- 1Department of Medical Oncology, Key Laboratory of Cancer Prevention and Intervention, Ministry of Education, The Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, China
- 2Cancer Institute, Key Laboratory of Cancer Prevention and Intervention, Ministry of Education, The Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, China
The adenomatous polyposis coli (APC) gene, known as tumor suppressor gene, has the two promoters 1A and 1B. Researches on APC have usually focused on its loss-of-function variants causing familial adenomatous polyposis. Hypermethylation, however, which is one of the key epigenetic alterations of the APC CpG sequence, is also associated with carcinogenesis in various cancers. Accumulating studies have successively explored the role of APC hypermethylation in gastrointestinal (GI) tumors, such as in esophageal, colorectal, gastric, pancreatic, and hepatic cancer. In sporadic colorectal cancer, the hypermethylation of CpG island in APC is even considered as one of the primary causative factors. In this review, we systematically summarized the distribution of APC gene methylation in various GI tumors, and attempted to provide an improved general understanding of DNA methylation in GI tumors. In addition, we included a robust overview of demethylating agents available for both basic and clinical researches. Finally, we elaborated our findings and perspectives on the overall situation of APC gene methylation in GI tumors, aiming to explore the potential research directions and clinical values.
Introduction
The adenomatous polyposis coli (APC) gene is a tumor suppressor gene located in the human chromosome region 5q21–22 (1, 2). As a housekeeping gene, it consists of 15 exons and encodes a 300 kDa protein composed of 2,843 amino acids, which plays a vital role in cellular proliferation, migration, DNA repair, and chromosomal segregation (3). It was initially identified as the pathogenic gene in familial adenomatous polyposis (FAP), an autosomal dominant hereditary disease with numerous adenomatous polyps all over the colorectum (4, 5). Subsequently, it was shown to play an important role in other tumors, such as gastric and pancreatic cancers (6–9). One of the best-known mechanisms of APC is the regulation of the Wnt/β-catenin signaling pathway (10). Without a Wnt signal, β-catenin in the cell is firstly phosphorylated by the destruction complex composed by APC, glycogen synthase kinase 3β (GSK3β), Axin and casein kinase 1 (CK1), then ubiquitinated by β-TrCP200, and finally targeted for proteosomal degradation. In this process, the T cell factor/lymphoid enhancer factor (TCF/LEF) in the nucleus bands with the Groucho co-repressor protein to repress target genes. When the Wnt protein binds with the Frizzled (Fzd) receptor and low-density lipoprotein receptor related protein (LRP) co-receptor, they form a complex to activate the Dishevelled (Dsh) protein. Dsh and Axin bind together to help the formation of Wnt–Frz–LRP5/6 complex, which leads to a disassembled destruction complex, resulting in the stabilization and accumulation of β-catenin. This translocates into the nucleus and binds to a member of the TCF/LEF transcription factor family, activating the expression of many genes, such as the S-phase regulators c-myc and cyclin D1 to promote proliferation (11).
Mutations in APC, such as frameshift mutations, nonsense mutations or splice variants can induce premature stop codons and lead to the production of truncated APC proteins (12). In addition, certain other mechanisms can reduce the expression of APC, such as the hypermethylation of APC promoters (13–18), high expression of EZH2 (19, 20) and YY1 (21), or low expression of CDX2 (22, 23), CEBPZ (24) and USF1/2 (25). All of these effects can lower the levels of destruction complex, causing the high β-catenin content to translocate into the nucleus and cause tumorigenesis (shown in Figure 1).
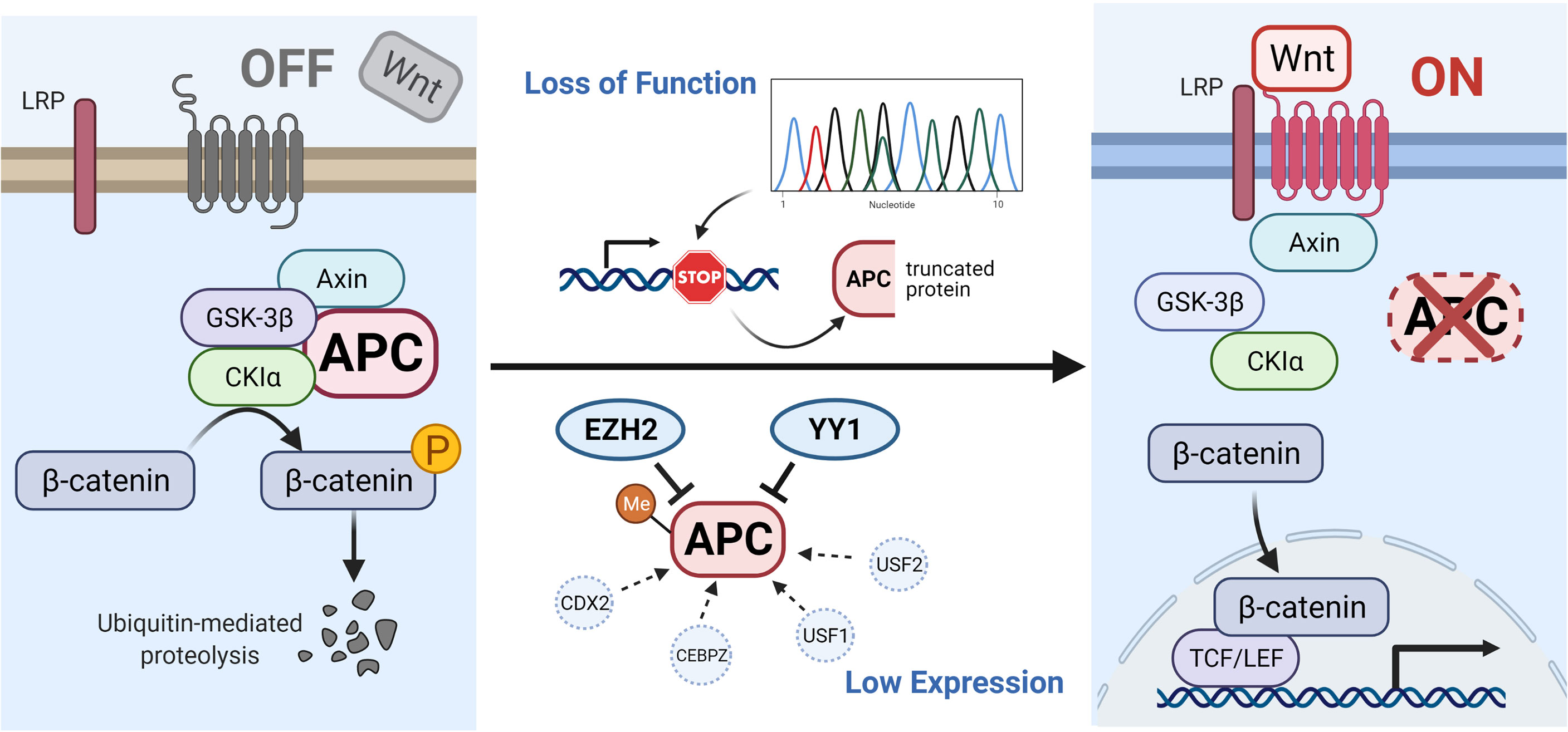
Figure 1 Illustration of APC gene-related pathways and interactive genes. In the absence of a Wnt signal (Wnt/β-catenin signaling pathway is inactive state, see left), the phosphorylated β-catenin can be formed by the destruction complex and degraded by ubiquitin-mediated proteolysis. Protein truncation generated by mutation in the APC gene or decreased level of APC protein influenced by other factors (such as hypermethylated APC promoter, improved expression of EZH2 and YY1, or suppressed expression of CDX2, CEBPZ, and USF1/2) can activate the canonical pathway (Wnt signaling active, see right). When the destruction complex disassembles, the β-catenin fails to be degraded and accumulates in the cytoplasm, translocates into the nucleus, and binds to TCF/LEF transcription factor family, causing the abnormal expression of downstream genes. The illustration was created with BioRender.com.
In this review, we will focus on the impacts of hypermethylation of APC promoters on the development of gastrointestinal cancers. Two promoters have been identified in APC, termed as promoter 1A and promoter 1B. The latter is the most distal promoter of the APC gene [(GRCh37/Hg19) 112,043,008–112,043,594, 586 bp]. Promoter 1A is located at approximately 30.4 kb in the downstream region [(GRCh37/Hg19) 112,072,720–112,073,585, 865 bp], as shown as Figure 2 (26, 27). Through alternative splicing, promoter 1A produces transcript 1A, and promoter 1B produces three transcripts, namely 1B1, 1B2, and 1B3 (28). Both promoters generate transcripts in all organs in the digestive tract, while the expression level of each transcript varies between body organs, thereby 1A and 1B have different importance (28). For example, compared with 1A, transcripts from APC 1B are predominant in normal colonic mucosae but not in the cerebrum (29–31).
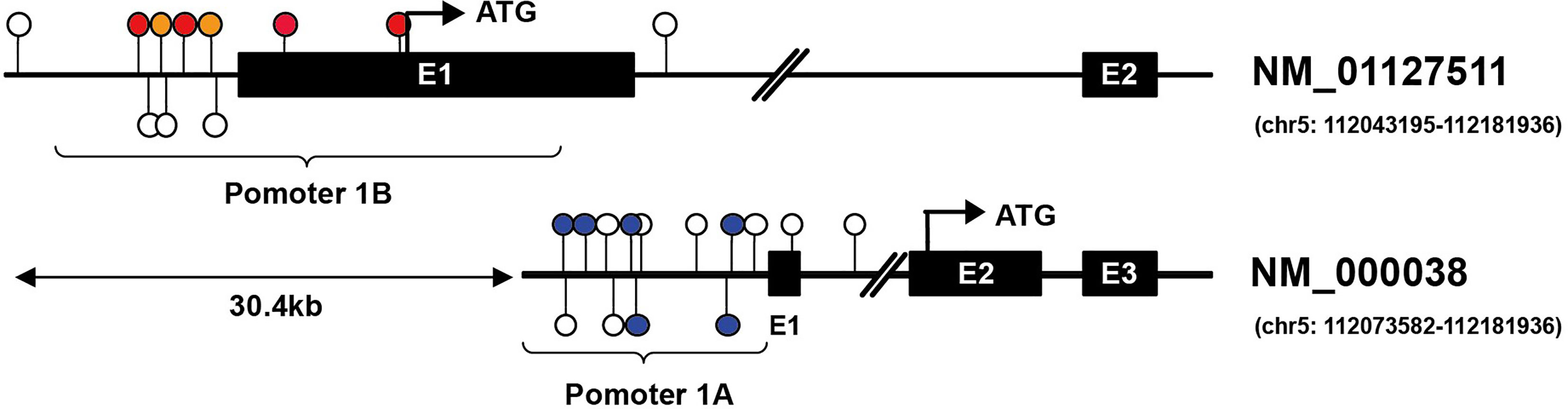
Figure 2 Illustration of CG locus in APC promoters in gastric and hepatic cancers. Promoter 1A (chr5: 112072720–112073585, GRCh37/hg19) is located at approximately 30.4 kb in the downstream region of promoter 1B (chr5: 112043008–112043594, GRCh37/hg19) of the APC gene. The colored lollipops denote statistically significant hypermethylated CG loci associated with prognosis, while the blue color indicates good prognosis, and the orange color means poor prognosis in gastric cancer; the red indicates poor prognosis in hepatic cancer [the colored lollipops referring to CG locus from left to right are cg04011030, cg18315896, cg08934600, cg18536802, cg08512345, cg25922032 (promoter 1B); cg00577935, cg14511739, cg14479889, cg16970232, cg23938220, cg02511809 (promoter 1A)]. Illustrator of biological sequences (IBS) was used to create the illustration.
The process of DNA methylation is a common but a crucial event of epigenetic regulation, which frequently occurs in the CG-rich DNA sequence, the CpG island (32). Saxonov et al. determined that promoters of 72% of DNA were rich in CpG dinucleotides (33). CpG islands comprise organized CpG clusters with more than 50% content of guanine and cytosine (G + C) (34). In promoters, CpG islands generally remain in hypomethylated or unmethylated status to ensure that the gene expression machinery can normally access the promoters of target genes. As outlined in Figure 3, the balance between methylation and demethylation is maintained by enzymes with opposite functions, with the DNA methyltransferase (DNMT) and ten-eleven translocation (TET) enzymes playing major roles (35). The three described members of DNMT are DNMT1, DNMT3a, and DNMT3b, which could transform cytosine to 5-methylcytosine through promoting the covalent transfer of methyl groups to cytosine in the DNA sequence (36). Altered DNMT level or function could impact the DNA methylation status, which was observed in tumors with APC hypermethylation (37). On the contrary, TET enzyme recruitment boosts DNA demethylation, which accomplishes the demethylation role by oxidizing 5-methylcytosine (5mC) to 5-hydroxymethylcytosine and (5hmC) and 5-formylcytosine (5fC)/5-carboxycytosine (5caC) (35, 38). The measurement of methylation status can be performed by diverse techniques. The most widely used method in the studies cited in our review for detecting DNA methylation was methylation-specific PCR (MSP) followed by MethyLight assay, in addition to other methods such as methylation-sensitive single-strand conformation analysis, Illumina Infinium Human Methylation 450 BeadChip array, methylation specific-melting curve analysis (MS-MCA), and so on. The hypermethylation of APC promoter was found to be prevalent in most gastrointestinal (GI) cancers; it was detected in tumor tissues and even the corresponding serum of cancer patients, whereas it was generally less ubiquitous or absent in non-tumor tissues (39). The relationship between cancer and hypermethylation in APC CpG islands, however, has been shown to differ from cancer to cancer. In some GI cancers, APC hypermethylation could even be a potential indicator of early diagnosis or prognostic prediction (39, 40), while in others, it might be relative to tumorigenesis.
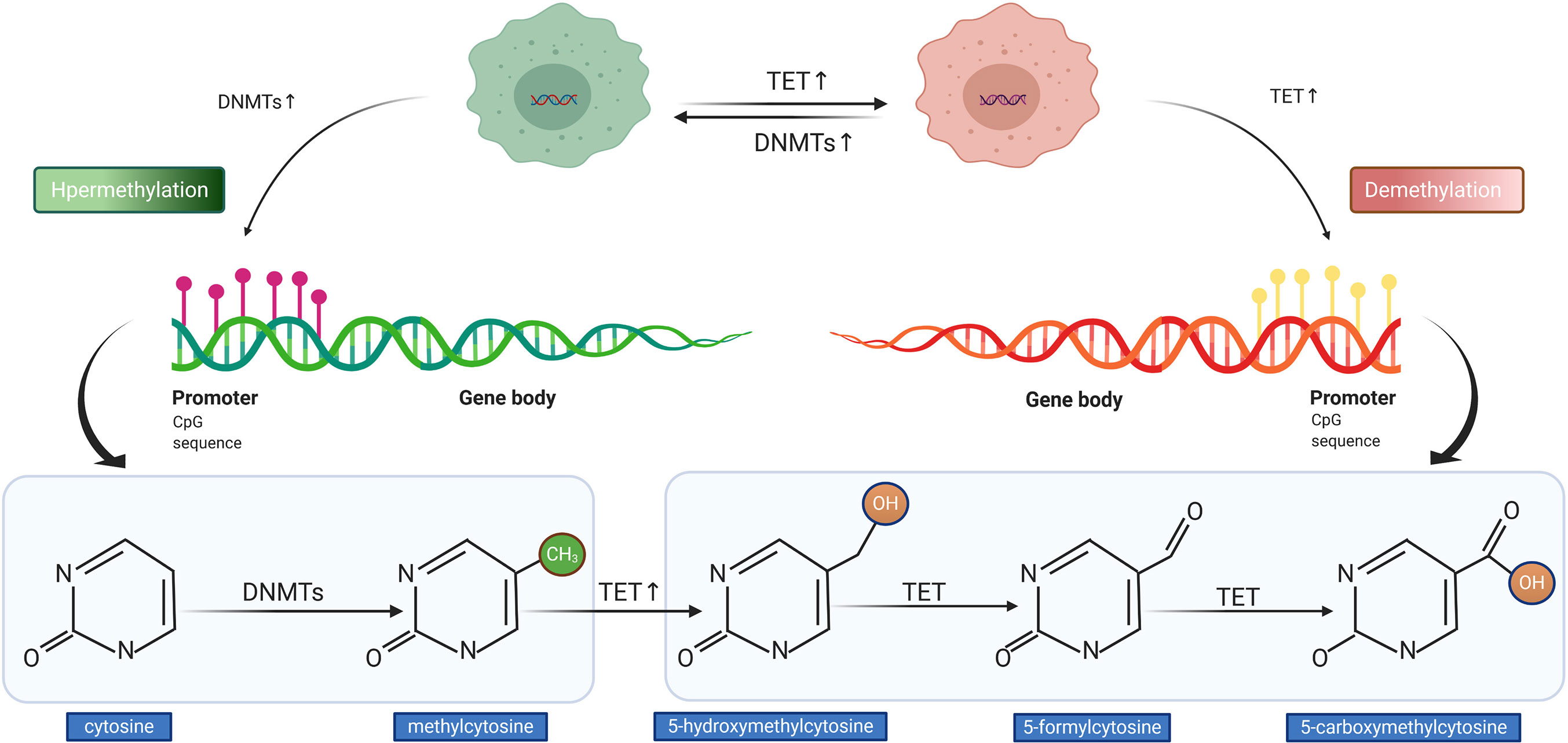
Figure 3 Illustration of DNA hypermethylation and demethylation patterns. The DNA methyltransferase (DNMT) mediates the hypermethylation of DNA promoter (left). The ten-eleven translocation (TET) enzyme conducts the demethylation of DNA promoter (right).
Although the studies dedicated to the role of hypermethylation of APC gene in digestive cancer have been thorough, a comprehensive knowledge of the mechanism is still not available. In this review, we attempt to expound an outline on the distribution of APC hypermethylation in GI cancers and promising demethylating agents that have been experimentally and clinically verified. We also discuss the underlying tumorigenesis mechanism and clinical value of APC hypermethylation.
APC Methylation in Different Gastrointestinal Cancers
Most gastrointestinal tumors were shown to feature APC methylation, while data for duodenal cancer and cholecystic cancer are rare. Therefore, we focus on the introduction and analysis of the relationship between APC methylation and tumors of the digestive organs, except the duodenum and the cholecyst.
Esophageal Cancer
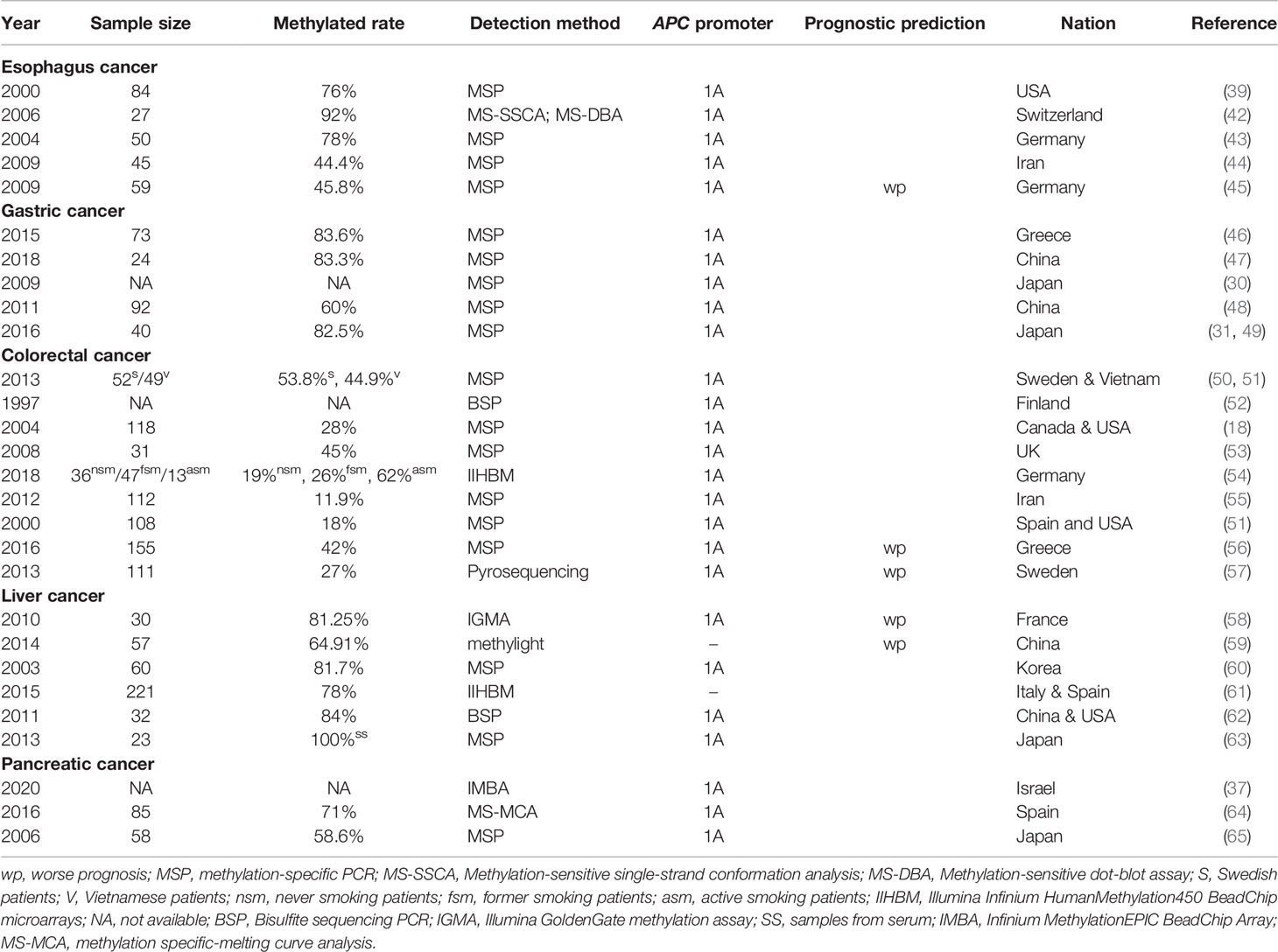
There are two major subtypes of esophageal cancer: adenocarcinoma and squamous cell carcinoma. The pathogenic mechanism of APC reported for other tumors was mainly through truncated proteins caused by APC mutations, but these causative mutations reported for esophageal cancer have been rare (41). Surprisingly, hypermethylation in the APC promoter was detected both in tissue from esophagus adenocarcinoma and squamous cell carcinoma without any radiotherapy or chemotherapy, which occurred in esophageal tissues in 68–92% of patients with esophageal adenocarcinoma, in 44–50% of patients with esophageal squamous cell carcinoma, but not in matching normal esophageal tissues (39, 42–44), as shown specifically in Table 1.
The hypermethylation of APC promoter was also observed in the plasma of patients suggested as hypermethylation positive for tumor tissue (39, 41, 45). Researchers found that the detection rate of APC methylation was higher in the blood of patients who had undergone surgery but featured residual tumor than that of patients with radical resection (45). Moreover, the hypermethylation level in the plasma increased with cancer stage (39, 41, 45). For patients with recurrent lesions, it was found that whether they had been plasma DNA-methylated or not, methylated APC could be detected in their plasma. It was further proposed that a high level of circulating methylated APC DNA and presence of tumor tissues was statistically significantly associated with reduced patient survival (39, 44).
According to these results, it is not difficult to conclude that the appearance of plasma DNA methylation may point to a more advanced tumor stage. Hypermethylated APC DNA can thus be a candidate biomarker to predict the tumor stage and provide prognosis. In addition, the detection of methylated APC in peripheral blood after surgery may provide crucial information about obvious residual tumors, potentially acting as a molecular “R0” marker (45).
Gastric Cancer
Gastric cancer (GC) is one of the cancers with the highest mortality worldwide (66). Compared with patients without GC, the incidence of APC hypermethylation was reported as higher in the tissues and blood of patients with GC (67), which was 60 to 83.6% in these patients (46, 47, 49, 68) (listed in Table 1). The role of APC methylation in gastric cancer is more controversial than in other gastrointestinal tumors. In 2009, Hosoya et al. proposed that APC promoter 1A hypermethylation acted as a passenger in human gastric carcinogenesis (30). In contrast, the work of Balgkouranidou et al. showed that patients with APC methylation had lower survival, and the incidence of death was significantly higher in patients with methylated than those with unmethylated APC promoter status (46). Nevertheless, both studies indicated that the promoter 1A of APC gene was frequently methylated, while promoter 1B was unmethylated (30). In a different study, the promoter 1B of patients with primary GC was also completely in unmethylated status, while the underlying mechanism remained unknown (49).
Infection by Helicobacter pylori (Hp) was known as a risk factor for the development of benign lesions into GC, while the underlying mechanism has not been elucidated. Furthermore, Hp infection was reported to induce APC methylation (48). In addition to Hp, the inducible role of Epstein–Barr virus (EBV) in APC methylation was also demonstrated (68, 69). Therefore, patients with Hp or EBV infection should consider corresponding actively anti-infective treatment and as soon as possible.
Colorectal Cancer
It is widely established that hereditary colorectal cancer (CRC) and sporadic CRC are the two main subtypes of colorectal cancer. Germline APC mutations are typically described as the cause of FAP, which is one of the hereditary forms of CRC (70). Concerning the role of APC in sporadic CRC, with the gradual deepening of researches on colorectal cancer, various studies have shown that the aberrant methylation of APC gene has a non-negligible impact on the occurrence and development of sporadic CRC (53, 54). The APC promoter methylation rate in CRC mucosa was detected in the range of 11.9 to 62% in different populations (18, 50, 51, 54, 55) (listed in Table 1). It was reported that hypermethylation occurred in multistage CRC tumorigenesis (53). According to certain studies, the heaviest CpG methylation occurred in colorectal cancer tissue, while it was slighter in adenoma and absent in normal colonic mucosa (40, 52). APC methylation was also measured in CRC patients at early or advanced stages (56).
As mutation appeared to be more common in cancer tissue, subsequent studies also measured the hypermethylation rate in APC promoter with or without mutation and found that hypermethylation was present in 20 of 108 (18%) tumors lacking APC mutation (51). In samples with mutant APC, however, only 2% demonstrated hypermethylated status. The remarkable finding was that, in FAP patients with certain germline mutations, hypermethylation was not detected at all (51). Thus, mutations in APC may antagonize the methylation of the APC promoter.
In a substantial proportion of sporadic CRCs, hypermethylation in the APC promoter can cause transcriptional silencing (40, 53). As a result of inactivation of APC transcription, transcripts expressed from 1A are significantly decreased, therefore it is quite likely that the Wnt signaling pathway may be activated, further causing cancer development in cells with hypermethylated APC.
APC methylation was also related to worse prognoses (56, 57). It was demonstrated that, independently of tumor stage, the survival outcome in methylation-negative CRC patients was better than that in methylation-positive patients (56).
Liver Cancer
Known as one of the leading causes of death from cancer, hepatocellular carcinoma (HCC) has aroused growing attention in recent years. The progression of HCC was frequently related to epigenetic alterations of genes including APC (58–60). The APC promoter was reported to be hypermethylated in about 64.91 to 84% liver cancer patients (listed in Table 1), which was significantly higher than the non-HCC group (58–62), and a strong association was detected between hypermethylation and the risk of suffering from HCC (71). A relevant study specifically examined the DNA methylation status of the sense and antisense strand and demonstrated the occurrence of aberrant methylation in both, while methylation in the sense strand had a significantly higher specificity of 79% to distinguish HCC tissues from non-HCC tissues. Moreover, aberrant APC methylation was also detected in the serum of HCC patients (63), and serum AFP level and APC methylation status were proposed to be two independent markers for HCC. Methylation of the sense strand of APC was detected in 40% of patients with negative serum AFP (62). Consequently, the combination of serum AFP level and APC methylation may be a reliable approach to screen for HCC.
There is a level of controversy in researches on the relationship between APC methylation and the prognosis of patients with HCC. Augusto Villanueva et al. proposed that, in samples from tumor tissue, APC methylation was an indicator of lower survival in HCC patients (61). However, other researches have suggested that a higher degree of CpG island methylation of APC promoter in tumor tissue was not a definitive marker of lower overall survival rate (58, 59). In vascular circulation samples, higher APC methylation was related to vascular invasion (63). Based on these varied results, more in-depth research will be needed to clarify the relationship between APC methylation and HCC.
Pancreatic Cancer
One of the most aggressive cancers is pancreatic cancer (PC). Compared with other GI cancers, less attention has been paid on the correlation between APC promoter methylation and pancreatic cancer. A number of early studies found that the frequency of APC promoter CpG island hypermethylation was quite distinguished and was detected in 58.6 to 71% of patients with pancreatic cancer (64, 65) (shown in Table 1). Neuroendocrine pancreatic cancer, but not the major population of pancreatic cancer, also exhibits hypermethylation in APC promoter 1A. The overexpression of DNMT1 was detected in the tissue with hypermethylated APC, which might account for this phenomenon (37). Results of He et al. showed that DNMT was regulated by transcriptional factor GLI, and APC methylation in PC might be promoted by DNMT upregulation (72).
Data From Public Databases
MethSurv is a web tool to perform multivariable survival analysis based on CpG methylation patterns and includes 7,358 methylomes from 25 different human cancers. It utilizes methylome data from Illumina Infinium HumanMethylation450 (HM450K). The microarray data is sourced from The Cancer Genome Atlas and GDAC Firehose, and the Cox proportional-hazards model is used to develop an interactive web interface for survival analysis. MethSurv enables survival analysis for a single CpG located in or around the proximity of a query gene. What is more, the clinical characteristics and optional browsing of top biomarkers for each cancer type are also available (73).
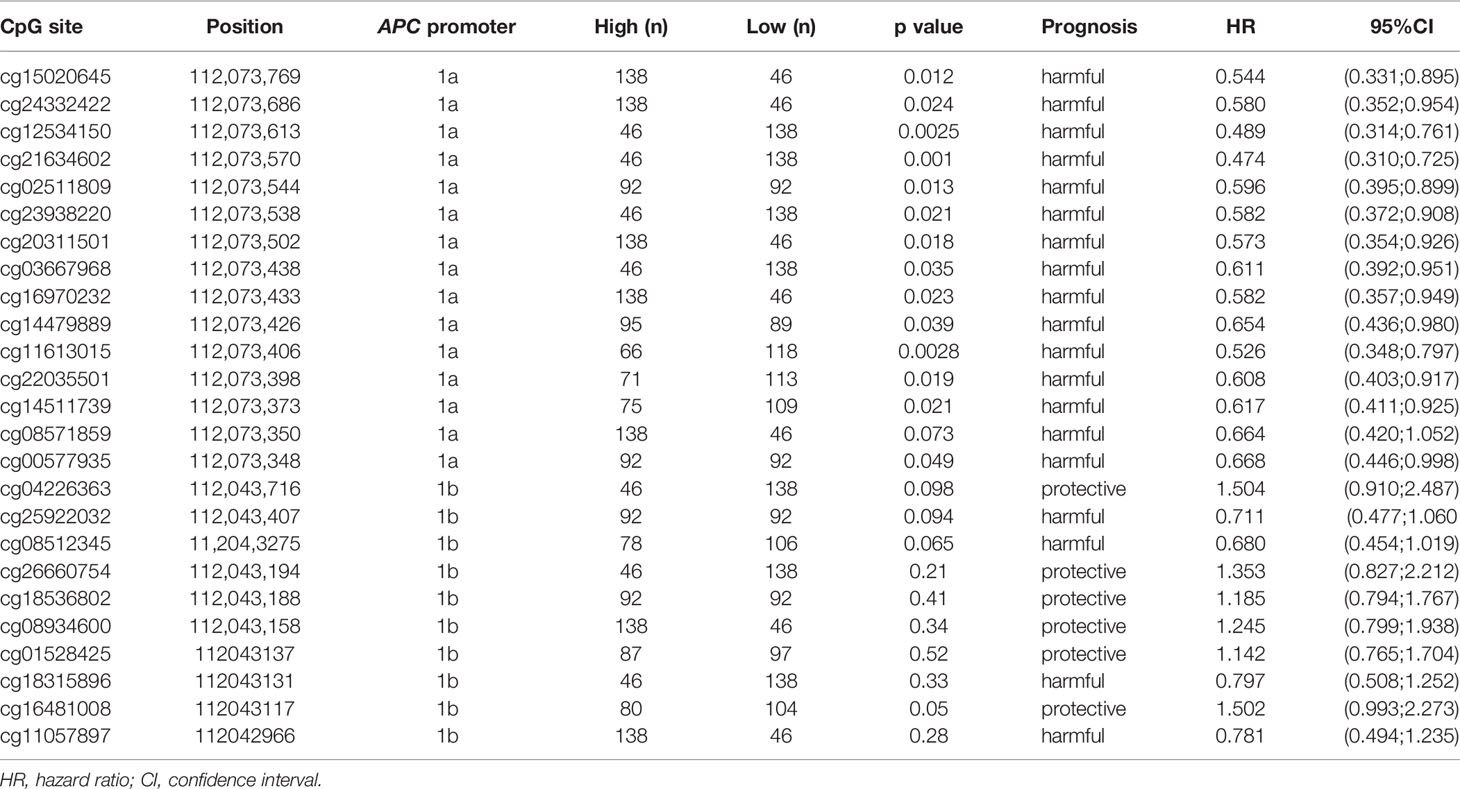
Upon analyzing the data downloaded from MethSurv, it is found that the level of methylation in APC promoters may be concerned with the prognosis and clinical stage of gastrointestinal cancers. In gastric cancer, for instance, the hypermethylation of CG locus in APC promoter 1A indicates good prognosis and has negative correlation with clinical stage, while the partial CG locus (red lollipops in Figure 2) in APC promoter 1B suggests that hypermethylation is closely associated with poor prognosis. Hypermethylation in some CG loci (orange lollipops in Figure 2) of APC promoter 1B in hepatic cancer prompts less median survival time, and the DNA methylation level in cg08512345 appears positively correlated with clinical stage. However, unlike in gastric cancer, most of the hypermethylated CG loci located in promoter 1A in pancreatic cancer demonstrate poor prognosis and are also positively correlated with clinical stage (except cg21634602), as summarized in Table 2. As opposed to gastric, hepatic, and pancreatic cancer, we find that, for esophageal cancer and colorectal cancer, whether in promoter 1A or 1B, the correlations between CG locus methylation level and prognosis or clinical stage are insignificant. Therefore, continued exploration is required to establish the exact role of CG locus hypermethylation in different cancers of the gastrointestinal tract.
Drug Resistance Related to DNA Methylation
Resistance to therapeutic drugs is a significant challenge affecting survival and prognosis for patients receiving medical treatment. Relevant studies have shown that, when a certain gene was abnormally methylated, the abnormal expression of that gene and resistance to anti-tumor drugs would develop (74–77). Shen et al. reported that decreased expression of the tumor suppressor gene KANK1 could increase paclitaxel resistance in non-small cell lung cancer (75). In CRC, NKX6.1 gene was demonstrated to participate in the regulation of chemotherapy sensitivity for its hypermethylation (78).
Limited research has focused on the relationship between drug-resistance and APC methylation. While studying the association between the methylation of multiple genes and sensitivity to pyrimidine synthesis inhibitors, such as cyclocytidine and AraC, researchers found that APC methylation was linked to cell sensitivity to this kind of drug (79). Some studies only exhibited a directed correlation between APC expression level and drug resistance (74, 80). The loss-of-function of APC was able to decrease sensitivity to doxorubicin (DOX) of triple negative breast cancer (TNBC) in the study of Stefanski et al. (80). Through cooperating with MDM2 (murine double minute 2 gene), APC might be involved in drug resistance in ovarian cancer (74). It is well-established that APC is abnormally methylated in both breast cancer (81, 82) and ovarian cancer (83, 84), therefore it cannot be ruled out that APC might affect cancer cell responsiveness to anti-tumor drugs through gene silencing induced by promoter methylation. However, specific experiments are needed to prove this postulation.
Potential Agents Exhibiting a Demethylating Effect
DNA hypermethylation may lead to gene silencing in the promoter region, while demethylation can reactivate gene expression (85–87). Relative agents used in modifying the hypermethylated status of different genes have emerged over the years, with the two most widely applied demethylating drugs 5-azacytidine (azacytidine) and 5-aza-2′-deoxycytidine (decitabine, DAC) (88–91). For example, in patients with myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML), azacytidine and DAC were used to demethylate tumor suppressor genes like CDH1 and CDNNA1, in order to achieve therapeutic effects (92–94).
Apart from MDS and AML, researches on demethylation agents also made some progress in certain solid tumors (95–98). On the one hand, ongoing clinical trials of these drugs to reverse the methylation status in genes have shown varying efficacy and promising results (93, 99). On the other hand, basic research also became intensive. In liver cancer, for example, arsenic trioxide (ATO), an ancient chemotherapy drug, could repress the NF-κB pathway through miR-148a demethylation, and further reverse the resistance of tumor cell (97). Moreover, DAC was able to improve the sensitivity of gastric cancer to irinotecan cisplatin through the demethylation of promoter CpG islands of RUNX3 and PYCARD (98).
In addition to common drugs like DAC, researches on further new agents gradually emerged (16, 100, 101), and some of them could even exhibit a demethylating effect on the APC gene. Selenite treatment was demonstrated to reduce levels of DNMT1 and reactivate methylation-silenced gene expression in prostate cancer cells (16). In another study, genistein was capable of competitively binding with DNMT1, which inhibited the interaction of DNMT1 and APC DNA to protect APC from methylation. By treating breast cancer cells presenting hypermethylated APC with genistein, the methylation-induced silencing in APC was reversed. Natural DNMT inhibitors, such as epigallocatechin-3-gallate (EGCG) and curcumin, also exerted inhibitory effects by competitively binding with DNMT1 (101, 102).
In pancreatic cancer, APC hypermethylation was inferred to result from DNMT upregulation (37, 72), thus agents targeting DNMT might be possible therapeutic drugs for PC. The expression of DNMT was not demonstrated in other GI cancers, therefore it remains uncertain whether the anti-DNMT drug effect potentially occurs in these cancers.
Conclusions and Perspectives
In the relevant literature, APC hypermethylation was found tumor-specific in most gastrointestinal cancers, and was suggested as a potential alternative tumorigenesis mechanism for GI cancers. In some GI cancers, APC hypermethylation may serve as a candidate molecular marker for early disease screening, clinical stage, prognostic prediction and drug efficacy. Nowadays, anti-cancer treatment is gradually moving toward precise and individualized treatment, and APC methylation can be used as a novel target for treating malignant tumors. A rising number of agents are found to reverse the hypermethylated status of APC, which can be a promising anti-cancer method, while more research is necessary to establish such effective therapeutic alternatives. The future of CRISPR Cas9 and other gene editing methods to achieve demethylation can also have significant importance for cancer therapy.
Author Contributions
LZ was mainly responsible for writing conception, literature searching, and drafting of the manuscript. XL participated in the collection of literatures and revision of article. YY was responsible for the revision of the manuscript. CD provided professional revision for the article. MY participated in the conception and revision of the article. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key R&D Program of China (2017YFC0908200), the National Natural Science Foundation of China (81872481), and Traditional Chinese Medicine (Integrated Chinese and Western Medicine) Key Discipline Construction Project of Zhejiang Province (2017-XK-A40).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Bodmer WF, Bailey CJ, Bodmer J, Bussey HJR, Ellis A, Gorman P, et al. Localization of the Gene for Familial Adenomatous Polyposis on Chromosome 5. Nature (1988) 328(6131):614–6. doi: 10.1038/328614a0
2. Kinzler KW, Nilbert MC, Su LK, Vogelstein B, Bryan TM, Levy DB, et al. Identification of FAP Locus Genes From Chromosome 5q21. Science (1991) 253(5020):661–5. doi: 10.1126/science.1651562
3. Senda T, Shimomura A, Iizuka-Kogo A. Adenomatous Polyposis Coli (Apc) Tumor Suppressor Gene as a Multifunctional Gene. Anat Sci Int (2005) 80(3):121–31. doi: 10.1111/j.1447-073x.2005.00106.x
4. Potter JD. Colorectal Cancer: Molecules and Populations. JNCI J Natl Cancer Inst (1999) 91(11):916–32. doi: 10.1093/jnci/91.11.916
5. Goss KH, Groden J. Biology of the Adenomatous Polyposis Coli Tumor Suppressor. J Clin Oncol (2000) 18(9):1967–79. doi: 10.1200/JCO.2000.18.9.1967
6. Rubinstein JC, Khan SA, Christison-Lagay ER, Cha C. APC Mutational Patterns in Gastric Adenocarcinoma are Enriched for Missense Variants With Associated Decreased Survival. Genes Chromosom Cancer (2020) 59(1):64–8. doi: 10.1002/gcc.22792
7. Cole JM, Simmons K, Prosperi JR. Effect of Adenomatous Polyposis Coli Loss on Tumorigenic Potential in Pancreatic Ductal Adenocarcinoma. Cells (2019) 8(9):1–12. doi: 10.3390/cells8091084
8. Cullingworth J, Hooper ML, Harrison DJ, Mason JO, Sirard C, Patek CE, et al. Carcinogen-Induced Pancreatic Lesions in the Mouse: Effect of Smad4 and Apc Genotypes. Oncogene (2002) 21(30):4696–701. doi: 10.1038/sj.onc.1205673
9. Kuo TL, Weng CC, Kuo KK, Chen CY, Wu DC, Hung WC, et al. APC Haploinsufficiency Coupled With p53 Loss Sufficiently Induces Mucinous Cystic Neoplasms and Invasive Pancreatic Carcinoma in Mice. Oncogene (2016) 35(17):2223–34. doi: 10.1038/onc.2015.284
10. Stefanski CD, Prosperi JR. Wnt-Independent and Wnt-Dependent Effects of APC Loss on the Chemotherapeutic Response. Int J Mol Sci (2020) 21(21):7844. doi: 10.3390/ijms21217844
11. Zhan T, Rindtorff N, Boutros M. Wnt Signaling in Cancer. Oncogene (2017) 36(11):1461–73. doi: 10.1038/onc.2016.304
12. Friedl W, Aretz S. Familial Adenomatous Polyposis: Experience From a Study of 1164 Unrelated German Polyposis Patients. Hered Cancer Clin Pract (2005) 3(3):95–114. doi: 10.1186/1897-4287-3-3-95
13. Ignatov A, Bischoff J, Ignatov T, Schwarzenau C, Krebs T, Kuester D, et al. APC Promoter Hypermethylation is an Early Event in Endometrial Tumorigenesis. Cancer Sci (2010) 101(2):321–7. doi: 10.1111/j.1349-7006.2009.01397.x
14. Sievers S, Fritzsch C, Lehnhardt M, Zahn S, Kutzner N, Kuhnen C, et al. Hypermethylation of the APC Promoter But Lack of APC Mutations in Myxoid/Round-Cell Liposarcoma. Int J Cancer (2006) 119(10):2347–52. doi: 10.1002/ijc.22117
15. Fu X, Li J, Li K, Tian X, Zhang Y. Hypermethylation of APC Promoter 1A is Associated With Moderate Activation of Wnt Signalling Pathway in a Subset of Colorectal Serrated Adenomas. Histopathology (2009) 55(5):554–63. doi: 10.1111/j.1365-2559.2009.03411.x
16. Xiang N, Zhao R, Song G, Zhong W. Selenite Reactivates Silenced Genes by Modifying DNA Methylation and Histones in Prostate Cancer Cells. Carcinogenesis (2008) 29(11):2175–81. doi: 10.1093/carcin/bgn179
17. Yegnasubramanian S, Kowalski J, Gonzalgo ML, Zahurak M, Piantadosi S, Walsh PC, et al. Hypermethylation of CpG Islands in Primary and Metastatic Human Prostate Cancer. Cancer Res (2004) 64(6):1975–86. doi: 10.1158/0008-5472.CAN-03-3972
18. Arnold CN, Goel A, Niedzwiecki D, Dowell JM, Wasserman L, Compton C, et al. APC Promoter Hypermethylation Contributes to the Loss of APC Expression in Colorectal Cancers With Allelic Loss on 5q1. Cancer Biol Ther (2004) 3(10):960–4. doi: 10.4161/cbt.3.10.1113
19. Fu X, Lin J, Qin F, Yang Z, Ding Y, Zhang Y, et al. LncAPC Drives Wnt/β-Catenin Activation and Liver TIC Self-Renewal Through EZH2 Mediated APC Transcriptional Inhibition. Mol Carcinog (2018) 57(3):408–18. doi: 10.1002/mc.22764
20. Liu Y, Sun J, Yu J, Ge W, Xiao X, Dai S, et al. Lncrna CACS15 Accelerates the Malignant Progression of Ovarian Cancer Through Stimulating EZH2-induced Inhibition of APC. Am J Transl Res (2019) 11(10):6561–8.
21. Yang Y, Zhou L, Lu L, Wang L, Li X, Jiang P, et al. A Novel miR-193a-5p-YY1-APC Regulatory Axis in Human Endometrioid Endometrial Adenocarcinoma. Oncogene (2013) 32(29):3432–42. doi: 10.1038/onc.2012.360
22. Olsen AK, Coskun M, Bzorek M, Kristensen MH, Danielsen ET, Jørgensen S, et al. Regulation of APC and AXIN2 Expression by Intestinal Tumor Suppressor CDX2 in Colon Cancer Cells. Carcinogenesis (2013) 34(6):1361–9. doi: 10.1093/carcin/bgt037
23. Coskun M, Olsen AK, Bzorek M, Holck S, Engel UH, Nielsen OH, et al. Involvement of CDX2 in the Cross Talk Between TNF-α and Wnt Signaling Pathway in the Colon Cancer Cell Line Caco-2. Carcinogenesis (2014) 35(5):1185–92. doi: 10.1093/carcin/bgu037
24. Deng G, Song GA, Pong E, Sleisenger M, Kim YS. Promoter Methylation Inhibits APC Gene Expression by Causing Changes in Chromatin Conformation and Interfering With the Binding of Transcription Factor CCAAT-Binding Factor. Cancer Res (2004) 64(8):2692–8. doi: 10.1158/0008-5472.CAN-03-3000
25. Jaiswal AS, Narayan S. Upstream Stimulating Factor-1 (USF1) and USF2 Bind to and Activate the Promoter of the Adenomatous Polyposis Coil (APC) Tumor Suppressor Gene. J Cell Biochem (2001) 81(2):262–77. doi: 10.1002/1097-4644(20010501)81:2<262::AID-JCB1041>3.0.CO;2-R
26. Pavicic W, Nieminen TT, Gylling A, Pursiheimo JP, Laiho A, Gyenesei A, et al. Promoter-Specific Alterations of APC are a Rare Cause for Mutation-Negative Familial Adenomatous Polyposis. Genes Chromosom Cancer (2014) 53(10):857–64. doi: 10.1002/gcc.22197
27. Kadiyska TK, Todorov TP, Bichev SN, Vazharova RV, Nossikoff AV, Savov AS, et al. APC Promoter 1B Deletion in Familial Polyposis-Implications for Mutation-Negative Families. Clin Genet (2014) 85(5):452–7. doi: 10.1111/cge.12210
28. Horii A, Nakatsuru S, Ichii S, Nagase H, Nakamura Y. Multiple Forms of the APC Gene Transcripts and Their Tissue-Specific Expression. Hum Mol Genet (1993) 2(3):283–7. doi: 10.1093/hmg/2.3.283
29. Bardos J, Sulekova Z, Ballhausen WG. Novel Exon Connections of the Brain-Specific (BS) Exon of the Adenomatous Polyposis Coli Gene. Int J Cancer (1997) 73(1):137–42. doi: 10.1002/(SICI)1097-0215(19970926)73:1<137::AID-IJC21>3.0.CO;2-C
30. Hosoya K, Yamashita S, Ando T, Nakajima T, Itoh F, Ushijima T. Adenomatous Polyposis Coli 1A is Likely to be Methylated as a Passenger in Human Gastric Carcinogenesis. Cancer Lett (2009) 285(2):182–9. doi: 10.1016/j.canlet.2009.05.016
31. Li J, Woods SL, Healey S, Beesley J, Chen X, Lee JS, et al. Point Mutations in Exon 1B of APC Reveal Gastric Adenocarcinoma and Proximal Polyposis of the Stomach as a Familial Adenomatous Polyposis Variant. Am J Hum Genet (2016) 98(5):830–42. doi: 10.1016/j.ajhg.2016.03.001
32. Romero-Garcia S, Prado-Garcia H, Carlos-Reyes A. Role of DNA Methylation in the Resistance to Therapy in Solid Tumors. Front Oncol (2020) 10(August):1152. doi: 10.3389/fonc.2020.01152
33. Saxonov S, Berg P. Brutlag Dl. A Genome-Wide Analysis of CpG Dinucleotides in the Human Genome Distinguishes Two Distinct Classes of Promoters. Proc Natl Acad Sci U S A (2006) 103(5):1412–7. doi: 10.1073/pnas.0510310103
34. Gardiner-Garden M, Frommer M. Cpg Islands in Vertebrate Genomes. J Mol Biol (1987) 196(2):261–82. doi: 10.1016/0022-2836(87)90689-9
35. Skvortsova K, Stirzaker C, Taberlay P. The DNA Methylation Landscape in Cancer. Blewitt M, Editor. Essays Biochem (2019) 63(6):797–811. doi: 10.1042/EBC20190037
36. Goyal R, Reinhardt R, Jeltsch A. Accuracy of DNA Methylation Pattern Preservation by the Dnmt1 Methyltransferase. Nucleic Acids Res (2006) 34(4):1182–8. doi: 10.1093/nar/gkl002
37. Tirosh A, Killian JK, Petersen D, Zhu YJ, Walker RL, Blau JE, et al. Distinct DNA Methylation Signatures in Neuroendocrine Tumors Specific for Primary Site and Inherited Predisposition. J Clin Endocrinol Metab (2020) 105(10):3285–94. doi: 10.1210/clinem/dgaa477
38. Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J, et al. Genome-Wide Regulation of 5hmC, 5mC, and Gene Expression by Tet1 Hydroxylase in Mouse Embryonic Stem Cells. Mol Cell (2011) 42(4):451–64. doi: 10.1016/j.molcel.2011.04.005
39. Kawakami K, Brabender J, Lord RV, Groshen S, Greenwald BD, Krasna MJ, et al. Hypermethylated APC DNA in Plasma and Prognosis of Patients With Esophageal Adenocarcinoma. J Natl Cancer Inst (2000) 92(22):1805–11. doi: 10.1093/jnci/92.22.1805
40. Usadel H, Brabender J, Danenberg KD, Jerónimo C, Harden S, Engles J, et al. Quantitative Adenomatous Polyposis Coli Promoter Methylation Analysis in Tumor Tissue, Serum, and Plasma DNA of Patients With Lung Cancer. Cancer Res (2002) 62(2):371–5.
41. Powell SM, Papadopoulos N, Kinzler KW, Smolinski KN, Meltzer SJ. APC Gene Mutations in the Mutation Cluster Region are Rare in Esophageal Cancers. Gastroenterology (1994) 107(6):1759–63. doi: 10.1016/0016-5085(94)90818-4
42. Clément G, Braunschweig R, Pasquier N, Bosman FT, Benhattar J. Methylation of APC, TIMP3, and TERT: A New Predictive Marker to Distinguish Barrett’s Oesophagus Patients At Risk for Malignant Transformation. J Pathol (2006) 208(1):100–7. doi: 10.1002/path.1884
43. Sarbia M, Geddert H, Klump B, Kiel S, Iskender E, Gabbert HE. Hypermethylation of Tumor Suppressor Genes (p16INK4A, P14 ARF and APC) in Adenocarcinomas of the Upper Gastrointestinal Tract. Int J Cancer (2004) 111(2):224–8. doi: 10.1002/ijc.20212
44. Zare M, Jazii FR, Alivand MR, Nasseri NK, Malekzadeh R, Yazdanbod M. Qualitative Analysis of Adenomatous Polyposis Coli Promoter: Hypermethylation, Engagement and Effects on Survival of Patients With Esophageal Cancer in a High Risk Region of the World, a Potential Molecular Marker. BMC Cancer (2009) 9:1–12. doi: 10.1186/1471-2407-9-24
45. Hoffmann A-C, Vallböhmer D, Prenzel K, Metzger R, Heitmann M, Neiss S, et al. Methylated DAPK and APC Promoter DNA Detection in Peripheral Blood is Significantly Associated With Apparent Residual Tumor and Outcome. J Cancer Res Clin Oncol (2009) 135(9):1231–7. doi: 10.1007/s00432-009-0564-x
46. Balgkouranidou I, Matthaios D, Karayiannakis A, Bolanaki H, Michailidis P, Xenidis N, et al. Prognostic Role of APC and RASSF1A Promoter Methylation Status in Cell Free Circulating DNA of Operable Gastric Cancer Patients. Mutat Res Mol Mech Mutagen (2015) 778:46–51. doi: 10.1016/j.mrfmmm.2015.05.002
47. Wang Z, Ye Y, Liu D, Yang X, Wang F. Hypermethylation of Multiple Wnt Antagonist Genes in Gastric Neoplasia: Is H Pylori Infection Blasting Fuse? Med (United States) (2018) 97(52):e13734. doi: 10.1097/MD.0000000000013734
48. Wang Z-K, Liu J, Liu C, Wang F-Y, Chen C-Y, Zhang X-H. Hypermethylation of Adenomatous Polyposis Coli Gene Promoter is Associated With Novel Wnt Signaling Pathway in Gastric Adenomas. J Gastroenterol Hepatol . (2012) 27(10):1629–34. doi: 10.1111/j.1440-1746.2012.07219.x
49. Tsuchiya T, Tamura G, Sato K, Endoh Y, Sakata K, Jin Z, et al. Distinct Methylation Patterns of Two APC Gene Promoters in Normal and Cancerous Gastric Epithelia. Oncogene (2000) 19(32):3642–6. doi: 10.1038/sj.onc.1203704
50. Dimberg J, Hong TT, Skarstedt M, Löfgren S, Zar N, Matussek A. Analysis of APC and IGFBP7 Promoter Gene Methylation in Swedish and Vietnamese Colorectal Cancer Patients. Oncol Lett (2013) 5(1):25–30. doi: 10.3892/ol.2012.967
51. Esteller M, Sparks A, Toyota M, Sanchez-Cespedes M, Capella G, Peinado MA, et al. Analysis of Adenomatous Polyposis Coli Promoter Hypermethylation in Human Cancer. Cancer Res (2000) 60(16):4366–71.
52. Hiltunen MO, Alhonen L, Koistinaho J, Myöhänen S, Pääkkönen M, Marin S, et al. Hypermethylation of the APC (Adenomatous Polyposis Coli) Gene Promoter Region in Human Colorectal Carcinoma. Int J Cancer (1997) 70(6):644–8. doi: 10.1002/(SICI)1097-0215(19970317)70:6<644::AID-IJC3>3.0.CO;2-V
53. Segditsas S, Sieber OM, Rowan A, Setien F, Neale K, Phillips RKS, et al. Promoter Hypermethylation Leads to Decreased APC mRNA Expression in Familial Polyposis and Sporadic Colorectal Tumours, But Does Not Substitute for Truncating Mutations. Exp Mol Pathol (2008) 85(3):201–6. doi: 10.1016/j.yexmp.2008.09.006
54. Barrow TM, Klett H, Toth R, Böhm J, Gigic B, Habermann N, et al. Smoking is Associated With Hypermethylation of the APC 1A Promoter in Colorectal Cancer: The ColoCare Study. J Pathol (2017) 243(3):366–75. doi: 10.1002/path.4955
55. Naghibalhossaini F, Zamani M, Mokarram P, Khalili I, Rasti M, Mostafavi-pour Z. Epigenetic and Genetic Analysis of WNT Signaling Pathway in Sporadic Colorectal Cancer Patients From Iran. Mol Biol Rep (2012) 39(5):6171–8. doi: 10.1007/s11033-011-1434-6
56. Matthaios D, Balgkouranidou I, Karayiannakis A, Bolanaki H, Xenidis N, Amarantidis K, et al. Methylation Status of the APC and RASSF1A Promoter in Cell-Free Circulating DNA and its Prognostic Role in Patients With Colorectal Cancer. Oncol Lett (2016) 12(1):748–56. doi: 10.3892/ol.2016.4649
57. Nilsson TK, Löf-Öhlin ZM, Sun X-F. DNA Methylation of the P14arf, RASSF1A and APC1A Genes as an Independent Prognostic Factor in Colorectal Cancer Patients. Int J Oncol (2013) 42(1):127–33. doi: 10.3892/ijo.2012.1682
58. Hernandez-Vargas H, Lambert M-P, Le Calvez-Kelm F, Gouysse G, McKay-Chopin S, Tavtigian SV, et al. Hepatocellular Carcinoma Displays Distinct Dna Methylation Signatures With Potential as Clinical Predictors. Feil R, editor. PloS One (2010) 5(3):e9749. doi: 10.1371/journal.pone.0009749
59. Xu B, Nie Y, Liu X, Feng S, Yang Z, Wang Z, et al. Quantitative Analysis of APC Promoter Methylation in Hepatocellular Carcinoma and its Prognostic Implications. Oncol Lett (2014) 7(5):1683–8. doi: 10.3892/ol.2014.1951
60. Lee S, Lee HJ, Kim JH, Lee HS, Jang JJ, Kang GH. Aberrant CpG Island Hypermethylation Along Multistep Hepatocarcinogenesis. Am J Pathol (2003) 163(4):1371–8. doi: 10.1016/S0002-9440(10)63495-5
61. Villanueva A, Portela A, Sayols S, Battiston C, Hoshida Y, Méndez-González J, et al. DNA Methylation-Based Prognosis and Epidrivers in Hepatocellular Carcinoma. Hepatology (2015) 61(6):1945–56. doi: 10.1002/hep.27732
62. Jain S, Chang T-T, Hamilton JP, Lin SY, Lin Y-J, Evans AA, et al. Methylation of the CpG Sites Only on the Sense Strand of the APC Gene Is Specific for Hepatocellular Carcinoma. Califano J, editor. PloS One (2011) 6(11):e26799. doi: 10.1371/journal.pone.0026799
63. Nishida N, Arizumi T, Takita M, Nagai T, Kitai S, Yada N, et al. Quantification of Tumor DNA in Serum and Vascular Invasion of Human Hepatocellular Carcinoma. Oncology (2013) 84(SUPPL.1):82–7. doi: 10.1159/000345895
64. Ginesta MM, Diaz-Riascos ZV, Busquets J, Pelaez N, Serrano T, Peinado MÀ, et al. APC Promoter is Frequently Methylated in Pancreatic Juice of Patients With Pancreatic Carcinomas or Periampullary Tumors. Oncol Lett (2016) 12(3):2210–6. doi: 10.3892/ol.2016.4868
65. Peng D-F, Kanai Y, Sawada M, Ushijima S, Hiraoka N, Kitazawa S, et al. DNA Methylation of Multiple Tumor-Related Genes in Association With Overexpression of DNA Methyltransferase 1 (DNMT1) During Multistage Carcinogenesis of the Pancreas. Carcinogenesis (2006) 27(6):1160–8. doi: 10.1093/carcin/bgi361
66. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2018) 68(6):394–424. doi: 10.3322/caac.21492
67. Zhou X, Jiao D, Dou M, Zhang W, Hua H, Chen J, et al. Association of APC Gene Promoter Methylation and the Risk of Gastric Cancer. Med (Baltimore) (2020) 99(16):e19828. doi: 10.1097/MD.0000000000019828
68. Geddert H, Zur HA, Gabbert HE, Sarbia M. EBV-Infection in Cardiac and non-Cardiac Gastric Adenocarcinomas is Associated With Promoter Methylation of p16, p14 and APC, But Not Hmlh1. Cell Oncol (2011) 34(3):209–14. doi: 10.1007/s13402-011-0028-6
69. Zhao Z, Liu W, Liu J, Wang J, Luo B. The Effect of EBV on WIF1, NLK, and APC Gene Methylation and Expression in Gastric Carcinoma and Nasopharyngeal Cancer. J Med Virol (2017) 89(10):1844–51. doi: 10.1002/jmv.24863
70. Lagarde A, Rouleau E, Ferrari A, Noguchi T, Qiu J, Briaux A, et al. Germline APC Mutation Spectrum Derived From 863 Genomic Variations Identified Through a 15-Year Medical Genetics Service to French Patients With FAP. J Med Genet (2010) 47(10):721–2. doi: 10.1136/jmg.2010.078964
71. Liu M, Cui L-H, Li C-C, Zhang L. Association of APC, GSTP1 and SOCS1 Promoter Methylation With the Risk of Hepatocellular Carcinoma. Eur J Cancer Prev (2015) 24(6):470–83. doi: 10.1097/CEJ.0000000000000121
72. He SS, Wang F, Yang LJ, Guo CY, Wan R, Ke AW, et al. Expression of DNMT1 and DNMT3a are Regulated by GLI1 in Human Pancreatic Cancer. PloS One (2011) 6(11):1–9. doi: 10.1371/journal.pone.0027684
73. Modhukur V, Iljasenko T, Metsalu T, Lokk K, Laisk-Podar T, Vilo J. Methsurv: A Web Tool to Perform Multivariable Survival Analysis Using DNA Methylation Data. Epigenomics (2018) 10(3):277–88. doi: 10.2217/epi-2017-0118
74. Yin F, Liu X, Li D, Wang Q, Zhang W, Li L. Tumor Suppressor Genes Associated With Drug Resistance in Ovarian Cancer (Review). Oncol Rep (2013) 30(1):3–10. doi: 10.3892/or.2013.2446
75. Pu J, Shen J, Zhong Z, Yanling M, Gao J. KANK1 Regulates Paclitaxel Resistance in Lung Adenocarcinoma A549 Cells. Artif Cells Nanomed Biotechnol (2020) 48(1):639–47. doi: 10.1080/21691401.2020.1728287
76. Jank P, Gehlhaar C, Bianca L, Caterina F, Andreas S, Karn T, et al. MGMT Promoter Methylation in Triple Negative Breast Cancer of the GeparSixto Trial. Banerjee SK, editor. PloS One (2020) 15(8):e0238021. doi: 10.1371/journal.pone.0238021
77. Rosell R, Monzo M, Brate AO, Taron M. Translational Oncogenomics: Toward Rational Therapeutic Decision-Making. Curr Opin Oncol (2002) 14(2):171–9. doi: 10.1097/00001622-200203000-00006
78. Chung HH, Te LC, Hu JM, YC C, YW L, Shih YL. NKX6.1 Represses Tumorigenesis, Metastasis, and Chemoresistance in Colorectal Cancer. Int J Mol Sci (2020) 21(14):1–20. doi: 10.3390/ijms21145106
79. Sasaki S, Kobunai T, Kitayama J, Hirokazu N. DNA Methylation and Sensitivity to Antimetabolites in Cancer Cell Lines. Oncol Rep (2008) 19(2):407–12. doi: 10.3892/or.19.2.407
80. Stefanski CD, Keffler K. APC Loss Affects DNA Damage Repair Causing Doxorubicin Resistance in Breast Cancer Cells. Neoplasia (2019) 21(12):1143–50. doi: 10.1016/j.neo.2019.09.002
81. Suman M, Dugué PA, Wong EM, Joo JE, Hopper JL, Dumont TN, et al. Association of Variably Methylated Tumour DNA Regions With Overall Survival for Invasive Lobular Breast Cancer. Clin Epigenet (2021) 13(1):1–16. doi: 10.1186/s13148-020-00975-6
82. Saelee P, Pongtheerat T. Apc Promoter Hypermethylation as a Prognostic Marker in Breast Cancer Patients. Asian Pac J Cancer Prev (2020) 21(12):3627–32. doi: 10.31557/APJCP.2020.21.12.3627
83. Bhagat R, Chadaga S, Premalata CS, Ramesh G, Ramesh C, Pallavi VR, et al. Aberrant Promoter Methylation of the RASSF1A and APC Genes in Epithelial Ovarian Carcinoma Development. Cell Oncol (2012) 35(6):473–9. doi: 10.1007/s13402-012-0106-4
84. Al-Shabanah OA, Hafez MM, Hassan ZK, Sayed-Ahmed MM, Abozeed WN, Alsheikh A, et al. Methylation of SFRPs and APC Genes in Ovarian Cancer Infected With High Risk Human Papillomavirus. Asian Pac J Cancer Prev (2014) 15(6):2719–25. doi: 10.7314/APJCP.2014.15.6.2719
85. Takeshima H, Yoda Y, Wakabayashi M, Hattori N, Yamashita S, Ushijima T. Low-Dose DNA Demethylating Therapy Induces Reprogramming of Diverse Cancer-Related Pathways At the Single-Cell Level. Clin Epigenet (2020) 12(1):142. doi: 10.1186/s13148-020-00937-y
86. Song C, Wang L, Wu X, Wang K, Xie D, Xiao Q, et al. PML Recruits TET2 to Regulate DNA Modification and Cell Proliferation in Response to Chemotherapeutic Agent. Cancer Res (2018) 78(10):2475–89. doi: 10.1158/0008-5472.CAN-17-3091
87. Agrawal K, Das V, Táborská N, Gurský J, Džubák P, Hajdúch M. Differential Regulation of Methylation-Regulating Enzymes by Senescent Stromal Cells Drives Colorectal Cancer Cell Response to DNA-Demethylating Epi-Drugs. Stem Cells Int (2018) 2018::6013728. doi: 10.1155/2018/6013728
88. Fan J-X, Deng R-H, Wang H, Liu X-H, Wang X-N, Qin R, et al. Epigenetics-Based Tumor Cells Pyroptosis for Enhancing the Immunological Effect of Chemotherapeutic Nanocarriers. Nano Lett (2019) 19(11):8049–58. doi: 10.1021/acs.nanolett.9b03245
89. Kuehl PJ, Tellez CS, Grimes MJ, March TH, Tessema M, Revelli DA, et al. 5-Azacytidine Inhaled Dry Powder Formulation Profoundly Improves Pharmacokinetics and Efficacy for Lung Cancer Therapy Through Genome Reprogramming. Br J Cancer (2020) 122(8):1194–204. doi: 10.1038/s41416-020-0765-2
90. Appel CK, Scheff NN, Viet CT, Schmidt BL, Heegaard A. Decitabine Attenuates Nociceptive Behavior in a Murine Model of Bone Cancer Pain. Pain (2019) 160(3):619–31. doi: 10.1097/j.pain.0000000000001442
91. Tomar T, De JS, NG A, RL H, GJ M, HG K, et al. Genome-Wide Methylation Profiling of Ovarian Cancer Patient-Derived Xenografts Treated With the Demethylating Agent Decitabine Identifies Novel Epigenetically Regulated Genes and Pathways. Genome Med (2016) 8(1):1–15. doi: 10.1186/s13073-016-0361-5
92. Quintás-Cardama A, Santos FPS, Garcia-Manero G. Therapy With Azanucleosides for Myelodysplastic Syndromes. Nat Rev Clin Oncol (2010) 7(8):433–44. doi: 10.1038/nrclinonc.2010.87
93. Savona MR, Odenike O, Amrein PC, Steensma DP, DeZern AE, Michaelis LC, et al. An Oral Fixed-Dose Combination of Decitabine and Cedazuridine in Myelodysplastic Syndromes: A Multicentre, Open-Label, Dose-Escalation, Phase 1 Study. Lancet Haematol (2019) 6(4):e194–203. doi: 10.1016/S2352-3026(19)30030-4
94. Lee V, Wang J, Zahurak M, Gootjes E, Verheul HM, Parkinson R, et al. A Phase I Trial of a Guadecitabine (SGI-110) and Irinotecan in Metastatic Colorectal Cancer Patients Previously Exposed to Irinotecan. Clin Cancer Res (2018) 24(24):6160–7. doi: 10.1158/1078-0432.CCR-18-0421
95. do Amaral GCLS, Planello AC, Borgato G, de Lima DG, Guimarães GN, Marques MR, et al. 5-Aza-CdR Promotes Partial MGMT Demethylation and Modifies Expression of Different Genes in Oral Squamous Cell Carcinoma. Oral Surg Oral Med Oral Pathol Oral Radiol (2019) 127(5):425–32. doi: 10.1016/j.oooo.2019.01.006
96. Hung C-S, Wang S-C, Yen Y-T, Lee T-H, Wen W-C, Lin R-K. Hypermethylation of CCND2 in Lung and Breast Cancer Is a Potential Biomarker and Drug Target. Int J Mol Sci (2018) 19(10):3096. doi: 10.3390/ijms19103096
97. Wang Y, Jiang F, Jiao K, Ju L, Liu Q, Li Y, et al. De-Methylation of miR-148a by Arsenic Trioxide Enhances Sensitivity to Chemotherapy Via Inhibiting the NF-κb Pathway and CSC Like Properties. Exp Cell Res (2020) 386(2):111739. doi: 10.1016/j.yexcr.2019.111739
98. Moro H, Hattori N, Nakamura Y, Kimura K, Imai T, Maeda M, et al. Epigenetic Priming Sensitizes Gastric Cancer Cells to Irinotecan and Cisplatin by Restoring Multiple Pathways. Gastric Cancer (2020) 23(1):105–15. doi: 10.1007/s10120-019-01010-1
99. Linnekamp JF, Butter R, Spijker R, Medema JP, van Laarhoven HWM. Clinical and Biological Effects of Demethylating Agents on Solid Tumours – A Systematic Review. Cancer Treat Rev (2017) 54:10–23. doi: 10.1016/j.ctrv.2017.01.004
100. Zwergel C, Schnekenburger M, Sarno F, Battistelli C, Manara MC, Stazi G, et al. Identification of a Novel Quinoline-Based DNA Demethylating Compound Highly Potent in Cancer Cells. Clin Epigenet (2019) 11(1):68. doi: 10.1186/s13148-019-0663-8
101. Assumpção JHM, Takeda AAS, Sforcin JM, Rainho CA. Effects of Propolis and Phenolic Acids on Triple-Negative Breast Cancer Cell Lines: Potential Involvement of Epigenetic Mechanisms. Molecules (2020) 25(6):1289. doi: 10.3390/molecules25061289
Keywords: adenomatous polyposis coli, promoter, methylation, gastrointestinal cancer, CpG island
Citation: Zhu L, Li X, Yuan Y, Dong C and Yang M (2021) APC Promoter Methylation in Gastrointestinal Cancer. Front. Oncol. 11:653222. doi: 10.3389/fonc.2021.653222
Received: 14 January 2021; Accepted: 30 March 2021;
Published: 23 April 2021.
Edited by:
Shama Prasada Kabekkodu, Manipal Academy of Higher Education, IndiaReviewed by:
Vaibhav Shukla, NIMS University, IndiaDil Afroze, Sher-I-Kashmir Institute of Medical Sciences, India
Copyright © 2021 Zhu, Li, Yuan, Dong and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Caixia Dong, ZG9uZ2NhaXhpYTE5ODJAemp1LmVkdS5jbg==; Mengyuan Yang, eWFuZ215MjAxN0B6anUuZWR1LmNu
†These authors have contributed equally to this work
 Lila Zhu
Lila Zhu Xinyu Li
Xinyu Li Ying Yuan
Ying Yuan Caixia Dong
Caixia Dong Mengyuan Yang
Mengyuan Yang