
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol. , 21 April 2021
Sec. Molecular and Cellular Oncology
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.659661
This article is part of the Research Topic Cancer Evolution: from biological insights to therapeutic opportunities View all 30 articles
 Yuanyuan Qin†
Yuanyuan Qin† Hong Yuan†
Hong Yuan† Xu Chen†
Xu Chen† Xinyi Yang
Xinyi Yang Zhengcao Xing
Zhengcao Xing Yajie Shen
Yajie Shen Wanying Dong
Wanying Dong Siming An
Siming An Yitao Qi*
Yitao Qi* Hongmei Wu*
Hongmei Wu*Breast cancer has the highest incidence among cancers and is the most frequent cause of death in women worldwide. The detailed mechanism of the pathogenesis of breast cancer has not been fully elucidated, and there remains a lack of effective treatment methods for the disease. SUMOylation covalently conjugates a large amount of cellular proteins, and affects their cellular localization and biological activity to participate in numerous cellular processes. SUMOylation is an important process and imbalance of SUMOylation results in the progression of human diseases. Increasing evidence shows that numerous SUMOylated proteins are involved in the occurrence and development of breast cancer. This review summarizes a series of studies on protein SUMOylation in breast cancer in recent years. The study of SUMOylated proteins provides a comprehensive understanding of the pathophysiology of breast cancer and provides evolving therapeutic strategies for the treatment of breast cancer.
Breast cancer is the most common type of cancer (1). Approximately 1.2 million women worldwide suffer from breast cancer every year in the world, and about one-half of these patients die within 10 years of diagnosis (2). According to the latest cancer data released by the National Cancer Center of China and the American Cancer Society in 2019, breast cancer ranks first among all new cancer diagnoses in women and second in terms of mortality, accounting for 15-30% of deaths from newly diagnosed cancers (3–5). It is estimated that the incidence and mortality of breast cancer will increase over the next 5-10 years (6). Furthermore, the morbidity of breast cancer is highest in Europe, North America, New Zealand, and Australia, and its mortality is highest in sub-Saharan Africa and some Asian countries (1, 7). These data suggest that breast cancer is still a global public health problem.
Breast cancer is a heterogeneous disease that can be classified into four subtypes according to histological features, including luminal A, luminal B, human epidermal growth factor receptor 2 (HER2)-positive and triple-negative breast cancer (TNBC). Luminal A and luminal B tumors are mostly ER-positive, and the difference between them is that luminal A tumors are low grade tumors, while luminal B tumors are high grade tumors. HER2-positive tumors exhibit overexpression of ERBB2 genes (1, 8). TNBC is a heterogeneous and aggressive form of breast cancer in which the cells do not express estrogen receptor α (ERα), progesterone receptor (PR), or HER2. TNBC accounts for 15% of breast carcinomas and 70-80% of basal-like breast cancers, and it is refractory to therapy (9, 10). Breast cancer is often accompanied with gene mutations, which are mainly divided into two types-functional gain mutations of oncogenes and functional loss mutations of tumor suppressor genes. BRCA1 and BRCA2 mutation play an important role in genetic susceptibility of breast cancer progression. Exon 4 and intron 3 of TP53 gene are frequently mutated in breast cancer, especially in TNBC. Most breast cancer cases have nothing to do with high penetrance mutations such as BRCA1, BRCA2, and TP53. Genes with low penetrance such as androgen receptor (AR), checkpoint kinase 2 (CHEK2), E-cadherin, Nijmegen breakage syndrome 1 (NBS1), RAD50, BRCA1 interacting protein C-terminal helicase 1 (BRIP1), and partner and localizer of BRCA2 (PALB2) are frequently mutated in the general population and play an important roles in the occurrence of breast cancer (11).
At present, the therapeutics of breast cancer mainly focus on surgery (12), radiation therapy (13), chemotherapy (14), endocrine therapy (15) and targeted therapy (16). Surgery is the most significant treatment (17), and remains the most accurate staging method for non-metastatic malignancies (12). Radiation therapy reduces local recurrence and breast cancer mortality after breast conservation after mastectomy (18), and radiation therapy after mastectomy is the standard of care for advanced breast cancer (13, 17). Chemotherapy is one of the main methods to improve survival and prognosis of patients through destroy cancer cells that have spread to various parts of the body. Current chemotherapeutic agents for breast cancer include alkylating agent cyclophosphamide (19), antimetabolic agent gemcitabine (14), anthracycline agent doxorubicin and paclitaxel agent paclitaxel (20). The advantage of endocrine therapy is that there are fewer adverse reactions and long drug maintenance. At the same time, endocrine therapy generally results in drug resistance, which is an urgent problem that needs to be solved. Targeted therapy can kill tumor cells efficiently and selectively with less adverse effects than chemotherapy. The drugs currently used in breast cancer include receptor tyrosine kinase inhibitor lapatinib, HER2 monoclonal antibody trastuzumab, mTOR inhibitor everolimus and the CDK4/6 inhibitor palbociclib (16). Although there are numerous studies on the treatment of breast cancer, many problems still need to be resolved.
SUMO proteins, including SUMO1, SUMO2 and SUMO3, constitute a class of proteins with a molecular weight of approximately 11 KD that have similar structure with ubiquitin. The mature SUMO proteins are activated by E1, which is composed of two subunits, SAE1/Aos1 and SAE2/Uba2. Subsequently, SUMOs are transferred to the Ubc9, the single E2, and finally conjugated to the specific lysine residues of the substrate protein with the help of E3 ligases, which includes members of the protein inhibitor of activated STAT (PIAS) family, Ran binding protein 2 (RanBP2), and a few other E3 ligases. SUMOylation is a dynamic and reversible process, and the modification is reversible because of the regulation of SUMO/sentrin-specific protease (SENP), which deconjugates attached molecules from substrates and is required for the maturation of SUMO proteins (Figure 1). The well-known protease families include Ulp1 and Ulp2 in yeast and SENPs (SENP1-3 and SENP5-7) in mammals, and they are involved in embryonic development and human diseases (21, 22). SUMOylation regulates a variety of biological processes including cell division, DNA replication and repair, signal transduction and cell metabolism (23). Typical SUMOylation is observed during cellular activities because a rapid modification of even a small portion of targets is sufficient to produce significant functional changes (24).
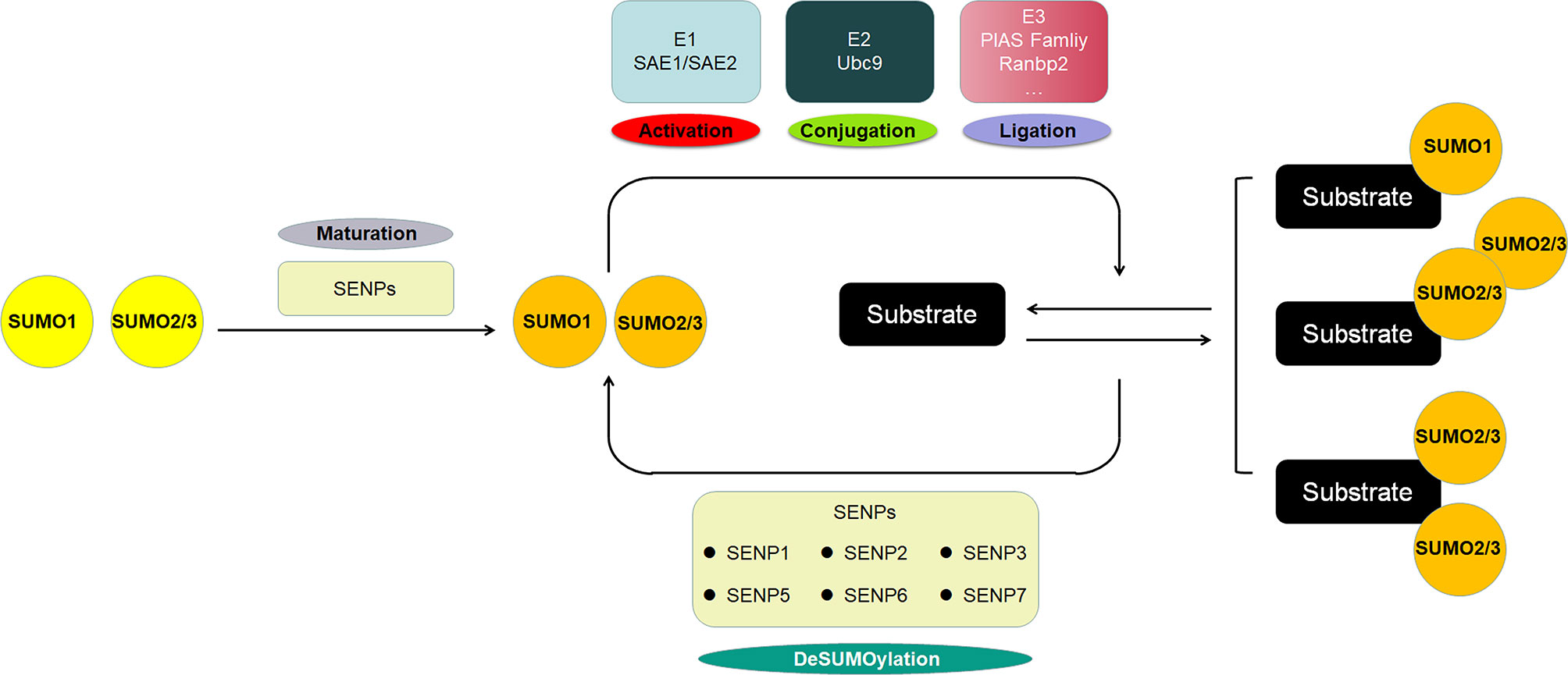
Figure 1 The scheme of SUMOylation pathway. The SUMO protein precursor is cleaved and matured by SENP, then activated by E1, transferred to E2, and ultimately ligated to the target protein by E3. SUMO1 modification is usually conjugated as a monomer, whereas SUMO2/3 modification is often form poly-chain. The SENP family deconjugates the SUMO protein from the substrate to deSUMOylate target protein.
In recent years, additional studies have shown that SUMOylation and its pathways are associated with human diseases (25–28). Various types of stress induce the upregulated SUMOylation, making SUMOylation a critical mechanism to protect cells from stress-induced apoptosis or cell death (29, 30). The low survival rate of patients with hepatocellular carcinoma is reported to be related to the overexpression of SUMO2 and E1 enzyme Uba2 subunits (31). The overexpression of E2 enzyme Ubc9 was found in human lung and neck cancers (32). The E3 enzyme PIAS3 was overexpressed in prostate, lung, colon, brain and breast cancers (33). Many recent reports have shown that SENPs and other SUMO-related proteins regulate the occurrence and development of breast cancer by modulating protein modifications (23). This suggests that SUMOylation is likely to play an important role in regulating breast cancer. In this review, we focus on the linkage between breast cancer and SUMOylation pathway to explore the role of SUMOylation in the occurrence and development of breast cancer.
SAE2 is a SUMO-activating enzyme (E1) in mammals (23). It was found that the global SUMO-2/3 modification was increased, but the SUMO2/3 modification of SAE2 was decreased in highly metastatic breast cancer cells (34). These two seemingly contradictory conclusions can be explained as follows: SAE2 can be SUMOylated at the K236 site, which alters its enzymatic activity and inhibits SUMO transfer from E1 to Ubc9; as a result, the decrease in SAE2 SUMOylation enhances global SUMOylation to some extent (35). Furthermore, SAE2 is required for Myc-dependent tumor growth in mice, and the analysis of gene expression in Myc-high human breast cancer suggests that low expression levels of SAE1 and SAE2 are correlated with longer metastasis-free survival of breast cancer patients (36). These results indicated that SAE1 and SAE2 may inhibit the development of tumor metastasis in breast cancer with high Myc expression (Table 1).
Ubc9 is the only SUMO-conjugating enzyme (23). It was observed that the expression of Ubc9 is 5.7-fold higher in breast cancer tissues, and ectopic expression of Ubc9 promotes tumor growth and invasion in an animal model (51, 52). In addition, it was found that Ubc9 positively regulates Bcl2, a well-known tumor promoter, indicating that Ubc9 may play a tumor promoter role in breast cancer development (53). Additional results showed that Ubc9 was downregulated by the tumor suppressor miR-30e and upregulated by cell division cycle 2 (Cdc2) in breast cancer (51, 54). Moreover, Ubc9 gene variants have been shown to be associated with the risk of grade 1 breast cancer (55). Recently, it was shown that the expression and activity of Ubc9 played a critical role in breast tumorigenesis and responded to anticancer drugs. It was reported that ERα and NF-Y bound directly to the proximal promoter of Ubc9 and were essential for the in vivo expression of Ubc9 through transcriptional regulation (41, 56), and the overexpression of Ubc9 increased ERα-mediated transcriptional activity via enhanced SUMOylation in MCF-7 breast cancer cells, suggesting a possible synergy between Ubc9 and the promoting factor during breast cancer development (57). These findings contribute to a better understanding of Ubc9 regulation in breast cancer cells and indicate that Ubc9 is a potential therapeutic target in breast cancer.
PIAS1 is a SUMO-ligating enzyme (58). Some reports have shown that PIAS1 is highly expressed in breast cancer and regulates breast cancer tumorigenesis (59). It was found that PIAS1 can enhance the expression of breast cancer signature genes, including ESR1 and CCND2, and the oncogene AIB1 (59, 60). However, PIAS1 can also cooperate with TNFγ to regulate SnoN SUMOylation and suppress the EMT, inhibiting the growth and invasion of MDA-MB-231 cell-derived organoids (44, 61). The role of PIAS1 in breast cancer may be a double-edged sword, and further investigations are required to clarify its regulatory mechanism.
Previous studies showed that SENP1 is highly expressed in human prostate cancer cells (62), lung cancer and colon cancer tissues (63, 64). SENP1 is also upregulated in TNBC tissues, and depletion of SENP1 attenuates TNBC cell proliferation and migration, tumor growth and metastasis (65). SENP1 may function by deSUMOylating related substrates. For example, a study found that SENP1 can deSUMOylate HIF-1α to enhance HIF-1α stabilization and ultimately promote breast cancer metastasis (66). Furthermore, SENP1 can deSUMOylate and regulate the protein activity and oncogenic function of the isomerase Pin1, which is an important regulator of cellular processes involving Pro-directed phosphorylation in breast cancer (67). These results suggest a critical role for SENP1 in TNBC cell proliferation, breast cancer formation and migration.
SENP2 plays important roles in embryonic development (21) and myogenesis (22), and reversing SUMOylation of potassium channels may present a novel approach for treating SUDEP (sudden unexplained death in epilepsy) patients (25–27). SENP2 has been reported to play a crucial role in hepatocellular carcinoma (HCC) cell growth by modulating β-catenin stability (68). Moreover, SENP2 functions as a suppressor in bladder cancer metastasis partially by inhibiting the expression of MMP13 (69). Considering these reports, some studies have focused on the correlation between SENP2 and breast cancer. One study showed that SENP2 significantly represses estrogen-dependent and estrogen-independent proliferation of MCF-7 cells and revealed a novel property of SENP2 as a typical transcription coregulator (70). The polymorphic SENP2 genes examined to date cannot be used as independent markers of breast cancer, but studies using these forms may be useful in identifying a set of clinical markers helpful for breast cancer diagnosis and treatment (71). However, no in-depth studies on the specific role and mechanism of SENP2 in the development and progression of breast cancer have been reported, and further studies need to be performed to determine the critical role of SENP2 in breast cancer.
Several studies have focused on the relationship between SENP5 and the development of breast cancer. One study showed that SENP5 silencing inhibits breast cancer cell growth, proliferation, migration and invasion by regulating the expression level of TGFβRI (48). Recently, another study showed that the expression levels of SENP5 were negatively correlated with the survival of breast cancer patients, and suggested SENP5 as a unique prognostic biomarker (72). These results were consistent with other findings demonstrating that SENP5 silencing reduced cell migration and invasion and indicating some interplay between TGFβRI and SENP5 (48). These results suggest that SENP5 acts as a tumor promoter in breast cancer development.
Different SENP7 isoforms, including SENP7S and SENP7L, have a different subcellular localizations and biological functions (73). SENP7S and SENP7L are two isoforms that have been intensively studied. SENP7S is mainly located in the cytoplasm and is highly expressed in mammary epithelia but expressed at low levels in precancerous ductal carcinoma and is lost in invasive breast cancer (73). Another study revealed that deletion of SENP7S can directly enhance the tumorigenicity of MCF-10-2A cells. Mechanistically, SENP7 loss enhances the SUMOylation of β-catenin, and SUMOylated β-catenin is transported to the nucleus and promotes the transcription of oncogenes, including c-Myc, cyclin D, and Aurora kinase, ultimately promoting metastasis and invasion of breast cancer (38).
In contrast, SENP7L is highly expressed, promotes aberrant proliferation and initiates the EMT and invasion of breast cancer (73). It was reported that the SUMOylation of HP1α promoted HP1α localization to promoters and subsequently silenced genes. SENP7L deSUMOylated HP1α to release the inhibition of downstream genes and ultimately promote the proliferation and invasion of breast cancer cells (74, 75). Breast cancer patients who express low SENP7L exhibit higher survival rates after chemotherapy than patients who express high SENP7L (74). These results indicated that SENP7S acts as a tumor suppressor but that SENP7L plays a tumor-promoting role in breast cancer.
α-Catenin is an essential protein in adherent junctions, critical for the maintenance of cellular adhesion and polarity (76, 77), and has been recognized as a novel tumor suppressor gene (37, 78). α-Catenin plays a role in two different ways. In the traditional way, loss of α-Catenin specifically causes the loss of intracellular adhesion in E-cadherin-expressing breast cancer cells and induces further resistance to anoikis (79, 80). Another way is the adherent junction-independent pathway in which α-catenin suppresses E-cadherin-negative basal-like breast cancer by inhibiting NF-κB signaling (81). It was reported that α-catenin is SUMOylated, and SUMOylation stabilizes its interaction with IκBα, inhibiting the expression of NF-κB target genes (37, 79). A survival analysis showed a significant association between abnormal α-catenin expression and poor survival of breast cancer patients (82). In conclusion, α-catenin plays a significant role in breast cancer development, and its abnormal expression is associated with severe symptoms of breast cancer.
AMPK is an anabolic pathway inhibitor found in all eukaryotes that controls fatty sugar and lipid metabolism processes (39, 83). Furthermore, the expression levels of AMPK are upregulated in TNBC and can be regarded as biomarkers for TNBC (84). Many targets are regulated by AMPK. Phosphorylated AMPK inactivates the serine/threonine protein kinase Akt, which is involved in tumor progression, thereby inhibiting anoikis and impairing autophagy, ultimately inhibiting anchorage-independent growth and metastasis (85). Furthermore, some reports revealed that the LKB1-AMPK axis governs the mTORC1 pathway to regulate tumor growth (83). AMPKα1 can be SUMOylated, and its SUMOylation inhibits the response of AMPK towards mTORC1 signaling, suggesting that suppression of AMPKα1 SUMOylation can be applied to regulate AMPK activation and thus suppress breast cancer cell growth (39). These findings indicate that the SUMOylation of AMPKα1 can be a potential target for the treatment of breast cancer.
It was reported that women with BRCA1 germline mutations usually develop TNBC (86). Researchers have identified a consensus SUMO modification site localized in the amino-terminal region of BRCA1. In contrast to the SUMO mutation in this potential SUMO-acceptor site of the BRCA1 protein, the wild-type BRCA1 protein can bind to the unique SUMO E2 Ubc9 (87). It seems that BRCA1 may be SUMOylated; however, research has shown that SUMO1 binds to the SUMO-binding motifs in BRCA1 and represses BRCA1-mediated transcription by recruiting HDAC in a SUMO-independent manner (88). Taken together, these results indicate that BRCA1 SUMOylation needs further investigation and that BRCA1 may regulate breast cancer in a SUMO-dependent and SUMO-independent manner.
The transcription factor FOXM1 (forkhead box protein M1) is a critical regulator governing cell cycle pathway essential for mitosis, DNA replication, and cell proliferation (40). There are three main subtypes of FOXM1, of which FOXM1B is closely related to tumor growth and metastasis (89). FOXM1B can be SUMOylated on the K463 residue, and SUMOylation of FOXM1B is mediated by PIASy, and this SUMOylation is deconjugated by SENP2. SUMOylation of FOXM1B is necessary for its transcriptional activity, thus promoting the expression of its target gene JNK1 and repressing the expression of its targets MiR-200b/c and p21, ultimately promoting MCF-7 cell proliferation (40). Therefore, follow-up studies can be initiated directed toward the regulatory action of the SUMOylation of FOXM1B to explore strategies for the treatment of breast cancer.
Forkhead box protein P3 (FOXP3) is involved in regulatory T (Treg) cell development and inhibits tumorigenicity by downregulating oncogenes such as HER2/ErbB2 in breast cancer (89). Tumor immunotherapy has been successfully applied in the clinic. The role of Treg cells in immune suppression is well defined, and FOXP3 is a pivotal marker of Treg cells with immunosuppressive functions. In TNBC, the increase in FOXP3-positive Treg cells is associated with an improved survival rate (90). Furthermore, FOXP3 is modified by phosphorylation and acetylation, and the removal of phosphate and acetyl groups from FOXP3 results in the attenuated transcriptional activity of Ubc9 (41). Increasing evidence has shown that FOXP3 mainly binds to the FOX response element in the proximal promoter region to activate the transcription of the Ubc9 gene. These results suggested that FOXP3 may have physiological functions as a novel regulator in global SUMOylation and in other post-translational modification systems in breast cancer.
Myc is an oncogenic transcription factor and is frequently dysregulated in human cancers. The overexpression of Myc may contribute to the acquired drug resistance in ER-positive breast cancer. One of the possible mechanisms is that Myc can positively regulate HSP111, which is an estrogen-responsive gene and is associated with the poor prognosis for patients (91). It has been reported that Myc may be SUMOylated by SUMO1 and deSUMOylated by SENP1. Myc SUMOylation can regulate its stabilization. Furthermore, PIAS1 can enhance the stability of Myc and promote Myc-driven tumorigenesis by recruiting JNK1 to phosphorylate Myc at S62 (92). SAE2 inhibition switches the Myc transcriptional subprogram from promoting to suppressing activity. SAE2 is necessary for Myc-dependent tumor growth in mice, and a gene expression analysis of human breast cancer with high Myc showed that lower SAE1 and SAE2 abundance in tumors is associated with a longer survival period without metastasis (36). Therefore, suppression of SUMOylation may be worthy of study, and inhibition of Myc SUMOylation is a potential treatment for Myc-driven breast cancer.
The NF-κB essential modulator (NEMO) is a key activator of NF-κB signaling and IL6 secretion (93, 94). Once the IL6 receptor is activated, its downstream protein STAT3 is phosphorylated, and phosphorylated STAT3 binds to the PML promoter to activate PML expression (94). Several studies have shown that NEMO is regulated by SUMOylation, and the inhibition of NEMO SUMOylation suppresses the activation of NF-κB signaling in cells (95). Doxorubicin is a chemical drug commonly used for the treatment of breast cancer. However, breast cancer often develops treatment resistance, which leads to the recurrence and poor prognosis of the disease. Researchers found that SENP2 overexpression sensitized drug-resistant breast cancer cells to doxorubicin therapy. Mechanistically, the overexpression of SENP2 deconjugates the SUMOs of NEMO and inhibits NF-kB activation, especially in drug-resistant breast cancer cells. More importantly, when treated with an NF-κB pathway activator, the SENP2 overexpression-induced sensitivity of drug-resistant breast cancer cells to doxorubicin was eliminated (42). Taken together, these results suggest SENP2 activators may be used to treat doxorubicin-sensitive breast cancer patients, although this finding needs to be confirmed in clinical trials.
PES1 is a component of the nucleolar PeBoW complex (consisting of Pes1, Bop1 and WDR12) and is highly expressed in several kinds of cancers, including breast cancer (43, 96). PES1 promotes breast cancer development through multiple pathways. PES1 can directly bind to telomerase reverse transcriptase (TRET) and promote the formation of the TRET and TR complex, resulting in enhanced telomerase activity and telomere elongation, and inhibited cell senescence (96). In addition, PES1 can increase the expression of tumor-promoting ERα and decrease the expression of tumor-suppressive ERβ (97). Some studies demonstrated that in breast cancer cells, PES1 can be SUMOylated on K517, stabilizing PES1, which then promotes ERα transcription and inhibits ERα ubiquitination (42). Hence, the ultimate effect of PES1 SUMOylation is the acceleration of cell proliferation and cell cycle progression.
The promyelocytic leukemia (PML) protein is highly expressed in TNBC. A report showed that PML inhibition led to cell proliferation arrest and senescence by downregulating Myc and PIM1 kinase, followed by the subsequent accumulation of p27 (98). Another study found that PML can promote the expression of SOX9 and thus enable breast cancer cells to acquire cancer-initiating cells (CIC) properties (99). PML can be modified by SUMO1, SUMO2, and SUMO3 at K65, K160, and K490, respectively (100). Previous studies have shown that global SUMO2/3 modification is enhanced in metastatic breast cancer. Consistently, the upregulation of PML SUMO-2/3 modification has been observed in metastatic breast cancer cells (34). Hence, it is likely that the upregulation of the PML SUMO-2/3 modification may result in the increased metastatic capacity of breast cells, a supposition that requires further investigation.
Smurf2 (Smad ubiquitination regulatory factor 2) is a HECT (homology to E6 carboxy-terminus domain)-containing ubiquitin E3 ligase that mediates substrate proteins for ubiquitination and degradation via the proteasome pathway (101). Smurf2 is expressed at low levels in breast cancer, especially in TNBC, and acts as a tumor suppressor. Smurf2 is located in the nucleus in normal cells but exhibits significant cytoplasmic sequestration in breast cancer cells (102, 103). Smurf2 can be SUMOylated at K29 and K369, and its SUMOylation contributes to the downregulation of TGFβ signaling and inhibits the EMT in breast cancer cells. Mechanistic studies showed that the SUMO E3 ligase PIAS3 maintains breast cancer organoids through Smurf2 SUMOylation under noninvasive conditions (101). Collectively, these findings identify a novel role for PIAS3-mediated Smurf2 SUMOylation in the suppression of breast cancer cell invasion (46). These findings identify Smurf2 SUMOylation as a novel biomarker and suggest the regulation of Smurf2 SUMOylation as a targeted approach to breast cancer therapy.
Signal transducer and transcriptional activator (STAT) proteins, in particular STAT3 and STAT5, are continuously activated in many human cancers and are related to dysregulated cell proliferation and apoptosis (104). Drug-targeted activation of STAT3 and STAT5 has been an active subject of cancer studies. System XC-, a cysteine/glutamate antiporter, contributes to the redox balance and facilitates the adaptation of aggressive cancer cells to increased levels of ROS (reactive oxygen species) (105). Protein xCT is the main impact factor in system XC- (105), and insufficient xCT expression potentially blocks cancer cell proliferation and metastasis (106). Previous studies have shown that the acute blockade of STAT3 and STAT5 with SH-4-54, a small-molecule inhibitor targeting the SH2 domains of these two proteins, can increase xCT expression and thus improve system XC- activity in breast cancer cells. However, current studies have shown that the chronic treatment of SH-4-54 followed by the cloning and selection of resistant MDA-MB-231 cells leads to the opposite effects (107). In resistant MDA-MB-231 cells, chronic treatment with SH-4-54 downregulates constitutive STAT3 phosphorylation and thus increases intracellular ROS levels, resulting in the deSUMOylation of STAT5 and the subsequent phosphorylation of STAT5. Activated STAT5 leads to a reduction in xCT mRNA and protein, which eventually abrogates cell growth and migration (47). Further studies addressing the relationship between STAT3 and STAT5 SUMOylation and the development of breast cancer are still needed.
Transcription factor activator protein-2 (TFAP-2) activates transcription through GC-rich DNA sequences (108) and is important for cell proliferation and migration and xenograft outgrowth (49). Many solid cancers have an amplified CD44+/CD24- cancer stem cell (CSC) population that is relatively chemically resistant and leads to recurrence and metastasis. A durable response requires the development of therapeutics specific to CSCs (109). Recent evidence suggests that inhibiting the SUMOylation pathway inhibits tumor growth and invasion. It has been reported that in basal breast cancer, the inhibition of the SUMO pathway suppresses the expression of MMP14 and CD44, accompanied by decreased cell invasiveness and loss of CSC function (110). Another report showed that TFAP2A mediates SUMO pathway inhibition in breast cancer, indicating that TFAP2A may act as an upstream regulator of the SUMO pathway to regulate global SUMO modification in tumor cells and thus influence the cellular phenotype (111). Another study showed that SUMOylation of TFAP2A is necessary to maintain basal breast cancer phenotypes (49), suggesting that there may be a mutual regulatory relationship between TFAP2A and the SUMO pathway.
TP53 (p53), a well-known tumor suppressor, is a critical transcription factor that regulates the expression of numerous target genes to induce cell cycle arrest, apoptosis, senescence, and other anti-proliferative outcomes (50). p53 is the most frequently mutated gene in breast cancer. The incidence of mutations depends on the molecular subtype of breast cancer, most common in the TN subtype and least in the Luminal A subtype (112). A research showed that p53 was SUMOylated by SUMO1 (50), another research found that p53 was conjugated with SUMO2/3 (113). Both SUMOylation promotes p53 bind to the target gene promoter, thereby enhances p53-mediated transcription. Meanwhile, it is found that SENP1 abolished SUMOylation of p53 and promotes cancer cell proliferation (50). These studies indicated that SUMO and SENP1 dynamically regulate the SUMOylation of p53, involving the progression of breast cancer.
SUMOylation has been studied since its discovery, and the understanding of its biochemistry and enzymological mechanisms has been advanced. SUMOylation is an important factor in the regulation of intracellular protein function, and the functional activity of other proteins can also be regulated by various other mechanisms. Abnormal SUMOylation levels lead to the occurrence and development of various human diseases. Numerous important transcription factors have been reported to be SUMOylated during the development of breast cancer (Figure 2), indicating that SUMOylation affects the occurrence and development of breast cancer.
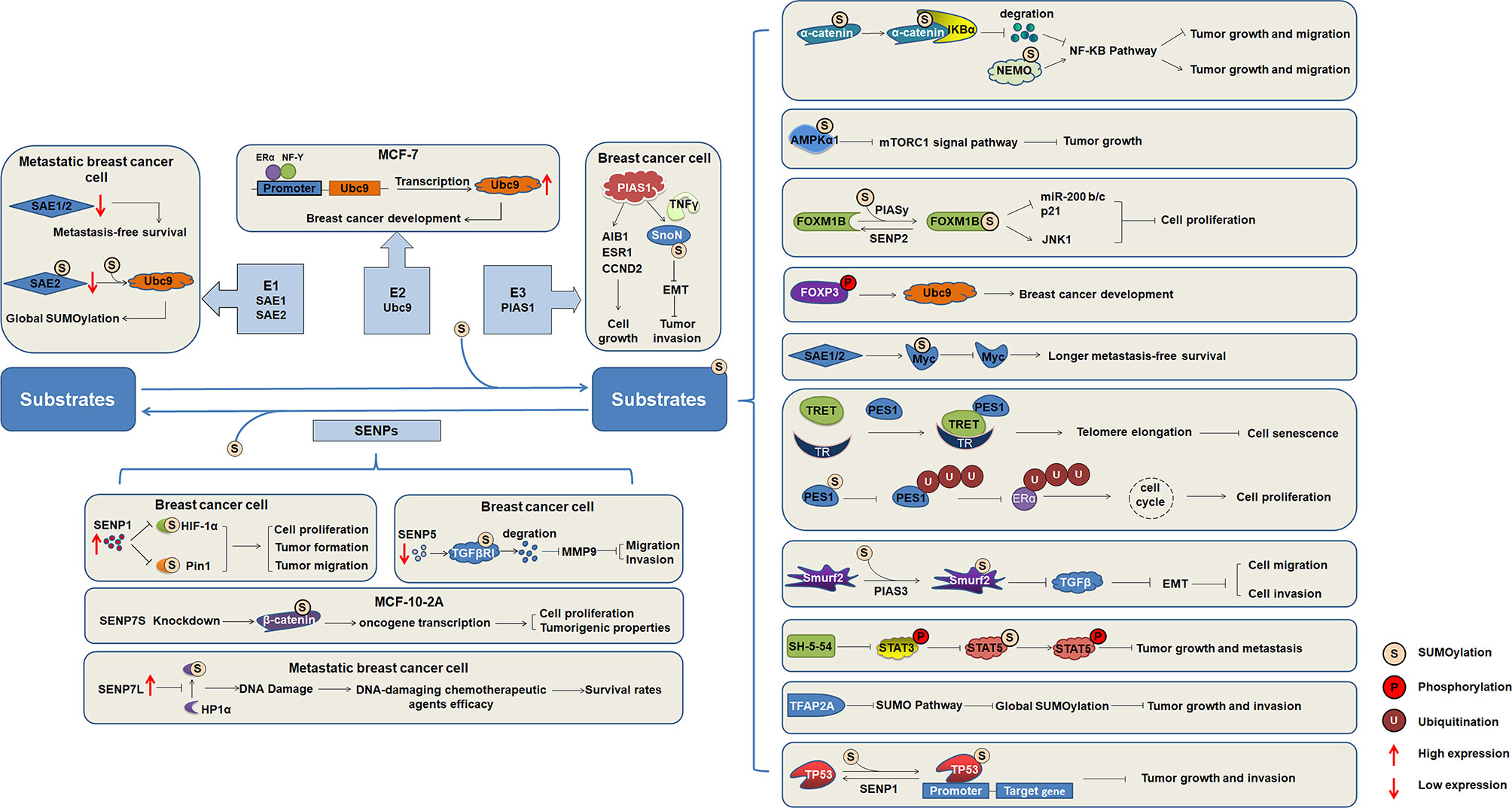
Figure 2 The regulatory mechanism of substrates SUMOylation in the occurrence and development of breast cancer. SUMO activating enzymes E1, conjugating enzyme Ubc9, and ligating enzyme PIAS1, as well as SENP1, SENP5, and SENP7 regulate the progression of breast cancer. Increasing number of target proteins are SUMOylated, and SUMOylation of substrates regulate the function and involved in the occurrence and development of breast cancer.
Global SUMOylation is greatly upregulated in metastatic breast cancer cells compared with nonmetastatic control cells. Substrates identified with altered SUMOylation levels are involved in the cell cycle, migration, inflammation, and glycolysis, suggesting that perturbations of SUMOylation might contribute to cancer metastasis by affecting one or more of these biological processes (34). Dysregulation of SUMOylation plays a critical role in the metastasis of breast cancer. Although the molecular details of how SUMOylation affects breast cancer progression and metastasis are not well understood, accumulating evidence has suggested that targeting the SUMOylation pathway may be a strategy for targeting breast cancer. Therefore, further studies into the mechanism of SUMO modification in the process of gene transcription regulation are necessary for providing new ideas and methods for the prevention and treatment of breast cancer and important references for clarifying the pathogenesis of other nuclear receptor-related tumors.
YTQ and HW conceived the idea for the review. YYQ, HY, and XC performed the retrieval and collection of relevant literatures. ZX, XY, YS, WD, and SA provided suggestions. YYQ, HY, XC, YTQ, and HW wrote the paper. YYQ designed and prepared the figures. All authors contributed to the article and approved the submitted version.
This research was funded by the National Natural Science Foundation of China (81671294 and 81870241 to YTQ), and the Fundamental Research Funds for the Central Universities (GK201903066 to HW).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Harbeck N, Gnant M. Breast cancer. Lancet (2017) 389(10074):1134–50. doi: 10.1016/S0140-6736(16)31891-8
2. Leyrer CM, Berriochoa CA, Agrawal S, Donaldson A, Calhoun BC, Shah C, et al. Predictive factors on outcomes in metaplastic breast cancer. Breast Cancer Res Treat (2017) 165(3):499–504. doi: 10.1007/s10549-017-4367-5
3. Chen WQ, Zheng RS, Baade PD, Zhang SW, Zeng HM, Bray F, et al. Cancer Statistics in China, 2015. Ca-a Cancer J Clin (2016) 66(2):115–32. doi: 10.3322/caac.21338
4. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. Ca-a Cancer J Clin (2019) 69(1):7–34. doi: 10.3322/caac.21551
5. Medina MA, Oza G, Sharma A, Arriaga LG, Hernandez Hernandez JM, Rotello VM, et al. Triple-Negative Breast Cancer: A Review of Conventional and Advanced Therapeutic Strategies. Int J Environ Res Public Health (2020) 17(6):2078. doi: 10.3390/ijerph17062078
6. Anastasiadi Z, Lianos GD, Ignatiadou E, Harissis HV, Mitsis M. Breast cancer in young women: an overview. Updates Surg (2017) 69(3):313–7. doi: 10.1007/s13304-017-0424-1
7. Torre LA, Islami F, Siegel RL, Ward EM, Jemal A. Global Cancer in Women: Burden and Trends. Cancer Epidemiol Biomarkers Prev (2017) 26(4):444–57. doi: 10.1158/1055-9965.EPI-16-0858
8. Kennecke H, Yerushalmi R, Woods R, Cheang MC, Voduc D, Speers CH, et al. Metastatic behavior of breast cancer subtypes. J Clin Oncol (2010) 28(20):3271–7. doi: 10.1200/JCO.2009.25.9820
9. Badve S, Dabbs DJ, Schnitt SJ, Baehner FL, Decker T, Eusebi V, et al. Basal-like and triple-negative breast cancers: a critical review with an emphasis on the implications for pathologists and oncologists. Mod Pathol (2011) 24(2):157–67. doi: 10.1038/modpathol.2010.200
10. Brenton JD, Carey LA, Ahmed AA, Caldas C. Molecular classification and molecular forecasting of breast cancer: ready for clinical application? J Clin Oncol (2005) 23(29):7350–60. doi: 10.1200/JCO.2005.03.3845
11. Sheikh A, Hussain SA, Ghori Q, Naeem N, Fazil A, Giri S, et al. The spectrum of genetic mutations in breast cancer. Asian Pac J Cancer Prev (2015) 16(6):2177–85. doi: 10.7314/APJCP.2015.16.6.2177
12. Matsen CB, Neumayer LA. Breast cancer: a review for the general surgeon. JAMA Surg (2013) 148(10):971–9. doi: 10.1001/jamasurg.2013.3393
13. Castaneda SA, Strasser J. Updates in the Treatment of Breast Cancer with Radiotherapy. Surg Oncol Clin N Am (2017) 26(3):371–82. doi: 10.1016/j.soc.2017.01.013
14. Hassan MS, Ansari J, Spooner D, Hussain SA. Chemotherapy for breast cancer (Review). Oncol Rep (2010) 24(5):1121–31. doi: 10.3892/or_00000963
15. Zelnak AB, O’Regan RM. Optimizing Endocrine Therapy for Breast Cancer. J Natl Compr Canc Netw (2015) 13(8):e56–64. doi: 10.6004/jnccn.2015.0125
16. Gu G, Dustin D, Fuqua SA. Targeted therapy for breast cancer and molecular mechanisms of resistance to treatment. Curr Opin Pharmacol (2016) 31:97–103. doi: 10.1016/j.coph.2016.11.005
17. Veronesi U, Boyle P, Goldhirsch A, Orecchia R, Viale G. Breast cancer. Lancet (2005) 365(9472):1727–41. doi: 10.1016/S0140-6736(05)66546-4
18. Boyages J. Radiation therapy and early breast cancer: current controversies. Med J Aust (2017) 207(5):216–22. doi: 10.5694/mja16.01020
19. Bocci G, Tuccori M, Emmenegger U, Liguori V, Falcone A, Kerbel RS, et al. Cyclophosphamide-methotrexate ‘metronomic’ chemotherapy for the palliative treatment of metastatic breast cancer. A comparative pharmacoeconomic evaluation. Ann Oncol (2005) 16(8):1243–52. doi: 10.1093/annonc/mdi240
20. McArthur HL, Hudis CA. Breast cancer chemotherapy. Cancer J (2007) 13(3):141–7. doi: 10.1097/PPO.0b013e318074dc6f
21. Kang X, Qi Y, Zuo Y, Wang Q, Zou Y, Schwartz RJ, et al. SUMO-specific protease 2 is essential for suppression of polycomb group protein-mediated gene silencing during embryonic development. Mol Cell (2010) 38(2):191–201. doi: 10.1016/j.molcel.2010.03.005
22. Qi Y, Zuo Y, Yeh ET, Cheng J. An essential role of small ubiquitin-like modifier (SUMO)-specific Protease 2 in myostatin expression and myogenesis. J Biol Chem (2014) 289(6):3288–93. doi: 10.1074/jbc.M113.518282
23. Chang HM, Yeh ETH. SUMO: From Bench to Bedside. Physiol Rev (2020) 100(4):1599–619. doi: 10.1152/physrev.00025.2019
24. Kumar A, Zhang KY. Advances in the development of SUMO specific protease (SENP) inhibitors. Comput Struct Biotechnol J (2015) 13:204–11. doi: 10.1016/j.csbj.2015.03.001
25. Qi Y, Wang J, Bomben VC, Li DP, Chen SR, Sun H, et al. Hyper-SUMOylation of the Kv7 potassium channel diminishes the M-current leading to seizures and sudden death. Neuron (2014) 83(5):1159–71. doi: 10.1016/j.neuron.2014.07.042
26. Wu H, Chen X, Cheng J, Qi Y. SUMOylation and Potassium Channels: Links to Epilepsy and Sudden Death. Adv Protein Chem Struct Biol (2016) 103:295–321. doi: 10.1016/bs.apcsb.2015.11.009
27. Chen X, Zhang S, Huang J, Dong W, Xiao H, Shao H, et al. Hyper-SUMOylation of K(+) Channels in Sudden Unexplained Death in Epilepsy: Isolation and Primary Culture of Dissociated Hippocampal Neurons from Newborn Mice for Subcellular Localization. Methods Mol Biol (2018) 1684:63–71. doi: 10.1007/978-1-4939-7362-0_6
28. Zhao B, Zhang Z, Chen X, Shen Y, Qin Y, Yang X, et al. The important roles of protein SUMOylation in the occurrence and development of leukemia and clinical implications. J Cell Physiol (2021) 236(5):3466–80. doi: 10.1002/jcp.30143
29. Guo C, Hildick KL, Luo J, Dearden L, Wilkinson KA, Henley JM. SENP3-mediated deSUMOylation of dynamin-related protein 1 promotes cell death following ischaemia. EMBO J (2013) 32(11):1514–28. doi: 10.1038/emboj.2013.65
30. Saitoh H, Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J Biol Chem (2000) 275(9):6252–8. doi: 10.1074/jbc.275.9.6252
31. Rabellino A, Andreani C, Scaglioni PP. The Role of PIAS SUMO E3-Ligases in Cancer. Cancer Res (2017) 77(7):1542–7. doi: 10.1158/0008-5472.CAN-16-2958
32. Bayer P, Arndt A, Metzger S, Mahajan R, Melchior F, Jaenicke R, et al. Structure determination of the small ubiquitin-related modifier SUMO-1. J Mol Biol (1998) 280(2):275–86. doi: 10.1006/jmbi.1998.1839
33. Wang L, Banerjee S. Differential PIAS3 expression in human malignancy. Oncol Rep (2004) 11(6):1319–24. doi: 10.3892/or.11.6.1319
34. Subramonian D, Raghunayakula S, Olsen JV, Beningo KA, Paschen W, Zhang XD. Analysis of changes in SUMO-2/3 modification during breast cancer progression and metastasis. J Proteome Res (2014) 13(9):3905–18. doi: 10.1021/pr500119a
35. Truong K, Lee TD, Chen Y. Small ubiquitin-like modifier (SUMO) modification of E1 Cys domain inhibits E1 Cys domain enzymatic activity. J Biol Chem (2012) 287(19):15154–63. doi: 10.1074/jbc.M112.353789
36. Kessler JD, Kahle KT, Sun TT, Meerbrey KL, Schlabach MR, Schmitt EM, et al. A SUMOylation-Dependent Transcriptional Subprogram Is Required for Myc-Driven Tumorigenesis. Science (2012) 335(6066):348–53 doi: 10.1126/science.1212728.
37. Chen H, Xu Z, Li X, Yang Y, Li B, Li Y, et al. alpha-catenin SUMOylation increases IkappaBalpha stability and inhibits breast cancer progression. Oncogenesis (2018) 7(3):28. doi: 10.1038/s41389-018-0037-7
38. Karami S, Lin FM, Kumar S, Bahnassy S, Thangavel H, Quttina M, et al. Novel SUMO-Protease SENP7S Regulates beta-catenin Signaling and Mammary Epithelial Cell Transformation. Sci Rep (2017) 7:46477. doi: 10.1038/srep46477
39. Yan Y, Ollila S, Wong IP, Vallenius T, Palvimo JJ, Vaahtomeri K, et al. SUMOylation of AMPKalpha1 by PIAS4 specifically regulates mTORC1 signalling. Nat Commun (2015) 6:8979. doi: 10.1038/ncomms9979
40. Wang CM, Liu R, Wang L, Nascimento L, Brennan VC, Yang WH. SUMOylation of FOXM1B alters its transcriptional activity on regulation of MiR-200 family and JNK1 in MCF7 human breast cancer cells. Int J Mol Sci (2014) 15(6):10233–51. doi: 10.3390/ijms150610233
41. Wang CM, Yang WH, Liu R, Wang L, Yang WH. FOXP3 Activates SUMO-Conjugating UBC9 Gene in MCF7 Breast Cancer Cells. Int J Mol Sci (2018) 19(7):2036. doi: 10.3390/ijms19072036
42. Gao X, Wu Y, Qiao L, Feng X. SENP2 suppresses NF-kappaB activation and sensitizes breast cancer cells to doxorubicin. Eur J Pharmacol (2019) 854:179–86. doi: 10.1016/j.ejphar.2019.03.051
43. Li S, Wang M, Qu X, Xu Z, Yang Y, Su Q, et al. SUMOylation of PES1 upregulates its stability and function via inhibiting its ubiquitination. Oncotarget (2016) 7(31):50522–34. doi: 10.18632/oncotarget.10494
44. Chanda A, Chan A, Deng L, Kornaga EN, Enwere EK, Morris DG, et al. Identification of the SUMO E3 ligase PIAS1 as a potential survival biomarker in breast cancer. PloS One (2017) 12(5):e0177639. doi: 10.1371/journal.pone.0177639
45. Chanda A, Ikeuchi Y, Karve K, Sarkar A, Chandhoke AS, Deng L, et al. PIAS1 and TIF1gamma collaborate to promote SnoN SUMOylation and suppression of epithelial-mesenchymal transition. Cell Death Differ (2021) 28(1):267–82. doi: 10.1038/s41418-020-00611-z
46. Chandhoke AS, Chanda A, Karve K, Deng L, Bonni S. The PIAS3-Smurf2 sumoylation pathway suppresses breast cancer organoid invasiveness. Oncotarget (2017) 8(13):21001–14. doi: 10.18632/oncotarget.15471
47. Linher-Melville K, Nashed MG, Ungard RG, Haftchenary S, Rosa DA, Gunning PT, et al. Chronic Inhibition of STAT3/STAT5 in Treatment-Resistant Human Breast Cancer Cell Subtypes: Convergence on the ROS/SUMO Pathway and Its Effects on xCT Expression and System xc- Activity. PloS One (2016) 11(8):e0161202. doi: 10.1371/journal.pone.0161202
48. Cashman R, Cohen H, Ben-Hamo R, Zilberberg A, Efroni S. SENP5 mediates breast cancer invasion via a TGFbetaRI SUMOylation cascade. Oncotarget (2014) 5(4):1071–82. doi: 10.18632/oncotarget.1783
49. Bogachek MV, Chen Y, Kulak MV, Woodfield GW, Cyr AR, Park JM, et al. Sumoylation pathway is required to maintain the basal breast cancer subtype. Cancer Cell (2014) 25(6):748–61. doi: 10.1016/j.ccr.2014.04.008
50. Chauhan KM, Chen Y, Chen Y, Liu AT, Sun XX, Dai MS. The SUMO-specific protease SENP1 deSUMOylates p53 and regulates its activity. J Cell Biochem (2021) 122(2):189–97. doi: 10.1002/jcb.29838
51. Wu F, Zhu S, Ding Y, Beck WT, Mo YY. MicroRNA-mediated regulation of Ubc9 expression in cancer cells. Clin Cancer Res (2009) 15(5):1550–7. doi: 10.1158/1078-0432.CCR-08-0820
52. Zhu S, Sachdeva M, Wu F, Lu Z, Mo YY. Ubc9 promotes breast cell invasion and metastasis in a sumoylation-independent manner. Oncogene (2010) 29(12):1763–72. doi: 10.1038/onc.2009.459
53. Mo YY, Yu Y, Theodosiou E, Ee PL, Beck WT. A role for Ubc9 in tumorigenesis. Oncogene (2005) 24(16):2677–83. doi: 10.1038/sj.onc.1208210
54. Tomasi ML, Tomasi I, Ramani K, Pascale RM, Xu J, Giordano P, et al. S-adenosyl methionine regulates ubiquitin-conjugating enzyme 9 protein expression and sumoylation in murine liver and human cancers. Hepatology (2012) 56(3):982–93. doi: 10.1002/hep.25701
55. Dunnebier T, Bermejo JL, Haas S, Fischer HP, Pierl CB, Justenhoven C, et al. Polymorphisms in the UBC9 and PIAS3 genes of the SUMO-conjugating system and breast cancer risk. Breast Cancer Res Treat (2010) 121(1):185–94. doi: 10.1007/s10549-009-0530-y
56. Ying S, Dunnebier T, Si J, Hamann U. Estrogen receptor alpha and nuclear factor Y coordinately regulate the transcription of the SUMO-conjugating UBC9 gene in MCF-7 breast cancer cells. PloS One (2013) 8(9):e75695. doi: 10.1371/journal.pone.0075695
57. Sentis S, Le Romancer M, Bianchin C, Rostan MC, Corbo L. Sumoylation of the estrogen receptor alpha hinge region regulates its transcriptional activity. Mol Endocrinol (2005) 19(11):2671–84. doi: 10.1210/me.2005-0042
58. Kahyo T, Nishida T, Yasuda H. Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol Cell (2001) 8(3):713–8. doi: 10.1016/S1097-2765(01)00349-5
59. Liu B, Tahk S, Yee KM, Yang R, Yang Y, Mackie R, et al. PIAS1 regulates breast tumorigenesis through selective epigenetic gene silencing. PloS One (2014) 9(2):e89464. doi: 10.1371/journal.pone.0089464
60. Li S, Yang C, Hong Y, Bi H, Zhao F, Liu Y, et al. The transcriptional activity of co-activator AIB1 is regulated by the SUMO E3 ligase PIAS1. Biol Cell (2012) 104(5):287–96. doi: 10.1111/boc.201100116
61. Chanda A, Ikeuchi Y, Karve K, Sarkar A, Chandhoke AS, Deng L, et al. PIAS1 and TIF1gamma collaborate to promote SnoN SUMOylation and suppression of epithelial-mesenchymal transition. Cell Death Differ (2021) 28(1):267–82. doi: 10.1038/s41418-020-0599-8
62. Cheng J, Bawa T, Lee P, Gong L, Yeh ET. Role of desumoylation in the development of prostate cancer. Neoplasia (2006) 8(8):667–76. doi: 10.1593/neo.06445
63. Wang RT, Zhi XY, Zhang Y, Zhang J. Inhibition of SENP1 induces radiosensitization in lung cancer cells. Exp Ther Med (2013) 6(4):1054–8. doi: 10.3892/etm.2013.1259
64. Xu Y, Li J, Zuo Y, Deng J, Wang LS, Chen GQ. SUMO-specific protease 1 regulates the in vitro and in vivo growth of colon cancer cells with the upregulated expression of CDK inhibitors. Cancer Lett (2011) 309(1):78–84. doi: 10.1016/j.canlet.2011.05.019
65. Wang Z, Jin J, Zhang J, Wang L, Cao J. Depletion of SENP1 suppresses the proliferation and invasion of triple-negative breast cancer cells. Oncol Rep (2016) 36(4):2071–8. doi: 10.3892/or.2016.5036
66. Jia Y, Guo Y, Jin Q, Qu H, Qi D, Song P, et al. A SUMOylation-dependent HIF-1alpha/CLDN6 negative feedback mitigates hypoxia-induced breast cancer metastasis. J Exp Clin Cancer Res (2020) 39(1):42. doi: 10.1186/s13046-020-01547-5
67. Chen CH, Chang CC, Lee TH, Luo M, Huang P, Liao PH, et al. SENP1 deSUMOylates and regulates Pin1 protein activity and cellular function. Cancer Res (2013) 73(13):3951–62. doi: 10.1158/0008-5472.CAN-12-4360
68. Goeres J, Chan PK, Mukhopadhyay D, Zhang H, Raught B, Matunis MJ. The SUMO-specific isopeptidase SENP2 associates dynamically with nuclear pore complexes through interactions with karyopherins and the Nup107-160 nucleoporin subcomplex. Mol Biol Cell (2011) 22(24):4868–82. doi: 10.1091/mbc.e10-12-0953
69. Tan MY, Mu XY, Liu B, Wang Y, Bao ED, Qiu JX, et al. SUMO-specific protease 2 suppresses cell migration and invasion through inhibiting the expression of MMP13 in bladder cancer cells. Cell Physiol Biochem (2013) 32(3):542–8. doi: 10.1159/000354458
70. Nait Achour T, Sentis S, Teyssier C, Philippat A, Lucas A, Corbo L, et al. Transcriptional repression of estrogen receptor alpha signaling by SENP2 in breast cancer cells. Mol Endocrinol (2014) 28(2):183–96. doi: 10.1210/me.2013-1376
71. Mirecka A, Morawiec Z, Wozniak K. Genetic Polymorphism of SUMO-Specific Cysteine Proteases - SENP1 and SENP2 in Breast Cancer. Pathol Oncol Res (2016) 22(4):817–23. doi: 10.1007/s12253-016-0064-7
72. Jin ZL, Pei H, Xu YH, Yu J, Deng T. The SUMO-specific protease SENP5 controls DNA damage response and promotes tumorigenesis in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci (2016) 20(17):3566–73.
73. Cai J, Wei X, Zhang G, Sui Y, Zhuang J, Liu Z, et al. Association of SENPs single-nucleotide polymorphism and breast cancer in Chinese population. Med (Baltimore) (2019) 98(6):e14168. doi: 10.1097/MD.0000000000014168
74. Lin FM, Kumar S, Ren J, Karami S, Bahnassy S, Li Y, et al. SUMOylation of HP1alpha supports association with ncRNA to define responsiveness of breast cancer cells to chemotherapy. Oncotarget (2016) 7(21):30336–49. doi: 10.18632/oncotarget.8733
75. Bawa-Khalfe T, Lu LS, Zuo Y, Huang C, Dere R, Lin FM, et al. Differential expression of SUMO-specific protease 7 variants regulates epithelial-mesenchymal transition. Proc Natl Acad Sci USA (2012) 109(43):17466–71. doi: 10.1073/pnas.1209378109
76. Buckley CD, Tan J, Anderson KL, Hanein D, Volkmann N, Weis WI, et al. Cell adhesion. The minimal cadherin-catenin complex binds to actin filaments under force. Science (2014) 346(6209):1254211. doi: 10.1126/science.1254211
77. Harris TJ, Tepass U. Adherens junctions: from molecules to morphogenesis. Nat Rev Mol Cell Biol (2010) 11(7):502–14. doi: 10.1038/nrm2927
78. Hollestelle A, Elstrodt F, Timmermans M, Sieuwerts AM, Klijn JG, Foekens JA, et al. Four human breast cancer cell lines with biallelic inactivating alpha-catenin gene mutations. Breast Cancer Res Treat (2010) 122(1):125–33. doi: 10.1007/s10549-009-0545-4
79. Piao HL, Yuan Y, Wang M, Sun Y, Liang H, Ma L. alpha-catenin acts as a tumour suppressor in E-cadherin-negative basal-like breast cancer by inhibiting NF-kappaB signalling. Nat Cell Biol (2014) 16(3):245–54. doi: 10.1038/ncb2909
80. de Groot JS, Ratze MA, van Amersfoort M, Eisemann T, Vlug EJ, Niklaas MT, et al. alphaE-catenin is a candidate tumor suppressor for the development of E-cadherin-expressing lobular-type breast cancer. J Pathol (2018) 245(4):456–67. doi: 10.1002/path.5099
81. Sun Y, Zhang J, Ma L. alpha-catenin. A tumor suppressor beyond adherens junctions. Cell Cycle (2014) 13(15):2334–9. doi: 10.4161/cc.29765
82. Nakopoulou L, Gakiopoulou-Givalou H, Karayiannakis AJ, Giannopoulou I, Keramopoulos A, Davaris P, et al. Abnormal alpha-catenin expression in invasive breast cancer correlates with poor patient survival. Histopathology (2002) 40(6):536–46. doi: 10.1046/j.1365-2559.2002.01392.x
83. Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer (2009) 9(8):563–75. doi: 10.1038/nrc2676
84. Huang X, Li X, Xie X, Ye F, Chen B, Song C, et al. High expressions of LDHA and AMPK as prognostic biomarkers for breast cancer. Breast (2016) 30:39–46. doi: 10.1016/j.breast.2016.08.014
85. Saha M, Kumar S, Bukhari S, Balaji SA, Kumar P, Hindupur SK, et al. AMPK-Akt Double-Negative Feedback Loop in Breast Cancer Cells Regulates Their Adaptation to Matrix Deprivation. Cancer Res (2018) 78(6):1497–510. doi: 10.1158/0008-5472.CAN-17-2090
87. Xu J, Watkins T, Reddy A, Reddy ES, Rao VN. A novel mechanism whereby BRCA1/1a/1b fine tunes the dynamic complex interplay between SUMO-dependent/independent activities of Ubc9 on E2-induced ERalpha activation/repression and degradation in breast cancer cells. Int J Oncol (2009) 34(4):939–49. doi: 10.3892/ijo_00000220
88. Park MA, Seok YJ, Jeong G, Lee JS. SUMO1 negatively regulates BRCA1-mediated transcription, via modulation of promoter occupancy. Nucleic Acids Res (2008) 36(1):263–83. doi: 10.1093/nar/gkm969
89. Barger CJ, Branick C, Chee L, Karpf AR. Pan-Cancer Analyses Reveal Genomic Features of FOXM1 Overexpression in Cancer. Cancers (Basel) (2019) 11(2). doi: 10.3390/cancers11020251
90. Kalaw E, Lim M, Kutasovic JR, Sokolova A, Taege L, Johnstone K, et al. Metaplastic breast cancers frequently express immune checkpoint markers FOXP3 and PD-L1. Br J Cancer (2020) 123(11):1665–72. doi: 10.1038/s41416-020-01065-3
91. Fallah Y, Brundage J, Allegakoen P, Shajahan-Haq AN. MYC-Driven Pathways in Breast Cancer Subtypes. Biomolecules (2017) 7(3):53. doi: 10.3390/biom7030053
92. Chen Y, Sun XX, Sears RC, Dai MS. Writing and erasing MYC ubiquitination and SUMOylation. Genes Dis (2019) 6(4):359–71. doi: 10.1016/j.gendis.2019.05.006
93. Maubach G, Schmadicke AC, Naumann M. NEMO Links Nuclear Factor-kappaB to Human Diseases: (Trends Mol Med. 23, 1138-1155; 2017). Trends Mol Med (2018) 24(7):654. doi: 10.1016/j.molmed.2018.01.009
94. Elsarraj HS, Valdez KE, Hong Y, Grimm SL, Ricci LR, Fan F, et al. NEMO, a Transcriptional Target of Estrogen and Progesterone, Is Linked to Tumor Suppressor PML in Breast Cancer. Cancer Res (2017) 77(14):3802–13. doi: 10.1158/0008-5472.CAN-16-2794
95. Chen H, Xu Z, Li X, Yang Y, Li B, Li Y, et al. α-catenin SUMOylation increases IκBα stability and inhibits breast cancer progression. Oncogenesis (2018) 7(3):28. doi: 10.1038/s41389-018-0037-7
96. Cheng L, Yuan B, Ying S, Niu C, Mai H, Guan X, et al. PES1 is a critical component of telomerase assembly and regulates cellular senescence. Sci Adv (2019) 5(5):eaav1090. doi: 10.1126/sciadv.aav1090
97. Cheng L, Li J, Han Y, Lin J, Niu C, Zhou Z, et al. PES1 promotes breast cancer by differentially regulating ERalpha and ERbeta. J Clin Invest (2012) 122(8):2857–70. doi: 10.1172/JCI62676
98. Arreal L, Piva M, Fernandez S, Revandkar A, Schaub-Clerigue A, Villanueva J, et al. Targeting PML in triple negative breast cancer elicits growth suppression and senescence. Cell Death Differ (2020) 27(4):1186–99. doi: 10.1038/s41418-019-0407-5
99. Martin-Martin N, Piva M, Urosevic J, Aldaz P, Sutherland JD, Fernandez-Ruiz S, et al. Stratification and therapeutic potential of PML in metastatic breast cancer. Nat Commun (2016) 7:12595 doi: 10.1038/ncomms12595.
100. Maroui MA, Kheddache-Atmane S, El Asmi F, Dianoux L, Aubry M, Chelbi-Alix MK. Requirement of PML SUMO interacting motif for RNF4- or arsenic trioxide-induced degradation of nuclear PML isoforms. PloS One (2012) 7(9):e44949. doi: 10.1371/journal.pone.0044949
101. Chandhoke AS, Karve K, Dadakhujaev S, Netherton S, Deng L, Bonni S. The ubiquitin ligase Smurf2 suppresses TGFbeta-induced epithelial-mesenchymal transition in a sumoylation-regulated manner. Cell Death Differ (2016) 23(5):876–88. doi: 10.1038/cdd.2015.152
102. Emanuelli A, Manikoth Ayyathan D, Koganti P, Shah PA, Apel-Sarid L, Paolini B, et al. Altered Expression and Localization of Tumor Suppressive E3 Ubiquitin Ligase SMURF2 in Human Prostate and Breast Cancer. Cancers (Basel) (2019) 11(4):506. doi: 10.3390/cancers11040556
103. Liu X, Gu X, Sun L, Flowers AB, Rademaker AW, Zhou Y, et al. Downregulation of Smurf2, a tumor-suppressive ubiquitin ligase, in triple-negative breast cancers: involvement of the RB-microRNA axis. BMC Cancer (2014) 14:57. doi: 10.1186/1471-2407-14-57
104. Yu H, Jove R. The STATs of cancer–new molecular targets come of age. Nat Rev Cancer (2004) 4(2):97–105. doi: 10.1038/nrc1275
105. Ishimoto T, Nagano O, Yae T, Tamada M, Motohara T, Oshima H, et al. CD44 variant regulates redox status in cancer cells by stabilizing the xCT subunit of system xc(-) and thereby promotes tumor growth. Cancer Cell (2011) 19(3):387–400. doi: 10.1016/j.ccr.2011.01.038
106. Chen RS, Song YM, Zhou ZY, Tong T, Li Y, Fu M, et al. Disruption of xCT inhibits cancer cell metastasis via the caveolin-1/beta-catenin pathway. Oncogene (2009) 28(4):599–609. doi: 10.1038/onc.2008.414
107. Xiong A, Yang Z, Shen Y, Zhou J, Shen Q. Transcription Factor STAT3 as a Novel Molecular Target for Cancer Prevention. Cancers (Basel) (2014) 6(2):926–57. doi: 10.3390/cancers6020926
108. Paonessa F, Foti D, Costa V, Chiefari E, Brunetti G, Leone F, et al. Activator protein-2 overexpression accounts for increased insulin receptor expression in human breast cancer. Cancer Res (2006) 66(10):5085–93. doi: 10.1158/0008-5472.CAN-05-3678
109. Bogachek MV, Park JM, De Andrade JP, Kulak MV, White JR, Wu T, et al. A novel animal model for locally advanced breast cancer. Ann Surg Oncol (2015) 22(3):866–73. doi: 10.1245/s10434-014-4174-8
110. Park SY, Lee HE, Li H, Shipitsin M, Gelman R, Polyak K. Heterogeneity for stem cell-related markers according to tumor subtype and histologic stage in breast cancer. Clin Cancer Res (2010) 16(3):876–87. doi: 10.1158/1078-0432.CCR-09-1532
111. Bogachek MV, Park JM, De Andrade JP, Lorenzen AW, Kulak MV, White JR, et al. Inhibiting the SUMO Pathway Represses the Cancer Stem Cell Population in Breast and Colorectal Carcinomas. Stem Cell Rep (2016) 7(6):1140–51. doi: 10.1016/j.stemcr.2016.11.001
112. Duffy MJ, Synnott NC, Crown J. Mutant p53 in breast cancer: potential as a therapeutic target and biomarker. Breast Cancer Res Treat (2018) 170(2):213–9. doi: 10.1007/s10549-018-4753-7
Keywords: SUMOylation, sentrin-specific protease, ubiquitin-proteasome system, breast cancer, cancer progression
Citation: Qin Y, Yuan H, Chen X, Yang X, Xing Z, Shen Y, Dong W, An S, Qi Y and Wu H (2021) SUMOylation Wrestles With the Occurrence and Development of Breast Cancer. Front. Oncol. 11:659661. doi: 10.3389/fonc.2021.659661
Received: 28 January 2021; Accepted: 02 March 2021;
Published: 21 April 2021.
Edited by:
Andrew Davis, Washington University in St. Louis, United StatesCopyright © 2021 Qin, Yuan, Chen, Yang, Xing, Shen, Dong, An, Qi and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yitao Qi, cWl5aXRhb0Bzbm51LmVkdS5jbg==; Hongmei Wu, aHE4NDc5QHNubnUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.