- Shanghai Institute of Hematology, State Key Laboratory of Medical Genomics, National Research Centre for Translational Medicine at Shanghai, Ruijin Hospital Affiliated to Shanghai Jiao Tong University School of Medicine, Shanghai, China
The clinically ideal time point and optimal approach for the assessment of measurable residual disease (MRD) in patients with acute myeloid leukemia (AML) are still inconclusive. We investigated the clinical value of multiparameter flow cytometry-based MRD (MFC MRD) after induction (n = 492) and two cycles of consolidation (n = 421). The latter time point was proved as a superior indicator with independent prognostic significance for both relapse-free survival (RFS, HR = 3.635, 95% CI: 2.433–5.431, P <0.001) and overall survival (OS: HR = 3.511, 95% CI: 2.191–5.626, P <0.001). Furthermore, several representative molecular MRD markers were compared with the MFC MRD. Both approaches can establish prognostic value in patients with NPM1 mutations, and FLT3, C-KIT, or N-RAS mutations involved in kinase-related signaling pathways, while the combination of both techniques further refined the risk stratification. The detection of RUNX1–RUNX1T1 fusion transcripts achieved a considerable net reclassification improvement in predicting the prognosis. Conversely, for patients with biallelic CEBPA or DNMT3A mutations, only the MFC method was recommended due to the poor prognostic discriminability in tracking mutant transcripts. In conclusion, this study demonstrated that the MFC MRD after two consolidation cycles independently predicted clinical outcomes, and the integration of MFC and molecular MRD should depend on different types of AML-related genetic lesions.
Introduction
Acute myeloid leukemia (AML) is a group of hematological malignant disorders characterized by the high heterogeneity in clinical manifestation, genetic abnormalities, and prognosis (1). Via the treatment modalities as exemplified by cytotoxic drugs or molecular targeted therapies, a high complete remission (CR) rate could be achieved, however, a substantial proportion of AML patients will relapse due to residual leukemic cells below the detection threshold of traditional morphologic methods. To achieve a long term remission and survival, post-remission therapies should be administered not only based on pre-treatment parameters including age, cytogenetics, and a limited set of molecular genetic markers, but also on several post-treatment factors, among which the detection of residual disease is of utmost importance (2, 3).
Measurable residual disease (MRD; also named minimal residual disease) detected in AML patients with hematological complete remission after treatment has been suggested as a powerful prognostic indicator (4, 5). In general clinical practice, MRD is reliably monitored using the two most common methods, multiparameter flow cytometry-based MRD (MFC MRD) and polymerase chain reaction (PCR)-based MRD (Gene MRD), quantifying MRD in AML by virtue of either immunophenotype or molecular abnormalities of leukemic cells. However, until now, issues on when and how to apply the evaluation of MRD in daily clinical practice are still controversial.
Numerous studies have highlighted the prognostic impact of MFC MRD assessment at diverse time points in AML patients, as exemplified by post-induction (PI), post-consolidation (PC), before and after hematopoietic stem cell transplantation (HSCT) (6–9), whereas the clinically ideal time point for MRD assessment is inconclusive. On the other side, the existing two common approaches differ in sensitivity and applicability. The MFC MRD can be monitored in the majority of AML patients with rapid turnaround time, but an established threshold with considerably high sensitivity and reproducibility is still an unmet clinical need (7, 10). Conversely, although the PCR technique is highly sensitive, its application is limited to a fraction of AML patients who harbored specific genetic aberrations suitable for MRD detection, including RUNX1–RUNX1T1, CBFB–MYH11, and PML–RARα fusions, and NPM1 mutations (5, 11–16). Notably, the next-generation sequencing (NGS) has introduced novel molecular markers for MRD assessment, and reliable criteria for their routine application in the clinical setting are under exploration. Consequently, how to compare and integrate current procedures for MRD monitoring is of great clinical significance.
In this study, we evaluated the prognostic value of the MFC MRD after induction and two consolidation cycles in AML to identify the optimal time point for MRD measurement. Furthermore, the tracking of molecular MRD on a series of AML-related gene alterations was compared with the MFC MRD, highlighting the necessity of combining both MFC and molecular techniques to establish an integrated methodology for MRD monitoring.
Materials and Methods
Patients and Treatment
From January 2011 to June 2018, a total of 833 consecutive newly diagnosed AML (non-M3) patients treated in Ruijin Hospital were enrolled. Among which, the majority of patients diagnosed after 2015 participated in one of three phase II/III clinical trials, which were registered at the Chinese Clinical Trial Registry (www.chictr.org.cn: ChiCTR-OPC-15006085; ChiCTR-OIC-16007764; ChiCTR-OIN-16008955).
Young patients (<60 years) were given standard intensive “3 + 7” IA/DA-based regimens as initial induction, which contained idarubicin/daunorubicin (10–12 mg/m2/45–60 mg/m2, D1–3) and cytarabine (100–200 mg/m2 D1–7). When CR was achieved, four cycles of high-dose cytarabine (2 g/m2) were delivered as consolidation. Elderly patients (≥60 years) were evaluated for the fitness by the treating physician. Fit patients received the same induction regimens as young patients but reduced the consolidation to two cycles of high-dose cytarabine. Unfit patients received either low dose “3 + 7” regimens, demethylation agents, or other less intensive therapies at the discretion of the physician. Treatment protocols of the three clinical trials are provided in Supplementary Methods.
Multiparameter Flow Cytometry
Bone marrow aspirate samples were obtained at diagnosis and before the first and third cycles of consolidation chemotherapy, which were processed through the standard procedure of our institution (17). The MFC MRD was monitored by using the 10-color flow cytometry, and monoclonal antibodies against 21 antigens, including stem cell and progenitor markers (CD34, CD38, CD45, CD117, CD123, and HLA-DR), myelomonocytic markers (CD13, CD11b, CD14, CD15, CD33, MPO, and CD64), and lymphoid lineage markers (CD2, cyCD3, CD4, CD7, CD19, cyCD79a, TdT, and CD56) were utilized. Identical antibody-fluorochrome combinations at diagnosis and during the follow-up period were utilized for tracking established LAIPs and newly emerging aberrant immunophenotypes. For statistical analyses, the “LAIP‐based different‐from‐normal approach” was applied. Detailed definitions concerning MFC are provided in Supplementary Methods.
Molecular Events
Molecular alterations of AML in this study were selected according to the established laboratory developed tests (LDTs) at Shanghai Institute of Hematology based on our previous work conducted in a large AML cohort, in which gene mutations and fusions showing significant prognostic value were then tested in daily clinical routine (18). Genetic alterations including FLT3-ITD/TKD, KMT2A-PTD, NPM1, NRAS, CKIT, CEBPA, DNMT3A, IDH1, and IDH2 were detected by PCR and Sanger sequencing. RUNX1–RUNX1T1, CBFβ–MYH11, and KMT2A-related fusion genes were detected via reverse transcription (RT)-PCR strategy as previously reported (19). The level of RUNX1–RUNX1T1 transcripts was measured by quantitative real-time PCR (qPCR), and a >3-log reduction compared with the baseline level at diagnosis was defined as molecular MRD negativity according to the published literature (14, 20).
Statistical Analyses
Kaplan–Meier and hazard ratio analyses were used to calculate and compare the relapse-free survival (RFS) and overall survival (OS). The Cox proportional hazard regression model was applied for the multivariate analysis of independent factors for RFS and OS. To investigate the prognostic accuracy of MRD status by the MFC and molecular methods, the net reclassification improvement (NRI) (21) was used to measure the net gain in risk reclassification between different techniques for MRD monitoring. All of the statistical procedures mentioned above were carried out using the R (version 4.0.0) and the SPSS (version 26.0) software packages.
Results
Characteristics of Patients and Their Associations With MFC MRD Status
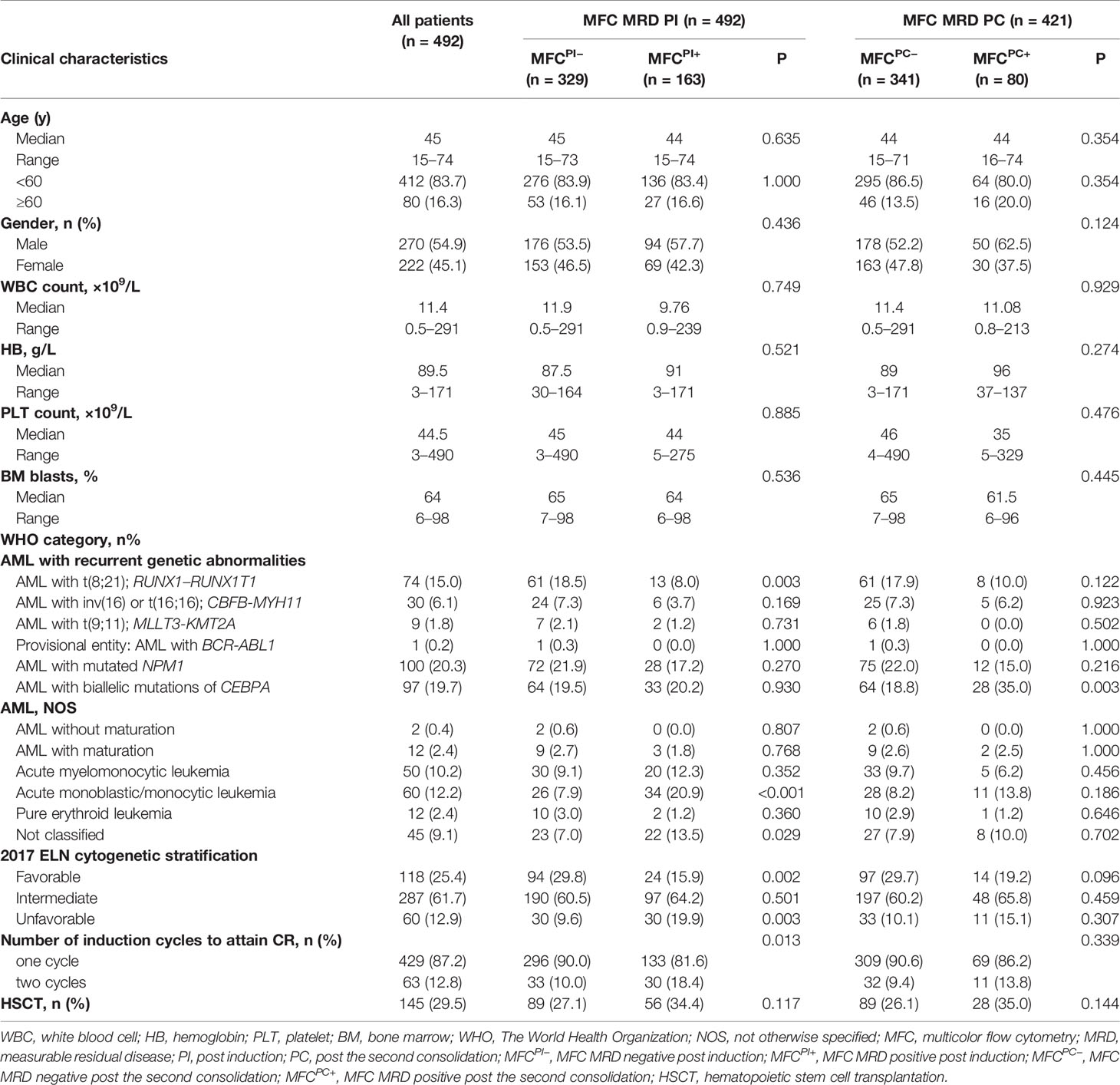
The patient flow diagram is depicted in Figure S1. Among the 833 AML patients, 639 (76.7%) achieved CR after induction chemotherapy, of whom 587 (91.9%) patients had specific LAIPs at diagnosis that were suitable for MRD monitoring by flow cytometry. The genetic alterations of AML patients stratified by LAIPs are shown in Table S1, and the most frequent LAIPs are summarized in Table S2. A higher frequency of RUNX1–RUNX1TI (P = 0.031) and biallelic CEBPA (BiCEBPA, P = 0.001) mutations, but lower frequency of CBFβ–MYH11 (P = 0.009) were observed in patients who had LAIPs at diagnosis. MRD analysis by MFC was available in 492 patients after induction, among which, 24 patients chose HSCT as consolidation, 41 patients relapsed, and six patients died. Consequently, 421 patients who were treated with chemotherapy only remained in CR and received MRD monitoring after two cycles of consolidation therapy. By comparing different cut-off levels including 0, 0.01, 0.035, 0.1, and 1% to distinguish MRD+ from MRD- patients, the cut-off of 0.1% was proved to be most relevant to prognosis, displaying significant disparities in both RFS and OS (Figure 1, Figure S2). Therefore, the level of ≥0.1% was considered as MRD positive in this study. As shown in Table 1, when one to two cycles of induction chemotherapy were completed, 329 (66.9%) of the 492 patients were classified into MFC MRD negative group, termed as MFCPI–. Moreover, 340 (81%) of 421 achieved MFC MRD negativity after two cycles of consolidation chemotherapy, termed as MFCPC–. There were no significant differences in age, gender, peripheral blood count, and BM blasts between MFC– and MFC+ at both time points. Patients in the MFCPI+ group were less likely to carry RUNX1–RUNX1T1 (P = 0.003), and those with MFCPC+ were more likely to harbor biallelic CEBPA (BiCEBPA) mutations at diagnosis (P = 0.003). In cytogenetic risk stratification, favorable cytogenetic abnormalities were less common in patients with MFCPI+ (P = 0.002), while a higher frequency of unfavorable risk was seen in the MFCPI+ group (P = 0.003). Besides, patients who required two induction cycles to attain CR were more likely to have MFCPI+ (P = 0.013). The follow-up of all patients ended in April 2020, with a median follow-up time of 45 (range 1–108) months.
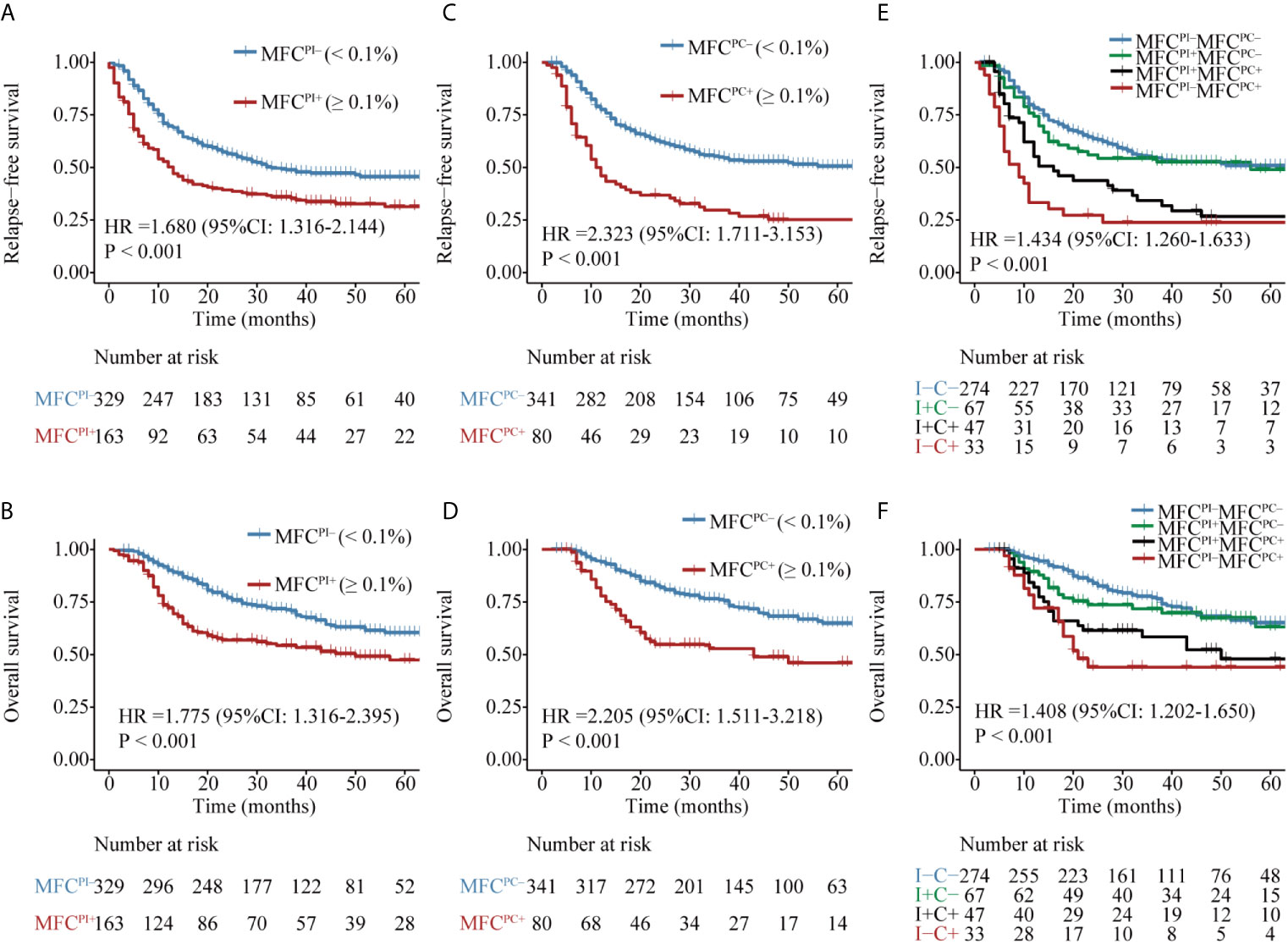
Figure 1 Kaplan–Meier curves for probability of relapse-free survival (RFS) and overall survival (OS) in AML patients. (A, B) RFS and OS based on MFC MRD status post induction. (C, D) RFS and OS based on MFC MRD status post two consolidation cycles. (E, F) RFS and OS based on MFC MRD status in the combination of both time points.
Prognostic Significance of MFC MRD at Different Time Points
Patients with MFCPI– (median MRD, 0; range, 0–0.09%) had a significantly favorable RFS and OS than those with MFCPI+ (median MRD, 0.43%; range, 0.1–11.4%) (Figures 1A, B and Table S3). Similarly, patients in the MFCPC– group (median MRD, 0; range, 0–0.09%) had a better prognosis than those whose MFC MRD status was positive (median MRD, 0.27%; range, 0.1–4.81%) (Figures 1C, D and Table S3). Of note, the status of MFCPC seemed to provide a better discrimination ability for both short- and long-term survival than that of MFCPI.
The dynamics of MFC MRD status after induction and the second cycle of consolidation therapy in the 421 patients who experienced MFC MRD monitoring at both time points were integratively evaluated. Patients were stratified into four groups based on the MFC MRD status at the two checkpoints. As shown in Figures 1E, F, there was no difference in the distribution of RFS and OS between MFCPI–MFCPC– and MFCPI+MFCPC– patients (P = 0.787), and between patients with MFCPI+MFCPC+ and MFCPI-MFCPC+ (P = 0.408), while both groups conferred an inferior prognosis compared to those with MFCPC–. Intriguingly, the prognostic impact of the MFCPC MRD status in both MFCPI– and MFCPI+ patients was significant (Figures 1E, F and Table S3). The prognostic value of the MFCPC MRD status was also observed in young and old AML patients, respectively (Figures S3, S4), and in the ELN low and intermediate cytogenetic risk group, respectively (Figures S5, S6), while it was not significant in the high-risk group (Figure S7).
The Post-Consolidation MFC MRD Was an Independent Prognostic Factor
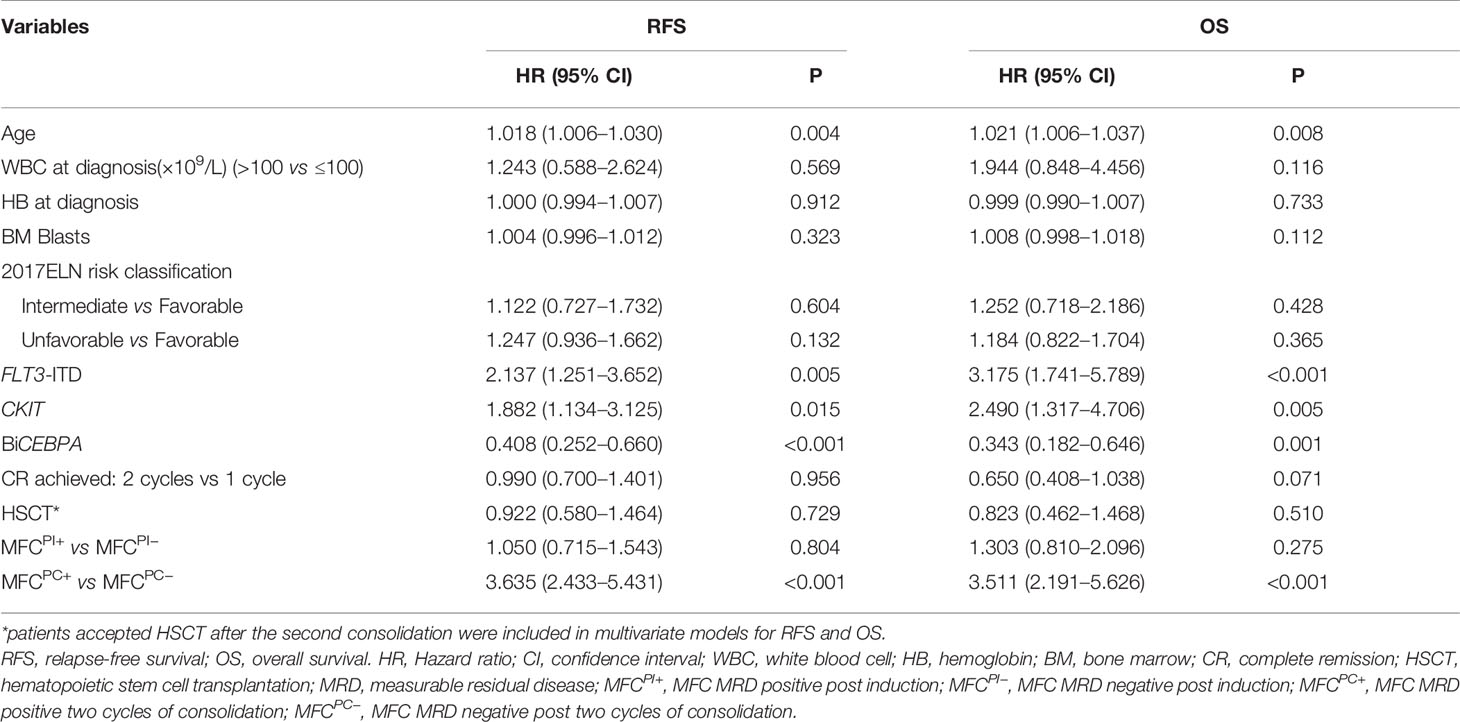
Age, gender, WBC count, hemoglobin (Hb), platelet count, BM blasts, cytogenetic risk stratification, recurrent genetic alterations at diagnosis, number of induction cycles to CR, HSCT, MFCPI, and MFCPC MRD status were included in the univariate analysis for both RFS and OS (Table S4), in which factors with P <0.1 were entered into the multivariable Cox analysis. As shown in Table 2, after adjusting the impact of well-established prognostic indicators such as age, cytogenetic risk stratification, FLT3-ITD, CKIT, and BiCEBPA mutations at diagnosis, the post-consolidation MFC MRD status was independently associated with both RFS and OS of AML patients (RFS: HR = 3.635, 95% CI: 2.433–5.431, P <0.001; OS: HR = 3.511, 95% CI: 2.191–5.626, P <0.001). In the ELN intermediate cytogenetic risk group, the MFCPC MRD was also an independent prognostic factor for both RFS and OS (Tables S5, S6).
Comparison of MRD Assessment by Different Detection Modalities
The clinical utility of molecular MRD and MFC MRD in diverse types of genetic abnormalities was compared in patients with MRD monitoring by both methods after two consolidation cycles. A series of gene markers either of fusions or mutations with certain incidences were selected and described as follows according to gene type and function.
Firstly, the two approaches were compared in 50 patients who harbored the RUNX1–RUNX1T1 fusion gene at diagnosis. Both MFC MRD status (median RFS, 33 vs 5 months, P = 0.008, median OS, NR vs 12 months, P <0.001) (Figures 2A, B) and molecular MRD status (median RFS, NR vs 11 months, P = 0.003; median OS, NR, P = 0.012) Figures 2C, D could distinguish patients with a relatively favorable outcome from those with an increased risk of relapse and mortality, while the presence of both molecular and MFC MRD indicated the worst prognosis (Figures 2E, F). Of note, the NRI of molecular MRD in 2-year RFS and OS was 21.9 and 15.5%, respectively, compared with MFC MRD (Figure 3), and more importantly, there was no improvement when two methods were combined.
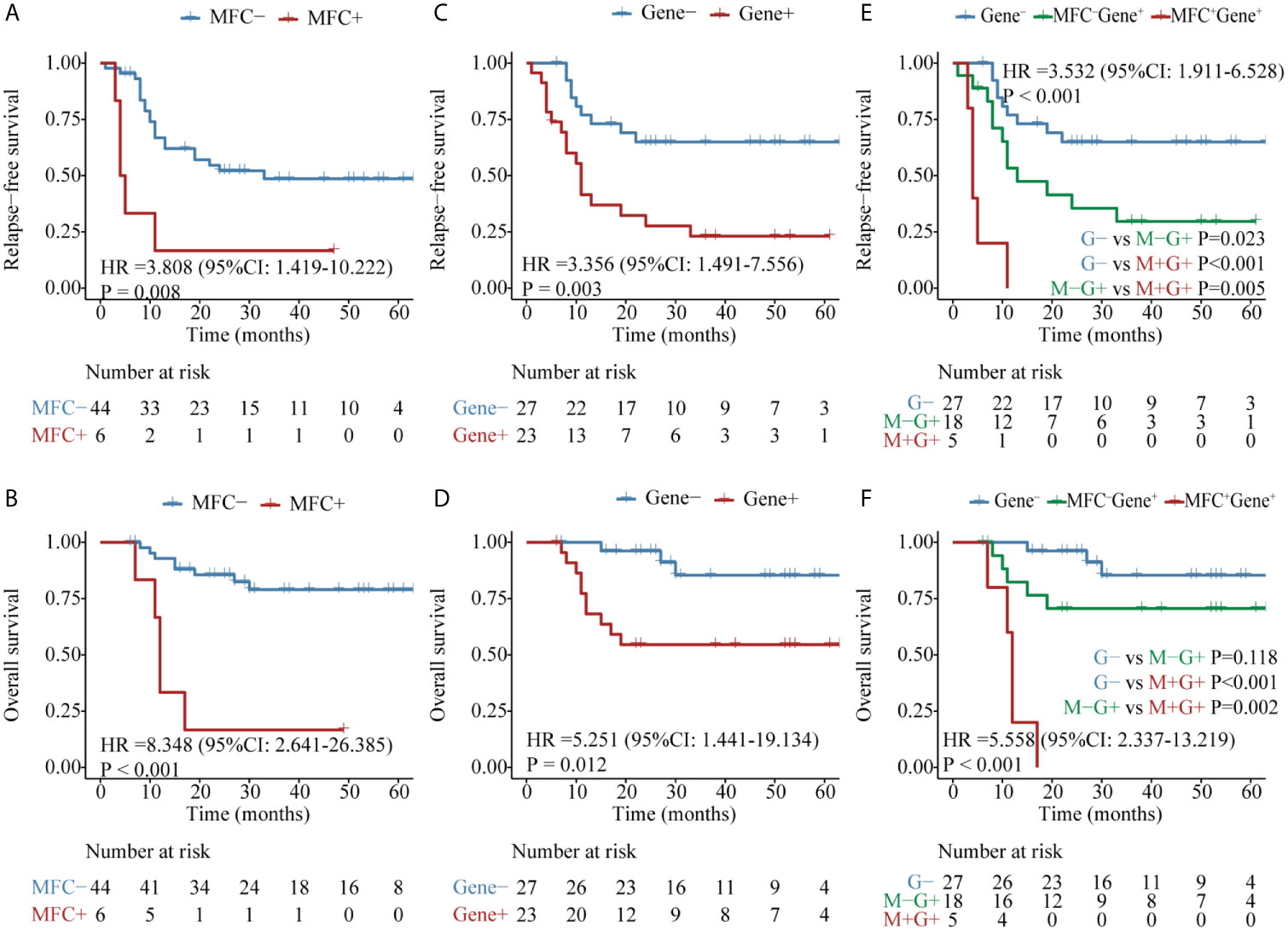
Figure 2 Kaplan–Meier curves for probability of relapse-free survival (RFS) and overall survival (OS) in AML patients with RUNX1–RUNX1T1 fusion gene. (A, B) RFS and OS based on MFC MRD status post two cycles of consolidation. (C, D) RFS and OS based on the quantification of RUNX1–RUNX1T1 transcript levels post two cycles of consolidation. (E, F) RFS and OS based on the integration of the two MRD monitoring methods.
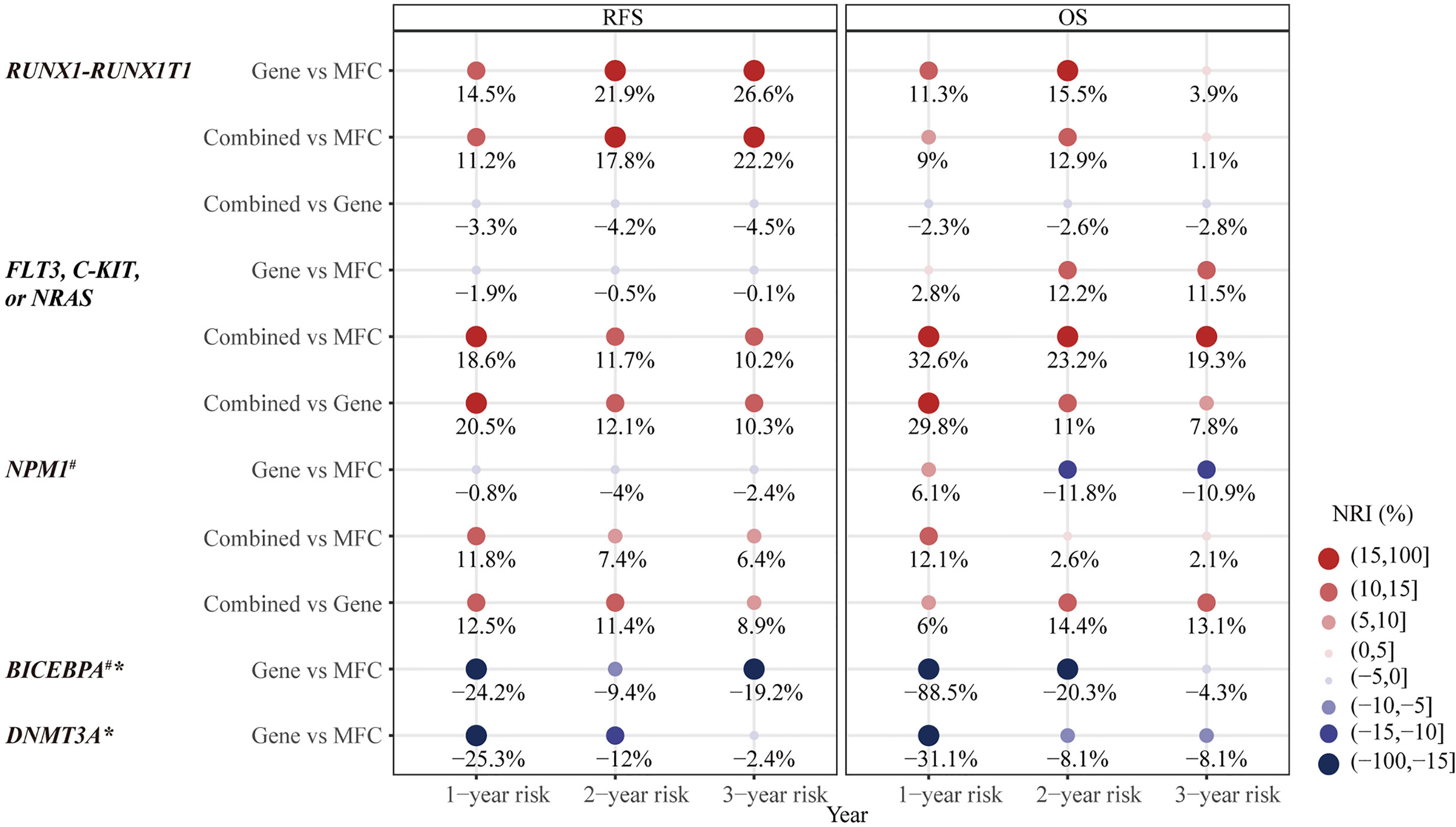
Figure 3 The category-based net reclassification improvement (NRI) of prediction on relapse-free survival (RFS) and overall survival (OS) by MRD status in different types of AML-related genetic lesions. #Notice, there were less than three people in Gene+ group. *no significant difference in prognosis predicted by Gene MRD and therefore no combination of MFC MRD and Gene MRD. NRI, net reclassification improvement; MFC, multicolor flow cytometry; MRD, measurable residual disease; Combined MRD, MRD positivity detected by either MFC or molecular method; RFS, relapse-free survival; OS, overall survival.
The second panel of genes was involved in activated signaling pathways, and 77 patients with FLT3, C-KIT, or N-RAS mutations at diagnosis were evaluated. Both MFC MRD status (median, 50 vs 10 months, P = 0.017) (Figure 4A) and molecular MRD status (median, 50 vs 7 months, P <0.001) (Figure 4C) had a significant prognostic impact on RFS. Gene MRD positivity conferred a significantly worse OS (median, NR vs 12 months, P <0.001, Figure 4D), while the presence of MFC MRD was borderline associated with an inferior OS (median, NR vs 43 months, P = 0.101, Figure 4B). Patients with MFC–Gene– had longer RFS and OS than those in either MFC+Gene– or MFC–Gene+ group (P <0.001, P = 0.002 for RFS; P <0.001, P = 0.029 for OS, respectively, Figures 4E, F). The NRI showed positive gains in reclassification when combing MFC and Gene MRD together, with 11.7 and 23.2% improved value in the prediction of 2-year RFS and OS, respectively, compared with MFC MRD status, and the improvement was 12.1 and 11%, respectively, compared with molecular MRD method (Figure 3).
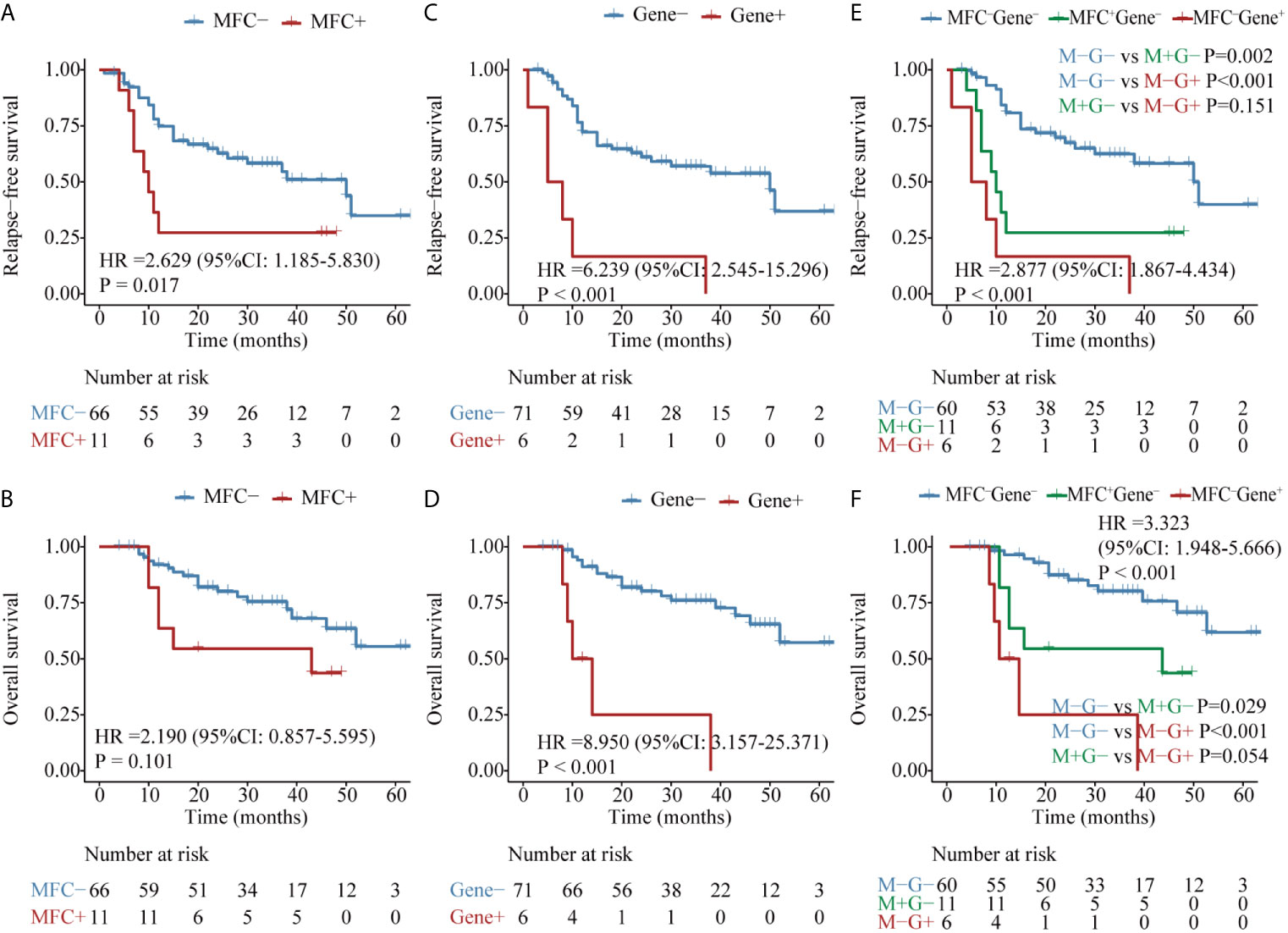
Figure 4 Kaplan–Meier curves for probability of relapse-free survival (RFS) and overall survival (OS) in AML patients with kinase-related signaling pathway mutations. (A, B) RFS and OS based on MFC MRD status post two consolidation cycles. (C, D) RFS and OS based on FLT3, C-KIT, or N-RAS mutations status by post two consolidation cycles. (E, F) RFS and OS based on MFC MRD status and Gene MRD status post two consolidation cycles.
Then, in 55 AML patients with NPM1 mutations, the MFC MRD status after the second consolidation exerted a significant prognostic impact on both RFS (median RFS, 32 vs 10 months, P = 0.036) and OS (median OS, NR vs 19 months, P = 0.028) (Figures 5A, B). Only two patients harbored NPM1 mutations post consolidation, and both experienced relapse within 10 months, one of whom succumbed to the disease, resulting in the shorter duration of both RFS (median RFS,30 vs 8 months, P = 0.010) and OS (median OS, NR vs 8 months, P = 0.090) compared with patients who had a clearance of NPM1 mutations (Figures 5C, D). Patients with MFC–Gene– obtained significantly longer RFS and OS than those with either MFC or Gene MRD positive (all P <0.05, Figures 5E, F). The integration of both assays yielded 7.4 and 11.4% NRI for 2-year RFS compared to the MRD status evaluated by MFC and NPM1 mutations, respectively, and 14.4% for 2-year OS compared to Gene MRD (Figure 3).
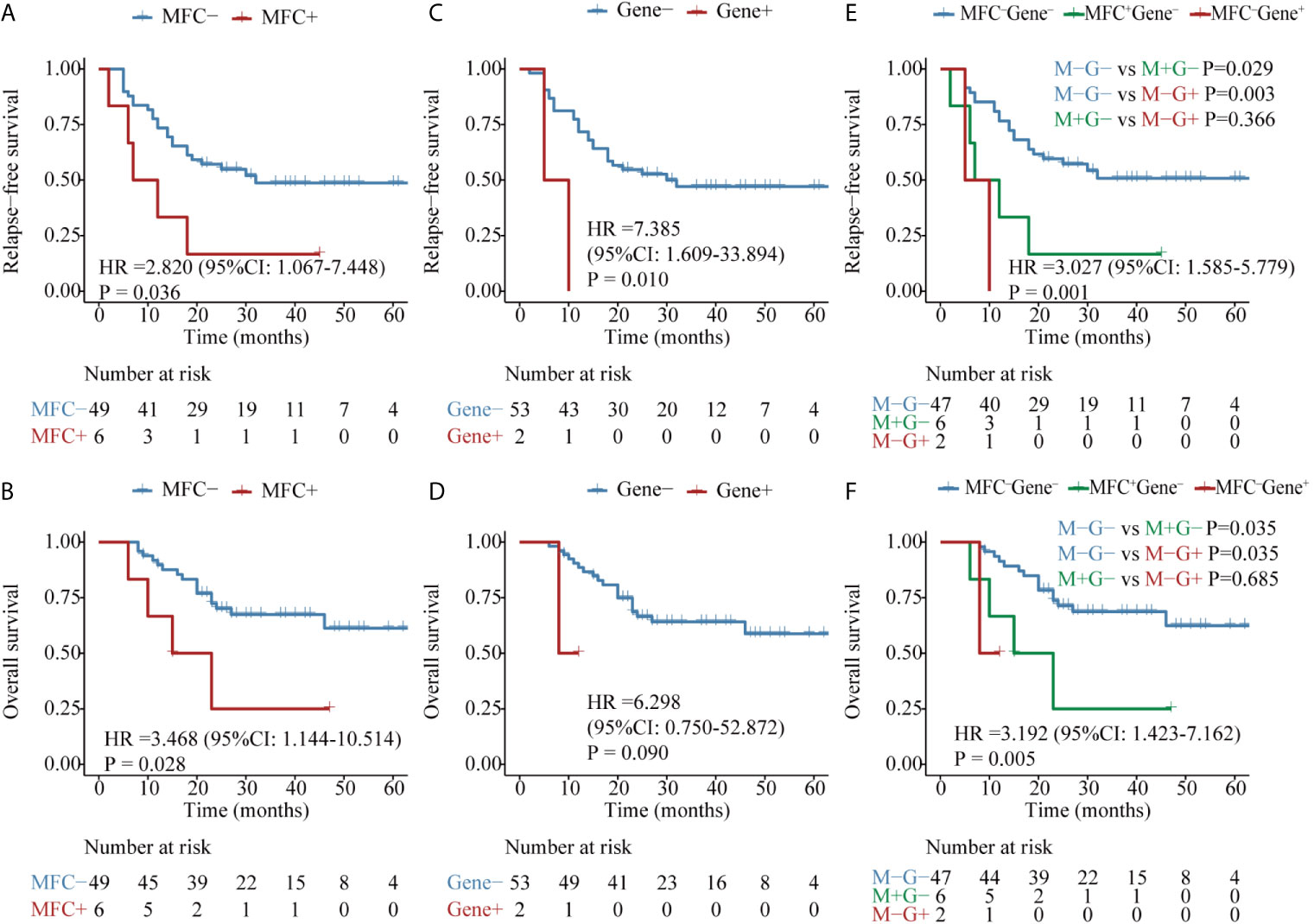
Figure 5 Kaplan–Meier curves for probability of relapse-free survival (RFS) and overall survival (OS) in AML patients with NPM1 mutations. (A, B) RFS and OS based on MFC MRD status post-consolidation. (C, D) RFS and OS based on NPM1 mutations status post two consolidation cycles. (E, F) RFS and OS based on MFC MRD status and Gene MRD status post two consolidation cycles.
Patients with BiCEBPA mutations were reported to be sensitive to standard chemotherapy. Consistently, there were only six patients in the MFC MRD+ group and 1 patient in the Gene MRD+ group after two cycles of consolidation. MFC MRD positivity tended to predict a worse RFS (P = 0.013, Figure S8A) but did not impact OS (P = 0.745, Figure S8B). The only patient with Gene MRD+ experienced a long-term survival, therefore no significant differences were observed in RFS and OS between different Gene MRD groups (Figures S8C, D).
For 25 patients carrying mutations in DNMT3A, the elimination of MFC MRD was significantly associated with longer RFS (P = 0.011) and OS (P = 0.049) (Figures S9A, B), while no significant differences were observed in RFS (P = 0.902) and OS (P = 0.596) between patients with detectable DNMT3A mutations and those whose molecular MRD turned into negative (Figures S9C, D).
Discussion
There is now mounting evidence that the identification of residual disease is of paramount importance in refining risk reclassification and informing therapeutic intervention for AML patients after the achievement of morphological remission (16, 22, 23). However, no consensus has been reached on the ideal time point and the optimal methodology for MRD evaluation, highlighting the need to establish standardized analysis and reporting procedures so as to improve the accuracy and reproducibility of MRD monitoring.
Our results indicated that the MFC MRD status after two consolidation cycles had a greater impact on the subsequent relapse and inferior outcome than that measured after induction. Notably, the same conclusion could be drawn when restricting the analysis to different patient subgroups (young vs. elderly patients, and ELN cytogenetic low- or intermediate-risk patients). While for high-risk patients, the MFC MRD status at both time points failed to forecast the prognosis, which merits further exploration considering that adverse molecular markers, e.g., TP53, ASXL1, and RUNX1 were not included in our study. Our results are consistent with previous studies that recommended MRD tracking after consolidation (9, 13, 24), although others favored the post-induction time point (25). It should be mentioned that the controversial interpretation of the prognostic value of MRD in different studies may be attributed to the number of induction and consolidation courses completed at the time of MFC MRD monitoring, and the modality and intensity of induction regimens, as reported by Minetto et al. for fludarabine plus high dose cytarabine-based induction, an earlier timepoint of MRD assessment may provide the most significant information on outcome (26).
The dynamics of sequential MRD monitoring demonstrated that patients who had detectable MRD after induction but entered MRD negativity after the second consolidation showed the same prognosis as those with a negative MRD at both time points. In contrast, the initial clearance of MRD did not guarantee a persistent remission and long-term survival, as exemplified by the dismal prognosis of patients whose MRD was eliminated early and subsequently converted into positive after consolidation. More importantly, the achievement of MFC MRD negativity after two consolidation cycles was an independent predictor for both RFS and OS, emphasizing the need to introduce new therapeutic modalities such as HSCT and targeted therapies to eradicate residual malignant cells when MFC MRD was positive at this checkpoint.
In addition to abnormal immunophenotypes detected by MFC, MRD could be reliably measured through genetic aberrations expressed in leukemic cells. We compared the Gene MRD and MFC MRD after the second consolidation in several molecular groups representative of different genetic etiology and biological function. Gene fusions involving transcription factors such as RUNX1–RUNX1T1 represent a specific subtype of AML. Although significantly diverse prognostic groups could be distinguished by both MFC and molecular MRD, the latter methodology was superior in terms of the NRI. Indeed, a less than 3 log reduction in the RUNX1–RUNX1T1 transcript levels was proved to be an independent adverse prognostic factor (23, 27).
Mutations in kinase-related signaling pathways including FLT3, CKIT, or N-RAS mutations usually occur at a later stage and are more likely to be eliminated by cytotoxic chemotherapy. The clearance of MRD confirmed by either MFC or sequencing-based approach conferred a favorable clinical outcome, and the combination of both methods showed greater discriminative ability. Similar results could be observed in patients with NPM1 mutations. Although only two patients harbored NPM1 mutations post consolidation, both displayed a dismal prognosis, which was in concordance with the widely appreciated role of NPM1 mutations in MRD testing (28, 29). Remarkably, the MFC MRD can provide complementary prognostic value.
Biallelic CEBPA mutations have been recognized as a favorable prognostic marker of AML (18, 30, 31). However, since only one patient was in the Gene MRD+ group after consolidation, the tracking of molecular MRD showed limited predictive power as reported in prior studies (32, 33). Likewise, the continuous presence of mutant transcripts in the epigenetic modifier gene DNMT3A did not exert any adverse impact on prognosis. Consistently, recent researches regarded DNMT3A, TET2, and ASXL1 (DTA) mutations as age-related clonal hematopoiesis and their persistent existence post remission was unable to forecast an increased risk of relapse (3, 34).
Despite the high performance of MRD monitoring by MFC and PCR-based assays, a proportion of AML patients lack a traceable MRD marker. So far, routine clinical practices of MRD tracking have dealt with only a small proportion of typical genetic anomalies in AML (5, 35). The overexpression of WT1 can be observed in more than 80% of AML patients, which may be an alternative PCR-based MRD testing since the quantification of WT1 expression after treatment has been proved to have significant prognostic value (36). In addition, the integration of WT1-based MRD and MFC MRD may improve the prediction of outcome in AML, although the limited sensitivity and specificity to some extent hamper the wide application of MRD monitoring based on WT1 expression (37–39). Encouragingly, the NGS technology holds great potential for the widely applicable MRD tracking as nearly all AML patients harbored at least one mutation at diagnosis. Growing evidence has proved the prognostic value of NGS-based MRD, either in the CR or pre-transplant stage, which can provide additional information on changes of variant allele frequency as well as clonal evolution during the follow-up period (3, 40–42). However, these NGS-based studies often integrated dozens to hundreds of genetic abnormalities without uniform design, technical and reporting standards, which may ignore the heterogeneity of molecular anomalies and their utility for MRD tracking in AML (40). It is noteworthy that the predictive value of molecular MRD varies in different categories of mutations as manifested in this study, and the optimal threshold of NGS-based MRD needs to be explored in a flexible and genotype-oriented way.
In summary, our study indicated that a positive MFC MRD after two consolidation cycles was an independent risk factor, and the comparison of molecular and MFC MRD in patients with different types of recurrent mutations lends support to the clinical implementation of NGS-based MRD assessment. Due to the limitation of technologies, a few germline mutations could affect the explanation of molecular MRD. Hence, how to integrate various detection methodologies and establish standard-of-care guidelines for MRD testing warrants further refinements in large prospective studies.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by Ruijin Hospital Ethics Committee Shanghai Jiao Tong University School of Medicine. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Author Contributions
F-JL, W-YC, X-JL, and S-YW contributed equally to this work. F-JL, W-YC, S-YW, and T-YJ collected clinical data. Y-MZ designed the experiments. Y-MZ and T-TM performed the experiments. F-JL and X-JL analyzed the data. F-JL and W-YC wrote the manuscript. YS conceived the overall study and revised the paper. All authors contributed to the article and approved the submitted version.
Funding
This work was supported in part by the National Natural Science Foundation of China (no. 81770141), the National Key R&D Program of China (no. 2016YFE0202800), and Shanghai Municipal Education Commission-Gaofeng Clinical Medicine Grant Support (no. 20161406).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank the patients and their families for their support of this initiative.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.677833/full#supplementary-material
References
1. Short NJ, Rytting ME, Cortes JE. Acute Myeloid Leukaemia. Lancet (2018) 392(10147):593–606. doi: 10.1016/s0140-6736(18)31041-9
2. Grimwade D, Freeman SD. Defining Minimal Residual Disease in Acute Myeloid Leukemia: Which Platforms are Ready for “Prime Time”? Blood (2014) 124(23):3345–55. doi: 10.1182/blood-2014-05-577593
3. Jongen-Lavrencic M, Grob T, Hanekamp D, Kavelaars FG, Al Hinai A, Zeilemaker A, et al. Molecular Minimal Residual Disease in Acute Myeloid Leukemia. N Engl J Med (2018) 378(13):1189–99. doi: 10.1056/NEJMoa1716863
4. Hourigan CS, Gale RP, Gormley NJ, Ossenkoppele GJ, Walter RB. Measurable Residual Disease Testing in Acute Myeloid Leukaemia. Leukemia (2017) 31(7):1482–90. doi: 10.1038/leu.2017.113
5. Schuurhuis GJ, Heuser M, Freeman S, Béné MC, Buccisano F, Cloos J, et al. Minimal/Measurable Residual Disease in AML: A Consensus Document From the European Leukemianet MRD Working Party. Blood (2018) 131(12):1275–91. doi: 10.1182/blood-2017-09-801498
6. Al-Mawali A, Gillis D, Lewis I. The Use of Receiver Operating Characteristic Analysis for Detection of Minimal Residual Disease Using Five-Color Multiparameter Flow Cytometry in Acute Myeloid Leukemia Identifies Patients With High Risk of Relapse. Cytometry B Clin Cytom (2009) 76(2):91–101. doi: 10.1002/cyto.b.20444
7. Kern W, Voskova D, Schoch C, Hiddemann W, Schnittger S, Haferlach T. Determination of Relapse Risk Based on Assessment of Minimal Residual Disease During Complete Remission by Multiparameter Flow Cytometry in Unselected Patients With Acute Myeloid Leukemia. Blood (2004) 104(10):3078–85. doi: 10.1182/blood-2004-03-1036
8. Venditti A, Buccisano F, Del Poeta G, Maurillo L, Tamburini A, Cox C, et al. Level of Minimal Residual Disease After Consolidation Therapy Predicts Outcome in Acute Myeloid Leukemia. Blood (2000) 96(12):3948–52. doi: 10.1182/blood.V96.12.3948
9. Buccisano F, Maurillo L, Gattei V, Del Poeta G, Del Principe MI, Cox MC, et al. The Kinetics of Reduction of Minimal Residual Disease Impacts on Duration of Response and Survival of Patients With Acute Myeloid Leukemia. Leukemia (2006) 20(10):1783–9. doi: 10.1038/sj.leu.2404313
10. Döhner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Büchner T, et al. Diagnosis and Management of AML in Adults: 2017 ELN Recommendations From an International Expert Panel. Blood (2017) 129(4):424–47. doi: 10.1182/blood-2016-08-733196
11. Grimwade D, Jovanovic JV, Hills RK, Nugent EA, Patel Y, Flora R, et al. Prospective Minimal Residual Disease Monitoring to Predict Relapse of Acute Promyelocytic Leukemia and to Direct Pre-Emptive Arsenic Trioxide Therapy. J Clin Oncol (2009) 27(22):3650–8. doi: 10.1200/jco.2008.20.1533
12. Shayegi N, Kramer M, Bornhäuser M, Schaich M, Schetelig J, Platzbecker U, et al. The Level of Residual Disease Based on Mutant NPM1 is an Independent Prognostic Factor for Relapse and Survival in AML. Blood (2013) 122(1):83–92. doi: 10.1182/blood-2012-10-461749
13. Ivey A, Hills RK, Simpson MA, Jovanovic JV, Gilkes A, Grech A, et al. Assessment of Minimal Residual Disease in Standard-Risk AML. N Engl J Med (2016) 374(5):422–33. doi: 10.1056/NEJMoa1507471
14. Rücker FG, Agrawal M, Corbacioglu A, Weber D, Kapp-Schwoerer S, Gaidzik VI, et al. Measurable Residual Disease Monitoring in Acute Myeloid Leukemia With T (8)(Q22;Q22.1): Results From the AML Study Group. Blood (2019) 134(19):1608–18. doi: 10.1182/blood.2019001425
15. Duan W, Liu X, Jia J, Wang J, Gong L, Jiang Q, et al. The Loss or Absence of Minimal Residual Disease of <0·1% At Any Time After Two Cycles of Consolidation Chemotherapy in CBFB-MYH11-Positive Acute Myeloid Leukaemia Indicates Poor Prognosis. Br J Haematol (2021) 192(2):265–71. doi: 10.1111/bjh.16745
16. Puckrin R, Atenafu EG, Claudio JO, Chan S, Gupta V, Maze D, et al. Measurable Residual Disease Monitoring Provides Insufficient Lead-Time to Prevent Morphologic Relapse in the Majority of Patients With Core-Binding Factor Acute Myeloid Leukemia. Haematologica (2021) 106(1):56–63. doi: 10.3324/haematol.2019.235721
17. Weng XQ, Shen Y, Sheng Y, Chen B, Wang JH, Li JM, et al. Prognostic Significance of Monitoring Leukemia-Associated Immunophenotypes by Eight-Color Flow Cytometry in Adult B-Acute Lymphoblastic Leukemia. Blood Cancer J (2013) 3(8):e133. doi: 10.1038/bcj.2013.31
18. Shen Y, Zhu YM, Fan X, Shi JY, Wang QR, Yan XJ, et al. Gene Mutation Patterns and Their Prognostic Impact in a Cohort of 1185 Patients With Acute Myeloid Leukemia. Blood (2011) 118(20):5593–603. doi: 10.1182/blood-2011-03-343988
19. Shen Y, Fu YK, Zhu YM, Lou YJ, Gu ZH, Shi JY, et al. Mutations of Epigenetic Modifier Genes as a Poor Prognostic Factor in Acute Promyelocytic Leukemia Under Treatment With All-Trans Retinoic Acid and Arsenic Trioxide. EBioMedicine (2015) 2(6):563–71. doi: 10.1016/j.ebiom.2015.04.006
20. Jourdan E, Boissel N, Chevret S, Delabesse E, Renneville A, Cornillet P, et al. Prospective Evaluation of Gene Mutations and Minimal Residual Disease in Patients With Core Binding Factor Acute Myeloid Leukemia. Blood (2013) 121(12):2213–23. doi: 10.1182/blood-2012-10-462879
21. Pencina MJ, D’Agostino RBSr., D’Agostino RB Jr, Vasan RS. Evaluating the Added Predictive Ability of a New Marker: From Area Under the ROC Curve to Reclassification and Beyond. Stat Med (2008) 27(2):157–72; discussion 207-12. doi: 10.1002/sim.2929
22. Forghieri F, Comoli P, Marasca R, Potenza L, Luppi M. Minimal/Measurable Residual Disease Monitoring in NPM1-Mutated Acute Myeloid Leukemia: A Clinical Viewpoint and Perspectives. Int J Mol Sci (2018) 19(11):3492. doi: 10.3390/ijms19113492
23. Yin JA, O’Brien MA, Hills RK, Daly SB, Wheatley K, Burnett AK. Minimal Residual Disease Monitoring by Quantitative RT-PCR in Core Binding Factor AML Allows Risk Stratification and Predicts Relapse: Results of the United Kingdom MRC AML-15 Trial. Blood (2012) 120(14):2826–35. doi: 10.1182/blood-2012-06-435669
24. Terwijn M, van Putten WL, Kelder A, van der Velden VH, Brooimans RA, Pabst T, et al. High Prognostic Impact of Flow Cytometric Minimal Residual Disease Detection in Acute Myeloid Leukemia: Data From the HOVON/SAKK AML 42A Study. J Clin Oncol (2013) 31(31):3889–97. doi: 10.1200/jco.2012.45.9628
25. Freeman SD, Hills RK, Virgo P, Khan N, Couzens S, Dillon R, et al. Measurable Residual Disease At Induction Redefines Partial Response in Acute Myeloid Leukemia and Stratifies Outcomes in Patients At Standard Risk Without NPM1 Mutations. J Clin Oncol (2018) 36(15):1486–97. doi: 10.1200/jco.2017.76.3425
26. Minetto P, Guolo F, Clavio M, Kunkl A, Colombo N, Carminati E, et al. Early Minimal Residual Disease Assessment After AML Induction With Fludarabine, Cytarabine and Idarubicin (FLAI) Provides the Most Useful Prognostic Information. Br J Haematol (2019) 184(3):457–60. doi: 10.1111/bjh.15106
27. Wang Y, Wu DP, Liu QF, Qin YZ, Wang JB, Xu LP, et al. In Adults With T (8)AML, Posttransplant RUNX1/RUNX1T1-Based MRD Monitoring, Rather Than C-KIT Mutations, Allows Further Risk Stratification. Blood (2014) 124(12):1880–6. doi: 10.1182/blood-2014-03-563403
28. Dillon R, Hills R, Freeman S, Potter N, Jovanovic J, Ivey A, et al. Molecular MRD Status and Outcome After Transplantation in NPM1-Mutated AML. Blood (2020) 135(9):680–8. doi: 10.1182/blood.2019002959
29. Balsat M, Renneville A, Thomas X, de Botton S, Caillot D, Marceau A, et al. Postinduction Minimal Residual Disease Predicts Outcome and Benefit From Allogeneic Stem Cell Transplantation in Acute Myeloid Leukemia With NPM1 Mutation: A Study by the Acute Leukemia French Association Group. J Clin Oncol (2017) 35(2):185–93. doi: 10.1200/jco.2016.67.1875
30. Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 Revision to the World Health Organization Classification of Myeloid Neoplasms and Acute Leukemia. Blood (2016) 127(20):2391–405. doi: 10.1182/blood-2016-03-643544
31. Taskesen E, Bullinger L, Corbacioglu A, Sanders MA, Erpelinck CA, Wouters BJ, et al. Prognostic Impact, Concurrent Genetic Mutations, and Gene Expression Features of AML With CEBPA Mutations in a Cohort of 1182 Cytogenetically Normal AML Patients: Further Evidence for CEBPA Double Mutant AML as a Distinctive Disease Entity. Blood (2011) 117(8):2469–75. doi: 10.1182/blood-2010-09-307280
32. Smith LL, Pearce D, Smith ML, Jenner M, Lister TA, Bonnet D, et al. Development of a Quantitative Real-Time Polymerase Chain Reaction Method for Monitoring CEBPA Mutations in Normal Karyotype Acute Myeloid Leukaemia. Br J Haematol (2006) 133(1):103–5. doi: 10.1111/j.1365-2141.2006.06001.x
33. Cruz NM, Mencia-Trinchant N, Hassane DC, Guzman ML. Minimal Residual Disease in Acute Myelogenous Leukemia. Int J Lab Hematol (2017) 39 Suppl 1(Suppl 1):53–60. doi: 10.1111/ijlh.12670
34. Zink F, Stacey SN, Norddahl GL, Frigge ML, Magnusson OT, Jonsdottir I, et al. Clonal Hematopoiesis, With and Without Candidate Driver Mutations, is Common in the Elderly. Blood (2017) 130(6):742–52. doi: 10.1182/blood-2017-02-769869
35. Voso MT, Ottone T, Lavorgna S, Venditti A, Maurillo L, Lo-Coco F, et al. MRD in AML: The Role of New Techniques. Front Oncol (2019) 9:655. doi: 10.3389/fonc.2019.00655
36. Cilloni D, Renneville A, Hermitte F, Hills RK, Daly S, Jovanovic JV, et al. Real-Time Quantitative Polymerase Chain Reaction Detection of Minimal Residual Disease by Standardized WT1 Assay to Enhance Risk Stratification in Acute Myeloid Leukemia: A European Leukemianet Study. J Clin Oncol (2009) 27(31):5195–201. doi: 10.1200/jco.2009.22.4865
37. Malagola M, Skert C, Borlenghi E, Chiarini M, Cattaneo C, Morello E, et al. Postremission Sequential Monitoring of Minimal Residual Disease by WT1 Q-PCR and Multiparametric Flow Cytometry Assessment Predicts Relapse and May Help to Address Risk-Adapted Therapy in Acute Myeloid Leukemia Patients. Cancer Med (2016) 5(2):265–74. doi: 10.1002/cam4.593
38. Rossi G, Minervini MM, Melillo L, di Nardo F, de Waure C, Scalzulli PR, et al. Predictive Role of Minimal Residual Disease and Log Clearance in Acute Myeloid Leukemia: A Comparison Between Multiparameter Flow Cytometry and Wilm’s Tumor 1 Levels. Ann Hematol (2014) 93(7):1149–57. doi: 10.1007/s00277-014-2029-9
39. Marani C, Clavio M, Grasso R, Colombo N, Guolo F, Kunkl A, et al. Integrating Post Induction WT1 Quantification and Flow-Cytometry Results Improves Minimal Residual Disease Stratification in Acute Myeloid Leukemia. Leuk Res (2013) 37(12):1606–11. doi: 10.1016/j.leukres.2013.07.005
40. Ghannam J, Dillon LW, Hourigan CS. Next-Generation Sequencing for Measurable Residual Disease Detection in Acute Myeloid Leukaemia. Br J Haematol (2020) 188(1):77–85. doi: 10.1111/bjh.16362
41. Klco JM, Miller CA, Griffith M, Petti A, Spencer DH, Ketkar-Kulkarni S, et al. Association Between Mutation Clearance After Induction Therapy and Outcomes in Acute Myeloid Leukemia. JAMA (2015) 314(8):811–22. doi: 10.1001/jama.2015.9643
Keywords: acute myeloid leukemia, measurable residual disease, molecular markers, multiparameter flow cytometry, prognosis
Citation: Liu F-J, Cheng W-Y, Lin X-J, Wang S-Y, Jiang T-Y, Ma T-T, Zhu Y-M and Shen Y (2021) Measurable Residual Disease Detected by Multiparameter Flow Cytometry and Sequencing Improves Prediction of Relapse and Survival in Acute Myeloid Leukemia. Front. Oncol. 11:677833. doi: 10.3389/fonc.2021.677833
Received: 08 March 2021; Accepted: 26 April 2021;
Published: 20 May 2021.
Edited by:
Claudio Cerchione, Istituto Scientifico Romagnolo per lo Studio e il Trattamento dei Tumori (IRCCS), ItalyReviewed by:
Monica Bocchia, University of Siena, ItalyFabio Guolo, San Martino Hospital (IRCCS), Italy
Copyright © 2021 Liu, Cheng, Lin, Wang, Jiang, Ma, Zhu and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yang Shen, eWFuZ19zaGVuQHNqdHUuZWR1LmNu
†These authors have contributed equally to this work
 Fu-Jia Liu
Fu-Jia Liu Wen-Yan Cheng†
Wen-Yan Cheng†