
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol., 10 September 2021
Sec. Cancer Immunity and Immunotherapy
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.661043
This article is part of the Research TopicTumor-Associated Antigens and Their Autoantibodies: From Discovering to Clinical UtilizationView all 19 articles
 Huimin Wang1,2,3,4,5†
Huimin Wang1,2,3,4,5† Xiaoang Yang1†
Xiaoang Yang1† Guiying Sun3,4,5
Guiying Sun3,4,5 Qian Yang3,4,5
Qian Yang3,4,5 Chi Cui1,3,4
Chi Cui1,3,4 Xiao Wang1,3,4
Xiao Wang1,3,4 Hua Ye3,4,5
Hua Ye3,4,5 Liping Dai1,3,4
Liping Dai1,3,4 Jianxiang Shi1,3,4
Jianxiang Shi1,3,4 Jianying Zhang1,3,4
Jianying Zhang1,3,4 Peng Wang1,3,4,5*
Peng Wang1,3,4,5*The study aims to explore the diagnostic value of anti-GNA11 autoantibody in esophageal squamous cell carcinoma (ESCC) from multiple levels. Autoantibody against GNA11 with the highest diagnostic performance was screened out from the customized protein microarray. A total of 486 subjects including ESCC patients and matched normal controls were recruited in the verification and validation phases by using enzyme-linked immunosorbent assay (ELISA). Western blotting analysis was used to verify the ELISA results. Immunohistochemistry (IHC) was used to evaluate GNA11 expression in ESCC tissues and para-tumor tissues. In addition, a bioinformatics approach was adopted to investigate the mRNA expression of GNA11 in ESCC. Results indicated that the level of anti-GNA11 autoantibody in ESCC patients was significantly higher than that in the normal controls, and it can be used to distinguish ESCC patients from normal individuals in clinical subgroups (p < 0.05), as revealed by both ELISA and Western blotting. The receiver operating characteristic (ROC) curve analysis showed that anti-GNA11 autoantibody could distinguish ESCC patients from normal controls with an area under the ROC curve (AUC) of 0.653, sensitivity of 10.96%, and specificity of 98.63% in the verification cohort and with an AUC of 0.751, sensitivity of 38.24%, and specificity of 88.82% in the validation cohort. IHC manifested that the expression of GNA11 can differentiate ESCC tissues with para-tumor tissues (p < 0.05), but it cannot be used to differentiate different pathological grades and clinical stages (p > 0.05). The mRNA expression of GNA11 in ESCC patients and normal controls was different with a bioinformatics mining with The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) data in Gene Expression Profiling Interactive Analysis (GEPIA). In summary, anti-GNA11 autoantibody has the potential to be a new serological marker in the diagnosis of ESCC.
Esophageal cancer (EC) is one of the common malignant tumors that threaten the health of human beings, with its incidence ranking seventh and cancer-related death ranking sixth worldwide (1). Studies estimated that there were 572,000 new cases and 508,600 deaths of EC around the world in 2018 (2). The incidence of EC in China is among the top 5 in the world (1). It was estimated that there were 246,000 new cases and 188,000 deaths of EC in 2015, which was the fourth leading cause of cancer-related death in China (3). Esophageal squamous cell carcinoma (ESCC) accounts for more than 90% of EC in China, which is a severe healthcare burden (4).
The 5-year overall survival rate of EC is less than 20%, and the prognosis is inferior (5, 6). The poor prognosis of EC is mainly due to the lack of clinical symptoms in patients in the early stage and the paucity of reliable non-invasive testing methods. However, relevant studies pointed out that early diagnosis of EC can significantly improve its poor prognosis, with a 5-year survival rate reaching as high as 80%–90% (7, 8). The standard diagnostic methods for EC screening include endoscopy and pathological biopsy, which are expensive and invasive. Consequently, a critically unmet need in the diagnosis and management of EC is identifying and developing novel non-invasive biomarkers that can complement the traditional diagnostic methods. Researchers have suggested that autoantibodies against tumor-associated antigens (TAAs) could be used as diagnostic biomarkers for the early diagnosis of cancer. These anti-TAA autoantibodies are stable in the circulating blood and can be produced as early as several years before the appearance of clinical symptoms (9–12). Currently, there are a lot of autoantibodies against TAAs that have been used in the early diagnosis of ESCC, such as FOXP3 (13), Fascin (14), Ezrin (15), STIP1 (16), LY6K (17), and MMP7 (18). However, the sensitivity and specificity of these anti-TAA autoantibodies cannot meet the needs of the clinical diagnosis of ESCC as biomarkers. Hence, it is of great importance to identify additional biomarkers with high specificity and sensitivity for the diagnosis of ESCC.
Heterotrimeric G proteins are widely expressed in all eukaryotic cells consisting of three subunits: alpha, beta, and gamma. GNA11 is the alpha subunit of G protein and is involved in a variety of transmembrane signaling systems. Our previous study indicated that GNA11 was involved in the development of hepatocellular carcinoma (HCC) and might be a potential biomarker in HCC detection (19). Our previous study also found that anti-GNA11 autoantibody had the potential to diagnose ESCC when we established a diagnostic model of an autoantibody panel to screen ESCC (20). GNA11 gene was frequently mutated in the conventional esophageal adenocarcinoma, and the mutation was related to critical cellular pathways including PI3K, RAS, and MAPK, which suggested that GNA11 mutation might be tightly linked to the occurrence of EC (21). In the current study, in order to evaluate the potential of anti-GNA11 autoantibody in the diagnosis of ESCC, the level of anti-GNA11 autoantibody in sera of ESCC patients and matched normal controls was detected by ELISA, and the protein level and mRNA level of GNA11 were further explored by immunohistochemistry (IHC) and bioinformatics. The overall design of the current study is shown in Figure 1.
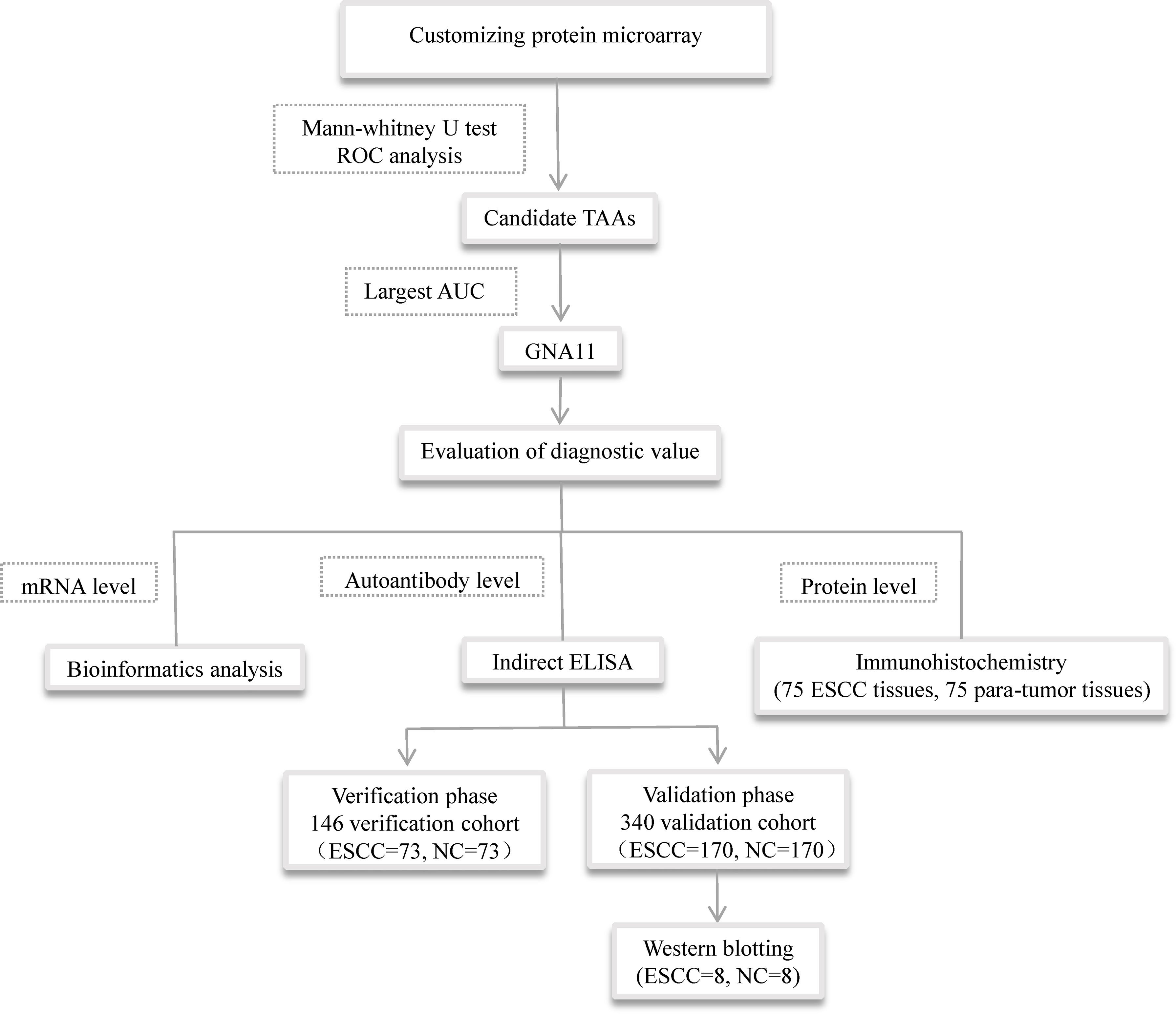
Figure 1 The overall design of the study. TAAs, tumor-associated antigens; ELISA, enzyme-linked immunosorbent assay.
Serum samples of 333 ESCC patients and 293 normal controls were used in the study. From July 2017 to October 2018, sera from ESCC patients were collected from a third-level grade A hospital in Henan Province, China. Normal control sera were derived from the biological specimen bank in Henan Key Laboratory of Tumor Epidemiology. All patients underwent pathological examination to confirm that they have not received any treatment prior to collecting blood, and none of the controls had autoimmune diseases or tumor-related diseases. Sera from 90 ESCC patients and 50 normal controls were selected for protein microarray assay, and 486 sera from 243 ESCC patients and 243 normal controls were used for ELISA. Table 1 shows the detailed clinical information of all participants. The peripheral blood of all subjects under fasting state was collected at 5 ml and placed into vacuum tube without anticoagulants. The collected whole blood samples were centrifuged at 3,000 rpm for 10 min after standing for 1 h at room temperature and then stored at −80°C. This study has been approved by the Ethics Committee of Zhengzhou University, and all the subjects had signed informed consent.
The study was authorized by Guangzhou Bochong Biotechnology Co., Ltd., to make focused array protein microarray including 154 recombinant human proteins or protein fragments (CDI lab), which included 143 proteins or protein fragments encoded by 138 cancer driver genes and 11 proteins with high diagnostic value in our previous studies containing IMP1, IMP2, IMP3, CyclinB1, c-Myc, CIP2A/p90, RalA, YWHAZ, RBM39, and two fragments of Survivin. Protein microarray was used to detect the expression levels of corresponding autoantibodies in 140 serums to screen out significant TAAbs associated with ESCC. To eliminate the bias brought by the difference of background values, signal-to-noise ratio (SNR) was defined as F median/B median, where F532 Median refers to the median of the foreground value of signal points in the 532-nm channel and B532 Median refers to the median of the background value of signal points in the 532-nm channel. The detailed experimental procedure was performed exactly as the standard protocol described in our previous study (22).
The level of anti-GNA11 autoantibody in sera of ESCC patients and normal controls was detected by ELISA. Purified recombinant protein GNA11 was purchased from the LD Biopharma Company (San Diego, USA). Horseradish peroxidase (HRP)-conjugated mouse anti-human IgG (Wuhan Aoko Biotechnology Co. Ltd.) was used as the secondary antibody. Each ELISA plate included eight standard concentrations of 10, 20, 50, 100, 150, 200, 250, and 300 ng/ml of IgG (Solarbio); a positive control; a negative control; and a blank control, which enabled the stability of all the plates. The detailed operation of ELISA was described in our previous study (23). The optical density (OD) of each well was measured at 450 and 620 nm by using a microplate reader (Thermo Fisher Scientific).
The positive and negative sera of anti-GNA11 autoantibody found by ELISA were randomly selected and detected by Western blotting to verify the immunoreactivity of the sera. The detailed procedure of Western blotting was described in our previous study (24). Briefly, purified recombinant protein GNA11 was electrophoresed by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gel and then transferred onto nitrocellulose membrane. Selected serum samples diluted at 1:100 and HRP-conjugated mouse anti-human IgG antibody with a dilution of 1:5,000 were utilized as the primary antibody and the secondary antibody, respectively. The positive reaction signal was obtained by Azure Biosystems with chemiluminescence (C300–C600) according to the manufacturer’s instructions.
Immunohistochemical analysis was performed to detect the expression level of GNA11 protein in 75 ESCC tissues and 75 corresponding para-tumor tissues. Tissue microarray (TMA; CGT No. HEso-Squ150CS-02) and the specific operations were provided by Shanghai Outdo Biotech Co. Ltd. Mouse monoclonal anti-GNA11 antibody was used as the first antibody. Biotin-labeled secondary antibody and diaminobenzidine (DAB) were used as detecting reagents. All the TMA results were evaluated by two independent pathologists without knowing the tumor stage of the TMA sections. Ten fields were randomly selected under the microscope for each microarray, and the score was calculated according to the percentage of positive cells (area score) and staining intensity (color score). There were five scoring conditions for the percentage of stained cells in a cell count, for example, score 0 (less than 10%), 1 (10%–25%), 2 (26%–50%), 3 (51%–75%), and 4 (more than 75%). Staining intensity incorporated four different evaluation criteria: negative, score 0; faint yellow, score 1; brown yellow, score 2; and medium brown, score 3. The final score was attained by multiplying the score of the percentage of positive cells by the staining intensity score, which was negative score 0, weakly positive score 1–4, positive score 5–8, and strongly positive score 9–12.
The expression of GNA11 at the mRNA level was further explored in Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn/). The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) normal data were chosen to be compared in esophageal carcinoma (ESCA) patients about the difference of GNA11 expression at the mRNA level. RNA sequencing expression data of ESCC were according to TCGA dataset. And the use of GEPIA was precisely based on the report from literature (25).
IBM statistical software (version 25.0) and GraphPad Prism 8.0 were utilized in the study. All statistical analysis processes were two-tailed tests, and the test level was set as α = 0.05. The non-parametric test was applied to compare the difference of anti-GNA11 autoantibody between ESCC patients and normal controls. The frequency of anti-GNA11 autoantibody in different clinical parameters and demographic characteristics in all ESCC patients was analyzed by chi-square test. One-way ANOVA was used to analyze IHC scores in different tissue types. The receiver operating characteristic (ROC) curve was performed to estimate the diagnostic ability of the biomarker in different groups and clinical subgroups. At the same time, Youden’s index, positive likelihood ratio (PLR), negative likelihood ratio (NLR), positive predictive value (PPV), negative predictive value (NPV), and accuracy were also calculated to appraise the better ability of anti-GNA11 autoantibody in differentiating ESCC patients from normal controls. The area under the ROC curve (AUC) values of different clinical subgroups were analyzed by DeLong test. The cutoff value was set as mean value plus one SD of the normal control OD values (mean ± SD).
In the present study, 86 sera from ESCC patients and 50 control sera were finally selected to evaluate the autoantibody levels with protein microarray. Interestingly, five candidate anti-TAA autoantibodies corresponding to P53, PTEN, GNA11, GNAS, and SRSF2 were identified by the Mann–Whitney U test and ROC analysis (20). In the ESCC group and normal control group, the positive rates of five anti-TAA autoantibodies ranged from 16.28% to 34.88% and 8.16% to 16.33%, respectively. The AUC values of five autoantibodies in diagnosing ESCC ranged from 0.606 to 0.682. The AUC value of anti-GNA11 autoantibody was the highest (0.682, 95% CI: 0.588–0.776) (Figure 2B) with sensitivity of 17.44% and specificity of 91.84%. The positive rate of anti-GNA11 autoantibody in the ESCC group and normal control group was 17.44% and 8.16%, respectively. The scatter plot showed that the level of anti-GNA11 autoantibody in the ESCC group was obviously higher than that in the normal control group (p < 0.05) (Figure 2A). It indicated that anti-GNA11 autoantibody has the potential to distinguish ESCC patients from normal controls. To verify the hypothesis, the performance of anti-GNA11 autoantibody was further explored in cohorts with a large sample size.

Figure 2 The expression level and diagnostic value of anti-GNA11 autoantibody in ESCC patients and normal controls in screening phase. (A) The scatter plot depicts the level of anti-GNA11 autoantibody in ESCC group and normal control group. (B) The receiver operating characteristic curve of anti-GNA11 autoantibody in the screening stage. ESCC, esophageal squamous cell carcinoma; NC, normal controls; SNR, signal-to-noise ratio.
To further validate the efficacy of anti-GNA11 autoantibody in the immunodiagnosis of ESCC, sera from 243 ESCC patients and 243 normal controls were used in ELISA. All 486 serum samples were divided into two cohorts according to the ratio of 3:7. In the verification cohort (73 ESCC vs. 73 NC), a higher expression level of anti-GNA11 autoantibody was observed in ESCC patients (mean ± SD: 0.301 ± 0.054) compared with normal controls (mean ± SD: 0.277 ± 0.049) (Figure 3A); and anti-GNA11 autoantibody can distinguish 10.96% of ESCC patients with an AUC of 0.653 at the specificity of 98.63% (Figure 3B). A validation cohort of larger sample size (170 ESCC vs. 170 NC) was used to further confirm the diagnostic value of anti-GNA11 autoantibody. In this validation cohort, the level of anti-GNA11 autoantibody was distinctly higher in the ESCC group (mean ± SD: 0.345 ± 0.068) than that in the normal control group (mean ± SD: 0.281 ± 0.079) (Figure 3C), and the AUC value was as high as 0.751 with sensitivity of 38.24% and specificity of 88.82% (Figure 3D). The detailed results are shown in Table 2.
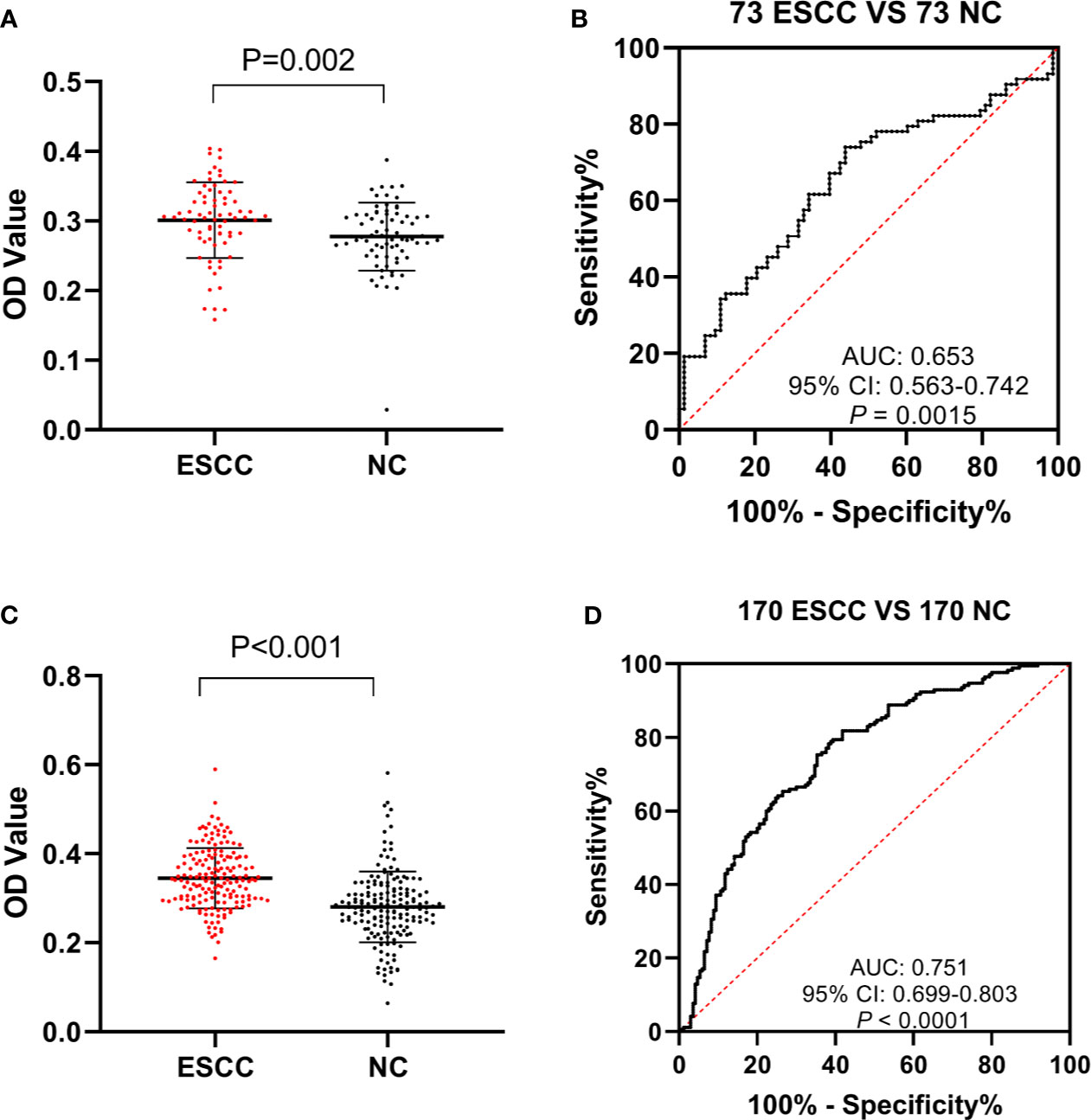
Figure 3 The expression level and diagnostic value of anti-GNA11 autoantibody in the phases of verification and validation. (A, C) The expression level of anti-GNA11 autoantibody in ESCC patients and normal controls in verification cohort and validation phase. (B, D) ROC analysis of anti-GNA11 autoantibody in the verification and validation phase. ESCC, esophageal squamous cell carcinoma; NC, normal controls; ROC, receiver operating characteristic.
Additionally, eight ESCC and eight normal control sera were randomly selected from ESCC patients and normal controls for Western blotting analysis to further confirm the results of ELISA. The molecular weight of GNA11 protein is 45.8 kDa. Results indicated that six of eight ESCC sera showed positive bands with molecular weight near 45 kDa, and none of the eight control sera showed a positive band in the corresponding 45 kDa position. The results of Western blotting are shown in Figure 4 and were consistent with those of ELISA.
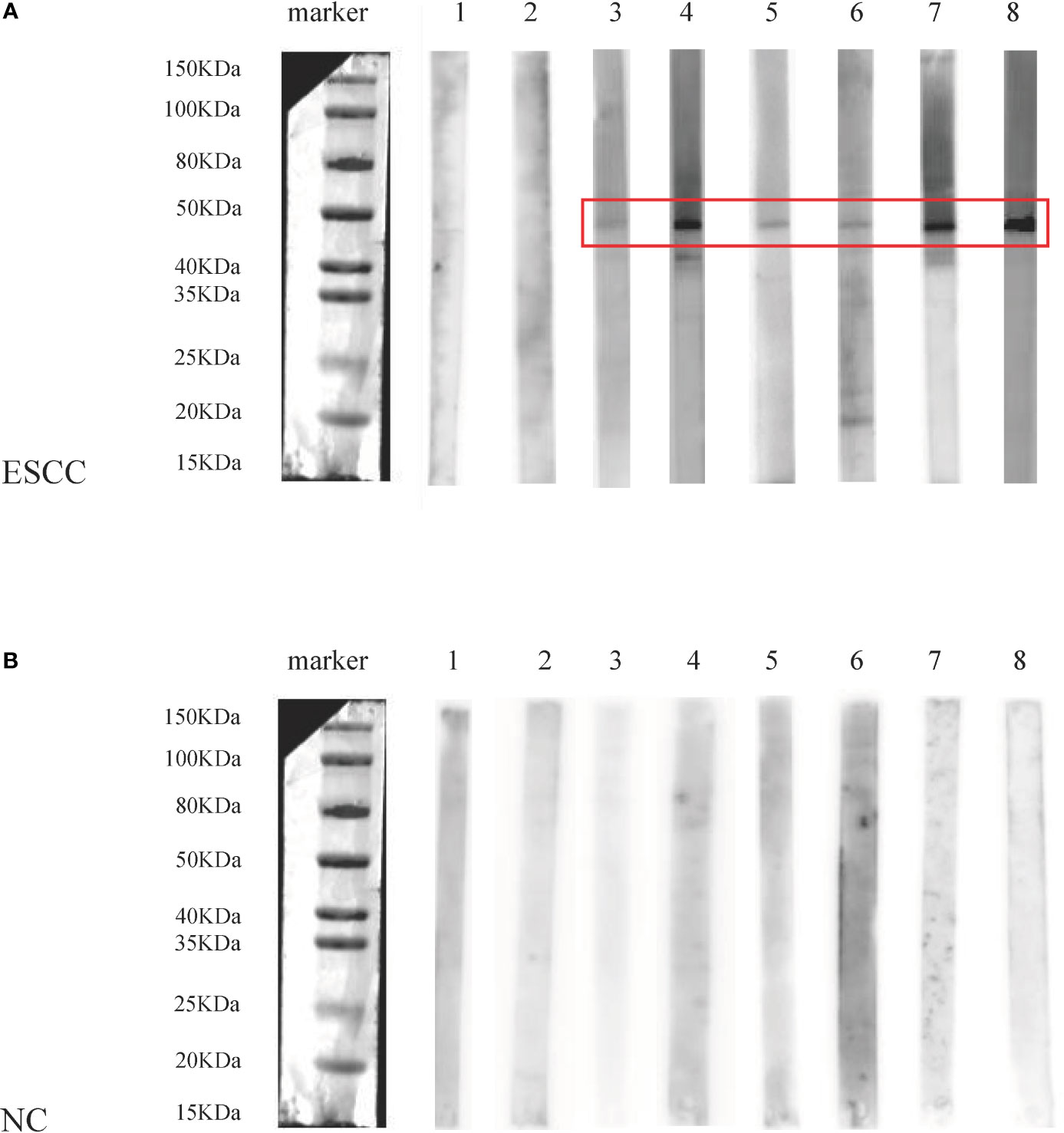
Figure 4 The analysis of anti-GNA11 autoantibody by Western blotting in sera of eight ESCC patients and eight normal controls. ESCC, esophageal squamous cell carcinoma; NC, normal controls. (A) The results of Western blotting in sera of 8 ESCC patients, of which No. 1-2 was negative, and No. 3-8 showed a positive band near 45 kDa. (B) The results of Western blotting in 8 control sera were negative, and there was no positive band near 45 kDa.
The performance of anti-GNA11 autoantibody in clinical variables (including lymphatic metastasis, differentiation, distance metastasis, TNM stage, family tumor history, gender, and age) was further explored. Anti-GNA11 autoantibody could significantly distinguish ESCC patients from normal controls in every subgroup (p < 0.05) (Figures 5A–N). The AUC values of clinical subgroups ranged from 0.661 to 0.774 (p < 0.05). The maximum AUC of 0.774 was observed among female ESCC patients (Figure 5L), and the minimum AUC of 0.661 was detected in ESCC patients with TNM stage III and IV (Figure 5H). The AUC values in comparison groups regarding lymphatic metastasis, differentiation, distance metastasis, TNM stage, family tumor history, and age was not significantly different (p > 0.05), yet it presented a marginal difference in gender (p = 0.05). In addition, there were no differences in the positive rates of anti-GNA11 autoantibody in all subgroups (p > 0.05) (Table 3).
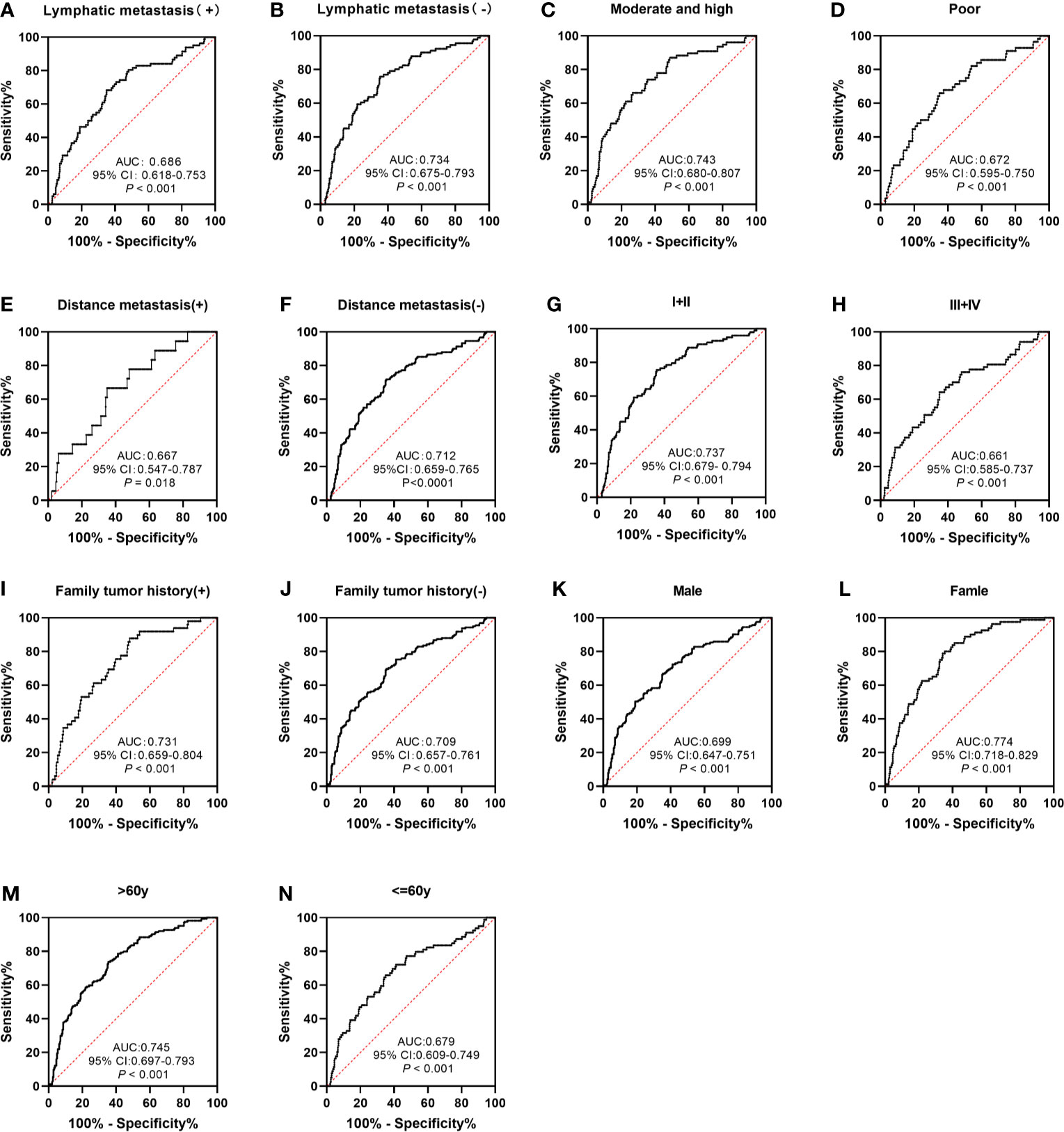
Figure 5 The performance of anti-GNA11 autoantibody according to different clinical variables of ESCC patients. ESCC, esophageal squamous cell carcinoma.
As shown in Figure 6, the mRNA level of GNA11 was higher (fold change > 1.4, p < 0.05) in ESCC patients compared with normal controls based on TCGA and GTEx data in GEPIA, which was consistent with the difference in serum level of anti-GNA11 autoantibody between ESCC patients and normal controls.
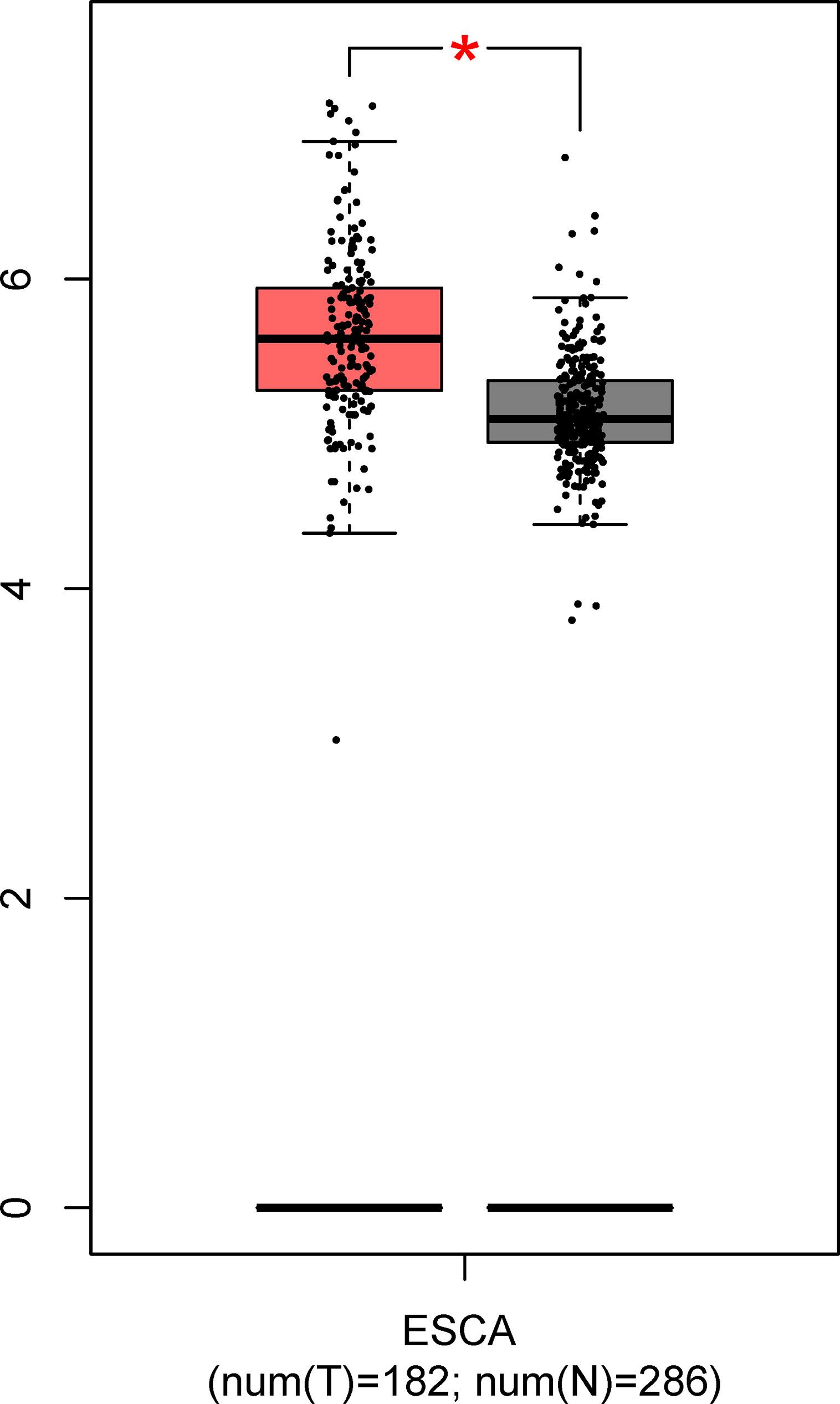
Figure 6 The expression of GNA11 at mRNA level based on TCGA and GTEx data in GEPIA. The red box represents the esophageal carcinoma group, and the gray box chart indicates the normal control group. ESCA, esophageal carcinoma; TCGA, The Cancer Genome Atlas; GTEx, Genotype-Tissue Expression; GEPIA, Gene Expression Profiling Interactive Analysis. *p < 0.05.
Furthermore, the expression of GNA11 at the protein level was investigated in the Human Protein Altas (HPA; https://www.proteinatlas.org/). However, there was no prior evidence on the expression of GNA11 at protein in ESCC tissues (Supplementary Figure 1). Therefore, IHC was employed to further verify the expression of GNA11 at the protein level in ESCC tissues and para-tumor tissues. TMA slides including 75 ESCC tissue samples and corresponding para-tumor tissues were commercially acquired for this study. The clinical information of 75 ESCC patients including American Joint Committee on Cancer (AJCC) clinical stage and pathological grade was analyzed to explore the association between GNA11 expression and tumor stage. The expression level of GNA11 in tumor tissues and para-tumor tissues was evaluated by staining intensity (color score) and the percentage of positive cells (area score).
Results showed that GNA11 was overexpressed in ESCC tissues compared with para-tumor tissues (final score 1.57 vs. final score 0.14, p < 0.05) (Table 4). The color scores in ESCC tissues were significantly higher than those in para-tumor tissues, whereas the area scores were similar in different groups. In addition, the expression of GNA11 protein was distinctly higher in ESCC patients of pathological grade III compared with pathological grade I and II patients in color score (p < 0.05). The area score showed boundary difference in ESCC patients of clinical stage 1–2 compared with clinical stage 3–4 (p = 0.05). And the difference in final score was only observed in ESCC patients of pathological grade I compared with pathological grade III (p < 0.05). The color score and final score in ESCC patients with clinical stage 3–4 were also higher than those in clinical stage 1–2 patients (p < 0.05). Figure 7 includes representative immunostaining for GNA11 protein in ESCC tissues and corresponding para-tumor tissues.
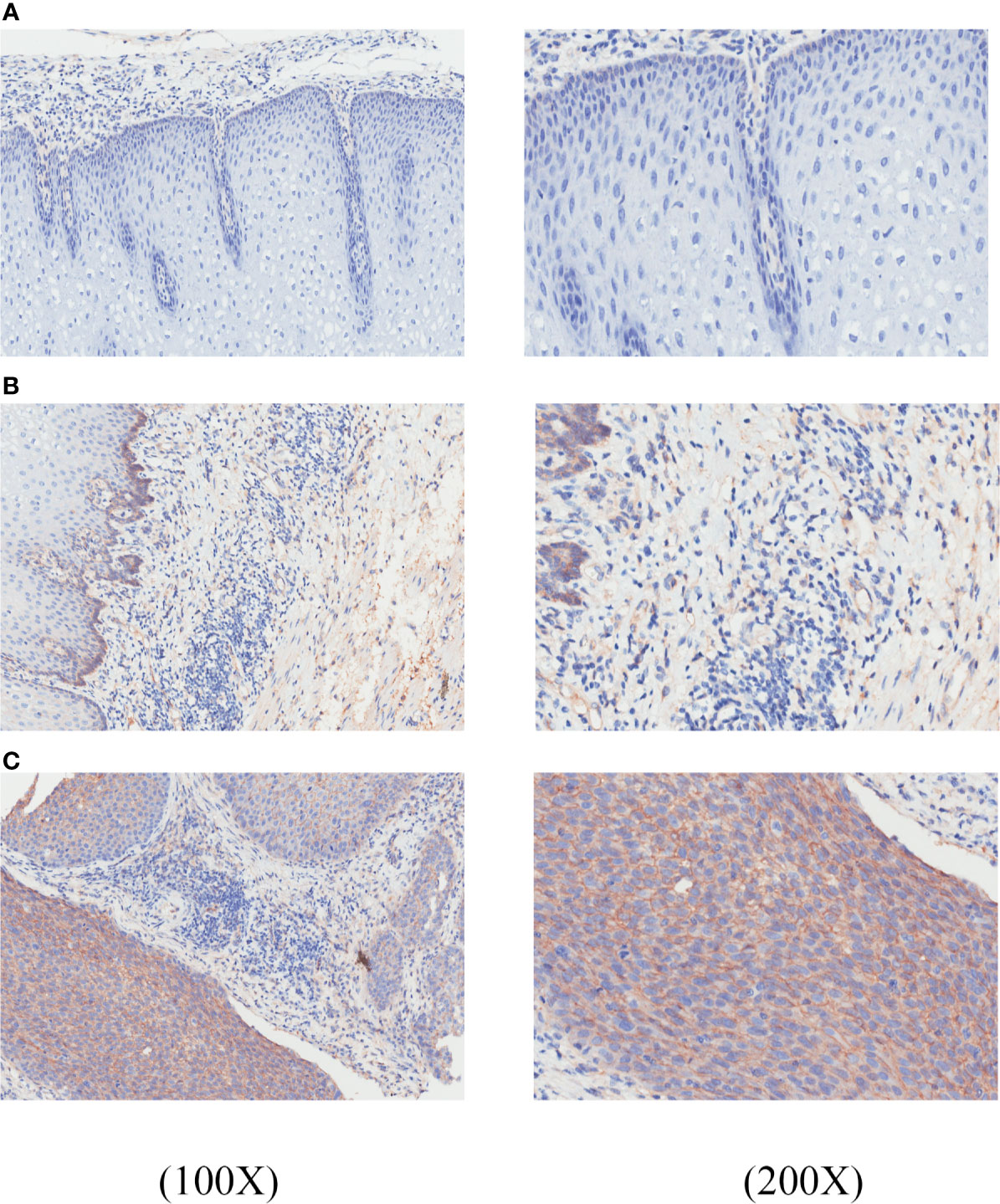
Figure 7 GNA11 is overexpressed in ESCC tissues compared with corresponding para-tumor tissues by immunohistochemistry. The histopathology of these tissue samples was tested by hematoxylin and eosin (H&E) staining. (A) A representative negative expression of GNA11 protein in para-tumor tissues at ×100 and ×200 magnification. (B) Weak positive expression of GNA11 protein in representative para-tumor tissues at ×100 and ×200 magnification. (C) Positive expression of GNA11 protein in representative ESCC tissues at ×100 and ×200 magnification. ESCC, esophageal squamous cell carcinoma.
Currently, the discovery of reliable biomarkers is still an important goal for the diagnosis of ESCC. However, only a few biomarkers had been used in the clinical detection of ESCC due to the limitation in study population or diagnostic value. So far, only a few tumor-associated proteins have been approved to be used as biomarkers in cancer (26). Therefore, it is necessary to find autoantibodies to TAAs as effective and reliable biomarkers. Protein microarray is an emerging high-throughput technique for protein detection and analysis with the merits of low sample consumption, simultaneous detection of multiple proteins, automation, and high sensitivity (27, 28). It has been widely used to detect protein biomarkers in a variety of cancers such as lung cancer, gastric cancer, and colorectal cancer (29–31). In the current study, the protein microarray containing 154 recombinant proteins was customized to screen out potential TAAs as biomarkers for the diagnosis of ESCC by using ROC analysis and Wilcoxon test. Through an extensive screening and validation, anti-GNA11 autoantibody was finally identified as a potential biomarker for the diagnosis of ESCC. Our study demonstrated that the positive rate of anti-GNA11 autoantibody was 38.24% (65/170), which was significantly higher than that in sera of normal controls 11.18% (19/170) (p < 0.05). Besides, our team further explored the performance of anti-GNA11 autoantibody as a biomarker in different subgroups including lymphatic metastasis, differentiation, distance metastasis, family tumor history, TNM stage, gender, and age. However, there was no obvious difference to distinguish these subgroups. Similar results had been seen in other studies about ESCC detection. It was reported that autoantibodies to p53, NY-ESO-1, matrix metalloproteinase-7 (MMP-7), heat shock protein 70 (Hsp70), and peroxiredoxin VI (Prx VI) had no difference in the detection of early-stage and late-stage ESCC patients (32). Chen et al. indicated that tumor-associated autoantibodies against Fascin also showed no difference in histological grade or TNM stage (14). It can be inferred that anti-GNA11 autoantibody may have a relation to the occurrence of ESCC rather than the progression from results above. Through bioinformatics analysis, our team also found that the mRNA level of GNA11 in ESCC is significantly higher than that in the normal control group. IHC analysis further confirmed that GNA11 protein was overall more abundant in ESCC tissues compared with para-tumor tissues. Multiple expression level analyses showed that anti-GNA11 autoantibody is elevated in ESCC patients.
GNA11 involves signaling pathways associated with cell survival including PI3K, RAS, and MAPK; and mutations of the corresponding genes could give the cancer cells a selective growth advantage that develop in a restricted nutritional condition (33). Mutations in GNA11 will lead to malformations and overgrowth of capillary in capillary malformation patients (34). Some studies have demonstrated that GNAQ/11 mutation occurs at the early stage of uveal melanoma and clonal expansion is possible only after GNAQ/11 mutation happens in normal cells (35, 36). Another study indicated that hotspot mutations of Gαq and Gα11 (R183 and Q209) that occurred in uveal melanoma acting as cancer driver genes will destroy the function of GTPase in the MAPK/MEK/ERK pathway, and the implementation of GNAQ/GNA11 mutation analysis in clinical diagnosis processes might be helpful in making treatment decisions (37). GNA* (GNAS, GNAQ, or GNA11) aberrations in gastrointestinal excluding appendiceal and colorectal cancer account for 7.3% in all GNA* mutated tumors, and there is a trend toward lower median overall survival in patients with GNA* mutated tumors compared with GNA* wild-type tumors (38). Gα11 is coupled to gonadotropin-releasing hormone receptor (GnRHR) and also involved in negatively regulating the growth of human breast epithelial cells through GnRH–GnRHR system (39). Studies suggested that the methylation of GNA11 resulted in reduced mRNA expression in breast cancer, which was of great help in the growth of human breast cancer cells (40). It was reported that 80% uveal melanomas have GNAQ or GNA11 mutations, which were potential drivers of MAPK activation; and a randomized phase II trial of selumetinib, a selective MEK inhibitor, has shown prospective therapeutic effect for uveal melanomas (41, 42). Consistent with the above studies, we also detected elevated levels of anti-GNA11 autoantibody in sera of ESCC patients, which may indicate changes in GNA11 gene during the original initiation of ESCC.
The present study reports the potential value of anti-GNA11 autoantibody in the diagnosis of ESCC patients from multiple levels. Anti-GNA11 autoantibody was initially screened out by protein microarray, and the performance of the autoantibody was further verified in two stages to obtain stable and reliable results. From multiple level analyses, we systematically confirmed that anti-GNA11 autoantibody is elevated in ESCC patients and may be a novel marker for the diagnosis of ESCC patients. However, some other studies on GNA11 mainly focus on the influence of its mutation on melanoma, and there are few studies working on GNA11 in ESCC. We believe that our subsequent studies on the mechanism of GNA11 in the occurrence and development of ESCC will fill this gap and provide more data to support our current findings.
In summary, this study has investigated the expression of GNA11 in ESCC from multiple levels. The results have demonstrated that anti-GNA11 autoantibody can be used as a potential biomarker in the detection of ESCC patients.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The studies involving human participants were reviewed and approved by the Institution Review Board of Zhengzhou University. The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
PW, XY, and JZ conceived the design of the current study. HW and GS conducted experiments and drafted the manuscript. QY and CC participated in the data analysis. HY, XW, JS, and LD helped to draft the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by The Funded Project of International Training of High-level talents in Henan Province, Zhengzhou Major Project for Collaborative Innovation (18XTZX12007) and the Major Project of Science and Technology in Henan Province (161100311400).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors thank Dr. Ying Peng for his helpful suggestions and comments on this manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.661043/full#supplementary-material
Supplementary Figure 1 | The expression of GNA11 protein in the tissues of different organs.
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2018) 68(6):394–424. doi: 10.3322/caac.21492
2. Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Pineros M, et al. Estimating the Global Cancer Incidence and Mortality in 2018: GLOBOCAN Sources and Methods. Int J Cancer (2019) 144(8):1941–53. doi: 10.1002/ijc.31937
3. Zheng RS, Sun KX SWZ. Report of Cancer Epidemiology in China, 2015. Chin J Oncol (2019) 41:19–28. doi: 10.3760/cma.j.issn.0253-3766.2019.01.005
4. Cao X. X. S. Incidence and Trend of Esophageal Cancer. Chin J Clin Oncol (2016) 43(21):932–6. doi: 10.3969/j.issn.1000-8179.2016.21.984
5. Enzinger PC, Mayer RJ. Esophageal Cancer. N Engl J Med (2003) 349(23):2241–52. doi: 10.1056/NEJMra035010
6. Kim T, Grobmyer SR, Smith R, Ben-David K, Ang D, Vogel SB, et al. Esophageal Cancer–the Five Year Survivors. J Surg Oncol (2011) 103(2):179–83. doi: 10.1002/jso.21784
7. Wang GQ, Jiao GG, Chang FB, Fang WH, Song JX, Lu N, et al. Long-Term Results of Operation for 420 Patients With Early Squamous Cell Esophageal Carcinoma Discovered by Screening. Ann Thorac Surg (2004) 77(5):1740–4. doi: 10.1016/j.athoracsur.2003.10.098
8. Law S, Wong J. The Current Management of Esophageal Cancer. Adv Surg (2007) 41:93–119. doi: 10.1016/j.yasu.2007.05.007
9. Tan EM. Autoantibodies as Reporters Identifying Aberrant Cellular Mechanisms in Tumorigenesis. J Clin Invest (2001) 108(10):1411–5. doi: 10.1172/jci14451
10. Tan EM, Zhang J. Autoantibodies to Tumor-Associated Antigens: Reporters From the Immune System. Immunol Rev (2008) 222:328–40. doi: 10.1111/j.1600-065X.2008.00611.x
11. Macdonald IK, Parsy-Kowalska CB, Chapman CJ. Autoantibodies: Opportunities for Early Cancer Detection. Trends Cancer (2017) 3(3):198–213. doi: 10.1016/j.trecan.2017.02.003
12. Anderson KS, LaBaer J. The Sentinel Within: Exploiting the Immune System for Cancer Biomarkers. J Proteome Res (2005) 4(4):1123–33. doi: 10.1021/pr0500814
13. Ye L, Guan S, Zhang C, Lee KH, Sun S, Wei J, et al. Circulating Autoantibody to FOXP3 may be a Potential Biomarker for Esophageal Squamous Cell Carcinoma. Tumour Biol (2013) 34(3):1873–7. doi: 10.1007/s13277-013-0729-8
14. Chen WX, Hong XB, Hong CQ, Liu M, Li L, Huang LS, et al. Tumor-Associated Autoantibodies Against Fascin as a Novel Diagnostic Biomarker for Esophageal Squamous Cell Carcinoma. Clinics Res Hepatol Gastroenterol (2017) 41(3):327–32. doi: 10.1016/j.clinre.2016.10.011
15. Li L, Liu M, Lin JB, Hong XB, Chen WX, Guo H, et al. Diagnostic Value of Autoantibodies Against Ezrin in Esophageal Squamous Cell Carcinoma. Dis Markers (2017) 2017:2534648. doi: 10.1155/2017/2534648
16. Xu YW, Liu CT, Huang XY, Huang LS, Luo YH, Hong CQ, et al. Serum Autoantibodies Against STIP1 as a Potential Biomarker in the Diagnosis of Esophageal Squamous Cell Carcinoma. Dis Markers (2017) 2017:5384091. doi: 10.1155/2017/5384091
17. Zhang B, Zhang ZL, Zhang XF, Gao X, Kernstine KH, Zhong L. Serological Antibodies Against LY6K as a Diagnostic Biomarker in Esophageal Squamous Cell Carcinoma. Biomarkers (2012) 17(4):372–8. doi: 10.3109/1354750x.2012.680609
18. Zhou JH, Zhang B, Kernstine KH, Zhong L. Autoantibodies Against MMP-7 as a Novel Diagnostic Biomarker in Esophageal Squamous Cell Carcinoma. World J Gastroenterol (2011) 17(10):1373–8. doi: 10.3748/wjg.v17.i10.1373
19. Wang K, Li M, Qin J, Sun G, Dai L, Wang P, et al. Serological Biomarkers for Early Detection of Hepatocellular Carcinoma: A Focus on Autoantibodies Against Tumor-Associated Antigens Encoded by Cancer Driver Genes. Cancers (Basel) (2020) 12(5):1271. doi: 10.3390/cancers12051271
20. Sun G, Ye H, Wang X, Cheng L, Ren P, Shi J, et al. Identification of Novel Autoantibodies Based on the Protein Chip Encoded by Cancer-Driving Genes in Detection of Esophageal Squamous Cell Carcinoma. Oncoimmunology (2020) 9(1):1814515. doi: 10.1080/2162402X.2020.1814515
21. van Nistelrooij AM, van Marion R, Biermann K, Spaander MC, van Lanschot JJ, Wijnhoven BP, et al. Early Onset Esophageal Adenocarcinoma: A Distinct Molecular Entity? Oncoscience (2016) 3(1):42–8. doi: 10.18632/oncoscience.290
22. Ma Y, Wang X, Qiu C, Qin J, Wang K, Sun G, et al. Using Protein Microarray to Identify and Evaluate Autoantibodies to Tumor-Associated Antigens in Ovarian Cancer. Cancer Sci (2020) 112(2):537–49. doi: 10.1111/cas.14732
23. Wang S, Qin J, Ye H, Wang K, Shi J, Ma Y, et al. Using a Panel of Multiple Tumor-Associated Antigens to Enhance Autoantibody Detection for Immunodiagnosis of Gastric Cancer. Oncoimmunology (2018) 7(8):e1452582. doi: 10.1080/2162402x.2018.1452582
24. Wang P, Song C, Xie W, Ye H, Wang K, Dai L, et al. Evaluation of Diagnostic Value in Using a Panel of Multiple Tumor-Associated Antigens for Immunodiagnosis of Cancer. J Immunol Res (2014) 2014:512540. doi: 10.1155/2014/512540
25. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A Web Server for Cancer and Normal Gene Expression Profiling and Interactive Analyses. Nucleic Acids Res (2017) 45(W1):W98–102. doi: 10.1093/nar/gkx247
26. Polanski M. Anderson NL. A List of Candidate Cancer Biomarkers for Targeted Proteomics. Biomark Insights (2007) 1:1–48. doi: 10.1177/117727190600100001
27. Mitchell P. A Perspective on Protein Microarrays. Nat Biotechnol (2002) 20(3):225–9. doi: 10.1038/nbt0302-225
28. Rosenberg JM, Utz PJ. Protein Microarrays: A New Tool for the Study of Autoantibodies in Immunodeficiency. Front Immunol (2015) 6:138. doi: 10.3389/fimmu.2015.00138
29. Pan J, Song G, Chen D, Li Y, Liu S, Hu S, et al. Identification of Serological Biomarkers for Early Diagnosis of Lung Cancer Using a Protein Array-Based Approach. Mol Cell Proteomics (2017) 16(12):2069–78. doi: 10.1074/mcp.RA117.000212
30. Yang L, Wang J, Li J, Zhang H, Guo S, Yan M, et al. Identification of Serum Biomarkers for Gastric Cancer Diagnosis Using a Human Proteome Microarray. Mol Cell Proteomics (2016) 15(2):614–23. doi: 10.1074/mcp.M115.051250
31. Babel I, Barderas R, Diaz-Uriarte R, Martinez-Torrecuadrada JL, Sanchez-Carbayo M, Casal JI. Identification of Tumor-Associated Autoantigens for the Diagnosis of Colorectal Cancer in Serum Using High Density Protein Microarrays. Mol Cell Proteomics (2009) 8(10):2382–95. doi: 10.1074/mcp.M800596-MCP200
32. Xu YW, Peng YH, Chen B, Wu ZY, Wu JY, Shen JH, et al. Autoantibodies as Potential Biomarkers for the Early Detection of Esophageal Squamous Cell Carcinoma. Am J Gastroenterol (2014) 109(1):36–45. doi: 10.1038/ajg.2013.384
33. Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW. Cancer Genome Landscapes. Sci (New York NY) (2013) 339(6127):1546–58. doi: 10.1126/science.1235122
34. Couto JA, Ayturk UM, Konczyk DJ, Goss JA, Huang AY, Hann S, et al. A Somatic GNA11 Mutation Is Associated With Extremity Capillary Malformation and Overgrowth. Angiogenesis (2017) 20(3):303–6. doi: 10.1007/s10456-016-9538-1
35. Robertson AG, Shih J, Yau C, Gibb EA, Oba J, Mungall KL, et al. Integrative Analysis Identifies Four Molecular and Clinical Subsets in Uveal Melanoma. Cancer Cell (2018) 33(1):151. doi: 10.1016/j.ccell.2017.12.013
36. Field MG, Durante MA, Anbunathan H, Cai LZ, Decatur CL, Bowcock AM, et al. Punctuated Evolution of Canonical Genomic Aberrations in Uveal Melanoma. Nat Commun (2018) 9(1):116. doi: 10.1038/s41467-017-02428-w
37. Schneider B, Riedel K, Zhivov A, Huehns M, Zettl H, Guthoff RF, et al. Frequent and Yet Unreported GNAQ and GNA11 Mutations are Found in Uveal Melanomas. Pathol Oncol Res (2019) 25(4):1319–25. doi: 10.1007/s12253-017-0371-7
38. Parish AJ, Nguyen V, Goodman AM, Murugesan K, Frampton GM, Kurzrock R. GNAS. GNAQ, and GNA11 Alterations in Patients With Diverse Cancers. Cancer (2018) 124(20):4080–9. doi: 10.1002/cncr.31724
39. Hsieh KP, Martin TF. Thyrotropin-Releasing Hormone and Gonadotropin-Releasing Hormone Receptors Activate Phospholipase C by Coupling to the Guanosine Triphosphate-Binding Proteins Gq and G11. Mol Endocrinol (Baltimore Md) (1992) 6(10):1673–81. doi: 10.1210/mend.6.10.1333052
40. Asada K, Miyamoto K, Fukutomi T, Tsuda H, Yagi Y, Wakazono K, et al. Reduced Expression of GNA11 and Silencing of MCT1 in Human Breast Cancers. Oncology (2003) 64(4):380–8. doi: 10.1159/000070297
41. Carvajal RD, Sosman JA, Quevedo JF, Milhem MM, Joshua AM, Kudchadkar RR, et al. Effect of Selumetinib vs Chemotherapy on Progression-Free Survival in Uveal Melanoma: A Randomized Clinical Trial. JAMA (2014) 311(23):2397–405. doi: 10.1001/jama.2014.6096
Keywords: GNA11, tumor-associated antigen, autoantibody, immunodiagnosis, esophageal squamous cell carcinoma
Citation: Wang H, Yang X, Sun G, Yang Q, Cui C, Wang X, Ye H, Dai L, Shi J, Zhang J and Wang P (2021) Identification and Evaluation of Autoantibody to a Novel Tumor-Associated Antigen GNA11 as a Biomarker in Esophageal Squamous Cell Carcinoma. Front. Oncol. 11:661043. doi: 10.3389/fonc.2021.661043
Received: 30 January 2021; Accepted: 18 August 2021;
Published: 10 September 2021.
Edited by:
Wei Wei, Institute for Systems Biology (ISB), United StatesReviewed by:
Pierre Busson, Centre National de la Recherche Scientifique (CNRS), FranceCopyright © 2021 Wang, Yang, Sun, Yang, Cui, Wang, Ye, Dai, Shi, Zhang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peng Wang, d2FuZ3BlbmcxNjU4QGhvdG1haWwuY29t
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.