
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol. , 29 April 2021
Sec. Cancer Immunity and Immunotherapy
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.672677
This article is part of the Research Topic The Clinical Application of Neoantigens View all 11 articles
Immunotherapy has significantly improved the clinical outcome of patients with cancer. However, the immune response rate varies greatly, possibly due to lack of effective biomarkers that can be used to distinguish responders from non-responders. Recently, clinical studies have associated high tumor neoantigen burden (TNB) with improved outcomes in patients treated with immunotherapy. Therefore, TNB has emerged as a biomarker for immunotherapy and other types of therapy. In the present review, the potential application of TNB as a biomarker was evaluated. The methods of neoantigen prediction were summarized and the mechanisms involved in TNB were investigated. The impact of high TNB and increased number of infiltrating immune cells on the efficacy of immunotherapy was also addressed. Finally, the future challenges of TNB were discussed.
Tumor immunotherapy aims to control tumor development by activating the immune system to attack tumor cells. By selecting appropriate antigens, notably neoantigens produced by tumor-specific mutations, an effective tumor-specific immune response can be mounted, and immune tolerance can be minimized (1). Non-synonymous somatic mutations will produce altered peptides, among which, some are processed and presented by the major histocompatibility complex (MHC) in order to generate neoantigens. These molecules are the key factors required for successful immunotherapy, including immune checkpoint inhibitors (ICIs), personalized tumor vaccines and adoptive T cell transfer immunotherapy (2–4). These strategies have shown promise in the treatment of solid tumors (5–7).
A higher number of DNA mutations are associated with higher number of candidate peptides, and results in an increased probability of successfully presented neoantigens (8). The response to immunotherapy correlates with tumor mutation burden (TMB) and mainly with the number of mutations in the coding region of the genome (exome) of the tumor cells. It is usually reported as the number of mutations present in a megabase of the genomic region by whole-exome sequencing or large-scale next-generation sequencing (9–12). Similarly, the tumor neoantigen burden (TNB) is defined by the number of neoantigens per megabase in the genome region (13, 14). Notably, TMB has become a biomarker for immunotherapy, assuming that higher TMB will increase the probability of tumor neoantigens and specific T-cell responses (15).
However, the role of TMB in immunotherapy remains controversial (16–18), since not all mutations produce neoantigens. Only a limited number of mutations can be properly processed, presented on the surface of the MHC complex and recognized by T cells (19). The TMB noted in pediatric tumors is considerably low (20). However, in certain tumors, such as pediatric medulloblastoma or acute lymphoblastic leukemia, which exhibit minimal mutational burden, a strong anti-tumor immune response can be induced by high-quality neoantigens (21, 22).
TMB generates neoantigens and causes tumor immunogenicity. This biomarker can be used as a valuable estimate of TNB to a certain extent. A positive correlation has been noted between TMB and TNB. However, TNB is directly used for neoantigen evaluation and may be considered an improved biomarker for immunotherapy compared with TMB (23–25). High TNB was associated with durable progression-free survival (PFS) in patients with non-small cell lung cancer (NSCLC) treated with programmed death 1 (PD-1) inhibitors (26). In addition, TNB correlated with clinical benefit in patients with metastatic melanoma treated with cytotoxic T-lymphocyte-associated protein 4 (CTLA4) inhibitors (27). Similarly, a phase I/II trial performed in patients with stage IV melanoma demonstrated that their clinical benefit was associated with a proposed immune activation signatures score. Among the score items, high TMB and predicted TNB were significantly associated with improved PFS and overall survival (28). The present review investigated the application of TNB as a biomarker in immunotherapy and other therapies and provided an in-depth discussion of the mechanisms, clinical application and challenges of this biomarker.
In general, in silico analysis on genome sequencing can aid the selection of immunogenic neoantigen peptides. Neoantigen prediction is usually performed prior to selecting immunogenic neoantigens to reduce the burden of immunogenicity testing by decreasing the number of candidate peptides. This is a necessary step in developing personalized immunotherapy. Several important steps are involved in neoantigen selection, including intracellular processing and transportation, the stability and affinity of peptide-MHC complex binding, the diversity of T cell receptors (TCR) and the recognition by TCR. In addition, the difference between the prediction algorithms is also important. Neopepsee is a neoantigen prediction algorithm that automatically extracts mutated peptide sequences and expression levels, and combines multiple immunogenic features to construct a machine-learning classifier (29). The application of deep learning to the determination of large human leukocyte antigen (HLA) peptides and genomic data sets from various tumors can aid the development of a computational model for neoantigen prediction (30). An additional prediction algorithm can determine the priority of neoantigens and discover immune characteristics in cancer immunotherapy by the classification of human neoantigen/neopeptide data into three categories based on different mutation positions (anchor mutation, MHC-contacting position and TCR-contacting position) (31).
Several computational pipelines have been developed for neoantigen prediction. However, the majority of them are based on peptide affinity with MHC (32–34). Furthermore, neoantigen prediction can be performed by prioritizing predicted peptides based on mutant allele expression, mutation clonality, MHC presentation, and T cell recognition, either alone or in combination (35–38). A Cauchy-Schwarz index of neoantigens score was proposed and the effects of both clonality and MHC binding affinity were included in order to accurately determine the concentration of neoantigens in truncal mutations (39). An additional prediction model was developed by integrating peptide presentation and recognition into antigenic determinant immunogenicity via the use of specific parameters.
To establish a global neoantigen prediction algorithm standard, several institutions established the tumor epitope selection alliance, which is a bioinformatics consortium with scientists from well-respected neoantigen research groups. These institutions independently mine the open database of tumor sequencing, predict potential neoantigens and rank candidate peptides. Different predictions may be collected and cross-matched to reach a final optimized consensus. This integration incorporates aspects of binding affinity, tumor abundance, stability and peptide identification in addition to antigen presentation. Therefore, higher precision would be expected (40).
Any form of genomic instability, including single -nucleotide variation (SNV), frameshift mutations, splicing variations or chromosome rearrangement, may result in TNB. The genomic instability can result from abnormalities in either DNA replication or mismatch repair (MMR) (41). The high-fidelity process of DNA replication requires replicative DNA polymerases, exonucleolytic proofreading and MMR. Abnormalities that may occur in any of these parts contribute to genetic instability. Inactivation of DNA polymerase leads to excessive mutations, such as ultra-hypermutated phenotype. Defective MMR (dMMR) leads to microsatellite instability (MSI), which is an ultra-hypervariable phenotype of short repetitive DNA sequences and SNV. Following exposure to either exogenous (smoking, ultraviolet radiation, chemicals, ionizing radiation) or endogenous (reactive oxygen species, endocrine abnormalities) mutagens, dMMR/MSI facilitates carcinogenicity and paradoxically increases TNB, which in turn enhances immunogenicity (42–46).
Although TNB is a biomarker of immunotherapy, current knowledge regarding its function is limited. Primarily, TNB analysis was performed on SNV (47). However, other genetic aberrations may produce comparable or even more immunogenic neoantigens (23). For example, neoantigens were found from a data set of gene fusion-positive tumors (48). Splice variants are also sources of neoantigens. High expression of PD-1 and programmed death-ligand 1 (PD-L1) were observed in tumors with splice variants (49). In addition, a new class of neoantigens was discovered, which was derived from intra- or inter-chromosomal rearrangements (50).
Various studies have analyzed the frequency of specific somatic mutations in multiple types of cancers, which demonstrated that the frequency of non-synonymous mutations varied greatly, ranging from ~0.001/Mb to higher than 400/Mb. The mutation frequency was very prominent in melanoma and lung cancer (20, 51). Notably, Samra Turajlic et al. analyzed and compared the counts of Insertion-and-deletion-derived tumor-specific neoantigens in pan-cancer, both SNV-derived neoantigens and frameshift indel-derived neoantigens in the study showed that melanoma, lung cancer, bladder cancer, colon cancer, and head and neck cancer with high TNB (47). Mutations in lung cancers can be attributed to direct DNA damage from cigarette smoke carcinogens. A significant dose-response association of smoking history with genetic alterations has been noted in advanced non-small-cell lung adenocarcinoma with regards to cancer-associated pathways and their corresponding mutant antigens (52, 53). Ultraviolet stimulation is the main factor leading to high TNB in melanoma (54). In other common cancer types, such as colorectal or endometrial cancer, which harbor DNA polymerase epsilon mutations, increased TNB is attributed to endogenous mutations (55, 56). However, in certain tumors, such as bladder cancer, the mechanisms underlying the formation of TNB are complex, including the apolipoprotein B mRNA editing enzyme catalytic polypeptide family, smoking, viral infection and genetic fusions (57).
A previous study dissected the genetic heterogeneity during the evolution of a primary osteosarcoma tumor to its metastatic variant. Metastases exhibited higher TNB compared with primary tumors, possibly due to the accumulating mutations in DNA damage response genes (58). Different mutational landscapes exist between the primary and metastatic sites and in the subclones noted inside different regions of a tumor (intra-tumoral heterogeneity; ITH). The presence of ITH suggests that the tumor cannot elicit equal immunity. Patients with tumors exhibiting high TNB and low ITH are more likely to benefit from immunotherapy (59, 60).
Neoantigens alone are not sufficient to mount an effective immune response and tumor-infiltrating lymphocytes (TILs) are also required for this process (61, 62). High TNB can promote the recognition and activation of T cells, which in turn increase TILs and improve the immune response of cancer patients to cancer cells (Figure 1). Colorectal tumors with dMMR exhibit neoantigen-stimulated lymphocyte infiltration and increased levels of inflammatory cytokines. In the absence of high TMB, high TNB alone correlates with the inflammatory microenvironment (63). Moreover, lung adenocarcinoma patients with high TMB presented with enhanced infiltration of activated CD4+ and CD8+ T cells, while the mutations detected could accurately predict the increased TNB and T cell infiltration. In addition, TNB was significantly associated with the expression levels of M1 polarized macrophage genes, namely PD-1, PD-L1, interferon-γ (IFNγ), Granzyme B FAS ligand and other immune-associated genes (64). The correlation between TNB and TILs has been verified in multiple studies (64–66).
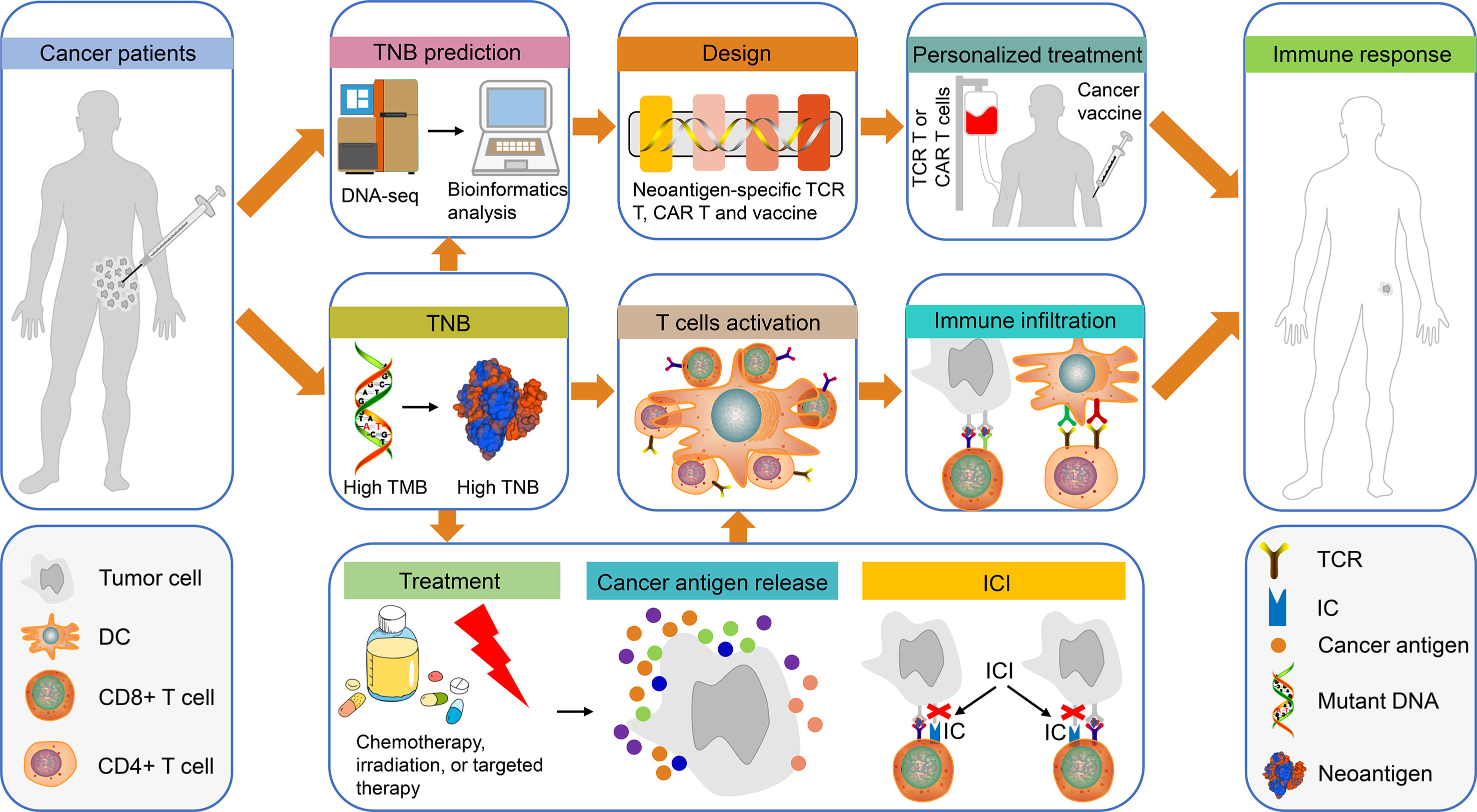
Figure 1 The use of TNB in personalized immunotherapy for patients with cancer. Tumor tissues from patients with cancer were obtained for DNA sequencing and bioinformatics prediction of TNB. Subsequently, the TCR-T, CAR-T and the cancer vaccines were designed based on TNB for personalized treatment. Finally, the patients exhibited an immune response to cancer cells. In addition, high TMB in patients with cancer can produce high TNB, which will promote T cell activation and immune cell infiltration, thereby causing an immune response in these patients. It is interesting to note that chemotherapy, radiotherapy or targeted therapy can promote the release of cancer antigens in patients with high TNB, which in turn can enhance the therapeutic effects of ICI-based treatment. This type of treatment will activate T cells, increase immune infiltration and produce immune responses. TNB, tumor neoantigen burden; TCR-T, neoantigen-specific T cell receptor engineered-T cell; CAR-T, chimeric antigen receptor-T cell; ICI, immune checkpoint inhibitor.
High TNB correlates with the abundance of TCR clonality and the infiltration of activated CD4+ and CD8+ T cells (50, 67). This may be mediated by the elevated expression of chemokines induced by IFN-γ, such as chemokine (C-X-C motif) ligand (CXCL) 9, and by the recruitment of T cells or myeloid dendritic cells (45, 68, 69). In the majority of the cases, high TNB can predict inflammatory microenvironment and optimal immune response. The infiltration of different immune cell subgroups is commonly noted in a special spatial compartment termed tertiary lymphoid structure (TLS). The mechanism by which TLS responds to the tumor microenvironment is actively studied. A previous report indicated that transforming growth factor β1 induced co-expression of CXCL13 and CD103 in CD8+ T cells, providing a potential link between CD8+ T cell activation and B cell migration (70, 71).
The use of individualized neoantigen vaccines and neoantigen-specific T cell therapy is actively explored. This topic has been well described in previous review articles (72–76) and will not be covered in the present review. It should be noted that individualized vaccines against a single neoantigen demonstrated limited efficacy. The use of a complete tumor lysate vaccine or a personalized vaccine containing multiple neoantigens can improve patient outcomes (6, 77). The dendritic cells were pulsed with oxidized autologous whole-tumor cell lysate, which was proved as an effective vaccine in patients with ovarian cancer. This vaccine amplified T cell responses against recognized neoepitopes and elicited de novo responses for previously unrecognized neoepitopes (78). An additional study tested the efficacy of an adenoviral vaccine consisting of multiple neoantigens. This vaccine facilitated T cell infiltration and expanded the breadth and efficacy of the TCR repertoire following ICI treatment (79). Tumor cell lysates differ in their efficiency as vaccines. A direct comparison of 2 autologous tumor cell lysate (with different TMB) vaccines demonstrated that the lysates with lower TMB inhibited tumor growth more efficiently. Thus, it may be considered that the neoantigen quality outranked the quantity (80).
Radiotherapy or chemotherapy can facilitate immunotherapy and is possibly attributed to increased exposure of neoantigens (81) (Figure 1). In addition to direct tumoricidal effects, radiotherapy converts the irradiated tumor cells into an in situ vaccine (82, 83). In locally advanced rectal cancer, neoadjuvant chemoradiotherapy induced new neoantigen epitopes and altered the immune function of the hosts (84). Similarly, patients with relapsed anal squamous cell carcinoma exhibited high TNB following radiochemotherapy and indicated objective responses to PD-1 inhibitors (85). In bladder cancer, dual poly (ADP-ribose) polymerase and PD/PD-L1 inhibition is used to improve disease prognosis (86). The standard treatment for high-grade serous ovarian carcinoma is surgery and/or chemotherapy. However, only dismal results are obtained, whereas in a subgroup of patients harboring high TNB, an improved prognosis was achieved. Moreover, an additional study demonstrated that TNB could be used to determine the prognosis of patients with clear cell renal cell carcinoma who received either surgery alone or surgery combined with adjuvant therapies (87–89).
The more important aspect is that pediatric tumors exhibit low TMB at diagnosis, whereas the levels of this biomarker increase when the tumor is exposed to chemoradiotherapy, resulting in neoantigen targets (90, 91). The majority of the pediatric tumors have less TIL and low MHC expression. In addition, the immune system of the children is immature. Consequently, the current immunotherapy alone is not sufficient to treat pediatric tumors efficiently. The combination of immunotherapy with conventional radio- and chemotherapy can achieve an improved survival benefit (92, 93).
High TNB produces neoantigens, contributing to an inflammatory microenvironment, which ultimately leads to improved outcomes following ICI therapy (Figure 1). A previous study performed in patients with NSCLC, who exhibited high TMB or genetic defects in the DNA repair pathway, demonstrated that they benefited from ICI treatment. At least in one responder, neoantigen-specific CD8+ T cell responses paralleled with tumor regression (26). It has also been shown that patients with melanoma, who are treated with CTLA4 inhibitors, demonstrated a significant association of TMB, TNB and cytolytic marker expression with clinical benefit (27). Recently, a model of immunotherapy score (ITS) mutation was proposed for predicting the response of patients with melanoma to ICI treatment. Patients with high TMB and TNB exhibited higher ITS scores and immunotherapy sensitive features (94).
A scoring system based on neoantigen concentration combined with clonality and MHC binding affinity predicted responses to ICIs and the prognosis of patients with melanoma, lung and kidney cancers (39). Neoantigen concentration levels were prognostic factors for patients with melanoma and chronic lymphocytic leukemia treated with ICIs (29). However, paradoxical results were reported, indicating inferior PFS with high TNB in patients with multiple myeloma. The possible explanation for these findings was that the disease progression caused reduced efficiency of T cell recruitment (24).
The diversity and clonality of neoantigen-responsive TCR are also potential biomarkers for immunotherapy (95, 96). Additional research studies in this topic will aid the successful application of immunotherapy.
The rapid development of immunology and bioinformatics has enabled the successful prediction of neoantigens. However, the standard pipeline for neoantigen prediction and the optimized cut-off value for TNB are unknown, since it is an emerging biomarker. In addition, the presence of specific mutations causing ITH should be taken into consideration (97). Heterogeneity exists not only locally but also between primary lesions and their successive metastases (98). Neoantigen prediction is currently hindered by the difficulty in exploring the entire tumor through a partial biopsy (99).
Several obstacles hinder the patient immune response, such as the loss of HLA (100–103). Failure of successful HLA presentation renders the candidate neoantigens ineffective in vivo. Therefore, TNB alone cannot accurately predict the immune response. Currently, the personalized detection of circulating tumor DNA is considered a powerful tool for the dynamic monitoring of TNB (16, 80, 104). However, the clinical application of TNB is still limited. Previous evidence has shown that the quality of neoantigens may be more important, since high-quality neoantigens can confer higher immunogenicity (105). According to our opinion, the real-time status of the high-quality neoantigen burden can monitor the treatment response more effectively. The construction of a neoantigen vaccine library and a neoantigen-responsive T cell receptor repertoire can provide a more comprehensive and personalized antitumor treatment. Future studies should focus on assessing the quality of TNB.
CW and PW conceptualized the study. PW wrote and edited the manuscript. YC helped revise the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by Clinical Research Incubation Project, West China Hospital, Sichuan University (2020HXFH056).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Hu Z, Ott PA, Wu CJ. Towards Personalized, Tumour-Specific, Therapeutic Vaccines for Cancer. Nat Rev Immunol (2018) 18:168–82. doi: 10.1038/nri.2017.131
2. Tran E, Turcotte S, Gros A, Robbins PF, Lu YC, Dudley ME, et al. Cancer Immunotherapy Based on Mutation-Specific CD4+ T Cells in a Patient With Epithelial Cancer. Science (2014) 344:641–5. doi: 10.1126/science.1251102
3. Gubin MM, Zhang X, Schuster H, Caron E, Ward JP, Noguchi T, et al. Checkpoint Blockade Cancer Immunotherapy Targets Tumour-Specific Mutant Antigens. Nature (2014) 515:577–81. doi: 10.1038/nature13988
4. Hinrichs CS, Rosenberg SA. Exploiting the Curative Potential of Adoptive T-cell Therapy for Cancer. Immunol Rev (2014) 257:56–71. doi: 10.1111/imr.12132
5. Rosenberg SA, Restifo NP. Adoptive Cell Transfer as Personalized Immunotherapy for Human Cancer. Science (2015) 348:62–8. doi: 10.1126/science.aaa4967
6. Ott PA, Hu Z, Keskin DB, Shukla SA, Sun J, Bozym DJ, et al. An Immunogenic Personal Neoantigen Vaccine for Patients With Melanoma. Nature (2017) 547:217–21. doi: 10.1038/nature22991
7. Schumacher TN, Schreiber RD. Neoantigens in Cancer Immunotherapy. Science (2015) 348:69–74. doi: 10.1126/science.aaa4971
8. Chalmers ZR, Connelly CF, Fabrizio D, Gay L, Ali SM, Ennis R, et al. Analysis of 100,000 Human Cancer Genomes Reveals the Landscape of Tumor Mutational Burden. Genome Med (2017) 9:34. doi: 10.1186/s13073-017-0424-2
9. Puccini A, Poorman K, Salem ME, Soldato D, Seeber A, Goldberg RM, et al. Comprehensive Genomic Profiling of Gastroenteropancreatic Neuroendocrine Neoplasms (GEP-Nens). Clin Cancer Res (2020) 26:5943–51. doi: 10.1158/1078-0432.ccr-20-1804
10. Tian Y, Xu J, Chu Q, Duan J, Zhang J, Bai H, et al. A Novel Tumor Mutational Burden Estimation Model as a Predictive and Prognostic Biomarker in NSCLC Patients. BMC Med (2020) 18:232. doi: 10.1186/s12916-020-01694-8
11. Mosele F, Remon J, Mateo J, Westphalen CB, Barlesi F, Lolkema MP, et al. Recommendations for the Use of Next-Generation Sequencing (NGS) for Patients With Metastatic Cancers: A Report From the ESMO Precision Medicine Working Group. Ann Oncol (2020) 31:1491–505. doi: 10.1016/j.annonc.2020.07.014
12. Yang H, Sun L, Guan A, Yin H, Liu M, Mao X, et al. Unique TP53 Neoantigen and the Immune Microenvironment in Long-Term Survivors of Hepatocellular Carcinoma. Cancer Immunol Immunother (2020) 70:667–77. doi: 10.1007/s00262-020-02711-8
13. Feng K, Liu Y, Zhao Y, Yang Q, Dong L, Liu J, et al. Efficacy and Biomarker Analysis of Nivolumab Plus Gemcitabine and Cisplatin in Patients With Unresectable or Metastatic Biliary Tract Cancers: Results From a Phase II Study. J Immunother Cancer (2020) 8:e000367. doi: 10.1136/jitc-2019-000367
14. Wang Q, Douglass J, Hwang MS, Hsiue EH, Mog BJ, Zhang M, et al. Direct Detection and Quantification of Neoantigens. Cancer Immunol Res (2019) 7:1748–54. doi: 10.1158/2326-6066.cir-19-0107
15. Ward JP, Gubin MM, Schreiber RD. The Role of Neoantigens in Naturally Occurring and Therapeutically Induced Immune Responses to Cancer. Adv Immunol (2016) 130:25–74. doi: 10.1016/bs.ai.2016.01.001
16. Wood MA, Weeder BR, David JK, Nellore A, Thompson RF. Burden of Tumor Mutations, Neoepitopes, and Other Variants are Weak Predictors of Cancer Immunotherapy Response and Overall Survival. Genome Med (2020) 12:33. doi: 10.1186/s13073-020-00729-2
17. McDermott DF, Huseni MA, Atkins MB, Motzer RJ, Rini BI, Escudier B, et al. Clinical Activity and Molecular Correlates of Response to Atezolizumab Alone or in Combination With Bevacizumab Versus Sunitinib in Renal Cell Carcinoma. Nat Med (2018) 24:749–57. doi: 10.1038/s41591-018-0053-3
18. Jia Q, Wu W, Wang Y, Alexander PB, Sun C, Gong Z, et al. Local Mutational Diversity Drives Intratumoral Immune Heterogeneity in non-Small Cell Lung Cancer. Nat Commun (2018) 9:5361. doi: 10.1038/s41467-018-07767-w
19. Coulie PG, Van den Eynde BJ, van der Bruggen P, Boon T. Tumour Antigens Recognized by T Lymphocytes: At the Core of Cancer Immunotherapy. Nat Rev Cancer (2014) 14:135–46. doi: 10.1038/nrc3670
20. Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational Heterogeneity in Cancer and the Search for New Cancer-Associated Genes. Nature (2013) 499:214–8. doi: 10.1038/nature12213
21. Blaeschke F, Paul MC, Schuhmann MU, Rabsteyn A, Schroeder C, Casadei N, et al. Low Mutational Load in Pediatric Medulloblastoma Still Translates Into Neoantigens as Targets for Specific T-cell Immunotherapy. Cytotherapy (2019) 21:973–86. doi: 10.1016/j.jcyt.2019.06.009
22. Zamora AE, Crawford JC, Allen EK, Guo XJ, Bakke J, Carter RA, et al. Pediatric Patients With Acute Lymphoblastic Leukemia Generate Abundant and Functional Neoantigen-Specific CD8(+) T Cell Responses. Sci Transl Med (2019) 11:eaat8549. doi: 10.1126/scitranslmed.aat8549
23. Chae YK, Viveiros P, Lopes G, Sukhadia B, Sheikh MM, Saravia D, et al. Clinical and Immunological Implications of Frameshift Mutations in Lung Cancer. J Thorac Oncol (2019) 14:1807–17. doi: 10.1016/j.jtho.2019.06.016
24. Miller A, Asmann Y, Cattaneo L, Braggio E, Keats J, Auclair D, et al. High Somatic Mutation and Neoantigen Burden are Correlated With Decreased Progression-Free Survival in Multiple Myeloma. Blood Cancer J (2017) 7:e612. doi: 10.1038/bcj.2017.94
25. Yarchoan M, Johnson BA3, Lutz ER, Laheru DA, Jaffee EM. Targeting Neoantigens to Augment Antitumour Immunity. Nat Rev Cancer (2017) 17:209–22. doi: 10.1038/nrc.2016.154
26. Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer Immunology. Mutational Landscape Determines Sensitivity to PD-1 Blockade in non-Small Cell Lung Cancer. Science (2015) 348:124–8. doi: 10.1126/science.aaa1348
27. Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, et al. Genomic Correlates of Response to CTLA-4 Blockade in Metastatic Melanoma. Science (2015) 350:207–11. doi: 10.1126/science.aad0095
28. Lauss M, Donia M, Harbst K, Andersen R, Mitra S, Rosengren F, et al. Mutational and Putative Neoantigen Load Predict Clinical Benefit of Adoptive T Cell Therapy in Melanoma. Nat Commun (2017) 8:1738. doi: 10.1038/s41467-017-01460-0
29. Kim S, Kim HS, Kim E, Lee MG, Shin EC, Paik S, et al. Neopepsee: Accurate Genome-Level Prediction of Neoantigens by Harnessing Sequence and Amino Acid Immunogenicity Information. Ann Oncol (2018) 29:1030–6. doi: 10.1093/annonc/mdy022
30. Bulik-Sullivan B, Busby J, Palmer CD, Davis MJ, Murphy T, Clark A, et al. Deep Learning Using Tumor HLA Peptide Mass Spectrometry Datasets Improves Neoantigen Identification. Nat Biotechnol (2018) 37:55–63. doi: 10.1038/nbt.4313
31. Bai P, Li Y, Zhou Q, Xia J, Wei PC, Deng H, et al. Immune-Based Mutation Classification Enables Neoantigen Prioritization and Immune Feature Discovery in Cancer Immunotherapy. Oncoimmunology (2021) 10:1868130. doi: 10.1080/2162402x.2020.1868130
32. Wu J, Wang W, Zhang J, Zhou B, Zhao W, Su Z, et al. Deephlapan: A Deep Learning Approach for Neoantigen Prediction Considering Both HLA-Peptide Binding and Immunogenicity. Front Immunol (2019) 10:2559. doi: 10.3389/fimmu.2019.02559
33. Shao XM, Bhattacharya R, Huang J, Sivakumar IKA, Tokheim C, Zheng L, et al. High-Throughput Prediction of MHC Class I and II Neoantigens With Mhcnuggets. Cancer Immunol Res (2020) 8:396–408. doi: 10.1158/2326-6066.cir-19-0464
34. Liu G, Li D, Li Z, Qiu S, Li W, Chao CC, et al. Pssmhcpan: A Novel PSSM-based Software for Predicting Class I peptide-HLA Binding Affinity. GigaScience (2017) 6:1–11. doi: 10.1093/gigascience/gix017
35. Hu Z, Anandappa AJ, Sun J, Kim J, Leet DE, Bozym DJ, et al. A Cloning and Expression System to Probe T-cell Receptor Specificity and Assess Functional Avidity to Neoantigens. Blood (2018) 132:1911–21. doi: 10.1182/blood-2018-04-843763
36. Chen F, Zou Z, Du J, Su S, Shao J, Meng F, et al. Neoantigen Identification Strategies Enable Personalized Immunotherapy in Refractory Solid Tumors. J Clin Invest (2019) 129:2056–70. doi: 10.1172/jci99538
37. Zhou C, Wei Z, Zhang Z, Zhang B, Zhu C, Chen K, et al. pTuneos: Prioritizing Tumor Neoantigens From Next-Generation Sequencing Data. Genome Med (2019) 11:67. doi: 10.1186/s13073-019-0679-x
38. Hundal J, Kiwala S, McMichael J, Miller CA, Xia H, Wollam AT, et al. Pvactools: A Computational Toolkit to Identify and Visualize Cancer Neoantigens. Cancer Immunol Res (2020) 8:409–20. doi: 10.1158/2326-6066.cir-19-0401
39. Lu T, Wang S, Xu L, Zhou Q, Singla N, Gao J, et al. Tumor Neoantigenicity Assessment With CSiN Score Incorporates Clonality and Immunogenicity to Predict Immunotherapy Outcomes. Sci Immunol (2020) 5:eaaz3199. doi: 10.1126/sciimmunol.aaz3199
40. Wells DK, van Buuren MM, Dang KK, Hubbard-Lucey VM, Sheehan KCF, Campbell KM, et al. Key Parameters of Tumor Epitope Immunogenicity Revealed Through a Consortium Approach Improve Neoantigen Prediction. Cell (2020) 183:818–34.e13. doi: 10.1016/j.cell.2020.09.015
41. Mardis ER. Neoantigens and Genome Instability: Impact on Immunogenomic Phenotypes and Immunotherapy Response. Genome Med (2019) 11:71. doi: 10.1186/s13073-019-0684-0
42. Green AR, Aleskandarany MA, Ali R, Hodgson EG, Atabani S, De Souza K, et al. Clinical Impact of Tumor Dna Repair Expression and T-cell Infiltration in Breast Cancers. Cancer Immunol Res (2017) 5:292–9. doi: 10.1158/2326-6066.cir-16-0195
43. Baretti M, Le DT. DNA Mismatch Repair in Cancer. Pharmacol Ther (2018) 189:45–62. doi: 10.1016/j.pharmthera.2018.04.004
44. Germano G, Lamba S, Rospo G, Barault L, Magrì A, Maione F, et al. Inactivation of DNA Repair Triggers Neoantigen Generation and Impairs Tumour Growth. Nature (2017) 552:116–20. doi: 10.1038/nature24673
45. Chae YK, Anker JF, Oh MS, Bais P, Namburi S, Agte S, et al. Mutations in DNA Repair Genes are Associated With Increased Neoantigen Burden and a Distinct Immunophenotype in Lung Squamous Cell Carcinoma. Sci Rep (2019) 9:3235. doi: 10.1038/s41598-019-39594-4
46. Ballhausen A, Przybilla MJ, Jendrusch M, Haupt S, Pfaffendorf E, Seidler F, et al. The Shared Frameshift Mutation Landscape of Microsatellite-Unstable Cancers Suggests Immunoediting During Tumor Evolution. Nat Commun (2020) 11:4740. doi: 10.1038/s41467-020-18514-5
47. Turajlic S, Litchfield K, Xu H, Rosenthal R, McGranahan N, Reading JL, et al. Insertion-and-Deletion-Derived Tumour-Specific Neoantigens and the Immunogenic Phenotype: A Pan-Cancer Analysis. Lancet Oncol (2017) 18:1009–21. doi: 10.1016/s1470-2045(17)30516-8
48. Yang W, Lee KW, Srivastava RM, Kuo F, Krishna C, Chowell D, et al. Immunogenic Neoantigens Derived From Gene Fusions Stimulate T Cell Responses. Nat Med (2019) 25:767–75. doi: 10.1038/s41591-019-0434-2
49. Jayasinghe RG, Cao S, Gao Q, Wendl MC, Vo NS, Reynolds SM, et al. Systematic Analysis of Splice-Site-Creating Mutations in Cancer. Cell Rep (2018) 23:270–81.e3. doi: 10.1016/j.celrep.2018.03.052
50. Mansfield AS, Peikert T, Smadbeck JB, Udell JBM, Garcia-Rivera E, Elsbernd L, et al. Neoantigenic Potential of Complex Chromosomal Rearrangements in Mesothelioma. J Thorac Oncol (2019) 14:276–87. doi: 10.1016/j.jtho.2018.10.001
51. Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of Mutational Processes in Human Cancer. Nature (2013) 500:415–21. doi: 10.1038/nature12477
52. Pfeifer GP, Denissenko MF, Olivier M, Tretyakova N, Hecht SS. Hainaut P. Tobacco Smoke Carcinogens, DNA Damage and p53 Mutations in Smoking-Associated Cancers. Oncogene (2002) 21:7435–51. doi: 10.1038/sj.onc.1205803
53. Wang X, Ricciuti B, Nguyen T, Li X, Rabin MS, Awad MM, et al. Association Between Smoking History and Tumor Mutation Burden in Advanced non-Small Cell Lung Cancer. Cancer Res (2021). doi: 10.1158/0008-5472.can-20-3991
54. Pham TV, Boichard A, Goodman A, Riviere P, Yeerna H, Tamayo P, et al. Role of Ultraviolet Mutational Signature Versus Tumor Mutation Burden in Predicting Response to Immunotherapy. Mol Oncol (2020) 14:1680–94. doi: 10.1002/1878-0261.12748
55. Castellucci E, He T, Goldstein DY, Halmos B, Chuy J. Dna Polymerase ϵ Deficiency Leading to an Ultramutator Phenotype: A Novel Clinically Relevant Entity. Oncologist (2017) 22:497–502. doi: 10.1634/theoncologist.2017-0034
56. Temko D, Van Gool IC, Rayner E, Glaire M, Makino S, Brown M, et al. Somatic POLE Exonuclease Domain Mutations are Early Events in Sporadic Endometrial and Colorectal Carcinogenesis, Determining Driver Mutational Landscape, Clonal Neoantigen Burden and Immune Response. J Pathol (2018) 245:283–96. doi: 10.1002/path.5081
57. Glaser AP, Fantini D, Shilatifard A, Schaeffer EM, Meeks JJ. The Evolving Genomic Landscape of Urothelial Carcinoma. Nat Rev Urol (2017) 14:215–29. doi: 10.1038/nrurol.2017.11
58. Wang D, Niu X, Wang Z, Song CL, Huang Z, Chen KN, et al. Multiregion Sequencing Reveals the Genetic Heterogeneity and Evolutionary History of Osteosarcoma and Matched Pulmonary Metastases. Cancer Res (2019) 79:7–20. doi: 10.1158/0008-5472.can-18-1086
59. McGranahan N, Furness AJ, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, et al. Clonal Neoantigens Elicit T Cell Immunoreactivity and Sensitivity to Immune Checkpoint Blockade. Science (2016) 351:1463–9. doi: 10.1126/science.aaf1490
60. Wolf Y, Bartok O, Patkar S, Eli GB, Cohen S, Litchfield K, et al. Uvb-Induced Tumor Heterogeneity Diminishes Immune Response in Melanoma. Cell (2019) 179:219–35.e21. doi: 10.1016/j.cell.2019.08.032
61. Mariathasan S, Turley SJ, Nickles D, Castiglioni A, Yuen K, Wang Y, et al. Tgfβ Attenuates Tumour Response to PD-L1 Blockade by Contributing to Exclusion of T Cells. Nature (2018) 554:544–8. doi: 10.1038/nature25501
62. Riaz N, Havel JJ, Makarov V, Desrichard A, Urba WJ, Sims JS, et al. Tumor and Microenvironment Evolution During Immunotherapy With Nivolumab. Cell (2017) 171:934–49.e16. doi: 10.1016/j.cell.2017.09.028
63. Willis JA, Reyes-Uribe L, Chang K, Lipkin SM, Vilar E. Immune Activation in Mismatch Repair-Deficient Carcinogenesis: More Than Just Mutational Rate. Clin Cancer Res (2020) 26:11–7. doi: 10.1158/1078-0432.ccr-18-0856
64. Chae YK, Anker JF, Bais P, Namburi S, Giles FJ, Chuang JH. Mutations in DNA Repair Genes are Associated With Increased Neo-Antigen Load and Activated T Cell Infiltration in Lung Adenocarcinoma. Oncotarget (2018) 9:7949–60. doi: 10.18632/oncotarget.23742
65. Maby P, Tougeron D, Hamieh M, Mlecnik B, Kora H, Bindea G, et al. Correlation Between Density of CD8+ T-Cell Infiltrate in Microsatellite Unstable Colorectal Cancers and Frameshift Mutations: A Rationale for Personalized Immunotherapy. Cancer Res (2015) 75:3446–55. doi: 10.1158/0008-5472.can-14-3051
66. Choudhury NJ, Kiyotani K, Yap KL, Campanile A, Antic T, Yew PY, et al. Low T-cell Receptor Diversity, High Somatic Mutation Burden, and High Neoantigen Load as Predictors of Clinical Outcome in Muscle-invasive Bladder Cancer. Eur Urol Focus (2016) 2:445–52. doi: 10.1016/j.euf.2015.09.007
67. Sneddon S, Rive CM, Ma S, Dick IM, Allcock RJN, Brown SD, et al. Identification of a CD8+ T-Cell Response to a Predicted Neoantigen in Malignant Mesothelioma. Oncoimmunology (2020) 9:1684713. doi: 10.1080/2162402x.2019.1684713
68. Tokunaga R, Zhang W, Naseem M, Puccini A, Berger MD, Soni S, et al. Cxcl9, CXCL10, CXCL11/CXCR3 Axis for Immune Activation - A Target for Novel Cancer Therapy. Cancer Treat Rev (2018) 63:40–7. doi: 10.1016/j.ctrv.2017.11.007
69. Church SE, Jensen SM, Antony PA, Restifo NP, Fox BA. Tumor-Specific CD4+ T Cells Maintain Effector and Memory Tumor-Specific CD8+ T Cells. Eur J Immunol (2014) 44:69–79. doi: 10.1002/eji.201343718
70. Workel HH, Lubbers JM, Arnold R, Prins TM, van der Vlies P, de Lange K, et al. A Transcriptionally Distinct CXCL13(+)CD103(+)CD8(+) T-Cell Population is Associated With B-cell Recruitment and Neoantigen Load in Human Cancer. Cancer Immunol Res (2019) 7:784–96. doi: 10.1158/2326-6066.cir-18-0517
71. Pfannstiel C, Strissel PL, Chiappinelli KB, Sikic D, Wach S, Wirtz RM, et al. The Tumor Immune Microenvironment Drives a Prognostic Relevance That Correlates With Bladder Cancer Subtypes. Cancer Immunol Res (2019) 7:923–38. doi: 10.1158/2326-6066.cir-18-0758
72. Blass E, Ott PA. Advances in the Development of Personalized Neoantigen-Based Therapeutic Cancer Vaccines. Nat Rev Clin Oncol (2021) 18:215–29. doi: 10.1038/s41571-020-00460-2
73. Villadangos JA. Antigen-Specific Impairment of Adoptive T-cell Therapy Against Cancer: Players, Mechanisms, Solutions and a Hypothesis. Immunol Rev (2016) 272:169–82. doi: 10.1111/imr.12433
74. Harari A, Graciotti M, Bassani-Sternberg M, Kandalaft LE. Antitumour Dendritic Cell Vaccination in a Priming and Boosting Approach. Nat Rev Drug Discovery (2020) 19:635–52. doi: 10.1038/s41573-020-0074-8
75. Bethune MT, Joglekar AV. Personalized T Cell-Mediated Cancer Immunotherapy: Progress and Challenges. Curr Opin Biotechnol (2017) 48:142–52. doi: 10.1016/j.copbio.2017.03.024
76. Li L, Goedegebuure SP, Gillanders WE. Preclinical and Clinical Development of Neoantigen Vaccines. Ann Oncol (2017) 28:xii11–7. doi: 10.1093/annonc/mdx681
77. Johanns TM, Miller CA, Liu CJ, Perrin RJ, Bender D, Kobayashi DK, et al. Detection of Neoantigen-Specific T Cells Following a Personalized Vaccine in a Patient With Glioblastoma. Oncoimmunology (2019) 8:e1561106. doi: 10.1080/2162402x.2018.1561106
78. Tanyi JL, Bobisse S, Ophir E, Tuyaerts S, Roberti A, Genolet R, et al. Personalized Cancer Vaccine Effectively Mobilizes Antitumor T Cell Immunity in Ovarian Cancer. Sci Transl Med (2018) 10:eaao5931. doi: 10.1126/scitranslmed.aao5931
79. D’Alise AM, Leoni G, Cotugno G, Troise F, Langone F, Fichera I, et al. Adenoviral Vaccine Targeting Multiple Neoantigens as Strategy to Eradicate Large Tumors Combined With Checkpoint Blockade. Nat Commun (2019) 10:2688. doi: 10.1038/s41467-019-10594-2
80. Salewski I, Gladbach YS, Kuntoff S, Irmscher N, Hahn O, Junghanss C, et al. In Vivo Vaccination With Cell Line-Derived Whole Tumor Lysates: Neoantigen Quality, Not Quantity Matters. J Transl Med (2020) 18:402. doi: 10.1186/s12967-020-02570-y
81. Lhuillier C, Rudqvist NP, Elemento O, Formenti SC, Demaria S. Radiation Therapy and Anti-Tumor Immunity: Exposing Immunogenic Mutations to the Immune System. Genome Med (2019) 11:40. doi: 10.1186/s13073-019-0653-7
82. Formenti SC, Demaria S. Radiation Therapy to Convert the Tumor Into an in Situ Vaccine. Int J Radiat Oncol Biol Phys (2012) 84:879–80. doi: 10.1016/j.ijrobp.2012.06.020
83. Wilkins A, McDonald F, Harrington K, Melcher A. Radiotherapy Enhances Responses of Lung Cancer to CTLA-4 Blockade. J Immunother Cancer (2019) 7:64. doi: 10.1186/s40425-019-0542-z
84. Ji D, Yi H, Zhang D, Zhan T, Li Z, Li M, et al. Somatic Mutations and Immune Alternation in Rectal Cancer Following Neoadjuvant Chemoradiotherapy. Cancer Immunol Res (2018) 6:1401–16. doi: 10.1158/2326-6066.cir-17-0630
85. Mouw KW, Cleary JM, Reardon B, Pike J, Braunstein LZ, Kim J, et al. Genomic Evolution After Chemoradiotherapy in Anal Squamous Cell Carcinoma. Clin Cancer Res (2017) 23:3214–22. doi: 10.1158/1078-0432.ccr-16-2017
86. Criscuolo D, Morra F, Giannella R, Visconti R, Cerrato A, Celetti A. New Combinatorial Strategies to Improve the PARP Inhibitors Efficacy in the Urothelial Bladder Cancer Treatment. J Exp Clin Cancer Res (2019) 38:91. doi: 10.1186/s13046-019-1089-z
87. Ren Y, Cherukuri Y, Wickland DP, Sarangi V, Tian S, Carter JM, et al. HLA Class-I and class-II Restricted Neoantigen Loads Predict Overall Survival in Breast Cancer. Oncoimmunology (2020) 9:1744947. doi: 10.1080/2162402x.2020.1744947
88. Matsushita H, Hasegawa K, Oda K, Yamamoto S, Asada K, Karasaki T, et al. Neoantigen Load and HLA-Class I Expression Identify a Subgroup of Tumors With a T-cell-inflamed Phenotype and Favorable Prognosis in Homologous Recombination-Proficient High-Grade Serous Ovarian Carcinoma. J Immunother Cancer (2020) 8:e000375. doi: 10.1136/jitc-2019-000375
89. Matsushita H, Sato Y, Karasaki T, Nakagawa T, Kume H, Ogawa S, et al. Neoantigen Load, Antigen Presentation Machinery, and Immune Signatures Determine Prognosis in Clear Cell Renal Cell Carcinoma. Cancer Immunol Res (2016) 4:463–71. doi: 10.1158/2326-6066.cir-15-0225
90. Schramm A, Köster J, Assenov Y, Althoff K, Peifer M, Mahlow E, et al. Mutational Dynamics Between Primary and Relapse Neuroblastomas. Nat Genet (2015) 47:872–7. doi: 10.1038/ng.3349
91. Eleveld TF, Oldridge DA, Bernard V, Koster J, Colmet Daage L, Diskin SJ, et al. Relapsed Neuroblastomas Show Frequent RAS-MAPK Pathway Mutations. Nat Genet (2015) 47:864–71. doi: 10.1038/ng.3333
92. Majzner RG, Heitzeneder S, Mackall CL. Harnessing the Immunotherapy Revolution for the Treatment of Childhood Cancers. Cancer Cell (2017) 31:476–85. doi: 10.1016/j.ccell.2017.03.002
93. Park JA, Cheung NV. Limitations and Opportunities for Immune Checkpoint Inhibitors in Pediatric Malignancies. Cancer Treat Rev (2017) 58:22–33. doi: 10.1016/j.ctrv.2017.05.006
94. Jiang J, Ding Y, Wu M, Chen Y, Lyu X, Lu J, et al. Integrated Genomic Analysis Identifies a Genetic Mutation Model Predicting Response to Immune Checkpoint Inhibitors in Melanoma. Cancer Med (2020) 9:8498–518. doi: 10.1002/cam4.3481
95. Simnica D, Akyüz N, Schliffke S, Mohme M, VW L, Mährle T, et al. T Cell Receptor Next-Generation Sequencing Reveals Cancer-Associated Repertoire Metrics and Reconstitution After Chemotherapy in Patients With Hematological and Solid Tumors. Oncoimmunology (2019) 8:e1644110. doi: 10.1080/2162402x.2019.1644110
96. Aversa I, Malanga D, Fiume G, Palmieri C. Molecular T-Cell Repertoire Analysis as Source of Prognostic and Predictive Biomarkers for Checkpoint Blockade Immunotherapy. Int J Mol Sci (2020) 21:2378. doi: 10.3390/ijms21072378
97. Dao T, Klatt MG, Korontsvit T, Mun SS, Guzman S, Mattar M, et al. Impact of Tumor Heterogeneity and Microenvironment in Identifying Neoantigens in a Patient With Ovarian Cancer. Cancer Immunol Immunother (2020) 70:1189–202. doi: 10.1007/s00262-020-02764-9
98. Jiang T, Cheng R, Pan Y, Zhang H, He Y, Su C, et al. Heterogeneity of Neoantigen Landscape Between Primary Lesions and Their Matched Metastases in Lung Cancer. Transl Lung Cancer Res (2020) 9:246–56. doi: 10.21037/tlcr.2020.03.03
99. Fennemann FL, de Vries IJM, Figdor CG, Verdoes M. Attacking Tumors From All Sides: Personalized Multiplex Vaccines to Tackle Intratumor Heterogeneity. Front Immunol (2019) 10:824. doi: 10.3389/fimmu.2019.00824
100. McGranahan N, Rosenthal R, Hiley CT, Rowan AJ, Watkins TBK, Wilson GA, et al. Allele-Specific HLA Loss and Immune Escape in Lung Cancer Evolution. Cell (2017) 171:1259–71.e11. doi: 10.1016/j.cell.2017.10.001
101. Gong Z, Jia Q, Chen J, Diao X, Gao J, Wang X, et al. Impaired Cytolytic Activity and Loss of Clonal Neoantigens in Elderly Patients With Lung Adenocarcinoma. J Thorac Oncol (2019) 14:857–66. doi: 10.1016/j.jtho.2019.01.024
102. Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and Genetic Properties of Tumors Associated With Local Immune Cytolytic Activity. Cell (2015) 160:48–61. doi: 10.1016/j.cell.2014.12.033
103. Sade-Feldman M, Jiao YJ, Chen JH, Rooney MS, Barzily-Rokni M, Eliane JP, et al. Resistance to Checkpoint Blockade Therapy Through Inactivation of Antigen Presentation. Nat Commun (2017) 8:1136. doi: 10.1038/s41467-017-01062-w
104. Jia Q, Chiu L, Wu S, Bai J, Peng L, Zheng L, et al. Tracking Neoantigens by Personalized Circulating Tumor Dna Sequencing During Checkpoint Blockade Immunotherapy in Non-Small Cell Lung Cancer. Advanced Sci (Weinheim Baden-Wurttemberg Germany) (2020) 7:1903410. doi: 10.1002/advs.201903410
Keywords: tumor neoantigen burden, biomarker, immunotherapy, immune response, tumor mutation burden
Citation: Wang P, Chen Y and Wang C (2021) Beyond Tumor Mutation Burden: Tumor Neoantigen Burden as a Biomarker for Immunotherapy and Other Types of Therapy. Front. Oncol. 11:672677. doi: 10.3389/fonc.2021.672677
Received: 26 February 2021; Accepted: 07 April 2021;
Published: 29 April 2021.
Edited by:
Min Cheng, Weifang Medical University, ChinaReviewed by:
Scott Moerdler, Rutgers Cancer Institute of New Jersey, United StatesCopyright © 2021 Wang, Chen and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chun Wang, d2FuZ2NodW5Ac2N1LmVkdS5jbg==
†ORCID: Peipei Wang, orcid.org/0000-0001-7201-4928
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.