
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol. , 22 April 2021
Sec. Thoracic Oncology
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.671799
This article is part of the Research Topic Management of Large Cell Neuroendocrine Carcinoma View all 9 articles
Pulmonary large cell neuroendocrine carcinoma (LCNEC) is an aggressive neoplasm with poor prognosis. Histologic diagnosis of LCNEC is not always straightforward. In particular, it is challenging to distinguish small cell lung carcinoma (SCLC) or poorly differentiated carcinoma from LCNEC. However, histological classification for LCNEC as well as their therapeutic management has not changed much for decades. Recently, genomic and transcriptomic analyses have revealed different molecular subtypes raising hopes for more personalized treatment. Two main molecular subtypes of LCNEC have been identified by studies using next generation sequencing, namely type I with TP53 and STK11/KEAP1 alterations, alternatively called as non-SCLC type, and type II with TP53 and RB1 alterations, alternatively called as SCLC type. However, there is still no easy way to classify LCNEC subtypes at the actual clinical level. In this review, we have discussed histological diagnosis along with the genomic studies and molecular-based treatment for LCNEC.
Neuroendocrine tumors (NET) of the lung account for approximately 20% of all lung cancers with a majority being represented by small cell lung cancer (SCLC) (1–3). Large cell neuroendocrine carcinoma (LCNEC) is rare, accounting for 1–3% of all lung cancer cases (4, 5). The increasing incidence has been reported recently, with a rate raised from 0.26 in 2004 to 0.39 per 100,000 people in 2015 (5–7). Most LCNEC patients are male and their median age is 66 (5–7). There is a high frequency of cigarette smoking history, up to 98% (4, 6). LCNEC usually occurs in the lung periphery. Median survival time from diagnosis is 9.7 months and 54.6% of patients have stage IV LCNEC at the time of diagnosis (7). LCNEC is a unique tumor that shows immunohistochemical and morphological traits of both SCLC and non-small cell lung carcinoma (NSCLC). Although LCNEC is currently categorized into NSCLC, due to its overlapping features of SCLC, treatment protocols for NSCLC and SCLC are used depending on the situations. Also with the rarity of the disease, no standard treatment for LCNEC has been currently developed. Recently, two main molecular subtypes of LCNEC have been identified by next generation sequencing (NGS) studies, namely type I with TP53 and STK11/KEAP1 alterations (NSCLC type), and type II with TP53 and RB1 alterations (SCLC type) (8). These subtypes are currently considered as relevant for prognosis and selecting the therapeutic regimen. In this review, we will first introduce current histopathological features of LCNEC and its criteria to distinguish from other histological types. And then, we will focus on the molecular subtypes and their influence on pathological diagnosis. The emphasis will be done on immunohistochemical subgrouping added by the concise review of studies about treatment for LCNEC based on molecular subtype.
The World Health Organization (WHO) 2015 classification of malignant pulmonary neuroendocrine tumors includes four major histological types: low-grade typical carcinoid, intermediate-grade atypical carcinoid, high-grade SCLC and LCNEC (1). There are tumorlets and diffuse idiopathic pulmonary neuroendocrine cell hyperplasia (DIPNECH) in the category of neuroendocrine cell tumors, but both are currently considered to be the pre-invasive benign lesions. All of these neoplasms show various levels of organoid growth patterns (rosette, palisading, trabeculae, ribbons, festoons, lobular nests), they are distinguished conventionally based on hematoxylin and eosin (H&E) features such as mitotic rate, presence of necrosis, and cytologic details including the presence of large nucleoli and abundant cytoplasm (Table 1, Figure 1). LCNEC is defined as a NSCLC with neuroendocrine morphology and the expression of neuroendocrine markers (1). LCNEC cells are typically more than three times diameter of small lymphocytes and exhibit abundant cytoplasm, nucleoli or often with vesicular chromatin. The use of immunohistochemistry (IHC) for the purpose of detecting neuroendocrine feature, synaptophysin, chromogranin, and CD56, is mandatory.
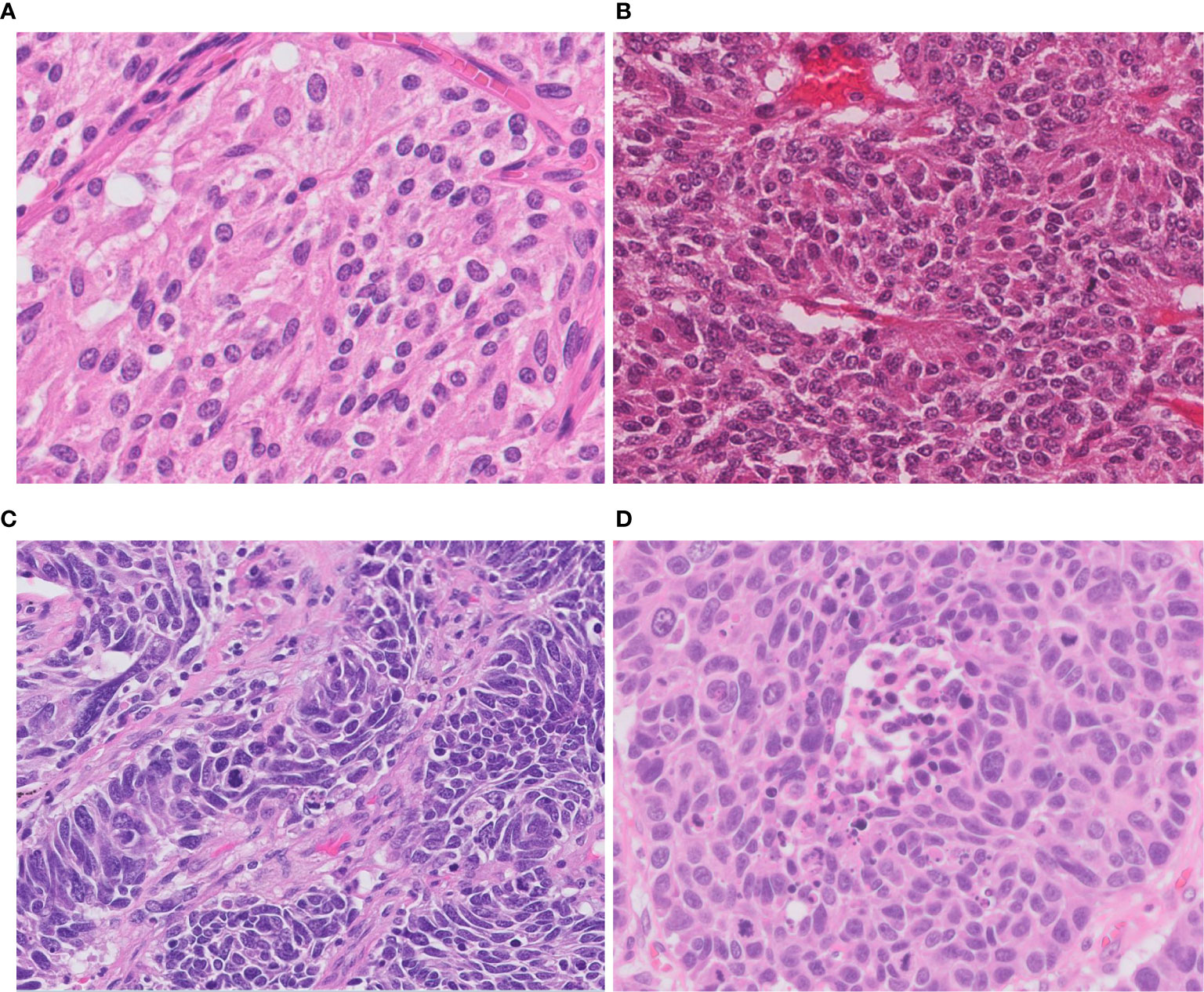
Figure 1 Morphology of neuroendocrine tumors of the lung. (A) Typical carcinoid shows solid nests with zellballen patterns; the tumor cells are uniform with a moderate amount of eosinophilic cytoplasm. (B) Atypical carcinoid with rosette formation. (C) SCLC showing sheets of small cells with scant cytoplasm, finely granular chromatin, and mitoses. (D) LCNEC displays organoid nesting and palisading patterns; tumor cells have abundant eosinophilic cytoplasm, coarsely granular chromatin, and prominent nucleoli. Magnification: ×40.
The primary differential diagnosis for LCNEC include SCLC and other types of NSCLC. LCNEC and SCLC are differentially diagnosed based on cytological appearance including prominent nucleoli, vesicular to clumped versus finely granular chromatin, cell size, and more abundant cytoplasm in LCNEC. NSCLC can be differentiated from LCNEC by absence of neuroendocrine morphology and IHC expression. Basaloid squamous cell carcinoma shows nested growth pattern with peripheral palisading and large areas of necrosis, mimicking LCNEC. Although p40 is useful squamous epithelial marker to distinguish LCNEC from basaloid squamous cell carcinoma, it should be noted that some LCNEC may also show focal staining (<10% tumor cells labeling) (9). The diffuse and strong p40 positivity is characteristic for basaloid squamous cell carcinoma.
The difficulty of distinguishing between LCNEC and SCLC is that there are cases showing borderline features (Figure 2A). Use of IHC to distinguish LCNEC and SCLC is with limited benefits. There are currently no entirely sensitive and specific IHC markers to separate LCNEC and SCLC. Some markers including CK7, 8, 18, and 19 were reported to be significantly weaker in SCLC than in LCNEC. In SCLC, these cytokeratins typically show dot-like cytoplasmic staining (10–12), however, similar staining patterns can also be seen in LCNEC. One of the potential useful markers is napsin A. Based on the literature, focal and weak staining can be seen in up to 15% of LCNECs, whereas SCLC is consistently negative for this marker (13, 14). Therefore, there are significant interobserver variability.
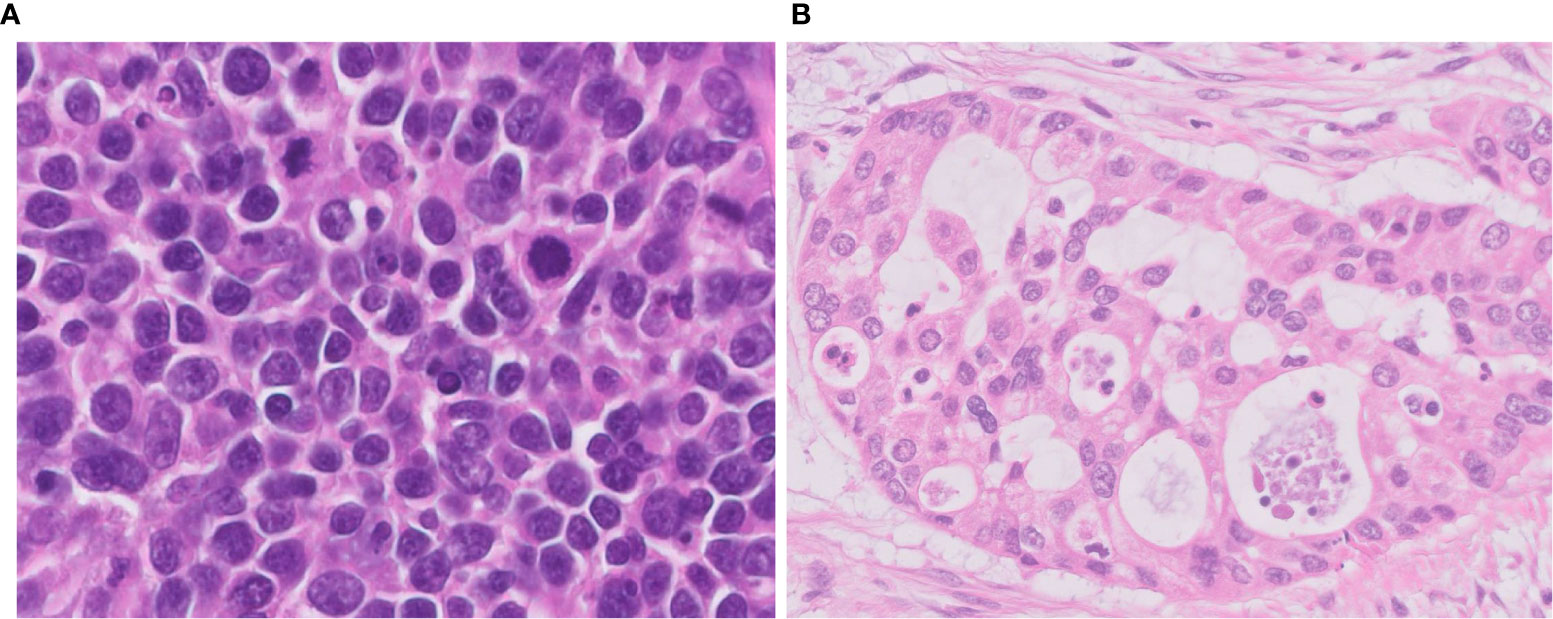
Figure 2 Difficulties in differential diagnosis of LCNEC. (A) This tumor is LCNEC that needs to be differentiated from SCLC; cell size is larger than that of SCLC. (B) This LCNEC needs to be differentiated from adenocarcinoma; the tumor shows pseudoglandular structures forming cribriform pattern. Magnification: ×40.
One of the important pathological judgements outside the pulmonary NETs is distinguishing some types of poorly differentiated NSCLC, which can be difficult to distinguish solely by H&E observation (Figure 2B). Usually, it is possible to distinguish between NSCLC and pulmonary NETs by the presence or absence of neuroendocrine marker expression (synaptophysin, chromogranin A, and CD56). However, it is important to know that neuroendocrine markers are not always clear to separate those two types since 15-23% of high-grade neuroendocrine carcinomas are stained negative even with a combination of all three markers and nearly 15% of NSCLCs are positive for one of these neuroendocrine markers (15, 16). Identifying neuroendocrine morphology along with confirming diffuse staining for neuroendocrine markers occupying the majority of the whole tumor area are critical (Figure 3).
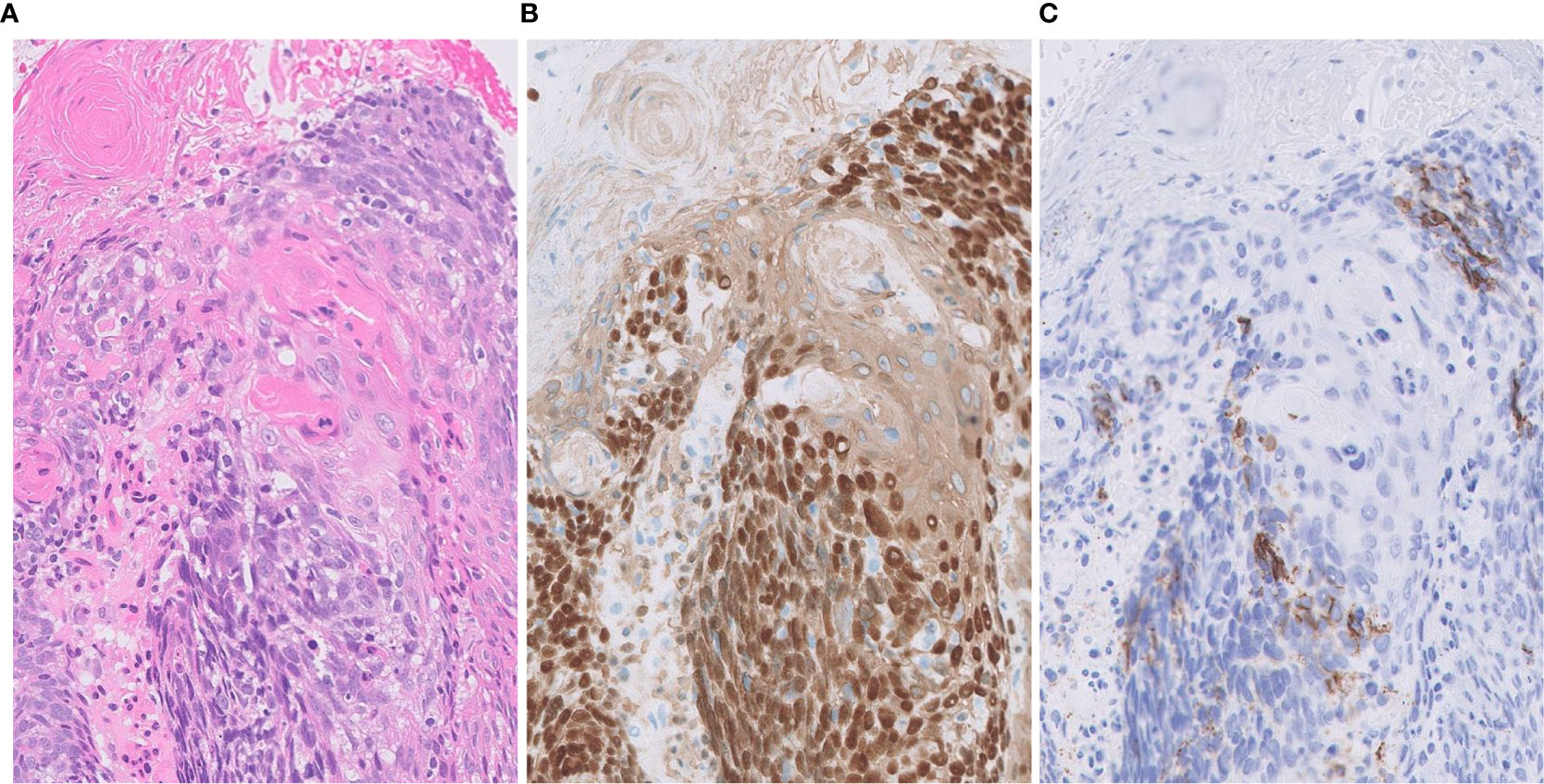
Figure 3 Focal expression of CD56 in squamous cell carcinoma. (A) Tumor cells form solid nests with occasional keratinization. (B) Diffusely positivity for p40 (nuclear) and CK14 (cytoplasmic). (C) Focal expression of neuroendocrine marker CD56. Magnification: ×20.
Recently, a new category of tumors called SMARCA4-deficient undifferentiated thoracic tumor has been reported. These tumors show undifferentiated round cells or rhabdoid morphology (17, 18). Although these tumors do not show neuroendocrine architecture, they are diffusely positive for synaptophysin in 70% of cases (17). They also show geographic necrosis, high mitotic and Ki-67 rate. Therefore, it should be also noted that in a crushed biopsy, these features may closely mimic LCNEC. Other immunostaining is useful to distinguish between the two in such cases. Typical immunohistochemical features for SMARCA4-deficient undifferentiated thoracic tumor is the lack of claudin 4 expression and low or absent keratin immunostaining, in addition to the loss of SMARCA4 (BRG-1) (17–20).
Recently, several new immunohistochemical markers, such as Insulinoma-associated protein 1 (INSM1) and Achaete-scute homolog-1 (ASCL1) have been reported as neuroendocrine markers with nuclear expression (16, 21–27). INSM1 has been reported to have excellent sensitivity and specificity for the diagnosis of lung NETs (26, 27), and no cases of NSCLC have reported to be positive for ASCL1 immunostaining so far (16). An important benefit of these markers is their staining localization. Nuclear staining is suggested optimal for lung NET diagnosis since many specimens of these tumors obtained by transbronchial biopsy show strong crush artifacts gotten at the time of sampling (Figure 4) and, for that occasion, evaluation of cytoplasmic (synoptophysin, chromagranin A) and membranous (CD56) immunoexpression is difficult.
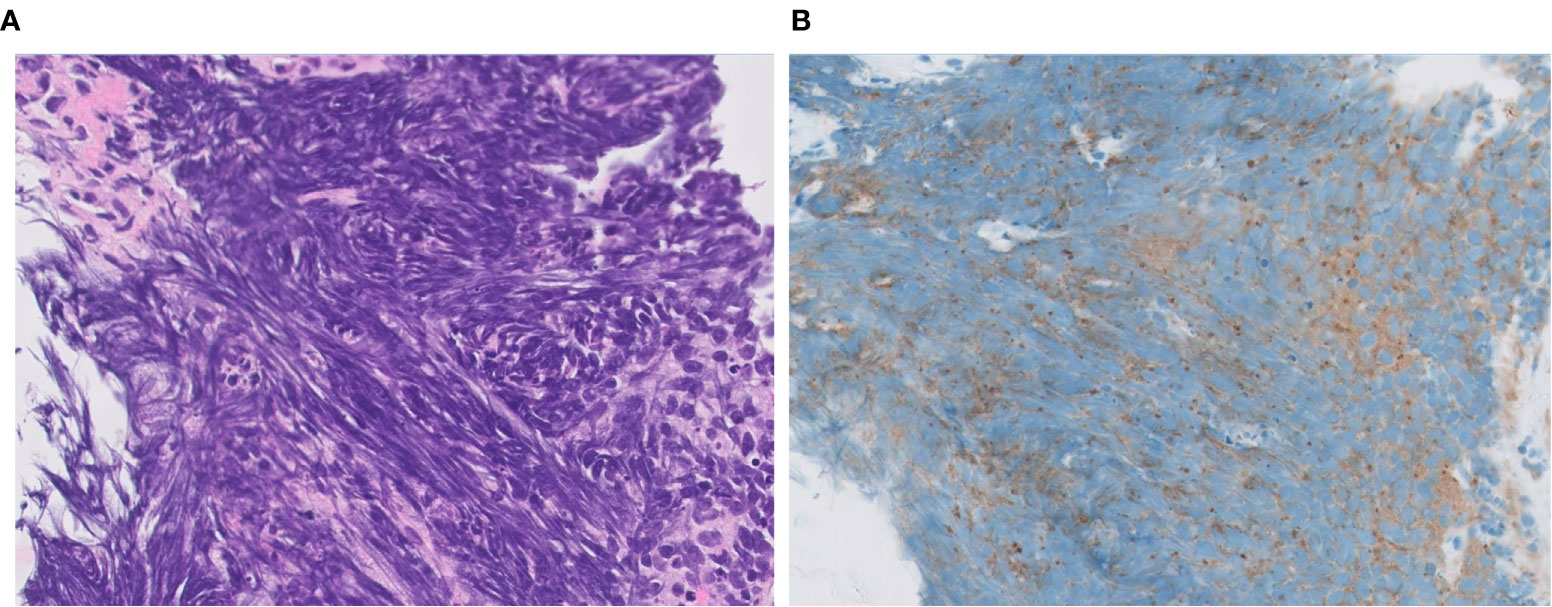
Figure 4 Extensive crush artifact in small cell lung carcinoma. SCLC is often crushed during biopsy, which affects appearance on routine staining (A, H&E) and immunohistochemistry (B, synaptophysin). Magnification: ×40.
Extrapulmonary NETs are graded as G1, G2, or G3 based on mitotic count, Ki-67 labeling index (LI) and presence of necrosis (28). Because Ki-67 LI is overlapped among different grades in pulmonary NETs without clear cutoff, the main utility of this biomarker is to distinguish the carcinoids from high grade pulmonary NETs (29). Ki-67 LI can be used to separate LCNEC from typical and atypical carcinoid tumors where LCNEC typically display LI >40% whereas that of carcinoid tumors is < 20% (9, 29) (Figure 5). Ki-67 LI has been proposed as a prognostic factor in surgically resected specimens of typical and atypical carcinoids, with cutoff values ranging from 2.5% to 5.8% (29). Our group showed that scoring hot sports is mandatory for survival prediction in NSCLC (30). However, studies exploring prognostic utility of Ki-67 in LCNEC could not find statistically significant correlation between LI and overall survival and disease-free survival (31, 32). Additional well-powered studies with standardized scoring approach are needed to elucidate a role of Ki-67 as diagnostic and/or prognostic biomarker in LCNEC.
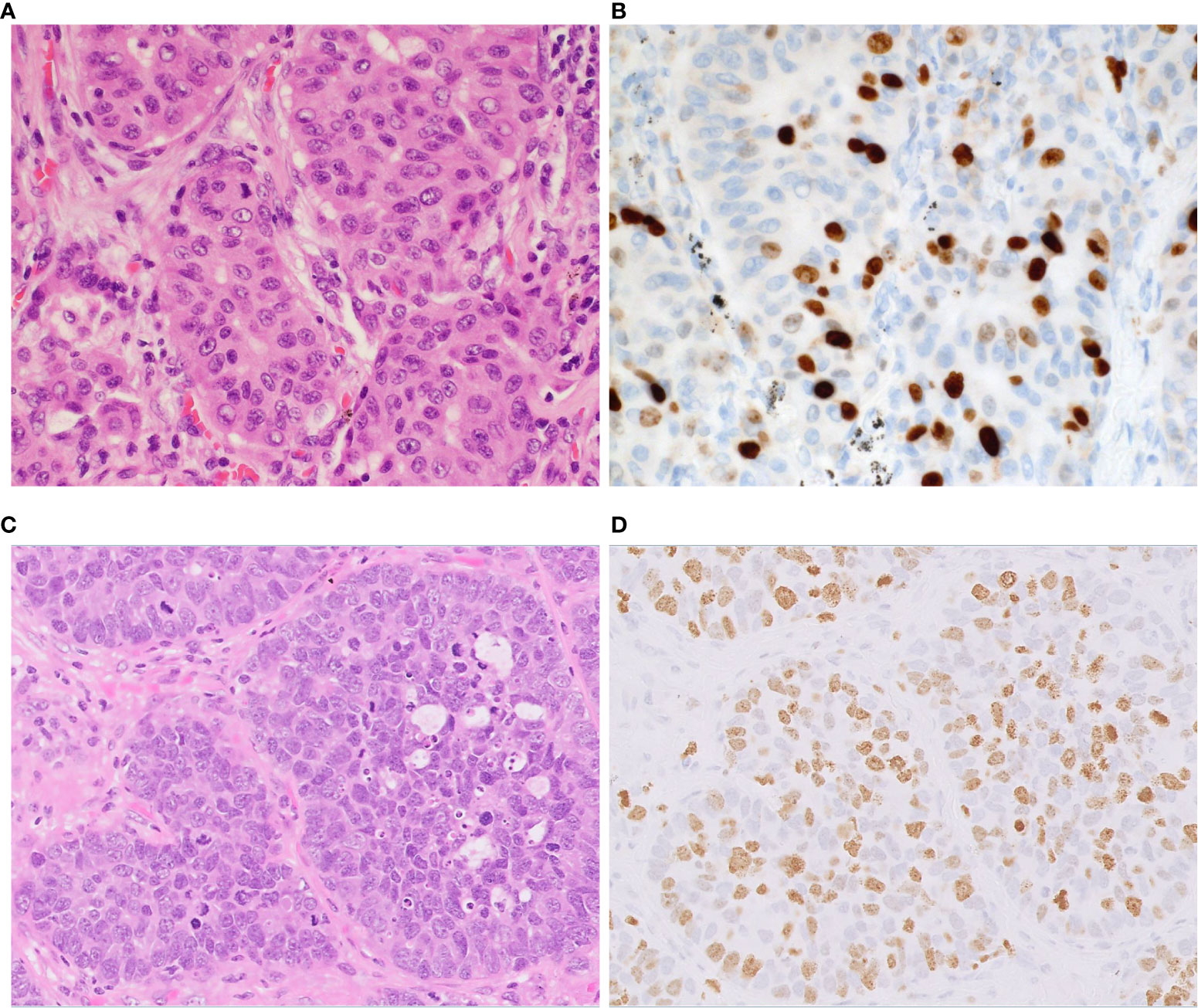
Figure 5 Proliferation index in atypical carcinoid vs. LCNEC. Representative examples of atypical carcinoid (A, B) and LCNEC (C, D). (A) The tumor shows nests of carcinoid tumor cells. (B) The Ki-67 proliferative index is approximately 20%. (C) The tumor shows nests and palisading patterns of LCNEC tumor cells. (D) The Ki-67 proliferative index is approximately 60%. Magnification: ×40.
The histological diagnosis of LCNEC is challenging in small biopsy specimens unless a full-developed morphology consistent with LCNEC diagnosis is disclosed. The major reason is tissue heterogeneity of the tumor due to admixture of other patterns (32). Therefore, WHO does not recommend rendering pathological diagnosis of LCNEC on the small biopsies, instead the diagnosis should be limited to surgically removed specimens. However, considering the unignorable frequency of advance disease status at the time of presentation and the importance of histological diagnosis to select the best therapeutic regimen, the improvement of accurate diagnosis of LCNEC for the small biopsies such as transbronchial lung biopsy, needle biopsy, and cryobiopsy is important. There indeed are several cases showing convincing morphological and IHC features to make diagnosis of LCNEC (Figure 6). Accumulating evidence to prove diagnostic feasibility for biopsy samples with combination of H&E and IHC panels is mandatory. For those occasions, molecular classifiers including new IHC markers described in below sections may be useful.
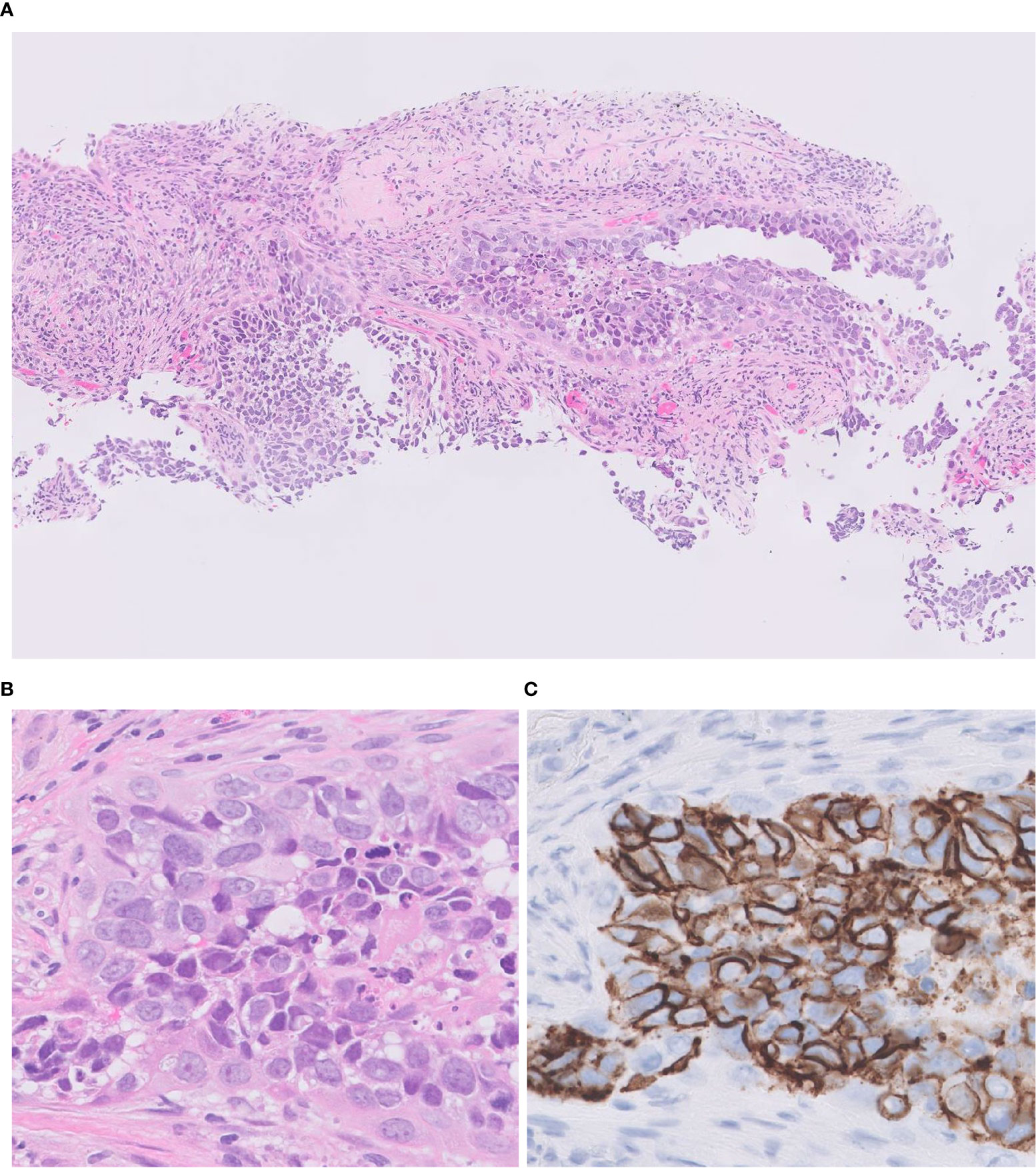
Figure 6 Transbronchial lung biopsy of LCNEC. A small-sized specimen featuring tumor cells with abundant eosinophilic cytoplasm, coarsely granular chromatin, and prominent nucleoli. H&E (A, B); CD 56 (C) Magnification: ×10 (A); ×40 (B, C).
Genetic background of LCNEC has been widely studied, particularly, with a reference to molecular signature of NSCLC. The common molecular alterations are shown in Table 2. Among tumor suppressor genes altered in LCNEC, the TP53 mutation is the most common (64-92%), followed by the RB1 mutation (19-42%) (Table 2) (8, 9, 33–44).
The most common alterations in driver genes are KRAS mutations (4-24%), with a much less frequency of EGFR, ALK, RET, and absence of others driver mutations associated with NSCLC, such as BRAF and ROS1 (9, 34, 39, 46). Genomic mutations associated with the PI3K-AKT-mTOR pathway are found in LCNECs, including PTEN (4–5%), PIK3CA (3%), AKT2 (4%), PICTOR (5%), mTOR: (1%), NF1 (5%), INSR (3%), TSC2 (2%), and PRCTR (2%) (34, 45). Amplifications in NK2 homeobox 1 (NKX2-1, also known as TTF-1; 10–20%), v-myc avian myelocytomatosis viral oncogene homolog gene (MYC; 5-13%), v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog gene (MYCL1; 12%), SRY-box 2 gene (SOX2; 11%), fibroblast growth factor receptor 1 (FGFR1; 4–7%), insulin receptor substrate 2 (IRS2; 3–4%) and v-myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog gene (MYCN; 2%) have also been identified. Cyclin-dependent kinase inhibitor 2A (CDKN2A) deletions were reported with a frequency of 4% to 8% (8, 9, 45).
In addition to gene mutations, chromosomal alterations have been reported in LCNEC; in particular, alterations greater than 10 Mb, losses of 1p, 3p, 4p, 4q, 5q, 8p, 10q, 13q, 17p and gains of 3q, 5p, 8q, 18q, were found to be much more frequent in LCNECs as compared to pulmonary carcinoid tumors (47).
George et al. reported the existence of two LCNEC genomic subtypes with specific transcriptional patterns, which they categorized as type I (NSCLC type) with TP53 and STK11/KEAP1 alterations and type II (SCLC type) with TP53 and RB1 alterations, which is introduced in the upcoming 2021 WHO classification (8). Although type I LCNEC shares genomic alterations with pulmonary adenocarcinomas and squamous cell carcinomas, no transcriptional relationship was found, and it was divided into transcriptional subgroups (ASCL1high/DLL3high/NOTCHlow) with similarity to SCLC. TP53 and RB1 are tumor suppressor genes, and mutations have been reported in LCNEC as well as in SCLC. One of the hallmarks of SCLC is bi-allelic alterations in TP53 and RB1 (48, 49). While type II LCNECs reveals genetic resemblance to SCLC, these tumors are markedly different from SCLC transcriptional subgroups (ASCL1low/DLL3low/NOTCHhigh). Therefore, the distinction between type I and type II LCNECs from SCLC is important to be able to evaluate the response of patients to treatment options.
Serine/threonine kinase 11 (STK11) encodes liver kinase B1 (LKB1) and is a commonly altered tumor suppressor that frequently occurs in NSCLC (50). LKB1 directly phosphorylates and activates adenosine monophosphate-activated protein kinase (AMPK) (51). In response to energetic stress, AMPK alters the cellular metabolism to restore nicotinamide adenine dinucleotide phosphate (NADPH) concentrations (52). It also regulates the activity of mTOR. Under energetic stress, the LKB1-AMPK axis plays a critical role in modulating cell growth and proliferation to maintain adequate ATP and NADPH levels (53).
Kelch-like ECH-associated protein 1 (KEAP1) forms a protein complex and ubiquitinates the N-terminal domain of NRF2, an oxidative stress-responsive transcription factor (54). Oxidative stress induces the oxidation of KEAP1 at key cysteine residues which causes a conformational change in KEAP1 releasing NRF2, resulting in translocation and nuclear accumulation of NRF2. In the nucleus, NRF2 forms a heterodimer with its partner sMAF (v-Maf avian musculoaponeurotic fibrosarcoma oncogene homolog) and binds to antioxidant responsive element (ARE) sequences to regulate the transcription of target genes (55). A major NRF2 transcriptional target is NADPH (56, 57). KEAP1 is not only a tumor suppressor gene, but also a metastasis suppressor gene (58). KEAP1 mutations co-occur with mutations in STK11, which have also been associated with poor response to immune checkpoint blockade in lung adenocarcinoma (59–61). Therefore, STK11/KEAP1 mutations can be expected as predictive biomarkers for anti-PD-1/PD-L1 therapy.
ASCL1 is a transcription factor and is selectively expressed in normal fetal pulmonary neuroendocrine cells. ASCL1 is highly expressed in SCLC and LCNEC, where it acts to maintain neuroendocrine features (62). The Notch pathway likewise plays an important role in the developing respiratory system and regulates neuroendocrine versus epithelial cell fate decisions (63). Notch genes encode single transmembrane receptors that mediate short-range communication between cells (64, 65). When Notch binds to its ligand (delta-like ligands: DLL1, DLL3 and DLL4, jagged ligands: JAG1 and JAG2) expressed on adjacent cells, Notch receptors (Notch1–4) release the Notch intracellular domain (NICD). NICD activates transcription of HES1 (hairy and enhancer of split 1 and HEY1 [hairy and enhancer of split-related protein 1]), which encodes transcriptional repressors of ASCL1 (66). On the other hand, DLL3 is a transcriptional target of ASCL1 (62, 67). And unlike other Notch ligands (DLL1, DLL4, JAG1 and JAG2), DLL3 without the conserved N-terminal module of agonistic Notch ligands can antagonize DLL1-Notch signaling (68). Thus, ASCL1 both activates Notch signaling and is repressed by it. DLL3 predominantly localizes to the Golgi apparatus, where it retains other Notch members and redirects them to endosomes for degradation (69). Some DLL3 is expressed on the cell surface, which is not expressed in normal lung tissue (70). Thus, DLL3 protein has emerged as a very promising drug target.
We reviewed the frequency of mutations associated with molecular subtypes of LCNEC. PubMed was searched for papers with keywords: lung, LCNEC and molecular/genetic alterations. Initially, we retrieved 94 papers. After excluding review papers and case reports, the remaining 17 studies containing information on alterations for TP53, RB1, STK11, KEAP1, NOTCH, ASCL1 and DLL3 were analyzed (8, 9, 24, 33–44, 71, 72). A summary of these studies is shown in Table 3 (raw data provided in Supplemental Table 1).
The most common alterations were TP53, followed by RB1 and STK11 (Table 3). No alterations were found in DLL3 and ASCL1. All alterations of TP53, RB1, STK11 and KEAP1 were examined in 205 cases, of which 26 cases could be classified as type I LCNEC (TP53 and STK11/KEAP1 alterations), and 71 cases could be classified into type II LCNECs (TP53 and RB1 alterations).
Chemotherapy treatment for LCNEC remains a subject of debate. In patients with advanced LCNEC, the chemotherapy regimens used in SCLC are still the standard of treatment, but results are not satisfactory (73). The type I (with TP53 and STK11/KEAP1 alterations) and type II LCNECs (with TP53 and RB1 alterations) may have a heterogeneous response to chemotherapy. Derks et al. reported patients with LCNEC tumors that carry a wild-type RB1 gene or express the RB1 protein do better with NSCLC type chemotherapy (platinum-gemcitabine or paclitaxel) than with SCLC type chemotherapy (platinum, etoposide). In contrast, no difference was observed in LCNEC cases with the RB1 mutation (35). Another study found that patients with NSCLC-like LCNEC treated with NSCLC-gemcitabine/taxane-platinum regimen had significantly shorter progression-free survival and overall survival than those treated with SCLC-etoposide-platinum regimen (44). It is not entirely clear why the above studies produced conflicting results. Baseline characteristics of the patients and combination therapy with irradiation could be contributing factors. Future studies in larger cohorts are needed to establish optimal protocols of chemotherapy in patients with different molecular subtypes of LCNEC.
The first antibody-drug conjugate in which DLL3 was investigated as a therapeutic target in SCLC patients is rovalpituzumab-tesirine (Rova-T) (74). Rova-T demonstrated encouraging single-agent antitumor activity with a manageable safety profile in a phase 1 trial. Unfortunately, modest clinical activity with associated toxicities led to the discontinuation of Rova-T in later studies (75). Two other novel DLL3-targeted therapies are anti-DLL3/CD3 bispecific antibodies (AMG 757) and DLL3-binding chimeric antigen receptor-modified T cells (AMG 119) (76, 77). Clinical evaluation of these therapies in SCLC patients is ongoing (NCT03319940 and NCT03392064). Since type I LCNECs show high neuroendocrine expression (ASCL1high/DLL3high/NOTCHlow) similar to SCLC, such tumors may also be susceptible to this agent.
Recently, it was reported that the high tumor mutational burden is related to better efficacy of immunotherapy (78, 79). The tumor mutational burden is high in LCNEC (>8 mutations/Mb) and was shown to be related to PD-L1 expression (8, 9, 80). Moreover, some LCNEC cases with negative PD-L1 expression but high tumor mutation burden may respond to immunotherapy (81, 82). PD-L1 expression was reported in 10–22% of studied LCNEC cases (80, 83–85). Although PD-L1 expression is known to be distinctly higher in NSCLCs as compared to SCLCs, Hermans et al. reported PD-L1 expression was equal in RB1 mutates (SCLC-like) and RB1 wildtype (NSCLC-like) cases. None of the seven STK11-mutated samples in this study harbored PD-L1 expression. The co-occurrence of KEAP1 and STK11 mutations has also been reported to be associated with poor response to immune checkpoint inhibition (13, 14, 47). Therefore, the effect of immunotherapeutic treatment might be reduced in type I LCNECs. George et al. also reported type II LCNECs exhibited a pattern of gene expression with upregulation of immune related pathways, which may impact the response of patients to immunotherapy. Clinical trials of immunotherapy in LCNEC patients are ongoing (NCT03591731, NCT02939651), but it will be necessary to consider other factors, including gene expression, in the future.
Rekhtman et al. reported that at least one alteration potentially targetable by investigational agents was present in 65% of LCNECs (30/45), being more common in RB1 wild type LCNEC than in RB1 mutated LCNEC (84% vs. 50%, respectively). Several case reports described a good response to EGFR-tyrosine kinase inhibitor (gefitinib/icotinib) in LCNEC with an activating EGFR gene mutation (exon 19) (86–88). Two case reports demonstrated that treatment with crizotinib of LCNEC with EML4-ALK rearrangement was ineffective or achieved only partial response (89, 90). On the other hand, a case of LCNEC with PLB1-ALK rearrangements reported to be sensitive to crizotinib (91). We anticipate that future studies with larger cohorts of LCNEC patients will produce more consistent results on the effect of the above targeted treatments.
The PI3K-AKT-mTOR pathway has been reported to be overactivated in lung NETs (85). Phase II clinical trials of mTOR inhibitors (everolimus) in low-to-intermediate NETs (including lung) showed encouraging results (92). Moreover, everolimus in combination with chemotherapy (carboplatin and paclitaxel) has been reported to be effective for patients with metastatic LCNEC (93). Therefore, evaluation of gene mutations in the PI3K-AKT-mTOR pathway may be useful for a targeted treatment in LCNEC.
Poly-ADP ribose polymerase (PARP) inhibitors are also being studied in combination with chemotherapy in SCLC (94). Recently, coiled-coil-domain containing 6 (CCDC6) has been indicated as a prognostic biomarker, which is also predictive of a possible response to treatment with PARP inhibitors in NSCLC (95). The CCDC6 levels are modulated by deubiquitinase ubiquitin specific protease 7 (USP7) (96). Malapelle et al. has reported that the immunostaining of pulmonary NET, including LCNEC, showed the intensity of CCDC6 staining correlated with the levels of USP7 expression (71). Moreover, the inhibition of USP7 by P5091 accelerated the degradation of CCDC6 versus control in cycloheximide treated SCLC cells in vitro and sensitized the cells to PARP inhibitors alone and in combination with cisplatin (71). This suggests that CCDC6 and USP7 have a predictive value for the clinical usage of USP7 inhibitors in combination with PARP inhibitors in SCLC. This can be expected to be effective in LCNEC, and further research in larger cohorts is desired in the future.
The enhancer of zeste homolog 2 (EZH2) is a histone methyltransferase that forms the polycomb repressive complex 2 (PRC2) (97). In SCLC, EZH2 is upregulated upon inactivation of the E2F/Rb pathway and leads to aberrant methylation of its target (98). Poirier et al. reported that inhibition of EZH2 suppressed tumor growth in vivo and in vitro in both SCLC cell lines and patient derived xenograft mouse models (99). Clinical trials of EZH2 inhibitors in SCLC patients are currently ongoing (NCT03460977). Noteworthy, high EZH2 expression in SCLC and LCNEC was reported in studies using IHC (100, 101). Therefore, EZH2 inhibition may provide new therapeutic perspectives for LCNEC as well as SCLC.
Although there have been remarkable scientific updates such as molecular subtyping, pathological diagnosis of LCNEC has not changed much since the release of the 2004 WHO classification (102). The diagnostic yield is limited to surgical material while small biopsies are frequently insufficient and unreliable for a definite diagnosis. However, the majority of LCNEC cases are found at the late stage of the disease progression, which makes small biopsies the common modality for tissue sampling. To improve the treatment of LCNEC, the diagnostic approach to this entity by the use of molecular classifiers or, ideally, by its cheap and robust IHC alternative has been long anticipated. There has been a large debate over whether patients with LCNEC should be treated as NSCLC or SCLC. At present, just a few clear solutions have been established through clinical trials. With this background, the recent conceptual separation of LCNEC into two different subtypes, NSCLC-like type I and SCLC-like type II, is hoped to be a promising stratification to provide better therapeutic options to patients.
In their seminal paper, Rekhtman et al. genotyped 45 LCNECs and found 56% of tumors displayed NSCLC-like molecular features, characterized by KRAS or KEAP1 mutations alone or concurrently with TP53 mutations (9). Remaining 40% of LCNECs exhibited a SCLC-like genomic profile, characterized by RB1/TP53 co-alteration. Additionally, less common molecular alterations seen almost exclusively in the NSCL-like LCNEC involved the BRAF, MAP2K1, ERBB2, and CDKN2A genes, and those seen exclusively in the SCLC-like LCNEC included MYCL1 amplification and PTEN mutations.
In the same study, SCLC-like LCNEC had higher Ki-67 rates and a spectrum of morphologic features closer to SCLC than NSCL-like LCNEC (9). Other groups also addressed a proliferation index detected by Ki-67 IHC for stratification of LCNEC. Milione et al. reported that LCNECs with co-mutation of TP53 and RB1 (SCLC-like) were significantly enriched in cases with a Ki-67 ≧55%, while the tumors with KRAS mutations were enriched in cases with Ki-67 <55% (39). Such findings along with molecular data suggest that there is an overlap between the two subtypes of LCNEC, and proliferation alone cannot predict genomic features.
A continuing interest in surrogate biomarkers to substitute genetic testing (i.e., in a context of LCNEC, to render molecular subtyping) is explained by the high costs of genotyping. For instance, compared to NGS, immunostaining is a simple method that can be performed in virtually any pathology laboratory. Several IHC biomarkers have been evaluated with this regard. RB1 immunoexpression was considered as a promising biomarker alternative to molecular subtyping, however, Derks et al. reported that RB1 expression was completely lost not only in almost all RB1-mutated LCNECs, but also in 47% of the wild-type cases (35).
The subtyping of LCNEC may be better accomplished by considering the biomarkers directly connected to the therapeutic targets. One such candidate is an expression of DLL3. Evaluating the effectiveness of DLL3 expression as a therapeutic biomarker, ideally by immunostaining, may provide a pivotal progress in the field.
Saunders et al. reported that immunohistochemical expression of the DLL3 protein was completely negative in normal lung parenchyma, but was observed in 65% of LCNEC cases (103). Several studies have reported a positive correlation between DLL3 and ASCL1 IHC expression in SCLC (21, 23). Hermans et al. has reported that DLL3 H-score and ASCL1 H-score were correlated in LCNEC (24). They demonstrated that DLL3 is highly expressed in STK11- and KEAP1-mutated type I subtype and in TP53 wild-type tumors, as well as in tumors positive for ASCL1 and more than two neuroendocrine markers (24). Moreover, they did not find any relationship between DLL3 expression and RB1 mutation status or Rb immunostaining. Brcic et al. analyzed high-grade pulmonary NETs and cell cultures using different DLL3 antibodies. They found no correlation between the expression of TP53 and RB1 and DLL3 expression in LCNEC (104).
On DLL3 IHC staining, the majority of tumors show cytoplasmic/membranous staining (Figure 7), but perinuclear dot-like staining has also been reported (24). So far, it is unclear whether the pattern of staining predicts a response to DLL3-targeted therapy. These studies (24, 104) have some limitations (e.g., size of cohorts, NGS not performed in all patients, cut-off value, etc.), therefore further research is needed to verify utility of DLL3 immunostaining for adoption in practical use.
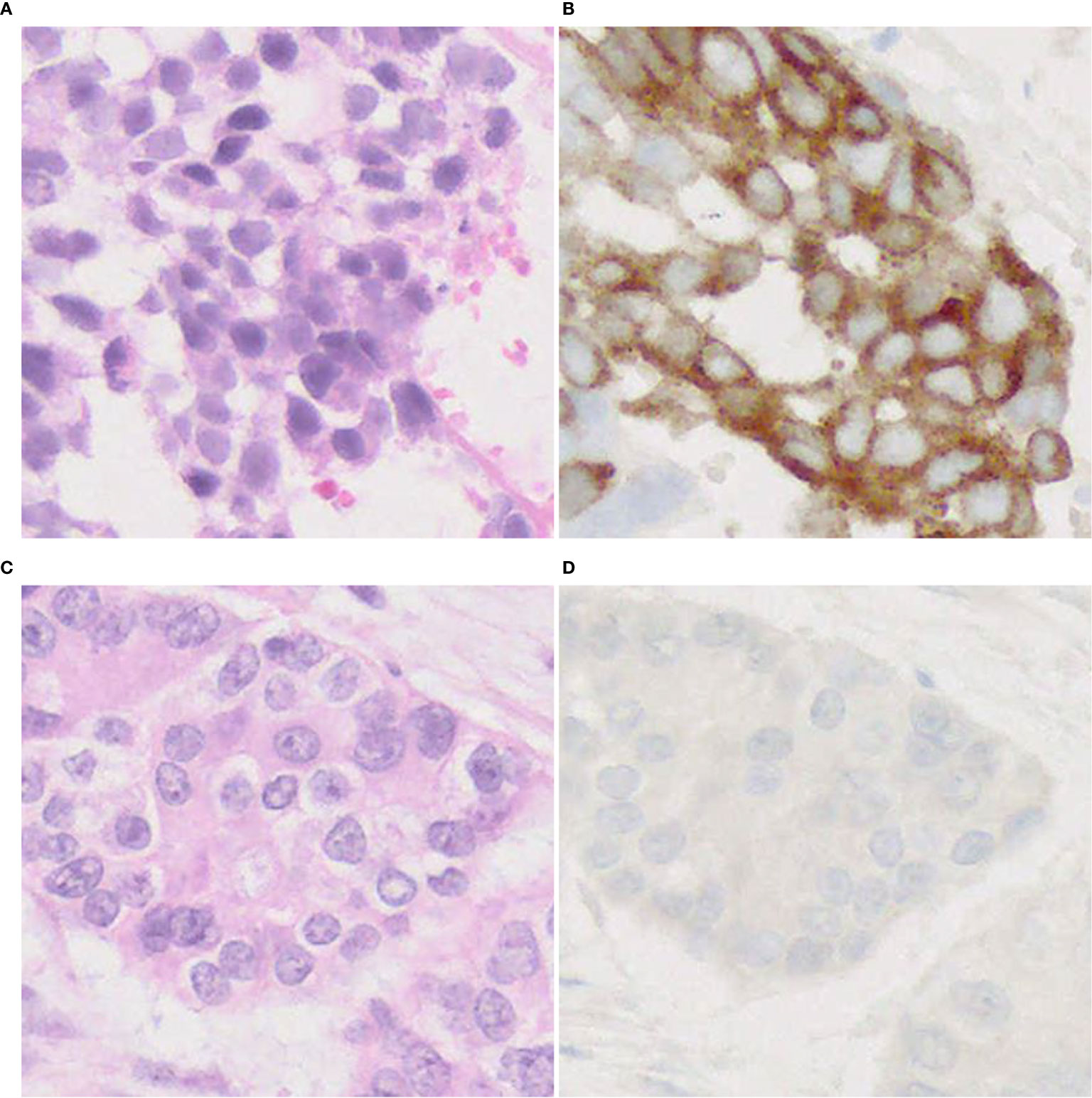
Figure 7 DLL3 immunophenotype of LCNEC. Representative cases with positive (A, B) and negative (C, D) expression of DLL3. Note a cytoplasmic pattern of immunostaining (B). Magnification: ×40.
We have done a review of 205 published LCNEC cases stratified by molecular subtype limited to alterations in four genes (TP53, RB1, STK11 and KEAP1). There were 108 cases (52.7%) that were not identified as either type I or type II LCNECs (Table 4). This suggests that the molecular spectrum of LCNEC is more heterogeneous and complex than expected.
Carcinoid tumors have distinct epidemiology and molecular pathogenesis from LCNEC. Rekhtman et al. reported carcinoid-like LCNEC, which was characterized by MEN1 alterations and low overall mutation burden (9). Very recently, a new entity of pulmonary carcinoids named supra-carcinoids has been discovered in an integrative genomic analysis study. Supra-carcinoids appeared morphologically as atypical carcinoids, but their molecular signature corresponded to the molecular cluster of LCNEC (40, 105). In addition to sharing the molecular features of LCNEC, supra-carcinoids also showed worse prognosis, similar to survival rates in LCNEC – 10-year overall survival of 33% and 19%, respectively (vs. 59% in conventional atypical carcinoid) (105, 106). However, histological and clinical characteristics of carcinoid-like LCNEC and supra-carcinoids are yet to be defined. This suggests the possibility of molecular link between pulmonary carcinoids and LCNEC.
It is obvious that comprehensive molecular analysis in well-defined large cohorts yields additional genotypic, phenotypic, and prognostic signatures within a family of lung NETs. We may predict that new types/subtypes of LCNEC other than the recently introduced types I–II will enter the classification scheme in the future.
With the rising impact of molecular pathology, the interest for reliable biomarkers grows, which can help to subclassify LCNECs and also enable personalized treatment for patients. In this review, we have discussed recent LCNEC genomic studies and treatments for LCNEC based on molecular subtype. To summarize, type I (NSCLC-like) LCNEC can be expected to respond to a DLL3 inhibitor, and type II (SCLC-like) can be expected to respond to immunotherapy. Classification of LCNECs will become important in choosing treatments. However, according to our literature review, there are many LCNECs that belong to groups other than these two categories. Moreover, there is no easy way to classify LCNEC subtypes at the clinical level. In the future, it will be necessary to study specific treatment and classification methods, and examine indicators for determining the efficacy of such methods.
MY: study design, literature review, article writing, and revision. KS: study design, literature review. AB: article writing, article revision. JF: study design, article writing, article revision. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors thank Mr. Ethan Okoshi, Department of Pathology, Nagasaki University Graduate School of Biomedical Sciences, for assistance with English language editing.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.671799/full#supplementary-material
1. Travis WD, Brambilla E, Burke AP, Marx A, Nicholson AG. Who Classification of Tumours of the Lung, Pleura, Thymus and Heart. 4th ed. Lyon: World Health Organization (2015).
2. Travis WD. Advances in Neuroendocrine Lung Tumors. Ann Oncol (2010) 21:vii65–71. doi: 10.1093/annonc/mdq380
3. Gustafsson BI, Kidd M, Chan A, Malfertheiner MV, Modlin IM. Bronchopulmonary Neuroendocrine Tumors. Cancer (2008) 113:5–21. doi: 10.1002/cncr.23542
4. Takei H, Asamura H, Maeshima A, Suzuki K, Kondo H, Niki T, et al. Large Cell Neuroendocrine Carcinoma of the Lung: A Clinicopathologic Study of Eighty-Seven Cases. J Thoracic Cardiovasc Surg (2002) 124:285–92. doi: 10.1067/mtc.2002.122523
5. Derks JL, Hendriks LE, Buikhuisen WA, Groen HJM, Thunnissen E, Suylen R-J, et al. Clinical Features of Large Cell Neuroendocrine Carcinoma: A Population-Based Overview. Eur Respir J (2016) 47:615–24. doi: 10.1183/13993003.00618-2015
6. Asamura H, Kameya T, Matsuno Y, Noguchi M, Tada H, Ishikawa Y, et al. Neuroendocrine Neoplasms of the Lung: A Prognostic Spectrum. JCO (2006) 24:70–6. doi: 10.1200/JCO.2005.04.1202
7. Kinslow CJ, May MS, Saqi A, Shu CA, Chaudhary KR, Wang TJC, et al. Large-Cell Neuroendocrine Carcinoma of the Lung: A Population-Based Study. Clin Lung Cancer (2020) 21:e99–e113. doi: 10.1016/j.cllc.2019.07.011
8. George J, Walter V, Peifer M, Alexandrov LB, Seidel D, Leenders F, et al. Integrative Genomic Profiling of Large-Cell Neuroendocrine Carcinomas Reveals Distinct Subtypes of High-Grade Neuroendocrine Lung Tumors. Nat Commun (2018) 9:1048. doi: 10.1038/s41467-018-03099-x
9. Rekhtman N, Pietanza MC, Hellmann MD, Naidoo J, Arora A, Won H, et al. Next-Generation Sequencing of Pulmonary Large Cell Neuroendocrine Carcinoma Reveals Small Cell Carcinoma–Like and Non–Small Cell Carcinoma–Like Subsets. Clin Cancer Res (2016) 22:3618–29. doi: 10.1158/1078-0432.CCR-15-2946
10. Nagashio R, Sato Y, Matsumoto T, Kageyama T, Satoh Y, Ryuge S, et al. Significant High Expression of Cytokeratins 7, 8, 18, 19 in Pulmonary Large Cell Neuroendocrine Carcinomas, Compared to Small Cell Lung Carcinomas. Pathol Int (2010) 60:71–7. doi: 10.1111/j.1440-1827.2009.02487.x
11. Nitadori J, Ishii G, Tsuta K, Yokose T, Murata Y, Kodama T, et al. Immunohistochemical Differential Diagnosis Between Large Cell Neuroendocrine Carcinoma and Small Cell Carcinoma by Tissue Microarray Analysis With a Large Antibody Panel. Am J Clin Pathol (2006) 125:682–92. doi: 10.1309/DT6BJ698LDX2NGGX
12. Sun L, Sakurai S, Sano T, Hironaka M, Kawashima O, Nakajima T. High-Grade Neuroendocrine Carcinoma of the Lung: Comparative Clinicopathological Study of Large Cell Neuroendocrine Carcinoma and Small Cell Lung Carcinoma. Pathol Int (2009) 59:522–9. doi: 10.1111/j.1440-1827.2009.02402.x
13. Baine MK, Sinard JH, Cai G, Homer RJ. A Semiquantitative Scoring System may Allow Biopsy Diagnosis of Pulmonary Large Cell Neuroendocrine Carcinoma. Am J Clin Pathol (2020) 153:165–74. doi: 10.1093/ajcp/aqz149
14. Rekhtman N, Pietanza CM, Sabari J, Montecalvo J, Wang H, Habeeb O, et al. Pulmonary Large Cell Neuroendocrine Carcinoma With Adenocarcinoma-Like Features: Napsin a Expression and Genomic Alterations. Mod Pathol (2018) 31:111–21. doi: 10.1038/modpathol.2017.110
15. Ionescu DN, Treaba D, Gilks CB, Leung S, Renouf D, Laskin J, et al. Nonsmall Cell Lung Carcinoma With Neuroendocrine Differentiation—an Entity of No Clinical or Prognostic Significance. Am J Surg Pathol (2007) 31:26–32. doi: 10.1097/01.pas.0000213319.04919.97
16. Ye B, Cappel J, Findeis-Hosey J, McMahon L, Yang Q, Xiao G-Q, et al. Hash1 is a Specific Immunohistochemical Marker for Lung Neuroendocrine Tumors. Hum Pathol (2016) 48:142–7. doi: 10.1016/j.humpath.2015.09.019
17. Rekhtman N, Montecalvo J, Chang JC, Alex D, Ptashkin RN, Ai N, et al. Smarca4-Deficient Thoracic Sarcomatoid Tumors Represent Primarily Smoking-Related Undifferentiated Carcinomas Rather Than Primary Thoracic Sarcomas. J Thoracic Oncol (2020) 15:231–47. doi: 10.1016/j.jtho.2019.10.023
18. Yoshida A, Kobayashi E, Kubo T, Kodaira M, Motoi T, Motoi N, et al. Clinicopathological and Molecular Characterization of SMARCA4-Deficient Thoracic Sarcomas With Comparison to Potentially Related Entities. Mod Pathol (2017) 30:797–809. doi: 10.1038/modpathol.2017.11
19. Sesboue C, Le Loarer F. Swi/Snf-Deficient Thoraco-Pulmonary Neoplasms. Semin Diagn Pathol [Preprint] (2021). Available at: https://www.sciencedirect.com/science/article/abs/pii/S074025702030109X.
20. Fukuoka J, Fujii T, Shih JH, Dracheva T, Meerzaman D, Player A, et al. Chromatin Remodeling Factors and BRM/BRG1 Expression as Prognostic Indicators in Non-Small Cell Lung Cancer. Clin Cancer Res (2004) 10:4314–24. doi: 10.1158/1078-0432.CCR-03-0489
21. Baine MK, Hsieh M-S, Lai WV, Egger JV, Jungbluth AA, Daneshbod Y, et al. Sclc Subtypes Defined by ASCL1, Neurod1, POU2F3, and YAP1: A Comprehensive Immunohistochemical and Histopathologic Characterization. J Thoracic Oncol (2020) 15:1823–35. doi: 10.1016/j.jtho.2020.09.009
22. Du E. Ttf-1 Expression is Specific for Lung Primary in Typical and Atypical Carcinoids: TTF-1-Positive Carcinoids are Predominantly in Peripheral Location. Hum Pathol (2004) 35:825–31. doi: 10.1016/j.humpath.2004.02.016
23. Furuta M, Sakakibara-Konishi J, Kikuchi H, Yokouchi H, Nishihara H, Minemura H, et al. Analysis of DLL3 and ASCL1 in Surgically Resected Small Cell Lung Cancer (Hot1702). Oncol (2019) 24:1172–9. doi: 10.1634/theoncologist.2018-0676
24. Hermans BCM, Derks JL, Thunnissen E, van Suylen RJ, den Bakker MA, Groen HJM, et al. Dll3 Expression in Large Cell Neuroendocrine Carcinoma (LCNEC) and Association With Molecular Subtypes and Neuroendocrine Profile. Lung Cancer (2019) 138:102–8. doi: 10.1016/j.lungcan.2019.10.010
25. Kontogianni K. Cd56: A Useful Tool for the Diagnosis of Small Cell Lung Carcinomas on Biopsies With Extensive Crush Artefact. J Clin Pathol (2005) 58:978–80. doi: 10.1136/jcp.2004.023044
26. Rooper LM, Sharma R, Li QK, Illei PB, Westra WH. Insm1 Demonstrates Superior Performance to the Individual and Combined Use of Synaptophysin, Chromogranin and CD56 for Diagnosing Neuroendocrine Tumors of the Thoracic Cavity. Am J Surg Pathol (2017) 41:1561–9. doi: 10.1097/PAS.0000000000000916
27. Viswanathan K, Siddiqui MT, Borczuk AC. Insulinoma-Associated Protein 1 is a Sensitive and Specific Marker for Lung Neuroendocrine Tumors in Cytologic and Surgical Specimens. J Am Soc Cytopathol (2019) 8:299–308. doi: 10.1016/j.jasc.2019.06.002
28. Rindi G, Klimstra DS, Abedi-Ardekani B, Asa SL, Bosman FT, Brambilla E, et al. A Common Classification Framework for Neuroendocrine Neoplasms: An International Agency for Research on Cancer (IARC) and World Health Organization (Who) Expert Consensus Proposal. Mod Pathol (2018) 31:1770–86. doi: 10.1038/s41379-018-0110-y
29. Pelosi G, Rindi G, Travis WD, Papotti M. Ki-67 Antigen in Lung Neuroendocrine Tumors: Unraveling a Role in Clinical Practice. J Thoracic Oncol (2014) 9:273–84. doi: 10.1097/JTO.0000000000000092
30. Tabata K, Tanaka T, Hayashi T, Hori T, Nunomura S, Yonezawa S, et al. Ki-67 is a Strong Prognostic Marker of Non-Small Cell Lung Cancer When Tissue Heterogeneity is Considered. BMC Clin Pathol (2014) 14:23. doi: 10.1186/1472-6890-14-23
31. Walts AE, Mirocha JM, Marchevsky AM. Challenges in Ki-67 Assessments in Pulmonary Large-Cell Neuroendocrine Carcinomas*. Histopathology (2021) 78:699–709. doi: 10.1111/his.14277
32. Igarashi T, Jiang S-X, Kameya T, Asamura H, Sato Y, Nagai K, et al. Divergent Cyclin B1 Expression and Rb/P16/Cyclin D1 Pathway Aberrations Among Pulmonary Neuroendocrine Tumors. Mod Pathol (2004) 17:1259–67. doi: 10.1038/modpathol.3800176
33. Karlsson A, Brunnström H, Lindquist KE, Jirström K, Jönsson M, Rosengren F, et al. Mutational and Gene Fusion Analyses of Primary Large Cell and Large Cell Neuroendocrine Lung Cancer. Oncotarget (2015) 6:22028–37. doi: 10.18632/oncotarget.4314
34. Miyoshi T, Umemura S, Matsumura Y, Mimaki S, Tada S, Makinoshima H, et al. Genomic Profiling of Large-Cell Neuroendocrine Carcinoma of the Lung. Clin Cancer Res (2017) 23:757–65. doi: 10.1158/1078-0432.CCR-16-0355
35. Derks JL, Leblay N, Thunnissen E, van Suylen RJ, den Bakker M, Groen HJM, et al. Molecular Subtypes of Pulmonary Large-Cell Neuroendocrine Carcinoma Predict Chemotherapy Treatment Outcome. Clin Cancer Res (2018) 24:33–42. doi: 10.1158/1078-0432.CCR-17-1921
36. Hermans BCM, Derks J, Groen HJM, Stigt JA, van Suylen RJ, Hillen LM, et al. Large Cell Neuroendocrine Carcinoma With a Solitary Brain Metastasis and Low Ki-67: A Unique Subtype. Endocrine Connections (2019)8:1600–6. doi: 10.1530/EC-19-0372
37. Karlsson A, Brunnström H, Micke P, Veerla S, Mattsson J, La Fleur L, et al. Gene Expression Profiling of Large Cell Lung Cancer Links Transcriptional Phenotypes to the New Histological WHO 2015 Classification. J Thoracic Oncol (2017) 12:1257–67. doi: 10.1016/j.jtho.2017.05.008
38. Meder L, König K, Ozretić L, Schultheis AM, Ueckeroth F, Ade CP, et al. NOTCH , ASCL1 , P53 and RB Alterations Define an Alternative Pathway Driving Neuroendocrine and Small Cell Lung Carcinomas. Int J Cancer (2016) 138:927–38. doi: 10.1002/ijc.29835
39. Milione M, Maisonneuve P, Grillo F, Mangogna A, Centonze G, Prinzi N, et al. Ki-67 Index of 55% Distinguishes Two Groups of Bronchopulmonary Pure and Composite Large Cell Neuroendocrine Carcinomas With Distinct Prognosis. Neuroendocrinology [Preprint] (2020). Available at: https://www.karger.com/Article/Purchase/508376.
40. Simbolo M, Barbi S, Fassan M, Mafficini A, Ali G, Vicentini C, et al. Gene Expression Profiling of Lung Atypical Carcinoids and Large Cell Neuroendocrine Carcinomas Identifies Three Transcriptomic Subtypes With Specific Genomic Alterations. J Thoracic Oncol (2019) 14:1651–61. doi: 10.1016/j.jtho.2019.05.003
41. Simbolo M, Mafficini A, Sikora KO, Fassan M, Barbi S, Corbo V, et al. Lung Neuroendocrine Tumours: Deep Sequencing of the Four World Health Organization Histotypes Reveals Chromatin-Remodelling Genes as Major Players and a Prognostic Role for TERT, RB1, MEN1 and KMT2D: Molecular Landscape of Lung Neuroendocrine Tumours. J Pathol (2017) 241:488–500. doi: 10.1002/path.4853
42. Vollbrecht C, Werner R, Walter RFH, Christoph DC, Heukamp LC, Peifer M, et al. Mutational Analysis of Pulmonary Tumours With Neuroendocrine Features Using Targeted Massive Parallel Sequencing: A Comparison of a Neglected Tumour Group. Br J Cancer (2015) 113:1704–11. doi: 10.1038/bjc.2015.397
43. Zhou Z, Zhu L, Niu X, Shen S, Zhao Y, Zhang J, et al. Comparison of Genomic Landscapes of Large Cell Neuroendocrine Carcinoma, Small Cell Lung Carcinoma, and Large Cell Carcinoma. Thorac Cancer (2019) 10:839–47. doi: 10.1111/1759-7714.13011
44. Zhuo M, Guan Y, Yang X, Hong L, Wang Y, Li Z, et al. The Prognostic and Therapeutic Role of Genomic Subtyping by Sequencing Tumor or Cell-Free DNA in Pulmonary Large-Cell Neuroendocrine Carcinoma. Clin Cancer Res (2020) 26:892–901. doi: 10.1158/1078-0432.CCR-19-0556
45. Derks JL, Leblay N, Lantuejoul S, Dingemans A-MC, Speel E-JM, Fernandez-Cuesta L. New Insights Into the Molecular Characteristics of Pulmonary Carcinoids and Large Cell Neuroendocrine Carcinomas, and the Impact on Their Clinical Management. J Thoracic Oncol (2018) 13:752–66. doi: 10.1016/j.jtho.2018.02.002
46. Hiroshima K, Mino-Kenudson M. Update on Large Cell Neuroendocrine Carcinoma. Transl Lung Cancer Res (2017) 6:530–9. doi: 10.21037/tlcr.2017.06.12
47. Swarts DRA, Ramaekers FCS, Speel E-JM. Molecular and Cellular Biology of Neuroendocrine Lung Tumors: Evidence for Separate Biological Entities. Biochim Biophys Acta (BBA) Rev Cancer (2012) 1826:255–71. doi: 10.1016/j.bbcan.2012.05.001
48. George J, Lim JS, Jang SJ, Cun Y, Ozretić L, Kong G, et al. Comprehensive Genomic Profiles of Small Cell Lung Cancer. Nature (2015) 524:47–53. doi: 10.1038/nature14664
49. Peifer M, Fernández-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, et al. Integrative Genome Analyses Identify Key Somatic Driver Mutations of Small-Cell Lung Cancer. Nat Genet (2012) 44:1104–10. doi: 10.1038/ng.2396
50. Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, et al. Somatic Mutations Affect Key Pathways in Lung Adenocarcinoma. Nature (2008) 455:1069–75. doi: 10.1038/nature07423
51. Shaw RJ, Kosmatka M, Bardeesy N, Hurley RL, Witters LA, DePinho RA, et al. The Tumor Suppressor LKB1 Kinase Directly Activates AMP-Activated Kinase and Regulates Apoptosis in Response to Energy Stress. Proc Natl Acad Sci (2004) 101:3329–35. doi: 10.1073/pnas.0308061100
52. Jeon S-M, Chandel NS, Hay N. Ampk Regulates Nadph Homeostasis to Promote Tumour Cell Survival During Energy Stress. Nature (2012) 485:661–5. doi: 10.1038/nature11066
53. Lamming DW. Sabatini DM. A Central Role for Mtor in Lipid Homeostasis. Cell Metab (2013) 18:465–9. doi: 10.1016/j.cmet.2013.08.002
54. Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, et al. The Selective Autophagy Substrate P62 Activates the Stress Responsive Transcription Factor Nrf2 Through Inactivation of Keap1. Nat Cell Biol (2010) 12:213–23. doi: 10.1038/ncb2021
55. Best SA, Sutherland KD. “Keaping” a Lid on Lung Cancer: The Keap1-Nrf2 Pathway. Cell Cycle (2018) 17:1696–707. doi: 10.1080/15384101.2018.1496756
56. Best SA, De Souza DP, Kersbergen A, Policheni AN, Dayalan S, Tull D, et al. Synergy Between the KEAP1/NRF2 and PI3K Pathways Drives non-Small-Cell Lung Cancer With an Altered Immune Microenvironment. Cell Metab (2018) 27:935–43.e4. doi: 10.1016/j.cmet.2018.02.006
57. Romero R, Sayin VI, Davidson SM, Bauer MR, Singh SX, LeBoeuf SE, et al. Keap1 Loss Promotes Kras-Driven Lung Cancer and Results in Dependence on Glutaminolysis. Nat Med (2017) 23:1362–8. doi: 10.1038/nm.4407
58. Lignitto L, LeBoeuf SE, Homer H, Jiang S, Askenazi M, Karakousi TR, et al. Nrf2 Activation Promotes Lung Cancer Metastasis by Inhibiting the Degradation of Bach1. Cell (2019) 178:316–29.e18. doi: 10.1016/j.cell.2019.06.003
59. Galan-Cobo A, Sitthideatphaiboon P, Qu X, Poteete A, Pisegna MA, Tong P, et al. LKB1 and KEAP1/NRF2 Pathways Cooperatively Promote Metabolic Reprogramming With Enhanced Glutamine Dependence in KRAS -Mutant Lung Adenocarcinoma. Cancer Res (2019) 79:3251–67. doi: 10.1158/0008-5472.CAN-18-3527
60. Papillon-Cavanagh S, Doshi P, Dobrin R, Szustakowski J, Walsh AM. STK11 and KEAP1 Mutations as Prognostic Biomarkers in an Observational Real-World Lung Adenocarcinoma Cohort. ESMO Open (2020) 5:e000706. doi: 10.1136/esmoopen-2020-000706
61. Skoulidis F, Goldberg ME, Greenawalt DM, Hellmann MD, Awad MM, Gainor JF, et al. Stk11/Lkb1 Mutations and PD-1 Inhibitor Resistance in KRAS -Mutant Lung Adenocarcinoma. Cancer Discovery (2018) 8:822–35. doi: 10.1158/2159-8290.CD-18-0099
62. Borges M, Linnoila RI, van de Velde HJK, Chen H, Nelkin BD, Mabry M, et al. An Achaete-Scute Homologue Essential for Neuroendocrine Differentiation in the Lung. Nature (1997) 386:852–5. doi: 10.1038/386852a0
63. Morimoto M, Nishinakamura R, Saga Y, Kopan R. Different Assemblies of Notch Receptors Coordinate the Distribution of the Major Bronchial Clara, Ciliated and Neuroendocrine Cells. Development (2012) 139:4365–73. doi: 10.1242/dev.083840
64. Meurette O, Mehlen P. Notch Signaling in the Tumor Microenvironment. Cancer Cell (2018) 34:536–48. doi: 10.1016/j.ccell.2018.07.009
65. Guruharsha KG, Kankel MW, Artavanis-Tsakonas S. The Notch Signalling System: Recent Insights Into the Complexity of a Conserved Pathway. Nat Rev Genet (2012) 13:654–66. doi: 10.1038/nrg3272
66. Vasconcelos FF, Castro DS. Transcriptional Control of Vertebrate Neurogenesis by the Proneural Factor Ascl1. Front Cell Neurosci (2014) 8:412. doi: 10.3389/fncel.2014.00412
67. Henke RM, Meredith DM, Borromeo MD, Savage TK, Johnson JE. Ascl1 and Neurog2 Form Novel Complexes and Regulate Delta-Like3 (Dll3) Expression in the Neural Tube. Dev Biol (2009) 328:529–40. doi: 10.1016/j.ydbio.2009.01.007
68. Katoh M, Katoh M. Precision Medicine for Human Cancers With Notch Signaling Dysregulation (Review). Int J Mol Med (2019) 45:279–97. doi: 10.3892/ijmm.2019.4418
69. Dylla SJ. Toppling High-Grade Pulmonary Neuroendocrine Tumors With a DLL3-Targeted Trojan Horse. Mol Cell Oncol (2016) 3:e1101515. doi: 10.1080/23723556.2015.1101515
70. Gazdar AF, Bunn PA, Minna JD. Small-Cell Lung Cancer: What We Know, What We Need to Know and the Path Forward. Nat Rev Cancer (2017) 17:725–37. doi: 10.1038/nrc.2017.87
71. Malapelle U, Morra F, Ilardi G, Visconti R, Merolla F, Cerrato A, et al. Usp7 Inhibitors, Downregulating CCDC6, Sensitize Lung Neuroendocrine Cancer Cells to PARP-Inhibitor Drugs. Lung Cancer (2017) 107:41–9. doi: 10.1016/j.lungcan.2016.06.015
72. Peng W-X, Shibata T, Katoh H, Kokubu A, Matsuno Y, Asamura H, et al. Array-Based Comparative Genomic Hybridization Analysis of High-Grade Neuroendocrine Tumors of the Lung. Cancer Sci (2005) 96:661–7. doi: 10.1111/j.1349-7006.2005.00092.x
73. Lo Russo G, Pusceddu S, Proto C, Macerelli M, Signorelli D, Vitali M, et al. Treatment of Lung Large Cell Neuroendocrine Carcinoma. Tumor Biol (2016) 37:7047–57. doi: 10.1007/s13277-016-5003-4
74. Rudin CM, Pietanza MC, Bauer TM, Ready N, Morgensztern D, Glisson BS, et al. Rovalpituzumab Tesirine, a DLL3-Targeted Antibody-Drug Conjugate, in Recurrent Small-Cell Lung Cancer: A First-in-Human, First-in-Class, Open-Label, Phase 1 Study. Lancet Oncol (2017) 18:42–51. doi: 10.1016/S1470-2045(16)30565-4
75. Morgensztern D, Besse B, Greillier L, Santana-Davila R, Ready N, Hann CL, et al. Efficacy and Safety of Rovalpituzumab Tesirine in Third-Line and Beyond Patients With Dll3-Expressing, Relapsed/Refractory Small-Cell Lung Cancer: Results From the Phase Ii TRINITY Study. Clin Cancer Res (2019) 25:6958–66. doi: 10.1158/1078-0432.CCR-19-1133
76. Giffin MJ, Cooke K, Lobenhofer EK, Estrada J, Zhan J, Deegen P, et al. AMG 757, a Half-Life Extended, DLL3-Targeted Bispecific T-Cell Engager, Shows High Potency and Sensitivity in Preclinical Models of Small-Cell Lung Cancer. Clin Cancer Res (2020) 1078-0432:CCR-20–2845. doi: 10.1158/1078-0432.CCR-20-2845
77. Owen DH, Giffin MJ, Bailis JM, Smit M-AD, Carbone DP, He K. Dll3: An Emerging Target in Small Cell Lung Cancer. J Hematol Oncol (2019) 12:61. doi: 10.1186/s13045-019-0745-2
78. Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer Immunology. Mutational Landscape Determines Sensitivity to PD-1 Blockade in Non-Small Cell Lung Cancer. Science (2015) 348:124–8. doi: 10.1126/science.aaa1348
79. Cao D, Xu H, Xu X, Guo T, Ge W. High Tumor Mutation Burden Predicts Better Efficacy of Immunotherapy: A Pooled Analysis of 103078 Cancer Patients. OncoImmunology (2019) 8:e1629258. doi: 10.1080/2162402X.2019.1629258
80. Kim HS, Lee JH, Nam SJ, Ock C-Y, Moon J-W, Yoo CW, et al. Association of PD-L1 Expression With Tumor-Infiltrating Immune Cells and Mutation Burden in High-Grade Neuroendocrine Carcinoma of the Lung. J Thoracic Oncol (2018) 13:636–48. doi: 10.1016/j.jtho.2018.01.008
81. Wang VE, Urisman A, Albacker L, Ali S, Miller V, Aggarwal R, et al. Checkpoint Inhibitor is Active Against Large Cell Neuroendocrine Carcinoma With High Tumor Mutation Burden. J Immunother Cancer (2017) 5:75. doi: 10.1186/s40425-017-0281-y
82. Zhang X, Sun Y, Miao Y, Xu S. Immune Checkpoint Inhibitor Therapy Achieved Complete Response for Drug-Sensitive Egfr/Alk Mutation-Negative Metastatic Pulmonary Large-Cell Neuroendocrine Carcinoma With High Tumor Mutation Burden: A Case Report. Onco Targets Ther (2020) 13:8245–50. doi: 10.2147/OTT.S259893
83. Eichhorn F, Harms A, Warth A, Muley T, Winter H, Eichhorn ME. Pd-L1 Expression in Large Cell Neuroendocrine Carcinoma of the Lung. Lung Cancer (2018) 118:76–82. doi: 10.1016/j.lungcan.2018.02.003
84. Hermans BCM, Derks JL, Thunnissen E, van Suylen RJ, den Bakker MA, Groen HJM, et al. Prevalence and Prognostic Value of PD-L1 Expression in Molecular Subtypes of Metastatic Large Cell Neuroendocrine Carcinoma (Lcnec). Lung Cancer (2019) 130:179–86. doi: 10.1016/j.lungcan.2019.02.022
85. Tsuruoka K, Horinouchi H, Goto Y, Kanda S, Fujiwara Y, Nokihara H, et al. Pd-L1 Expression in Neuroendocrine Tumors of the Lung. Lung Cancer (2017) 108:115–20. doi: 10.1016/j.lungcan.2017.03.006
86. Wang Y, Shen YH, Ma S, Zhou J. A Marked Response to Icotinib in a Patient With Large Cell Neuroendocrine Carcinoma Harboring an EGFR Mutation: A Case Report. Oncol Lett (2015) 10:1575–8. doi: 10.3892/ol.2015.3405
87. Aroldi F, Bertocchi P, Meriggi F, Abeni C, Ogliosi C, Rota L, et al. Tyrosine Kinase Inhibitors in EGFR-Mutated Large-Cell Neuroendocrine Carcinoma of the Lung a Case Report. Case Rep Oncol (2014) 7:478–83. doi: 10.1159/000365413
88. De Pas TM, Giovannini M, Manzotti M, Trifirò G, Toffalorio F, Catania C, et al. Large-Cell Neuroendocrine Carcinoma of the Lung Harboring EGFR Mutation and Responding to Gefitinib. JCO (2011) 29:e819–22. doi: 10.1200/JCO.2011.36.2251
89. Zheng Q, Zheng M, Jin Y, Shen X, Shan L, Shen L, et al. Alk-Rearrangement Neuroendocrine Carcinoma of the Lung: A Comprehensive Study of a Rare Case Series and Review of Literature. OTT (2018) Volume 11:4991–8. doi: 10.2147/OTT.S172124
90. Omachi N, Shimizu S, Kawaguchi T, Tezuka K, Kanazu M, Tamiya A, et al. Atagi S. A Case of Large-Cell Neuroendocrine Carcinoma Harboring an EML4–ALK Rearrangement With Resistance to the ALK Inhibitor Crizotinib. J Thoracic Oncol (2014) 9:e40–2. doi: 10.1097/JTO.0000000000000103
91. Wang S, Wu X, Zhao J, Chen H, Zhang Z, Wang M, et al. Next-Generation Sequencing Identified a Novel Crizotinib-Sensitive PLB1-ALK Rearrangement in Lung Large-Cell Neuroendocrine Carcinoma. Clin Lung Cancer [Preprint] (2020). Available at: https://www.clinical-lung-cancer.com/article/S1525-7304(20)30166-2/fulltext.
92. Yao JC, Phan AT, Chang DZ, Wolff RA, Hess K, Gupta S, et al. Efficacy of RAD001 (Everolimus) and Octreotide LAR in Advanced Low- to Intermediate-Grade Neuroendocrine Tumors: Results of a Phase Ii Study. JCO (2008) 26:4311–8. doi: 10.1200/JCO.2008.16.7858
93. Christopoulos P, Engel-Riedel W, Grohé C, Kropf-Sanchen C, von Pawel J, Gütz S, et al. Everolimus With Paclitaxel and Carboplatin as First-Line Treatment for Metastatic Large-Cell Neuroendocrine Lung Carcinoma: A Multicenter Phase Ii Trial. Ann Oncol (2017) 28:1898–902. doi: 10.1093/annonc/mdx268
94. Owonikoko TK, Dahlberg SE, Sica GL, Wagner LI, Wade JL, Srkalovic G, et al. Randomized Phase Ii Trial of Cisplatin and Etoposide in Combination With Veliparib or Placebo for Extensive-Stage Small-Cell Lung Cancer: ECOG-ACRIN 2511 Study. JCO (2019) 37:222–9. doi: 10.1200/JCO.18.00264
95. Morra F, Luise C, Visconti R, Staibano S, Merolla F, Ilardi G, et al. New Therapeutic Perspectives in CCDC6 Deficient Lung Cancer Cells: CCDC6 as Predictive Biomarker of Drug Resistance in NSCLC. Int J Cancer (2015) 136:2146–57. doi: 10.1002/ijc.29263
96. Morra F, Luise C, Merolla F, Poser I, Visconti R, Ilardi G, et al. FBXW7 and USP7 Regulate CCDC6 Turnover During the Cell Cycle and Affect Cancer Drugs Susceptibility in NSCLC. Oncotarget (2015) 6:12697–709. doi: 10.18632/oncotarget.3708
97. Kondo Y. Targeting Histone Methyltransferase EZH2 as Cancer Treatment. J Biochem (2014) 156:249–57. doi: 10.1093/jb/mvu054
98. Coe BP, Thu KL, Aviel-Ronen S, Vucic EA, Gazdar AF, Lam S, et al. Genomic Deregulation of the E2F/Rb Pathway Leads to Activation of the Oncogene EZH2 in Small Cell Lung Cancer. PloS One (2013) 8:e71670. doi: 10.1371/journal.pone.0071670
99. Poirier JT, Gardner EE, Connis N, Moreira AL, de Stanchina E, Hann CL, et al. Dna Methylation in Small Cell Lung Cancer Defines Distinct Disease Subtypes and Correlates With High Expression of EZH2. Oncogene (2015) 34:5869–78. doi: 10.1038/onc.2015.38
100. Findeis-Hosey JJ, Huang J, Li F, Yang Q, McMahon LA, Xu H. High-Grade Neuroendocrine Carcinomas of the Lung Highly Express Enhancer of Zeste Homolog 2, But Carcinoids Do Not. Hum Pathol (2011) 42:867–72. doi: 10.1016/j.humpath.2010.09.019
101. Saito M, Saito K, Shiraishi K, Maeda D, Suzuki H, Minamiya Y, et al. Identification of Candidate Responders for Anti-PD-L1/PD-1 Immunotherapy, Rova-T Therapy, or EZH2 Inhibitory Therapy in Small-Cell Lung Cancer. Mol Clin onc (2017) 8:310–4. doi: 10.3892/mco.2017.1536
102. Travis WD MD. Pathology & Genetics of Tumours of the Lung, Thymus and Heart. 1st ed. Lyon: Intl Agency for Research on Cancer (2004).
103. Saunders LR, Bankovich AJ, Anderson WC, Aujay MA, Bheddah S, Black K, et al. A DLL3-Targeted Antibody-Drug Conjugate Eradicates High-Grade Pulmonary Neuroendocrine Tumor-Initiating Cells in Vivo. Sci Transl Med (2015) 7:302ra136–302ra136. doi: 10.1126/scitranslmed.aac9459
104. Brcic L, Kuchler C, Eidenhammer S, Pabst D, Quehenberger F, Gazdar AF, et al. Comparison of Four Dll3 Antibodies Performance in High Grade Neuroendocrine Lung Tumor Samples and Cell Cultures. Diagn Pathol (2019) 14:47. doi: 10.1186/s13000-019-0827-z
105. Alcala N, Leblay N, Gabriel AAG, Mangiante L, Hervas D, Giffon T, et al. Integrative and Comparative Genomic Analyses Identify Clinically Relevant Pulmonary Carcinoid Groups and Unveil the Supra-Carcinoids. Nat Commun (2019) 10:3407. doi: 10.1038/s41467-019-11276-9
Keywords: large cell neuroendocrine carcinoma, lung, DLL3, ASCL1, RB1, TP53
Citation: Yoshimura M, Seki K, Bychkov A and Fukuoka J (2021) Molecular Pathology of Pulmonary Large Cell Neuroendocrine Carcinoma: Novel Concepts and Treatments. Front. Oncol. 11:671799. doi: 10.3389/fonc.2021.671799
Received: 24 February 2021; Accepted: 31 March 2021;
Published: 22 April 2021.
Edited by:
Takefumi Komiya, Parkview Health, United StatesReviewed by:
Janaki Deepak, University of Maryland, Baltimore, United StatesCopyright © 2021 Yoshimura, Seki, Bychkov and Fukuoka. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junya Fukuoka, ZnVrdW9rYWpAbmFnYXNha2ktdS5hYy5qcA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.