- 1Department of Urology, Taizhou Central Hospital (Taizhou University Hospital), Taizhou, China
- 2Department of Clinical Laboratory, Taizhou Central Hospital (Taizhou University Hospital), Taizhou, China
- 3Department of Urology, Maoming People’s Hospital, Maoming, China
- 4Department of Hepatobiliary Surgery, Taizhou Central Hospital (Taizhou University Hospital), Taizhou, China
Cholangiocarcinoma (CCA), a high mortality malignant carcinoma characterized by advanced disease and frequent recurrence, constitutes a major challenge for treatment and prognosis. AT-rich interaction domain 1A (ARID1A) variation is a distinct genetic entity in CCA, getting mounting concerns recently. Here, we comprehensively reviewed the clinical significance and molecular mechanisms of ARID1A alterations in CCA. Based on the independent data derived from 29 relevant studies, the variation rate of ARID1A in intrahepatic and extrahepatic CCA is reported at 6.9–68.2% and 5–55%, respectively. Most of the included studies (28/29, 96.6%) suggest that ARID1A serves as a tumor suppressor in CCA. ARID1A variation may be an important prognostic indicator to predict disease mortality, metastasis, and recurrence in patients with CCA. Multifactorial molecular mechanisms are involved in the relationship between ARID1A variations and the pathogenesis and pathophysiology of CCA, including disruption of the cell cycle, chromatin remodeling, oxidative stress damage, DNA hypermethylation, and the interaction of multiple genes being affected. This review describes that ARID1A variation might be a potential diagnostic and prognostic biomarker for CCA. Future diagnoses and treatments targeting ARID1A hint towards a precision medicine strategy in the management of CCA.
Introduction
Cholangiocarcinoma (CCA) is the second most frequently diagnosed primary liver malignancy, accounting for 10–20% of primary hepatic carcinomas, and representing 3% of all gastrointestinal tumors (1). Accounting to the anatomic location of biliary tree involvement, CCA can be classified into intrahepatic (ICC, 6–8% of CCA) and extrahepatic (ECC, including perihilar and distal CCA) (2). CCA is a highly malignant neoplasm resulting from the malignant transformation of the epithelium of the biliary tract, of which approximately 90% are adenocarcinoma (3). The incidence and mortality rates of CCA has steadily increased in recent decades. The rising incidence is considered to be related to the burden of hepatitis B and hepatitis C virus infection (4). In addition, primary sclerosing cholangitis, obesity, diabetes, inflammatory bowel disease, non-alcoholic fatty liver diseases, and liver fluke infestation are also the key risk factors for CCA development (5). As reported, only 10–15% of CCA patients are eligible for curative surgery (6). Due to the high rates of disease recurrence (50–60%), the 5-year overall survival rates are only 30% even after surgery (7). Furthermore, locally advanced and distant metastasis terrifically contribute to the high mortality of CCA patients. Therefore, it is necessary to look for potential biomarkers that could help with identifying the disease prognosis.
Heterogeneity in cancer aggressiveness and prognosis of CCA may be driven by the differential alteration of the genetic variations. It is reported that genes encoding components of the SWItch/Sucrose Non-Fermenting (SWI/SNF) chromatin-remodeling complex may be one of the most commonly mutated genes in multiple malignancies. SWI/SNF chromatin remodeling complex is involved in transcription and DNA replication and repair (8). The AT-rich interaction domain 1A (ARID1A) is the most frequently mutated SWI/SNF gene across a broad spectrum of human cancers, which facilitates access of proteins to DNA (9, 10). ARID1A is presumed to be a tumor suppressor based on loss-of-function mutational profiles observed in many cancers (11), including gastrointestinal cancers. It was reported that ARID1A variations were present in 18.7% of gastric, 13.7% of hepatocellular, 9.4% of colorectal, and 3.6% of pancreatic cancers (12). Most of the ARID1A variations are inactivating alterations, leading to loss of ARID1A protein expression (13). In recent years, accumulated evidence indicates that ARID1A variation is associated with clinicopathologic features of CCA (14, 15). ARID1A variations were identified in 7.2 to 36% of ICC and 5 to 12.3% of extrahepatic CCA (16).
At present, the exact role of ARID1A on the prognosis and clinicopathologic features of CCA is still controversial among different clinical studies. It was suggested that ARID1A had dual roles in both oncogenicity and tumor suppression in CCA. Based on the published data, most studies indicated ARID1A may be a tumor suppressor gene. For example, Yang et al. (17) demonstrated that low expression of ARID1A was associated with worse prognosis in intrahepatic CCA than those with high expression. However, a recent study developed by Bi et al. (18) indicated that high expression of ARID1A might be correlated with worse prognosis in intrahepatic CCA patients than those with low expression. The effect of ARID1A in CCA is currently inconclusive. In this review, we aim to summarize all the evidence on the association between ARID1A variations or expression and CCA development, as well as open up currently known mechanisms therefore to facilitate clinical understanding of the role of ARID1A in CCA.
Alteration Of ARID1A In CCA Among Different Relevant Studies
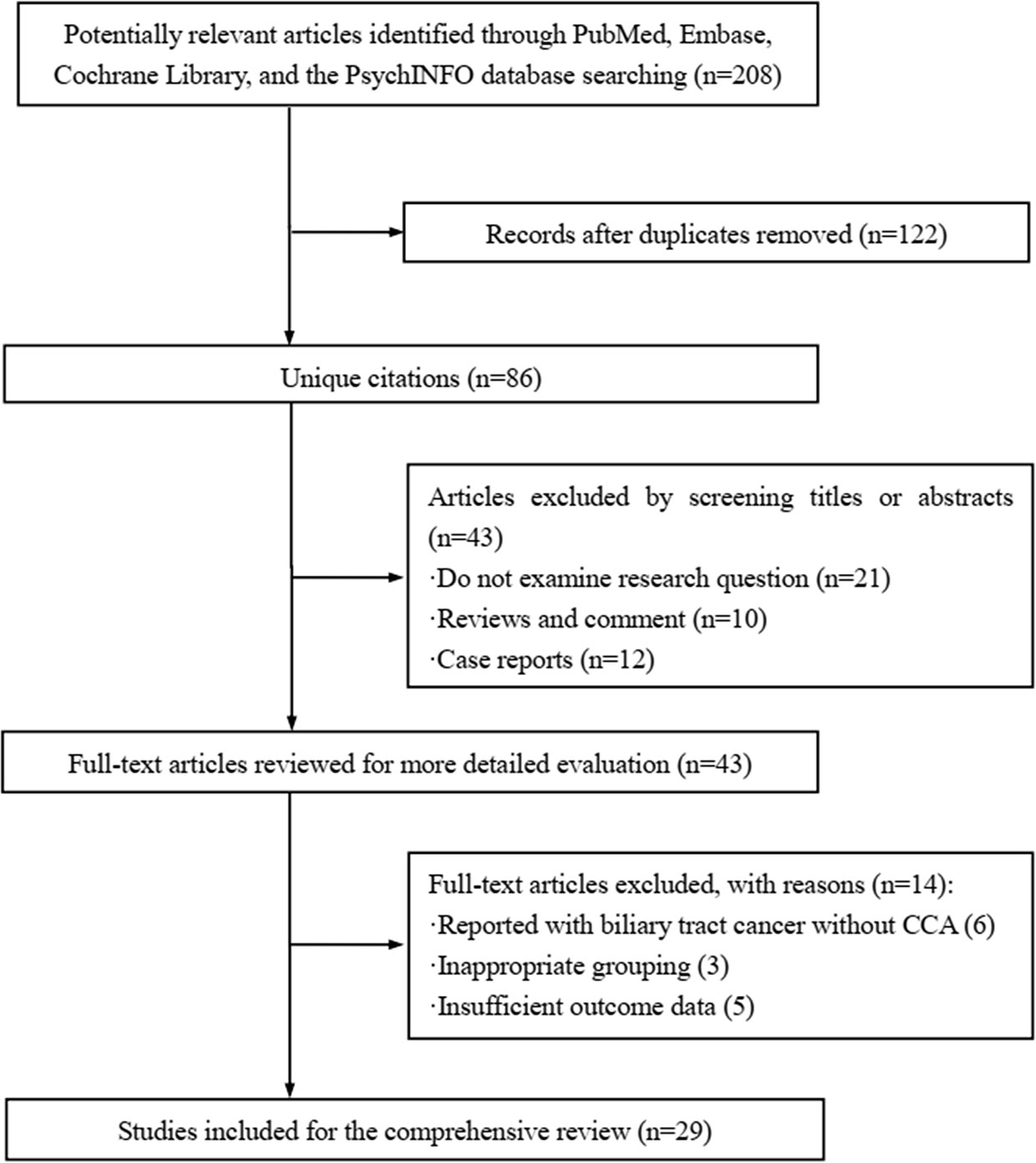
Four databases, i.e., MEDLINE (PubMed), EMBASE (OVID), Cochrane Library, and PsychINFO were systematically searched to screening the related studies prior to April 1, 2021. Only studies reporting with English language were included. The searching strategy employed for identifying the eligible studies in PubMed databases was: ((((((((((((((“Cholangiocarcinoma”[Mesh]) OR (Cholangiocarcinomas)) OR (Cholangiocellular Carcinoma)) OR (Carcinoma, Cholangiocellular)) OR (Carcinomas, Cholangiocellular)) OR (Cholangiocellular Carcinomas)) OR (Extrahepatic Cholangiocarcinoma)) OR (Cholangiocarcinoma, Extrahepatic)) OR (Cholangiocarcinomas, Extrahepatic)) OR (Extrahepatic Cholangiocarcinomas)) OR (Intrahepatic Cholangiocarcinoma)) OR (Cholangiocarcinoma, Intrahepatic)) OR (Cholangiocarcinomas, Intrahepatic)) OR (Intrahepatic Cholangiocarcinomas)) AND ((((((((((((((ARID1A) OR (B120)) OR (BAF250)) OR (BAF250a)) OR (BM029)) OR (C1orf4)) OR (CSS2)) OR (ELD)) OR (MRD14)) OR (OSA1)) OR (P270)) OR (SMARCF1)) OR (hELD)) OR (hOSA1)). Additional studies were detected by manual inspection of reference lists in the relevant publications. The following information was extracted based on a data collection form, including the first authors’ names of the included studies, publication year, country, type of cholangiocarcinoma, ARID1A variations presented with cases and percentage, effect of ARID1A on CCA, and clinical implications or biological functioning of ARID1A.
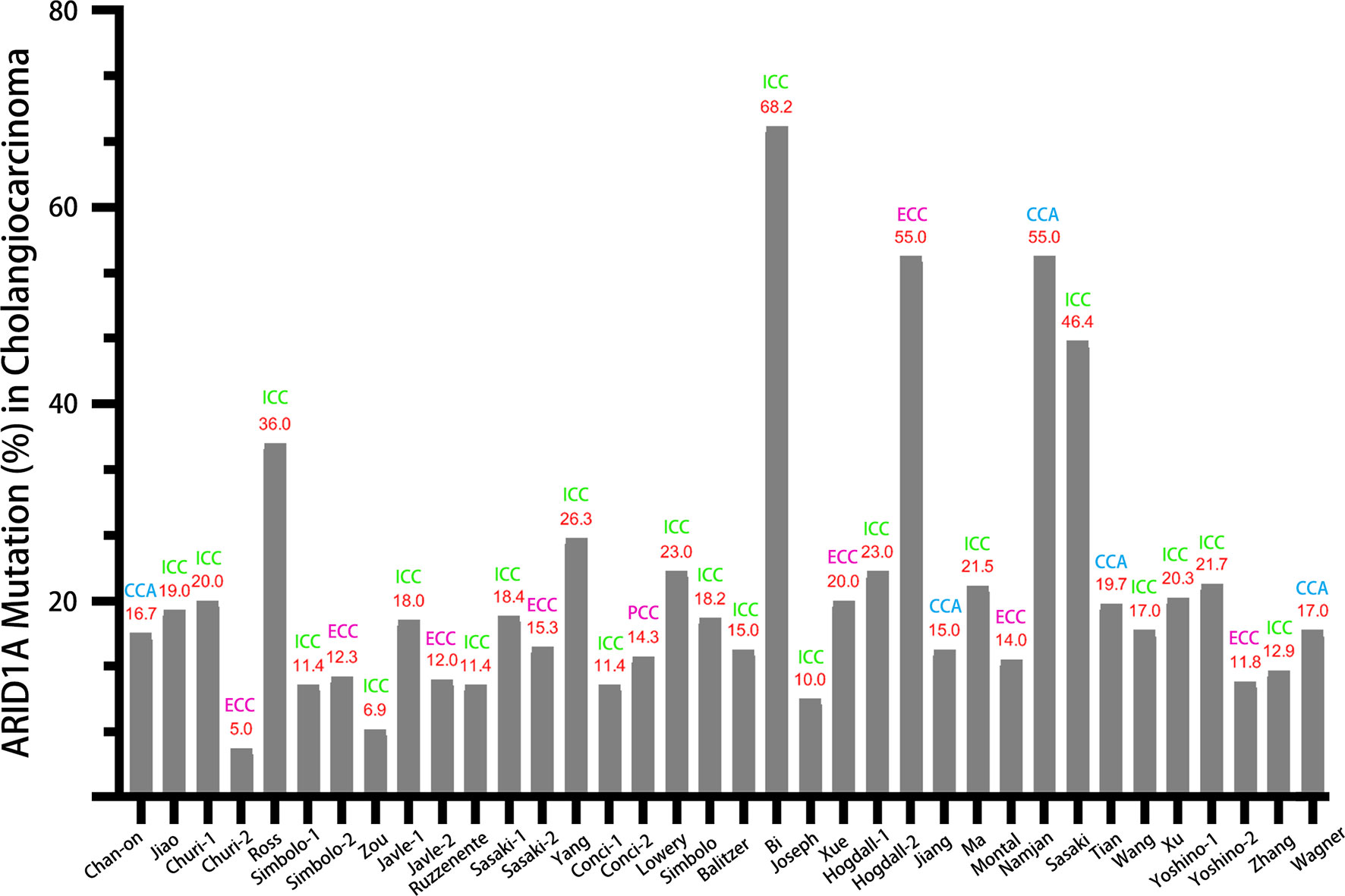
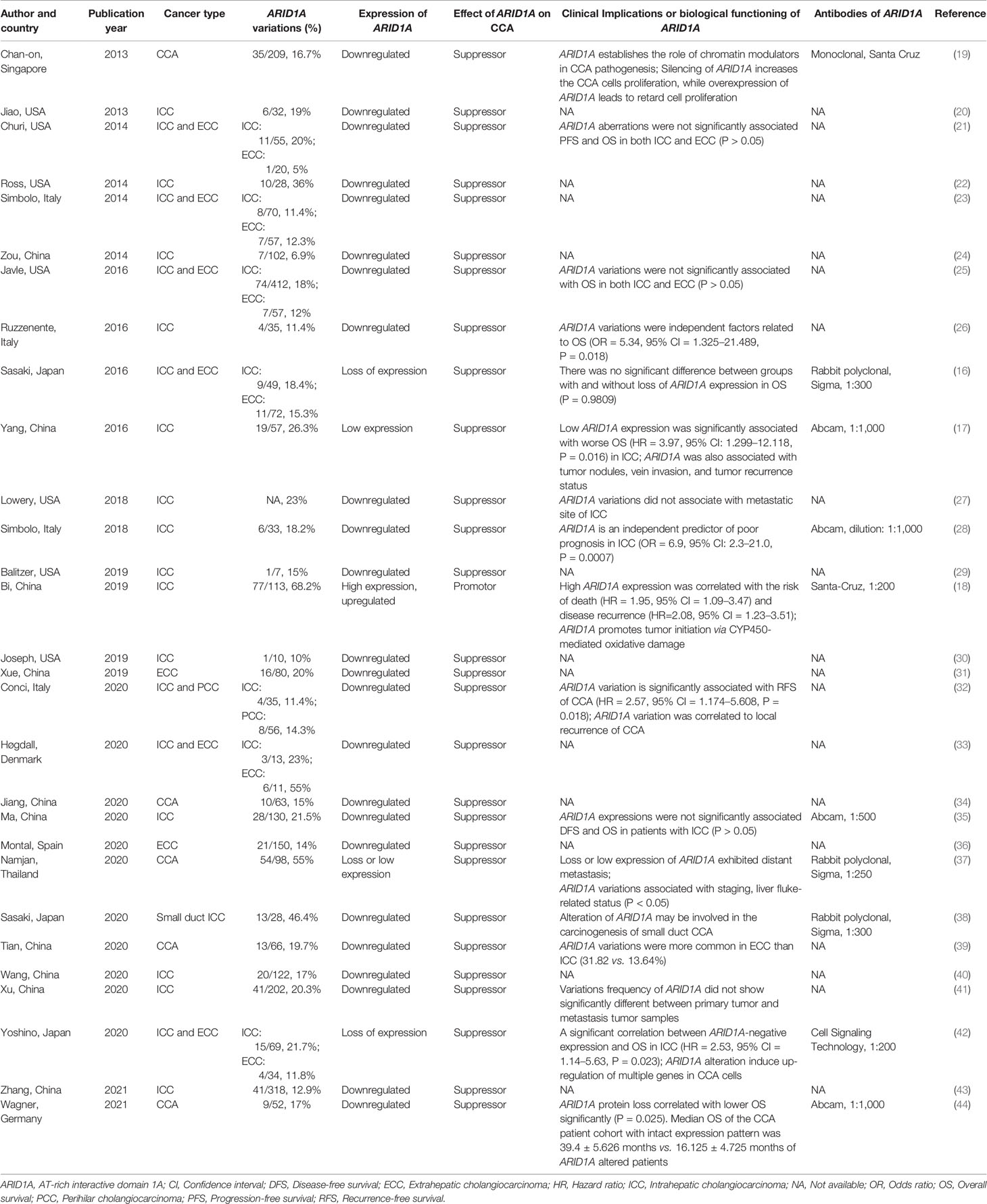
Figure 1 displayed the search flowchart for identifying the eligible studies reporting ARID1A variants and CCA. In the initial database search, 208 publications were detected, of which 82 from MEDLINE, 47 from EMBASE, 41 from the Cochrane Library, and 38 from the PsychINFO database. Finally, 29 eligible studies (16–44) with a total of 2,945 subjects were included. The publication years of the included studies ranged from 2013 to 2021. Seven studies were conducted in Europe, 15 studies in Asia, and 7 studies in America. The cancer type in those eligible studies included CCA (reported in 5 studies), ICC (reported in 22 studies), and ECC (reported in 9 studies). The sample size ranged from 7 to 412 patients. ARID1A variations or expressions were determined by various methods, including targeted sequencing study, immunohistochemistry (IHC), tissue microarrays, western-blot, quantitative real-time reverse transcription-polymerase chain reaction, and chromatin immunoprecipitation. The anti-ARID1A antibodies were inconsistent among different studies, including different antigenic determinant (monoclonal or polyclonal), various antibody manufacturer (i.e., Santa Cruz, Sigma, Abcam, and Cell Signaling Technology), diverse dilution rate (ranged from 1:1,000 to 1:200), and the different cut off that used for defining “positive vs. negative” samples. As shown in Figure 2, the frequency of ARID1A variations among the 29 studies ranged from 5 to 68.2%. Only one study reported that ARID1A variants might serve as a cancer-promoting gene in CCA, while the remaining 28 included studies indicated ARID1A variants might be a suppressor in CCA development and progression. Four studies have provided the data of the type of ARID1A variants in CCA. There are two types of ARID1A variants, including mutant and wild type (not mutated, the opposite of mutant type), while a mutant one is defined as loss of ARID1A expression or low expression of ARID1A. In Namjan et al.’s study (37), the authors reported that reduction of ARID1A expression and/or somatic mutation was associated with CCA progression. They also found that the truncation mutations (92%) of ARID1A were significantly associated with loss of ARID1A expression in CCA. In a cohort of 209 CCA cases (19), Chan-on et al. suggested the tumor-suppressive functions for ARID1A in CCA pathogenesis and further identified a total of 35 non-synonymous ARID1A somatic mutations, including 17 indels, 14 non-sense mutations, 3 missense mutations, and 1 splice-site mutation. Jiao et al. (20) reported that ARID1A variations occurred in 6 of 32 CCA patients (19%) and the types of mutations, including 3 non-sense mutations, 1 frameshift insertion-deletion, and 2 missense mutations. ARID1A variations in Simbolo et al.’s study (28) were found at 18.2% (6/33), the mutation types including 2 missense, 3 non-sense, and 1 frameshift. The characteristics of the 29 included studies are summarized in Table 1.
Since the variation rates, clinical implications, and the biological functioning of ARID1A are different among the 29 eligible studies, we perform an in-depth review on these studies as follows.
ARID1A Expressions and Variations in CCA
The Variation Rate of ARID1A in CCA
Comprehensive genome analysis is a useful tool for identifying various oncogene variations, especially in those genes encoding the chromatin remodeling factors. ARID1A is a part of the SWI/SNF chromatin-remodeling complex. SWI/SNF complexes are the most frequently mutated epigenetic regulators, controlling gene expression and cellular differentiation (45). Mounting evidence suggests ARID1A variation is commonly associated with multiple cancers, including gynecologic cancers, urothelial carcinoma, and gastrointestinal cancer (46, 47), with a mutational rate from 3.6 to 45.2% depending on different tumor types (48). In recent decades, more and more studies (49, 50) have found that there is a close relationship between ARID1A variant and the clinicopathologic features of CCA. It was reported that ARID1A gene variations were detected in 7.2–36% of ICC and 5–12.3% of ECC (16). However, no related review article has published focusing on this issue.
In this comprehensive review, we have summarized all the evidence relating to the ARID1A variation in CCA. Based on data from the 29 included studies, the variation rate of ARID1A varies greatly, ranging from 5 to 68.2%. As displayed in Figure 2, the variation rate of ARID1A in CCA (without distinguishing cancer type), ICC, and ECC was 15–55%, 6.9–68.2%, and 5–55%, respectively. According to previous reports, the highest ARID1A variant rate was found in ovarian clear cell carcinoma, up to 46–57% (42). And the ARID1A variation ratio was identified in about 10–20% of hepatobiliary and pancreatic cancers. Referred to the present systematic review, the variation rate of ARID1A was up to over 50% in some studies regardless of any type of CCA. Tian et al. (39) compared the ratio of ARID1A variations in the different types of CCA and found that ARID1A variations were more common in ECC than ICC (31.82 vs. 13.64%). The above results indicate that there might be a pivotal pathogenic link between the high frequency of ARID1A variations and CCA development.
The differences in ARID1A mutation rates and expression level that observed between different studies could be due to multiple factors, including distinct demographic characteristics (sample size, race, and regions), different types (ICC, ECC, or combined) and disease states (early or advanced CCA), specific anti-ARID1A antibodies and measurements for assessing the ARID1A expression (immunohistochemistry, targeted sequencing analysis, tissue microarrays, western blot, quantitative real-time reverse transcription PCR, and chromatin immunoprecipitation), and various co-present or targeted proteins being affected by ARID1A. Based on this evidence, an international multicenter with a large sample size and well-designed study is still needed to better illuminate the relationship between ARID1A mutations and CCA development, which is crucial for future strategies of CCA treatment.
Of note, except for one study reporting the significantly high level of ARID1A in CCA tissues, the remaining 28 studies (96.6%) suggested that the expression of ARID1A in CCA was low or absent. In other words, only one study concluded that ARID1A plays an oncogenic role, while the remaining studies suggest that ARID1A serves as a tumor suppressor and this is consistent with the ARID1A functions in other cancer types, including ovarian clear cell, gastric, pancreatic, colon, breast, lung, bladder, and renal cancer (51).
Based on the current evidence, ARID1A variation is expected to solely play a tumor suppressor in the tumorigenesis of multiple cancers, including CCA. Intriguingly, however, a previous retrospective study conducted by Bi et al. showed that the ARID1A variant might also serve as a cancer promotor in CCA development (18). They found that ARID1A was highly expressed in ICC tumor tissues, showing a total of 68% (77/113) tumor tissues presented with positive immunohistochemical staining in ICC. However, the majority of the clinical studies showed that decreased/absent ARID1A correlated with worse stage and prognosis in patients with ICC (17). This opposite effect of ARID1A is also detected in liver cancer. Sun et al. demonstrated that ARID1A had context-dependent tumor-suppressive and oncogenic roles in the liver cancers (52). The author showed that ARID1A was required for initial tumor development and to be inhibitory of hepatocellular carcinoma metastatic potential, indicating the role of ARID1A in oncogenesis was dependent on tissue context. Bi et al. (18) believed that the dual roles of ARID1A in CCA might be due to different sample sizes and recommended using multiple prognostic factors to avoid inconsistencies.
Prognostic Significance of ARID1A in CCA
Though a high frequency of ARID1A variations is observed in the CCA, the prognostic value of ARID1A in CCA is still controversial. Ruzzenente et al. (26) recruited 35 patients with CCA and found that the ARID1A variations rate was 11.4%. The authors also found that ARID1A variation was an independent factor for the overall survival (OS) in CCA (OR = 5.34, 95% CI = 1.325–21.489, P = 0.018). The median overall survival of ICC in mutation and wild type of ARID1A was 14 and 52 months, respectively (P = 0.012). Yang et al. (17) reported that ARID1A variation was detected in 19/57 (26.3%) ICC patients and found that low ARID1A expression was dramatically correlated with worse OS (HR = 3.97, 95% CI: 1.299–12.118, P = 0.016). In addition, they also found that ARID1A was associated with tumor nodules, vein invasion, and tumor recurrence status in CCA. Conci et al. (32) indicated that ARID1A variation was remarkably associated with recurrence-free survival (RFS) of CCA (HR = 2.57, 95% CI = 1.174–5.608, P = 0.018). They further found that ARID1A variation was associated with a local recurrence in 43% of cases. A study (28) developed in Italy revealed that ARID1A variation was recorded in 18.2% and indicated that ARID1A variations are an independent predictor of poor prognosis in ICC (OR = 6.9, 95% CI: 2.3–21.0, P = 0.0007). In a study in Thailand (37), the authors found that loss or low expression of ARID1A was liable to distant metastasis in CCA. They also observed that ARID1A variations were associated with staging and liver fluke-related status (P < 0.05). Sasaki et al. (38) reported that the variations rate of ARID1A was up to 46.4% in small duct ICC and found that alteration of ARID1A might be involved in the carcinogenesis of CCA. Another study conducted by Sasaki et al. (52) indicated that ARID1A alteration was correlated with the degree of ductal plate malformation (DPM)-pattern of CCA. Yoshino et al. (42) showed a significant correlation between ARID1A-negative expression and OS in ICC (HR = 2.53, 95% CI = 1.14–5.63, P = 0.023). Wagner et al. demonstrated that suppressor ARID1A protein loss correlated with lower OS significantly loss of ARID1A protein expression is significantly correlated with lower OS when compared to the intact expression pattern (16.125 ± 4.725 vs. 39.4 ± 5.626 months, P = 0.025). The above studies that confirmed the prognostic value of ARID1A variations in CCA were correlated to the ARID1A deficiency or low expression. However, Bi et al. (18) also found that ARID1A alteration was associated with the risk of death (HR = 1.95, 95% CI = 1.09–3.47) as well as disease recurrence (HR = 2.08, 95% CI = 1.23–3.51), but such prognostic effects were based on high expression of ARID1A. Notably, an opposite effect played by ARID1A in CCA was found in Bi et al.’s study when compared to the remainder 28 included studies. Since Bi et al. have employed immunohistochemistry (IHC) to evaluate ARID1A variations rather than the sequencing analysis which was commonly used by other investigators, experimental and technical challenges (i.e. antibody clone and specificity, quality of IHC performance, etc.) might play role in the inconsistent effect of ARID1A identified in different studies. For example, in each of the seven studies conducted by IHC (16, 18, 28, 35, 37, 38, 42, 44), a different anti-ARID1A antibody is used, including the antibody’s clone, manufacturer, dilution rate, the IHC scoring, and the cut-off values. These factors could be potential reasons behind the different results obtained from each of these studies, especially the one by Bi and colleagues which is the only study that shows an oncogenic potential for ARID1A. The above data suggested that ARID1A variations regardless of low or high expression was correlated with the prognostic significance in CCA.
However, other studies have not found any correlation between ARID1A variation or expression level and disease prognoses in the CCA. Churi et al. (21) have identified 20 and 5% of variation for ARID1A in ICC and ECC, respectively. They have not found any significant correlation between ARID1A aberration and PFS or OS in either of ECC or ICC patients. In line with these findings, Javle et al. (25) have also found that ARID1A variations were not significantly associated with OS in both ICC and ECC patients (P > 0.05). Lowery et al. (27) demonstrated that ARID1A variations did not correlate with the metastatic site of ICC despite a high variant rate of 23%. Three studies developed in Asia also did not support a positive relationship between ARID1A variation and CCA prognosis. A study (16) in Japan revealed that there was no significant difference between groups with and without loss of ARID1A expression and OS in CCA patients (P = 0.9809). Similarly, Ma et al. (35) also demonstrated that ARID1A expressions were not significantly associated with DFS and OS in patients with ICC (P > 0.05). Xu et al. (41) reported the ARID1A variations rate was up to 20.3%, but the variant frequency of ARID1A did not show a significant difference between primary tumor and metastasis tumor samples. Based on these results, a significant correlation was not achieved between the ARID1A variations and the survival as well as the metastasis of CCA in these studies. A possible explanation for this phenomenon may be due to the low rate of ARID1A variations in CCA in some studies. For example, the ARID1A variations rate in ECC of Churi et al.’s study (21) was reported at only 5%, and the authors found that ARID1A variations were not significantly associated with PFS and OS in ECC (all P > 0.05). On the contrary, those studies reported the ARID1A variations rate over 20% were more likely to more likely to identify a poor prognostic significance for ARID1A variations in CCA (17, 18, 37, 42).
Collectively, ARID1A variations might be an important prognostic indicator that can predict disease mortality, metastasis, and recurrence in CCA patients, which also suggests that ARID1A could play important roles in the CCA progression and worth more attentions. Of note, since several studies have not supported such a prognostic value for ARID1A further studies with larger sample sizes are warranted to validate the prognostic values of ARID1A variations in CCA.
Biological Functions of ARID1A and Its Pathological Impact on CCA
Since a causal relationship between ARID1A alteration and CCA is suggestive from many clinical studies, a better understanding of the biological functions of ARID1A and its underlying mechanisms in CCA development is profound for the investigators. ARID1A is a driver gene encoding the DNA-binding subunit of the SWI/SNF chromatin-remodeling complexes. ARID1A provides specificity for SWI/SNF complex, facilitating proteinprotein or proteinDNA molecule interactions. Knockdown of ARID1A gene could induce dysregulation of cell cycle arrest thus enhance tumorigenesis (53). Inactivation of ARID1A might activate cell cycle progression, leading to an uncontrolled cellular proliferation in cancer cells. ARID1A commonly exerts the tumor-suppressive functions in CCA as well as multiple cancers. It was suggested that frequent variations in ARID1A related to perturbation in chromatin remodeling and chromosome organization might participate in the carcinogenesis and progression in CCA (54). Chan-on et al. (19) reported that ARID1A played a role of chromatin modulator in CCA pathogenesis, showing that silencing of ARID1A enhanced the CCA cells proliferation and upregulation of ARID1A causes disruption of cell proliferation. Yoshino et al. (42) indicated that ARID1A alteration could induce up-regulation of multiple genes (i.e., ALDH1A1, Aldehyde dehydrogenase 1A1, a potent cancer stem cell marker) in CCA cells. ARID1A-knockout in CCA cells lines promotes migration, invasion, and sphere formation activity, which might be correlated to transcriptional suppression of ALDH1A1 expression with decreasing histone H3K27 acetylation. A study conducted by Sasaki et al. (16) suggests that loss of ARID1A expression might be an early event in CCA development which presents a novel molecular pathway that is characterized by non-papillary and tubular adenocarcinoma. A precursor lesion with loss of ARID1A expression might cause a premalignant lesion of cholangiocarcinoma. TP53, also known as P53, is one of the most frequent genetic variants in human cancers, which plays a key role in the control of the cell cycle, apoptosis, and DNA repair (55). Alteration of PT53 is a prognostic biomarker for cancer due to its biological function of carcinogenesis. A meta-analysis indicated that TP53 might be a pivotal prognostic factor for the OS of patients with ECC (56). Both TP53 and ARID1A are frequently mutated in patients with CCA for their chromatin remodeling function. Interestingly, many researchers have found that TP53 and ARID1A variations appeared simultaneously in CCA (57). It was reported that ARID1A and P53 collaborated to prevent tumorigenesis by transcriptional activation of the tumor-inhibiting downstream genes (48). Therefore, the prognostic value and the molecular biological effect of ARID1A in CCA might partially depend on the alteration of TP53. Sasaki et al. (58) developed a study related to the combined hepatocellular-cholangiocarcinoma (cHC-CC) and found that the effect of ARID1A variations on the clinicopathological significance of cHC-CC might be correlated to oxidative stress and alpha-fetoprotein (AFP)-positivity. Farshidfar et al. (59) showed that ARID1A exhibited DNA hypermethylation and decreased expression of the isocitrate dehydrogenase (IDH) in the mutant subtype of CCA.
In a more recent study (60) recruited of 412 intrahepatic CCA, Boerner et al. found that both ARID1A (20%) and TP53 (17%) were among the most common oncogenic alterations in CCA. In line with this finding, Zhang et al. (43) also reported that both TP53 and ARID1A were among the most frequently mutated genes in intrahepatic CCA. In addition to TP53, ARID1A variations can co-occur and probably interact with multiple other genes (i.e., ALDH1A1, Beclin-1, BAP1, and PBRM1) which could be involved in CCA development. For example, Jiao et al. (20) demonstrated that genes involved in chromatin remodeling (including BAP1, ARID1A, and PBRM1), which was considered as the frequently targeted pathway in CCA, were somatically altered in almost half of the intrahepatic CCA cases. In 2013, Chan-on et al. (19) identified ARID1A and BAP1 as two new genes mutated in CCA. They further found that these two genes showed typical features of tumor suppressors and their mutations were mainly truncating and were scattered throughout the entire gene. However, we should also note that ARID1A might play an independent role in CCA development. Because some other studies [i.e. Sasaki et al. (16)] have failed to find CCA harboring both ARID1A and KRAS mutations (another frequently mutated gene in CCA), whereas these tumors had lost ARID1A expression suggesting that loss of ARID1A expression might represent an alternative (to ARID1A genomic variations) mechanism for ARID1A-driven carcinogenesis in CCA, and this could also be an alternative to KRAS mutations-driven CCA development. In line with this, Namjan et al. (37) have not been able to identify the co-occurrence of ARID1A and TP53/KRAS mutations in some CCA cases.
In summary, ARID1A variations appear to play important roles in the CCA tumorigenesis and progression. As shown in Figure 3, this schematic diagram summarizes that multifactorial mechanisms that are potentially involved in the ARID1A-driven CCA development, including opposing functions in cell cycle arrest, chromatin remodeling and chromosome organization, oxidative stress damage, DNA hypermethylation, downregulation of IDH, and the interaction of multiple genes (i.e., TP53, ALDH1A1, and Beclin-1 target) that enhance cellular proliferation and anti-apoptotic processes. However, further comprehensive researches are still warranted to better elucidate the underlying mechanisms of CCA development initiated by ARID1A variations.
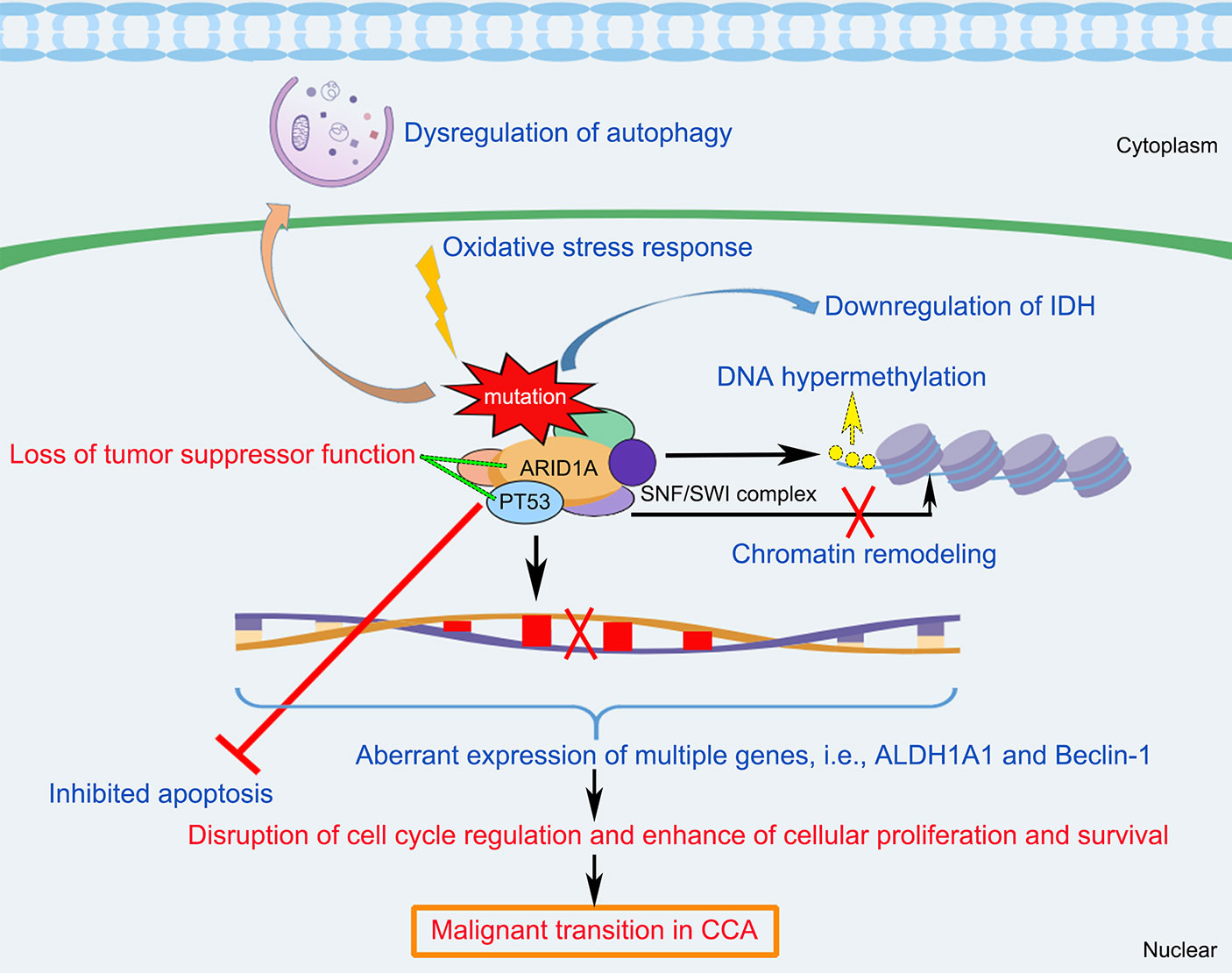
Figure 3 Schematic diagram of the molecular mechanisms underlying ARID1A variant and the pathogenesis and pathophysiology of CCA. ARID1A alteration associated with disruption of cell cycle regulation, chromatin remodeling, oxidative stress damage, DNA hypermethylation, downregulation of IDH, and the interaction of multiple genes (i.e., PT53, ALDH1A1, and Beclin-1), resulting in inhibition of apoptosis, dysregulation of autophagy, and enhance of cellular proliferation and survival, which cause loss of tumor suppressor functioning of ARID1A and induce malignant transition in CCA. ALDH1A1, Aldehyde dehydrogenase 1A1; ARID1A, AT-rich interaction domain 1A; IDH, isocitrate dehydrogenase; SWI/SNF, SWItch/Sucrose Non-Fermenting.
Limitations and Perspectives
To our knowledge, this is the first study for conducting a comprehensive review to summarize all the evidence of the relationship between ARID1A variations and CCA at both clinical and biological levels. However, some inherent limitations should be noted. First, the variation rate of ARID1A in CCA diverse in different studies, ranging from 5 to 68.2%. Different cancer types and stages, study design, sample size, geographical areas, gender, and age could all be partly responsible for this heterogeneity. Second, the prognostic significance of ARID1A in CCA is still controversial among the included studies. Nevertheless, we do not perform a meta-analysis for these studies due to due to unavailability of sufficient relevant data (i.e., OS, PFS, RFS, DFS, and metastatic site) in most of the studies. Third, the molecular mechanisms for the involvement of ARID1A variants in CCA tumorigenesis remain to be explored. In addition to ARID1A alteration, some other common contributors in cancer development, such as tumor microenvironment and tumor immunity, might also play roles in the pathomechanism of CCA, which should be further investigated.
Conclusions
This review has shown ARID1A variations in CCA cases are diverse in different studies, ranging from 5 to 68.2%. A higher ARID1A variation rate has been identified in the ICC subgroup when compared to the ECC subgroup (6.9–68.2% vs. 5–55%). Excepting for one study indicating the carcinogenic functions for ARID1A, the remaining 28 included studies suggest that ARID1A is a tumor suppressor in CCA. Though more studies show that loss or low ARID1A expression is significantly correlated with worse survival and recurrence in CCA, several studies do not support these prognostic values of ARID1A. Multifactorial mechanisms are involved in the pathogenesis of CCA caused by ARID1A variations. More investigations are necessary to validate the prognostic significance of ARID1A variations and to identify its molecular mechanism in CCA development.
Author Contributions
SZ, YX, and WW contributed to conceive and design the study. YW and PW performed the systematic searching. HJ and JZ extracted the data. SZ and JZ wrote the manuscript. YX and WW supervised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Welfare Technology Applied Research Project of Zhejiang Province (LGC20H200004), Reagent Project of Taizhou City, Zhejiang Province (ID: 1901ky38), the High-level Hospital Construction Research Project of Maoming People’s Hospital, and the Technology Planning Project of Taizhou City, Zhejiang Province (ID: 20ywb40).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Bergquist A, von Seth E. Epidemiology of Cholangiocarcinoma. Best Pract Res Clin Gastroenterol (2015) 29(2):221–32. doi: 10.1016/j.bpg.2015.02.003
2. Entezari P, Riaz A. Intrahepatic Cholangiocarcinoma. Semin Intervent Radiol (2020) 37(5):475–83. doi: 10.1055/s-0040-1719188
3. Fostea RM, Fontana E, Torga G, Arkenau HT. Recent Progress in the Systemic Treatment of Advanced/Metastatic Cholangiocarcinoma. Cancers (Basel) (2020) 12(9):2599. doi: 10.3390/cancers12092599
4. Weber SM, Ribero D, O’Reilly EM, Kokudo N, Miyazaki M, Pawlik TM. Intrahepatic Cholangiocarcinoma: Expert Consensus Statement. HPB (Oxford) (2015) 17(8):669–80. doi: 10.1111/hpb.12441
5. Petrick JL, Thistle JE, Zeleniuch-Jacquotte A, Zhang X, Wactawski-Wende J, Van Dyke AL, et al. Body Mass Index, Diabetes and Intrahepatic Cholangiocarcinoma Risk: The Liver Cancer Pooling Project and Meta-Analysis. Am J Gastroenterol (2018) 113(10):1494–505. doi: 10.1038/s41395-018-0207-4
6. Nathan H, Pawlik TM, Wolfgang CL, Choti MA, Cameron JL, Schulick RD. Trends in Survival After Surgery for Cholangiocarcinoma: A 30-Year Population-Based SEER Database Analysis. J Gastrointest Surg (2007) 11(11):1488–96; discussion 1496-7. doi: 10.1007/s11605-007-0282-0
7. Boscoe AN, Rolland C, Kelley RK. Frequency and Prognostic Significance of Isocitrate Dehydrogenase 1 Mutations in Cholangiocarcinoma: A Systematic Literature Review. J Gastrointest Oncol (2019) 10(4):751–65. doi: 10.21037/jgo.2019.03.10
8. Wilson BG, Roberts CW. SWI/SNF Nucleosome Remodellers and Cancer. Nat Rev Cancer (2011) 11(7):481–92. doi: 10.1038/nrc3068
9. Kadoch C, Crabtree GR. Reversible Disruption of mSWI/SNF (BAF) Complexes by the SS18-SSX Oncogenic Fusion in Synovial Sarcoma. Cell (2013) 153(1):71–85. doi: 10.1016/j.cell.2013.02.036
10. Wang X, Nagl NG, Wilsker D, Van Scoy M, Pacchione S, Yaciuk P, et al. Two Related ARID Family Proteins Are Alternative Subunits of Human SWI/SNF Complexes. Biochem J (2004) 383(Pt 2):319–25. doi: 10.1042/BJ20040524
11. Hohmann AF, Vakoc CR. A Rationale to Target the SWI/SNF Complex for Cancer Therapy. Trends Genet (2014) 30(8):356–63. doi: 10.1016/j.tig.2014.05.001
12. Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J, et al. Proteomic and Bioinformatic Analysis of Mammalian SWI/SNF Complexes Identifies Extensive Roles in Human Malignancy. Nat Genet (2013) 45(6):592–601. doi: 10.1038/ng.2628
13. Wiegand KC, Shah SP, Al-Agha OM, Zhao Y, Tse K, Zeng T, et al. ARID1A Mutations in Endometriosis-Associated Ovarian Carcinomas. N Engl J Med (2010) 363(16):1532–43. doi: 10.1056/NEJMoa1008433
14. Sasaki M, Sato Y, Nakanuma Y. Cholangiolocellular Carcinoma With “Ductal Plate Malformation” Pattern may Be Characterized by ARID1A Genetic Alterations. Am J Surg Pathol (2019) 43(3):352–60. doi: 10.1097/PAS.0000000000001201
15. Shiao MS, Chiablaem K, Charoensawan V, Ngamphaiboon N, Jinawath N. Emergence of Intrahepatic Cholangiocarcinoma: How High-Throughput Technologies Expedite the Solutions for a Rare Cancer Type. Front Genet (2018) 9:309. doi: 10.3389/fgene.2018.00309
16. Sasaki M, Nitta T, Sato Y, Nakanuma Y. Loss of ARID1A Expression Presents a Novel Pathway of Carcinogenesis in Biliary Carcinomas. Am J Clin Pathol (2016) 145(6):815–25. doi: 10.1093/ajcp/aqw071
17. Yang SZ, Wang AQ, Du J JT, Yu WW, Liu Q, Wu YF, et al. Low Expression of ARID1A Correlates With Poor Prognosis in Intrahepatic Cholangiocarcinoma. World J Gastroenterol (2016) 22(25):5814–21. doi: 10.3748/wjg.v22.i25.5814
18. Bi C, Liu M, Rong W, Wu F, Zhang Y, Lin S, et al. High Beclin-1 and ARID1A Expression Corelates With Poor Survival and High Recurrence in Intrahepatic Cholangiocarcinoma: A Histopathological Retrospective Study. BMC Cancer (2019) 19(1):213. doi: 10.1186/s12885-019-5429-3
19. Chan-On W, Nairismagi ML, Ong CK, Lim WK, Dima S, Pairojkul C, et al. Exome Sequencing Identifies Distinct Mutational Patterns in Liver Fluke-Related and Non-Infection-Related Bile Duct Cancers. Nat Genet (2013) 45(12):1474–8. doi: 10.1038/ng.2806
20. Jiao Y, Pawlik TM, Anders RA, Selaru FM, Streppel MM, Lucas DJ, et al. Exome Sequencing Identifies Frequent Inactivating Mutations in BAP1, ARID1A and PBRM1 in Intrahepatic Cholangiocarcinomas. Nat Genet (2013) 45(12):1470–3. doi: 10.1038/ng.2813
21. Churi CR, Shroff R, Wang Y, Rashid A, Kang HC, Weatherly J, et al. Mutation Profiling in Cholangiocarcinoma: Prognostic and Therapeutic Implications. PloS One (2014) 9(12):e115383. doi: 10.1371/journal.pone.0115383
22. Ross JS, Wang K, Gay L, Al-Rohil R, Rand JV, Jones DM, et al. New Routes to Targeted Therapy of Intrahepatic Cholangiocarcinomas Revealed by Next-Generation Sequencing. Oncologist (2014) 19(3):235–42. doi: 10.1634/theoncologist.2013-0352
23. Simbolo M, Fassan M, Ruzzenente A, Mafficini A, Wood LD, Corbo V, et al. Multigene Mutational Profiling of Cholangiocarcinomas Identifies Actionable Molecular Subgroups. Oncotarget (2014) 5(9):2839–52. doi: 10.18632/oncotarget.1943
24. Zou S, Li J, Zhou H, Frech C, Jiang X, Chu JS, et al. Mutational Landscape of Intrahepatic Cholangiocarcinoma. Nat Commun (2014) 5:5696. doi: 10.1038/ncomms6696
25. Javle M, Bekaii-Saab T, Jain A, Wang Y, Kelley RK, Wang K, et al. Biliary Cancer: Utility of Next-Generation Sequencing for Clinical Management. Cancer-Am Cancer Soc (2016) 122(24):3838–47. doi: 10.1002/cncr.30254
26. Ruzzenente A, Fassan M, Conci S, Simbolo M, Lawlor RT, Pedrazzani C, et al. Cholangiocarcinoma Heterogeneity Revealed by Multigene Mutational Profiling: Clinical and Prognostic Relevance in Surgically Resected Patients. Ann Surg Oncol (2016) 23(5):1699–707. doi: 10.1245/s10434-015-5046-6
27. Lowery MA, Ptashkin R, Jordan E, Berger MF, Zehir A, Capanu M, et al. Comprehensive Molecular Profiling of Intrahepatic and Extrahepatic Cholangiocarcinomas: Potential Targets for Intervention. Clin Cancer Res (2018) 24(17):4154–61. doi: 10.1158/1078-0432.CCR-18-0078
28. Simbolo M, Vicentini C, Ruzzenente A, Brunelli M, Conci S, Fassan M, et al. Genetic Alterations Analysis in Prognostic Stratified Groups Identified TP53 and ARID1A as Poor Clinical Performance Markers in Intrahepatic Cholangiocarcinoma. Sci Rep (2018) 8(1):7119. doi: 10.1038/s41598-018-25669-1
29. Balitzer D, Joseph NM, Ferrell L, Shafizadeh N, Jain D, Zhang X, et al. Immunohistochemical and Molecular Features of Cholangiolocellular Carcinoma Are Similar to Well-Differentiated Intrahepatic Cholangiocarcinoma. Mod Pathol (2019) 32(10):1486–94. doi: 10.1038/s41379-019-0290-0
30. Joseph NM, Tsokos CG, Umetsu SE, Shain AH, Kelley RK, Onodera C, et al. Genomic Profiling of Combined Hepatocellular-Cholangiocarcinoma Reveals Similar Genetics to Hepatocellular Carcinoma. J Pathol (2019) 248(2):164–78. doi: 10.1002/path.5243
31. Xue L, Guo C, Zhang K, Jiang H, Pang F, Dou Y, et al. Comprehensive Molecular Profiling of Extrahepatic Cholangiocarcinoma in Chinese Population and Potential Targets for Clinical Practice. Hepatobiliary Surg Nutr (2019) 8(6):615–22. doi: 10.21037/hbsn.2019.08.05
32. Conci S, Ruzzenente A, Simbolo M, Bagante F, Rusev B, Isa G, et al. Multigene Mutational Profiling of Biliary Tract Cancer Is Related to the Pattern of Recurrence in Surgically Resected Patients. Updates Surg (2020) 72(1):119–28. doi: 10.1007/s13304-020-00718-5
33. Hogdall D, Larsen OF, Linnemann D, Svenstrup PT, Hogdall EV. Exome Sequencing of 22 Genes Using Tissue From Patients With Biliary Tract Cancer. APMIS (2020) 128(1):3–9. doi: 10.1111/apm.13003
34. Jiang G, Zhang W, Wang T, Ding S, Shi X, Zhang S, et al. Characteristics of Genomic Alterations in Chinese Cholangiocarcinoma Patients. Jpn J Clin Oncol (2020) 50(10):1117–25. doi: 10.1093/jjco/hyaa088
35. Ma B, Meng H, Tian Y, Wang Y, Song T, Zhang T, et al. Distinct Clinical and Prognostic Implication of IDH1/2 Mutation and Other Most Frequent Mutations in Large Duct and Small Duct Subtypes of Intrahepatic Cholangiocarcinoma. BMC Cancer (2020) 20(1):318. doi: 10.1186/s12885-020-06804-6
36. Montal R, Sia D, Montironi C, Leow WQ, Esteban-Fabro R, Pinyol R, et al. Molecular Classification and Therapeutic Targets in Extrahepatic Cholangiocarcinoma. J Hepatol (2020) 73(2):315–27. doi: 10.1016/j.jhep.2020.03.008
37. Namjan A, Techasen A, Loilome W, Sa-Ngaimwibool P, Jusakul A. ARID1A Alterations and Their Clinical Significance in Cholangiocarcinoma. PeerJ (2020) 8:e10464. doi: 10.7717/peerj.10464
38. Sasaki M, Sato Y, Nakanuma Y. Bile Duct Adenoma may be a Precursor Lesion of Small Duct Type Intrahepatic Cholangiocarcinoma. Histopathology (2021) 78(2):310–20. doi: 10.1111/his.14222
39. Tian W, Hu W, Shi X, Liu P, Ma X, Zhao W, et al. Comprehensive Genomic Profile of Cholangiocarcinomas in China. Oncol Lett (2020) 19(4):3101–10. doi: 10.3892/ol.2020.11429
40. Wang L, Zhu H, Zhao Y, Pan Q, Mao A, Zhu W, et al. Comprehensive Molecular Profiling of Intrahepatic Cholangiocarcinoma in the Chinese Population and Therapeutic Experience. J Transl Med (2020) 18(1):273. doi: 10.1186/s12967-020-02437-2
41. Xu SF, Guo Y, Zhang X, Zhu XD, Fan N, Zhang ZL, et al. Somatic Mutation Profiling of Intrahepatic Cholangiocarcinoma: Comparison Between Primary and Metastasis Tumor Tissues. J Oncol (2020) 2020:5675020. doi: 10.1155/2020/5675020
42. Yoshino J, Akiyama Y, Shimada S, Ogura T, Ogawa K, Ono H, et al. Loss of ARID1A Induces a Stemness Gene ALDH1A1 Expression With Histone Acetylation in the Malignant Subtype of Cholangiocarcinoma. Carcinogenesis (2020) 41(6):734–42. doi: 10.1093/carcin/bgz179
43. Zhang R, Li Q, Fu J, Jin Z, Su J, Zhang J, et al. Comprehensive Analysis of Genomic Mutation Signature and Tumor Mutation Burden for Prognosis of Intrahepatic Cholangiocarcinoma. BMC Cancer (2021) 21(1):112. doi: 10.1186/s12885-021-07788-7
44. Wagner B, Plum P, Apel K, Scherer M, Buchner D, Brinkmann S, et al. Protein-Loss of SWI/SNF-complex Core Subunits Influences Prognosis Dependent on Histological Subtypes of Intra- and Extrahepatic Cholangiocarcinoma. Oncol Lett (2021) 21(5):349. doi: 10.3892/ol.2021.12610
45. Centore RC, Sandoval GJ, Soares L, Kadoch C, Chan HM. Mammalian SWI/SNF Chromatin Remodeling Complexes: Emerging Mechanisms and Therapeutic Strategies. Trends Genet (2020) 36(12):936–50. doi: 10.1016/j.tig.2020.07.011
46. Mao TL, Shih I. The Roles of ARID1A in Gynecologic Cancer. J Gynecol Oncol (2013) 24(4):376–81. doi: 10.3802/jgo.2013.24.4.376
47. Goswami S, Chen Y, Anandhan S, Szabo PM, Basu S, Blando JM, et al. ARID1A Mutation Plus CXCL13 Expression Act as Combinatorial Biomarkers to Predict Responses to Immune Checkpoint Therapy in Mucc. Sci Transl Med (2020) 12(548):eabc4220. doi: 10.1126/scitranslmed.abc4220
48. Mathur R. ARID1A Loss in Cancer: Towards a Mechanistic Understanding. Pharmacol Ther (2018) 190:15–23. doi: 10.1016/j.pharmthera.2018.05.001
49. Yoon KA, Woo SM, Kim YH, Kong SY, Han SS, Park SJ, et al. Somatic Mutations From Whole Exome Sequencing Analysis of the Patients With Biliary Tract Cancer. Genomics Inform (2018) 16(4):e35. doi: 10.5808/GI.2018.16.4.e35
50. Ito T, Sakurai-Yageta M, Goto A, Pairojkul C, Yongvanit P, Murakami Y. Genomic and Transcriptional Alterations of Cholangiocarcinoma. J Hepatobiliary Pancreat Sci (2014) 21(6):380–7. doi: 10.1002/jhbp.67
51. Wu JN, Roberts CW. ARID1A Mutations in Cancer: Another Epigenetic Tumor Suppressor? Cancer Discov (2013) 3(1):35–43. doi: 10.1158/2159-8290.CD-12-0361
52. Sun X, Wang SC, Wei Y, Luo X, Jia Y, Li L, et al. Arid1a Has Context-Dependent Oncogenic and Tumor Suppressor Functions in Liver Cancer. Cancer Cell (2017) 32(5):574–589.e6. doi: 10.1016/j.ccell.2017.10.007
53. Nagl NJ, Patsialou A, Haines DS, Dallas PB, Beck GJ, Moran E. The p270 (ARID1A/SMARCF1) Subunit of Mammalian SWI/SNF-Related Complexes Is Essential for Normal Cell Cycle Arrest. Cancer Res (2005) 65(20):9236–44. doi: 10.1158/0008-5472.CAN-05-1225
54. Liu ZH, Lian BF, Dong QZ, Sun H, Wei JW, Sheng YY, et al. Whole-Exome Mutational and Transcriptional Landscapes of Combined Hepatocellular Cholangiocarcinoma and Intrahepatic Cholangiocarcinoma Reveal Molecular Diversity. Biochim Biophys Acta Mol Basis Dis (2018) 1864(6 Pt B):2360–8. doi: 10.1016/j.bbadis.2018.01.027
55. Pinto EM, Zambetti GP. What 20 Years of Research Has Taught Us About the TP53 p.R337H Mutation. Cancer-Am Cancer Soc (2020) 126(21):4678–86. doi: 10.1002/cncr.33143
56. Wang J, Wang X, Xie S, Yan Z, Li Z, Li Y, et al. p53 Status and Its Prognostic Role in Extrahepatic Bile Duct Cancer: A Meta-Analysis of Published Studies. Dig Dis Sci (2011) 56(3):655–62. doi: 10.1007/s10620-010-1352-9
57. Chaisaingmongkol J, Budhu A, Dang H, Rabibhadana S, Pupacdi B, Kwon SM, et al. Common Molecular Subtypes Among Asian Hepatocellular Carcinoma and Cholangiocarcinoma. Cancer Cell (2017) 32(1):57–70.e3. doi: 10.1016/j.ccell.2017.05.009
58. Sasaki M, Sato Y, Nakanuma Y. Mutational Landscape of Combined Hepatocellular Carcinoma and Cholangiocarcinoma, and Its Clinicopathological Significance. Histopathology (2017) 70(3):423–34. doi: 10.1111/his.13084
59. Farshidfar F, Zheng S, Gingras MC, Newton Y, Shih J, Robertson AG, et al. Integrative Genomic Analysis of Cholangiocarcinoma Identifies Distinct Idh-Mutant Molecular Profiles. Cell Rep (2017) 18(11):2780–94. doi: 10.1016/j.celrep.2017.02.033
Keywords: ARID1A variations, cholangiocarcinoma, biomarker, prognosis (carcinoma), pathogenesis
Citation: Zhao S, Xu Y, Wu W, Wang P, Wang Y, Jiang H and Zhu J (2021) ARID1A Variations in Cholangiocarcinoma: Clinical Significances and Molecular Mechanisms. Front. Oncol. 11:693295. doi: 10.3389/fonc.2021.693295
Received: 10 April 2021; Accepted: 03 June 2021;
Published: 25 June 2021.
Edited by:
Jiayi Wang, Shanghai Jiaotong University, ChinaReviewed by:
Steven de Jong, University Medical Center Groningen, NetherlandsReza Ghasemi, Washington University School of Medicine in St. Louis, United States
Copyright © 2021 Zhao, Xu, Wu, Wang, Wang, Jiang and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jie Zhu, emh1ajIxMDBAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Shankun Zhao
Shankun Zhao Youwen Xu2†
Youwen Xu2† Jie Zhu
Jie Zhu