
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol. , 15 October 2021
Sec. Molecular and Cellular Oncology
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.708765
 Khalid Otmani*
Khalid Otmani* Philippe Lewalle
Philippe LewalleMicroRNAs (miRNAs) are noncoding RNAs that have been identified as important posttranscriptional regulators of gene expression. miRNAs production is controlled at multiple levels, including transcriptional and posttranscriptional regulation. Extensive profiling studies have shown that the regulation of mature miRNAs expression plays a causal role in cancer development and progression. miRNAs have been identified to act as tumor suppressors (TS) or as oncogenes based on their modulating effect on the expression of their target genes. Upregulation of oncogenic miRNAs blocks TS genes and leads to tumor formation. In contrast, downregulation of miRNAs with TS function increases the translation of oncogenes. Several miRNAs exhibiting TS properties have been studied. In this review we focus on recent studies on the role of TS miRNAs in cancer cells and the tumor microenvironment (TME). Furthermore, we discuss how TS miRNA impacts the aggressiveness of cancer cells, with focus of the mechanism that regulate its expression. The study of the mechanisms of miRNA regulation in cancer cells and the TME may paved the way to understand its critical role in the development and progression of cancer and is likely to have important clinical implications in a near future. Finally, the potential roles of miRNAs as specific biomarkers for the diagnosis and the prognosis of cancer and the replacement of tumor suppressive miRNAs using miRNA mimics could be promising approaches for cancer therapy.
MicroRNAs (miRNAs) are small noncoding RNAs that regulate gene expression posttranscriptionally. These RNAs regulate the expression of their target genes by degrading mRNA transcripts or by inhibiting mRNA translation. Dysregulation of miRNA expression is closely associated with cancer initiation, progression, and metastasis. In particular, miRNA gene expression control is critical for the cellular response to environmental stresses, such as starvation, hypoxia, oxidative stress, and DNA damage, largely implicated in cancer. Based on their inhibition of a large variety of tumor-suppressive and oncogenic mRNAs, some upregulated miRNAs act as tumor-promoting oncomiRs undergoing gain of function in cancer development, and other downregulated miRNAs act as tumor suppressors (TS) undergoing loss of function in tumor growth. Numerous miRNAs have tumor suppressive roles in cancer, and their aberrant underexpression leads to abnormalities in cellular processes, such as an increase in apoptosis, enhanced cell growth, invasion and metastasis and decreased sensitivity to treatment through negative suppression of oncogene function.
Studies have shown that a single miRNA complex may bind to more than 200 target genes, which may have a variety of functions, such as transcription factors, receptors and transporters; therefore, it is very challenging to identify target transcripts and pathways that are regulated by specific miRNAs (1). Since microRNAs can target multiple genes, it is not surprising that they impact a variety of cellular pathways. Moreover, numerous miRNAs have dual effects depending on the tumor type. In some cancers, they act as tumor suppressors, while in other cancers, they behave as tumor activators. A global decrease in the expression of mature miRNA in cancer cells causally contributes to the pathogenesis of various cancers. A profiling study of 217 mammalian miRNAs from normal and human cancer samples showed that miRNA expression is globally suppressed in tumor cells compared to normal cells (2). Nevertheless, more specific alterations of individual miRNA expression are also apparent in tumors (3).
An important aspect of the evolution and progression of cancer is the crosstalk between cancer cells and the surrounding microenvironment. This communication has been shown to be mediated by microRNAs. The deregulation in the expression of TS miRNA is not confined to the cancer cell but extends to the tumor microenvironment (TME). The increase in TS miRNA levels in cancer-associated fibroblasts (CAFs) in the TME severely impairs tumor-supporting capacity by reducing proliferation and migration and impairing tumor expansion (4). In addition, downregulation of mature TS miRNA expression in cancer cells and the TME is highly regulated at multiple levels, including transcriptional and posttranscriptional regulation. A variety of posttranscriptional regulation occurs through the biogenesis machinery of miRNA, affecting the components of the miRNA processing machinery such as Dicer1 (5, 6).
In this review, we will first briefly summarize oncogenic network target by TS microRNAs in cancer cells and some of the recent advance in understanding the complex interplay between TS miRNA downregulation and the TME. Given that microRNAs are frequently dysregulated in cancer, an important question is how microRNAs are regulated. We will therefore mainly focus on important mechanisms of the regulation of TS miRNAs and their impaired biological functions in the initiation and progression of various cancers.
TS miRNAs prevent cancer initiation through the modulation of oncoproteins coding gene expression (7). Many studies have shown that downregulation of some miRNAs is correlated with cancer progression. However, only some TS miRNAs that are often downregulated in cancer cells induce a robust phenotype. This has been demonstrated in various miRNA families that have received substantial attention, including the known tumoral suppressors let-7, miR-15/16, miR-34, and miR-200. The tumor suppressive role of these miRNAs is demonstrated by the repression of their target oncogenic mRNA network, leading to an inhibition of tumorigenesis.
The miR-15/16 clusters function as TS miRNAs that is often deleted or downregulated in CLL (8). miR-15/16 are reported to be downregulated in various solid tumors, such as melanoma, bladder cancer, colorectal cancer, pituitary adenomas, and prostate carcinoma (9). The main target of miR-15/16 that has been identified is bcL2 (10), miR-15/16 triggers apoptosis by suppressing the expression of bcL2, but it also targets other oncogenes, such as cyclin D1, MCL1, CDC2, ETS1 and JUN, that are involved in cancer progression (9). More recently, ROR1 was discovered as a target of miR-15/16, and it was shown that when the level of ROR1 is lower, miR-15/16 expression is higher (11). ROR1 encodes an oncoembryonic surface protein expressed on the CLL cells of over 90% of patients, and ROR1 is a receptor for Wnt5a that promotes leukemia cell proliferation and survival (11). A causative relationship between p53 loss and a decrease in miR-15/16 in the development of CLL was demonstrated in a mouse model, suggesting that the p53-miR15/miR-16-MCL1 axis may contribute to aggressiveness and drug resistance in CLL cells (9). More recently some TS miRNA were reported to promote apoptosis by targeting Bcl2. miR-140 was shown to inhibits colorectal cancer progression and its liver metastasis by targeting BCL9 and BCL2 (12). Many studies described the regulation of Bcl-2 by miR-148a in various cancer. Recently it has been shown that the miR-148a targets Bcl-2 in patients with Non-Small Cell Lung Cancer (13). Several studies reported that miR-340 exert multiple significant effects in cancer cell by triggering apoptosis. miR-340 decreased the expression of Notch and Bcl2 and increased the expression the levels of BIM and Bax inducing cell apoptosis (14). In CRC miR-340 was shown to trigger apoptosis by targeting RLIP76 (15) and REV3L (16). Moreover, miR-340 mediated apoptosis in SGC-7901 cells (17), by increasing the level of apoptosis-related factors pro-caspase 3, cleaved-caspase 3, and Bax, but inhibited Bcl-2. In ovarian cancer (OC) miR-340 was reported to promote apoptosis by downregulation of NF-κB1 (18). miR-34a was recently shown to increase apoptosis by targeting SYT1 in human colon cancer (CC) (19).
The EMT mechanism is important for cancer progression, invasion and metastasis. The miR-200 family is an important TS miRNA family that is downregulated in various cancer types and plays an crucial role in the regulation of EMT mechanism (20). The EMT mechanism is regulated by a complex network, including many transcription factors, such SNAIl, TWIST and ZEB (20, 21). In this context, miR-200 inhibits EMT by directly inhibiting zinc-finger E-box-binding homeobox 1 (ZEB1) and SIP1, also known as ZEB2 (22). In addition, ZEB1 exerts negative feedback to regulate the expression of miR-200 by binding to its promoter, inducing reduced miR-200 expression (23). Several tumor profiling studies demonstrated the importance of the miR-200-ZEB1 axis as important for the control of the EMT process. Recently, it has been shown that the disruption of miR-200/Zeb1 is sufficient to induce an effect on EMT and tumor progression (24). Another factor that plays an important role in the regulation of EMT via miR-200 is the transforming growth factor β1 (TGFβ). TGFβ is important for the induction and maintenance of stable mesenchymal cells. TGFβ downregulates miR-200 by inducing reversible DNA methylation of the miR-200 loci (25). Moreover, the loss of let-7 levels in breast carcinoma initiates and maintains the oncostatin O (OSM)-induced EMT genetic program, and HMGA2 acts as a master switch in this event (26). The EMT transcription factor SNAI1 represses transcription of the TS miRNA Let-7 in cancer. SNAI1 bind let-7 family promoters to inhibit miRNA transcription (27).
One of the most important function of TS miRNAs in cancer is the ability to suppress the proliferation of cancer cells. In cancer development it is well known that Wnt/β-catenin pathway drives tumorigenesis and cancer progression (28). Emergence evidence has indicated that some TS miRNAs suppress the Wnt pathways. the increase level of miR-340 that act as TS miRNA inhibit the expression of Wnt and represses cell proliferation, while its inhibition induces proliferation (28). miR-340 inhibits the Wnt/β-catenin pathway by targeting LGR5 or FHL2 as well as the CTNNB1-mediated Notch signaling pathway, repressing cell proliferation. This mechanism occur by modulating the expression of β-catenin. Many miRNAs as the well-studied miR-200 were shown to target β-catenin (29). It was also shown that miR-19 inhibits cell proliferation in gastric cancer by targeting Myocyte enhancer factor 2D (MEF2D) (30). MEF2D is a transcription factor of the MEF2 family. MEF2D inhibition leads to repression of the Wnt pathway (30). Recently it was shown that miR-133a-5p suppresses Gastric cancer (GC) proliferation by targeting TCF (31). This gene belong to a group of transcription factors that play an important role in the Wnt pathways by recruiting the β-catenin to enhance the transcription of targeting genes, including some oncogene such c-Myc (31). A recent work showed that ectopic expression of miR-34b-3p that is downregulated in non-small-cell lung cancer (NSCLC) tumoral tissue and the cell lines A549 and H1299, represses cell proliferation and cell cycle progression and induces apoptosis of lung cancer cell lines A549 and H1299 by targeting CDK4 kinase, which is important for cell cycle G1 phase progression (32). miR-34a was recently shown to target SYT1 in human colon cancer. SYT1 confers proliferation advantages to cancer cells and promotes the survival of cancer stem cells. Therefore low levels of miR-34a in colon cancer tissues promote cell proliferation, migration and invasion (19).
The miR-34 family has been identified as a TS miRNA in various cancers by targeting many mRNA oncogenes (33). Low levels of miR-34 are observed in several cancers, including prostate cancer (34), colorectal cancer (35) breast cancer and lung carcinoma (36). Many studies have demonstrated that the aberrant expression of programmed cell death protein (PDL-1) allows cancer cells to evade the immune system and is associated with tumor aggressiveness (37). miR-34 was found to target PDL-1 in NSCLC cell lines (38) and acute myeloid leukemia (AML) (39). In NSCLC, the TS p53 represses PDL1 expression by miR-34, revealing another mechanism by which tumor immune escape is regulated by the p53/miR-34/PDL-1 axis. Given the wide range of oncogenes that are modulated by miR-34, miR-34 represents a novel potential therapeutic target for cancer treatment. The Let-7 miRNA family also acts as a TS miRNA and is silenced in various cancers. The human Let-7 family includes ten mature isoform sequences that are produced from 13 precursors (40). Loss of Let-7 expression is associated with the progression of several human cancers (41–45). Let-7 negatively targets the expression of a wide range of oncogenes, such as the RAS family, HMGA2 (46) and Myc (47). In addition, Let-7 has been reported to play crucial roles in metastasis by targeting HMGA2 in nasopharyngeal carcinoma (42), in breast cancer (44) and in oral cancer (48).
The expression of miRNAs is highly regulated at multiple levels, including transcriptional and posttranscriptional regulation. Posttranscriptional regulation occurs in the miRNA biogenesis machinery, which affects the components of miRNA processing, such Drosha, Dicer, Argonaute 2 and DGCR8 (Figure 1).
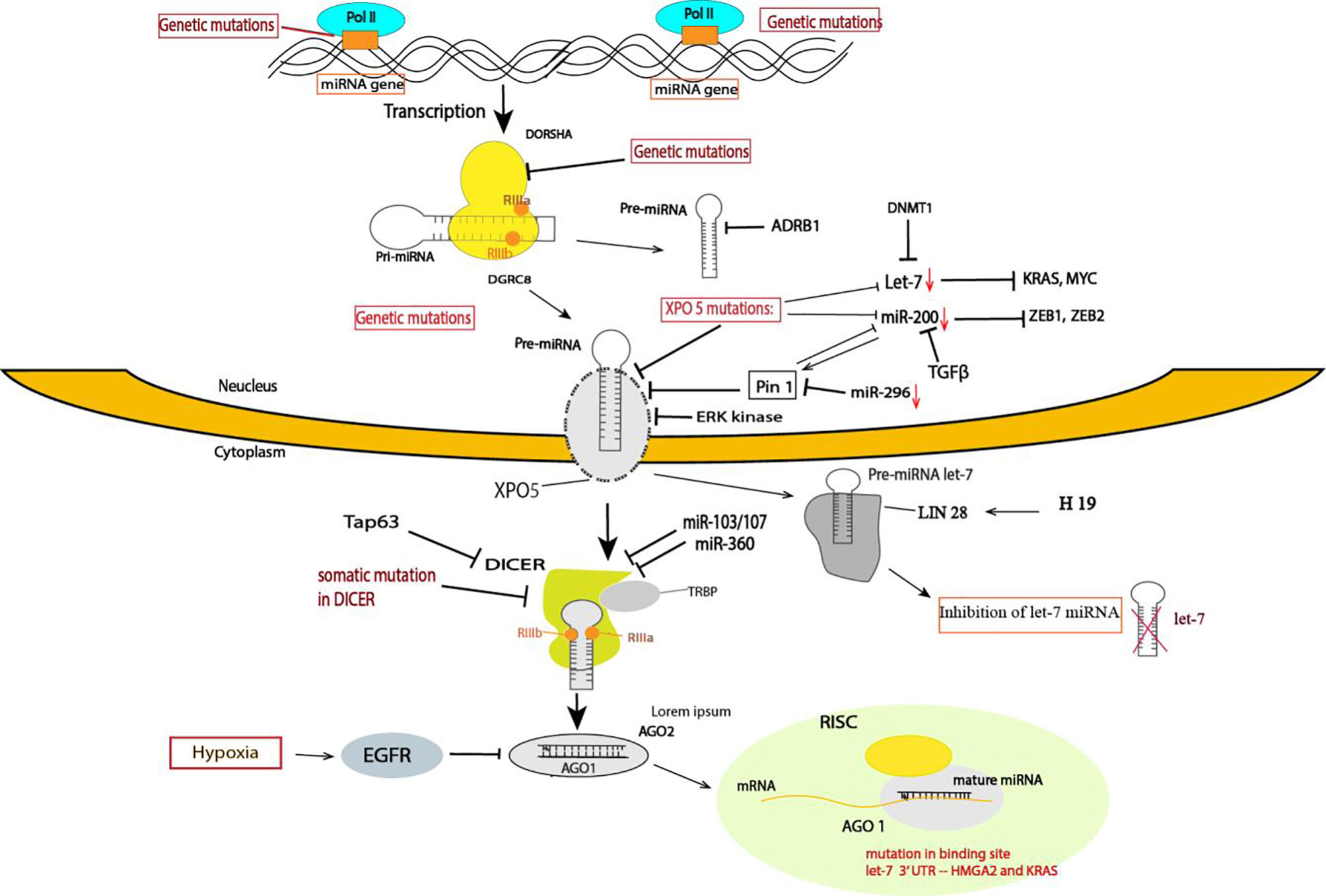
Figure 1 Summary of the mechanism of TS miRNA regulation in cancer. The mechanism controlling TS miRNAs occurs at different steps in cancer, including transcriptional and posttranscriptional regulation. A variety of posttranscriptional regulation occurs in the miRNA biogenesis machinery, which affects the components of miRNA processing. miRNA processing of some TS miRNAs is regulated at multiple levels: at the gene level, through genetic mutations in the miRNA gene and in other components of miRNA processing, such Dorsha, Dicer, and TRBP, and in the binding site of miRNA targets and exportin-5. In XPO mutations, several TS miRNAs are downregulated in cancer, such as miR-200 and let-7. The miRNA transcripts are altered at transcription by several oncogenes, such Myc and TGFB, or by epigenetic modification. LIN 28 proteins specifically block the processing of the pre-miRNA of Let-7. Several studies have highlighted that Dicer downregulation in cancer is caused by several factors, such as somatic mutations, Tap63, miR-360, and miR-103/107. EGFR suppresses the maturation of specific TS miRNAs through phosphorylation of AGO2 and prevents the association of AGO2 with Dicer.
The biogenesis of miRNA implies multiple steps. Similar to protein-encoding genes, miRNAs are transcribed by RNA polymerase II from independent miRNA-encoding genes or represent introns of protein-coding genes (49). Initially, miRNAs are transcribed to primary miRNAs (pri-miRNAs), and this transcript is further cleaved by Drosha and DGCR8, generating precursor miRNA (pre-miRNA). Pre-miRNA is exported from the nucleus to the cytoplasm by exportin-5 (XPO5), where it is processed by Dicer into mature miRNA, which forms what is called the miRNA-induced silencing complex (miRISC) containing the Argonaute 2 protein and glycine-tryptophan protein (GW182). The complex is formed where the miRNA and mRNA target interact (50).
Impaired miRNA biogenesis is one of the mechanisms for the global downregulation of miRNAs observed in various cancers. It was shown for the first time in leukemic cells that impaired miRNA maturation contributes to the downregulation of TS miRNAs (51). Another study confirmed that impaired miRNA biogenesis enhances cellular transformation and promotes tumorigenesis. The authors demonstrated that the global repression of miRNA maturation occurs by the expression of short hairpin RNAs (shRNAs) by cancer cells targeting three different components of the miRNA processing machinery, driving a substantial decrease in steady-state miRNA levels and a transformed phenotype (6). In line with this finding, downregulation of Drosha and Dicer, two key component proteins involved in the miRNA processing pathway, was reported in various cancers and is often associated with a poor clinical outcome (41, 42, 51, 52).
Dicer downregulation in cancer cells is induced by several mechanisms. For example, Tap63 is a transcription factor that activates the transcription of Dicer by directly binding to the promoter; low levels of Tap63 induce a downregulation of Dicer (53), and loss of Tap63 has been found to be associated with various cancers. The expression of Dicer can also be repressed by direct targeting of the 3’UTR by miR-103/107 (54) and miR-360 (55).
Nevertheless, Drosha and Dicer1 can also be expressed at higher levels in some cancers, such as in cervical cancer, prostate adenocarcinoma (56), AML (57) and in precursor lesions of lung adenocarcinoma (58). Moreover, a specific impairment of TS miR-15/16 family processing leads to its reduced levels. This defective processing is linked to the RNA-editing gene ADRB1, an RNA-specific deaminase that is known to modify pre-miRNA and interfere with miRNA processing (51). However, the variable expression of those enzymes in human cancer indicates a complex and likely a cancer-specific dependency of the miRNA biogenesis mechanism in cancer development.
In addition to those crucial enzymes, other miRNA processing pathway enzymes, such as AGO2 and TRABP2, have also been reported to be downregulated. Epidermal growth factor receptor (EGFR), which is an oncogene in human cancer, suppresses the maturation of specific TS miRNAs through phosphorylation of Argonaute 2 (AGO2), which prevents the association of AGO2 with Dicer and inhibits miRNA processing from precursor miRNAs to mature miRNAs in response to hypoxic stress (59).
The biogenesis of miRNA is regulated at diverse steps: transcription of miRNA genes, processing by Dicer and Drosha and transportation of pre-miRNAs to the cytoplasm by XPO5. Nuclear export of pre-miRNA by XPO5 is a crucial step in miRNA biogenesis. Recently, posttranscriptional modification of XPO5 or abnormal expression levels of XPO5 proteins leading to decreased mature miRNA production have been found to be associated with tumorigenesis. Importantly, the phosphorylation of the XPO5 protein by ERK kinase, followed by its cis/trans isomerization by prolyl isomerase (Pin), impairs the ability of XPO5 to transport pre-miRNA, inducing a downregulation of mature miRNA expression and leading to increased migration, cell proliferation and invasion in hepatocellular carcinoma (HCC) (60). A recent study showed that Pin1 downregulates the expression of TS miRNAs by modulating the phosphorylated serine-proline (pS-P) motif of the XPO5 protein (61). Downregulation of miRNA in cancer has a significantly negative correlation with Pin1 expression. In prostate cancer, miR-296 functions as a TS miRNA and induces an inhibition of prostate cancer cell proliferation by targeting Pin (62). It has also been shown recently that targeting Pin1 exerts potent antitumor activity by inhibiting tumor metastasis in pancreatic ductal carcinoma (63). Downregulation of the miR-200 family contributes to Pin1 overexpression and promotes tumorigenesis in breast cancer (55, 56). It was demonstrated that Pin1 inhibits the expression of miR-200b, which can act as a positive feedback loop of Pin1 overexpression in cancer (61).
LncRNAs are a type of a non-coding RNA which is longer than nucleotide in length (64). LncRNAs binds with miRNAs as a competitive endogenous RNA, which preventing miRNAs from binding to mRNA targets and consequently antagonized their functions (65, 66). LncRNAs was shown to play a crucial role in in multiple biological function, including apoptosis, cell proliferation and differentiation. Recently, lncRNAs have been reported as an important regulatory factors in tumorogeneisis (67–70). LncRNAs was shown in many cancers type to be enriched and target negatively the expression of TS miRNAs. In breast cancer steam cell (BCSCs) and breast tumor samples, lncRNA H19 was shown to be expressed in high level (45). The ectopic expression promotes breast cancer development. Conversely, silencing of lncRNA H19 represses the BCSC progression. Mechanistically, lncRNA H19 act as a molecular sponge to regulate the expression of miRNA Let-7. H19 increases the expression of LIN28 and in its turn bind the conserved terminal loop of precurssor miRNA-let-7 elements preventing DICER to cleave pre-Let-7 transcript, inhibiting the production of mature Let-7 miRNAs (45). LncRNA LINC01018 inhibits the progression of AML by targeting miR-499-5p to regulate PDCD4 (71). Moreover, lncRNA SNHG15 was reported to executed oncogenic proprieties in GC by impairing miR-506-5p expression (72). A very recent study has shown that LncRNA LINC01410 induced by Myc accelerates Glioma progression via decreasing the expression of miR-506-3p and Modulating Notch2 Expression to motivate Notch signaling pathway. It was also shown that LINC01410 promotes the progression of cholangiocarcinoma through modulating miR-124-3p/SMAD5 axis (73). Moreover, lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) has been shown to be overexpressed in MM and directly target the expression of miR-188-5p that serves as TS miRNA (74).
Considering that approximately half of the miRNA genes are usually found in the introns of coding genes or long noncoding genes, while the other miRNAs are derived from their own promoters (75), alteration of transcription factor in cancer cells induce a downregulation of TS miRNA in cancer cells.
The transcription of TS miRNAs is often repressed by oncogenic transcription factors. Myc is an oncogenic transcription factor that regulates the expression of various miRNAs. However, the main function of Myc is the repression of a wide repertoire of miRNAs that have a tumor suppressive characteristic, many of which have anti-proliferative and pro-apoptotic effects, such as miR-34, Let-7 and miR-15a/16-1 (76). Myc can also repress the expression of TS miRNA indirectly by activating the expression of LIN28A and LIN28B, which are required for repression of the Let-7 miRNA (77). Many studies have demonstrated a role of miRNAs in other transcription factor networks. For example, the miR-200 family regulates the expression of ZEB1 and ZEB2, which in turn represses the transcription of miR-200 genes, generating a complex feedback loop, and strongly regulates epithelial differentiation in many cancer types (22, 23).
Several studies has investigated the role of estrogen and its nuclear receptor (ER) in modulating miRNA expression in cancer especially in breast cancer cells. Estrogen is involved in various pathological processes through ER-mediated transcriptional gene regulation. One of the estrogen-regulated miRNAs, miR-34b, has been functionally identified and validated as a TS miRNA downregulated by estrogen (78). Estrogen repress the expression of miR-34b through the interaction of ER and p53 which serves as transcriptional factor of miR-34b (78). Another study has shown the identification of differential expression profile of miRNAs in breast cancer cell line MCF-7, in response to oestrogen treatment, miR-107, miR-570 and miR-618 were shown to be strongly downregulated (79). A very recent study demonstrated that miR-1291 targets the expression of ER and exert an antitumor effect. miR-1291 inhibits the proliferation capacity and metabolism status of pancreatic cancer cell line PANC-1 and the breast cancer cell line MDA-MB-231 by targeting the ER. miR-1921 form a regulatory axis with ER that controls the proliferation and cell metabolism by targeting the expression of Carnitine palmitoyltransferase 1C (CPT1C) in breast and pancreatic cancer (80). Moreover, miR-26a and miR-26b were identified as novel target of ER in breast cancer. ER stimulate Myc to suppress the expression of miR-26a/b mediating cell proliferation by targeting CHD1, GREB1 and KPNA2 (81).
The oncogenic protein Yes-associated protein (YAP) that is upregulated in cancer is reported to mediate the global miRNA suppression in tumors by regulating miRNA processing. YAP exerts its function through binding and sequestering DDX17 (DEADBox Helicase 17, also known as p72) away from Drosha and DGCR8 in a cell-density-dependent manner, reducing the processing of pri-miRNA to pre-miRNA by Microprocessor, thus limiting overall miRNA production. At higher cell density, YAP is inactivated by exclusion from the cell nucleus, thereby allowing p72 to associate with microprocessor and pri-miRNAs, resulting in enhanced miRNA biogenesis (82). The activation of YAP causes widespread miRNA suppression in cells and tumors resulting in the posttranscriptional induction of oncoproteins such Myc, suggesting an important role of miRNA biogenesis in YAP-induced tumorigenesis. Myc was shown to be responsible of suppression of miRNA biogenesis in many cancer type. However, the authors considered that suppression of a certain subset of miRNAs by YAP might lead to posttranscriptional enhancement of target genes important for tumorigenesis and growth (82). YAP was shown to be a target of some TS miRNAs in cancer such miR-591 and miR-200 BC (83, 84) and in HCC (85).
Downregulation of TS miRNA can also result from loss of TS transcription factors. However, transcription of miRNAs with TS effects is often activated by transcription factors that have their own important growth suppressive effects. p53 is a transcription factor with a TS function that plays a pivotal role in the regulation of apoptosis and cell cycle progression and governs the cellular response to DNA damage. Many TS miRNAs are under transcriptional control of p53, such as the miR-34 and miR-200 families. The miR-34 family, which is commonly deleted in human cancer, has been shown to be an important component of the p53 TS network (86, 87). The global expression analysis of miRNA levels after genotoxic stress that induces the upregulation of p53 has identified that miR-34 family members a, b, and c are strongly upregulated miRNAs (86). Similar to the known target set of miR-34 family-regulated genes, p53-responsive genes are highly implicated in apoptosis and cell cycle progression. Given the function of the wide range of target genes that are modulated by miR-34, it is clear that the p53 network suppresses tumor formation and is coordinated by transcriptional activation of miR-34 (83). Several other TS miRNAs are regulated by p53 miR-15/16, miR-107 and miR-145. Recently, a novel axis involving p53, miR-30a and ZEB2 was identified that controls tumor progression and metastasis in triple-negative breast cancer (88). The miR-30 member family was shown to act as a TS miRNA in breast cancer growth and metastasis (89). In particular, the expression of miR-30a (both miR-30a-5p and miR-30a-3p) was significantly altered in human breast cancer carrying the p53 gene alteration (88). Additionally, p53 regulates the expression of the component of miRNA biogenesis, either directly by binding to Drosha or indirectly (90). p53 interacts with Drosha through the association with DEAD-box- RNA helicase p68, thus activating the processing of pri-miRNA generating the precursor miRNA. Also the inactivation of transcriptionally p53 induces the decrease in the miRNa processing activity through the inhibition of the functional assembly between Drosha and p68 (90). The p53 family member p63 has been shown to activate the transcription of Dicer1 and miR-130b (53). Inactivated p63 by association with the mutant-p53 induces a downregulation of Dicer1 expression, resulting in low levels of mature miRNAs and increased metastasis (53).
Many tumor suppressors are mutated or repressed in cancer, such as PTEN and p53, resulting in loss of expression of miRNAs with antitumorigenic effects (91, 92). The function of p53 is deregulated in many cancer types, and it’s mutated in more than 50% of human cancers (93), resulting in an oncogenic functions known as a gain-of-function (GOF) activities promoting tumorigenesis (94, 95). At molecular level, GOF acquired by mutant-p53 forms show a dominant-negative (DN) effect either through the binding, sequestration, and the inactivation of tumor suppressor proteins (96), or through the binding to the gene promotor and enhancing its transcriptional regulation (97). However, mutant-p53 was described as transcriptional factor of some miRNAs (97). The widespread nature of mutant-p53 in cancer has suggested the relationship between mutant-p53 GOF activities and the deregulation of miRNA biogenesis observed in cancer (98). Mutant-p53 was shown to suppress the expression of miR-26a by disrupting p68-Drosha complex assembly (99). Another important work demonstrated that mutant-p53 proteins downregulates the expression of miRNAs not only at transcriptional but also at post-transcriptional level by direct binding and sequestering RNA helicases p72/82 through its N-terminal domain from microprosessor complex, hindering the association with Drosha/p72/82 and pri-miRNA and leading to the inhibition of the biogenesis of a subset of miRNA that positively regulated by p72, promoting cell survival and cell migration (100). Recently, in colorectal cancer (CRC) cell line SW480/OxR with mutant-p53, LINC00460-miR-149-5p/miR-150-5p-mutant p53 feedback loop was found to play an important role for oxaliplatin resistance in CRC. In this study the authors demonstrated that the upregulation of LINC00460 in CRC cell line promotes mutant-p53 expression through competitive binding of miR-149-5p/miR150-5p, and in turn mutant-p53 induces the expression of LINC00460, thus forming a positive feedback loop that drives oxaliplatin resistance in SW480/OxR cells (101).
MicroRNA-mediated gene silencing is considered one of the major epigenetic modifications that change the genomic structure and thereby influences gene expression without modifying the DNA sequence (102, 103). Increasing evidence supports the idea that miRNAs can also be deregulated in cancer by abnormal CpG methylation or histone modification. A signature of miRNAs hypermethylated in metastatic cell lines has been found in colon, melanoma, and head and neck cancers. miRNA-34, miR-148a and miR-9 were found to be associated with CpG island hypermethylation in metastatic cell lines in primary colon, oral, breast, lung, gastric head, neck, melanoma and pancreatic cancers (104, 105). miR-200 has been found to be downregulated in the stem-like cell fraction isolated from metastatic breast cancer, and it has been demonstrated that the miR-200c-141 cluster is silenced by CpG hypermethylation, whereas the miR-200b-200a-429 cluster is repressed through polycomb group-mediated histone modification (106). Recently, EZH2 a histone methyltransferase was shown to be responsible for the epigenetic regulation of TS miRNAs in HCC. EZH2 inhibit the expression of miR-200c promoting the hepatocarcinogenesis through increasing the expression of BMI1 (107). More recently, miR-22 that act as TS miRNA in wide variety of cancer such as colorectal cancer, gastric cancer, breast cancer, hepatocellular carcinoma and lung cancer, was shown to be subject to epigenetic regulation by methyl-CpG-binding protein 2 (MeCP2) in gastric cancer (108). MeCP2 silenced miR-22 by binding to their upstream element, including not only the promotor but also the distal enhancer (108). Several TS miRNAs have been found to be inactivated by hypermethylation in multiple myeloma, such as miR-188-5p (74), miR-34b/c and miR-203 (109). Very recently, it have been shown that the SWitch/Sucrose Non-Fermentable (SWI/SNF) complex, a member of the family of ATP-dependent chromatin remodeling complexes is involved in miRNAs expression regulation, SMARCA4-SWI/SNF complex binds to the miR-222 enhancer, increasing the expression of miR-222, who acts as TS miRna (110). SWI/SNF complex plays a relevant role in pathogenesis. SWI/SNF is one of the most frequently mutated epigenetic regulators in cancer (111).
miRNA genes throughout the genome are usually not disturbed, and SNPs in miRNA genes are significantly lower in comparison to the whole genome (112). Nevertheless, miRNA coding sequences are often amplified or deleted in cancer, similar to TS genes and protein coding oncogenes (113). Genomic alterations are critical in oncogenesis, and an increasing number of single nucleotide polymorphisms (SNPs), mutations and chromosomal translocations have been shown to be capable of driving cancer (114). In addition, genomic alteration of miRNA may be an important mechanism responsible for miRNA deregulation reported in several tumor types (115). A very recent work characterized and identified several significant mutations in miRNA gene sequences, and most of these mutations were typically observed in TS miRNAs (112). This finding coincides with previous work suggesting the pattern of loss-of-function mutations observed in suppressor genes (116). Moreover, a somatic mutation in Dicer promotes cell proliferation and regulates the expression of differentiation genes partially through silencing the TS miRNA let-7 family (117).
Moreover, it was shown that mutations in human Dicer are recurrent in several cancers (118, 119),, suggested by partial loss of function of the miRNA processing machinery in human tumors. Previously, it was shown that the Dicer1 single copy deletion in human cancers provides a relevant mechanism for impaired miRNA biogenesis, suggesting a broad role for Dicer1 as a tumor suppressor. A very recent report highlighted the importance of a Dicer1 mutation in anaplastic sarcomas of the kidney (120), and other recent publications have shown that Müllerian adenosarcoma tumorigenesis is characterized by a somatic Dicer1 mutation (121). A Dicer1 hotspot mutation has been shown to be involved in the dysregulation of hormone synthesis in ovarian Sertoli-Leydig patients (122). Sarcoma and lung cancer exhibit impaired miRNA processing without loss of the wild-type Dicer1 allele. Full deletion of Dicer1 induces inhibition of tumorigenesis (123), indicating that cancer cells need a certain minimum level of Dicer expression for tumorigenesis or cell survival. The presence of Dicer knockout cell lines, such as embryonic stem cells, indicates that this phenomenon may be cell type-specific (124).
Mutations in other components of the miRNA processing machinery occur, as in the TARBP2 miRNA processing component in colon cancer and in human ovarian cancer cell lines. Mutations have also been described for Dicer1 and Drosha, which show no correlation with those genes expression levels. However, the low expression of Dicer1 and Drosha is associated with a poor outcome in patients with ovarian cancer (125). A very recent published work demonstrated that a somatic Dicer1 RNase llla/b mutation hotspot previously studied in endometrial cancer (119) impedes 5’ pre-miRNA biogenesis and activity, implying significant changes in targets in a few specific miRNA families, including let-7 and miR-15/16. These Dicer1 RNase llla/b biallelic mutations are notably present in uterine cancer (126).
Disrupting the pairing between miRNA and mRNA targets by mutation is another type of oncogene activating event. Let-7 represses HMGA2, a chromosomal translocation previously associated with tumors, and disrupts the repression of HMGA2 by Let-7 (127).
A mutation in the gene encoding XPO5 induces a decrease in the export of TS miRNA in cancer. Two of the best examples are Let-7 and miR-200, which are downregulated in the XPO5 mutant. Importantly, impairment of TS miRNAs in the XPO5 mutant leads to an increase in their target oncogenes, such Myc and K-RAS for Let-7 and ZEB1 for miR-200 (128). A small but significant mutation in miRNA genes or in their biogenesis machinery can alter miRNA processing, which ultimately influences the level and function of mature miRNAs. This highlights the interplay between genetic variation in miRNA synthesis and posttranscriptional regulation of miRNA biogenesis.
The dynamic crosstalk between cancer cells and normal cells in the TME by the secretion of molecules such as cytokines, chemokines and growth factors has been largely studied, but other molecules, such miRNAs, are also involved in the regulation of several physiological intercellular dialogues and play a significant role in disease progression, including cancer. The TME is very heterogeneous and incorporates various cell types, such as immune cells, fibroblasts, and endothelial cells, and the extracellular matrix (ECM). miRNA can affect the tumor immune microenvirement by regulating the function of immune cells, which in turn modulates the progression of tumor cells (129). TS miRNAs show the ability to act on both stromal and tumor cells to inhibit cancer. Recently, in prostate cancer, it has been shown that the TS miR-15/16 is underexpressed in fibroblasts surrounding prostate tumors. Downregulation of those miRNAs in CAFs promotes the progression and growth of tumors through a reduction in Fgf-2 and repression of its receptor Fgfr1. Fgf-2 and its receptor Fgfr1 play a crucial role in both stromal and cancer cells in enhancing cancer cell survival (4). This suggests a molecular pathway in which both miR-15/16 and its target cooperate to promote tumor progression and invasiveness through concurrent activity on cancer and stromal cells. It has been recently shown that downregulation of exosomal miR-148b from CAFs promotes endometrial cancer cell invasion and metastasis. miR-148b acts as a TS by directly suppressing the expression of DNMT1, which plays a crucial role in enhancing cancer metastasis by inducing EMT (130) (Figure 2).
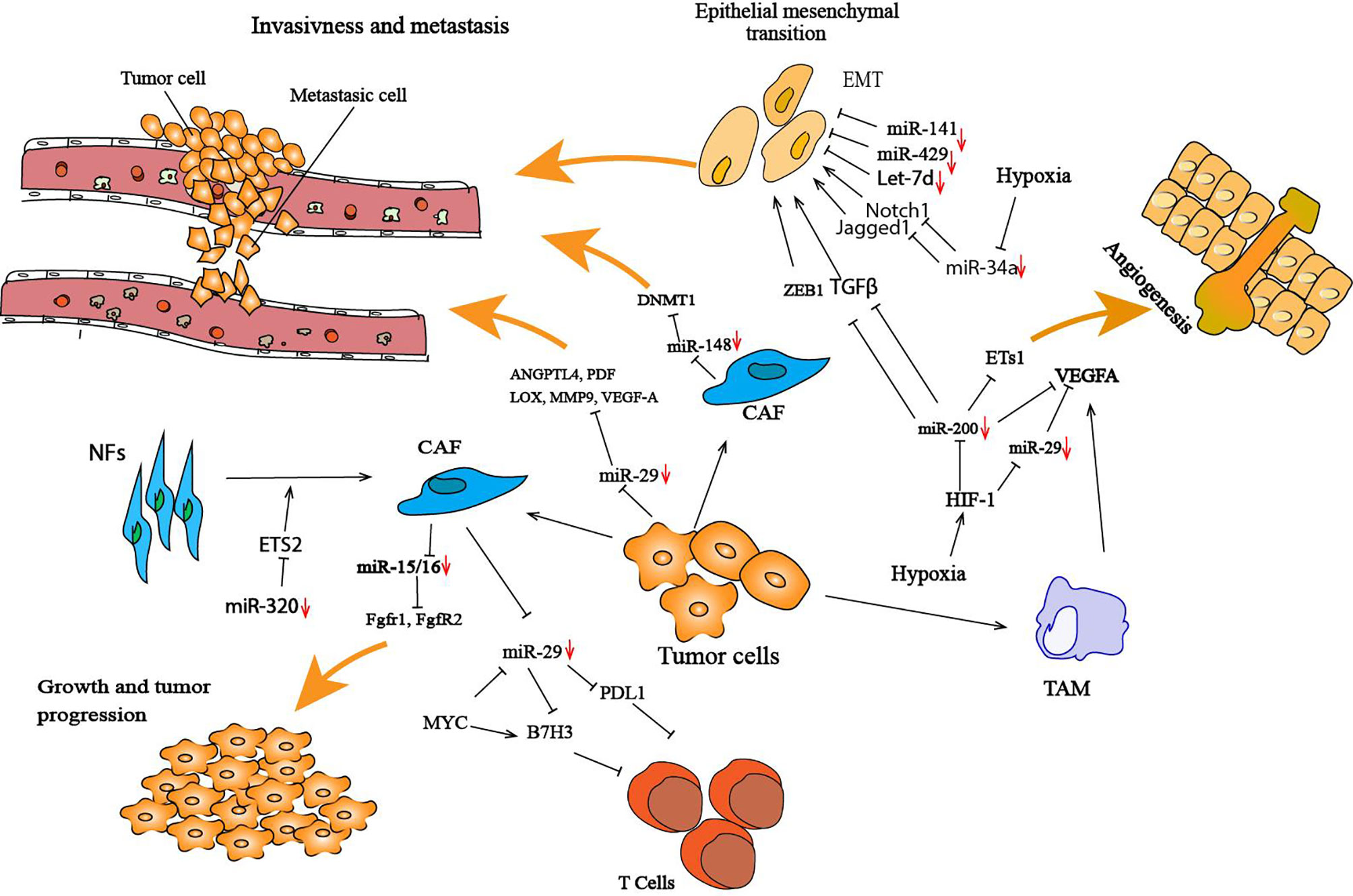
Figure 2 Illustration of cancer cells and the tumor microenvironment-deregulated TS miRNAs that lead to tumor development and progression. In the tumor microenvironment, miRNAs regulate the interaction between cancer cells and their surrounding microenvironment by targeting several genes involved in this interaction. Several TS miRNAs are deregulated in the tumor microenvironment; for example, miR-29 and mR-148 are both downregulated in CAFs, and this deregulation promotes cancer cell invasion and metastasis. These miRNAs have been found to negatively target several genes that are crucial in enhancing cancer metastasis, such DNMT1 for miR-148 and ANGPTL4, PDF, LOX, MMP9, VEGF-A for miR-29. Hypoxia in the tumor microenvironment has a protumorigenic role by altering the expression of TS miRNAs, such as miR-200, miR-34, miR-29 and miR-15/16. Inhibition of miR-200 can significantly increase the expression of Ets-1, resulting in the promotion of angiogenesis. HIF-1 is one of the most studied genes in hypoxia that plays a crucial role in the tumor microenvironment and has been associated with the downregulation of miRNAs. CAFs play an important role in cancer cells during initial cancer and metastasis processes. miR-320 is a TS miRNA that is known to play a part in transforming NFs to CAFs. miR-320 targets ETS2, a cancer-specific transcription factor, resulting in increased oncogenic secretome secretion, and this oncogenic secretome converts NFs to CAFs in the tumor microenvironment (97).
miRNA-29b, a TS miRNA, is downregulated in various types of cancer, including lymphoma, nasopharyngeal carcinoma, glioblastoma and osteosarcoma (87, 88). miRNA-29 possesses antitumoral effects, including inhibition of proliferation and migration. Moreover, miR-29b has been shown to alter the TME by affecting angiogenesis and collagen remodeling to inhibit metastasis and to induce antitumor epigenetic modulation. miR-29b forms a regulatory axis with GATA3 and acts as a TS by modulating the TME in breast cancer. Low expression of miR-29b increases metastasis and promotes a mesenchymal phenotype (127). Mechanistically, miR-29b inhibits metastasis by targeting various genes involved in angiogenesis and collagen remodeling, including ANGPTL4, platelet-derived growth factor (PDF), LOX, matrix metalloproteinase 9 (MMP9) and vascular endothelial growth factor-A (VEGF-A). miR-29 has been shown to directly inhibit the expression of B7-H3 (131). B7H3, known as an inhibitory immune checkpoint ligand (132), is highly expressed on the vascularization surrounding breast, lung and kidney tumors (133) and is responsible for the suppression of T cell function in several cancers (134). A recent study showed a triangular regulation between Myc-miR-29 and B7-H3 in medulloblastoma (MB) tumoral cells, in which Myc directly regulates the expression of B7-H3 and indirectly regulates the expression of miR-29. The upregulation of B7-H3 by Myc promotes MB angiogenesis and can be inhibited by miR-29 overexpression via miR-29-mediated B7-H3 downregulation (131) (Figure 2).
More important, TS miRNA have shown abilities to enhance immune response, by diminishing immune-suppressive mechanisms and or suppress the STAT3 pathway. Cancer cells may also downregulate some TS miRNA that target negatively the expression of immunosuppressive factors and inhibitory costimulatory molecules involved in the inhibition of effector T-cell activation. For example miR-322 that behaves as TS miRNA, suppress the expression of galectin-3 (135), while miR-181 block the biosynthesis of TGF-β receptor 1 and TGF-β receptor associated protein 1 (TGFBRAP1) suggesting that miR-181a may enhance immune response by suppressing the immunosuppressive TGF-β pathway (136).
Some circulating and intracellular miRNAs are dysregulated in cancer, and many studies have shown that the alteration of miRNAs in cancer cells modulates the TME. The alteration of the expression of miRNA profiles is induced not only by dysregulation in signaling pathways in different cancer types but also by general events such as hypoxia (137). Some studies have shown that miRNAs are associated with some signaling pathways in response to hypoxia and play an important role in hypoxic stress. miR-200 has been shown to be downregulated by hypoxia and hypoxia-inducible factor-1 (HIF-1), and this inhibition of expression induced high Ets-1 expression that promotes angiogenesis (138). HIF-1 is one of the most reported genes in hypoxia, which plays an important role in the TME and is associated with miRNA downregulation (137).
Another regulatory mechanism is when the TME alters not only the expression of miRNAs but also negatively modulates their function. Hypoxia attenuates VEGF-A repression by inducing the cytoplasmic translocation of the nuclear ribonucleoprotein L (hnRNP L), which binds the VEGF-A 3’UTR in CA-rich elements, thereby inhibiting miRNA silencing activity (139). The expression of VEGF-A by tumor-associated macrophages (TAMs) plays an important role in metastasis and tumor progression (139). This gene is targeted by several TS miRNAs, such miR-15/16 and miR-29, and downregulation of this miRNA by hypoxia induces an increase in VEGF-A that promotes angiogenesis and initiates tumor vascularization (140). Moreover, hypoxia increase the expression of EGFR, which impairs TS miRNA maturation (59). Another new mechanism was identified in which tumor hypoxia decreases TS miRNAs by inducing a downregulation of Dicer expression. In such events, hypoxia regulates the expression of Dicer by epigenetic mechanisms through inhibition of the oxygen-dependent trimethylated histone H3 lysine (H3K27me3) demethylases KDM6A/B (141). This downregulation of Dicer induces a decrease in miR-200 family processing, leading to an increased level of ZEB1 and activation of the EMT in breast cancer.
Several studies have shown that the quantification of some circulating miRNA levels in peripheral blood can be used to detect tumors and to predict progression, prognosis and response in many cancer types (2). miRNAs are more stable than mRNAs and exhibit tissue specificity, making them potential diagnostic and prognostic biomarkers. Downregulation of circulating TS miRNA is implicated in the progression of many cancers and is associated with a poor clinical outcome. Circulating miRNAs in human plasma or serum are protected from endogenous RNA activity by residing in macrovesicles, exosomes, or apoptotic bodies. These miRNAs can easily be quantified in human plasma or serum and can distinguish patients with cancer from healthy controls (142). The expression levels of some miRNAs, such as Let-7, miR-10b and miR-155, can distinguish patients with cancer from healthy donors in the majority of patients in a non-cancer-specific manner (143). This finding suggests that an individual cancer can exhibit a specific miRNA profile, which may help in discriminating cancer types. Another study demonstrated that some miRNAs can also detect stage I or stage II breast cancer patients at significant levels compared to healthy donors, making them attractive candidates for early cancer diagnosis (144). Changes in miRNA expression can also correlate with metastasis and can therefore be used as a prediction for distal metastasis. Downregulation of miR-361 has been implicated in the progression of lung cancer (145), breast cancer (146), glioma (147) and papillary thyroid carcinoma. Reduced miRNA-361 expression in tumoral tissue is associated with shorter overall survival in lung cancer (145) and colon cancer (148). The Let-7 miRNA family is repressed in many different types of cancer, including breast cancer (41), lung cancer (46), gastric cancer (43) and nasopharyngeal carcinoma (42). Lower levels of Let-7 are correlated with aggressiveness and a poor prognosis, while increased levels are correlated with a good prognosis and prolonged patient survival (44, 143, 149). Lower expression of the Let-7 family is detected in metastatic sites than in primary tumors in gastric and breast cancer (21, 43, 133). Increased levels of Let-7 in breast cancer are associated with lower liver and lung metastases, while its downregulation increases the number of metastases (41, 109). Recent studies have demonstrated that the miR-200 family can be used as a prognostic marker in various cancers (150, 151). Some miRNA families, due to their heterogeneous expression in different tissues and between different cancers, have variable predictive value for patient outcomes. A very recent meta-analysis showed that the miR-200 family may serve as a prognostic biomarker for cancer. It should be used in different tumor tissues based on different members of the miRNA family and requires specific interpretation (152, 153). Taken together, these data show that miRNAs can be useful specific cancer biomarkers to predict clinical outcome and can help in the discrimination of different cancer subtypes.
Globally, lower levels of TS miRNAs are causal agents in several cancers and are correlated with tumor progression and poor patient outcome. Owing to the ability of miRNAs to behave as TS, miRNAs have become potential candidates and provide an opportunity for new cancer therapy strategies. Several miRNA-based therapeutics have reached the preclinical stage, and miR-122 reached phase II trials for the treatment of hepatitis (154, 155). miR-34 was the first to undergo phase I clinical trials in oncology for the treatment of several solid and hematological malignancies (156). miRNA-based therapeutics can be divided into miRNA mimics related to the restoration of TS miRNA expression and inhibitors of miRNAs known as antagomiRs. Therapeutically replenishing TS miRNAs using miRNA mimics has been considered for the clinical modulation of miRNAs. Various studies have shown that the restoration of TS miRNA has significant antitumor activities. Several clinical studies using miR-34 mimics encapsulated in lipid nanoparticles have shown their great potential as therapeutic agents, and they are considered the most advanced miRNA therapeutics for cancer. A recent study showed that treatment with miR-34 encapsulated in lipids induced a significant decrease in NSCLC tumors in a mouse model (157). A subsequent study showed that a combination treatment with miR-34a and Let-7 using lipid nanoparticles resulted in a significant decrease in tumors, and combinatorial treatment with the EGFR inhibitor EROLINIB resulted in the strongest synergistic effect in inhibiting the proliferation of NSNLC (158). Another miRNA that has been used in preclinical studies is miR-200. In a lung cancer xenograft model, the treatment of tumors with miR-200c mimic increased cellular radiosensitivity by directly targeting, in addition to the repressor ZEB1, the genes of oxidative stress response sestrin1 (SESN1), NF-related factor 2 (NRF2) and peroxiredoxin 2 (PRDX2), thereby increasing the levels of reactive oxygen species (ROS) (159). Several TS miRNAs delivery systems are under development. miRNAs could emerge as a key technology for the treatment of cancer and other diseases if clinical trials using miRNAs confirm their preliminary encouraging success.
Cancer is an extremely complex disease that is driven by a combination of various aberrantly regulated processes, such as, deregulation of the epigenetic machinery. miRNAs involved in cancer are divided in two groups: the oncomirs that can induce tumor initiation and progression, and their expression are higher in cancer cells. The second group is TS miRNAs, that are downregulated in cancer cells, prevent cancer beginning and progression of cancer through suppressing the expression of various oncogenes. In this review we have highlighted recent advances in the understanding of the role of TS miRNAs, and how TS miRNAs affect the progression of cancer cells and modulate the TME by antagonizing processes that are crucial for cancer cells development. We also emphasized the most important mechanisms of the regulation that specially silence the expression of TS miRNA.
TS miRNA are downregulated in cancer tissues, upon re-expression they suppress various processes relevant to tumorigenesis, including growth and proliferation as well as invasion and metastasis. However, reducing TS miRNA expression in cancer cells, allow cancer progression by reducing apoptosis, inducing cell survival and suppressing tumor invasion and metastasis. In some instances, the overexpression of these miRNAs was shown to be effective in inhibiting cancer proliferation, increasing the sensitivity of cancer cells to apoptosis signals, and suppressing tumor invasion, metastasis and in vivo angiogenesis, by targeting the production of oncogene (127, 140).
Accordingly, the restauration of the expression of TS miRNA to inhibit cell proliferation and growth emerges as a new target strategy that could be used in many cancer types, given that TS miRNA play pivotal role in a wider range of human cancers. Importantly, many studies have shown the efficiency of combined treatments with current chemotherapies. Currently, there is emergence of several potential TS miRNAs that have been demonstrated to negatively target cancer cells stability in various pre-clinical cancer models. Further studies are required to evaluate the anti-cancer efficiency of the specific TS miRNA in multiple pre-clinical models and eventually in clinical trials.
Under the pressure of immune system, cancer cells develop multiples mechanism to evade attack by immune system. One of the mechanism used is the deregulation of the expression of miRNA in cancer cell and TME. Tumor cells aberrantly express a number of miRNAs, and this abnormal miRNA expression profile is characterized by the downregulation of TS miRNA and by the maintenance of an increased levels of oncogenic miRNA (oncomiR).
Generally, the expression of miRNA in cancer show a global downregulation (2), this downregulation affect especially TS miRNA maturation. Due to their role in the regulation of gene expression, the expression of mature miRNA is highly regulated at multiple levels including transcriptional and posttranscriptional regulation. A variety of posttranscriptional regulation occurs in biogenesis machinery of miRNA, which affect, the components of the miRNA processing machinery (Drosha, Dicer, argonote 2 or DGCR8), as example, impaired miRNA biogenesis enhance cellular transformation and promote tumorigenesis (6). Recent research has highlighted the role of genetic variation in miRNA genes and components of the biogenesis machinery in alteration of miRNA processing, which ultimately affects the levels of mature TS miRNAs in cancer. Deregulation of the expression of miRNAs can occur from aberrant transcription factor activity in cancer cells. Numerous Pol II–associated transcription factors are involved in the transcriptional control of miRNA genes. Some miRNAs are therefore under the control of tumor–suppressive or oncogenic transcription factors. Intriguingly, miRNAs have been shown in many studies to have a role in many transcription factor networks in cancer, representing a complex feedforward loop.
The activity of TS miRNA is not confined to the cancer cell but is extended to TME. In addition to these mechanism, cancer cells deregulate the expression of some miRNA in TME which creates an immunosuppressive microenvironment. TS miRNA have been shown to act directly by affecting cancer cell growth and viability but also indirectly by affecting angiogenesis and metastasis via secretion into the TME. These miRNAs regulate different stromal cells and exert their impact to recruit or promote the differentiation of suppressive immune cells, such Treg, CAF, TAMs and others that are involved in the TME (123). Reduced O2 availability (hypoxia) in the TME was shown to exert major regulatory effect in the expression of TS miRNA in TME.
As has been shown through this review, TS miRNAs have an important role as biomarkers in diagnosis and prognosis and have prominent clinical implications. Nevertheless, studies of miRNA-mediated interactions between cancer cells and stromal cells, specifically those focused on understanding the origin of miRNA alterations, are needed to improve targeted therapy.
KO, first author writer of the review. PL, supervision and correction of the work. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Palmero EI, de Campos SG, Campos M, de Souza NC, Guerreiro ID, Carvalho AL, et al. Mechanisms and Role of microRNA Deregulation in Cancer Onset and Progression. Genet Mol Biol (2011) 34(3):363–70. doi: 10.1590/S1415-47572011000300001
2. Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA Expression Profiles Classify Human Cancers. Nature (2005) 435(7043):834–8. doi: 10.1038/nature03702
3. Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA Expression Signature of Human Solid Tumors Defines Cancer Gene Targets. Proc Natl Acad Sci U S A (2005) 103(7):2257–61. doi: 10.1073/pnas.0510565103
4. Musumeci M, Coppola V, Addario A, Patrizii M, Maugeri-Saccà M, Memeo L, et al. Control of Tumor and Microenvironment Cross-Talk by miR-15a and miR-16 in Prostate Cancer. Oncogene (2011) 30(41):4231–42. doi: 10.1038/onc.2011.140
5. Ramírez-Moya J, Wert-Lamas L, Riesco-Eizaguirre G, Santisteban P. Impaired microRNA Processing by DICER1 Downregulation Endows Thyroid Cancer With Increased Aggressiveness. Oncogene (2019) 38(27):5486–99. doi: 10.1038/s41388-019-0804-8
6. Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA Processing Enhances Cellular Transformation and Tumorigenesis. Nat Genet (2007) 39(5):673–7. doi: 10.1038/ng2003
7. Bartel DP, Lee R, Feinbaum R. MicroRNAs : Genomics, Biogenesis, Mechanism, and Function Genomics : The miRNA Genes. Cell (2004) 116:281–97. doi: 10.1016/s0092-8674(04)00045-5
8. Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent Deletions and Down-Regulation of Micro-RNA Genes Mir15 and Mir16 at 13q14 in Chronic Lymphocytic Leukemia. Proc Natl Acad Sci USA (2002) 99(24):15524–9. doi: 10.1073/pnas.242606799
9. Pekarsky Y, Croce CM. Role of miR-15/16 in CLL. Cell Death Differ (2015) 22(1):6–11. doi: 10.1038/cdd.2014.87
10. Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, et al. miR-15 and miR-16 Induce Apoptosis by Targeting BCL2. Proc Natl Acad Sci USA (2005) 102(39):13944–9. doi: 10.1073/pnas.0506654102
11. Rassenti LZ, Balatti V, Ghia EM, Palamarchuk A, Tomasello L, Fadda P, et al. MicroRNA Dysregulation to Identify Therapeutic Target Combinations for Chronic Lymphocytic Leukemia. Proc Natl Acad Sci USA (2017) 114(40):10731–6. doi: 10.1073/pnas.1708264114
12. Liu D, Chen C, Cui M, Zhang H. miR-140-3p Inhibits Colorectal Cancer Progression and its Liver Metastasis by Targeting BCL9 and BCL2. Cancer Med (2021) 10(10):3358–72. doi: 10.1002/cam4.3840
13. Elnaggar GN, El-Hifnawi NM, Ismail A, Yahia M, Elshimy RAA. Micro RNA-148a Targets Bcl-2 in Patients With Non-Small Cell Lung Cancer. Asian Pacific J Cancer Prev (2021) 22(6):1949–55. doi: 10.31557/APJCP.2021.22.6.1949
14. Xie W, Qin W, Kang Y, Zhou Z, Qin A. MicroRNA-340 Inhibits Tumor Cell Proliferation and Induces Apoptosis in Endometrial Carcinoma Cell Line RL 95-2. Med Sci Monit (2016) 22:1540–6. doi: 10.12659/MSM.898121
15. Zhang LL, Xie FJ, Tang CH, Xu WR, Ding XS, Liang J. miR-340 Suppresses Tumor Growth and Enhances Chemosensitivity of Colorectal Cancer by Targeting RLIP76. Eur Rev Med Pharmacol Sci (2017) 21(12):2875–86.
16. Arivazhagan R, Lee J, Bayarsaikhan D, Kwak P, Son M, Byun K, et al. MicroRNA-340 Inhibits the Proliferation and Promotes the Apoptosis of Colon Cancer Cells by Modulating REV3L. Oncotarget (2018) 9(4):5155–68. doi: 10.18632/oncotarget.23703
17. Yu J, Wang R, Chen J, Wu J, Dang Z, Zhang Q, et al. miR-340 Inhibits Proliferation and Induces Apoptosis in Gastric Cancer Cell Line SGC-7901, Possibly via the AKT Pathway. Med Sci Monit (2017) 23:71–7. doi: 10.12659/MSM.898449
18. Li P, Sun Y, Liu Q. MicroRNA-340 Induces Apoptosis and Inhibits Metastasis of Ovarian Cancer Cells by Inactivation of NF-κb1. Cell Physiol Biochem (2016) 38(5):1915–27. doi: 10.1159/000445553
19. Lu H, Hao L, Yang H, Chen J, Liu J. miRNA-34a Suppresses Colon Carcinoma Proliferation and Induces Cell Apoptosis by Targeting SYT1. Int J Clin Exp Pathol (2019) 12(8):2887–97. doi: 10.1016/j.scitotenv.2019.136171
20. Lamouille S, Xu J, Derynck R. Molecular Mechanisms of Epithelial – Mesenchymal Transition. Nat Publ Gr (2014) 15(3):178–96. doi: 10.1038/nrm3758
21. Krebs AM, Mitschke J, Lasierra Losada M, Schmalhofer O, Boerries M, Busch H, et al. The EMT-Activator Zeb1 is a Key Factor for Cell Plasticity and Promotes Metastasis in Pancreatic Cancer. Nat Cell Biol (2017) 19(5):518–29. doi: 10.1038/ncb3513
22. Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 Family and miR-205 Regulate Epithelial to Mesenchymal Transition by Targeting ZEB1 and SIP1. Nat Cell Biol (2008) 10(5):593–601. doi: 10.1038/ncb1722
23. Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, et al. A Reciprocal Repression Between ZEB1 and Members of the miR-200 Family Promotes EMT and Invasion in Cancer Cells. EMBO Rep (2008) 9(6):582–9. doi: 10.1038/embor.2008.74
24. Title AC, Hong SJ, Pires ND, Hasenöhrl L, Godbersen S, Stokar-Regenscheit N, et al. Genetic Dissection of the miR-200–Zeb1 Axis Reveals its Importance in Tumor Differentiation and Invasion. Nat Commun (2018) 9(1):4671. doi: 10.1038/s41467-018-07130-z
25. Gregory PA, Bracken CP, Smith E, Bert AG, Wright JA, Roslan S, et al. An Autocrine TGF-β/ZEB/miR-200 Signaling Network Regulates Establishment and Maintenance of Epithelial-Mesenchymal Transition. Mol Biol Cell (2011) 22(10):1686–98. doi: 10.1091/mbc.E11-02-0103
26. Guo L, Chen C, Shi M, Wang F, Chen X, Diao F, et al. Circuits Initiate and Maintain Oncostatin M-Driven Epithelial – Mesenchymal Transition. (2013) 32(45):5272–82. doi: 10.1038/onc.2012.573
27. Wang H, Chirshev E, Hojo N, Suzuki T, Bertucci A, Pierce M, et al. The Epithelial–Mesenchymal Transcription Factor Snai1 Represses Transcription of the Tumor Suppressor miRNA Let-7 in Cancer. Cancers (Basel) (2021) 13(6):1–18. doi: 10.3390/cancers13061469
28. Mohammadi-Yeganeh S, Paryan M, Arefian E, Vasei M, Ghanbarian H, Mahdian R, et al. MicroRNA-340 Inhibits the Migration, Invasion, and Metastasis of Breast Cancer Cells by Targeting Wnt Pathway. Tumor Biol (2016) 37(7):8993–9000. doi: 10.1007/s13277-015-4513-9
29. Peng Y, Zhang X, Feng X, Fan X, Jin Z. The Crosstalk Between microRNAs and the Wnt/β-Catenin Signaling Pathway in Cancer. Oncotarget (2017) 8(8):14089–106. doi: 10.18632/oncotarget.12923
30. Pon JR, Marra MA. MEF2 Transcription Factors: Developmental Regulators and Emerging Cancer Genes. Oncotarget (2015) 7(3):2297–312. doi: 10.18632/oncotarget.6223
31. He MQ, Wan JF, Zeng HF, Tang YY, He MQ. miR-133a-5p Suppresses Gastric Cancer Through TCF4 Down-Regulation. J Gastrointest. Oncol (2021) 12(3):1007–19. doi: 10.21037/jgo-20-418
32. Feng H, Ge F, Du L, Zhang Z, Liu D. MiR - 34b - 3p Represses Cell Proliferation, Cell Cycle Progression and Cell Apoptosis in Non - Small - Cell Lung Cancer (NSCLC) by Targeting CDK4. J Cell Mol Med (2019) 23(8):5282–91. doi: 10.1111/jcmm.14404
33. Raver-shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, et al. Article Transcriptional Activation of miR-34a Contributes to P53-Mediated Apoptosis. Mol Cell (2007) 26(5):731–43. doi: 10.1016/j.molcel.2007.05.017
34. Liu C, Kelnar K, Liu B, Chen X, Calhoun-Davis T, Li H, et al. Identification of miR-34a as a Potent Inhibitor of Prostate Cancer Progenitor Cells and Metastasis by Directly Repressing CD44 HHS Public Access. Nat Med (2011) 17(2):211–5. doi: 10.1038/nm.2284
35. Rokavec M, Öner MG, Li H, Jackstadt R, Jiang L, Lodygin D, et al. Promotes EMT-Mediated Colorectal Cancer Invasion and Metastasis. J Clin Invest (2014) 124(4):1853–67. doi: 10.1172/JCI73531DS1
36. Okada N, Lin CP, Ribeiro MC, Biton A, Lai G, He X, et al. A Positive Feedback Between P53 and miR-34 miRNAs Mediates Tumor Suppression. Genes Dev (2014) 28(5):438–50. doi: 10.1101/gad.233585.113
37. Pardoll DM. The Blockade of Immune Checkpoints in Cancer Immunotherapy. Nat Rev Cancer (2016) 12: (4):252–64. doi: 10.1038/nrc3239.The
38. Cortez MA, Ivan C, Valdecanas D, Wang X, Peltier HJ, Ye Y, et al. PDL1 Regulation by P53 via miR-34. J Natl Cancer Inst (2016) 108(1):djv303. doi: 10.1093/jnci/djv303
39. Wang X, Li J, Dong K, Lin F, Long M, Ouyang Y, et al. Tumor Suppressor miR-34a Targets PD-L1 and Functions as a Potential Immunotherapeutic Target in Acute Myeloid Leukemia. Cell Signal (2015) 27(3):443–52. doi: 10.1016/j.cellsig.2014.12.003
40. Roush S, Slack FJ. The Let-7 Family of microRNAs. Trends Cell Biol (2008) 18(10):505–16. doi: 10.1016/j.tcb.2008.07.007
41. Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, et al. Let-7 Regulates Self Renewal and Tumorigenicity of Breast Cancer Cells. Cell (2007) 131(6):1109–23. doi: 10.1016/j.cell.2007.10.054
42. Wu A, Wu K, Li J, Mo Y, Lin Y, Wang Y, et al. Let-7a Inhibits Migration, Invasion and Epithelial-Mesenchymal Transition by Targeting HMGA2 in Nasopharyngeal Carcinoma. J Transl Med (2015) 13(1):1–13. doi: 10.1186/s12967-015-0462-8
43. Guo M, Zhao X, Yuan X, Jiang J, Li P. MiR-Let-7a Inhibits Cell Proliferation, Migration, and Invasion by Down-Regulating PKM2 in Cervical Cancer. Oncotarget (2017) 8(17):28226–36. doi: 10.18632/oncotarget.15999
44. Yang N, Kaur S, Volinia S, Greshock J, Lassus H, Hasegawa K, et al. MicroRNA Microarray Identifies Let-7i as a Novel Biomarker and Therapeutic Target in Human Epithelial Ovarian Cancer. Cancer Res (2008) 68(24):10307–14. doi: 10.1158/0008-5472.CAN-08-1954
45. Peng F, Li TT, Wang KL, Xiao GQ, Wang JH, Zhao HD, et al. H19/let-7/LIN28 Reciprocal Negative Regulatory Circuit Promotes Breast Cancer Stem Cell Maintenance. Cell Death Dis (2017) 8(1):e2569. doi: 10.1038/cddis.2016.438
46. Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, et al. RAS is Regulated by the Let-7 microRNA Family. Cell (2005) 120(5):635–47. doi: 10.1016/j.cell.2005.01.014
47. Worringer KA, Rand TA, Hayashi Y, Sami S, Takahashi K, Tanabe K, et al. The Let-7/LIN-41 Pathway Regulates Reprogramming to Human Induced Pluripotent Stem Cells by Controlling Expression of Prodifferentiation Genes. Cell Stem Cell (2014) 14(1):40–52. doi: 10.1016/j.stem.2013.11.001
48. Chang C, Hsu CC, Chang CH, Tsai LL, Chang YC, Lu SW, et al. Let-7d Functions as Novel Regulator of Epithelial-Mesenchymal Transition and Chemoresistant Property in Oral Cancer. Oncol Rep (2011) 110:1003–10. doi: 10.3892/or.2011.1360
49. Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, et al. MicroRNA Genes are Transcribed by RNA Polymerase II. EMBO J (2004) 23(20):4051–60. doi: 10.1038/sj.emboj.7600385
50. Krol J, Loedige I, Filipowicz W. The Widespread Regulation of microRNA Biogenesis, Function and Decay. Nat Rev Genet (2010) 11(9):597–610. doi: 10.1038/nrg2843
51. Allegra D, Bilan V, Garding A, Döhner H, Stilgenbauer S, Kuchenbauer F, et al. Defective DROSHA Processing Contributes to Downregulation of MiR-15/-16 in Chronic Lymphocytic Leukemia. Leukemia (2014) 28(1):98–107. doi: 10.1038/leu.2013.246
52. Torres A, Torres K, Paszkowski T, Jodłowska-Jędrych B, Radomański T, Książek A, et al. Major Regulators of microRNAs Biogenesis Dicer and Drosha are Down-Regulated in Endometrial Cancer. Tumor Biol (2011) 32(4):769–76. doi: 10.1007/s13277-011-0179-0
53. Su X, Chakravarti D, Cho MS, Liu L, Gi YJ, Lin YL, et al. TAp63 Suppresses Metastasis Through Coordinate Regulation of Dicer and miRNAs. Nature (2010) 467(7318):986–90. doi: 10.1038/nature09459
54. Martello G, Rosato A, Ferrari F, Manfrin A, Cordenonsi M, Dupont S, et al. A microRNA Targeting Dicer for Metastasis Control. Cell (2010) 141(7):1195–207. doi: 10.1016/j.cell.2010.05.017
55. Rupaimoole R, Ivan C, Yang D, Gharpure KM, Wu SY, Pecot CV, et al. Hypoxia-Upregulated microRNA-630 Targets Dicer, Leading to Increased Tumor Progression. Oncogene (2016) 35(33):4312–20. doi: 10.1038/onc.2015.492
56. Chiosea S, Jelezcova E, Chandran U, Acquafondata M, McHale T, Sobol RW, et al. Up-Regulation of Dicer, a Component of the microRNA Machinery, in Prostate Adenocarcinoma. Am J Pathol (2006) 169(5):1812–20. doi: 10.2353/ajpath.2006.060480
57. Martin MG, Payton JE, Link DC. Dicer and Outcomes in Patients With Acute Myeloid Leukemia (AML). Leuk Res (2009) 33(8):63110. doi: 10.1016/j.leukres.2009.02.003
58. Chiosea S, Jelezcova E, Chandran U, Luo J, Mantha G, Sobol RW, et al. Overexpression of Dicer in Precursor Lesions of Lung Adenocarcinoma. Cancer Res (2007) 67(5):2345–50. doi: 10.1158/0008-5472.CAN-06-3533
59. Shen J, Xia W, Khotskaya YB, Huo L, Nakanishi K, Lim SO, et al. EGFR Modulates microRNA Maturation in Response to Hypoxia Through Phosphorylation of AGO2. Nature (2013) 497(7449):383–7. doi: 10.1038/nature12080
60. Sun HL, Cui R, Zhou J, Teng KY, Hsiao YH, Nakanishi K, et al. ERK Activation Globally Downregulates miRNAs Through Phosphorylating Exportin-5. Cancer Cell (2016) 30(5):723–36. doi: 10.1016/j.ccell.2016.10.001
61. Li J, Pu W, Sun HL, Zhou JK, Fan X, Zheng Y, et al. Pin1 Impairs microRNA Biogenesis by Mediating Conformation Change of XPO5 in Hepatocellular Carcinoma. Cell Death Differ (2018) 25(9):1612–24. doi: 10.1038/s41418-018-0065-z
62. Lee KH, Lin FC, Hsu TI, Lin JT, Guo JH, Tsai CH, et al. MicroRNA-296-5p (miR-296-5p) Functions as a Tumor Suppressor in Prostate Cancer by Directly Targeting Pin1. Biochim Biophys Acta - Mol Cell Res (2014) 1843(9):2055–66. doi: 10.1016/j.bbamcr.2014.06.001
63. Chen L, Xu X, Wen X, Xu S, Wang L, Lu W, et al. Targeting PIN1 Exerts Potent Antitumor Activity in Pancreatic Ductal Carcinoma via inhibiting Tumor Metastasis. Cancer Sci (2019) 110(8):2442–55. doi: 10.1111/cas.14085
64. Tang X, Long Y, Xu L, Yan X. Lncrna Mort Inhibits Cancer Cell Proliferation and Promotes Apoptosis in Mantle Cell Lymphoma by Upregulating Mirna-16. Cancer Manage Res (2020) 12:2119–25. doi: 10.2147/CMAR.S233859
65. Li X, Chen N, Zhou L, Wang C, Wen X, Jia L, et al. Genome-Wide Target Interactome Profiling Reveals a Novel EEF1A1 Epigenetic Pathway for Oncogenic lncRNA MALAT1 in Breast Cancer. Am J Cancer Res (2019) 9(4):714–29.
66. Liang Y, Song X, Li Y, Chen B, Zhao W, Wang L, et al. LncRNA BCRT1 Promotes Breast Cancer Progression by Targeting miR-1303/PTBP3 Axis. Mol Cancer (2020) 19(1):1–20. doi: 10.1186/s12943-020-01206-5
67. Song W, Mei JZ, Zhang M. Long Noncoding RNA PlncRNA-1 Promotes Colorectal Cancer Cell Progression by Regulating the PI3K/Akt Signaling Pathway. Oncol Res (2018) 26(2):261–8. doi: 10.3727/096504017X15031557924132
68. Xu Y, Zhang G, Zou C, Zhang H, Gong Z, Wang W, et al. LncRNA MT1JP Suppresses Gastric Cancer Cell Proliferation and Migration Through MT1JP/MiR-214-3p/RUNX3 Axis. Cell Physiol Biochem (2018) 46(6):2445–59. doi: 10.1159/000489651
69. Zhang Y, Liu X, Li Q, Zhang Y. IncRNA LINC00460 Promoted Colorectal Cancer Cells Metastasis via miR-939-5p Sponging. Cancer Manage Res (2019) 11:1779–89. doi: 10.2147/CMAR.S192452
70. Ghafouri-Fard S, Gholipour M, Hussen BM, Taheri M. The Impact of Long Non-Coding RNAs in the Pathogenesis of Hepatocellular Carcinoma. Front Oncol (2021) 11:649107. doi: 10.3389/fonc.2021.649107
71. Zhou H, Shi P, Jia X, Xue Q. Long Non−Coding RNA LINC01018 Inhibits the Progression of Acute Myeloid Leukemia by Targeting Mir−499a−5p to Regulate PDCD4. Oncol Lett (2021) 22(1):1–11. doi: 10.3892/ol.2021.12802
72. Chen Z, Zhong T, Li T, Zhong J, Tang Y, Liu Z, et al. LncRNA SNHG15 Modulates Gastric Cancer Tumorigenesis by Impairing miR-506-5p Expression. Biosci Rep (2021) 41(7):1–10. doi: 10.1042/bsr20204177
73. Jiang T, Wang C, Zhu Y, Han H. LINC01410 Promotes Cell Proliferation and Migration of Cholangiocarcinoma Through Modulating miR-124-3p/SMAD5 Axis. J Gene Med (2020) 22(6):1–9. doi: 10.1002/jgm.3162
74. Liu H, Chi Z, Jin H, Yang W. MicroRNA miR-188-5p as a Mediator of Long Non-Coding RNA MALAT1 Regulates Cell Proliferation and Apoptosis in Multiple Myeloma. Bioengineered (2021) 12(1):1611–26. doi: 10.1080/21655979.2021.1920325
75. Kim VN, Han J, Siomi MC. Biogenesis of Small RNAs in Animals. Nat Rev Mol Cell Biol (2009) 10(2):126–39. doi: 10.1038/nrm2632
76. Bosia C, Osella M, Caselle M, El Baroudi M, Cora D. A Curated Database of miRNA Mediated Feed-Forward Loops Involving MYC as Master Regulator. PLoS One (2011) 6(3):17–8. doi: 10.1371/journal.pone.0014742
77. Chang TC, Zeitels LR, Hwang HW, Chivukula R, Wentzel EA, Dews M, et al. Lin-28B Transactivation is Necessary for Myc-Mediated Let-7 Repression and Proliferation. Proc Natl Acad Sci USA (2009) 106(9):3384–9. doi: 10.1073/pnas.0808300106
78. Lee YM, Lee JY, Ho CC, Hong QS, Yu SL, Tzeng CR, et al. miRNA-34b as a Tumor Suppressor in Estrogen-Dependent Growth of Breast Cancer Cells. Breast Cancer Res (2011) 13(6):R116. doi: 10.1186/bcr3059
79. Cicatiello L, et al. Estrogen Receptor α Controls a Gene Network in Luminal-Like Breast Cancer Cells Comprising Multiple Transcription Factors and microRNAs. Am J Pathol (2010) 176(5):2113–30. doi: 10.2353/ajpath.2010.090837
80. Chen Y, Zhou Y, Han F, Zhao Y, Tu M, Wang Y, et al. A Novel miR-1291-Errα-CPT1C Axis Modulates Tumor Cell Proliferation, Metabolism and Tumorigenesis. Theranostics (2020) 10(16):7193–210. doi: 10.7150/thno.44877
81. Tan S, Ding K, Li R, Zhang W, Li G, Kong X, et al. Identification of miR-26 as a Key Mediator of Estrogen Stimulated Cell Proliferation by Targeting CHD1, GREB1 and KPNA2. Breast Cancer Res (2014) 16(2):1–13. doi: 10.1186/bcr3644
82. Mori M, Triboulet R, Mohseni M, Schlegelmilch K, Shrestha K, Camargo FD, et al. Hippo Signaling Regulates Microprocessor and Links Cell-Density-Dependent Mirna Biogenesis to Cancer. Cell (2014) 156(5):893–906. doi: 10.1016/j.cell.2013.12.043
83. Huang X, Tang F, Weng Z, Zhou M, Zhang Q. MiR-591 Functions as Tumor Suppressor in Breast Cancer by Targeting TCF4 and Inhibits Hippo-YAP/TAZ Signaling Pathway. Cancer Cell Int (2019) 19(1):1–9. doi: 10.1186/s12935-019-0818-x
84. Yu SJ, Hu JY, Kuang XY, Luo JM, Hou YF, Di GH, et al. MicroRNA-200a Promotes Anoikis Resistance and Metastasis by Targeting YAP1 in Human Breast Cancer. Clin Cancer Res (2013) 19(6):1389–99. doi: 10.1158/1078-0432.CCR-12-1959
85. Wang Y, Cui M, Di Sun B, Liu FB, Zhang XD, Ye LH. MiR-506 Suppresses Proliferation of Hepatoma Cells Through Targeting YAP mRNA 3′UTR. Acta Pharmacol Sin (2014) 35(9):1207–14. doi: 10.1038/aps.2014.59
86. Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, et al. Transactivation of miR-34a by P53 Broadly Influences Gene Expression and Promotes Apoptosis. Mol Cell (2007) 26(5):745–52. doi: 10.1016/j.molcel.2007.05.010
87. He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, et al. A microRNA Component of the P53 Tumour Suppressor Network. Nature (2007) 447(7148):1130–4. doi: 10.1038/nature05939
88. Di Gennaro A, Damiano N, Brisotto N, Armellin N, Perin N, Zucchetto N, et al. A P53/miR-30a/ZEB2 Axis Controls Triple Negative Breast Cancer Aggressiveness. Cell Death Differ (2018) 25(12):2165–80. doi: 10.1038/s41418-018-0103-x
89. Zhang N, Wang X, Huo Q, Sun M, Cai C, Liu Z, et al. MicroRNA-30a Suppresses Breast Tumor Growth and Metastasis by Targeting Metadherin. Oncogene (2014) 33(24):3119–28. doi: 10.1038/onc.2013.286
90. Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA Processing by P53. Nature (2009) 460(7254):529–33. doi: 10.1038/nature08199
91. Sai X, Qin C, Wu Y, Zhao Y, Bian T. Downregulation of PTEN Mediates Bleomycin-Induced Premature Senescence in Lung Cancer Cells by Suppressing Autophagy. J Int Med Res (2020) 48(5):1–12. doi: 10.1177/0300060520923522
92. Donehower LA, Soussi T, Korkut A, Liu Y, Schultz A, Cardenas M, et al. Integrated Analysis of TP53 Gene and Pathway Alterations in The Cancer Genome Atlas. Cell Rep (2019) 28(5):1370–84.e5. doi: 10.1016/j.celrep.2019.07.001
93. Petitjean A, Achatz MIW, Borresen-Dale AL, Hainaut P, Olivier M. TP53 Mutations in Human Cancers: Functional Selection and Impact on Cancer Prognosis and Outcomes. Oncogene (2007) 26(15):2157–65. doi: 10.1038/sj.onc.1210302
94. Muller PAJ, Vousden KH. Mutant P53 in Cancer: New Functions and Therapeutic Opportunities. Cancer Cell (2014) 25(3):304–17. doi: 10.1016/j.ccr.2014.01.021
95. Donehower LA, Lozano G. 20 Years Studying P53 Functions in Genetically Engineered Mice. Nat Rev Cancer (2009) 11:831–41. doi: 10.1038/nrc2731
96. Donehower LA. Insights Into Wild-Type and Mutant P53 Functions Provided by Genetically Engineered Mice. Hum Mutat (2014) 35(6):715–27. doi: 10.1002/humu.22507
97. Gurtner A, Starace G, Norelli G, Piaggio G, Sacchi A, Bossi G. Mutant P53-Induced Up-Regulation of Mitogen-Activated Protein Kinase Kinase 3 Contributes to Gain of Function. J Biol Chem (2010) 285(19):14160–9. doi: 10.1074/jbc.M109.094813
98. Gurtner A, Falcone E, Garibaldi F, Piaggio G. Dysregulation of microRNA Biogenesis in Cancer: The Impact of Mutant P53 on Drosha Complex Activity. J Exp Clin Cancer Res (2016) 35(1):1–9. doi: 10.1186/s13046-016-0319-x
99. Jiang FZ, He YY, Wang HH, Zhang HL, Zhang J, Yan XF, et al. Mutant P53 Induces EZH2 Expression and Promotes Epithelial-Mesenchymal Transition by Disrupting P68-Drosha Complex Assembly and Attenuating miR-26a Processing. Oncotarget (2015) 6(42):44660–74. doi: 10.18632/oncotarget.6350
100. Garibaldi F, Falcone E, Trisciuoglio D, Colombo T, Lisek K, Walerych D, et al. Mutant P53 Inhibits miRNA Biogenesis by Interfering With the Microprocessor Complex. Oncogene (2016) 35(29):3760–70. doi: 10.1038/onc.2016.51
101. Meng X, Sun W, Yu J, Zhou Y, Gu Y, Han J, et al. LINC00460-miR-149-5p/miR-150-5p-Mutant P53 Feedback Loop Promotes Oxaliplatin Resistance in Colorectal Cancer. Mol Ther - Nucleic Acids (2020) 22:1004–15. doi: 10.1016/j.omtn.2020.10.018
102. Tsai K, Wu CW, Hu LY, Li SC, Liao YL, Lai CH, et al. Epigenetic Regulation of miR-34b and miR-129 Expression in Gastric Cancer. Int J Cancer (2011) 2610(11):2600–10. doi: 10.1002/ijc.25919
103. Ushijima T, Asada K. Aberrant DNA Methylation in Contrast with Mutations. Cancer Sci (2009) 101(2):300–5. doi: 10.1111/j.1349-7006.2009.01434.x
104. Suzuki H, Maruyama R, Yamamoto E, Kai M. DNA Methylation and microRNA Dysregulation in Cancer. Mol Oncol (2012) 6(6):567–78. doi: 10.1016/j.molonc.2012.07.007
105. Lujambio A, Calin GA, Villanueva A, Ropero S, Sánchez-Céspedes M, Blanco D, et al. A microRNA DNA Methylation Signature for Human Cancer Metastasis. Proc Natl Acad Sci USA (2008) 105(36):13556–61. doi: 10.1073/pnas.0803055105
106. Lim YY, Wright JA, Attema JL, Gregory PA, Bert AG, Smith E, et al. Epigenetic Modulation of the miR-200 Family is Associated With Transition to a Breast Cancer Stem-Celllike State. J Cell Sci (2013) 126(10):2256–66. doi: 10.1242/jcs.122275
107. Xu L, Lin J, Deng W, Luo W, Huang Y, Liu CQ, et al. EZH2 Facilitates BMI1-Dependent Hepatocarcinogenesis Through Epigenetically Silencing microRNA-200c. Oncogenesis (2020) 9(11):101. doi: 10.1038/s41389-020-00284-w
108. Tong D, Zhang J, Wang X, Li Q, Liu L, Lu A, et al. MiR-22, Regulated by MeCP2, Suppresses Gastric Cancer Cell Proliferation by Inducing a Deficiency in Endogenous S-Adenosylmethionine. Oncogenesis (2020) 9(11):99. doi: 10.1038/s41389-020-00281-z
109. Wong KY, Yim RLH, So CC, Jin DY, Liang R, Chim CS. Epigenetic Inactivation of the MIR34B/C in Multiple Myeloma. Blood (2011) 118(22):5901–4. doi: 10.1182/blood-2011-06-361022
110. Peinado P, Andrades A, Martorell-Marugán J, Haswell JR, Slack FJ, Carmona-Sáez P, et al. The SWI/SNF Complex Regulates the Expression of miR-222, a Tumor Suppressor microRNA in Lung Adenocarcinoma. Hum Mol Genet (2021) 00(00):1–9. doi: 10.1093/hmg/ddab187
111. Masliah-Planchon J, Bièche I, Guinebretière JM, Bourdeaut F, Delattre O. SWI/SNF Chromatin Remodeling and Human Malignancies. Annu Rev Pathol Mech Dis (2015) 10:145–71. doi: 10.1146/annurev-pathol-012414-040445
112. Galka-Marciniak P, Urbanek-Trzeciak MO, Nawrocka PM, Dutkiewicz A, Giefing M, Lewandowska MA, et al. Somatic Mutations in miRNA Genes in Lung Cancer—Potential Functional Consequences of Non-Coding Sequence Variants. Cancers (Basel) (2019) 11(6):1–26. doi: 10.3390/cancers11060793
113. Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, et al. MicroRNAs Exhibit High Frequency Genomic Alterations in Human Cancer. Proc Natl Acad Sci U S A (2006) 103(24):9136–41. doi: 10.1073/pnas.0508889103.
114. Albertson DG, Collins C, McCormick F, Gray JW. Chromosome Aberrations in Solid Tumors. Nat Genet (2003) 34(4):369–76. doi: 10.1038/ng1215
115. Czubak K, Lewandowska MA, Klonowska K, Roszkowski K, Kowalewski J, Figlerowicz M, et al. High Copy Number Variation of Cancer-Related microRNA Genes and Frequent Amplification of DICER1 and DROSHA in Lung Cancer. Oncotarget (2015) 6(27):23399–416. doi: 10.18632/oncotarget.4351
116. Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer Genome Landscapes. Sci (80-) (2013) 340(6127):1546–58. doi: 10.1126/science.1235122
117. Wang Y, Chen J, Yang W, Mo F, Senz J, Yap D, et al. The Oncogenic Roles of DICER1 RNase IIIb Domain Mutations in Ovarian Sertoli-Leydig Cell Tumors. Neoplasia (2015) 17(8):650–60. doi: 10.1016/j.neo.2015.08.003
118. Dennis DA, Ivanovich J, Priest JR, Gurnett CA, Dehner LP, Desruisseau D, et al. Germline DICER1 Mutations in Familial Pleuropulmonary Blastoma. Sci (80-) (2012) 127(5943):358–66. doi: 10.1016/j.jsbmb.2011.07.002.Identification
119. Chen J, Wang Y, McMonechy MK, Anglesio MS, Yang W, Senz J, et al. Recurrent DICER1 Hotspot Mutations in Endometrial Tumours and Their Impact on microRNA Biogenesis. J Pathol (2015) 237(2):215–25. doi: 10.1002/path.4569
120. Wu MK, Vujanic GM, Fahiminiya S, Watanabe N, Thorner PS, O'Sullivan MJ, et al. Anaplastic Sarcomas of the Kidney are Characterized by DICER1 Mutations. Mod Pathol (2018) 31(1):169–78. doi: 10.1038/modpathol.2017.100
121. Bean GR, Anderson J, Sangoi AR, Krings G, Garg K. DICER1 Mutations are Frequent in Müllerian Adenosarcomas and Are Independent of Rhabdomyosarcomatous Differentiation. Mod Pathol (2019) 32(2):280–9. doi: 10.1038/s41379-018-0132-5
122. Kato N, Kusumi T, Kamataki A, Tsunoda R, Fukase M, Kurose A. DICER1 Hotspot Mutations in Ovarian Sertoli-Leydig Cell Tumors: A Potential Association With Androgenic Effects. Hum Pathol (2017) 59:41–7. doi: 10.1016/j.humpath.2016.09.005
123. Kumar MS, Pester RE, Chen CY, Lane K, Chin C, Lu J, et al. Dicer1 Functions as a Haploinsufficient Tumor Suppressor. Genes Dev (2009) 23(23):2700–4. doi: 10.1101/gad.1848209
124. Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, et al. Dicer-Deficient Mouse Embryonic Stem Cells Are Defective in Differentiation and Centromeric Silencing. Genes Dev (2005) 19(4):489–501. doi: 10.1101/gad.1248505
125. Merritt WM, Lin YG, Han LY, Kamat AA, Spannuth WA, Schmandt R, et al. Dicer, Drosha, and Outcomes in Patients With Ovarian Cancer. New Engl J (2008) 359(25):2641–50. doi: 10.1056/NEJMoa0803785
126. Vedanayagam J, Chatila WK, Aksoy BA, Majumdar S, Skanderup AJ, Demir E, et al. Cancer-Associated Mutations in DICER1 RNase IIIa and IIIb Domains Exert Similar Effects on miRNA Biogenesis. Nat Commun (2019) 10(1):3682. doi: 10.1038/s41467-019-11610-1
127. Chou J, Lin JH, Brenot A, Kim JW, Provot S, Werb Z. GATA3 Suppresses Metastasis and Modulates the Tumour Microenvironment by Regulating microRNA-29b Expression. Nat Cell Biol (2013) 15(2):201–13. doi: 10.1038/ncb2672
128. Melo SA, Moutinho C, Ropero S, Calin GA, Rossi S, Spizzo R, et al. A Genetic Defect in Exportin-5 Traps Precursor MicroRNAs in the Nucleus of Cancer Cells. Cancer Cell (2010) 18(4):303–15. doi: 10.1016/j.ccr.2010.09.007
129. Xing Y, Ruan G, Ni H, Qin H, Chen S, Gu X, et al. Tumor Immune Microenvironment and Its Related miRNAs in Tumor Progression. Front Immunol (2021) 12:624725. doi: 10.3389/fimmu.2021.624725
130. Li BL, Lu W, Qu JJ, Ye L, Du GQ, Wan XP. Loss of Exosomal miR-148b From Cancer-Associated Fibroblasts Promotes Endometrial Cancer Cell Invasion and Cancer Metastasis. J Cell Physiol (2019) 234(3):2943–53. doi: 10.1002/jcp.27111
131. Purvis IJ, Avilala J, Guda MR, Venkataraman S, Vibhakar R, Tsung AJ, et al. Role of MYC-miR-29-B7-H3 in Medulloblastoma Growth and Angiogenesis. J Clin Med (2019) 8(8):1158. doi: 10.3390/jcm8081158
132. Gregorio A, Corrias MV, Castriconi R, Dondero A, Mosconi M, Gambini C, et al. Small Round Blue Cell Tumours: Diagnostic and Prognostic Usefulness of the Expression of B7-H3 Surface Molecule. Histopathology (2008) 53(1):73–80. doi: 10.1111/j.1365-2559.2008.03070.x
133. Mesri MJ, Gillette M, Carr SA, Paulovich AG, Smith RD, Rodland KK, et al. Identification and Characterization of Angiogenesis Targets Through Proteomic Profiling of Endothelial Cells in Human Cancer Tissues. PloS One (2013) 8(11). doi: 10.1371/journal.pone.0078885
134. Ma J, Ma P, Zhao C, Xue X, Han H, Liu C, et al. B7-H3 as a Promising Target for Cytotoxicity T Cell in Human Cancer Therapy. Oncotarget (2016) 7(20):29480–91. doi: 10.18632/oncotarget.8784
135. Bieg D, Sypniewski D, Nowak E, Bednarek I. MiR − 424 − 3p Suppresses Galectin − 3 Expression and Sensitizes Ovarian Cancer Cells to Cisplatin. Arch Gynecol Obstet (2019) 299(4):1077–87. doi: 10.1007/s00404-018-4999-7
136. Bhushan R, Grünhagen J, Becker J, Robinson PN, Ott CE, Knaus P. The International Journal of Biochemistry miR-181a Promotes Osteoblastic Differentiation Through Repression of TGF- β Signaling Molecules. Int J Biochem Cell Biol (2013) 45(3):696–705. doi: 10.1016/j.biocel.2012.12.008
137. Shen G, Li X, Jia YF, Piazza GA, Xi Y. Hypoxia-Regulated microRNAs in Human Cancer. Acta Pharmacol Sin (2013) 34(3):336–41. doi: 10.1038/aps.2012.195
138. Chan YC, Khanna S, Roy S, Sen CK. MiR-200b Targets Ets-1 and is Down-Regulated by Hypoxia to Induce Angiogenic Response of Endothelial Cells. J Biol Chem (2011) 286(3):2047–56. doi: 10.1074/jbc.M110.158790
139. Jafarifar F, Yao P, Fox PL. Repression of VEGFA by CA-Rich Element-Binding microRNAs Is Modulated by hnRNP L. EMBO J (2011) 30(7):1324–34. doi: 10.1038/emboj.2011.38
140. Hua Z, Lv Q, Ye W, Wong CK, Cai G, Gu D, et al. MiRNA-Directed Regulation of VEGF and Other Angiogenic Factors Under Hypoxia. PLoS One (2006) 1(1):e116. doi: 10.1371/journal.pone.0000116
141. Van Den Beucken T, Koch E, Chu K, Rupaimoole R, Prickaerts P, Adriaens M, et al. Europe PMC Funders Group Hypoxia Promotes Stem Cell Phenotypes and Poor Prognosis Through Epigenetic Regulation of DICER. (2015) 5:5203. doi: 10.1038/ncomms6203.Hypoxia
142. Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as Stable Blood-Based Markers for Cancer Detection. Proc Natl Acad Sci USA (2008) 105(30):10513–8. doi: 10.1073/pnas.0804549105
143. Shell S, Park SM, Radjabi AR, Schickel R, Kistner EO, Jewell DA, et al. Let-7 Expression Defines Two Differentiation Stages of Cancer. Proc Natl Acad Sci USA (2007) 104(27):11400–5. doi: 10.1073/pnas.0704372104
144. Tang Z, Ow GS, Thiery JP, Ivshina AV, Kuznetsov VA. Prognostic Biomarker and Predicts Molecular and Clinical Subclasses in High-Grade Serous Ovarian Carcinoma. Int J Cancer (2014) 318:306–18. doi: 10.1002/ijc.28371
145. Bottani M, Banfi G, Lombardi G. Circulating miRNAs as Diagnostic and Prognostic Biomarkers in Common Solid Tumors: Focus on Lung, Breast, Prostate Cancers, and Osteosarcoma. J Clin Med (2019) 8(10):1661. doi: 10.3390/jcm8101661
146. Han J, Yu J, Dai Y, Li J, Guo M, Song J, et al. Overexpression of miR-361-5p in Triple-Negative Breast Cancer (TNBC) Inhibits Migration and Invasion by Targeting RQCD1 and Inhibiting the EGFR/PI3K/Akt Pathway. Bosn. J Basic Med Sci (2019) 19(1):52–9. doi: 10.17305/BJBMS.2018.3399
147. Liu J, Yang J, Yu L, Rao C, Wang Q, Sun C, et al. MiR-361-5p Inhibits Glioma Migration and Invasion by Targeting SND1. Onco. Targets. Ther (2018) 11:5239–52. doi: 10.2147/OTT.S171539
148. Ma F, Song H, Guo B, Zhang Y, Zheng Y, Lin C, et al. MiR-361-5p Inhibits Colorectal and Gastric Cancer Growth and Metastasis by Targeting Staphylococcal Nuclease Domain Containing-1. Oncotarget (2015) 6(19):17404–16. doi: 10.18632/oncotarget.3744
149. Tang Z, Ow GS, Thiery JP, Ivshina AV, Kuznetsov VA. Meta-Analysis of Transcriptome Reveals Let-7b as an Unfavorable Prognostic Biomarker and Predicts Molecular and Clinical Subclasses in High-Grade Serous Ovarian Carcinoma. Int J Cancer (2014) 134(2):306–18. doi: 10.1002/ijc.28371
150. Saleeb R, Kim SS, Ding Q, Scorilas A, Lin S, Khella HW, et al. The miR-200 Family as Prognostic Markers in Clear Cell Renal Cell Carcinoma. Urol. Oncol Semin Orig Investig (2019) 37(12):955–63. doi: 10.1016/j.urolonc.2019.08.008
151. Yu C, Wan H, Shan R, Wen W, Li J, Luo D, et al. The Prognostic Value of the MiR-200 Family in Colorectal Cancer: A Meta-Analysis With 1882 Patients. J Cancer (2019) 10(17):4009–16. doi: 10.7150/jca.27529
152. Yuan L, Bing Z, Yan P, Li R, Wang C. Integrative Data Mining and Meta-Analysis to Investigate the Prognostic Role of microRNA-200 Family in Various Human Malignant Neoplasms : A Consideration on Heterogeneity. Gene (2019) 716:144025. doi: 10.1016/j.gene.2019.144025
153. Huang GL, Sun J, Lu Y, Liu Y, Cao H, Zhang H, et al. MiR-200 Family and Cancer: From a Meta-Analysis View. Mol Aspects Med (2019) 70:57–71. doi: 10.1016/j.mam.2019.09.005
154. Jopling CL, Yi MK, Lancaster AM, Lemon SM, Sarnow P. Molecular Biology: Modulation of Hepatitis C Virus RNA Abundance by a Liver-Specific MicroRNA. Sci (80-) (2005) 309(5740):1577–81. doi: 10.1126/science.1113329
155. Thibault PA, Huys A, Amador-Cañizares Y, Gailius JE, Pinel DE, Wilson JA. Regulation of Hepatitis C Virus Genome Replication by Xrn1 and MicroRNA-122 Binding to Individual Sites in the 5′ Untranslated Region. J Virol (2015) 89(12):6294–311. doi: 10.1128/jvi.03631-14
156. Rupaimoole R, Slack FJ. MicroRNA Therapeutics: Towards a New Era for the Management of Cancer and Other Diseases. Nat Rev Drug Discov (2017) 16(3):203–21. doi: 10.1038/nrd.2016.246
157. Wiggins JF, Ruffino L, Kelnar K, Omotola M, Patrawala L, Brown D, et al. Development of a Lung Cancer Therapeutic Based on the Tumor Suppressor microRNA-34. Cancer Res (2010) 70(14):5923–30. doi: 10.1158/0008-5472.CAN-10-0655
158. Stahlhut C, Slack FJ. Combinatorial Action of MicroRNAs Let-7 and miR-34 Effectively Synergizes With Erlotinib to Suppress Non-Small Cell Lung Cancer Cell Proliferation. Cell Cycle (2015) 14(13):2171–80. doi: 10.1080/15384101.2014.1003008
Keywords: cancer, microRNA, tumor suppressor miRNA, tumor microenvironment, functional regulation
Citation: Otmani K and Lewalle P (2021) Tumor Suppressor miRNA in Cancer Cells and the Tumor Microenvironment: Mechanism of Deregulation and Clinical Implications. Front. Oncol. 11:708765. doi: 10.3389/fonc.2021.708765
Received: 12 May 2021; Accepted: 27 September 2021;
Published: 15 October 2021.
Edited by:
Nan-Shan Chang, National Cheng Kung University, TaiwanReviewed by:
Giulia Piaggio, Regina Elena National Cancer Institute (IRCCS), ItalyCopyright © 2021 Otmani and Lewalle. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Khalid Otmani, b3RtYW5pX2toYWxpZDAwMUBob3RtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.