- Key Laboratory of Gastroenterology and Hepatology, Ministry of Health, Division of Gastroenterology and Hepatology, Shanghai Institute of Digestive Disease, Renji Hospital, School of Medicine, Shanghai Jiaotong University, Shanghai, China
Objective: Up to now, non-invasive diagnosis of laterally spreading tumor (LST) and prediction of adenoma recurrence after endoscopic resection of LSTs is inevitable. This study aimed to identify a microbial signature with clinical significance of diagnosing LSTs and predicting adenoma recurrence after LSTs colectomy.
Methods: We performed 16S rRNA sequencing in 24 mucosal samples, including 5 healthy controls (HC), 8 colorectal adenoma (CRA) patients, and 11 LST patients. The differentiating microbiota in fecal samples was quantified by qPCR in 475 cases with 113 HC, 208 CRA patients, 109 LST patients, and 45 colorectal cancer (CRC) patients. We identified differentially abundant taxa among cases and controls using linear discriminant analysis effect size analysis. ROC curve was used to evaluate diagnostic values of the bacterial candidates. Pairwise comparison of AUCs was performed by using the Delong’s test. The Mantel-Haenszel hazard models were performed to determine the effects of microbial compositions on recurrence free survival.
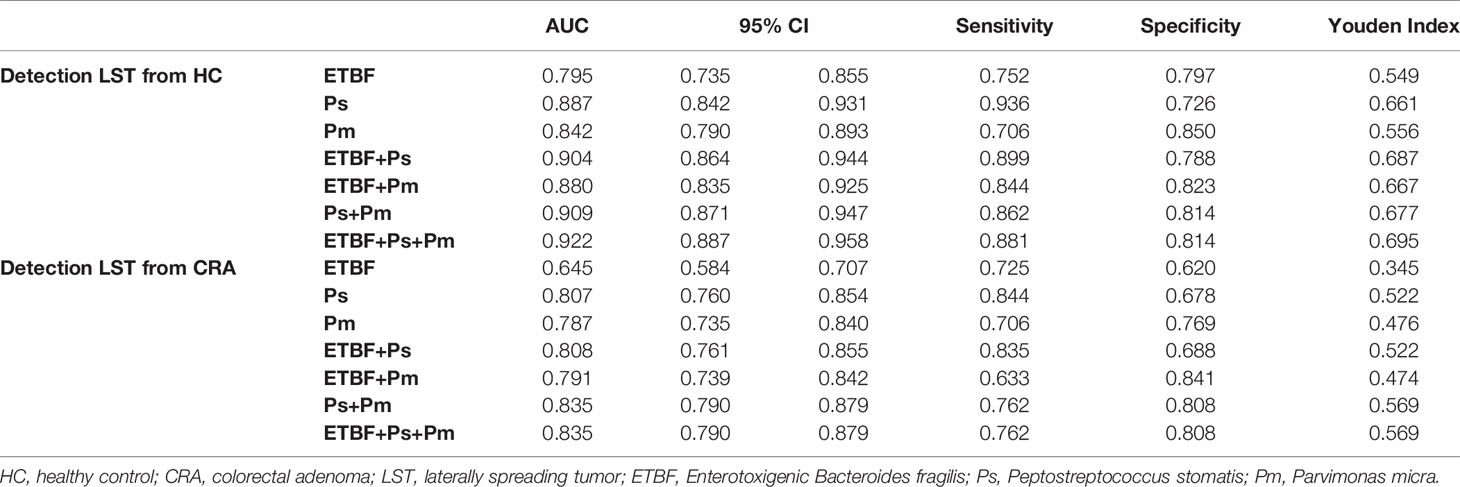
Results: The microbial dysbiosis of LST was characterized by relative high abundance of the genus Lactobacillus-Streptococcus and the species enterotoxigenic Bacteroides fragilis (ETBF)–Peptostreptococcus stomatis (P. stomatis)–Parvimonas micra (P. micra). The abundance of ETBF, P. stomatis, and P. micra were steadily increasing in LST and CRC groups. P. stomatis behaved stronger value on diagnosing LST than the other two bacteria (AUC 0.887, 95% CI 0.842–0.931). The combination of P. stomatis, P. micra, and ETBF (AUC 0.922, 95% CI 0.887–0.958) revealed strongest diagnostic power with 88.7% sensitivity and 81.4% specificity. ETBF, P. stomatis, and P. micra were associated with malignant LST (PP.stomatis = 0.0015, PP.micra = 0.0255, PETBF = 0.0169) and the abundance of IL-6. The high abundance of P. stomatis was related to the adenoma recurrence after LST resection (HR = 3.88, P = 0.008).
Conclusions: Fecal microbiome signature (ETBF–P. stomatis–P. micra) can diagnose LSTs with high accuracy. ETBF, P. stomatis, and P. micra were related to malignant LST and P. stomatis exhibited high predictive value on the adenoma recurrence after resection of LSTs. The fecal microbiome signature of LST may provide a noninvasive alternative to early detect LST and predict the adenoma recurrence risk after resections of LSTs.
Introduction
Globally, colorectal cancers (CRC) ranked second in mortality and third in incidence among cancers in 2020 (1). Nearly 90% of colorectal cancers develop in the adenoma-carcinoma sequence over 10 years (2, 3). Adenomas are crucial precursor lesions of CRC. To avoid the carcinogenesis progress, adenomas can be removed by colonoscopic resection. Laterally spreading tumors (LSTs) are well known as primary precursor lesions of CRC (4, 5). LSTs are featured by horizontally extending growth patterns and are at least 1cm in diameter (6). It is classified into two types: LST-Gs with granules or nodules on surfaces and LST-NGs featured with smooth surfaces without nodules or granules (6). LST-Gs are further categorized into nodular mixed (LST-G-M) and homogeneous (LST-G-H) subtypes. LST-NGs include pseudo-depressed (LST-NG-PD) and flat-elevated (LST-NG-FE) subtypes. The detection of LST is extremely difficult due to the special morphology and growth pattern. Thus, there is a need to sensitively detect LSTs to prevent the incidence of CRC. Moreover, adenoma recurrence after endoscopic resection of LSTs is frequent (7, 8). Despite the widespread use of endoscopic resection, few studies examined the risk factors for adenoma recurrence after LST resection. Many LST patients without adenoma recurrence were subjected to ultimately and frequently redundant surveillance procedures. Hence, a non-invasive, economic and convenient marker that can sensitively detect LSTs and predict the adenoma recurrence of LSTs yields to be explored.
The colorectal epithelium has a constant crosstalk with approximately 3 × 1013 gut microorganisms (9). In the past 5 years, the roles of gut microbiota in the carcinogenesis of CRC have received lots of attention. Human studies generally showed that the gut microbiota related to CRC patients was significantly different compared with healthy individuals. It showed higher species richness and increased abundant procarcinogenic taxa (such as Fusobacterium, Bacteroides, Porphyromonas, Escherichia) and lower abundance of potentially protective taxa (10–12). And these studies pinpoint a potential core series of carcinogenic microorganisms and provide scientific resource for developing fecal microbial indicators for the diagnosis of CRC. Existing studies have applied the abundance of multiple fungal, bacterial, viral species to differentiate CRC patients from HC and these noninvasive biomarkers exerted high sensitivity and specificity (12–17). Apart from microbial diagnostic applications, relations concerning clinical outcomes of CRC and bacterial biomarkers have raised the probability of prognostic bacterial markers. Numerous studies have disclosed the associations between CRC survival and tumoral bacterial abundance (18–20). The microbiota may be an effective, noninvasive, economic, and convenient biomarker for prognosis of CRC.
LST, an important precursor of CRC, was hardly recognized and resected for flat morphology during endoscopy. Exploring the microbiome signature of LSTs may inspire new approaches to identify or slow LSTs progressions. Current noninvasive screening tests including tumor markers and fecal occult blood test have low sensitivities for detecting LST. While microbiome signature for LSTs and noninvasive stool-based screening tests remain rare-reported. For widely screen and surveillance of LSTs, cost-effective, non-invasive biomarkers are urgently required, which are apparently beneficial to patients’ compliance.
In this study, we identified a microbiome signature with clinical significance to detect LSTs from healthy controls (HC) and colorectal adenoma (CRA). And we identified the microbiota with high accuracy on predicting the adenoma recurrence after colonoscopic resection of LSTs.
Materials and Methods
Study Design and Participants
This retrospective cohort study was carried out at Renji Hospital, School of Medicine, Shanghai Jiaotong University and approval for the study was obtained from the ethics committee of Renji Hospital, School of Medicine, Shanghai Jiaotong University. The study complied with the Helsinki Declaration of 1975. A total number of 475 patients was consisted with 113 HCs, 208 CRA patients, 109 LST patients, and 45 CRC patients (work flowchart see Figure 1). Excluding criteria were established to prevent potential gut microbial alternation. The exclusion criteria included: 1) with the upper gastrointestinal (GI) tracts surgery history; 2) with the history of hereditary non-polyposis CRC (Lynch syndrome), familial adenomatous polyposis (FAP), or Peutz-Jeghers syndrome or uncontrolled chronic metabolic disorder, including diabetes and hypertension; 3)with uninterested GI tract neoplasia history; 4) active GI tracts bleeding in recent six months; 5) using probiotics, antibiotics, immunosuppressor, nonsteroidal anti-inflammatory drugs at least 1 month before enrollment; 6)with eating habits changes in recent 1 month (17).
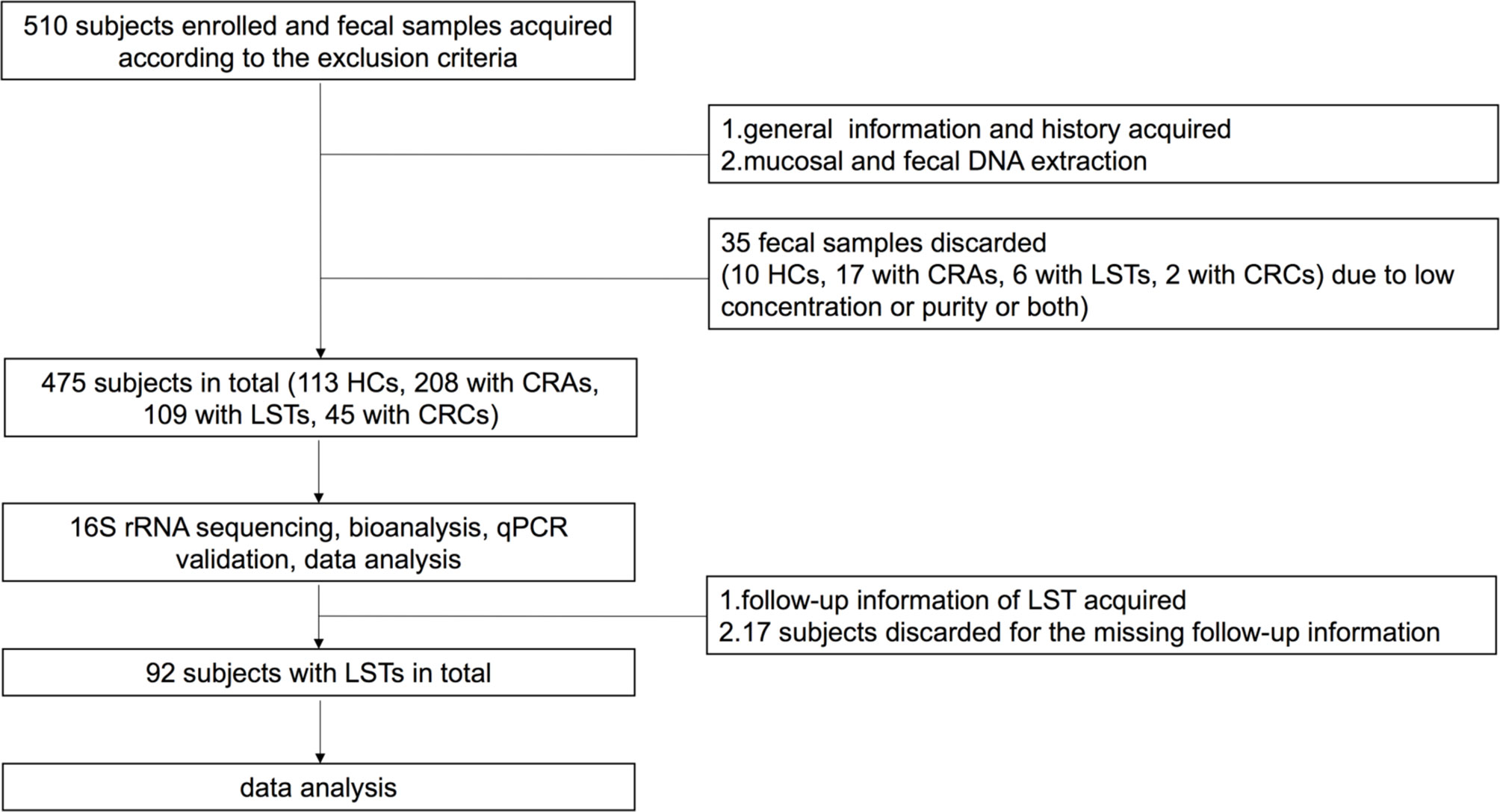
Figure 1 Work flowcharts. HC, healthy control; CRA, colorectal adenoma; LST, laterally spreading tumor; CRC, colorectal carcinoma; qPCR, quantitative polymerase chain reaction.
Enrolled subjects were divided into four groups: HC group, CRA group, LST group and CRC group and the clinical phenotype was defined by pathological and endoscopic diagnosis. Distal tumors were considered to be those in descending colon, sigmoid colon and rectum, while proximal tumors included those in the rest of colon. Conventional-type CRA include villous adenoma, tubular adenoma, tubulovillous adenoma. Serrated polyps include hyperplastic polyps and serrated adenoma. The early stage CRC (S0) as well as stage 0/pTis CRC was defined as adenoma with high-grade dysplasia and intramucosal carcinoma. Adenoma recurrence was endoscopically confirmed under surveillance in 3 to 36 months and was defined as detection of recurrent adenomas in patients without the recurrence evidence during any previous surveillance period. Late adenoma recurrence was described as recurrent adenoma at the site of previous resection at ≥12 months’ surveillance.
Stool and Mucosal Samples Collection
Before bowel preparation for endoscopy, all 475 stool samples (113 HC, 208 CRA patients, 109 LST patients, 45 CRC patients) were collected in the germ-free containments. All stool samples were temporarily preserved in −20°C and transferred to −80°C for storage within 48 h. All 24 mucosal samples (5 HC, 8 CRA patients, 11 LST patients) were collected in the special germ-free containment after endoscopic resection and were immediately moved to −80°C for storage.
DNA Extraction, 16S rRNA Gene Sequencing, and Quantitative Polymerase Chain Reaction
QIAamp DNA Tissue Mini Kit and QIAamp DNA Stool Mini Kit were used according to the instructions (Qiagen, Hilden, Germany). All extracts were preserved at −80°C before 16S rRNA sequencing and subsequent polymerase chain reaction (qPCR). We performed 16S rRNA sequencing in mucosal samples. The 16S rRNA V3-V4 region was sequenced on the Miseq platform (Illumia, San Diego, California, USA). We verify the potential microbiota by qPCR according to the sequencing results. The relative primers sequences were described in Supplementary Table 1. All SYBR-green based qPCR samples were in triplicates and average Ct value was calculated. The relative abundance of the target gut microbiota was based on the ΔCt value defined as the target Ct value subtracted Ct value for 16s rRNA (21, 22). 10 μl SYBR Green II was the qPCR reaction system using SYBR®Premix Ex TaqTMII (TliRNaseH Plus) of TAKARA cooperation. Stepone®plus by ABI company was used in all operations and configurations in 40 cycles of 95°C denaturation for 5 s, 60°C annealing and 30 s extension after pre-denaturation at 95°C for 30 s.
Statistical Analysis
The 16S rRNA sequencing statistics were analyzed by Quantitative Insights Into Microbial Ecology (QIIME2 V.2018.2). Basic bioinformatics analysis included reads splicing quality control and OTU clustering analysis. Reads splicing quality control is the original sequencing data using QIIME to filter the sequence quality according to the following criteria: 1) The two-terminal sequences obtained by MiSeq sequencing were spliced into one sequence. 2) The minimum overlap length is 10 bp and the maximum mismatch ratio allowed in the overlap region is 0.2. 3) Barcode, the sequence used to distinguish different samples, required the exact match. The maximum number of mismatches for the primer is 2. OTU was applied according to the similarity of the sequence. The specific OTU clustering steps were as following: 1) extracting the non-repetitive sequence in the optimized sequence; 2) performing OTU clustering under the criterion of 97% similarity. The OTU information have been uploaded on. We used the Bayesian algorithm to classify the OTU representative sequence in the domain, kingdom, phylum, class, order, genus, and species levels referring to the Silva database (http://www.arb-silva.de). To investigate whether the sample size of discovery cohort was sufficient, we performed Core curve analysis and Rarefaction curve analysis. With the sample size increasing, the Core curve tended to be horizontal, which indicated the number of core species did not change significantly. In Rarefaction curve analysis, as the number of sequencing samples increasing, the Shannon diversity indices tended to be flat with increasing number of sequencing samples. Rarefaction curve analysis indicated that the sequencing data were reasonable as well. The Core curve analysis and Rarefaction curve of mucosal samples indicated that our sample size was suitable for 16S rRNA sequencing (Supplementary Figure 1). Then we performed the entero-typing analysis on family, genus, and species levels and performed the distance heatmap, heatmap, cicros, and tertiary analysis to find the differentially taxa among groups. We identified differentially abundant species among cases and controls using linear discriminant analysis effect size (LEfSe) analysis. To explore the relationship between the targeted bacteria and other bacteria, we use the GMrepo bacteria database (https://gmrepo.humangut.info). The other statistical analysis involved Kruskal-Wallis test, Fisher’s exact test and partial Spearman’s rank correlation (PResiduals package).
All qPCR samples were in triplicates. All the relative abundance was normalized by the abundance of 16S rRNA. Average Ct value was calculated from the triplicates. The ΔCt value was defined as the target microbioma Ct value subtracted Ct value for 16S rRNA and the relative abundance of the target microbiota was based on ΔCt (21). Kruskal-Wallis test and Mann-Whitney U test were respectively used in comparison for nonparametric and continuous analysis. Receiver operating characteristic (ROC) curve evaluated the diagnostic value of bacterial candidates in differentiating LST from HC and CRA. And the ROC curve was used to establish the cut-off value maximizing the Youden index (J = Sensitivity + Specificity − 1). The Delong’s test was performed to pairwisely compare areas under ROC (AUCs). Recurrence-free survival (RFS) was described as the time from diagnosis to the colorectal adenoma recurrence in 36 months. Patients without adenoma recurrence were censored at date of last surveillance. Mantel-Haenszel hazard models determined the effects of microbial composition on RFS. All tests were done by SPSS statistical software (version 23.0, IBM Corp, Armonk, New York, 2012) and R statistical software (version 3.6.0, 2019).
Results
Bacteria Composition Was Significantly Different Between LSTs and Controls
A total number of 475 patients were divided into 113 HCs, 208 CRA patients, 109 LST patients, and 45 CRC patients (basic demographic features see Supplementary Table 2; Work flow chart see Figure 1). To examine the bacteria composition in LSTs, the 16S rRNA sequencing was performed in 24 mucosal samples (5 HCs, 8 CRA patients, 11 LST patients). Bacteria compositions in HCs, CRA patients, and LST patients were compared. In entero-typing analysis, it was found that bacteria were significantly different in family, genus, and species levels among the above three groups (Figure 2A), as well as in the distance heatmap analysis on genus level (Figure 2B). Lactobacsillus and Streptococcus were abundant on genus level in LST group compared to the other two groups (Figures 2C–E, Supplementary Figure 2). The potential differential bacterium patterns among the three groups was defined by Kruskal-Wallis test and LEfSe algorithm. Lactobacillus johnsonil (L. johnsonil) and Bacteroide fragilis were significantly abundant in LST group (Figures 3A, B). By analyzing the GMrepo bacteria database, 19 bacteria co-occurred with Bacteroide fragilis in colorectal neoplasms. We found only two bacteria showed the positive correction coefficient in both Pearson’s and Spearman’s analyses (Supplementary Table 3). The abundance of Peptostreptococcus stomatis (P. stomatis) and Parvimonas micra (P. micra) were found to be positively relevant to the abundance of Bacteroide fragilis (Figure 3C). Moreover, enterotoxigenic Bacteroides fragilis (ETBF) was reported to be driver bacteria in colorectal cancer development (23–27). Thus, we focused on verifying the abundance of ETBF, P. stomatis, P. micra, and L. johnsonil in cases.
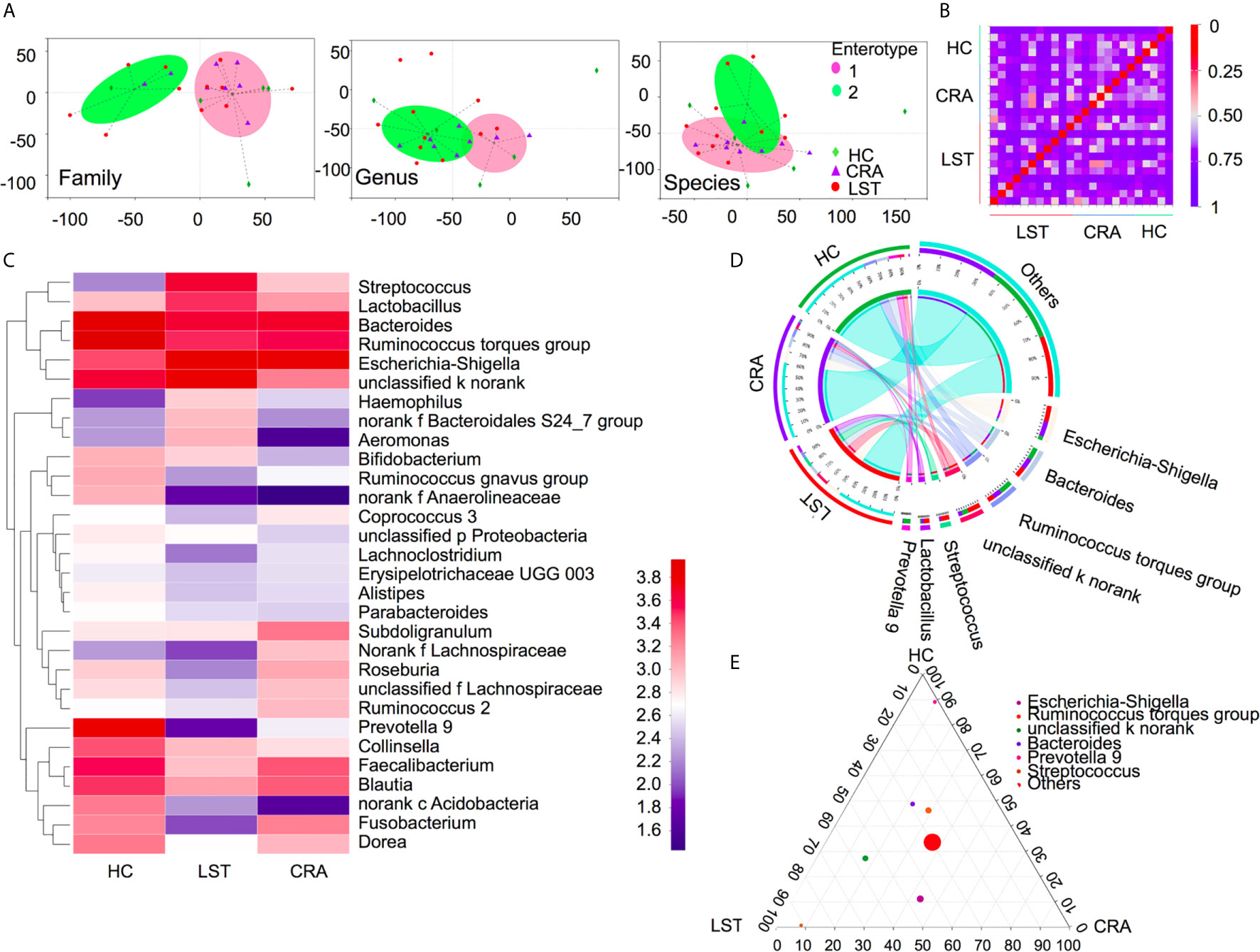
Figure 2 Variations of mucosal microbiota composition in LST. (A) Entero-typing analysis on family, genus and species groups in the HC (n = 5), CRA (n = 8), and LST group (n = 11). The green diamond represented the HC group. The purple triangle represented the CRA group and the red dot represented the LST group. The pink dot represented the enterotype 1 while the green dot represented the enterotype 2. (B) Distance heatmap in samples on genus level. (C) Heatmap in the three groups on genus level. (D) Cicros analysis on genus in three groups. (E) Tertiary analysis on genus level in three groups.
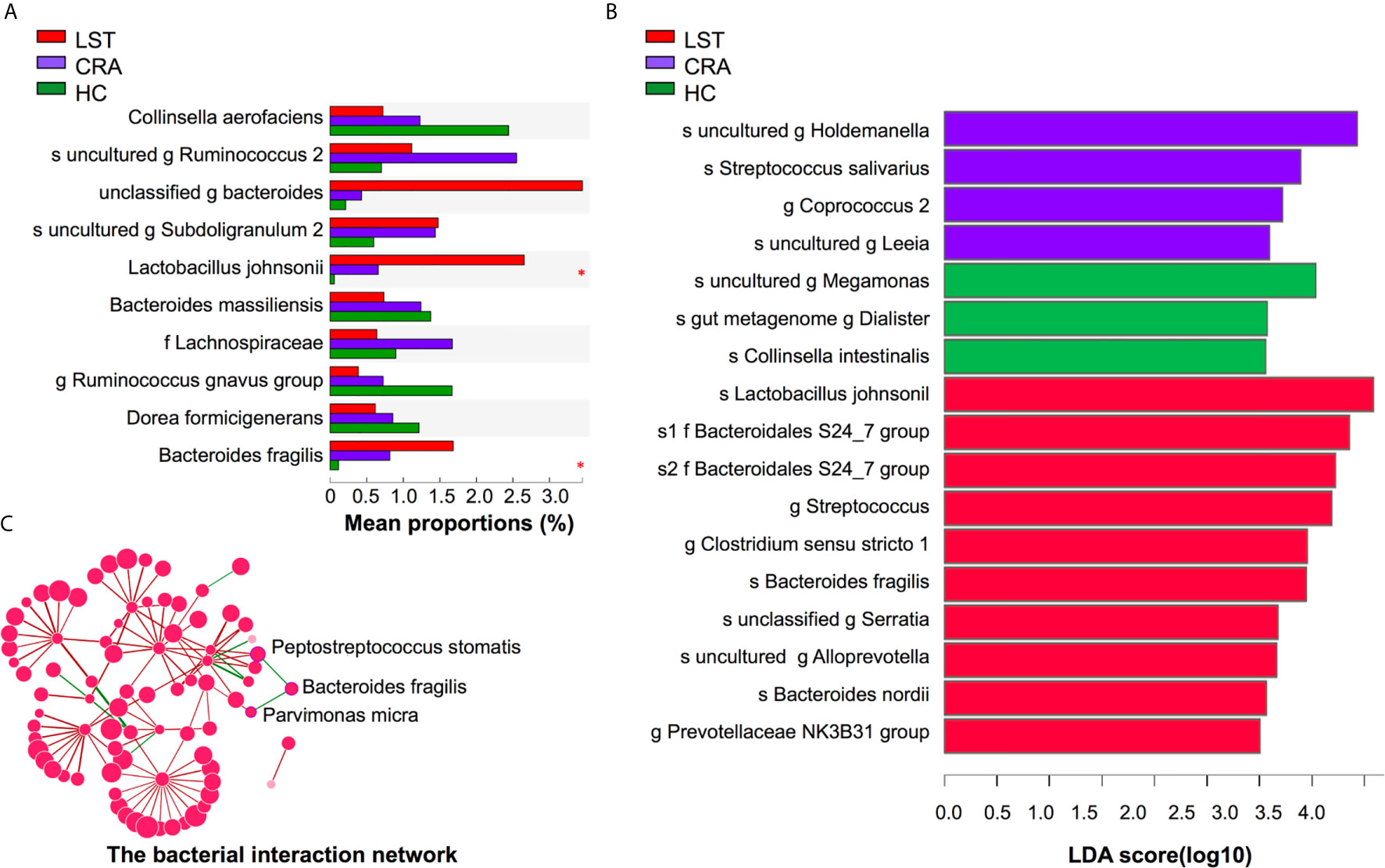
Figure 3 Variations of mucosal microbiota composition on species level in LST. (A) Kruskal-Wallis test analysis on species level in the three groups. The X axis represented the mean proportions (%) of species in three groups. Lactobacillus johnsonii and Bacteroides fragilis presented high abundance in the LST group. (B) LDA analysis on species level in the three groups. The X axis represented LDA score of species in three groups. Lactobacillus johnsonii and Bacteroides fragilis significantly over-presented in the LST group. (C) The interaction network among Bacteroides fragilis and other bacteria. The abundance of Bacteroides fragilis was positively related with the abundance of Peptostreptococcus stomatis and Parvimonas micra. LDA, linear discriminant analysis; HC, healthy control; CRA, colorectal adenoma; LST, laterally spreading tumor. *P < 0.05.
The Abundance of ETBF, P. stomatis, and P. micra were Increased in LST and CRC Groups
ETBF, P. stomatis, P. micra, and Lactobacillus johnsonil were also tested in fecal samples. Increased abundance of ETBF, P. stomatis, and P. micra in LST patients compared with HCs and CRA patients was observed. Moreover, there was an increase of the above microbiota in CRC patients over LST patients, as well (Figure 4A). Comparing LST patients with HCs, ETBF increased by 9.26 folds (P < 0.0001), P. stomatis increased by 14.79 folds (P < 0.0001), and P. micra increased by 10.17 folds (P < 0.0001). Comparing CRA patients with LST patients, ETBF increased by 2.95 folds (P < 0.0001), P. stomatis increased by 8.76 folds (P < 0.0001), and P. micra increased by 6.65 folds (P < 0.0001). Comparing LST patients with CRC patients, ETBF increased by 3.00 folds (P = 0.0327), P. stomatis increased by 15.31 folds (P = 0.0058), and P. micra increased by 6.41 folds (P = 0.0413) in CRC patients group. Further, no discrepancy was found for the relative abundance of three intestinal bacteria among distal and proximal colon for LSTs (Supplementary Figure 3A). In terms of topography and histopathology, no distinction was found between LST-G and LST-NG, as well as traditional adenoma type and serrated polyp type (Supplementary Figures 3B, C). In addition, the dysplasia degree of adenoma led to totally different prognosis and clinical decisions. Accordingly, the three bacteria in terms of dysplasia degree reported by pathologist were tested. The S0 LST had higher abundance of fecal bacteria (PP.stomatis = 0.0015, PP.micra= 0.0255, PETBF =0.0169) (Figure 4B). To further explore the relationship between inflammatory cytokines and the above three bacteria, we collected the clinical laboratory test results of TNF-α, IL-1β, IL-6, IL-8, IL-10 of 35 LST patients (Supplementary Table 4). The results showed that P. stomatis, P. micra, and ETBF significantly increased IL-6 secretion (Figure 4C) while the secretion of TNF-α, IL-1β, IL-8, IL-10 showed no significant relation with the above three bacteria (Supplementary Figure 4). Moreover, the abundance of P. stomatis (PP.stomatis = 0.0024) and P. micra (PP.micra =0.0077) positively correlated with the abundance of ETBF (Figure 4D).
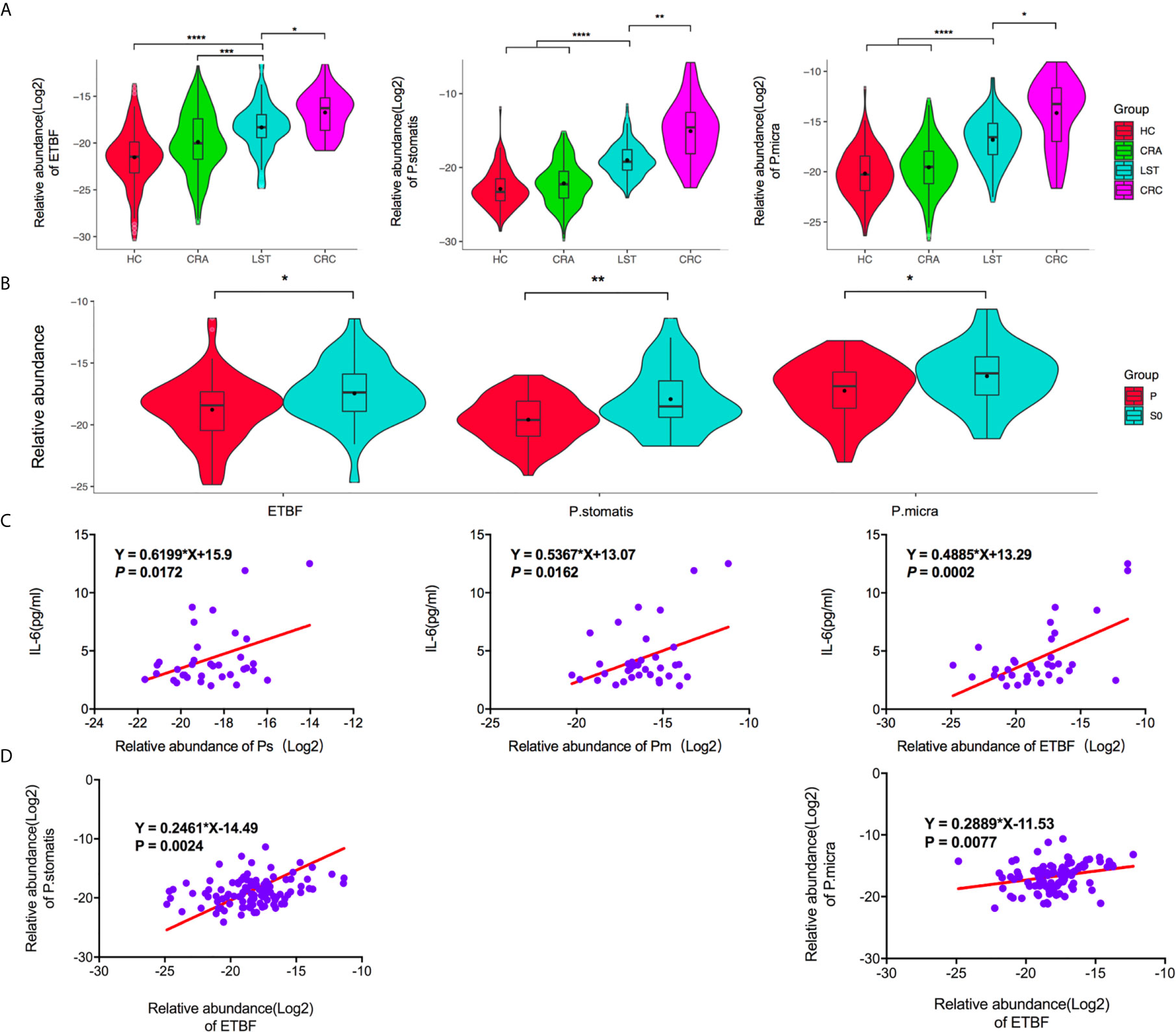
Figure 4 The fecal relative abundance of ETBF, P. stomatis, and P. micra among groups. (A) The fecal relative abundance of ETBF, P. stomatis, and P. micra was stepwise increased in the three groups. (B) The fecal high abundance of ETBF, P. stomatis and P. micra was relevant with S0 stage LSTs. (C) The expression of IL-6 was positively related with the abundance of the three bacteria. (D) The abundance of P. stomatis and P. micra increased with the increasing abundance of ETBF. HC, healthy control; CRA, colorectal adenoma; LST, laterally spreading tumor; CRC, colorectal carcinoma; ETBF, Enterotoxigenic Bacteroides fragilis; P. stomatis, Peptostreptococcus stomatis; P. micra, Parvimonas micra; P, LSTs with hyperplastic type or low-grade dysplasia; S0, LSTs with intramucosal carcinoma or high-grade dysplasia. *P < 0.05. **P < 0.01. ***P < 0.001. ****P < 0.0001.
As for Lactobacillus johnsonil, no significant changes have been revealed among CRA and LST groups (Supplementary Figure 5). Therefore, ETBF, P. stomatis, and P. micra were selected as non-invasive candidate biomarkers for detecting LSTs.
The Predictive Values of ETBF, P. stomatis, and P. micra for the Occurrence of LSTs
The relative abundance of the ETBF–P. stomatis–P. micra microbiome showed an increasing in the LST group as compared to HC group and CRA group as described above (Figure 4A). In comparison between HC group and LST group, the ROC curves were performed. In terms of AUCs, P. stomatis behaved stronger than the other two bacteria (AUCP.stomatis0.887, 95% CI 0.842–0.931) (Figure 5A). These three contributing markers were therefore united as an independent model. Combination of P. stomatis–P. micra and P. stomatis–P. micra-ETBF revealed stronger diagnostic potential over the single P. stomatis (PP.stomatis–P.micra = 0.0376, PP.stomatis–P.micra-ETBF=0.00930)(Figure 5B). The combination of the three bacteria showed the maximum AUC (AUC 0.922, 95% CI 0.887–0.958) and Youden index with 88.7% sensitivity and 81.4% specificity (Table 1).
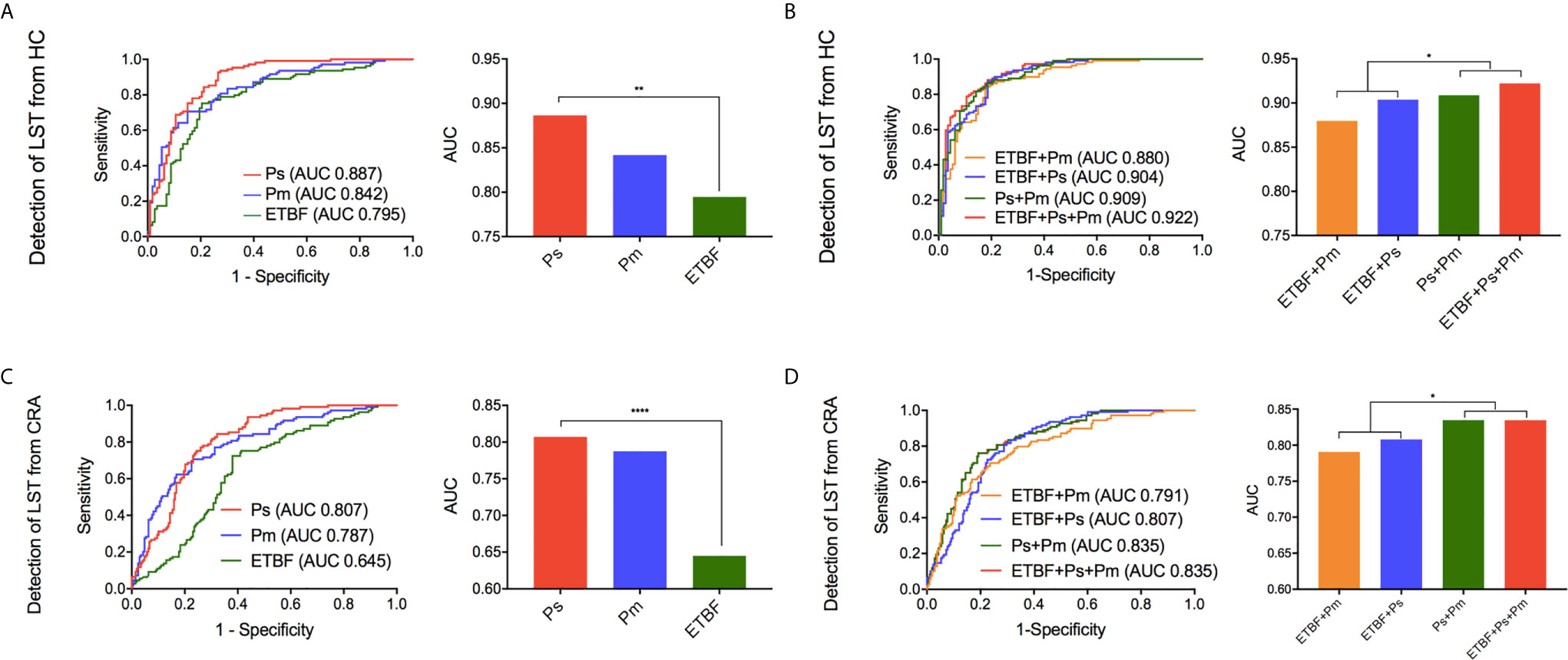
Figure 5 The diagnostic performance of marker ETBF, P. stomatis, P. micra, and combined test by ROC curve analysis. (A) Differentiating LST from HC, the ROC curves and AUCs (Delong’s test) of the three bacteria. (B) Differentiating LST from HC, the ROC curves and the AUCs (Delong’s test) of the promising diagnostic bacterial models. (C) Differentiating LST from CRA, the ROC curves and AUCs (Delong’s test) of the three bacteria. (D) Differentiating LST from CRA, the ROC curves and the AUCs (Delong’s test) of the promising diagnostic bacterial models. HC, healthy control; LST, laterally spreading tumor; ETBF, Enterotoxigenic Bacteroides fragilis; Ps, Peptostreptococcus stomatis; Pm, Parvimonas micra; AUC, area under the receiver-operating characteristic curve. *P < 0.05, **P < 0.01, ****P < 0.0001.
To distinguish LSTs from CRAs, the ROC curves were made. The result showed that P. stomatis exhibited higher AUCs than the other two bacteria (AUCP.stomatis0.807, 95% CI 0.760–0.854)(Figure 5C). Nonetheless, the diagnostic combination of P. stomatis–P.micra and P. stomatis–P. micra-ETBF showed higher AUCs than the single P. stomatis (PP.stomatis–P.micra = 0.0411, PP.stomatis–P.micra-ETBF = 0.0416) (Figure 5D). In terms of Youden index, the P. stomatis–P. micra marker was equal to the P. stomatis–P. micra–ETBF marker with 76.2% sensitivity and 80.8% specificity (Table 1).
P. stomatis Were Positively Related to Adenoma Recurrence After Resection of LSTs
Considering the relationship between signature bacteria and tumoral development of LST, the next step of this study aims to find if there were differentiations in the above three bacteria composition between recurrence or non-recurrence adenoma after resection of LSTs. After 36 months’ follow-ups, there were 23 LST patients with adenoma recurrence among 92 LST patients. To this end, the distribution of ETBF, P. stomatis, and P. micra among the recurrence group and non-recurrence group was first evaluated. The recurrence group exhibited a predominant abundance of P. stomatis (P < 0.0001). Besides, no difference of the other two bacteria between the two groups were detected (Figure 6A) and P. stomatis presented strong value on predicting the adenoma recurrence (AUC 0.800, 95% CI 0.678–0.915, P < 0.0001). Based on these results, the enrolled patients were divided into high abundance of P. stomatis group and low abundance of P. stomatis group. Univariate analysis showed the dysplasia type of LST and the abundance of P. stomatis were related to the adenoma recurrence, while gender, age, history of hypertension or diabetes mellitus, diameter, location, topography, histopathology futiled (Supplementary Table 5). The survival curve showed the S0 LSTs (HR = 2.78, P = 0.026) and high-abundance of P. stomatis (HR = 3.88, P = 0.008) were related to the adenoma recurrence event (Figure 6B). Furthermore, the abundance difference of P. stomatis in early-recurrence group and late-recurrence group was also tested to be no significant difference between the above two groups (Supplementary Figure 6).
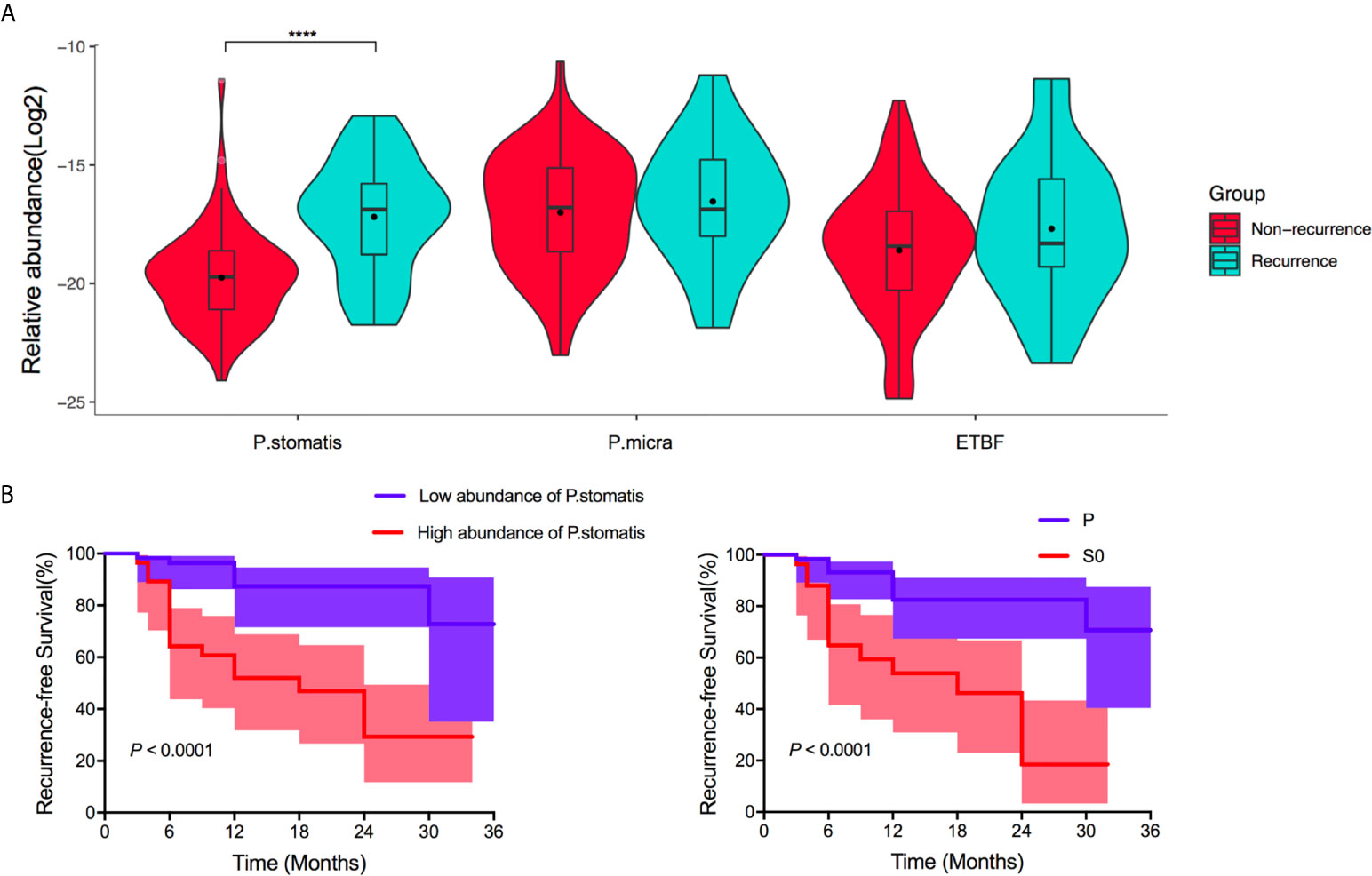
Figure 6 Performance of ETBF, P. stomatis, and P. micra on adenoma recurrence after resection of LST. (A) The fecal relative abundance of P. stomatis, P. micra, and ETBF among non-recurrence group and recurrence group. P. stomatis was related with adenoma recurrence. (B) Kaplan-Meier estimates for survival probability based on the abundance levels of P. stomatis in LST and histopathology of LST. High abundance of P. stomatis and S0 stage LST indicted adenoma recurrence. P. stomatis, Peptostreptococcus stomatis; P. micra, Parvimonas micra; ETBF, Enterotoxigenic Bacteroides fragilis; P, LSTs with hyperplastic type or llow-grade dysplasia; S0, LSTs with intramucosal carcinoma or high-grade dysplasia. ****P < 0.0001.
Discussion
Growing evidence revealed that alteration in gut microbiome relates to colorectal neoplasms (12, 28, 29). However, LST, as a principal precursor lesion of CRC, was never reported about the intestinal microbiota signature. Moreover, early detection of LSTs and early prediction of the risk of adenoma recurrence after LST resection were essential to prevent CRC. Herein, the gut microbiota signature of LST cohort by means of 16S rRNA gene sequencing was delineated in this study. The results demonstrate that LST microbial dysbiosis was characterized by relative high abundance of the genus Lactobacillus-Streptococcus and the species ETBF-P. stomatis–P. micra. Based on the microbial signature, fecal microbial biomarkers ETBF–P. stomatis–P. micra were defined as early noninvasive biomarkers of LST. It indicates microbiome may form a synergistic system resulting diseases. Moreover, P. stomatis behaved high accuracy on predicting adenoma recurrence after LST resections.
ETBF has been known as a contributor to many colonic diseases including colonic dysfunctions, intestinal inflammation, oncogenic transformation and colorectal precancerous and cancerous lesions (23–27). Combined action of IL-17 and ETBF on colonic epithelial cells suppressed T cell proliferation and promoted the differentiation of monocytic-myeloid-derived suppressor cells (MO-MDSCs) (26). Bacteroides fragilis-associated lncRNA1(BFAL1) is an important modulator of ETBF-induced carcinogenesis. Thus, for ETBF-induced CRC, BFAL1 can be a potential therapeutic target (27). The ETBF-bearing biofilms in colon biopsies from CRC patients susceptibility loci were recently reported to strongly suggest that ETBF plays a key role in CRC development (30). P.stomatis and P. micra are both part of the oral and gut commensal microbioma and gram-positive anaerobic bacterium (31, 32). These organisms have been concerned as contributing agents of many diseases (12, 13, 31, 33–35), including oral squamous cell carcinoma (33) and apical abscess (34). A study of 526 fecal shotgun metagenome data sets revealed seven enriched and core bacteria in CRC (28). The core set of bacteria included: ETBF; four oral bacteria of Fusobacterium nucleatum (F. nucleatum) (36, 37), Parvimonas micra, Prevotella intermedia (38), and Porphyromonas asaccharolytica (39); and two other bacteria, Thermanaerovibrio acidaminovorans (28) and Alistipes finegoldii (40). Yachida et al. (29) collected 616 stool samples and found these bacteria were observed in intramucosal carcinomas and multiple CRAs. Stage-specific analysis disclosed two elevated patterns of significant species: the first only increased in early stage, whereas the second one was elevated across early to later stages. The second pattern was characterized by F. nucleatum, Solobacterium moorei (38), P. stomatis, Peptostreptococcus anaerobius (41, 42), Lactobacillus sanfranciscensis, P. micra, and Gemella morbillorum (43). ETBF, P. stomatis, and P. micra were crucial to colorectal carcinogenesis while they were rarely reported to be biomarkers to detect precancerous lesions, especially in LSTs.
Many studies have applied the abundance of various bacteria to distinguish CRC patients from HCs. Among bacterial candidates, F. nucleatum appeared as a key marker either when being combined with other potential bacteria (13, 17, 44) or quantified alone (16, 21), specifically combined with Clostridium symbiosum (17). These findings provide sturdy support that a limited number of targeted bacterial markers may provide accurate stool-based noninvasive diagnostic value. In this study, the fecal abundance of the three bacteria ETBF–P. stomatis–P. micra displayed considerably high sensitivity and specificity in detecting LST, especially P. stomatis. While combinations of P. stomatis–P. micra–ETBF revealed higher diagnostic potential compared with the single biomarker P. stomatis and combined biomarkers of P. stomatis–P. micra, P. stomatis–ETBF, P. micra–ETBF. The wide application of the above bacterial markers in LST detection may be rather practical since qPCR detection of fecal bacterial DNA is apparently more cost-effective and reliable than strict endoscopic screening.
Adenoma recurrence after endoscopic resection of LSTs is frequent (7, 8, 45, 46). Despite the widespread use of endoscopic resection, few studies examined the risk factors of adenoma recurrence after standard therapy for LST. Cancerous LSTs were related with adenoma recurrence event after endoscopic therapy (47). Some studies showed that independent risk factors recurrent adenoma were lesion size >60 mm, lesion occupying 75% of the luminal circumference, high grade dysplasia of LSTs, adjunctive argon plasma coagulation and piecemeal resection (8, 47, 48). Furthermore, adenoma recurrence surveillance requests regular endoscopy screening. Many LST patients not developing adenoma recurrence may be subjected to ultimately unnecessary surveillance procedures. Inconvenient bowel preparation, well-trained technician and carefully installed devices before endoscopy are resource-consuming and could limit the use for population-wide screening. Based on the results of this study, it is worth noting that the relative high abundance of P. stomatis positively correlated with adenoma recurrence after LST resection. Therefore, application of the biomarker P. stomatis to predict adenoma recurrence was promising and feasible.
Furthermore, ETBF–P. stomatis–P. micra modulation, especially P. stomatis may be important for LST development, adenoma recurrence after LST resection and even colorectal carcinogenesis. It will require concerted efforts to translate it into a clinical product to find the appropriate interventional methods to manipulate the microbiota.
However, there is no totally clear answer about the role of ETBF–P. stomatis–P. micra in the development of LST or CRC. The process is intricate and influenced by environmental and genetic factors. Mechanisms include immune regulation, metabolism of dietary components, genotoxin production, and inflammation (49–51). There remained a question that what are the driver and passenger bacteria in LST development? As described, the three above bacteria increased IL-6 secretion in LST group. IL-6 is an inflammatory cytokine and plays important roles in microenviroment of colorectal cancers (52–55). IL-6 level was reported to be higher in CRC tissues compared with noncancerous tissues (52). Moreover, STAT3 activation through IL-6/IL-11 in cancer associated fibroblasts (CAFs) promoted CRC development and poor prognosis (53). IL-6 trans-signaling in a mouse model of CRC acted downstream of epidermal growth factor signaling in myeloid cells (54). It could be hypothesized that the three bacteria may induce inflammatory environment to accelerate the development of LSTs, while it deserves in-depth studies about this carcinogenesis process.
The strengths of the present study were as follows. First, we reported for the first time that the bacteria signature of LST. Second, we found fecal microbiome signature (ETBF–P. stomatis–P. micra) can effectively predict the occurrence of LSTs. Moreover, ETBF, P. stomatis, and P. micra were related to the S0 stage LST. Third, our study provided new evidence that P. stomatis exhibited the high accuracy on predicting the adenoma recurrence after endoscopic resection of LSTs.
Despite the clinical data for endoscopic resections was collected prospectively and all consecutive patients were enrolled, this study was limited by its retrospective design, as all results were only from the single tertiary center and could not be generalizable. In future, studies in large cohorts including need to be carried out to enroll more patients, which can derive the best diagnostic algorithm across populations.
Conclusion
In conclusion, we observed, for the first time, fecal microbiome signature (ETBF-P. stomatis–P. micra) can effectively predict the presence of LSTs. Moreover, ETBF, P. stomatis, and P. micra were related to malignant LST and P. stomatis exhibited the high accuracy on predicting the adenoma recurrence after endoscopic resection of LSTs. Thus, the signature bacteria of LST can provide a noninvasive method to early detect LST and predict the adenoma recurrence risk after resections.
Data Availability Statement
The 16S rRNA sequences were deposited at NCBI Sequence Read Archive repository with accession code SRP317984.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Committee of Renji Hospital, School of Medicine, Shanghai Jiaotong University. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
XS analyzed the data and wrote the manuscript. XS, JLL, JQL, YZ, XL, and QG conceived the study and collected samples. YC and XC performed the pathological diagnosis. YXC and J-YF revised the manuscript. J-YF designed and supervised the study. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by grants from the National Natural Science Foundation of China (81421001, 81530072, 81830081).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to all members of the group at the Shanghai Institute of Digestive Disease. This manuscript has been released as a pre-print at ResearchGate (XS, JLL, JQL, et al.).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.661048/full#supplementary-material
References
1. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2020. CA Cancer J Clin (2020) 70(1):7–30. doi: 10.3322/caac.21590
2. Cotton S, Sharp L, Little J. The Adenoma-Carcinoma Sequence and Prospects for the Prevention of Colorectal Neoplasia. Crit Rev Oncog (1996) 7(5-6):293–342. doi: 10.1615/critrevoncog.v7.i5-6.10
3. Lee-Six H, Olafsson S, Ellis P, Osborne RJ, Sanders MA, Moore L, et al. The Landscape of Somatic Mutation in Normal Colorectal Epithelial Cells. Nature (2019) 574(7779):532–7. doi: 10.1038/s41586-019-1672-7
4. Ishigaki T, Kudo SE, Miyachi H, Hayashi T, Minegishi Y, Toyoshima N, et al. Treatment Policy for Colonic Laterally Spreading Tumors Based on Each Clinicopathological Feature of 4 Subtypes: Actual Status of Pseudo-Depressed Type. Gastrointest Endosc (2020) 92(5):1083–94.e6. doi: 10.1016/j.gie.2020.04.033
5. Kumarasinghe MP, Bourke MJ, Brown I, Draganov PV, McLeod D, Streutker C, et al. Pathological Assessment of Endoscopic Resections of the Gastrointestinal Tract: A Comprehensive Clinicopathologic Review. Mod Pathol (2020) 33(6):986–1006. doi: 10.1038/s41379-019-0443-1
6. Kudo S. Endoscopic Mucosal Resection of Flat and Depressed Types of Early Colorectal Cancer. Endoscopy (1993) 25(7):455–61. doi: 10.1055/s-2007-1010367
7. Hassan C, Repici A, Sharma P, Correale L, Zullo A, Bretthauer M, et al. Efficacy and Safety of Endoscopic Resection of Large Colorectal Polyps: A Systematic Review and Meta-Analysis. Gut (2016) 65(5):806–20. doi: 10.1136/gutjnl-2014-308481
8. Moss A, Williams SJ, Hourigan LF, Brown G, Tam W, Singh R, et al. Long-Term Adenoma Recurrence Following Wide-Field Endoscopic Mucosal Resection (WF-EMR) for Advanced Colonic Mucosal Neoplasia is Infrequent: Results and Risk Factors in 1000 Cases From the Australian Colonic EMR (ACE) Study. Gut (2015) 64(1):57–65. doi: 10.1136/gutjnl-2013-305516
9. Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A Human Gut Microbial Gene Catalogue Established by Metagenomic Sequencing. Nature (2010) 464(7285):59–65. doi: 10.1038/nature08821
10. Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, et al. Fusobacterium Nucleatum Infection is Prevalent in Human Colorectal Carcinoma. Genome Res (2012) 22(2):299–306. doi: 10.1101/gr.126516.111
11. Feng Q, Liang S, Jia H, Stadlmayr A, Tang L, Lan Z, et al. Gut Microbiome Development Along the Colorectal Adenoma-Carcinoma Sequence. Nat Commun (2015) 6:6528. doi: 10.1038/ncomms7528
12. Yu J, Feng Q, Wong SH, Zhang D, Liang QY, Qin Y, et al. Metagenomic Analysis of Faecal Microbiome as a Tool Towards Targeted non-Invasive Biomarkers for Colorectal Cancer. Gut (2017) 66(1):70–8. doi: 10.1136/gutjnl-2015-309800
13. Baxter NT, Ruffin M, Rogers MA, Schloss PD. Microbiota-Based Model Improves the Sensitivity of Fecal Immunochemical Test for Detecting Colonic Lesions. Genome Med (2016) 8(1):37. doi: 10.1186/s13073-016-0290-3
14. Nakatsu G, Zhou H, Wu WKK, Wong SH, Coker OO, Dai Z, et al. Alterations in Enteric Virome Are Associated With Colorectal Cancer and Survival Outcomes. Gastroenterology (2018) 155(2):529–41.e5. doi: 10.1053/j.gastro.2018.04.018
15. Gao R, Kong C, Li H, Huang L, Qu X, Qin N, et al. Dysbiosis Signature of Mycobiota in Colon Polyp and Colorectal Cancer. Eur J Clin Microbiol Infect Dis (2017) 36(12):2457–68. doi: 10.1007/s10096-017-3085-6
16. Suehiro Y, Zhang Y, Hashimoto S, Takami T, Higaki S, Shindo Y, et al. Highly Sensitive Faecal DNA Testing of TWIST1 Methylation in Combination With Faecal Immunochemical Test for Haemoglobin is a Promising Marker for Detection of Colorectal Neoplasia. Ann Clin Biochem (2018) 55(1):59–68. doi: 10.1177/0004563217691064
17. Xie YH, Gao QY, Cai GX, Sun XM, Zou TH, Chen HM, et al. Fecal Clostridium Symbiosum for Noninvasive Detection of Early and Advanced Colorectal Cancer: Test and Validation Studies. EBioMedicine (2017) 25:32–40. doi: 10.1016/j.ebiom.2017.10.005
18. Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA, et al. Fusobacterium Nucleatum in Colorectal Carcinoma Tissue and Patient Prognosis. Gut (2016) 65(12):1973–80. doi: 10.1136/gutjnl-2015-310101
19. Wei Z, Cao S, Liu S, Yao Z, Sun T, Li Y, et al. Could Gut Microbiota Serve as Prognostic Biomarker Associated With Colorectal Cancer Patients’ Survival? A Pilot Study on Relevant Mechanism. Oncotarget (2016) 7(29):46158–72. doi: 10.18632/oncotarget.10064
20. Flanagan L, Schmid J, Ebert M, Soucek P, Kunicka T, Liska V, et al. Fusobacterium Nucleatum Associates With Stages of Colorectal Neoplasia Development, Colorectal Cancer and Disease Outcome. Eur J Clin Microbiol Infect Dis (2014) 33(8):1381–90. doi: 10.1007/s10096-014-2081-3
21. Wong SH, Kwong TNY, Chow TC, Luk AKC, Dai RZW, Nakatsu G, et al. Quantitation of Faecal Fusobacterium Improves Faecal Immunochemical Test in Detecting Advanced Colorectal Neoplasia. Gut (2017) 66(8):1441–8. doi: 10.1136/gutjnl-2016-312766
22. Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, et al. Fusobacterium Nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell (2017) 170(3):548–63 e16. doi: 10.1016/j.cell.2017.07.008
23. Zamani S, Taslimi R, Sarabi A, Jasemi S, Sechi LA, Feizabadi MM. Enterotoxigenic Bacteroides Fragilis: A Possible Etiological Candidate for Bacterially-Induced Colorectal Precancerous and Cancerous Lesions. Front Cell Infect Microbiol (2019) 9:449. doi: 10.3389/fcimb.2019.00449
24. Geis AL, Fan H, Wu X, Wu S, Huso DL, Wolfe JL, et al. Regulatory T-cell Response to Enterotoxigenic Bacteroides Fragilis Colonization Triggers IL17-Dependent Colon Carcinogenesis. Cancer Discov (2015) 5(10):1098–109. doi: 10.1158/2159-8290.CD-15-0447
25. Chung L, Thiele Orberg E, Geis AL, Chan JL, Fu K, DeStefano Shields CE, et al. Bacteroides Fragilis Toxin Coordinates a Pro-carcinogenic Inflammatory Cascade Via Targeting of Colonic Epithelial Cells. Cell Host Microbe (2018) 23(2):203–14.e5. doi: 10.1016/j.chom.2018.01.007
26. Thiele Orberg E, Fan H, Tam AJ, Dejea CM, Destefano Shields CE, Wu S, et al. The Myeloid Immune Signature of Enterotoxigenic Bacteroides Fragilis-Induced Murine Colon Tumorigenesis. Mucosal Immunol (2017) 10(2):421–33. doi: 10.1038/mi.2016.53
27. Bao Y, Tang J, Qian Y, Sun T, Chen H, Chen Z, et al. Long Noncoding RNA BFAL1 Mediates Enterotoxigenic Bacteroides Fragilis-Related Carcinogenesis in Colorectal Cancer Via the RHEB/mTOR Pathway. Cell Death Dis (2019) 10(9):675. doi: 10.1038/s41419-019-1925-2
28. Dai Z, Coker OO, Nakatsu G, Wu WKK, Zhao L, Chen Z, et al. Multi-Cohort Analysis of Colorectal Cancer Metagenome Identified Altered Bacteria Across Populations and Universal Bacterial Markers. Microbiome (2018) 6(1):70. doi: 10.1186/s40168-018-0451-2
29. Yachida S, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T, et al. Metagenomic and Metabolomic Analyses Reveal Distinct Stage-Specific Phenotypes of the Gut Microbiota in Colorectal Cancer. Nat Med (2019) 25(6):968–76. doi: 10.1038/s41591-019-0458-7
30. Dejea CM, Fathi P, Craig JM, Boleij A, Taddese R, Geis AL, et al. Patients With Familial Adenomatous Polyposis Harbor Colonic Biofilms Containing Tumorigenic Bacteria. Science (2018) 359(6375):592–7. doi: 10.1126/science.aah3648
31. Kistler JO, Pesaro M, Wade WG. Development and Pyrosequencing Analysis of an in-Vitro Oral Biofilm Model. BMC Microbiol (2015) 15:24. doi: 10.1186/s12866-015-0364-1
32. Shah MS, DeSantis TZ, Weinmaier T, McMurdie PJ, Cope JL, Altrichter A, et al. Leveraging Sequence-Based Faecal Microbial Community Survey Data to Identify a Composite Biomarker for Colorectal Cancer. Gut (2018) 67(5):882–91. doi: 10.1136/gutjnl-2016-313189
33. Zhang L, Liu Y, Zheng HJ, Zhang CP. The Oral Microbiota May Have Influence on Oral Cancer. Front Cell Infect Microbiol (2019) 9:476. doi: 10.3389/fcimb.2019.00476
34. Nobrega LM, Montagner F, Ribeiro AC, Mayer MA, Gomes BP. Molecular Identification of Cultivable Bacteria From Infected Root Canals Associated With Acute Apical Abscess. Braz Dent J (2016) 27(3):318–24. doi: 10.1590/0103-6440201600715
35. Drewes JL, White JR, Dejea CM, Fathi P, Iyadorai T, Vadivelu J, et al. High-Resolution Bacterial 16S rRNA Gene Profile Meta-Analysis and Biofilm Status Reveal Common Colorectal Cancer Consortia. NPJ Biofilms Microbiomes (2017) 3:34. doi: 10.1038/s41522-017-0040-3
36. Brennan CA, Garrett WS. Fusobacterium Nucleatum - Symbiont, Opportunist and Oncobacterium. Nat Rev Microbiol (2019) 17(3):156–66. doi: 10.1038/s41579-018-0129-6
37. Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, et al. Analysis of Fusobacterium Persistence and Antibiotic Response in Colorectal Cancer. Science (2017) 358(6369):1443–8. doi: 10.1126/science.aal5240
38. Yang Y, Cai Q, Shu XO, Steinwandel MD, Blot WJ, Zheng W, et al. Prospective Study of Oral Microbiome and Colorectal Cancer Risk in Low-Income and African American Populations. Int J Cancer (2019) 144(10):2381–9. doi: 10.1002/ijc.31941
39. Ai D, Pan H, Han R, Li X, Liu G, Xia LC. Using Decision Tree Aggregation With Random Forest Model to Identify Gut Microbes Associated With Colorectal Cancer. Genes (Basel) (2019) 10(2):112. doi: 10.3390/genes10020112
40. Yang Y, Jobin C. Novel Insights Into Microbiome in Colitis and Colorectal Cancer. Curr Opin Gastroenterol (2017) 33(6):422–7. doi: 10.1097/MOG.0000000000000399
41. Long X, Wong CC, Tong L, Chu ESH, Ho Szeto C, Go MYY, et al. Peptostreptococcus Anaerobius Promotes Colorectal Carcinogenesis and Modulates Tumour Immunity. Nat Microbiol (2019) 4(12):2319–30. doi: 10.1038/s41564-019-0541-3
42. Tsoi H, Chu ESH, Zhang X, Sheng J, Nakatsu G, Ng SC, et al. Peptostreptococcus Anaerobius Induces Intracellular Cholesterol Biosynthesis in Colon Cells to Induce Proliferation and Causes Dysplasia in Mice. Gastroenterology (2017) 152(6):1419–33.e5. doi: 10.1053/j.gastro.2017.01.009
43. Kwong TNY, Wang X, Nakatsu G, Chow TC, Tipoe T, Dai RZW, et al. Association Between Bacteremia From Specific Microbes and Subsequent Diagnosis of Colorectal Cancer. Gastroenterology (2018) 155(2):383–90.e8. doi: 10.1053/j.gastro.2018.04.028
44. Guo S, Li L, Xu B, Li M, Zeng Q, Xiao H, et al. A Simple and Novel Fecal Biomarker for Colorectal Cancer: Ratio of Fusobacterium Nucleatum to Probiotics Populations, Based on Their Antagonistic Effect. Clin Chem (2018) 64(9):1327–37. doi: 10.1373/clinchem.2018.289728
45. Li Y, Zhang Y, Chen Y, Wang Y, Dou L, Wang X, et al. Long-Term Outcomes of Endoscopic Treatment for Colorectal Laterally Spreading Tumor: A Large-Scale Multicenter Retrospective Study From China. Surg Endosc (2020) 35(2):736–44. doi: 10.1007/s00464-020-07440-8
46. Soliman H, Brieau B, Guillaumot MA, Leblanc S, Barret M, Camus M, et al. Invasive Pit Pattern, Macronodule and Depression are Predictive Factors of Submucosal Invasion in Colorectal Laterally Spreading Tumours From a Western Population. United Eur Gastroenterol J (2018) 6(10):1569–77. doi: 10.1177/2050640618804713
47. Tate DJ, Desomer L, Klein A, Brown G, Hourigan LF, Lee EY, et al. Adenoma Recurrence After Piecemeal Colonic EMR is Predictable: The Sydney EMR Recurrence Tool. Gastrointest Endosc (2017) 85(3):647–56.e6. doi: 10.1016/j.gie.2016.11.027
48. Emmanuel A, Lapa C, Ghosh A, Gulati S, Burt M, Hayee B, et al. Risk Factors for Early and Late Adenoma Recurrence After Advanced Colorectal Endoscopic Resection At an Expert Western Center. Gastrointest Endosc (2019) 90(1):127–36. doi: 10.1016/j.gie.2019.01.031
49. Lasry A, Zinger A, Ben-Neriah Y. Inflammatory Networks Underlying Colorectal Cancer. Nat Immunol (2016) 17(3):230–40. doi: 10.1038/ni.3384
50. Zhang FF, Cudhea F, Shan Z, Michaud DS, Imamura F, Eom H, et al. Preventable Cancer Burden Associated With Poor Diet in the United States. JNCI Cancer Spectr (2019) 3(2):pkz034. doi: 10.1093/jncics/pkz034
51. Tilg H, Adolph TE, Gerner RR, Moschen AR. The Intestinal Microbiota in Colorectal Cancer. Cancer Cell (2018) 33(6):954–64. doi: 10.1016/j.ccell.2018.03.004
52. Zeng J, Tang ZH, Liu S, Guo SS. Clinicopathological Significance of Overexpression of Interleukin-6 in Colorectal Cancer. World J Gastroenterol (2017) 23(10):1780–6. doi: 10.3748/wjg.v23.i10.1780
53. Heichler C, Scheibe K, Schmied A, Geppert CI, Schmid B, Wirtz S, et al. STAT3 Activation Through IL-6/IL-11 in Cancer-Associated Fibroblasts Promotes Colorectal Tumour Development and Correlates With Poor Prognosis. Gut (2020) 69(7):1269–82. doi: 10.1136/gutjnl-2019-319200
54. Schmidt S, Schumacher N, Schwarz J, Tangermann S, Kenner L, Schlederer M, et al. ADAM17 is Required for EGF-R-induced Intestinal Tumors Via IL-6 Trans-Signaling. J Exp Med (2018) 215(4):1205–25. doi: 10.1084/jem.20171696
Keywords: laterally spreading tumor, enterotoxigenic Bacteroides fragilis, Peptostreptococcus stomatis, Parvimonas micra, noninvasive biomarker, diagnosis and prognosis
Citation: Shen X, Li J, Li J, Zhang Y, Li X, Cui Y, Gao Q, Chen X, Chen Y and Fang J-Y (2021) Fecal Enterotoxigenic Bacteroides fragilis–Peptostreptococcus stomatis–Parvimonas micra Biomarker for Noninvasive Diagnosis and Prognosis of Colorectal Laterally Spreading Tumor. Front. Oncol. 11:661048. doi: 10.3389/fonc.2021.661048
Received: 30 January 2021; Accepted: 20 April 2021;
Published: 11 May 2021.
Edited by:
Zexian Liu, Sun Yat-sen University Cancer Center (SYSUCC), ChinaCopyright © 2021 Shen, Li, Li, Zhang, Li, Cui, Gao, Chen, Chen and Fang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yingxuan Chen, eWluZ3h1YW5jaGVuNzFAMTI2LmNvbQ==; Jing-Yuan Fang, amluZ3l1YW5mYW5nQHNqdHUuZWR1LmNu
 Xiaonan Shen
Xiaonan Shen Jialu Li
Jialu Li Yao Zhang
Yao Zhang Xiaobo Li
Xiaobo Li Yingxuan Chen
Yingxuan Chen Jing-Yuan Fang
Jing-Yuan Fang