- 1Laboratory Animal Center, College of Animal Science, Jilin University, Changchun, China
- 2Department of Hand Surgery, The First Hospital of Jilin University, Changchun, China
Background: Chrysin is a natural flavone that is present in honey and has exhibited anti-tumor properties. It has been widely studied as a therapeutic agent for the treatment of various types of cancers. The objectives of this present study were to elucidate how chrysin regulates non-coding RNA expression to exert anti-tumor effects in gastric cancer cells.
Methods: Through the use of RNA sequencing, we investigated the differential expression of mRNAs in gastric cancer cells treated with chrysin. Furthermore, COPB2, H19 and let-7a overexpression and knockdown were conducted. Other features, including cell growth, apoptosis, migration and invasion, were also analyzed. Knockout of the COPB2 gene was generated using the CRISPR/Cas9 system for tumor growth analysis in vivo.
Results: Our results identified COPB2 as a differentially expressed mRNA that is down-regulated following treatment with chrysin. Moreover, the results showed that chrysin can induce cellular apoptosis and inhibit cell migration and invasion. To further determine the underlying mechanism of COPB2 expression, we investigated the expression of the long non-coding RNA (lncRNA) H19 and microRNA let-7a. Our results showed that treatment with chrysin significantly increased let-7a expression and reduced the expression of H19 and COPB2. In addition, our results demonstrated that reduced expression of COPB2 markedly promotes cell apoptosis. Finally, in vivo data suggested that COPB2 expression is related to tumor growth.
Conclusions: This study suggests that chrysin exhibited anti-tumor effects through a H19/let-7a/COPB2 axis.
Introduction
Currently, gastric cancer (GC) is the third most common cause of human mortality among malignant cancers (1). Although surgical treatment for GC has led to increased survival rates, the diagnosis of GC needs to improve (2). There are several potential biomarkers of GC, including many genes and cell signaling pathways that are involved in GC development, such as BRCA2 and Ras-Raf-MAPK signaling (3, 4). Coatomer protein complex subunit beta 2 (COPB2) is a protein that functions to transport other proteins as vesicles from the endoplasmic reticulum to Golgi apparatus (5). Recently, numerous reports have indicated that COPB2 is abnormally expressed in colorectal cancer (CRC), cholangiocellular carcinoma and lung cancer (6–8). Previous studies have indicated that the reduction of COPB2 expression inhibited cell growth and induced apoptosis through the JNK/c-Jun signaling pathway in RKO and HCT116 cells (9). Moreover, miR4461 and miR335 were found to regulate COPB2 expression, which subsequently inhibited cell growth in CRC and lung cancer cells (10, 11).
Increasing evidence suggests that non-coding RNAs (ncRNAs), such as miRNAs, can be applied to the classification of GC (12). As tumor suppressors, the let-7 family is down-regulated in GC (13). Increased expression of let-7a has been shown to inhibit cell migration and invasion in prostate cancer (14). Compared to the loss of let-7a expression, the long non-coding RNA (lncRNA) H19 has been shown to be highly expressed in cancers, including GC (15). As a molecular sponge, H19 was found to be related to let-7 in the context of breast cancer stem cells (16). Previous reports have suggested that H19 expression suppressed endogenous let-7 while H19 mutant was not related to let-7 (17). Additionally, reduced expression of H19 induced cellular apoptosis and inhibited cell growth in HCC (18). However, there is little evidence to suggest that COPB2 expression is associated with lncRNAs and miRNAs in GC.
As a traditional Chinese medicine, chrysin is a natural flavone that has anti-cancer function (19). Previous reports have indicated that chrysin induces cellular apoptosis and inhibits tumor glycolysis in HCC (20). Moreover, chrysin has been shown to inhibit cell migration and invasion in melanoma cells (21). In this study, chrysin was used to treat GC cells and we screened differentially expressed genes using RNA-seq. Additionally, we created a COPB2 knockout (KO) cell line using the CRISPR/Cas9 system. Our findings indicate that chrysin can regulate COPB2 expression through let-7, which is antagonized by H19.
Materials and Methods
Cell Culture and Chrysin Treatment
Human GC cell lines (SGC7901, MKN45 and BGC823) and the human gastric epithelial cell line GES-1 were grown in Dulbecco’s modified Eagle’s (DMEM; Gibco) supplemented with 10% fetal bovine serum (Gibco), and cultured at 37°C in 5% CO2. The human gastric epithelial cell GES1 served as control. Experiments were conducted by treating GES1, SGC7901, MKN45 and BGC823 cells with 40 μM of Chrysin (Yuanye Bio-Technology, Shanghai) for 48 h when they reached 80 to 90% confluence.
RNA Isolation and RNA-Seq Analysis
GC cells were treated with 40 μM chrysin for 48 h, after which total RNA was extracted. To remove and purify ribosomal RNA (rRNA), we used the RiboZero Magnetic Gold Kit (Epidemiology, USA). Then, RNA-seq (Sangon Biotech, Shanghai, China) was carried out on HiSeq2500 (Illumina, USA) to analyze raw reads, which were quality controlled by FastQC. Using the HISAT2 software, the sequenced-reads were aligned to the reference sequence. The gene expression analysis and differential gene expression analysis were determined using DEGseq and DESeq program, respectively, in HISAT2 (qValue <0.05, Fold Change >2). Using clusterProfiler, the enrichment analysis, including Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway, of differential expressed genes was determined.
Knockdown and Overexpression of COPB2, H19 and let-7a
The pcDNA3.1 (GenePharma, China) vector served as the backbone for the overexpression construct of COPB2 (pcDNA3.1-COPB2) and H19 (pcDNA3.1-H19). The cells were cultured without FBS once they reached a confluence of 80% over 12–16 h. Next, the pcDNA3.1-COPB2 (2 μg), pcDNA3.1-H19 (2 μg) and Lipofectamine 2000 (Invitrogen, USA) were utilized for transfection. After incubating for 48 h, G418 (400 mg/ml, Invitrogen, USA) was added to GC cells. The clones were grown and picked after 14 days.
The siRNAs of COPB2 (si-COPB2) and H19 (si-H19) were obtained from RiboBio (Guangzhou, China). The target sequences of small interfering RNAs (siRNAs) are listed in Table S1. The mimics and inhibitors of let-7a-3p, miR29b-3p and miR675-3p were obtained from RiboBio. The GC cells were transfected with knockdown (siRNA), let-7a-3p mimics, and let-7a-3p inhibitor for 48 h. The nonspecific siRNA (si-Nc) was transfected into control cells.
The COPB2-KO was generated using the CRISPR/Cas9 system (px459, Addgene, USA). The single guide RNAs (sgRNAs) were designed as previously reported (22). The sequences of sgRNAs are listed in Table S2. Transfection was conducting using COPB2-KO (5 μg) and Lipofectamine 2000 (Invitrogen, USA) for 48 h. Next, puromycin helped select the positive clones. After 14 days, the clones (COPB2-KO) were grown and picked for subsequent western blot, qPCR and sequencing analysis.
DNA Methylation Analysis
The Bisulfite Sequencing PCR (BSP) protocol was carried out as previously described (23). Using the TIANamp Genomic DNA Kit (TIANGEN, Beijing, China), the DNA of SGC7901 and BGC823 cells were extracted. The DNA was modified using CpGenome™ Turbo Bisulfite Modification Kit (Millipore, USA). The differentially methylated regions (DMRs) of H19 were amplified using nested PCR. The products, which included 10 positive clones, were analyzed using the BiQ Analyzer software (http://biq-analyzer.bioinf.mpi-inf.mpg.de/tools/MethylationDiagrams/index.php). The primers of H19 DMR are listed in Table S3.
Gene Expression Analysis
The GC cells’ RNA was extracted and cDNAs were generated using the cDNA first-strand synthesis kit (TIANGEN, China). Gene expression analysis was conducted using quantitative real-time PCR (qPCR). The conditions for qPCR included heating to 94°C for 3 min, and then denaturation at 94°C for 10 s after 35 cycles. The annealing was carried out at 59°C for 15 s. The products were extended at 72°C for 30 s. The internal controls included GAPDH and U6 for genes and miRNAs, respectively. The primers for qPCR are listed in Table S4. The sequences of COPB2 exon 5 and exon 22 are listed in Table S5.
Cell Migration and Invasion Analysis
Wound healing assay was conducted to analyze cell migration of GC cells. In brief, 5 × 105 cells were cultured and seeded before treatment with chrysin, siRNAs, overexpression vectors, let-7a-3p mimics or let-7a-3p inhibitor. The cells were cultured after a scratched line was created with culture medium without serum. Cell migration was measured using the scratched area at 12, 24 and 48 h.
For cell invasion assays, GC cells (3 × 104) were cultured and seeded with 20 μl Matrigel prior to treatment with chrysin, siRNAs, overexpression vectors, let-7a-3p mimics and let-7a-inhibitor (BD Biosciences, USA). Next, 0.5 ml of medium, which contained 10% FBS, was added to the cells for 24 h. Then, 0.2% crystal violet dye (Solarbio, China) was used to stain cells after being fixed with 4% paraformaldehyde. The stained cells were assayed using the ImageJ software.
Cell Counting Kit-8 Assay
The GC cells (4 × 103) were cultured and seeded prior treatment with chrysin, siRNAs, overexpression vectors, let-7a-3p mimics and let-7a-3p inhibitors in order to conduct cell viability assay, as previously described (24). Then, Cell Counting Kit-8 (CCK-8) solution (10 μl, Dojindo, Kumamoto, Japan) was added to each well. After incubating for 2.5 h, the cells were measured using absorbance (OD) at 450 nm to analyze cell viability.
Cell Apoptosis Analysis
GC cells (1 × 106) were cultured prior to treatment with chrysin, siRNAs, overexpression vectors, let-7a-3p mimics and let-7a-inhibitor for detection of cellular apoptosis, as previously described (25). Then, Annexin V-FITC/PI reagent was added to cell to react for 30 min and flow cytometry (BD Biosciences, Franklin Lakes, NJ, USA) was used to detect fluorescent cells.
Western Blot Analysis
Total protein was extracted from GC cells using protein extraction buffer (Beyotime, China). Then, proteins were quantified utilizing the BCA protein assay kit (TIANGEN, Beijing, China). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was used to separate the proteins. After electrophoresis, proteins were transferred to the polyvinylidene difluoride (PVDF) membrane. The membrane was then incubated with primary antibodies, including anti-COPB2 (BETHYL, A304-522A-M-1, USA), anti-P53 (Abcam, ab131442, USA), anti-BAX (CST, D2E11, USA), anti-BCL2 (CST, D55G8, USA), anti-E-CADHERIN (Proteintech, 20874-1-AP,USA) and anti-GAPDH (Bioworld, AP0066, USA), overnight at 4°C. Subsequently, membranes were incubated with HRP-conjugated affiniPure goat antibodies IgG (BOSTER, China) for 1.5 h. The target bands were analyzed using ECL Super Signal (Pierce, USA).
Hematoxylin and Eosin (H&E) Staining
Tumor tissues from the control and chrysin groups were fixed in 4% paraformaldehyde for 48 h, embedded in paraffin wax and sliced into 5 μm sections. The slides were then stained with H&E and cancer cell infiltration was determined by observation under a light microscope.
Animals and Animal Care
For in vivo experiments, 17 female nude mice (6–8 weeks old) were utilized to determine the effect of chrysin treatment and COPB2 KO on tumor growth. The mice were acquired and grouped-housed in the Laboratory Animal Center of Jilin University. All mice were provided ad libitum access to standard rodent food and tap water within the laboratory cages, as well as under specific pathogen-free (SPF) conditions. The BGC823, pcDNA3.1-COPB2 and COPB2-KO cell lines (3 × 106) were subcutaneously injected into the left flank of each mouse, and tumors were observed after seven days. The tumor length (L) and width (W) were calculated as L × W2/2.
Statistical Analysis
An unpaired Student’s t-test was utilized in the present study. The SPSS 16.0 software (SPSS Inc., Chicago, IL, USA) helped conduct statistical analysis. All data was expressed as mean ± SD. A p-value of <0.05 was considered to be statistically significant. The website of http://ualcan.path.uab.edu/index.html was used for The Cancer Genome Atlas (TCGA) analysis. The TargetScan database was used to predict miRNA.
Results
Screen of Differentially Expressed Gene of Chrysin-Treated GC Cells
In order to analyze gene expression patterns of chrysin treatment in gastric cancer cells, we performed RNA-Seq. Overall, 20,010 genes were identified as core genes (Figure 1A). Compared to the control group, 380 genes were significantly up-regulated while 2,071 were significantly down-regulated (Figure 1B). Data from heatmap and KEGG pathway suggests that the differentially expressed genes have functions in cell death and growth (Figures 1C, D). In order to confirm this data, six genes (CAPN2, MXI1, HSPA9, RHBDD2, COPB2 and GABAPAPL1), which were related to cell growth and death, were further validated (Figure 1E). The qPCR results indicated that COPB2 expression was downregulated upon chrysin treatment in SGC7901 (Figure 1F) and MKN45 (Figure 1G) cells. These results indicate that COPB2 expression is regulated by chrysin in GC cells.
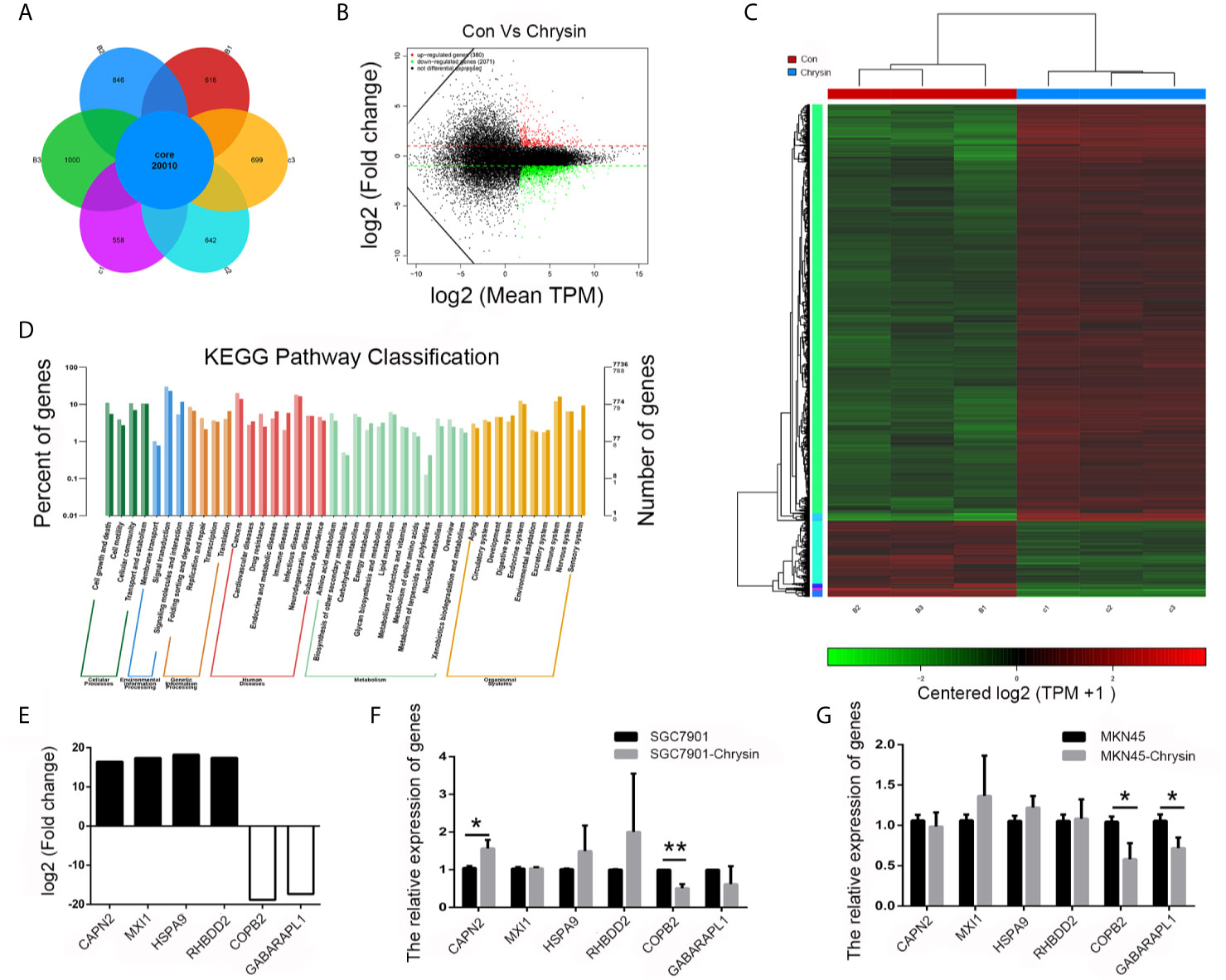
Figure 1 Screening of differentially expressed genes by RNA-seq. Analysis of differential expressed genes after chrysin treatment (A), Identification of different expressed genes (B). The heatmap was drawn to show the differentially expressed genes (C). KEGG pathway of the differentially expressed genes (D). The expression of log2 fold change in six genes (E). Relative expression of CAPN2, MXI1, HSPA9, RHBDD2, COPB2 and GABAPAPL1 were analyzed by qPCR after Chrysin treatment in SGC7901 (F) and MKN45 cells (G). The data are represented as the mean ± SD (n = 3). * (p <0.05) and ** (p <0.01) indicate statistically significant differences.
Chrysin Increased let-7a and Inhibited H19 and COPB2 Expression in GC Cells
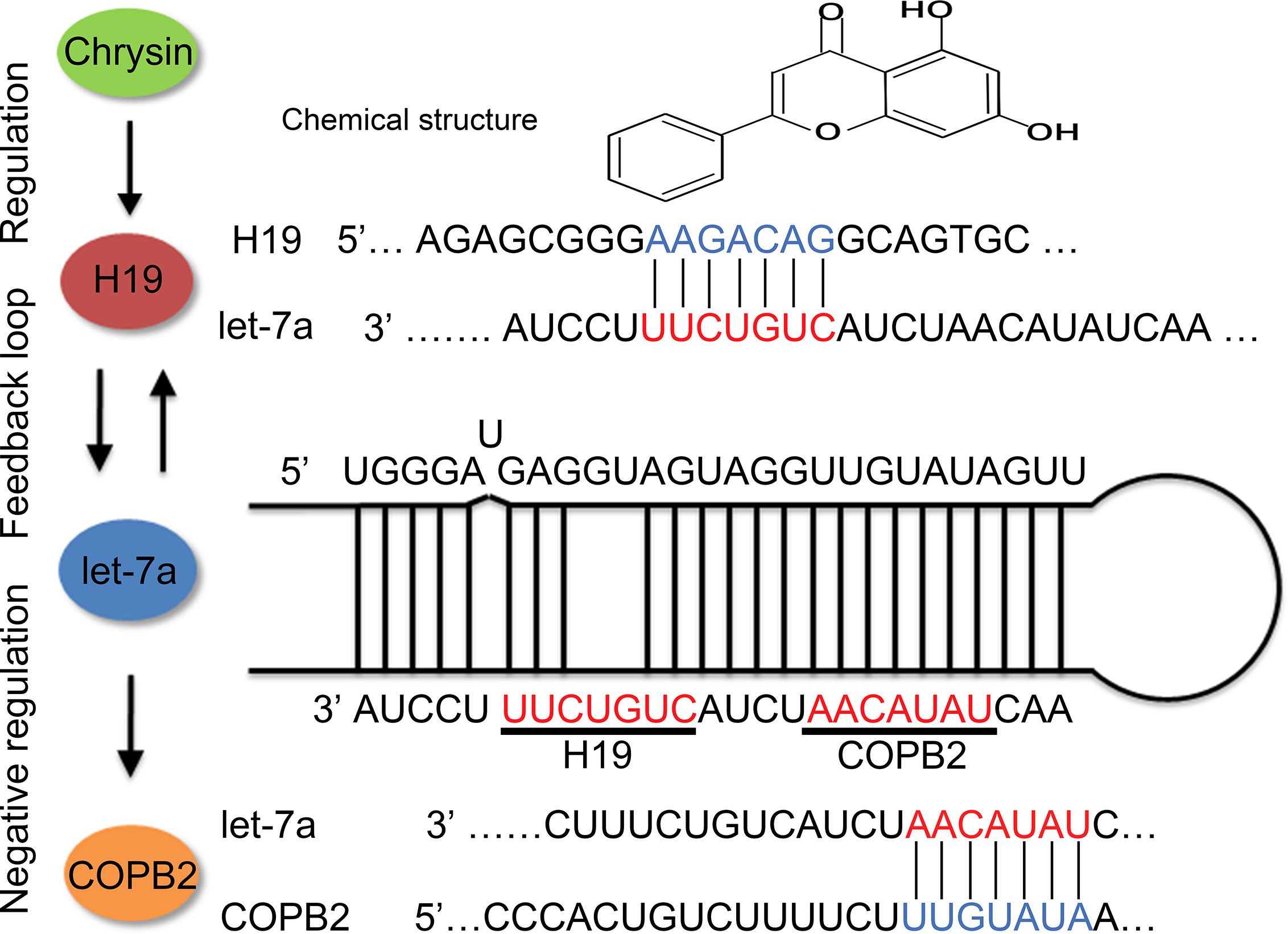
In order to further verify the expression levels of COPB2 in GC, we utilized the TCGA database. Results indicated increased expression of COPB2 in primary tumor of the stomach adenocarcinoma (STAD) patients (Figure S1A). Compared to GES1 cells, qPCR and western bolt results indicated increased expression of COPB2 in MKN45, SGC7901 and BGC823 cells (Figures 2A, B). To investigate which miRNAs were involved in COPB2 expression, bioinformatics analysis was performed. The database suggested that let-7a targets COPB2 (Figures 2C, S2). Furthermore, we analyzed let-7a levels in the TCGA database. Results indicated no differences between normal and tumor tissues (Figure S1B). However, qPCR results suggested that let-7a levels were reduced in GC cells, compared to GES1 cells (Figure 2D). Considering that let-7a is associated with expression of the lncRNA H19, we analyzed the expression pattern of H19. The TCGA database indicated increased expression of H19 among STAD patients (Figure S1C). The qPCR results indicated increased expression of H19 in GC cells (Figure 2E). DNA methylation results indicated the hypo-methylation profile of H19 DMR in GC cells (Figure S3). The cell growth was analyzed after chrysin treatment. The CCK8 results indicated that 40 μM was the optimal dose for subsequent experiments (Figure 2F). Moreover, qPCR results indicated increased expression of let-7a, as well as reduced expression of H19 after chrysin treatment in GC cells (Figure 2G). Besides, chrysin was able to induce cell apoptosis, as well as inhibit cell migration and invasion in GC cells (Figures 3, S4). These results indicate that chrysin has an anti-cancer role and regulates expression of COPB2, H19 and let-7a in GC cells.
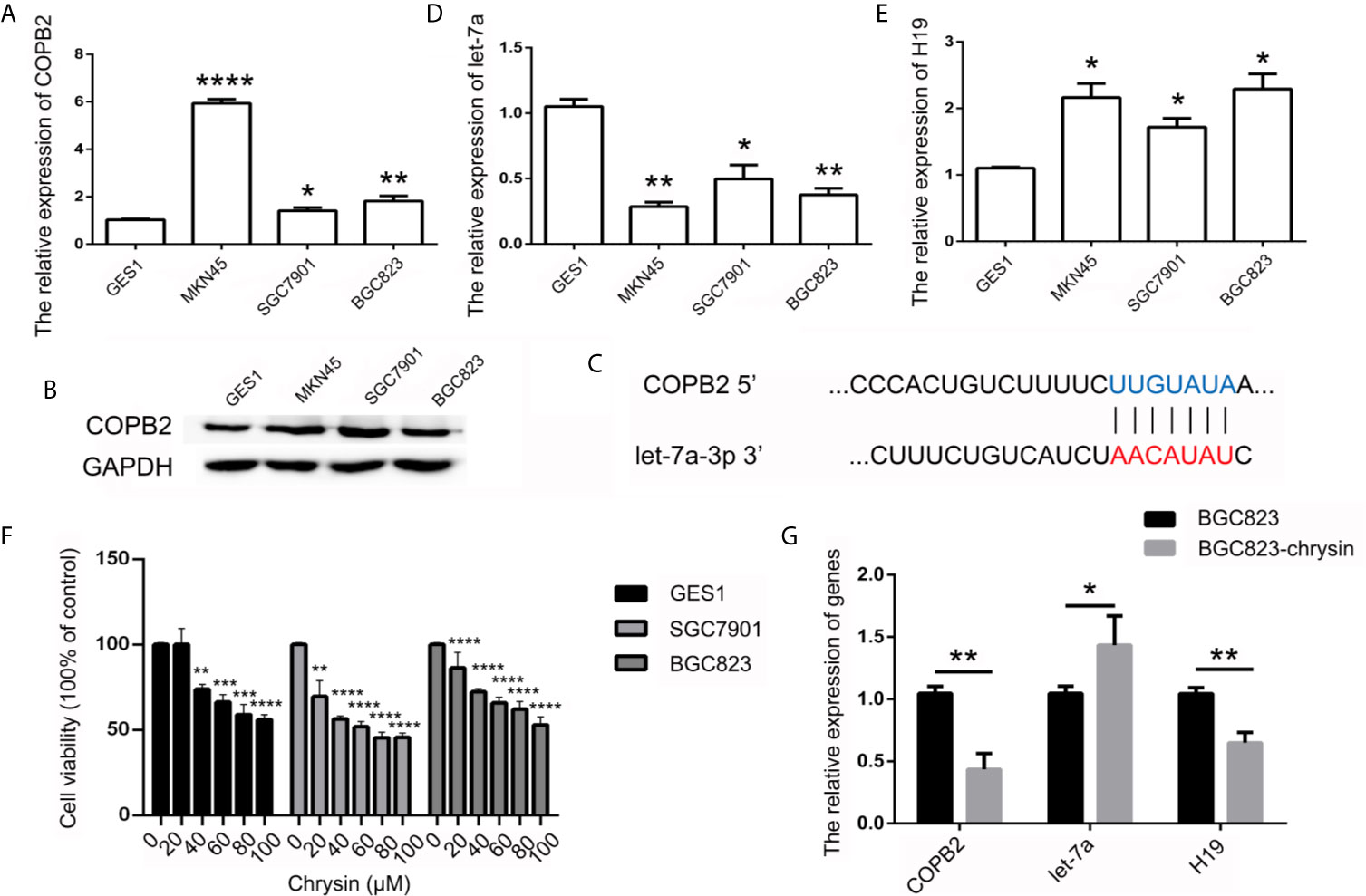
Figure 2 Analysis of COPB2, let-7a and H19 expression pattern. The expression of COPB2 in GC cells using qPCR (A) and western blot (B). Schematic representations of COPB2 and let-7a (C). Relative expression of let-7a in GC ells (D). Relative expression of H19 in GC ells (E). The cell growth was analyzed by CCK8 assay (F). Relative expression of COPB2, let-7a and H19 was analyzed by qPCR after chrysin treatment in GC cells (G). The data are represented as the mean ± SD (n = 3). * (p <0.05), ** (p <0.01), *** (p <0.001) and **** (p <0.0001) indicate statistically significant differences.
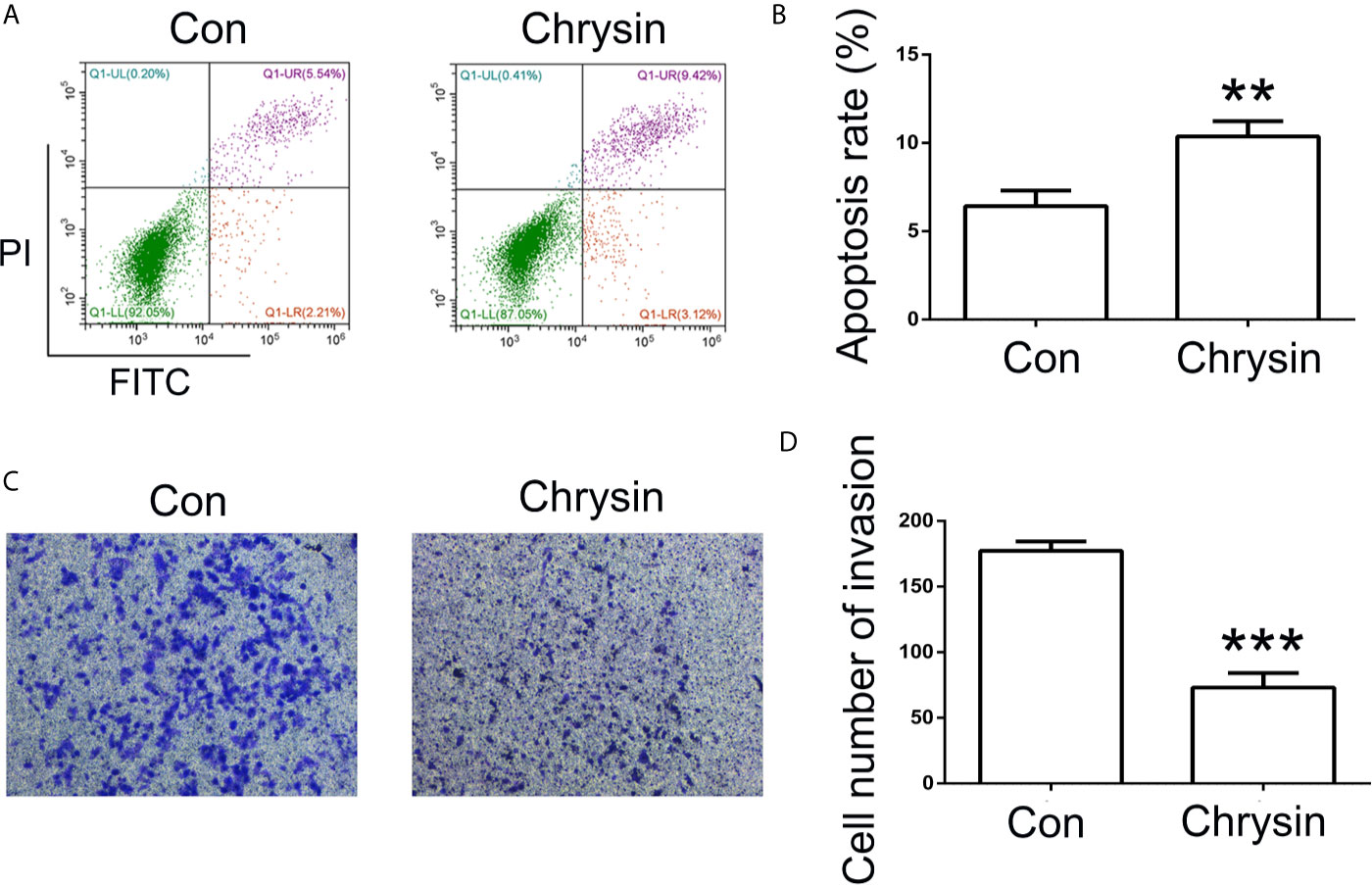
Figure 3 Analysis of cell apoptosis and invasion after chrysin treatment. The cell apoptosis was analyzed between Con and chrysin group (A). Statistical analysis of the percentage of cell apoptosis (B). The cell invasion was analyzed (C). Statistical analysis of the percentage of cell invasion (D). The data are represented as the mean ± SD (n = 3). ** (p <0.01) and *** (p <0.001) indicate statistically significant differences.
H19/let-7a Regulate COPB2 Expression
Considering that H19 has a role in let-7a expression, we analyzed the effect of H19 knockdown and overexpression in GC cells. The results indicated reduced expression of let-7a in the H19 overexpression group, as well as overexpression of let-7a in the H19 knockdown group (Figures 4A–C). In order to investigate the expression pattern of let-7a, we utilized miRNA mimics and inhibitors. The qPCR and western blot results demonstrated that let-7a mimics led to suppressed expression of COPB2 (Figures 4D–G). These results confirm that H19 acts as a sponge that competes with let-7a, thus regulating COPB2 expression.
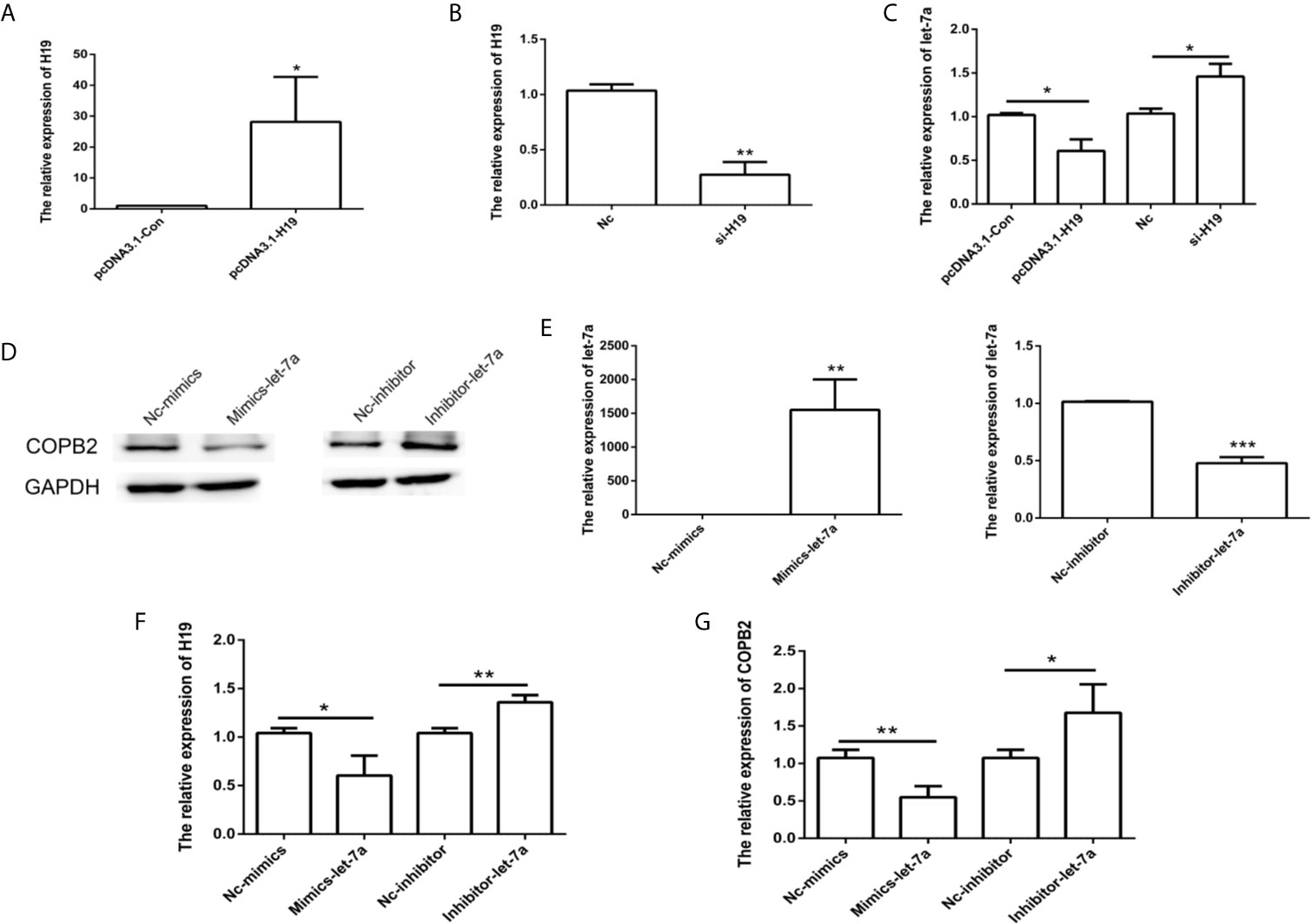
Figure 4 Analysis of COPB2 expression pattern through H19/let-7a in GC cells. Relative expression of H19 and let-7a in pcDNA3.1-Con, pcDNA3.1-H19, Nc and si-H19 group using qPCR (A–C). Expression of COPB2 protein in Nc-mimics, mimics-let-7a, Nc-inhibitor and inhibitor-let-7a group using Western blot (D). Relative expression of let-7a, H19 and COPB2 in Nc-mimics, mimics-let-7a, Nc-inhibitor and inhibitor-let-7a group using qPCR (E–G). The data are represented as the mean ± SD (n = 3). * (p <0.05), ** (p <0.01) and *** (p <0.001) indicate statistically significant differences.
Reduced Expression of COPB2 Induced Cell Apoptosis and Inhibited Cell Invasion
In order to analyze whether COPB2 expression has an effect on cell death and growth, we transfected knockdown and overexpression vector of COPB2 into GC cells. The CCK8 data demonstrates that reduced expression of COPB2 and increased expression of let-7a inhibits cell growth (Figure S5). Moreover, our results suggested that reduced expression of COPB2 induced cellular apoptosis (Figures 5A, B). In order to validate this, we analyzed markers of cell apoptosis. The results showed that increased expression of p53 was observed in the COPB2 knockdown group (Figure S6). In addition, COPB2 expression did not have an effect on cell migration in GC cells (Figures 5C, D). However, our data shows that reduced expression of COPB2 inhibited cell invasion (Figures 5E, F). Next, we investigated the effect of chrysin about cell migration and invasion in the COPB2 overexpression group. This result demonstrated that chrysin induced cell apoptosis and inhibited cell migration and invasion (Figures S7, S8). In order to validate this finding, the overexpression and knockdown of H19 and let-7a were used to analyze cell apoptosis. The results demonstrated that reduced expression of H19 and increased expression of let-7a induced cell apoptosis (Figure S9). Overall, these results suggest that COPB2 has a role in cell apoptosis and invasion.
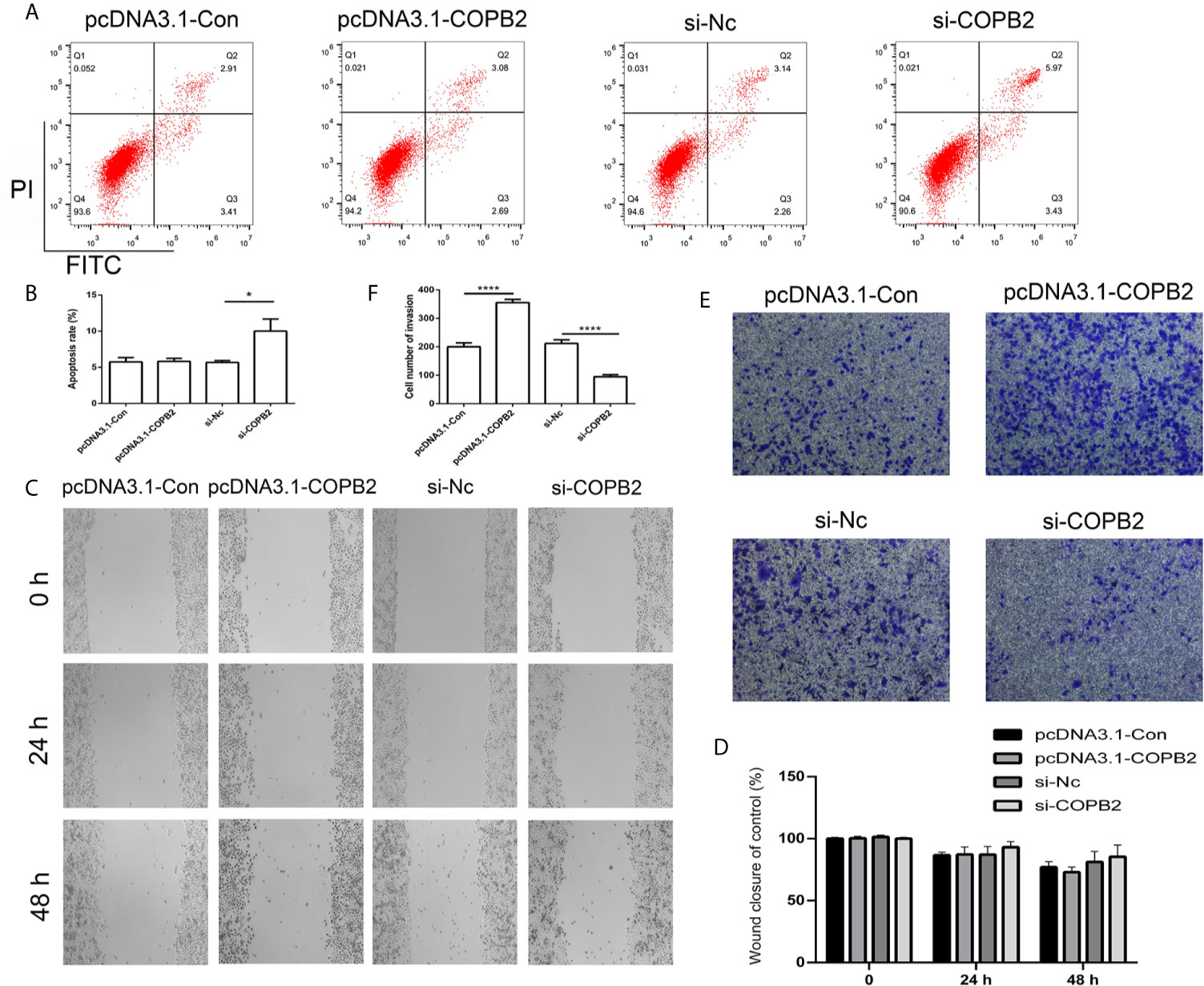
Figure 5 Analysis of cell apoptosis, migration and invasion after knockdown and overexpression of COPB2. Cell apoptosis was analyzed in the pcDNA3.1-Con, pcDNA3.1-COPB2, si-Nc, and si-COPB2 group (A, B). Cell migration was analyzed after si-COPB2 and pcDNA3.1-COPB2 were transfected (C, D). The cell invasion was analyzed in pcDNA3.1-Con, pcDNA3.1-COPB2, si-Nc, and si-COPB2 group (E, F). Statistical analysis of the percentage of cell invasion (D). The data are represented as the mean ± SD (n = 3). * (p <0.05) and **** (p <0.0001) indicate statistically significant differences.
Loss Expression of COPB2 Inhibited Tumor Growth In Vivo
In order to assess the anti-cancer effect of chrysin in vivo, we utilized nude mice. The BGC823 cells were injected into nude mice and after seven days, mice were treated with chrysin (20 mg/kg) for two weeks. The results showed that chrysin is able to inhibit tumor growth and COPB2 expression in vivo (Figures 6A–D). The H&E staining results confirm this data (Figure S10). Moreover, qPCR result indicated that chrysin inhibited the expression of H19, and increased let-7a in vivo (Figure S11). In order to determine the effect of loss expression of COPB2 in vivo, CRISPR/Cas9 system was used to edit the COPB2 exon 5 (Figure 6E). The qPCR data showed lower expression of COPB2 in the COPB2 KO group compared to the control group (Figure 6F). Moreover, chrysin treatment reduced COPB2 expression in overexpression and KO of COPB2 cells (Figure S12). To further confirm the effect of COPB2 expression, COPB2 KO cells were injected into nude mice. Results suggested that loss of expression of COPB2 inhibited tumor growth (Figures 6G, H). Overall, these results indicate that reduced expression of COPB2 leads to anti-tumor effects.
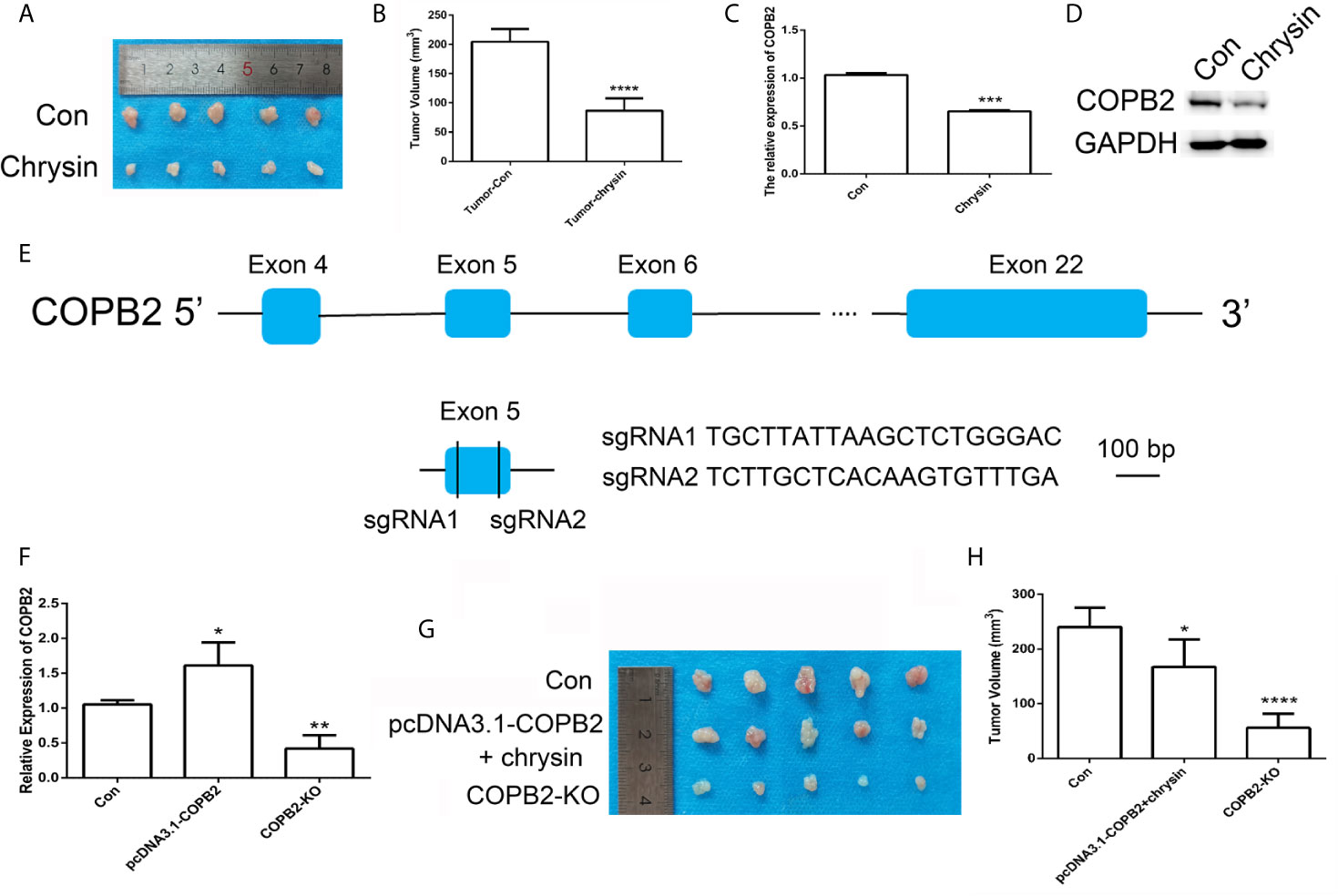
Figure 6 CRISPR/Cas9-mediated gene targeting of COPB2. Morphological observation of mouse tumor tissue (A). Analysis of tumor volume (B). The expression pattern of COPB2 in tumor of mice after chrysin treatment (C, D). Schematic diagram of sgRNAs targeting the COPB2 gene loci (E). The expression of COPB2 using qPCR (F). The tumor morphology (G) and volume (H). Blue indicated COPB2. * (p < 0.05), ** (p < 0.01), *** (p < 0.001) **** and (p < 0.0001) indicate statistically significant differences.
Discussion
Chrysin, a natural medicine, has anti-inflammatory and anti-cancer function and has been used to treat degenerative disorders and cancers in several Asian countries (26, 27). In this study, chrysin was used to treat GC cells in order to evaluate its effect on cellular apoptosis, growth, migration and invasion. Previous reports have indicated that chrysin induces cell apoptosis and inhibits cell growth, migration and invasion in glioblastoma cells (28), which was by validated by our results, which showed that chrysin has anti-cancer effects in SGC7901, MKN45 and BGC823 cells. Moreover, chrysin was found to increase expression of miR-9 and let-7a in GC cells, in accordance with previous data (29). Interestingly, our previous data suggested that chrysin inhibited cell migration and invasion in MKN45 cells through TET1, which regulates global DNA methylation (30). Previous reports have indicated abnormal DNA methylation in GC (31). Herein, our results suggested that H19 DMR is hypomethylated in SGC7901 and BGC823 cell lines, which is related to increased expression of H19. In addition, chrysin functions to regulate the expression of H19, let-7a and COPB2. Furthermore, chrysin inhibited cell invasion which was overexpression of COPB2.
Recently, lncRNAs and miRNAs have been demonstrated to have roles in development of different types of cancer, including HCC (18). There is evidence that reduced expression of lncRNA H19 leads to inhibition of tumor growth in breast cancer, bladder cancer and colorectal cancer (32). Moreover, H19, as a competitive endogenous RNA, is associated with miRNAs, such as miR-29 and let-7 (16, 33). Our results indicated that H19 and let-7a have competitive regulation in GC cells, which has also been confirmed in a previous report (17). Emerging evidence suggested that loss of expression of let-7 correlates with poor prognosis in various cancer (34). Our data showed that chrysin increased expression of let-7a and inhibited cell migration and invasion. Previous reports indicated that silencing of let-7, which targets MDM4, promotes cell proliferation, migration and invasion (35). Further, reduced expression of H19 and increased let-7a expression induced cell apoptosis in GC cells, which validated the previous report (36). These results suggested that the expression of H19 and let-7a is involved in cell apoptosis, growth and invasion of GC cells.
Previous reports indicated that let-7f targets HMGA2 in thyroid cancer (37). Compared to let-7f, IL-6 and CKIP-1 were reported to be targets of let-7a (38, 39). Our results suggest that let-7a targets COPB2, which leads to differential expression after chrysin treatment in GC cells. A previous study indicated that the expression of COPB2 is associated with cell growth, apoptosis, migration and invasion, functioning through a miR-216a manner, in lung cancer (8). Our results suggested that expression of COPB2 was regulated by H19/let-7 axis in GC cells (Figure 7). Moreover, reduced expression of COPB2 induced cellular apoptosis and inhibited cell growth in prostate cancer (40). Our data showed reduced expression of COPB2 increased p53 and E-cadherin expression. These results indicated that reduced expression of COPB2 induced cell apoptosis and inhibited invasion through H19/let-7a.
In order to confirm the effect of COPB2 in vivo, a xenograft model using nude mice was established. The in vivo results suggested that chrysin led to reduced expression of COPB2, which further confirms our data in GC cells. Furthermore, chrysin inhibited tumor growth in vivo, which is in accordance with a previous report in melanomas (41). These data indicate that chrysin can regulate COPB2 expression, which inhibits tumor growth in vivo. In order to further analyze the putative effect of COPB2, the KO and overexpression COPB2 cell line were injected into nude mice. As a powerful gene editing tool, the CRISPR/Cas9 system was wildly used in vitro and in vivo. Our results indicated that COPB2 KO suppressed tumor growth.
In summary, COPB2 is a differentially expressed gene that was identified after chrysin treatment in GC cells. Our data indicated that COPB2 is regulated by let-7a, which acts as a molecular sponge of H19. Moreover, reduced expression of COPB2 induced cellular apoptosis and inhibited cell growth and invasion. Therefore, this present study revealed that COPB2 is a potential molecular targeted therapy in GC.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by Laboratory Animal Center of Jilin University.
Author Contributions
DW designed the experiments and wrote the manuscript. LC, QL, HH, and CL performed cell experiment and gene expression analysis. TW and ZJ contributed reagents and materials. YG and CL carried out animal experiment. DW analyzed the data and prepared figures. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Jilin Health Commission Program under Grant 2020J05S, the Fundamental Research Funds for the Central Universities under Grant 2019JCKT-70, the Jilin Education Department Program under Grant JJKH20200950KJ, and the Jilin Scientific and Technological Development Program under Grant 20190103071JH.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.651644/full#supplementary-material
Supplementary Figure 1 | Analysis of COPB2, let-7a and H19 in TCGA database. Effect of COPB2 (A), let-7a (B) and H19 (C) expression on GC patient survival in TCGA database.
Supplementary Figure 2 | Screen of miRNA which regulated by COPB2.
Supplementary Figure 3 | The DNA methylation status of H19 DMR in MGC823 and SGC7901 using BSP.
Supplementary Figure 4 | Analysis of cell migration after chrysin treatment. The cell migration was analyzed between Con and chrysin group (A). Statistical analysis of the percentage of cell migration (B). **** (p < 0.0001) indicate statistically significant differences.
Supplementary Figure 5 | The CCK8 assay results in overexpression and knockdown of COPB2. The CCK8 assay in GES1 cells after chrysin treatment (A). The CCK8 assay in pcDNA3.1-Con, pcDNA3.1-H19, Nc and si-H19 group (B). The CCK8 assay in Nc-mimics, mimics-let-7a, Nc-inhibitor and inhibitor-let-7a group (C). The mRNA expression of COPB2 in the pcDNA3.1-Con, pcDNA3.1-COPB2, si-Nc, si-COPB2 group using qPCR (D). The CCK8 assay in pcDNA3.1-Con, pcDNA3.1-COPB2, si-Nc, and si-COPB2 group (E). ** (p < 0.01), *** (p < 0.001) and **** (p < 0.0001) indicate statistically significant differences.
Supplementary Figure 6 | Analysis of cell apoptosis markers. The protein expression of COPB2, BAX, BCL2, P53 and E-Cadherin were analyzed using western blot (A). The relative mRNA expression of BAX, BCL2 and TP53 were analyzed using qPCR (B). * (p < 0.05) and ** (p < 0.01) indicate statistically significant differences.
Supplementary Figure 7 | Cell apoptosis analysis. The cell apoptosis was analyzed after chrysin treatment in pcDNA3.1-COPB2 group (A, B). *** (p < 0.001) indicate statistically significant differences.
Supplementary Figure 8 | The cell migration and invasion analysis. The cell migration was analyzed after chrysin treatment in pcDNA3.1-COPB2 group (A, B). Cell invasion was analyzed after chrysin treatment in the pcDNA3.1-COPB2 group (C, D). * (p < 0.05) and ** (p < 0.01) indicate statistically significant differences.
Supplementary Figure 9 | Cell apoptosis and cell growth analysis. The cell apoptosis was analyzed in pcDNA3.1-Con, pcDNA3.1-H19, Nc and si-H19 group (A, B). Cell apoptosis was analyzed in the Nc-mimics, mimics-let-7a, Nc-inhibitor and inhibitor-let-7a group (C, D). ** (p < 0.01) and *** (p < 0.001) indicate statistically significant differences.
Supplementary Figure 10 | Histopathological observation of tumor tissue.
Supplementary Figure 11 | Analysis of H19 and let-7a levels after chrysin treatment using qPCR in vivo. * (p < 0.05) and **** (p < 0.0001) indicate statistically significant differences.
Supplementary Figure 12 | Analysis of chrysin treatment in overexpression and KO of COPB2. The genes expression in pcDNA3.1-COPB2 (A) and COPB2-KO (B) group after chrysin treatment using qPCR. The expression of COPB2 and P53 in pcDNA3.1-COPB2 (C) and COPB2-KO (D) group after chrysin treatment using western blot. * (p < 0.05) and ** (p < 0.01) indicate statistically significant differences.
References
1. Global Burden of Disease Cancer C, Fitzmaurice C, Dicker D, Pain A, Hamavid H, Moradi-Lakeh M, et al. The Global Burden of Cancer 2013. JAMA Oncol (2015) 1(4):505–27. doi: 10.1001/jamaoncol.2015.0735
2. Chia NY, Tan P. Molecular Classification of Gastric Cancer. Ann Oncol: Off J Eur Soc Med Oncol (2016) 27(5):763–9. doi: 10.1093/annonc/mdw040
3. Chen K, Yang D, Li X, Sun B, Song F, Cao W, et al. Mutational Landscape of Gastric Adenocarcinoma in Chinese: Implications for Prognosis and Therapy. Proc Natl Acad Sci USA (2015) 112(4):1107–12. doi: 10.1073/pnas.1422640112
4. Huang Y, Yuan K, Tang M, Yue J, Bao L, Wu S, et al. Melatonin Inhibiting the Survival of Human Gastric Cancer Cells Under ER Stress Involving Autophagy and Ras-Raf-MAPK Signalling. J Cell Mol Med (2020) 25(3):1480–92. doi: 10.1111/jcmm.16237
5. Beck R, Rawet M, Wieland FT, Cassel D. The COPI System: Molecular Mechanisms and Function. FEBS Lett (2009) 583(17):2701–9. doi: 10.1016/j.febslet.2009.07.032
6. Wang Y, Chai Z, Wang M, Jin Y, Yang A, Li M. COPB2 Suppresses Cell Proliferation and Induces Cell Cycle Arrest in Human Colon Cancer by Regulating Cell Cycle-Related Proteins. Exp Ther Med (2018) 15(1):777–84. doi: 10.3892/etm.2017.5506
7. Li ZS, Liu CH, Liu Z, Zhu CL, Huang Q. Downregulation of COPB2 by RNAi Inhibits Growth of Human Cholangiocellular Carcinoma Cells. Eur Rev Med Pharmacol Sci (2018) 22(4):985–92. doi: 10.26355/eurrev_201802_14380
8. Wang X, Shi J, Niu Z, Wang J, Zhang W. MiR-216a-3p Regulates the Proliferation, Apoptosis, Migration, and Invasion of Lung Cancer Cells Via Targeting COPB2. Biosci Biotechnol Biochem (2020) 84(10):2014–27. doi: 10.1080/09168451.2020.1783197
9. Wang Y, Xie G, Li M, Du J, Wang M. COPB2 Gene Silencing Inhibits Colorectal Cancer Cell Proliferation and Induces Apoptosis Via the JNK/c-Jun Signaling Pathway. PloS One (2020) 15(11):e0240106. doi: 10.1371/journal.pone.0240106
10. Chen HL, Li JJ, Jiang F, Shi WJ, Chang GY. MicroRNA-4461 Derived From Bone Marrow Mesenchymal Stem Cell Exosomes Inhibits Tumorigenesis by Downregulating COPB2 Expression in Colorectal Cancer. Biosci Biotechnol Biochem (2020) 84(2):338–46. doi: 10.1080/09168451.2019.1677452
11. Pu X, Jiang H, Li W, Xu L, Wang L, Shu Y. Upregulation of the Coatomer Protein Complex Subunit Beta 2 (COPB2) Gene Targets microRNA-335-3p in NCI-H1975 Lung Adenocarcinoma Cells to Promote Cell Proliferation and Migration. Med Sci Monitor: Int Med J Exp Clin Res (2020) 26:e918382. doi: 10.12659/MSM.918382
12. Alessandrini L, Manchi M, De Re V, Dolcetti R, Canzonieri V. Proposed Molecular and Mirna Classification of Gastric Cancer. Int J Mol Sci (2018) 19(6):1683. doi: 10.3390/ijms19061683
13. Hu W, Chen Y, Lv K. The Expression Profiles of MicroRNA Let-7a in Peripheral Blood Mononuclear Cells From Patients of Gastric Cancer With Neoadjuvant Chemotherapy. Clin Lab (2018) 64(5):835–9. doi: 10.7754/Clin.Lab.2017.171213
14. Tang G, Du R, Tang Z, Kuang Y. MiRNALet-7a Mediates Prostate Cancer PC-3 Cell Invasion, Migration by Inducing Epithelial-Mesenchymal Transition Through CCR7/MAPK Pathway. J Cell Biochem (2018) 119(4):3725–31. doi: 10.1002/jcb.26595
15. Chen JS, Wang YF, Zhang XQ, Lv JM, Li Y, Liu XX, et al. H19 Serves as a Diagnostic Biomarker and Up-Regulation of H19 Expression Contributes to Poor Prognosis in Patients With Gastric Cancer. Neoplasma (2016) 63(2):223–30. doi: 10.4149/207_150821N454
16. Peng F, Li TT, Wang KL, Xiao GQ, Wang JH, Zhao HD, et al. H19/let-7/LIN28 Reciprocal Negative Regulatory Circuit Promotes Breast Cancer Stem Cell Maintenance. Cell Death Dis (2017) 8(1):e2569. doi: 10.1038/cddis.2016.438
17. Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, et al. The Imprinted H19 lncRNA Antagonizes Let-7 Micrornas. Mol Cell (2013) 52(1):101–12. doi: 10.1016/j.molcel.2013.08.027
18. Zhong X, Huang S, Liu D, Jiang Z, Jin Q, Li C, et al. Galangin Promotes Cell Apoptosis Through Suppression of H19 Expression in Hepatocellular Carcinoma Cells. Cancer Med (2020) 9(15):5546–57. doi: 10.1002/cam4.3195
19. Xiang Y, Guo Z, Zhu P, Chen J, Huang Y. Traditional Chinese Medicine as a Cancer Treatment: Modern Perspectives of Ancient But Advanced Science. Cancer Med (2019) 8(5):1958–75. doi: 10.1002/cam4.2108
20. Xu D, Jin J, Yu H, Zhao Z, Ma D, Zhang C, et al. Chrysin Inhibited Tumor Glycolysis and Induced Apoptosis in Hepatocellular Carcinoma by Targeting Hexokinase-2. J Exp Clin Cancer Res: CR (2017) 36(1):44. doi: 10.1186/s13046-017-0514-4
21. Yufei Z, Yuqi W, Binyue H, Lingchen T, Xi C, Hoffelt D, et al. Chrysin Inhibits Melanoma Tumor Metastasis Via Interfering With the FOXM1/Beta-Catenin Signaling. J Agric Food Chem (2020) 68(35):9358–67. doi: 10.1021/acs.jafc.0c03123
22. Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science (2013) 339(6121):819–23. doi: 10.1126/science.1231143
23. Clark SJ, Harrison J, Paul CL, Frommer M. High Sensitivity Mapping of Methylated Cytosines. Nucleic Acids Res (1994) 22(15):2990–7. doi: 10.1093/nar/22.15.2990
24. Ding C, Li L, Yang T, Fan X, Wu G. Combined Application of Anti-VEGF and Anti-EGFR Attenuates the Growth and Angiogenesis of Colorectal Cancer Mainly Through Suppressing AKT and ERK Signaling in Mice Model. BMC Cancer (2016) 16(1):791. doi: 10.1186/s12885-016-2834-8
25. William-Faltaos S, Rouillard D, Lechat P, Bastian G. Cell Cycle Arrest and Apoptosis Induced by Oxaliplatin (L-OHP) on Four Human Cancer Cell Lines. Anticancer Res (2006) 26(3A):2093–9.
26. Naz S, Imran M, Rauf A, Orhan IE, Shariati MA, Iahtisham Ul H, et al. Chrysin: Pharmacological and Therapeutic Properties. Life Sci (2019) 235:116797. doi: 10.1016/j.lfs.2019.116797
27. Kasala ER, Bodduluru LN, Madana RM, AK V, Gogoi R, Barua CC. Chemopreventive and Therapeutic Potential of Chrysin in Cancer: Mechanistic Perspectives. Toxicol Lett (2015) 233(2):214–25. doi: 10.1016/j.toxlet.2015.01.008
28. Wang J, Wang H, Sun K, Wang X, Pan H, Zhu J, et al. Chrysin Suppresses Proliferation, Migration, and Invasion in Glioblastoma Cell Lines Via Mediating the ERK/Nrf2 Signaling Pathway. Drug Design Dev Ther (2018) 12:721–33. doi: 10.2147/DDDT.S160020
29. Mohammadian F, Pilehvar-Soltanahmadi Y, Zarghami F, Akbarzadeh A, Zarghami N. Upregulation of miR-9 and Let-7a by Nanoencapsulated Chrysin in Gastric Cancer Cells. Artif Cells Nanomed Biotechnol (2017) 45(6):1–6. doi: 10.1080/21691401.2016.1216854
30. Zhong X, Liu D, Jiang Z, Li C, Chen L, Xia Y, et al. Chrysin Induced Cell Apoptosis and Inhibited Invasion Through Regulation of TET1 Expression in Gastric Cancer Cells. OncoTarg Ther (2020) 13:3277–87. doi: 10.2147/OTT.S246031
31. Tahara T, Arisawa T. DNA Methylation as a Molecular Biomarker in Gastric Cancer. Epigenomics-Uk (2015) 7(3):475–86. doi: 10.2217/epi.15.4
32. Ghafouri-Fard S, Esmaeili M, Taheri M. H19 Lncrna: Roles in Tumorigenesis. Biomed Pharmacother Biomed Pharmacother (2020) 123:109774. doi: 10.1016/j.biopha.2019.109774
33. Lv M, Zhong Z, Huang M, Tian Q, Jiang R, Chen J. Lncrna H19 Regulates Epithelial-Mesenchymal Transition and Metastasis of Bladder Cancer by miR-29b-3p as Competing Endogenous RNA. Biochim Biophys Acta Mol Cell Res (2017) 1864(10):1887–99. doi: 10.1016/j.bbamcr.2017.08.001
34. Balzeau J, Menezes MR, Cao S, Hagan JP. The LIN28/let-7 Pathway in Cancer. Front Genet (2017) 8:31. doi: 10.3389/fgene.2017.00031
35. Zhang L, Wang K, Wu Q, Jin L, Lu H, Shi Y, et al. Let-7 Inhibits the Migration and Invasion of Extravillous Trophoblast Cell via Targeting MDM4. Mol Cell Probes (2019) 45:48–56. doi: 10.1016/j.mcp.2019.05.002
36. Zhu Y, Xu F. Up-Regulation of Let-7a Expression Induces Gastric Carcinoma Cell Apoptosis In Vitro. Chin Med Sci J Chung-kuo I Hsueh K’o Hsueh Tsa Chih (2017) 32(1):44–7. doi: 10.24920/j1001-9242.2007.006
37. Damanakis AI, Eckhardt S, Wunderlich A, Roth S, Wissniowski TT, Bartsch DK, et al. MicroRNAs Let7 Expression in Thyroid Cancer: Correlation With Their Deputed Targets HMGA2 and SLC5A5. J Cancer Res Clin Oncol (2016) 142(6):1213–20. doi: 10.1007/s00432-016-2138-z
38. Sun Y, Zhong L, He X, Wang S, Lai Y, Wu W, et al. Lncrna H19 Promotes Vascular Inflammation and Abdominal Aortic Aneurysm Formation by Functioning as a Competing Endogenous RNA. J Mol Cell Cardiol (2019) 131:66–81. doi: 10.1016/j.yjmcc.2019.04.004
39. Yang Z, Jiang X, Zhang J, Huang X, Zhang X, Wang J, et al. Let-7a Promotes Microglia M2 Polarization by Targeting CKIP-1 Following ICH. Immunol Lett (2018) 202:1–7. doi: 10.1016/j.imlet.2018.07.007
40. Mi YY, Yu ML, Zhang LF, Sun CY, Wei BB, Ding WH, et al. Copb2 Is Upregulated in Prostate Cancer and Regulates Pc-3 Cell Proliferation, Cell Cycle, and Apoptosis. Arch Med Res (2016) 47(6):411–8. doi: 10.1016/j.arcmed.2016.09.005
41. Sassi A, Maatouk M, El Gueder D, Bzeouich IM, Abdelkefi-Ben Hatira S, Jemni-Yacoub S, et al. Chrysin, a Natural and Biologically Active Flavonoid Suppresses Tumor Growth of Mouse B16F10 Melanoma Cells: In Vitro and In Vivo Study. Chem-Biol Interact (2018) 283:10–9. doi: 10.1016/j.cbi.2017.11.022
Keywords: H19, let-7a, COPB2, chrysin, gastric cancer
Citation: Chen L, Li Q, Jiang Z, Li C, Hu H, Wang T, Gao Y and Wang D (2021) Chrysin Induced Cell Apoptosis Through H19/let-7a/COPB2 Axis in Gastric Cancer Cells and Inhibited Tumor Growth. Front. Oncol. 11:651644. doi: 10.3389/fonc.2021.651644
Received: 10 January 2021; Accepted: 10 May 2021;
Published: 03 June 2021.
Edited by:
Yue Hou, Northeastern University, ChinaReviewed by:
Yi Qin, Fudan University, ChinaJiang Pi, Guangdong Medical University, China
Xiangyan Li, Changchun University of Chinese Medicine, China
Chunmeng Sun, China Pharmaceutical University, China
Copyright © 2021 Chen, Li, Jiang, Li, Hu, Wang, Gao and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dongxu Wang, d2FuZ19kb25nX3h1QGpsdS5lZHUuY24=
†These authors have contributed equally to this work
 Lin Chen
Lin Chen Qirong Li
Qirong Li Ziping Jiang
Ziping Jiang Chengshun Li
Chengshun Li Haobo Hu1
Haobo Hu1 Dongxu Wang
Dongxu Wang