- Department of Medical Ultrasound, Affiliated Hospital of Jiangsu University, Zhenjiang, China
Despite accumulating cell- or animal-based experiments providing the relationship between Gasdermin E (GSDME) and human diseases, especially in malignant cancers, no pan-cancer analysis about the function of GSMDE in cancer management can be available up to date. Our research, for the first time, explored the potential carcinogenic role of GSDME across 33 tumors from the public platform of TCGA (The cancer genome atlas) database. GSDME is highly expressed in most malignant cancers, and obvious relationship exists between GSDME level and survival prognosis of cancer patients. The expression of GSDME was statically associated with the cancer-associated fibroblast infiltration in diverse cancer types, such as BLCA, CHOL, GBM, KIRC, LIHC, MESO, STAD, and UCEC. Furthermore, pyroptosis, sensory perception of sound, and defense response to bacterium were involved in the functional mechanisms of GSDME expression from GO analysis. Last but not the least, in vitro experiments were also performed to identify GSDME-induced pyroptosis. Our first pan-cancer analysis of GSDME not only broadens the understanding of the carcinogenic roles of GSDME but also provides a promising therapeutic strategy for benefiting an increasing number of cancerous patients based on GSDME-induced pyroptosis.
Introduction
As ranking second among human diseases in the United States, malignant cancer has become a major threat to human life worldwide, which has raised great attention of scientists in different fields (1, 2). In view of the sophistication of carcinogenesis, it is extremely urgent to accomplish a pan-cancer analysis of any interested genes and exploit their association with potential molecular mechanisms and underlying clinical prognosis (3). Although great efforts have been devoted in early diagnosis of diverse kinds of cancers and the adoption of innovative approaches to decrease the mortality, the incidence of malignant cancer remains a major concern of human health (4, 5). Fortunately, with the advanced progress of genome-wide sequencing technology, an amount of functional genomics databases can be available from public platforms, such as The Cancer Genome Atlas (TCGA) project (6–9).
The Gasdermin E (GSDME), originally known as deafness, autosomal dominant 5 (DFNA5), was discovered on chromosome 7p15.3 with a peculiar form of autosomal dominant, progressive, sensorineural, and non-syndromic deafness (10). GSDME is one of the Gasdermin (GSDM) families, which possesses its nomenclature to the high-expression pattern along the gastrointestinal tract and skin (dermis) (11–16). It is interesting that GSDME can be cleaved by caspase-3 into pore-formation GSDME-N domain, as a result converting noninflammatory apoptosis to inflammatory pyroptosis in cancer cells (17). On one hand, GSDME is low expression in most cancers and decreased GSDME levels are also related to reduced survival prognosis, indicating that GSDME might function as a role of tumor suppressor (18, 19). However, on other hand, the inflammatory factors released during dysregulation of pyroptosis are widely associated with the tumorigenesis, as well as their drug resistance to chemotherapy (20–22). The function of GSDME in cancer has been increasingly prominent as the investigations has advanced. The structure and function research of GSDME has also been carried out from the aspects of physiology and pathology in various cancers; however, there is still no pan-cancer analysis about association between GSDME and different cancer types from public clinical information.
Our research, for the first time, utilized the TCGA database to perform a pan-cancer expression analysis of GSDME and included a series of relevant studies, including GSDME expression, clinical survival prognosis, immune cell infiltration, and potential signaling pathway, to explore the underlying mechanism in the tumorigenesis and tumor suppression across different cancer species. We strongly believe that the analysis of GSDME could assist in predicting the survival of cancer patients and further deepen our understanding of the individual management for cancer precision therapy.
Materials And Methods
Raw Data Acquisition and Processing
We entered DFNA5 into the “Gene_DE” module of TIMER2 website (http://timer.cistrome.org/). Then we scanned DFNA5 expression difference between malignant cancers and corresponding normal tissues for diverse cancers and certain subtypes. To solve the imbalance between the tumor and normal data, which can cause inefficiency in various differential analyses, TCGA and GTEx gene expression data were available from GEPIA (Gene Expression Profiling Interactive Analysis) web server (http://gepia.cancer-pku.cn/#analysis) that are re-computed from raw RNA-Seq data by the UCSC Xena project based on a uniform pipeline, thus allowing for the formation of the most comprehensive expression information (23). Herein, for specific cancers with limited normal and without normal tissues, we applied the “Expression analysis-BoxPlot” module and clicked “Match TCGA normal and GTEx data” module of GEPIA to analyze DFNA5 level between the malignant cancers and adjacent normal tissues.
Survival Analysis and Relationship With Clinical Stage
The overall survival (OS) and disease-free survival (DFS) information of GSDME within all TCGA cancers were obtained from the “Survival Map” module of GEPIA2. Cutoff-high (50%) and cutoff-low (50%) values were utilized to be thresholds to separate the high- and low-expression cases. In addition, the log-rank test was applied for the hypothesis test, and the survival curves were then graphed via the “Survival Analysis” module. Furthermore, the violin images of the GSDME in various pathological stages were also required from the “Pathological Stage Plot” module.
Genetic Alteration Analysis of GSDME
The genetic alteration features of DFNA5 were queried from the “TCGA Pan Cancer Atlas Studies” module of cBioPortal web (https://www.cbioportal.org/) (24, 25). The information of the alteration frequency, mutation type, and copy number alteration (CNA) were shown in the “Cancer Types Summary” module.
GSDME-Related Immune Infiltration Analysis
The immune infiltration cells of CD4+ T-cells and cancer-associated fibroblasts were chosen for immune infiltration evaluation by through TIMER, CIBERSORT, XCELL, EPIC, QUANTISEQ, MCPCOUNTER, and TIDE algorithms. Correlation (cor) values and P-values were also calculated by purity-adjusted Spearman’s rank correlation test.
GSDME-Related Gene Enrichment Analysis
The determined GSDME-binding proteins were downloaded from STRING (https://string-db.org/). We picked up the top 100 GSDME-binding genes from TCGA all cancers and adjacent normal tissues via “Similar Gene Detection” module. Then the “correlation analysis” module was used for achieving a pairwise gene Pearson correlation analysis of GSDME and these correlated genes. Furthermore, we applied the “Gene_Corr” module to image the heatmap information of these determined genes. Finally, these gene lists were uploaded into DAVID website (database for annotation, visualization, and integrated discovery) to perform the (gene ontology) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. The results with P <0.05 were considered as statistically significant, providing credibility for the data analysis.
Results
As graphed in Figure 1A, the expression level of GSDME in malignant cancers of cholangiocarcinoma (CHOL), glioblastoma multiforme (GBM), head and neck squamous cell carcinoma (HNSC), kidney renal papillary cell carcinoma (KIRP), liver hepatocellular carcinoma (LIHC), lung squamous cell carcinoma (LUSC), pheochromocytoma and paraganglioma (PCPG), stomach adenocarcinoma (STAD) is higher than their adjacent normal tissues (P<0.05). After including the normal cases from GTEx project as controls, our group proceeded to perform the expression analysis of GSDME level between the adjacent normal and cancer tissues of adrenocortical carcinoma (ACC), lymphoid neoplasm diffuse large B-cell lymphoma (DLBC), acute myeloid leukemia (LAML), brain lower grade glioma (LGG), ovarian cancer (OV), skin cutaneous melanoma (SKCM), testicular germ cell tumors (TGCT), sarcoma (SARC), thymoma (THYM), and uterine carcinosarcoma (UCS) (Figure 1B, P<0.05).
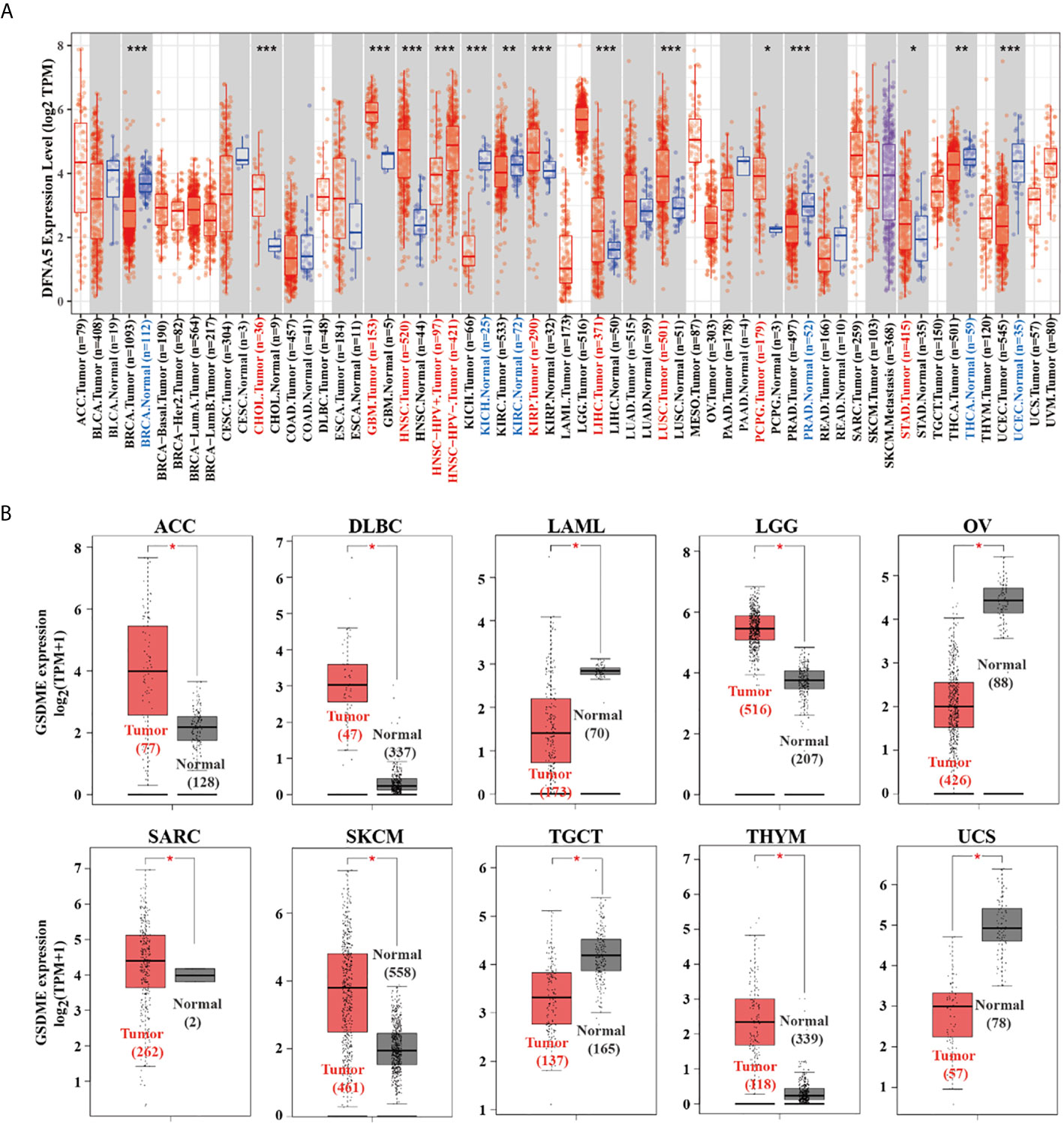
Figure 1 (A) The expression difference of the GSDME in diverse cancers or certain cancer subtypes was analyzed via TIMER2 algorithm. (B) For ACC, DLBC, LAML, LGG, OV, SARC, SKCM, TGCT, THYM, and UCS in the TCGA database, the adjacent normal tissues of the GTEx project were included as controls. The box plots were then graphed. *P < 0.05, **P < 0.01, ***P < 0.001.
Clinicopathological Stages and Prognostic Value of GSDME in Cancers
The “Pathological Stage Plot” module of GEPIA was used to analyze the relationship between GSDME expression and the pathological stages of different cancers, including BLCA, ESCA, KICH, KIRC, KIRP, READ, and UCEC (Figure 2A, all P value <0.05). We separated the cancer species into high- and low-expression groups, and then explored the association between GSDME level and the clinical prognosis of patients bearing various malignant cancers. As displayed in Figure 2B, high expression of GSDME was related to poorer prognosis of overall survival (OS) for malignancies, such as COAD (P=0.032), KIRC (P=0.0021), and KIRP (P=0.0081) across the TCGA database. Disease-free survival (DFS) analysis information revealed an association of high GSDME expression with poorer prognosis of COAD (P=0.0082) and KIRC (P=0.0012). Furthermore, low expression of the GSDME was also associated with poorer OS prognosis for ACC (P=0.026) and DFS prognosis for ACC (P=0.0019) and KIRP (P=0.0099). The abovementioned results suggested that GSDME expression is differentially correlated to the survival prognosis of patients bearing diverse cancers.
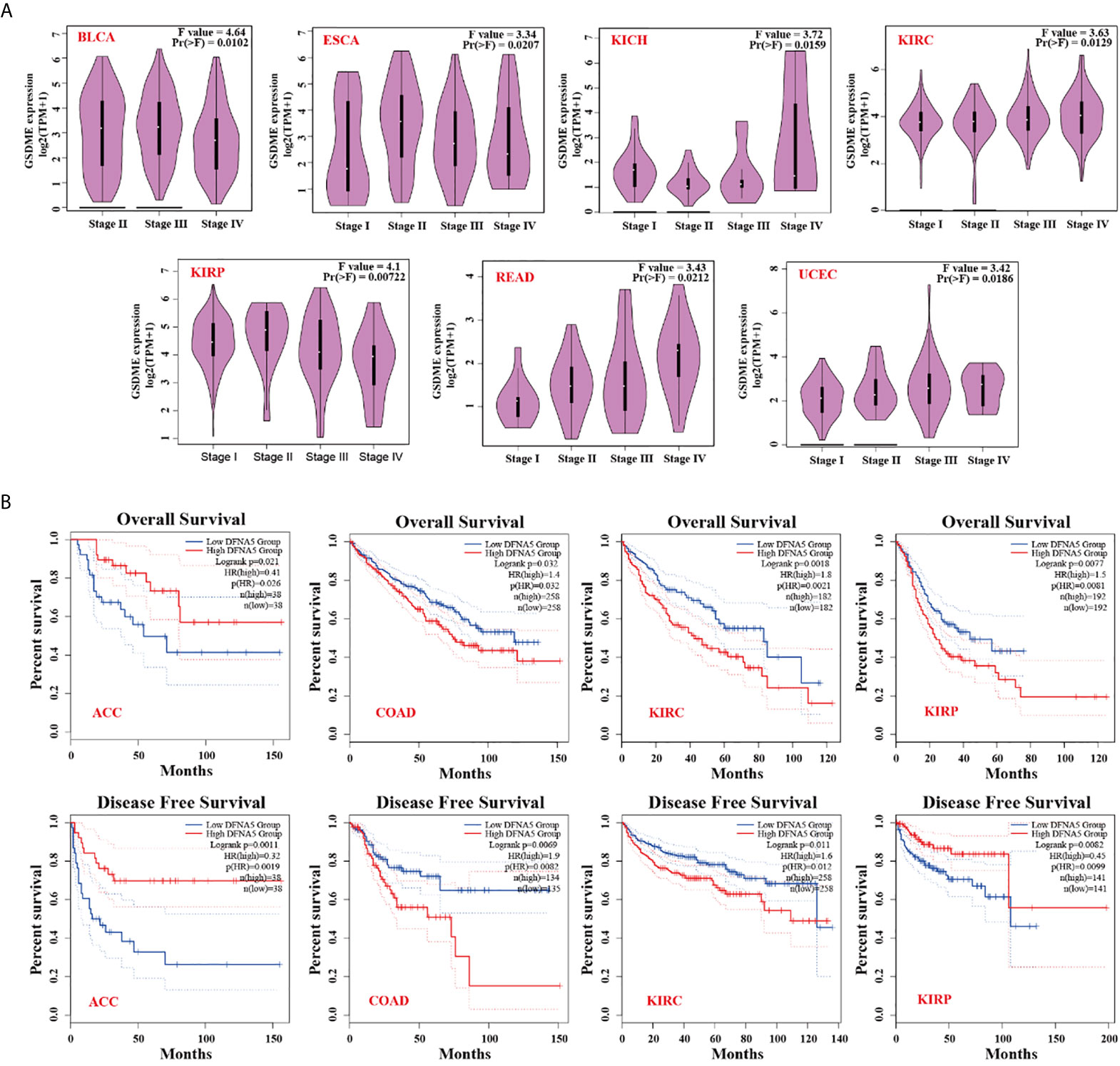
Figure 2 (A) Expression status of GSDME in various pathological stages (stage I, stage II, stage III, and stage IV) of BLCA, ESCA, KICH, KIRC, KIRP, READ, and UCEC. (B) Correlation of GSMDE expression with overall survival and disease-free survival across different TCGA cancers.
Genetic Alteration Analysis Information
We studied genetic alteration features of GSDME across the TCGA all cancers. The more than 5.5% mutation frequency of GSDME existed in patients with uterine corpus uterine corpus endometrial cancers. The “amplification” was the dominant type in the esophageal adenocarcinoma types (Figure 3A). Moreover, we investigated the possible relationship between the GSDME alteration and relevant survival prognosis. The graphs of Figure 3B indicated that CUEC cases with mutated GSDME revealed no association with disease-free survival (P=0.232), disease-specific survival (P=0.790), overall survival (P=0.558), and progression-free survival (P=0.368), as compared with species without GSDME alternation.
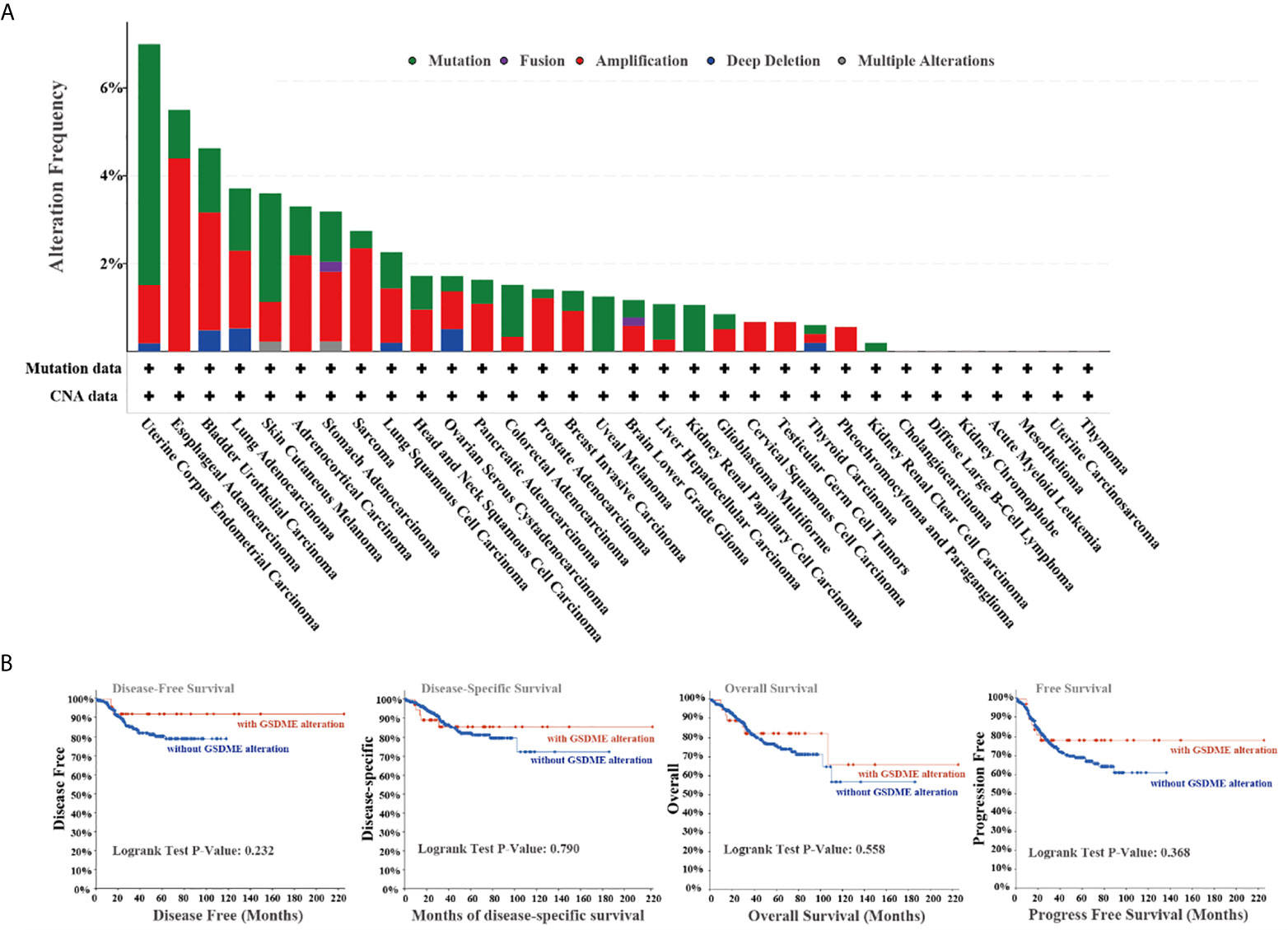
Figure 3 (A) Alteration characterizes of GSDME in diverse cancers in TCGA project. (B) The potential relationship between mutation status and disease-free, disease-specific, and progression-free survival from the cBioPortal tool.
Immune Infiltration Analysis
As important components of the tumor microenvironment, tumor-infiltrating immune cells were widely associated with the occurrence, development, or metastasis of malignant cancers (26, 27). Tumor-associated fibroblasts in the tumor microenvironment were shown to take part in regulating the function of different cancer-infiltrating immune cells (28, 29). We applied a series of algorithms to study the potential correlation of GSDME expression with the infiltration level of immune cells in different cancers form TCGA project. We found a positively correlated of GSDME expression with the estimated infiltration value of tumor-associated fibroblasts for BLCA, CHOL, KIRC, LIHC, STAD, and UCEC, and found a negative correlation for GBM and MESO (Figure 4A). The scatter plots of these cancers generated by one algorithm are graphed in Figure 4B. According to the TIDE algorithm, for instance, the expression of GSDME in BLCA is positively associated with the status of tumor-related fibroblasts infiltration (Figure 4B, cor =0.219, P = 2.29e-05).
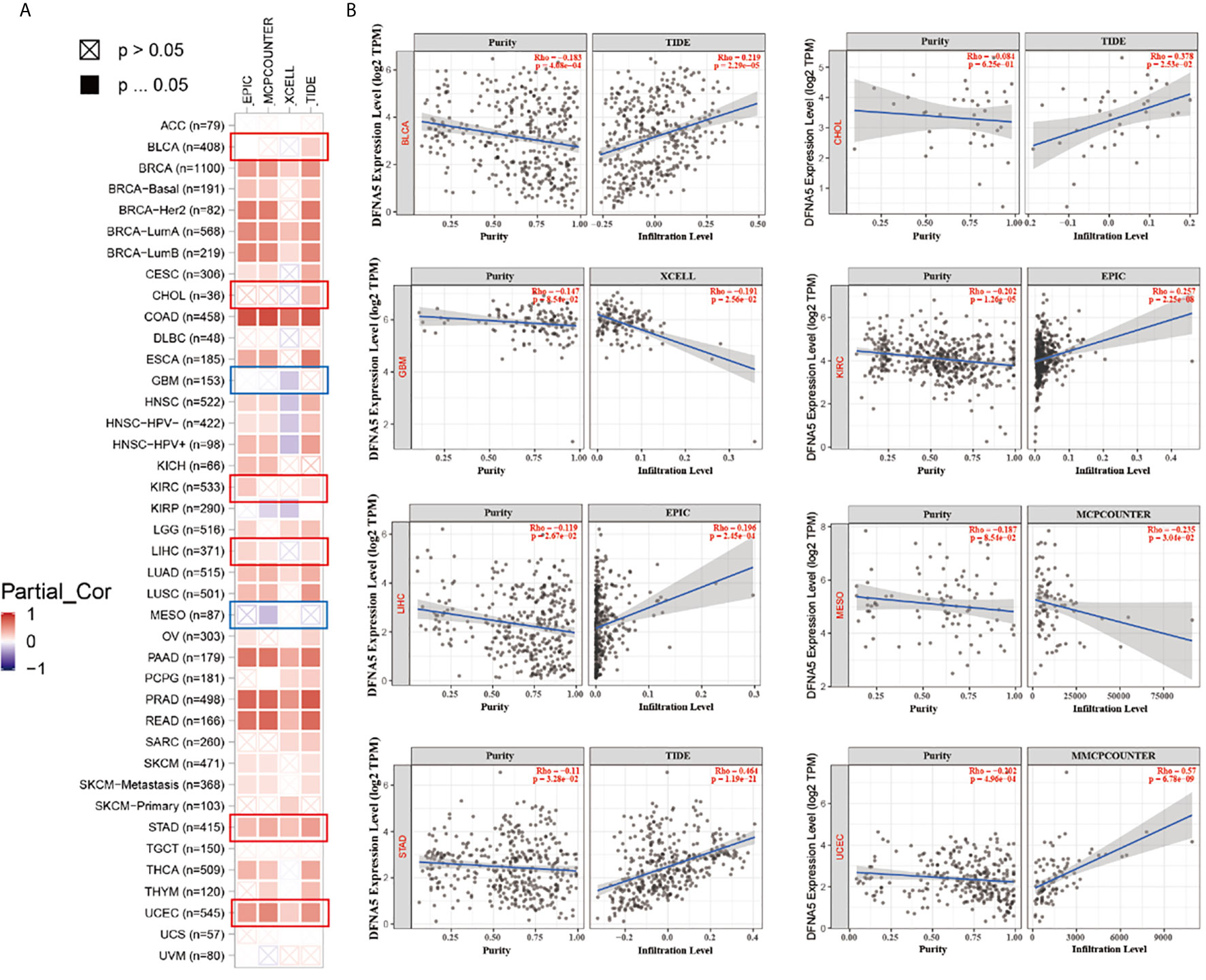
Figure 4 (A) The potential correlation between GSDME expression and immune cell infiltration of cancer-associated fibroblasts in all TCGA cancers. (B) The scatter plots of related cancers generated by one algorithm, including BLCA, CHOL, GBM, KIRC, LIHC, MESO, STAD and UCEC.
Enrichment Analysis and In Vitro Experiments
To deeply understand the potential mechanism of the GSDME underlying the carcinogenesis or antitumor therapy, our group further made great efforts to pick up the GSDME-related proteins and genes for subsequent enrichment analyses. We acquired the interaction network of 10 GSDME-related proteins, which have been supported by determined evidence (Figure 5A). The GSDME expression was statistically related to that of FEZ1 (R=0.51), GNA12 (R=0.54), MAP4 (R=0.53), SEPT7 (R=0.52), and TPST1 (R=0.51) genes (all P values <0.001) (Figure 5B). We employed the GEPIA2 tool to require the top 100 GSDME-binding genes (Figure 5C). The GO enrichment analysis revealed the majority of binding genes were associated with the cellular biology or pathways of pyroptosis, sensory perception of sound, sensory perception of mechanical stimulus, defense response to bacterium, defense response to other organism, and response to bacterium (Figure 5D). Unfortunately, any biological information related to KEGG has been found in our study. Moreover, it is very profound for us to perform in vitro experiments to verify GSDME expression for inducing pyroptosis. We used human MCF-7 cells with high expression of GSDME under the stimulation of apoptosis, MCF-7 cancer cells were characterized by cellular swelling, cytoplasmic membrane pore formation, and balloon bulging of membrane rupture (Figure 5E). Lactate dehydrogenase (LDH) and adenosine triphosphate (ATP) release are typical characterizes of pyroptotic induction, and we found that the release of LDH and ATP is significantly higher in GSDME expression MCF-7 cells as compared with 4T-1 cells, which can strongly support our points that GSDME expression is a crucial executor for pyroptosis occurrence (Figures S1, 2).
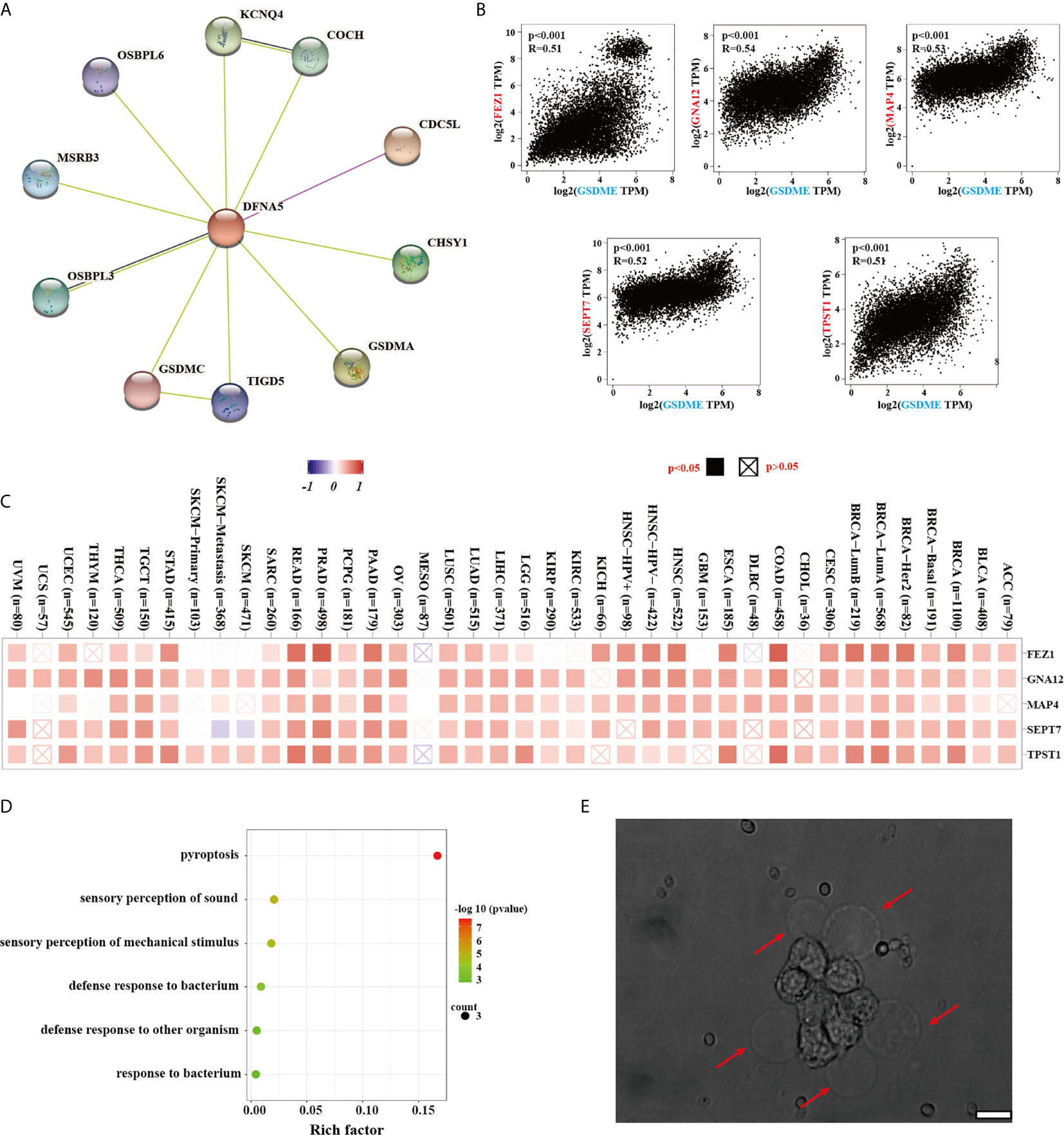
Figure 5 (A) The available determined GSDME-related proteins from the STRING website. (B) The correlation between GSDME and selected binding genes, such as PEZ1, GNA12, MAP4, SEPT7 TPST1, and TBL2 from the GEPIA2 tool. (C) The corresponding heatmap data of different cancer types are graphed in detail. (D) GO analysis was displayed based on the GSDME-binding genes. (E) The typical morphology of pyroptosis in MCF-7 cells after apoptotic stimuli. Error bar: 200 nm.
Discussion
Accumulating evidence has been documented the unique GSDME protein is involved in a series of biological processes in various cancer types, including ontogeny, homeostasis, and pathological processes. Emerging literatures have reported there is a closely relationship between GSDME expression and human diseases, particularly for malignant cancers (30). Unfortunately, the role of GSDME in the cancer pathogenesis and underlying molecular mechanism deserve further investigation. Through previous publications, it is unable to retrieve any available reports about a pan-cancer analysis of GSDME across all tumors. Thus, we carried out a comprehensively screen of GSDME expression, the genetic alteration, immune cell infiltration, and possible signal pathway.
Immunotherapy has completely revolutionized the management of tumor therapy, which restarts and maintains the tumor immune cycle to restore the normal anti-tumor immune activity (31). Unfortunately, currently, immunotherapy only benefits a subset of patients, and one of the reasons is that cancer cells may effectively evade the host immune system caused by inherently immunosuppressive microenvironment (32–36). GSDME is important pore-formation protein for pyroptosis induction, which could be cleaved by caspase-3 into N-terminal fragment of GSDME to squeeze into the lipid contents, form pores in the surface membrane, and drive the cell pyroptosis (37–39). Pyroptosis, an inflammatory programmed cell death (PCD) is triggered by Gasdermin proteins including GSDMA, GSDMB, GSDMC, GSDMD, and GSDME (16, 40–43). Gasdermin proteins have been documented to possess inherent toxic activity of the Gasdermin-N terminal fragments, which are frequently masked by inhibitory Gasdermin-C terminal fragments (19, 44, 45). Proteolytic cleavage the hinge loop between these terminals to unleash the necrotic Gasdermin-N fragments to squeeze the inner leaflet of membrane and form nanopores with a size of ~18 nm in the cellular plasma membrane (44, 46–48). Physical rupture of pyroptotic cells causes the leakage of inflammatory factors IL-18, IL-1β, and DAMPs to amplify the local and systemic inflammatory response, suggesting the immunogenic potential of pyroptosis (49, 50). Pore formation of pyroptosis may lead to the release of immune stimulants, including HMGB1, which can cause dendritic cell (DC) activation and, in turn, propagate antitumor T-cell activity. Moreover, cleaved GSDME can also permeate the mitochondria to positively feedback to the intrinsic apoptotic pathway (16, 51). Thus, therapeutic induction of pyroptosis in tumor cells can be of important value to stimulate a “cold” immunosuppressive microenvironment into “hot” immunogenic microenvironment, thereby eliciting high performance of inhibiting and eliminating malignant tumors (52).
However, despite lots of efforts have made in investigating and integrating information from available TCGA projects, there were still some limitations in the current investigation. First, although bioinformatic analysis offered us some significant insights of GSDME in cancers, it is necessary to conducted biological experiments in vitro or in vivo for checking on our findings and accelerating clinical application. Further mechanistic explorations will be helpful for clarifying the role of GSDME at the molecular levels. Second, posttranslational modifications are of great value in regulating the activity of intracellular signal transduction and regulatory factors, but post-translational modification data of GSDME is not available in these databases. Third, GSDME expression was found to be associated with cancer immunity and clinical survival prognosis; however, we were not sure that GSDME influenced clinical survival via definite signal pathway. Therefore, we are only just beginning to understand the molecular mechanisms of GSDME and its emerging role in cancer research, a great deal of work requires to be done to further broad the applications of GSDME in human cancers.
Conclusion
In summary, our pan-cancer analysis of GSDME not only broadens understanding of the carcinogenic roles of GSDME, but also provides a promising therapeutic strategy for benefiting an increasing number of cancer patients based on GSDME-induced pyroptosis.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
We greatly acknowledged the Fifth Phase “169 Project” Scientific Research Project of Zhenjiang City (YLJ201931), Zhenjiang Social Development Fund (SH2018035), Zhenjiang Key Research and Development Plan Fund (SH2016036), and the Doctoral Start-up Fund of Affiliated Hospital of Jiangsu University (jdfyRC2019005).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.706266/full#supplementary-material
Supplementary Figure 1 | The release of LDH in MCF-7 and 4T-1 cells under apoptotic stimuli. *P < 0.05; **P < 0.01; ***P < 0.001.
Supplementary Figure 2 | The release of ATP in MCF-7 and 4T-1 cells under apoptotic stimuli. *P < 0.05; **P < 0.01; ***P < 0.001
References
1. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2018. CA Cancer J Clin (2018) 60:277–300. doi: 10.3322/caac.20073
2. Martincorena I, Roshan A, Gerstung M, Ellis P, Van Loo P, McLaren S, et al. Tumor Evolution. High Burden and Pervasive Positive Selection of Somatic Mutations in Normal Human Skin. Science (2015) 348(6237):880–6. doi: 10.1126/science.aaa6806
3. Cui X, Zhang X, Liu M, Zhao C, Zhang N, Ren Y, et al. A Pan-Cancer Analysis of the Oncogenic Role of Staphylococcal Nuclease Domain-Containing Protein 1 (SND1) in Human Tumors. Genomics (2020) 112:3958–67. doi: 10.1016/j.ygeno.2020.06.044
4. Gao Q, Xie H, Zhan H, Li J, Liu Y, Huang W. Prognostic Values of Long Noncoding RNA GAS5 in Various Carcinomas: An Updated Systematic Review and Meta-Analysis. Front Physiol (2017) 8:814. doi: 10.3389/fphys.2017.00814
5. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer Incidence and Mortality Worldwide: Sources, Methods and Major Patterns in GLOBOCAN 2012. Int J Cancer (2015) 136:E359–86. doi: 10.1002/ijc.29210
6. Blum A, Wang P, Zenklusen J. Snapshot: TCGA-Analyzed Tumors. Cell (2018) 173:530. doi: 10.1016/j.cell.2018.03.059
7. Tomczak K, Czerwińska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): An Immeasurable Source of Knowledge. Contemp Oncol (Poznan Poland) (2015) 19:A68–77. doi: 10.5114/wo.2014.47136
8. Li S, Li J, Chen C, Zhang R, Wang K. Pan-Cancer Analysis of Long Non-Coding RNA NEAT1 in Various Cancers. Genes Dis (2018) 5(1):27–35. doi: 10.1016/j.gendis.2017.11.003
9. Sang C, Chao C, Wang M, Zhang Y, Luo G, Zhang X. Identification and Validation of Hub microRNAs Dysregulated in Esophageal Squamous Cell Carcinoma. Aging (2020) 12:9807–24. doi: 10.18632/aging.103245
10. Van Laer L, Huizing E, Verstreken M, van Zuijlen D, Wauters J, Bossuyt P, et al. Nonsyndromic Hearing Impairment Is Associated With a Mutation in DFNA5. Nat Genet (1998) 20:194–7. doi: 10.1038/2503
11. Saeki N, Kuwahara Y, Sasaki H, Satoh H, Shiroishi T. Gasdermin (Gsdm) Localizing to Mouse Chromosome 11 Is Predominantly Expressed in Upper Gastrointestinal Tract But Significantly Suppressed in Human Gastric Cancer Cells. Mamm Genome (2000) 11:718–24. doi: 10.1007/s003350010138
12. Tamura M, Tanaka S, Fujii T, Aoki A, Komiyama H, Ezawa K, et al. Members of a Novel Gene Family, Gsdm, Are Expressed Exclusively in the Epithelium of the Skin and Gastrointestinal Tract in a Highly Tissue-Specific Manner. Genomics (2007) 89:618–29. doi: 10.1016/j.ygeno.2007.01.003
13. Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, et al. BioGPS: An Extensible and Customizable Portal for Querying and Organizing Gene Annotation Resources. Genome Biol (2009) 10:R130. doi: 10.1186/gb-2009-10-11-r130
14. Bischoff A, Luijendijk M, Huygen P, van Duijnhoven G, De Leenheer E, Oudesluijs G, et al. A Novel Mutation Identified in the DFNA5 Gene in a Dutch Family: A Clinical and Genetic Evaluation. Audiol Neurootol (2004) 9:34–46. doi: 10.1159/000074185
15. Yu C, Meng X, Zhang S, Zhao G, Hu L, Kong X. A 3-Nucleotide Deletion in the Polypyrimidine Tract of Intron 7 of the DFNA5 Gene Causes Nonsyndromic Hearing Impairment in a Chinese Family. Genomics (2003) 82:575–9. doi: 10.1016/S0888-7543(03)00175-7
16. Rogers C, Fernandes-Alnemri T, Mayes L, Alnemri D, Cingolani G, Alnemri E. Cleavage of DFNA5 by Caspase-3 During Apoptosis Mediates Progression to Secondary Necrotic/Pyroptotic Cell Death. Nat Commun (2017) 8:14128. doi: 10.1038/ncomms14128
17. Zhang Z, Zhang Y, Xia S, Kong Q, Li S, Liu X, et al. Gasdermin E Suppresses Tumour Growth by Activating Anti-Tumour Imunity. Nature (2020) 579(7799):415–20. doi: 10.1038/s41586-020-2071-9
18. Ee Ck KB, Laer LV, Camp GV. DFNA5, a Gene Involved in Hearing Loss and Cancer: A Review. Ann Otol Rhinol Laryngol (2012) 121:197–207. doi: 10.1177/000348941212100310
19. Xia X, Wang X, Cheng Z, Qin W, Hu J. The Role of Pyroptosis in Cancer: Pro-Cancer or Pro-“Host”? Cell Death Dis (2019) 10. doi: 10.1038/s41419-019-1883-8
20. Trang H, Thi H, Suntaek, Hong. Inflammasome as a Therapeutic Target for Cancer Prevention and Treatment. J Cancer Prev (2017) 22:62–73. doi: 10.15430/JCP.2017.22.2.62
21. Zhou C, Fang J. The Role of Pyroptosis in Gastrointestinal Cancer and Immune Responses to Intestinal Microbial Infection. Biochim Biophys Acta Rev Cancer (2019) 1872:1–10. doi: 10.1016/j.bbcan.2019.05.001
22. Wei Z, Sun Y, Li G, Liu Z, Jiao S. Advances of Research in Cancer-Associated Inflammation and Tumor Microenvironments. Chin J Clin Oncol (2018) 11:1009. doi: 10.3389/fphar.2020.01009
23. Tang Z, Li C, Kang B, Gao G, Zhang Z. GEPIA: A Web Server for Cancer and Normal Gene Expression Profiling and Interactive Analyses. Nucleic Acids Res (2017) 45:W98–102. doi: 10.1093/nar/gkx247
24. Gao J, Aksoy B, Dogrusoz U, Dresdner G, Gross B, Sumer S, et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the Cbioportal. Sci Signaling (2013) 6:pl1. doi: 10.1126/scisignal.2004088
25. Cerami E, Gao J, Dogrusoz U, Gross B, Sumer S, Aksoy B, et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov (2012) 2:401–4. doi: 10.1158/2159-8290.CD-12-0095
26. Fridman W, Galon J, Dieunosjean M, Cremer I, Fisson S, Damotte D, et al. Immune Infiltration in Human Cancer: Prognostic Significance and Disease Control. Curr Top Microbiol Immunol (2011) 344:1. doi: 10.1007/82_2010_46
27. Steven A, Seliger B. The Role of Immune Escape and Immune Cell Infiltration in Breast Cancer. Breast Care (Basel Switzerland) (2018) 13:16–21. doi: 10.1159/000486585
28. Chen X, Song E. Turning Foes to Friends: Targeting Cancer-Associated Fibroblasts. Nat Rev Drug Discov (2019) 18:99–115. doi: 10.1038/s41573-018-0004-1
29. Kwa M, Herum K, Brakebusch C. Cancer-Associated Fibroblasts: How Do They Contribute to Metastasis? Clin Exp Metastasis (2019) 36:71–86. doi: 10.1007/s10585-019-09959-0
30. Wang Y, Liu X, Zhao R. Induction of Pyroptosis and Its Implications in Cancer Management. Front Oncol (2019) 9:971. doi: 10.3389/fonc.2019.00971
31. Rao G, Latha K, Ott M, Sabbagh A, Marisetty A, Ling X, et al. Anti-Pd-1 Induces M1 Polarization in the Glioma Microenvironment and Exerts Therapeutic Efficacy in the Absence of CD8 Cytotoxic T Cells. Clin Cancer Res (2020) 26:4699–712. doi: 10.1158/1078-0432.CCR-19-4110
32. Anker J, Naseem A, Mok H, Schaeffer A, Abdulkadir S, Thumbikat P. Multi-Faceted Immunomodulatory and Tissue-Tropic Clinical Bacterial Isolate Potentiates Prostate Cancer Immunotherapy. Nat Commun (2018) 9:1591. doi: 10.1038/s41467-018-03900-x
33. Peranzoni E, Lemoine J, Vimeux L, Feuillet V, Barrin S, Kantari-Mimoun C, et al. Macrophages Impede CD8 T Cells From Reaching Tumor Cells and Limit the Efficacy of Anti-PD-1 Treatment. Proc Natl Acad Sci USA (2018) 115:E4041–50. doi: 10.1073/pnas.1720948115
34. Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer Immunoediting: From Immunosurveillance to Tumor Escape. Nat Immunol (2002) 3:991–8. doi: 10.1038/ni1102-991
35. Schreiber RD, Old LJ, Smyth MJ. Cancer Immunoediting: Integrating Immunity’s Roles in Cancer Supression and Promotion. Science (2011) 331:1565–70. doi: 10.1126/science.1203486
36. Carreno B, Magrini V, Becker-Hapak M, Kaabinejadian S, Hundal J, Petti A, et al. Cancer Immunotherapy. A Dendritic Cell Vaccine Increases the Breadth and Diversity of Melanoma Neoantigen-Specific T Cells. Science (New York NY) (2015) 348:803–8. doi: 10.1126/science.aaa3828
37. Ruan J, Wang S, Wang J. Mechanism and Regulation of Pyroptosis-Mediated in Cancer Cell Death. Chemico-Biol Interact (2020) 323:109052. doi: 10.1016/j.cbi.2020.109052
38. Wang Y, Gao W, Shi X, Ding J, Liu W, He H, et al. Chemotherapy Drugs Induce Pyroptosis Through Caspase-3 Cleavage of a Gasdermin. Nature (2017) 547:99–103. doi: 10.1038/nature22393
39. Hu L, Chen M, Chen X, Zhao C, Fang Z, Wang H, et al. Chemotherapy-Induced Pyroptosis Is Mediated by BAK/BAX-Caspase-3-GSDME Pathway and Inhibited by 2-Bromopalmitate. Cell Death Dis (2020) 11:281. doi: 10.1038/s41419-020-2476-2
40. He W, Wan H, Hu L, Chen P, Wang X, Huang Z, et al. Gasdermin D Is an Executor of Pyroptosis and Required for Interleukin-1β Secretion. Cell Res (2015) 25:1285–98. doi: 10.1038/cr.2015.139
41. Holohan C, Van Schaeybroeck S, Longley D, Johnston P. Cancer Drug Resistance: An Evolving Paradigm. Nat Rev Cancer (2013) 13:714–26. doi: 10.1038/nrc3599
42. Ding J, Wang K, Liu W, She Y, Sun Q, Shi J, et al. Pore-Forming Activity and Structural Autoinhibition of the Gasdermin Family. Nature (2016) 535:111–6. doi: 10.1038/nature18590
43. Ding J, Wang K, Liu W, She Y, Sun Q, Shi J, et al. Erratum: Pore-forming Activity and Structural Autoinhibition of the Gasdermin Family. Nature (2016) 540:150. doi: 10.1038/nature20106
44. Liu X, Zhang Z, Ruan J, Pan Y, Magupalli VG, Wu H, et al. Inflammasome-Activated Gasdermin D Causes Pyroptosis by Forming Membrane Pores. Nature (2016) 535:153. doi: 10.1038/nature18629
45. Cookson B, Brennan M. Pro-Inflammatory Programmed Cell Death. Trends Microbiol (2001) 9:113–4. doi: 10.1016/S0966-842X(00)01936-3
46. Karmakar M, Minns M, Greenberg E, Diaz-Aponte J, Pestonjamasp K, Johnson J, et al. N-GSDMD Trafficking to Neutrophil Organelles Facilitates IL-1β Release Independently of Plasma Membrane Pores and Pyroptosis. Nat Commun (2020) 11:2212. doi: 10.1038/s41467-020-16043-9
47. Zhang X, Zhang P, An L, Sun N, Peng L, Tang W, et al. Miltirone Induces Cell Death in Hepatocellular Carcinoma Cell Through GSDME-dependent Pyroptosis. Acta Pharm Sinica B (2020) 10:1397–413. doi: 10.1016/j.apsb.2020.06.015
48. Aglietti RA, Estevez A, Gupta A, Ramirez MG, Dueber EC. Gsdmd p30 Elicited by Caspase-11 During Pyroptosis Forms Pores in Membranes. Proc Natl Acad Sci (2016) 113:7858–63. doi: 10.1073/pnas.1607769113
49. Jorgensen I, Miao E. Pyroptotic Cell Death Defends Against Intracellular Pathogens. Immunolo Rev (2015) 265:130–42. doi: 10.1111/imr.12287
50. Yu P, Wang H, Tian M, Li A, Chen X, Wang X, et al. Eukaryotic Elongation Factor-2 Kinase Regulates the Cross-Talk Between Autophagy and Pyroptosis in Doxorubicin-Treated Human Melanoma Cells In Vitro. Acta Pharmacol Sin (2019) 40:1237–44. doi: 10.1038/s41401-019-0222-z
51. Galluzzi L, Buqué A, Kepp O, Zitvogel L, Kroemer G. Immunogenic Cell Death in Cancer and Infectious Disease. Nat Rev Immunol (2016) 17:97. doi: 10.1038/nri.2016.107
Keywords: gasdermin E, pan-cancer analysis, immune infiltrate, survival analysis, pyroptosis
Citation: Zhang Z, Zhao S, Yang H, Chen Y, Feng H, An M and Chen B (2021) Prognostic and Immunological Role of Gasdermin E in Pan-Cancer Analysis. Front. Oncol. 11:706266. doi: 10.3389/fonc.2021.706266
Received: 07 May 2021; Accepted: 06 July 2021;
Published: 26 July 2021.
Edited by:
Xueli Zhang, Shanghai Jiao Tong University, ChinaReviewed by:
Leping Li, National Institute of Environmental Health Sciences, United StatesXin-Wu Cui, Huazhong University of Science and Technology, China
Copyright © 2021 Zhang, Zhao, Yang, Chen, Feng, An and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuangshuang Zhao, dWpzemhhb3NzQDE2My5jb20=; Baoding Chen, YWxwaGFsaWZlQDE2My5jb20=
 Zheng Zhang
Zheng Zhang Shuangshuang Zhao*
Shuangshuang Zhao*