- Department of Hematology, The First Affiliated Hospital of China Medical University, Shenyang, China
Background: ZNF384 rearrangements are found in 5-10% of B-cell acute lymphoblastic leukemia (B-ALL) and 48% of B cell/myeloid mixed phenotype acute leukemia (B/M MPAL). ZNF384-rearranged B-ALL is prone to lineage conversion after chemotherapy. TCF3 is the second most common rearrangement partner of ZNF384 in B-ALL (27.5%) and the most common partner in B/M MPAL (53.3%). TCF3-ZNF384 fusion is related to a poor steroid response and a high frequency of relapse. It is mostly reported in children and adolescents but rarely seen in adults.
Patients and Methods: Here, we report a rare case of adult common B-ALL with TCF3-ZNF384 fusion in which the patient relapsed after one cycle of consolidation chemotherapy. Relapsed leukemia cells from the bone marrow were cultured for 72 hours ex vivo, and a panel of 156 kinds of cytotoxic drugs, targeted therapy drugs, combination chemotherapy drugs, etc., was used for sensitivity screening. The literature on TCF3-ZNF384 fusion was reviewed, and reported cases with TCF3-ZNF384 fusion were summarized. Clinical characteristics were compared between B-ALL and MPAL with TCF3-ZNF384 fusion.
Results: The relapsed lymphoblasts showed moderate sensitivity to both acute myelocytic leukemia (AML) - and acute lymphoblastic leukemia (ALL)-directed combination chemotherapy schemes, as well as to multiple targeted therapeutic drugs. The hyper-CVAD (B) scheme showed synergistic effects with multiple targeted compounds and had the highest sensitivity. The patient chose the hyper-CVAD (B) scheme combined with sorafenib and achieved complete remission (CR), then consolidated with myeloid-directed homoharringtonine+cytarabine+daunorubicin (HAD) scheme and gained molecular CR. By reviewing the literature, we found that both the genomic landscapes and gene expression profiles of ZNF384-rearranged B-ALL and MPAL are similar and that both diseases have lineage plasticity. The gene expression profile in TCF3-ZNF384-positive patients shows enrichment of hematopoietic stem cell features. No significant differences in clinical characteristics were found between TCF3-ZNF384-positive ALL and MPAL.
Conclusion: TCF3-ZNF384-positive leukemia may be a distinct subtype of leukemia regardless of immunophenotype. Considering the frequent lineage switches and sensitivity to both ALL- and AML-directed schemes, a uniform strategy directed at both lymphoid and myeloid lineages or at hematopoietic stem cells may be better for TCF3-ZNF384-positive leukemia. Small molecule targeted therapies may be promising treatment options and deserve further investigation.
Introduction
Cytogenetic and molecular genetic information has great value in the diagnosis, treatment and prognostic evaluation of acute leukemia. For example, patients with the AML/ETO fusion are diagnosed with acute myeloid leukemia (AML) with AML/ETO regardless of the percentage of leukemia cells and are believed to have a good prognosis (1). ZNF384 rearrangement is found in 5-10% of B-cell acute lymphoblastic leukemia (B-ALL) (2–5) and 48% of B cell/myeloid mixed phenotype acute leukemia (B/M MPAL) (6). ZNF384-rearranged B-ALL often presents as CD10-negative, accompanied by aberrant expression of the myeloid markers CD13 and CD33 (5), and is prone to lineage conversion after chemotherapy. To date, 15 different rearrangement partners of ZNF384 have been found (7). Among them, TCF3 is the second most common partner in BCP-ALL (27.5%) (7) and the most common partner in B/M MPAL (53.3%) (6). B-ALL patients with TCF3-ZNF384 fusion have a poor steroid response and a high frequency of relapse (5). This scenario has mostly been reported in children and adolescents but rarely seen in adults. Liu, Y. F. sequenced samples from 177 adult B-ALL patients, and TCF3-ZNF384 fusion was not detected (4).
Here, we present the treatment process of a case of adult B-ALL with TCF3-ZNF384 fusion and in vitro drug sensitivity screening results. Through this case and literature review, we provide evidence that leukemia with TCF3-ZNF384 rearrangement is a distinct disease needing distinct treatments, and this idea deserves further investigation.
Case Presentation
A 41-year-old male was admitted to our hospital in November 2020 due to a fever that had been present for two weeks with a maximum temperature of 38.2°C. He had no relevant genetic family history. Physical examination showed multiple enlarged lymph nodes in the groin and axilla, which were firm and showed moderate activity, with the largest lymph node being approximately 2.5 cm in diameter. The blood tests showed a white blood cell (WBC) count of 150×109/L, a hemoglobin (HGB) level of 101 g/L and a platelet count of 17×109/L. Bone marrow aspiration was performed and revealed hypercellularity with 98.4% lymphoblasts. He was diagnosed as L2-type ALL according to FAB classification (Figure 1A). Flow cytometry showed malignant B lymphoblasts (P3 group, 92.1%) mainly positive for CD34, CD19, CD10, cCD79a, TdT, CD22, HLA-DR, CD58, and CD13; partially positive for CD38 and CD123; weakly positive for CD33; and negative for CD117, CD7, CD3, MPO, cCD3, CD56, CD15, CD79a, cIgM, CD25, CD20, surface Kappa and Lambda, in accordance with com-B-ALL (Figure 1B) (Experiment was conducted by CANTO II, 6 color flow cytometry, Becton, Dickinson and Company(BD) and all antibodies were purchased from BD company). Karyotype analysis showed that the patient had a 46,XY karyotype [20]. Quantitative polymerase chain reaction (qPCR) covering 56 commonly detected fusion genes in leukemia (listed in Supplementary Table 1) detected only the expression of WT1. Next generation RNA sequencing of bone marrow cells detected a rare fusion (TCF3-ZNF384), a FLT3 mutation c.2028C>A|p.Asn676Ly), a TCF3 mutation (c.1061delC|p.Ser354fs), a NOTCH2 mutation (c.1274A>G|p.Asn425Ser), a CARD11 mutation (c.2735G>A|p.Arg912Gln), and an SH2B3 mutation (c.1307A>G|p.Asp436Gly). The TCF3-ZNF384 fusion was further confirmed by qPCR and fluorescence in situ hybridization (FISH) using two break-apart probes for TCF3 and ZNF384 (Figures 1C, D) (Wuhan Kanglu Biotechnology Co., Ltd).
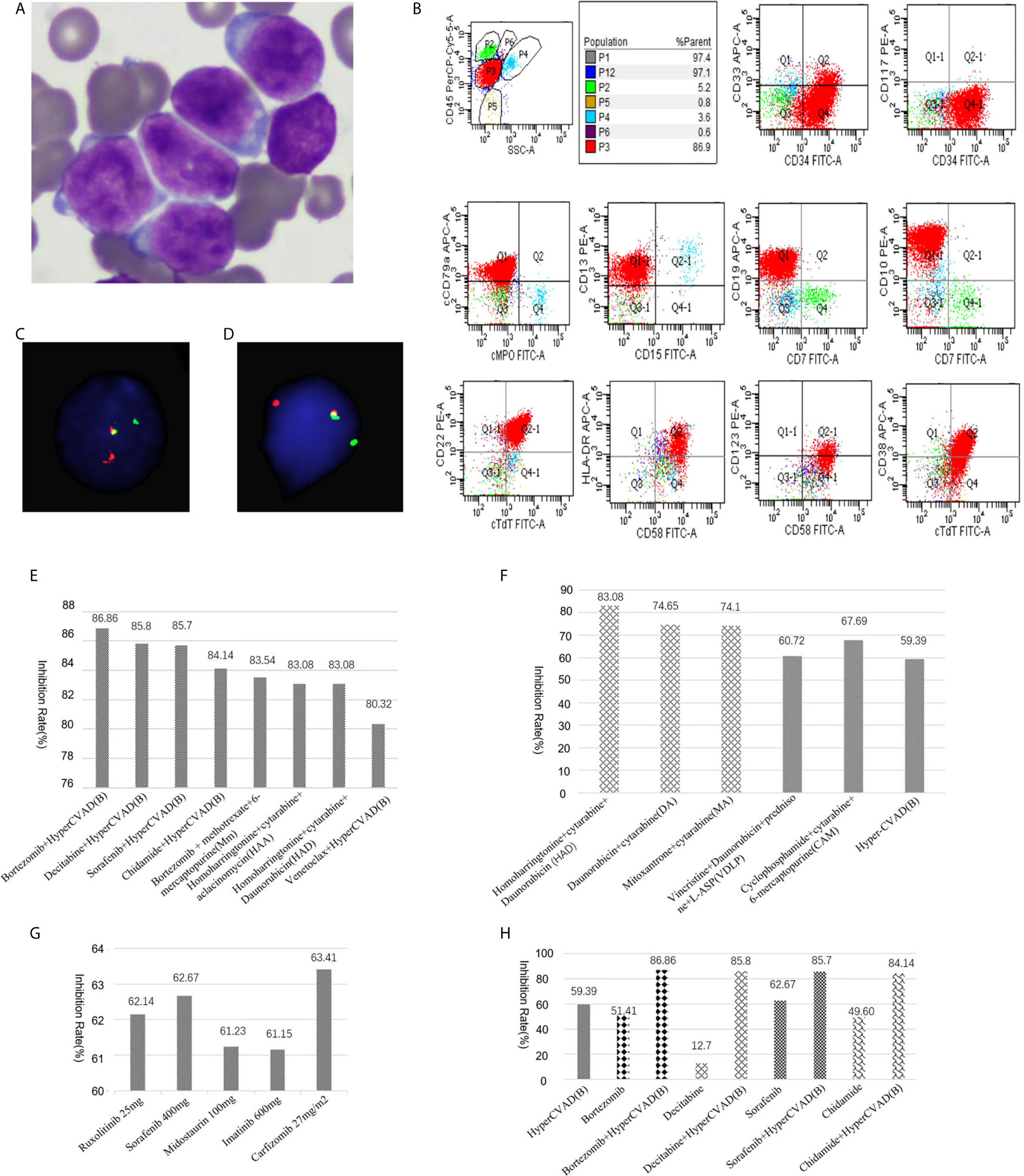
Figure 1 (A) Morphology of lymphoblasts at diagnosis (original magnification, × 1000). (B) Flow cytometry result. (C) FISH results using a break-apart probe for TCF3. Rearrangement of TCF3 was shown as one red and one green signal. (D) FISH results using a break-apart probe for ZNF384. (E) Drug sensitivity screening test in vitro with a panel of 156 kinds of cytotoxic drugs, molecular targeted therapy drugs, and combination chemotherapy regimens. Relative inhibition rates higher than 80% were listed. (F) Relapsed lymphoblasts showed moderate sensitivity to both an ALL scheme [VDLP, CAM, hyper-CVAD (B)] and AML schemes (HAD, DA, MA). (G) Relapsed lymphoblasts were sensitive to multiple targeted therapeutic drugs. (H) The hyper- CVAD (B) scheme showed multiple synergistic effects with multiple targeted compounds.
The patient received chemotherapy according to the JALSG-ALL202-O scheme (8). He achieved complete remission (CR) after induction chemotherapy but relapsed after one cycle of consolidation chemotherapy. Immunophenotyping revealed 29.9% lymphoblasts (mainly positive for CD34, CD19, CD10, CD58, and CD81dim; negative for CD20, CD38, CD123). Relapse leukemia cells from the bone marrow were cultured for 72 hours ex vivo, and a panel of 156 kinds of cytotoxic drugs, targeted therapy drugs, combination chemotherapy drugs, etc. (listed in Supplementary Table 2), was used for sensitivity screening. This service was provided by Hefei PreceDo Pharmaceuticals Co., Ltd. Relative inhibition rates higher than 80% were listed in Figure 1E. Interestingly, the lymphoblasts showed moderate sensitivity to both ALL schemes (VDLP, CAM, hyper-CVAD (B), etc.) and AML schemes (HAD, DA, MA, etc.) (Figure 1F), as well as to multiple targeted therapeutic drugs, such as the Jak2 inhibitor ruxolitinib, the tyrosine kinase inhibitors sorafenib, midostaurin, and imatinib, the proteasome inhibitors carfilzomib (Figure 1G). Furthermore, the hyper-CVAD (B) scheme showed multiple synergistic effects with multiple targeted compounds (bortezomib, decitabine, sorafenib and chidamide) (Figure 1H) and had the highest inhibition rates (Figure 1E). The patient chose the hyper-CVAD (B) scheme combined with sorafenib and gained CR with 0.15% minimal residual disease by flow cytometry (positive for CD19,CD34,CD10,CD123,CD81; negative for CD20,CD38) and 6.11% of TCF3-ZNF384 by Q-PCR after one cycle of treatment. Then the patient received a second cycle of consolidation chemotherapy with HAD scheme directing against the myeloid lineage (9, 10) and gained molecular CR with negative TCF3-ZNF384 by qPCR. Currently, the patient is waiting for stem cell transplantation.
Discussion
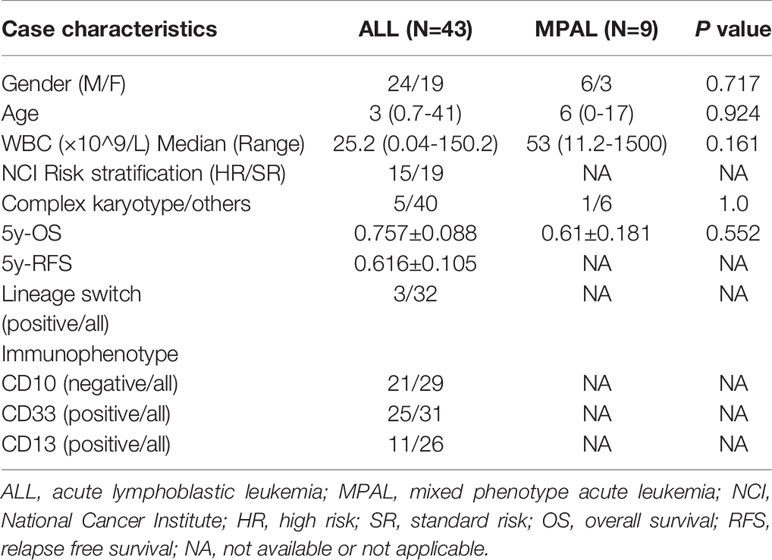
TCF3, also called E2A, can encode several transcription factors via alternative splicing and plays important roles in lymphopoiesis in B- and T-cell lineage (11). ZNF384, also called CIZ or NMP4, is a nucleocytoplasmic shuttling protein that associates with focal adhesions and regulates matrix metalloproteinase (MMP) expression (12). The TCF3-ZNF384 fusion can transform 3T3 cells, and it is the pathogenic basis of leukemia (13). As shown in Supplementary Table 3, most TCF3-ZNF384 fusions contain exon 11 or 13 of TCF3 and exon 2 or 3 of ZNF384, and such fusions retain the transactivation domain of TCF3 and the entire coding region of ZNF384. As such, both loss of function of TCF3 and transactivation of ZNF384 target genes may contribute to leukemogenesis; however, transactivation of ZNF384 is likely more important, as the gene expression profiles of ALL cases with TCF3-ZNF384 fusion are similar to those with other ZNF384 rearrangements but different from those with the TCF3-PBX1 fusion (5). The TCF3-ZNF384 fusion was first reported by Hunger, SP and Zhong, CH in 2002 in an abstract form (14). Later, it was reported to be present in multiple pediatric B-ALL and B/M MPAL cases. We reviewed the literature and found 43 cases of BCP-ALL (including this case) and 9 cases of MPAL with detailed clinical data, as shown in Supplementary Table 3 (4–6, 15–25). Only one adult B-ALL case has been reported. The case included a 21-year-old male who relapsed and died 15 months after diagnosis (23). Here, we report a second adult B-ALL patient with the TCF3-ZNF384 fusion. This patient also showed aberrant expression of myeloid marker CD13 and CD33. However CD10 was expressed in this patient, which accounted only a small part (8/29, Table 1) of patients.
Our case also showed early relapse, which implies poor prognosis. Interestingly, in vitro drug sensitivity screening using the relapsed leukemia cells showed sensitivity to both ALL- and AML-directed schemes. By reviewing the literature, we found that the genomic landscapes of B-ALL and MPAL with ZNF384 rearrangement were similar, and the gene expression profiles of ZNF384-rearranged B-ALL and MPAL cases were essentially indistinguishable (6). Under selective pressure, 10.7% (3/28) of TCF3-ZNF384 B-ALL patients showed lineage switching (Supplementary Table 3). Even without selective pressure, transplantation of sorted subpopulations of cells from a ZNF384-rearranged cell line showed propagation of immunophenotypic diversity (6). Moreover, the gene expression profile in TCF3-ZNF384-positive patients showed enrichment of hematopoietic stem cell features (5), and a case of twins with ZNF384-rearranged ALL indicated that fetal hematopoietic progenitor cells are the cell of origin of this disease (26). All of the above findings imply that leukemias with TCF3-ZNF384 fusion may be derived from hematopoietic stem and progenitor cells, and regardless of immunophenotype, they may have a similar pathogenic basis and clinical characteristics. To further support this hypothesis, we compared the clinical characteristics of TCF3-ZNF384-positive ALL and MPAL, as shown in Table 1. No statistically significant differences in clinical characteristics such as gender, age, WBC, karyotype and overall survival (OS) were found between the two groups. We further subclassified TCF3-ZNF384-rearranged ALL according to the expression of CD10, CD13 and CD33 and compared relapse-free survival (RFS) and OS. Still, we did not find any effects of the immunophenotype on prognosis (all P >0.05, data not shown). Even though more cases need to be accumulated, all above suggests that TCF3-ZNF384-positive leukemia may be a distinct subtype of leukemia regardless of immunophenotype. Our in vitro drug sensitivity screening further supported the above speculation and provided first-hand evidence that TCF3-ZNF384-rearranged ALL can be treated with both ALL- and AML-directed schemes. Considering the frequent lineage switches, a uniform strategy directed at both lymphoid and myeloid lineages or at hematopoietic stem cells may be better for TCF3-ZNF384-positive leukemia. This patient received lymphoid lineage directed scheme firstly and reached CR, then consolidated with myeloid lineage directed scheme and gained molecular CR, which provided a good model for the treatment of this disease. This idea was further supported by the fact that the two TCF3-ZNF384 rearranged MPAL patients who received therapy targeting both myeloid and lymphoid lineages had the longest OS among all 9 patients (6).
Notably, our results show moderate sensitivity of leukemia cells to multiple targeted therapeutic drugs, Jak2 inhibitor ruxolitinib, tyrosine kinases inhibitor midostaurin, sorafenib, imatinib and proteasome inhibitor carfilzomib. This phenomenon may be partly related to the multiple concurrent molecular mutations. The patient in this case showed a FLT3 mutation, which confers sensitivity to midostaurin and sorafenib. SH2B3 mutation is involved in abnormal activation of the Jak-stat signaling pathway, which provides a molecular mechanism explaining the sensitivity to Jak2 inhibitors in this patient (27). It in unknown whether leukemias with ZNF384 rearrangement without these mutations also show sensitivity to these small molecule compounds. In fact, ZNF384-rearranged leukemia shows overexpression of FLT3 and remarkable responsiveness to FLT3 kinase inhibitors even without FLT3 mutation (28). JAK/STAT-class and RAS/RAF/MAPK-class aberrations were found in 21% and 43% of ZNF384 rearrangement patients, respectively (29). Therefore, small molecule targeted therapies may be promising treatment options and deserve further investigation. This patient received tyrosine kinases inhibitor sorafenib combined with hyper-CVAD (B) scheme and achieved CR after only one cycle of treatment. This case provided a good model for individualized accurate treatment under the direction of drug sensitivity screening. Even though the ex-vivo approach may not always work as expected as the pharmacodynamics of each patient contribute to the drug sensitivity and toxicity, it did help guide the treatment of many disease, such as chronic myeloid leukemia (CML) (30), acute myeloid leukemia (AML) (31, 32) and ALL (33).
In conclusion, this study reports a rare case of adult B-ALL with TCF3-ZNF3 fusion that was characterized by early recurrence. The relapsed leukemia cells were sensitive to both ALL and AML schemes in vitro, which supports the viewpoint that TCF3-ZNF384-positive leukemia may be a distinct subtype of leukemia regardless of immunophenotype and should be treated with a uniform strategy directed at both lymphoid and myeloid lineages or at hematopoietic stem cells. Small molecule targeted therapies may be promising treatment options and deserve further investigation.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Materials. Further inquiries can be directed to the corresponding author.
Ethics Statement
Ethical review and approval were not required for the study on human participants in accordance with the local legislation and institutional requirements. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
XY and DC guided the treatment of this case. NL and LW drafted the manuscript and reviewed all related literature. XY and LW interpreted data and critically revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by the National Youth Top-notch Talent of Ten Thousand Talent Program (2014–253) and Translational Research Grant of HCRCH (2020ZKMB06).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to thank Hefei PreceDo Pharmaceuticals Co., Ltd for their work on drug sensitivity screening in vitro and thank the Precision Targeted Therapy Discovery Center, Institute of Technology Innovation, Precedo Pharmaceuticals Co., Ltd., Hefei Institutes of Physical Science, Chinese Academy of Sciences for their work on RNA-sequencing analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.709036/full#supplementary-material
References
1. Lin S, Mulloy JC, Goyama S. RUNX1-ETO Leukemia. Adv Exp Med Biol (2017) 962:151–73. doi: 10.1007/978-981-10-3233-2_11
2. Mullighan CG. How Advanced are We in Targeting Novel Subtypes of ALL? Best Pract Res Clin Haematol (2019) 32:101095. doi: 10.1016/j.beha.2019.101095
3. Yasuda T, Tsuzuki S, Kawazu M, Hayakawa F, Kojima S, Ueno T, et al. Recurrent DUX4 Fusions in B Cell Acute Lymphoblastic Leukemia of Adolescents and Young Adults. Nat Genet (2016) 48:569–74. doi: 10.1038/ng.3535
4. Liu YF, Wang BY, Zhang WN, Huang JY, Li BS, Zhang M, et al. Genomic Profiling of Adult and Pediatric B-Cell Acute Lymphoblastic Leukemia. EBioMedicine (2016) 8:173–83. doi: 10.1016/j.ebiom.2016.04.038
5. Hirabayashi S, Ohki K, Nakabayashi K, Ichikawa H, Momozawa Y, Okamura K, et al. ZNF384-Related Fusion Genes Define a Subgroup of Childhood B-Cell Precursor Acute Lymphoblastic Leukemia With a Characteristic Immunotype. Haematologica (2017) 102:118–29. doi: 10.3324/haematol.2016.151035
6. Alexander TB, Gu Z, Iacobucci I, Dickerson K, Choi JK, Xu B, et al. The Genetic Basis and Cell of Origin of Mixed Phenotype Acute Leukaemia. Nature (2018) 562:373–9. doi: 10.1038/s41586-018-0436-0
7. Hirabayashi S, Butler ER, Ohki K, Kiyokawa N, Bergmann AK, Moricke A, et al. Clinical Characteristics and Outcomes of B-ALL With ZNF384 Rearrangements: A Retrospective Analysis by the Ponte Di Legno Childhood ALL Working Group. Leukemia (2021). doi: 10.1038/s41375-021-01199-0
8. Sakura T, Hayakawa F, Sugiura I, Murayama T, Imai K, Usui N, et al. High-Dose Methotrexate Therapy Significantly Improved Survival of Adult Acute Lymphoblastic Leukemia: A Phase III Study by JALSG. Leukemia (2018) 32:626–32. doi: 10.1038/leu.2017.283
9. Qin T, Xu Z, Zhang Y, Lin Y, Ru K, Fang L, et al. Long-Term Outcomes of Homoharringtonine, Cytarabine, Daunorubicin or Idarubicin (HAD/HAI) as Induction Chemotherapy in De Novo Acute Myeloid Leukemia. Zhonghua Xue Ye Xue Za Zhi (2016) 37:94–9. doi: 10.3760/cma.j.issn.0253-2727.2016.02.002
10. Xiao Z, Xue H, Li R, Zhang L, Yu M, Hao Y. The Prognostic Significance of Leukemic Cells Clearance Kinetics Evaluation During the Initial Course of Induction Therapy With HAD (Homoharringtonine, Cytosine Arabinoside, Daunorubicin) in Patients With De Novo Acute Myeloid Leukemia. Am J Hematol (2008) 83:203–5. doi: 10.1002/ajh.21068
11. Aspland SE, Bendall HH, Murre C. The Role of E2A-PBX1 in Leukemogenesis. Oncogene (2001) 20:5708–17. doi: 10.1038/sj.onc.1204592
12. Nakamoto T, Yamagata T, Sakai R, Ogawa S, Honda H, Ueno H, et al. CIZ, a Zinc Finger Protein That Interacts With P130(Cas) and Activates the Expression of Matrix Metalloproteinases. Mol Cell Biol (2000) 20:1649–58. doi: 10.1128/mcb.20.5.1649-1658.2000
13. Corveleyn A, Janssen H, Martini A, Somers R, Cools J, Marynen P. Cellular Transformation of NIH3T3 Fibroblasts by CIZ/NMP4 Fusions. J Cell Biochem (2005) 94:1112–25. doi: 10.1002/jcb.20369
14. Hunger SPZC. Cryptic T (12,19) in ALL With Lineage Switch to AML Creates Two New E2A Fusion Proteins. Blood (2002) 100:529a.
15. Kapadia AB, Sreedharanunni S, Muhammed S, Karmakar I, Rana S, Sharma P, et al. Emergence of a Prominent Myeloid Clone in a ZNF384-Rearranged B-Cell Precursor Acute Lymphoblastic Leukaemia Post-Corticosteroid Pre-Phase Therapy. Pediatr Blood Cancer (2020) 67:e28513. doi: 10.1002/pbc.28513
16. Rowsey RA, Smoley SA, Williamson CM, Vasmatzis G, Smadbeck JB, Ning Y, et al. Characterization of TCF3 Rearrangements in Pediatric B-Lymphoblastic Leukemia/Lymphoma by Mate-Pair Sequencing (MPseq) Identifies Complex Genomic Rearrangements and a Novel TCF3/TEF Gene Fusion. Blood Cancer J (2019) 9:81. doi: 10.1038/s41408-019-0239-z
17. Nishimura A, Hasegawa D, Hirabayashi S, Kanabuchi S, Yamamoto K, Aiga S, et al. Very Late Relapse Cases of TCF3-ZNF384-Positive Acute Lymphoblastic Leukemia. Pediatr Blood Cancer (2019) 66:e27891. doi: 10.1002/pbc.27891
18. Bueno C, Ballerini P, Varela I, Menendez P, Bashford-Rogers R. Shared D-J Rearrangements Reveal Cell of Origin of TCF3-ZNF384 and PTPN11 Mutations in Monozygotic Twins With Concordant BCP-ALL. Blood (2020) 136:1108–11. doi: 10.1182/blood.2020006604
19. Marincevic-Zuniga Y, Dahlberg J, Nilsson S, Raine A, Nystedt S, Lindqvist CM, et al. Transcriptome Sequencing in Pediatric Acute Lymphoblastic Leukemia Identifies Fusion Genes Associated With Distinct DNA Methylation Profiles. J Hematol Oncol (2017) 10:148. doi: 10.1186/s13045-017-0515-y
20. Shago M, Abla O, Hitzler J, Weitzman S, Abdelhaleem M. Frequency and Outcome of Pediatric Acute Lymphoblastic Leukemia With ZNF384 Gene Rearrangements Including a Novel Translocation Resulting in an ARID1B/ZNF384 Gene Fusion. Pediatr Blood Cancer (2016) 63:1915–21. doi: 10.1002/pbc.26116
21. Zhong CH, Prima V, Liang X, Frye C, McGavran L, Meltesen L, et al. E2A-ZNF384 and NOL1-E2A Fusion Created by a Cryptic T(12;19)(P13.3; P13.3) in Acute Leukemia. Leukemia (2008) 22:723–9. doi: 10.1038/sj.leu.2405084
22. Barber KE, Harrison CJ, Broadfield ZJ, Stewart AR, Wright SL, Martineau M, et al. Molecular Cytogenetic Characterization of TCF3 (E2A)/19p13.3 Rearrangements in B-Cell Precursor Acute Lymphoblastic Leukemia. Genes Chromosomes Cancer (2007) 46:478–86. doi: 10.1002/gcc.20431
23. La Starza R, Aventin A, Crescenzi B, Gorello P, Specchia G, Cuneo A, et al. CIZ Gene Rearrangements in Acute Leukemia: Report of a Diagnostic FISH Assay and Clinical Features of Nine Patients. Leukemia (2005) 19:1696–9. doi: 10.1038/sj.leu.2403842
24. Qian M, Zhang H, Kham SK, Liu S, Jiang C, Zhao X, et al. Whole-Transcriptome Sequencing Identifies a Distinct Subtype of Acute Lymphoblastic Leukemia With Predominant Genomic Abnormalities of EP300 and CREBBP. Genome Res (2017) 27:185–95. doi: 10.1101/gr.209163.116
25. Oberley MJ, Gaynon PS, Bhojwani D, Pulsipher MA, Gardner RA, Hiemenz MC, et al. Myeloid Lineage Switch Following Chimeric Antigen Receptor T-Cell Therapy in a Patient With TCF3-ZNF384 Fusion-Positive B-Lymphoblastic Leukemia. Pediatr Blood Cancer (2018) 65:e27265. doi: 10.1002/pbc.27265
26. Bueno C, Tejedor JR, Bashford-Rogers R, Gonzalez-Silva L, Valdes-Mas R, Agraz-Doblas A, et al. Natural History and Cell of Origin of TC F3-ZN F384 and PTPN11 Mutations in Monozygotic Twins With Concordant BCP-ALL. Blood (2019) 134:900–5. doi: 10.1182/blood.2019000893
27. Cheng Y, Chikwava K, Wu C, Zhang H, Bhagat A, Pei D, et al. LNK/SH2B3 Regulates IL-7 Receptor Signaling in Normal and Malignant B-Progenitors. J Clin Invest (2016) 126:1267–81. doi: 10.1172/jci81468
28. Griffith M, Griffith OL, Krysiak K, Skidmore ZL, Christopher MJ, Klco JM, et al. Comprehensive Genomic Analysis Reveals FLT3 Activation and a Therapeutic Strategy for a Patient With Relapsed Adult B-Lymphoblastic Leukemia. Exp Hematol (2016) 44:603–13. doi: 10.1016/j.exphem.2016.04.011
29. Zaliova M, Stuchly J, Winkowska L, Musilova A, Fiser K, Slamova M, et al. Genomic Landscape of Pediatric B-Other Acute Lymphoblastic Leukemia in a Consecutive European Cohort. Haematologica (2019) 104:1396–406. doi: 10.3324/haematol.2018.204974
30. Wu J, Wang A, Li X, Chen C, Qi Z, Hu C, et al. Discovery and Characterization of a Novel Highly Potent and Selective Type II Native and Drug-Resistant V299L Mutant BCR-ABL Inhibitor (CHMFL-ABL-039) for Chronic Myeloid Leukemia (CML). Cancer Biol Ther (2019) 20:877–85. doi: 10.1080/15384047.2019.1579958
31. Liang X, Wang B, Chen C, Wang A, Hu C, Zou F, et al. Discovery of N-(4-(6-Acetamidopyrimidin-4-Yloxy)Phenyl)-2-(2-(Trifluoromethyl)Phenyl)Acetamide (CHMFL-FLT3-335) as a Potent FMS-Like Tyrosine Kinase 3 Internal Tandem Duplication (FLT3-ITD) Mutant Selective Inhibitor for Acute Myeloid Leukemia. J Med Chem (2019) 62:875–92. doi: 10.1021/acs.jmedchem.8b01594
32. Wang A, Hu C, Chen C, Liang X, Wang B, Zou F, et al. Selectively Targeting FLT3-ITD Mutants Over FLT3-Wt by a Novel Inhibitor for Acute Myeloid Leukemia. Haematologica (2021) 106:605–9. doi: 10.3324/haematol.2019.244186
Keywords: TCF3-ZNF384 fusion, acute lymphoblastic leukemia (ALL), mixed phenotype acute leukemia (MPAL), ZNF384 rearrangement, treatment
Citation: Lin N, Yan X, Cai D and Wang L (2021) Leukemia With TCF3-ZNF384 Rearrangement as a Distinct Subtype of Disease With Distinct Treatments: Perspectives From A Case Report and Literature Review. Front. Oncol. 11:709036. doi: 10.3389/fonc.2021.709036
Received: 13 May 2021; Accepted: 12 July 2021;
Published: 28 July 2021.
Edited by:
Michele Malagola, University of Brescia, ItalyReviewed by:
Michael Diamantidis, University Hospital of Larissa, GreeceSreejesh Sreedharanunni, Post Graduate Institute of Medical Education and Research (PGIMER), India
Copyright © 2021 Lin, Yan, Cai and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lei Wang, bGVpd2VuMDAwQHNpbmEuY29tLmNu
 Na Lin
Na Lin Xiaojing Yan
Xiaojing Yan