNA prediction (20). Recently, it has been an emerging interest in predicting individual outcomes as a result of genomic alterations (21). Machine learning is defined as the subfield of AI (artificial intelligence).
These methods can accelerate the miRNA studies. Similarly, Lee et al. designed a prediction model by means of ML and bioinformatics tools and based on the genomic study that predict late toxicity after radiotherapy (22).
Circulating MiRNA as Prognostic and Diagnostic Biomarkers
Alternations in miRNA regulation, i.e., upregulation or downregulation, during carcinogenesis, tumor progression, and metastatic transformation, have been reported in several investigations of tumors, including prostate (23–34). The association between miRNA activity and the steps of prostate cancer has been demonstrated (Figure 1) (5). Sylvestre et al. assessed the expression of miR-20a in the P.C. cell line and its correlation with apoptosis, showing that miRNA has a potential antiapoptotic role. Any inhibition of miR-20a resulted in increased cell death. However, its overexpression in doxorubicin-treated P.C. led to a 2-fold decrease in cell death and increased survival by 2 in that cell line (35). In a study by Agaoglu et al., plasma levels of miRNAs including miR-21, miR-141, and miR-221 were measured by real-time PCR in the plasma of fifty one pathologically confirmed P.C. patients in two subgroups, with localized/local advanced or metastatic P.C. and twenty healthy people (23). Agaoglu et al. report that amounts of two of these cancer-related miRNAs, i.e., miR-21 and miR-221, were significantly higher in patients than in normal people. These miRNAs had increased dramatically in metastatic patients than in patients with localized/local advanced disease; however, the results were most considerable for miR-141. The study finally concluded that among these miRNAs, miR-21, and miR-141 are the two most valuable measurements, the former functions better as a diagnostic tool, i.e., for discrimination of P.C. patients from healthy controls, and the latter serves better as a prognostic tool for determination of the stage of disease (23).
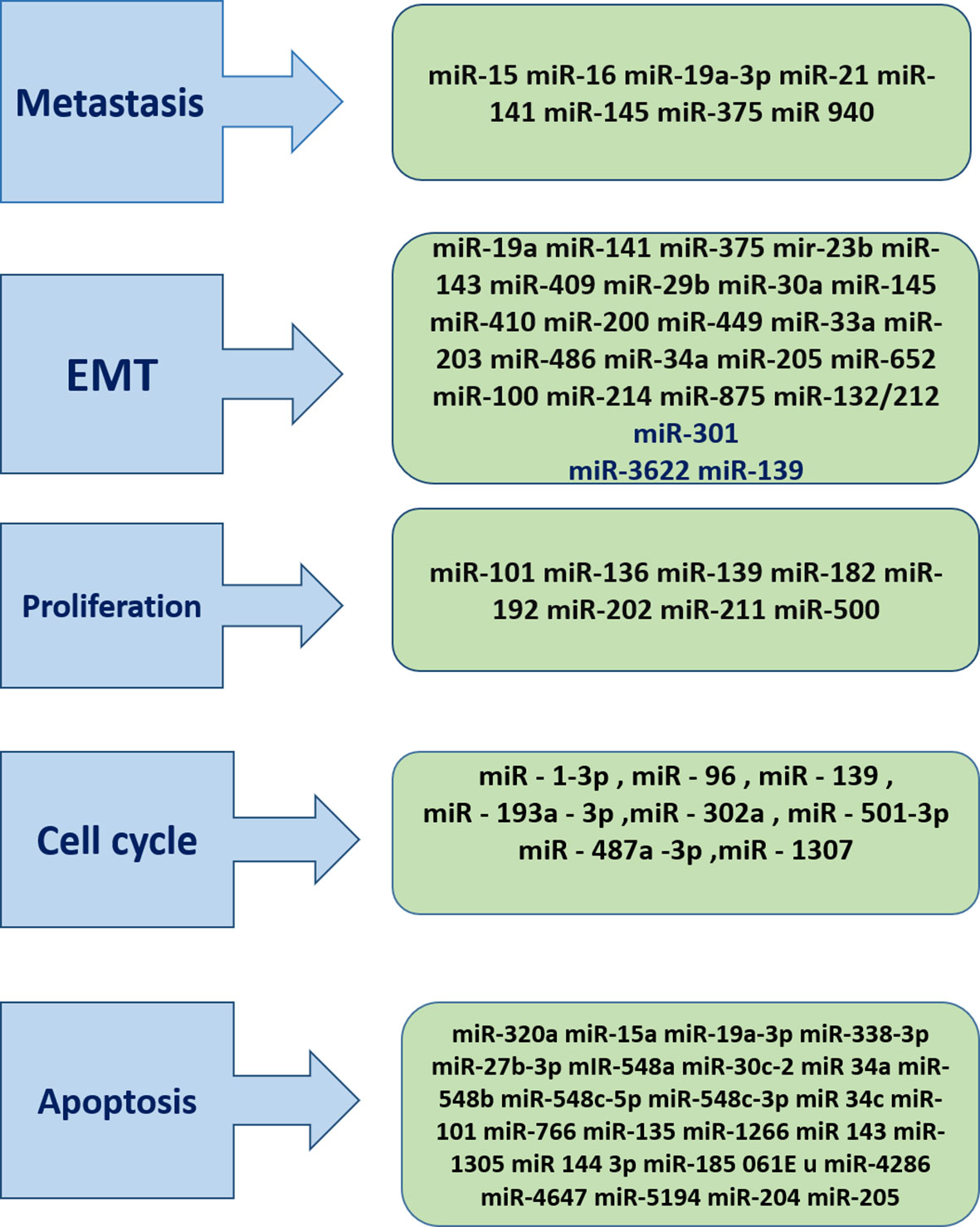
Figure 1 miRNAs Play critical role in cancer progression .As it has been described miRNAs are presented in different stage of cancer. Hence they can be target of the therapeutics and diagnostics approaches.
The assessment of circulating microRNAs in the autochthonous mouse models and patients with metastatic castration-resistant prostate cancer (mCRPC) by Selth et al. showed an alternation of mmu-miR-141, mmu-miR-298 and mmu-miR-375, mmu-miR-346 levels in both groups. The upregulation of the first three of these microRNAs in the cancerous tissue has been evident in P.C. compared with normal prostate tissue. It seems that by progression of disease, mmu-miR-141, mmu-miR-298, and mmu-miR-375 are released into the serum. Furthermore, there was a positive correlation between presence of hsa-miR-141 and hsa-miR-375 in the cancerous tissue and the possibility of biochemical relapse of P.C. suggests the role of hsa-miR-141 and hsa-miR-375 in prostate cancer pathophysiology (26).
Serum samples of about 600 participants including healthy people and patients with different types of cancers with different stages were evaluated by Lodes et al. in terms of miRNA expression patterns (24). All cancer samples were from patients with carcinoma of breast, male and female genitourinary system (prostate and ovary), gastrointestinal organ (colon), and the lung. Based on the results of Lodes et al., overexpression of different miRNAs was reported in the serum of patients with advanced-stage P.C. in comparison to serum of normal participants. They point out these upregulated miRNAs as “miR-16, -92a, -103, -107, -197, -34b, -328, -485-3p, -486-5p, -92b, -574-3p, -636, -640, -766, -885-5p”. Also, a modest elevation in the signal for miR-141 was recorded in patients suffering from stage III–IV PC consistent with a previous report by Mitchell et al. (24, 25). Plasma miR-141 represents various functions in different studies. Mitchell et al. showed its value as a diagnostic tool for discrimination of P.C. patients; however, Agaoglu et al. showed its role as a prognostic measurement as it can distinguish metastatic prostate cancer patients from other stages (23–25). In a study by Fredsøe et al., they assessed the expression of 92 selected miRNAs in plasma samples of 753 patients with different stages of P.C. and non-cancer controls. They found various dysregulations of miRNAs with the overlap of 59 dysregulated miRNAs between BPH versus advanced P.C. and localized P.C. versus advanced P.C. They also identified four miRNA diagnostic ratio models, named bCaP (miR-375*miR-33a-5p/miR-16-5p*miR-409-3p) (36).
Rajendiran et al. found that miR-940 serum levels were significantly higher in cancer patients, especially those with clinically advanced tumors. This study also showed that miR-940 in combination with PSA has a higher value than miR-940 alone for the diagnosis of prostate cancer (37).
Circulating MiRNAs as Predictive Biomarkers in Response to Therapy
Response assessment during treatment is essential to discriminate against the patient who benefits from treatment. Many efforts have been made; however, there is no agreement on the approach of choice. TaqMan Human MicroRNA Arrays and RT-PCR were used by Nguyen et al. to assess serum miRNAs of patients with different groups of P.C. including low-risk localized and high-risk localized disease and mCRPC to find the roles of these negatively regulated genes during carcinogenesis and tumor progression. They reported that P.C. has unique miRNA signatures even in its different subgroups. With disease progression from low-risk disease to mCRPC, increased serum levels of miR-375, -378, and 141 were observed (38). Considering previous reports supporting the results of Nguyen et al., they strongly suggest that the levels of miR-375 and miR-141 in the blood serum are two potential markers for follow-up and surveillance of patients with P.C. (38, 39). Newly diagnosed patients with metastatic hormone-sensitive prostate cancer were enrolled by Cheng et al. for measurement of the circulating miR-141, -200a-b, -210, and -375 by RT-PCR before and after (at 13 weeks) treatment (40). They showed the prognostic and predictive values of a new miRNA, miR-375, in the assessment of treatment of P.C. and confirmed the prognostic role of previously known miRNAs, miR-141 and miR-200a, in this disease. In another effort, Corcoran et al. assessed the value of miR-34a in the prediction of response to chemotherapy in patients with P.C. In their study, they enrolled P.C. patients with and without resistance to docetaxel and found that detection of miR-34a in exosomes is strongly correlated with response to the treatment (41). A study by Lin et al. found that the measurement of circulation levels of miR-200b and miR-20a is beneficial for predicting response to docetaxel in patients with P.C. (42). In a study by Benoist et al., miR-3687 was identified as a prognostic marker for response to enzalutamide in patients with mCRPC, and they confirmed the prognostic importance of miR-375 (43). The expression of five miRNAs (miR-93-5p, -125b-1-5p, -141-3p, -221-3p, and miR-375-3p) was assessed in 84 mCRPC patients in two groups treated with docetaxel and abiraterone by Zedan et al. They found that plasma levels of miR-141-3p and miR-375-3p can predict time to progression in mCRPC patients treated with docetaxel or abiraterone (44). Their results were a primary proof showing the potential benefit of circulating miRNAs as predictive biomarkers in response to therapy.
MiRNAs as Therapeutic Targets
Granulin (GRN) has a critical role in different cancers, and its products cause malignant transformation, induce metastases, and interfere with antiapoptotic processes (45, 46). Recent studies have shown that as GRN is upregulated, miR-107 gets downregulated, and there is an inverse correlation between them. A study by Wang et al. showed that miRNAs including miR-15/107 can potentially target GRN mRNA; i.e., by transfection of GRN protein into P.C. cells, different members of the miR-107 gene group suppress the level of this protein. Hypothetically, it is possible that the expression of GRN can be attenuated by positive manipulation of miR-15/107 gene group expression. It potentially reduces the malignant transformation and metastases and subsequently regulates the antiapoptotic processes of GRN productions (28). The oncogenic effects of overexpression of MiR-21 have been indicted in many human cancers. The effects of MiR-21 on the signal transducer and activator of transcription 3 (STAT3) pathway have been reported by Yang et al., revealing that “miR-21 can be upregulated by IFN and that manipulation of miR-21 expression by miR-21 knockdown can be employed to enhance IFN’s apoptotic action” (29).
Conclusions
Prostate cancer is one of the most frequently diagnosed cancers among men and the first leading cause of death worldwide. PSA is the only available standard method to assess patients’ response to treatment and follow-up. However, it has a lot of limitations, although there are several diagnostic and prognostic markers without consensus regarding them. Recently, dysregulations of miRNAs have been introduced as potential biomarkers for the diagnosis and assessment of prognosis of patients with P.C. and prediction of their response to treatment which can be found as chromosomal level or in the circulation. There are a lot of pitfalls in our knowledge about the molecular basis of P.C., and future investigations should be conducted to find out the role of dysregulations of miRNAs during carcinogenesis and progression of tumors to their advanced stages. The recent trends have also shown that the non-coding RNA analysis should also focus on the sequence composition, and several tools have been proposed for this aim, such as BioSeq-Analysis (47), Pse-in-One (48), and Pse-Analysis (49) (Figure 2).
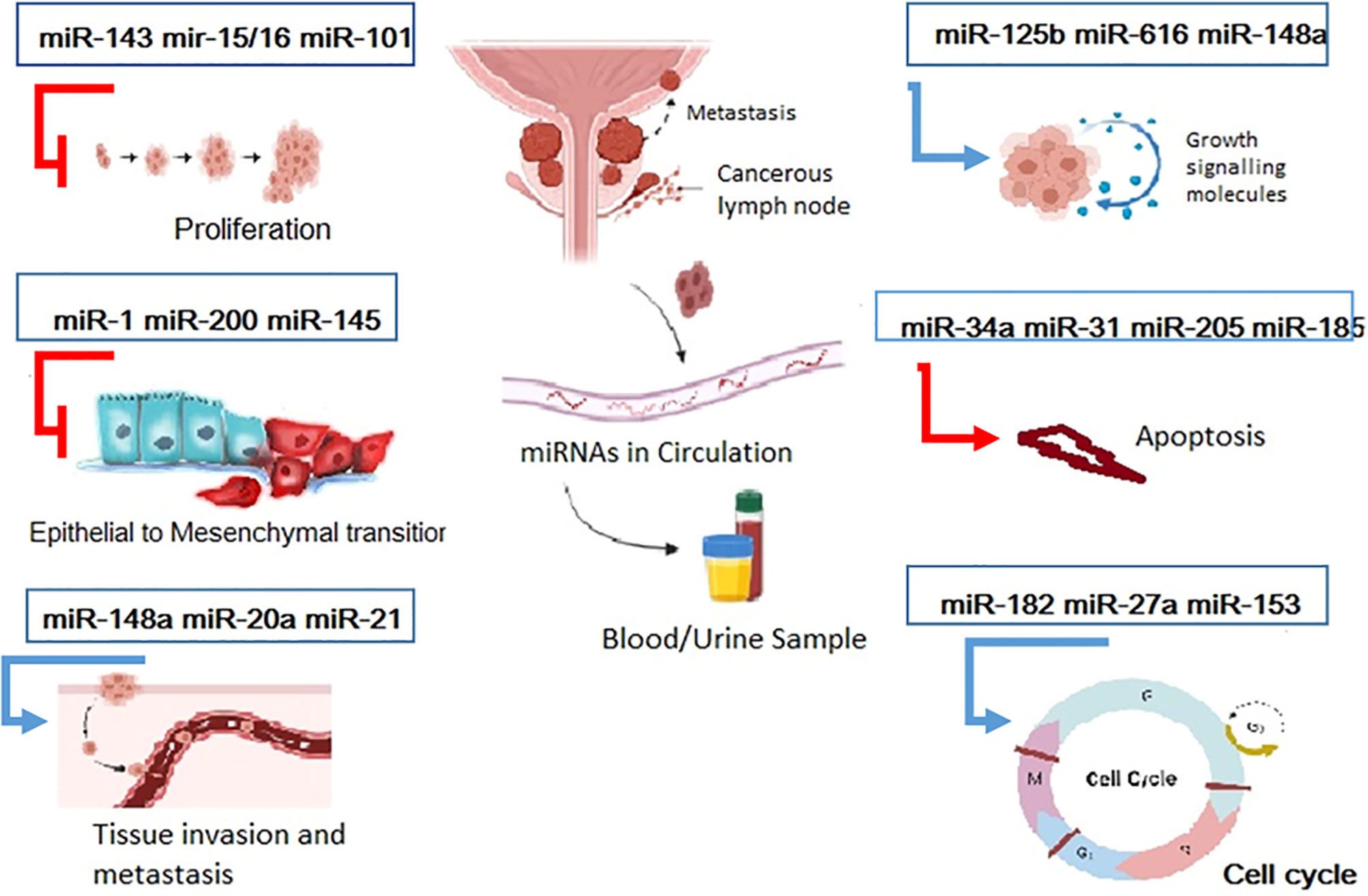
Figure 2 Overview of several mirNAs involved in both two different pathways for tumor progression and tumor suppression. Some miRNAs Promote cancer progression through induction of growth signals, tissue invasion and manipulation of cell cycle ,hence they called onco-miRNAs. On the contrary some miRNAs play protective role against cancer by inhibition of proliferation, EMT initiation of apoptosis. So this tumor suppressor mirs can be target as a therapeutic approaches.
Author Contributions
ES and AA conceived of the presented idea. SAJ, AF, and SSH developed the theory and performed the computations. GP and AM verified the methods. SMH and SM provided the initial draft of the manuscript. All authors discussed the results and contributed to the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors sincerely thank the Vasei Clinical Research Development Unit in Sabzevar University of Medical Sciences for providing advice and guidance in conducting this research.
References
1. US Preventive Services Task Force, Grossman DC, Curry SJ, Owens DK, Bibbins-Domingo K, Caughey AB, et al. Screening for Prostate Cancer: US Preventive Services Task Force Recommendation Statement. Jama (2018) 319(18):1901–13. doi: 10.1001/jama.2018.3710
2. Taghizadeh Kermani A, Hosseini S, Fanipakdel A, Joudi Mashhad M, Akhavan Rezayat K, Zardadi M, et al. A Randomized Clinical Trial on the Antitumoral Effects of Low Molecular Weight Heparin in the Treatment of Esophageal Cancer. J Cell Physiol (2018) 234(4):4191–9. doi: 10.1002/jcp.27177
3. Anceschi U, Tuderti G, Lugnani F, Biava PM, Malossini G, Luciani L, et al. Novel Diagnostic Biomarkers of Prostate Cancer: An Update. Curr Med Chem (2019) 26(6):1045–58. doi: 10.2174/0929867325666180914115416
4. Signore M, Alfonsi R, Federici G, Nanni S, Addario A, Bertuccini L, et al. Diagnostic and Prognostic Potential of the Proteomic Profiling of Serum-Derived Extracellular Vesicles in Prostate Cancer. Cell Death Dis (2021) 12:636. doi: 10.1038/s41419-021-03909-z
5. Cochetti G, Rossi de Vermandois JA, Maulà V, Giulietti M, Cecati M, Del Zingaro M, et al. Role of Mirnas in Prostate Cancer: Do We Really Know Everything? Urol Oncol (2020) 38(7):623–35. doi: 10.1016/j.urolonc.2020.03.007.
6. Cannistraci A, Federici G, Addario A, Di Pace AL, Grassi L, Muto G, et al. C-Met/Mir-130b Axis as Novel Mechanism and Biomarker for Castration Resistance State Acquisition. Oncogene (2017) 36(26):3718–28. doi: 10.1038/onc.2016.505
7. Wang J, Ni J, Beretov J, Thompson J, Graham P, Li Y, et al. Exosomal Micrornas as Liquid Biopsy Biomarkers in Prostate Cancer. Crit Rev Oncology/hematol (2020) 145:102860. doi: 10.1016/j.critrevonc.2019.102860
8. Sharma N, Baruah MM. The Microrna Signatures: Aberrantly Expressed Mirnas in Prostate Cancer. Clin Trans Oncol (2019) 21(2):126–44. doi: 10.1007/s12094-018-1910-8
9. Pastina P, Nardone V, Croci S, Battaglia G, Vanni F, Bellan C, et al. Anti-Cancer Activity of Dose-Fractioned Mpe+/– Bevacizumab Regimen Is Paralleled by Immune-Modulation in Advanced Squamous NSLC Patients. J Thorac Dis (2017) 9(9):3123. doi: 10.21037/jtd.2017.08.68
10. Babaei K, Shams S, Keymoradzadeh A, Vahidi S, Hamami P, Khaksar R, et al. An Insight of Micrornas Performance in Carcinogenesis and Tumorigenesis; an Overview of Cancer Therapy. Life Sci (2020) 240:117077. doi: 10.1016/j.lfs.2019.117077
11. Moya L, Meijer J, Schubert S, Matin F, Batra J. Assessment of Mir-98-5p, Mir-152-3p, Mir-326 and Mir-4289 Expression as Biomarker for Prostate Cancer Diagnosis. Int J Mol Sci (2019) 20(5):1154. doi: 10.3390/ijms20051154
12. Di Leva G, Garofalo M, Croce CM. Micrornas in Cancer. Annu Rev Pathology: Mech Dis (2014) 9:287–314. doi: 10.1146/annurev-pathol-012513-104715
13. Bryant RJ, Pawlowski T, Catto JW, Marsden G, Vessella RL, Rhees B, et al. Changes in Circulating Microrna Levels Associated With Prostate Cancer. Br J Cancer (2012) 106(4):768–74. doi: 10.1038/bjc.2011.595
14. Liu B, Fang L, Wang S, Wang X, Li H, Chou KC, et al. Identification of Microrna Precursor With the Degenerate K-Tuple or Kmer Strategy. J Theor Biol (2015) 385:153–9. doi: 10.1016/j.jtbi.2015.08.025
15. Zhang Y, Huang H, Zhang D, Qiu J, Yang J, Wang K, et al. A Review on Recent Computational Methods for Predicting Noncoding Rnas. BioMed Res International (2017) 2017:14. doi: 10.1155/2017/9139504
16. Liu B, Fang L, Liu F, Wang X, Chou KC. Imirna-Psedpc: Microrna Precursor Identification With a Pseudo Distance-Pair Composition Approach. J Biomol Struct Dyn (2016) 34(1):223–35. doi: 10.1080/07391102.2015.1014422
17. Chen J, Wang X, Liu B. Imirna-SSF: Improving the Identification of Microrna Precursors by Combining Negative Sets With Different Distributions. Sci Rep (2016) 6:19062. doi: 10.1038/srep19062
18. Liu B, Fang L, Chen J, Liu F, Wang X. Mirna-Dis: Microrna Precursor Identification Based on Distance Structure Status Pairs. Mol Biosyst (2015) 11(4):1194–204. doi: 10.1039/c5mb00050e
19. Liu B, Yang F, Chou K-C. 2L-Pirna: A Two-Layer Ensemble Classifier for Identifying Piwi-Interacting Rnas and Their Function. Mol Therapy-Nucleic Acids (2017) 7:267–77. doi: 10.1016/j.omtn.2017.04.008
20. Riffo-Campos Á., Riquelme I, Brebi-Mieville P. Tools for Sequence-Based Mirna Target Prediction: What to Choose? Int J Mol Sci (2016) 17(12):1987. doi: 10.3390/ijms17121987
21. Checcucci E, Autorino R, Cacciamani GE, Amparore D, De Cillis S, Piana A, et al. Artificial Intelligence and Neural Networks in Urology: Current Clinical Applications. Minerva Urol Nefrol (2020) 72(1):49–57. doi: 10.23736/S0393-2249.19.03613-0
22. Lee S, Kerns S, Ostrer H, Rosenstein B, Deasy JO, Oh JH, et al. Machine Learning on a Genome-Wide Association Study to Predict Late Genitourinary Toxicity After Prostate Radiation Therapy. Int J Radiat Oncol Biol Phys (2018) 101(1):128–35. doi: 10.1016/j.ijrobp.2018.01.054
23. Agaoglu FY, Kovancilar M, Dizdar Y, Darendeliler E, Holdenrieder S, Dalay N, et al. Investigation of Mir-21, Mir-141, and Mir-221 in Blood Circulation of Patients With Prostate Cancer. Tumor Biol (2011) 32(3):583–8. doi: 10.1007/s13277-011-0154-9
24. Lodes MJ, Caraballo M, Suciu D, Munro S, Kumar A, Anderson B. Detection of Cancer With Serum Mirnas on an Oligonucleotide Microarray. PloS One (2009) 4(7):e6229. doi: 10.1371/journal.pone.0006229
25. Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating Micrornas as Stable Blood-Based Markers for Cancer Detection. Proc Natl Acad Sci USA (2008) 105(30):10513–8. doi: 10.1073/pnas.0804549105
26. Selth LA, Townley S, Gillis JL, Ochnik AM, Murti K, Macfarlane RJ, et al. Discovery of Circulating Micrornas Associated With Human Prostate Cancer Using a Mouse Model of Disease. Int J Cancer (2012) 131(3):652–61. doi: 10.1002/ijc.26405
27. Shen J, Hruby GW, McKiernan JM, Gurvich I, Lipsky MJ, Benson MC, et al. Dysregulation of Circulating Micrornas and Prediction of Aggressive Prostate Cancer. Prostate (2012) 72(13):1469–77. doi: 10.1002/pros.22499
28. Wang WX, Kyprianou N, Wang X, Nelson PT. Dysregulation of the Mitogen Granulin in Human Cancer Through the Mir-15/107 Microrna Gene Group. Cancer Res (2010) 70(22):9137–42. doi: 10.1158/0008-5472.CAN-10-1684
29. Yang CH, Yue J, Fan M, Pfeffer LM. IFN Induces Mir-21 Through a Signal Transducer and Activator of Transcription 3-Dependent Pathway as a Suppressive Negative Feedback on IFN-Induced Apoptosis. Cancer Res (2010) 70(20):8108–16. doi: 10.1158/0008-5472.CAN-10-2579
30. Brase JC, Johannes M, Schlomm T, Fälth M, Haese A, Steuber T, et al. Circulating Mirnas Are Correlated With Tumor Progression in Prostate Cancer. Int J Cancer (2011) 128(3):608–16. doi: 10.1002/ijc.25376
31. McDonald AC, Vira M, Walter V, Shen J, Raman JD, Sanda MG, et al. Circulating Micrornas in Plasma Among Men With Low-Grade and High-Grade Prostate Cancer at Prostate Biopsy. Prostate (2019) 79(9):961–8. doi: 10.1002/pros.23803
32. Alhasan AH, Scott WH, Wu JJ, Feng G, Meeks JJ, Thaxton CS, et al. Circulating Microrna Signature for the Diagnosis of Very High-Risk Prostate Cancer. Proc Natl Acad Sci USA (2016) 113(38):10655–60. doi: 10.1073/pnas.1611596113
33. Mihelich BL, Maranville JC, Nolley R, Peehl DM, Nonn L. Elevated Serum Microrna Levels Associate With Absence of High-Grade Prostate Cancer in a Retrospective Cohort. PloS One (2015) 10(4):e0124245. doi: 10.1371/journal.pone.0124245
34. Moltzahn F, Olshen AB, Baehner L, Peek A, Fong L, Stöppler H, et al. Microfluidic-Based Multiplex Qrt-PCR Identifies Diagnostic and Prognostic Microrna Signatures in the Sera of Prostate Cancer Patients. Cancer Res (2011) 71(2):550–60. doi: 10.1158/0008-5472.CAN-10-1229
35. Sylvestre Y, De Guire V, Querido E, Mukhopadhyay UK, Bourdeau V, Major F, et al. An E2F/Mir-20a Autoregulatory Feedback Loop. J Biol Chem (2007) 282(4):2135–43. doi: 10.1074/jbc.M608939200
36. Fredsøe J, Rasmussen AKI, Mouritzen P, Bjerre MT, Østergren P, Fode M, et al. Profiling of Circulating Micrornas in Prostate Cancer Reveals Diagnostic Biomarker Potential. Diagnostics (Basel) (2020) 10(4):188. doi: 10.3390/diagnostics10040188
37. Rajendiran S, Maji S, Haddad A, Lotan Y, Nandy RR, Vishwanatha JK, et al. Microrna-940 as a Potential Serum Biomarker for Prostate Cancer. Front Oncol (2021) 11:628094. doi: 10.3389/fonc.2021.628094
38. Nguyen HC, Xie W, Yang M, Hsieh CL, Drouin S, Lee GS, et al. Expression Differences of Circulating Micrornas in Metastatic Castration Resistant Prostate Cancer and Low-Risk, Localized Prostate Cancer. Prostate (2013) 73(4):346–54. doi: 10.1002/pros.22572
39. Szczyrba J, Löprich E, Wach S, Jung V, Unteregger G, Barth S, et al. The Microrna Profile of Prostate Carcinoma Obtained by Deep Sequencing. Mol Cancer Res (2010) 8(4):529–38. doi: 10.1158/1541-7786.MCR-09-0443
40. Cheng HH, Plets M, Li H, Higano CS, Tangen CM, Agarwal N, et al. Circulating Micrornas and Treatment Response in the Phase II SWOG S0925 Study for Patients With New Metastatic Hormone-Sensitive Prostate Cancer. Prostate (2018) 78(2):121–7. doi: 10.1002/pros.23452
41. Corcoran C, Rani S, O’Driscoll L. Mir-34a Is an Intracellular and Exosomal Predictive Biomarker for Response to Docetaxel With Clinical Relevance to Prostate Cancer Progression. Prostate (2014) 74(13):1320–34. doi: 10.1002/pros.22848
42. Lin HM, Castillo L, Mahon KL, Chiam K, Lee BY, Nguyen Q, et al. Circulating Micrornas Are Associated With Docetaxel Chemotherapy Outcome in Castration-Resistant Prostate Cancer. Br J Of Cancer (2014) 110(10):2462–71. doi: 10.1038/bjc.2014.181
43. Benoist GE, van Oort M, Boerrigter E, Verhaegh GW, van Hooij O, Groen L, et al. Prognostic Value of Novel Liquid Biomarkers in Patients With Metastatic Castration-Resistant Prostate Cancer Treated With Enzalutamide: A Prospective Observational Study. Clin Chem (2020) 66(6):842–51. doi: 10.1093/clinchem/hvaa095
44. Zedan AH, Osther PJS, Assenholt J, Madsen JS, Hansen TF. Circulating Mir-141 and Mir-375 Are Associated With Treatment Outcome in Metastatic Castration Resistant Prostate Cancer. Sci Rep (2020) 10(1):227. doi: 10.1038/s41598-019-57101-7
45. Vachher M, Arora K, Burman A, Kumar B. NAMPT, GRN, and SERPINE1 Signature as Predictor of Disease Progression and Survival in Gliomas. J Cell Biochem (2020) 121(4):3010–23. doi: 10.1002/jcb.29560
46. Arechavaleta-Velasco F, Perez-Juarez CE, Gerton GL, Diaz-Cueto L. Progranulin and Its Biological Effects in Cancer. Med Oncol (2017) 34(12):194. doi: 10.1007/s12032-017-1054-7
47. Liu B. Bioseq-Analysis: A Platform for DNA, RNA and Protein Sequence Analysis Based on Machine Learning Approaches. Briefings Bioinf (2017). doi: 10.1093/bib/bbx165
48. Liu B, Liu F, Wang X, Chen J, Fang L, Chou KC. Pse-in-One: A Web Server for Generating Various Modes of Pseudo Components of DNA, RNA, and Protein Sequences. Nucleic Acids Res (2015) 43(W1):W65–71. doi: 10.1093/nar/gkv458
Keywords: Prostate cancer, miRNA, Circulating biomarker, Prognostic factor, Therapeutic targets
Citation: Samami E, Pourali G, Arabpour M, Fanipakdel A, Shahidsales S, Javadinia SA, Hassanian SM, Mohammadparast S and Avan A (2022) The Potential Diagnostic and Prognostic Value of Circulating MicroRNAs in the Assessment of Patients With Prostate Cancer: Rational and Progress. Front. Oncol. 11:716831. doi: 10.3389/fonc.2021.716831
Received: 29 May 2021; Accepted: 31 December 2021;
Published: 04 February 2022.
Edited by:
Ke-hung Tsui, Taipei Medical University, TaiwanReviewed by:
Gian Maria Busetto, University of Foggia, ItalyGiuseppe Simone, Hospital Physiotherapy Institutes (IRCCS), Italy
Copyright © 2022 Samami, Pourali, Arabpour, Fanipakdel, Shahidsales, Javadinia, Hassanian, Mohammadparast and Avan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amir Avan, YXZhbmFAbXVtcy5hYy5pcg==
 Elham Samami
Elham Samami Ghazaleh Pourali2,3
Ghazaleh Pourali2,3 Seyed Alireza Javadinia
Seyed Alireza Javadinia