
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol., 19 March 2021
Sec. Genitourinary Oncology
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.648968
This article is part of the Research TopicBiomarkers in Genitourinary Cancers, Volume IView all 21 articles
 Chunyu Zhang1
Chunyu Zhang1 Jiao Hu1
Jiao Hu1 Huihuang Li1
Huihuang Li1 Hongzhi Ma2
Hongzhi Ma2 Belaydi Othmane1
Belaydi Othmane1 Wenbiao Ren1,3
Wenbiao Ren1,3 Zhenglin Yi1
Zhenglin Yi1 Dongxu Qiu1
Dongxu Qiu1 Zhenyu Ou1
Zhenyu Ou1 Jinbo Chen1*
Jinbo Chen1* Xiongbing Zu1*
Xiongbing Zu1*Bladder cancer is one of the leading causes of cancer deaths worldwide. Early detection of lymph node metastasis of bladder cancer is essential to improve patients’ prognosis and overall survival. Current diagnostic methods are limited, so there is an urgent need for new specific biomarkers. Non-coding RNA and m6A have recently been reported to be abnormally expressed in bladder cancer related to lymph node metastasis. In this review, we tried to summarize the latest knowledge about biomarkers, which predict lymph node metastasis in bladder cancer and their mechanisms. In particular, we paid attention to the impact of non-coding RNA on lymphatic metastasis of bladder cancer and its specific molecular mechanisms, as well as some prediction models based on imaging, pathology, and biomolecules, in an effort to find more accurate diagnostic methods for future clinical application.
Bladder cancer (BCa) is the 10th most common cancer form, causing an estimated 549,000 new cases and 200,000 deaths in 2018. The incidence of BCa in men is four times that of women, and smoking is the most important risk factor for BCa in the population (1). More than 90% of bladder cancers are urothelial carcinoma, and the rest are squamous cell carcinoma and adenocarcinoma.
The most common metastatic manner of BCa is lymph node metastasis (LNM), which is more common in pelvic lymph nodes. LNM has a great influence on the prognosis and survival rate of BCa patients. For BCa patients with positive LNM, the 5-year CSS rate was 27.7%, which is significantly lower than that of patients without lymph node metastasis (2). CT or MRI is commonly used in clinical practice to diagnose pelvic LNM, but it is often difficult to accurately detect metastatic lymph nodes less than 6.8 mm in diameter (3). Many studies have recently reported the correlation between molecular markers and BCa metastasis, indicating a direct link between LNM and abnormal expression of specific biomarkers. Therefore, high-risk LNM patients can be diagnosed by detecting specific biomarkers to achieve early detection and early treatment, thereby achieving timely treatment and improving the survival rate.
Moreover, some predictive models, including imaging, pathology, and molecular markers, have been gradually developed and verified. In this review, we summarized the markers for LNM in BCa from different aspects, including genes, non-coding RNA, and some predictive models (Figure 1). The downstream genes of non-coding RNA are specifically listed here (Table 1). Generally, mechanisms for LNM in cancers mainly include cell proliferation, cell invasion and migration, inhibition of cell apoptosis, and chemosensitivity. Based on this, we also elaborated on the regulation mechanism of these biomarkers.
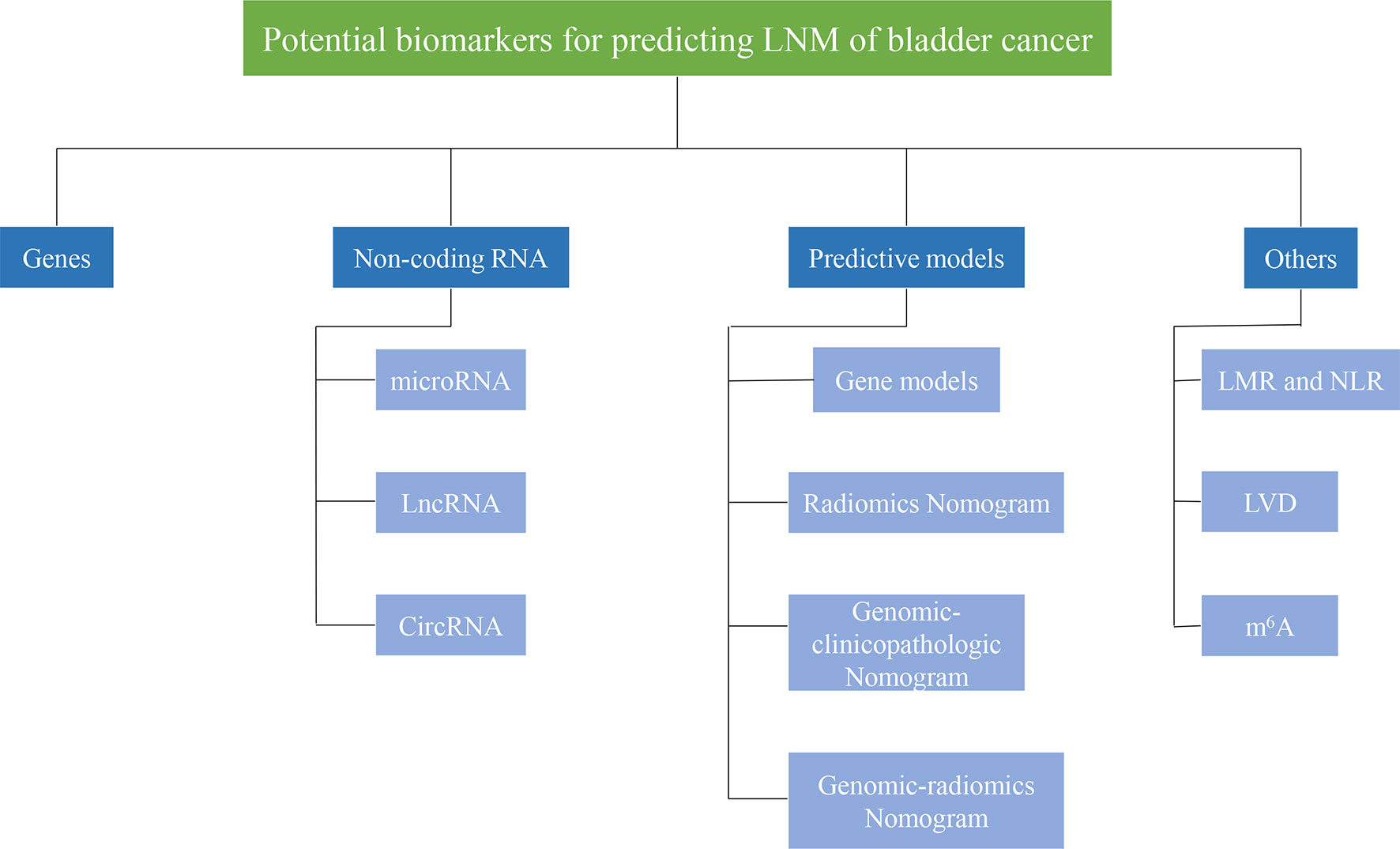
Figure 1 Potential biomarkers for predicting lymph node metastasis of bladder cancer. Biomarkers for predicting lymphatic metastasis of bladder cancer can be divided into different categories, such asgenes, non-coding RNA, prediction models.
There have been many studies on genes as markers for lymph node metastasis in bladder cancer. These genes act as oncogenes or tumor suppressor genes to influence the progression of cancer (Figure 2).
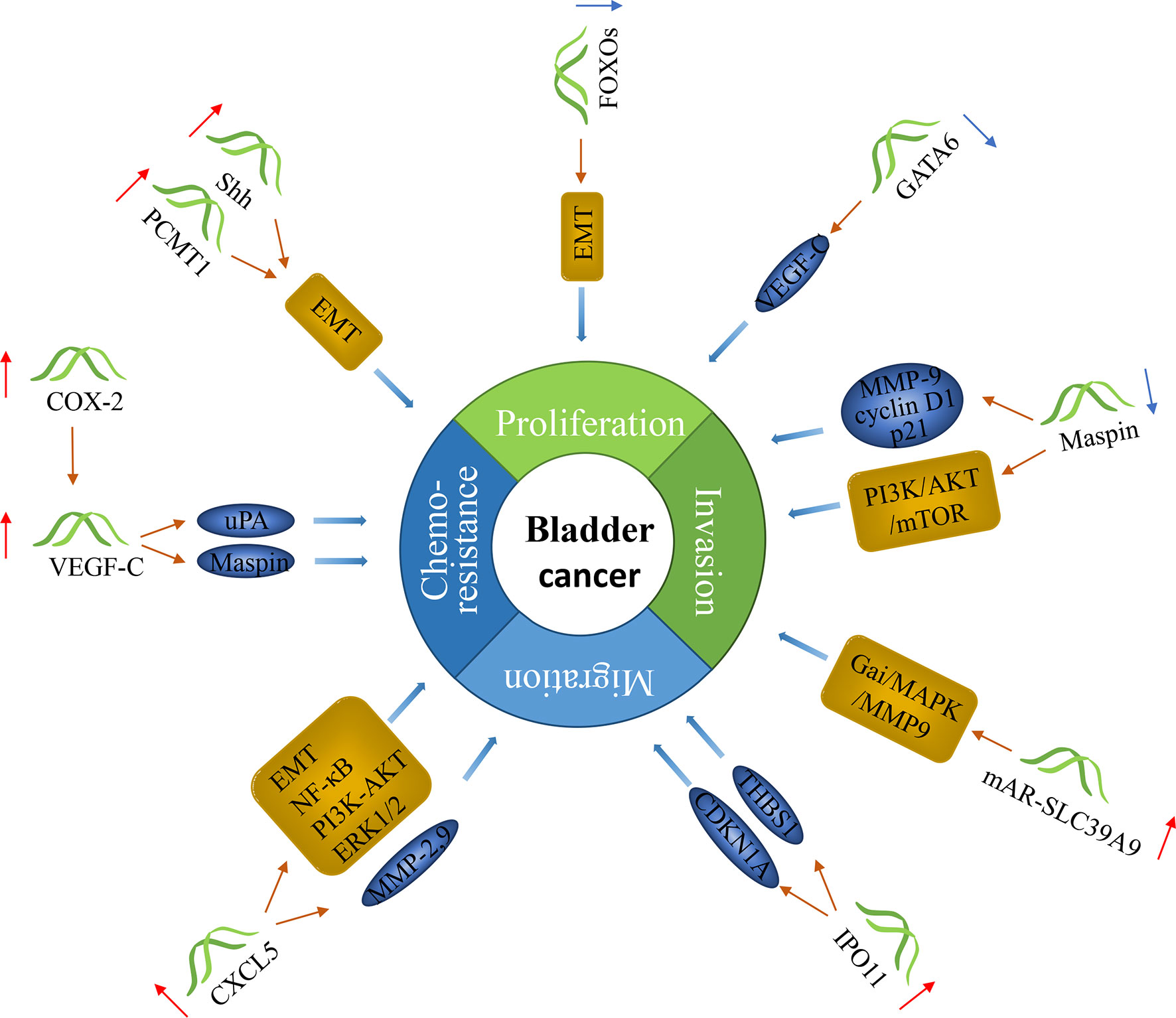
Figure 2 The molecular function of genes in bladder cancer with lymph node metastasis. Genes can predict lymph node metastasis in bladder cancer. Some of them can promote the progression of cancer, and some can inhibit it.
VEGF-C (vascular endothelial growth factor C) is the first discovered lymphangiogenesis factor. It contains the mature form of the VEGF homology region. Our team’s studies found that the expression of VEGF-C in BCa patients with LNM was significantly higher than that in BCa patients without LNM (57). Simultaneously, we also found that VEGF-C can promote proliferation, invasion, metastasis, and mitomycin C resistance of BCa cells. The mechanisms for that are thought to be related to the increased ratio of Bcl-2/Bax, inactivation of Caspase-3, and increased expression of MMP-9. Also, phosphorylated p38 MAPK and Akt, Keratin 8, Serpin B5, and Annexin A8 may be involved (58, 59). VEGF-C can promote the formation of tumor lymphatic vessels and the metastasis of tumor cells to regional lymph nodes. The combination of the activated VEGF-C and VEGFR-3 can induce phosphorylation of tyrosine kinase, causing the proliferation of lymphatic endothelial cells, thereby promoting the proliferation or expansion of lymphatic vessels (60). VEGF-C also positively affected primary tumor cells’ invasiveness since it changed the adhesion of tumor cells to the extracellular matrix, thereby providing the necessary environmental conditions for tumor cells to more easily transfer to the surrounding extracellular matrix. VEGF-C can stimulate lymphatic endothelial cells to release proteolytic enzymes, such as uPA, which facilitate the invasion and infiltration of cancer cells into the matrix, making cancer cells more easily detached from the original tissue (61). The up-regulation of VEGF-C may be the reason for BCa cells’ resistance to cisplatin, and the inhibition of VEGF-C reverses the resistance by increasing the expression level of maspin (62). Therefore, we suggest that VEGF-C and VEGFR-3 expression may serve as new indicators for early detection and diagnosis of BCa lymphatic metastasis in the future. Additionally, COX-2 may stimulate VEGF-C secretion to promote the formation of lymphatic vessels (63). COX-2, a subtype enzyme in the COX family, is an inducible enzyme. COX (Cyclooxygenase) is a rate-limiting enzyme in prostaglandin synthesis, which can catalyze arachidonic acid metabolites to prostaglandins. Previous studies have shown that COX-2 expression was significantly increased in BCa tissues and was associated with LNM (64).
Another well-known gene that functions as an oncogene in BCa is PCMT1. PCMTl gene is located at 6p22.3-6q24, about 60kb in length, and contains eight exons and seven introns. Studies have shown that the expression of PCMT1 in BCa tissue was higher than that in normal urothelial tissue, and its expression was significantly associated with LNM. PCMT1 regulated the migration and invasion of BCa cells by regulating the expression of epithelial-mesenchymal transition (EMT) related genes, such as E-cadherin, vimentin, Snail, and Slug (65). Sonic Hedgehog (Shh) also activated EMT to promote tumorigenicity and stemness in BCa (66). Shh is a member of the Hedgehog (HH) family. The study found that the expression of Shh protein was significantly correlated with LNM (67). Shh can promote the migration and invasion of BCa cells. The Shh pathway’s activation through the binding of the Shh ligand to the transmembrane protein Patched1 eliminates the inhibitory effect on smoothened (SMO). The activation of SMO produced a downstream signaling cascade that led to the nuclear translocation of the transcription factor Gli1, which further induce the transcription of target genes (68).
The overexpression of CXCL5 can promote the progression of BCa. CXCL5, known as epithelial-derived neutrophil-activating peptide 78 (ENA78), is a small (8-14 kDa) protein belonging to the CXC-type chemokine family. CXCL5 (chemokine C-X-C motif ligand 5) was expressed higher in BCa tissues than normal tissues, which was associated with LNM (69). It is also related to promoting mitomycin resistance by activating EMT and NF-κB pathway (70). Moreover, CXCL5 increased BCa cells proliferation, migration, and decreased cell apoptosis through Snail, PI3K-AKT, and ERK1/2 signaling pathways. In addition, CXCL5 combined with CXCR2 induces the expression of MMP-2 and MMP-9 and activates the PI3K/AKT signaling pathway (71, 72). Matrix metalloproteinases (MMPs) are a family of structurally related zinc-dependent endopeptidases that can substantially degrade all components of the extracellular matrix (ECM). MMP2, MMP7, and MMP9 are important members of the matrix metalloproteinase family. MMP-2 can physiologically degrade type IV collagen. Mohammad et al. (73) found that the higher the MMP-2 activity level in BCa, the higher the positive rate of LNM. MMP-7, also known as matrilysin, is the smallest MMP. It is produced by the tumor cells themselves, unlike other MMPs which are solely produced by stromal cells. Studies have shown that high expression of MMP-7 was significantly associated with LNM of BCa (74). Studies have shown that MMP-9 genes and proteins’ expression levels in urine and blood of patients with BCa were significantly increased (75). These genes can also decompose the extracellular matrix, make cancer cells easily pass through the extracellular matrix, and promote tumor metastasis.
In addition, Zhao et al. (76) identified a new oncogene candidate, IPO11, in BCa, which is located on chromosome 5q12. Importin-11, a 116 kD protein, is encoded by IPO11. It is a karyopherin family member, which mediates the nucleocytoplasmic transport of proteins and nucleic acids through the nuclear pore complexes. Studies have shown that IPO11 mRNA was highly expressed in invasive BCa cell lines. The overexpression of importin-11 was positively correlated with LNM. Importin-11 can promote BCa cells’ invasiveness, which may be related to the abnormal expression of CDKN1A and THBS1 (77). Presler et al. (78) found that SCD1 was overexpressed in BCa, which was related to LNM. SCD-1 (Stearoyl-CoA desaturase-1) can convert SFA (saturated fatty acids) to MUFA (monounsaturated fatty acids). It is located on chromosome 10q24.31. SCD inhibitors and SCD gene interference reduced the proliferation and invasion of BCa cells (79). FGFR3 (fibroblast growth factor receptor 3) stimulated SCD1 activity to promote tumor growth in BCa cells (80).
The studies of our team also found some new oncogenes. ISYNA1 (Inositol-3-phosphate synthase 1) was positively associated with tumor T stage and LNM of BCa patients. It is an important regulatory factor in promoting proliferation and inhibiting apoptosis in BCa cells (81). The high expression of mAR-SLC39A9 was directly associated with BCa pathological stage, pathological grade, and lymph node metastasis presence. It also increased BCa metastasis through Gαi/MAPK/MMP9 signaling (82).
Maspin (mammary sefine protease inhibitor) is an important member of the serin protease inhibitor (serpin) superfamily. It is located at 18q21.3-q23. Our team’s previous studies found that Maspin expression in BCa tissue was significantly down-regulated in comparison with normal tissues adjacent to the cancer and was related also to LNM. The negative correlation between the protein expression level and VEGF-C is statistically significant (83, 84). Maspin can inhibit the invasion of BCa cells, and its growth-inhibiting properties were related to its localization in cells. The surface-bound Maspin directly controlled the adhesion of BCa cells to the blood vessel wall (85). The combination of nuclear-localized maspin and chromatin can effectively prevent cell migration. Mapsin mainly promoted the development of BCa through DNA methylation and histone deacetylation to cause low expression of genes (86). Maspin modulated HDAC1 target genes, including cyclin D1, p21, MMP9, and vimentin (87). In our previous study, maspin could enhance Cisplatin chemosensitivity through the PI3K/AKT/mTOR signaling pathway in MIBC T24 and 5637 cell lines (88).
Another gene that functions as a tumor suppressor in BCa is GATA6. GATA6 (GATA-binding factor 6), a zinc-finger transcription factor, is located at 18q11.2. It regulates transcription cofactors and RNA polymerase II to the proximal promoter to regulate target genes’ transcription. Wang et al. (89) found that GATA6 decreased in BCa, and further decreased in patients with positive LNM. GATA6 was significantly down-regulated in BCa through frequent promoter methylation. GATA6 mainly inhibited LNM of BCa by regulating VEGF-C. Down-regulation of GATA6 promoted VEGF-C transcription, which promoted lymphangiogenesis, resulting in an increased lymphatic spread of BCa. This increased spread shows that it is of great significance to check the methylation status of the GATA6 promoter in the urine of BCa patients. The low expression of FOXOs was also associated with LNM in BCa (90). FOXO (Forkhead box class O) is the subgroup O of forkhead box (FOX) transcription factors, which has four members, FOXO1, FOXO3, FOXO4 and FOXO6. FOXOs have a highly conserved forkhead DNA binding domain. FOXOs can inhibit the invasion of BCa cells by down-regulating Twist2 and YB-1 and up-regulating E-cadherin (91).
MiRNA is a type of 21-23nt small RNA, which can complement mRNA and either silence it or degrade it. Most miRNAs are down-regulated in bladder cancer. Moreover, they inhibit the lymph node metastasis of bladder cancer (Figure 3).
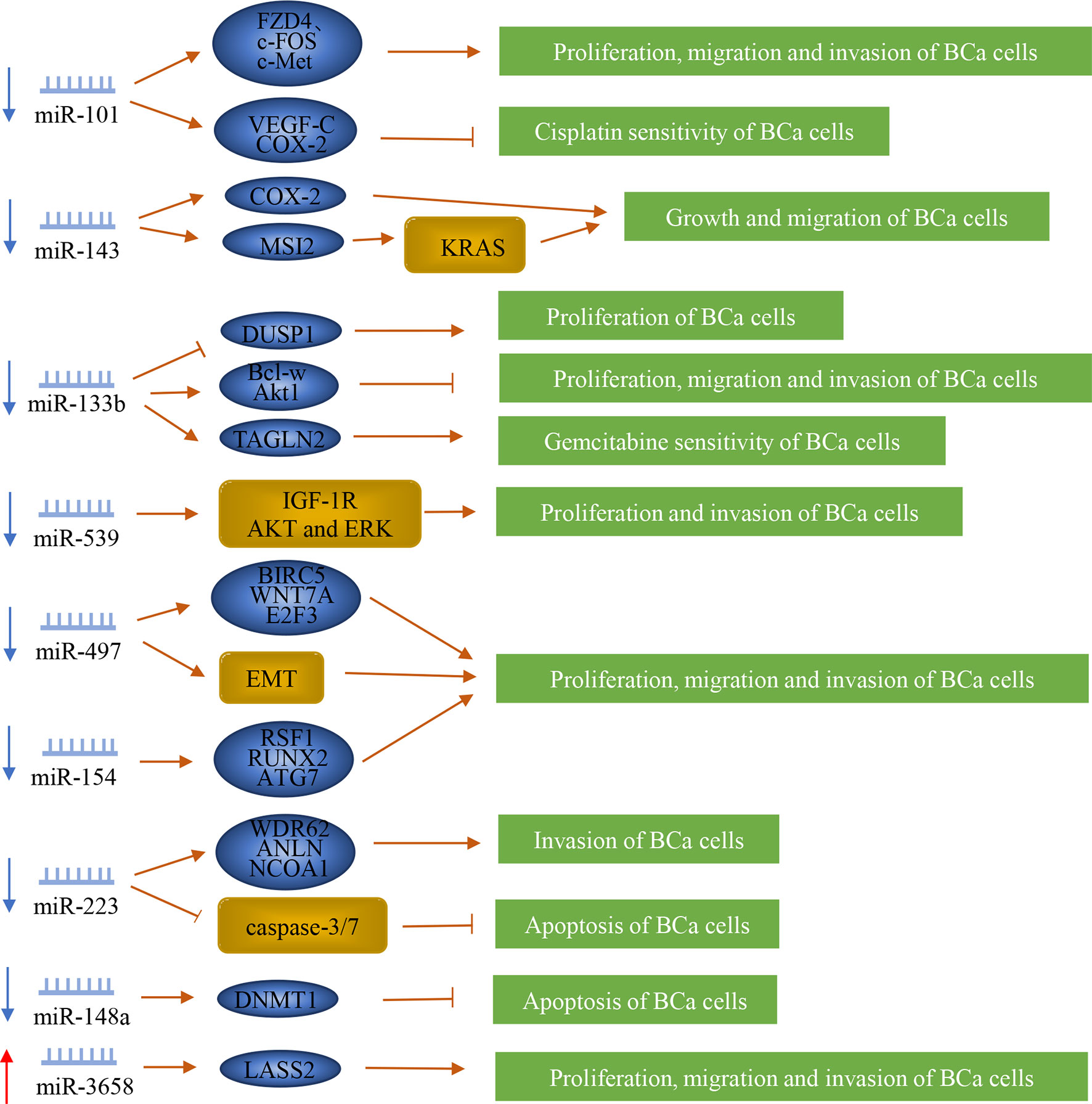
Figure 3 Regulation of microRNAs in bladder cancer patients with lymph node metastasis. MiRNAs play a vital role in the lymph node metastasis of bladder cancer. They can promote or inhibit the metastasis of bladder cancer by regulating downstream genes or proteins.
MiR-101 can suppress the progression of BCa. Studies have shown that the expression of miR-101 in BCa patients was down-regulated and significantly associated with LNM (92). Moreover, it can inhibit the proliferation, migration, and invasion of BCa cells by directly targeting FZD4 (frizzled class receptor 4), c-FOS, and c-Met (4–6). MiR-101 increased Cisplatin sensitivity by inhibiting the expression of VEGF-C and COX-2 in BCa cells (7, 8). MiR-143 also inhibited the growth and migration of BCa cells by targeting COX-2 (9). MiR-143 was reported to suppress the progression of BCa as well and it is located on chromosome 5q32. Liu et al. (93) found that miR-143 was down-expressed in the serum of BCa patients with LNM. It also directly affected the expression of MSI2 through its RNAi effect, which also effectively inhibited the KRAS network, thereby regulating BCa cells (10).
Another gene, miR-133b, is located on chromosome 6p12.2. Studies have shown that the expression level of miR-133b in BCa tissues is significantly reduced, which was significantly correlated with LNM (94). MiR-133b may inhibit the proliferation of BCa by up-regulating dual-specificity protein phosphatase 1 (DUSP1) (11). It inhibited angiogenesis and enhanced BCa cells’ chemosensitivity to Gemcitabine by targeting transgelin 2 (TAGLN2) (14). MiR-133b can regulate the proliferation, migration, and invasion of BCa cells by down-regulating Bcl-w, Akt1, and epidermal growth factor receptor along with its downstream effector protein (12, 13). Liao et al. (15) found that miR-539 was down-regulated in BCa, and was related to LNM. MiR-539 is located on chromosome 14q32.31, and it can inhibit the proliferation and invasion of BCa cells by directly targeting IGF-1R and inactivating the AKT and ERK signaling pathways.
MiR-497 is also known as a tumor suppressor in BCa, and it is located on chromosome 17p13.1. Studies have revealed that the expression of miR-497 in BCa tissue was lower than that of adjacent non-cancer tissues, and it was correlated with LNM (16). MiR-497 can inhibit the proliferation, migration, and invasion of BCa by up-regulating E-cadherin and down-regulating vimentin, α-smooth muscle actin, BIRC5, WNT7A, and E2F3 (16–18). Previous studies have found that miR-154 was significantly down-regulated in BCa tissues and was associated with LNM. MiR-154 is located in the human imprinted 14q32 domain. MiR-154 inhibited the proliferation, migration, and invasion of BCa cells by regulating the expression of RSF1, RUNX2, and ATG7 (19, 20). MiR-223 is located on chromosome Xq12. Sugita et al. (21) found that the expression level of miR-223 was significantly reduced in BCa tissues, which was related to LNM. MiR-223 inhibited cell invasion and promoted cell apoptosis in BCa via caspase-3/7 activation and negatively regulating WDR62 (WD repeat domain 62), ANLN, and nuclear receptor coactivator 1 (21–23). MiR-148a, with 68 nucleotide sequences, locates to 7p15.2, and is confirmed by Ma et al. (95) that its expression level in BCa tissue is lower than that of adjacent normal tissues, and that its low expression level is associated with advanced tumor progression and LNM. Also, Lombard et al. (24) found that miR-148a increased the apoptosis of BCa cells by reducing the expression of DNA methyltransferase 1 (DNMT1).
MiR-3658 is known as an oncogene in BCa. The expression of miR-3658 in BCa tissue was up-regulated, and its expression was significantly related to the lymph node infiltration, distant metastasis, and TNM stage (96). It can also promote cell proliferation, migration, and invasion by targeting LASS2 (25).
LncRNA is a non-coding RNA with a length of more than 200 nucleotides and is closely related to cancer occurrence and development. It can directly bind to proteins to block its functions or change its cellular location, regulate mRNA translation and act as a miRNA sponge. Most lncRNAs act as oncogenes to promote lymphatic metastasis of bladder cancer (Figure 4).
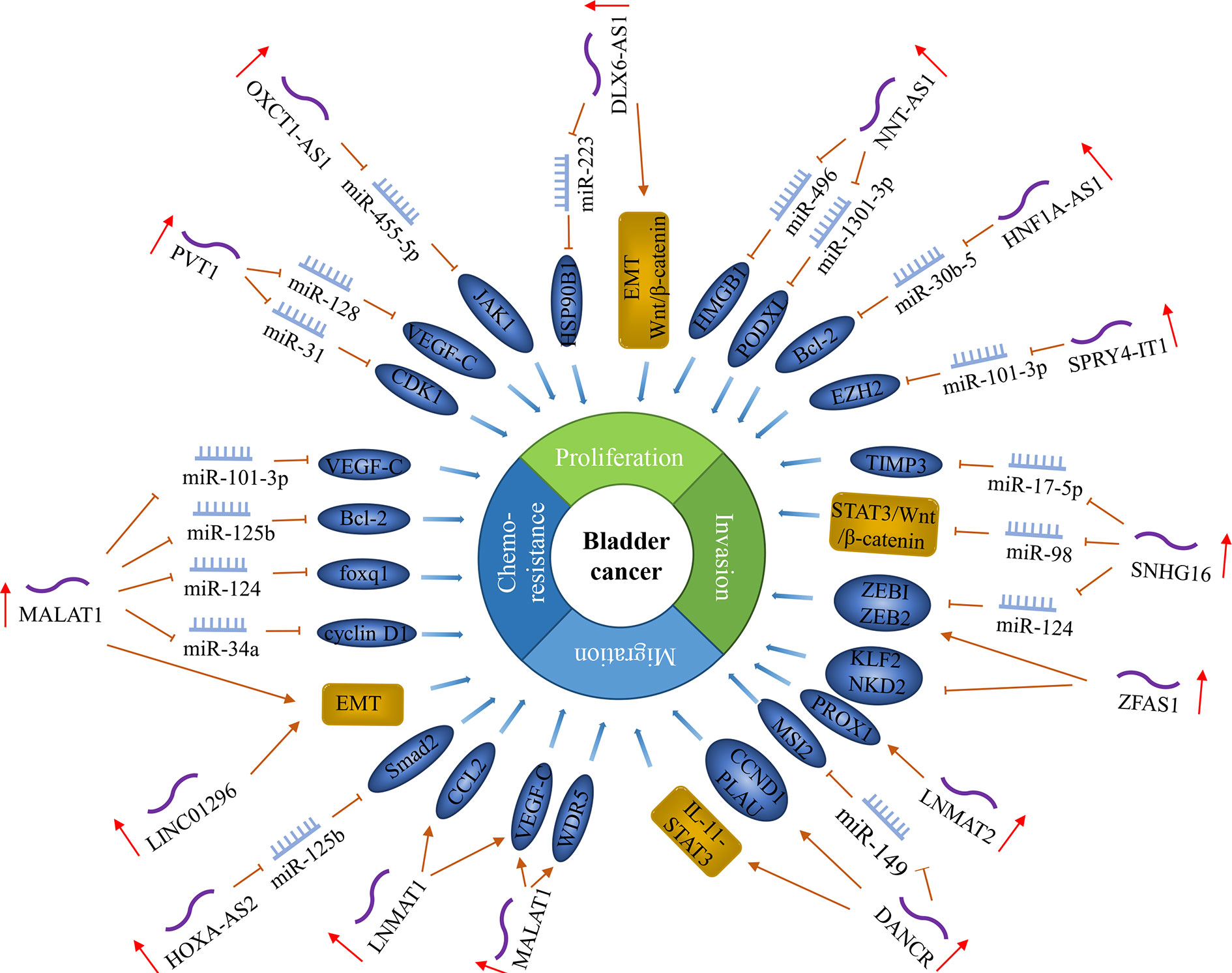
Figure 4 LncRNAs regulate lymph node metastasis in bladder cancer. In bladder cancer, the expression level of some lncRNAs is related to lymph node metastasis and regulates lymph node metastasis by regulating cancer cell proliferation, metastasis, invasion, and chemosensitivity.
Our team’s studies found several lncRNAs as oncogenes, such as MALAT1, PVT1, and OXCT1-AS1. The expression of MALAT1 was positively associated with LNM in BCa. It enhanced the Cisplatin resistance of the BCa cells by regulating the miR-101-3p/VEGF-C pathway (27, 97). MALAT1 promoted proliferation and invasion by miR-125b-Bcl-2/MMP-13, miR-124/foxq1 and microRNA-34a/cyclin D1 in BCa cells (28–30). It also up-regulated EMT-associated ZEB1, ZEB2, and Slug and downregulated E-cadherin levels (26). LncRNA PVT1 is located at 8q24, downstream of MYC. High PVT1 expression is associated with higher tumor stage and positive lymph node metastasis (98). PVT1 directly interacted with miR-128, reducing the binding of miR-128 to VEGF-C, thereby inhibiting the degradation of VEGFC mRNA by miR-128 (31). Moreover, PVT1 down-regulated miR-31 to enhance CDK1 expression and promote the proliferation, migration, and invasion of BCa cells (32). LncRNA OXCT1-AS1 (OXCT1 antisense RNA 1) is located on chromosome 5p13.1 and was also significantly up-regulated in BCa cell lines with LNM and was found to be inhibiting miR-455-5p in order to up-regulate the expression of JAK1, thus promoting the invasion of BCa (33).
Some lncRNAs regulate VEGF-C to promote the progression of BCa. BLACAT2 (bladder cancer-associated transcript 2) was significantly overexpressed in BCa patients with LNM. It combines with the VEGF-C promoter by forming triplexes to up-regulate VEGF-C expression, thereby promoting lymphangiogenesis and lymphatic metastasis. BLACAT2 directly interacted with WDR5 (the core component of the histone H3K4 methyltransferase complex) to epigenetically induce lymphangiogenesis and invasion (34). LNMAT1 (lymph node metastasis-associated Transcript 1) was significantly up-regulated in BCa with LNM. LNMAT1 recruited hnRNPL to the CCL2 promoter to activate CCL2 expression, resulting in increased H3K4 trimethylation, thereby ensuring hnRNPL binding and enhancing transcription. In addition, LNMAT1-induced CCL2 regulated the tumor microenvironment in BCa tissues through tumor-associated macrophages (TAMs) infiltration and VEGF-C upregulation, which ultimately led to lymphangiogenesis and lymphatic metastasis (35).
Several lncRNAs promote the progression of BCa by regulating ZEB1 and ZEB2. LncRNA SNHG16 (small nucleolar RNA host gene 16) is encoded by a 7571‐bp region at chromosome 17q25.1. Previous studies have found that SNHG16 was highly expressed in BCa tissues and was positively correlated with LNM (37). SNHG16 can regulate the proliferation, apoptosis, EMT, invasion, and migration of BCa by directly acting on the miR-17-5p/metalloproteinase 3 (TIMP3) axis, miR-200a-3p/ZEB1/ZEB2 axis, and miR-98/STAT3/Wnt/β-catenin pathway axis (36–38). LncRNA ZFAS1 (zinc finger antisense 1), located on the antisense strand of the ZNFX1 promoter region, is transcript antisense to the 5′- end of the gene zinc finger NFX1-type containing 1 (ZNFX1). Yang et al. (39) found that the expression level of ZFAS1 in BCa was increased and positively correlated with LNM. ZFAS1 can promote the proliferation, migration and invasion of BCa by down-regulating the expression of KLF2 and NKD2, and at the same time, up-regulating the expression of ZEB1 and ZEB2. It also promotes tumorigenesis of BCa through sponging miR-329 (99).
Also, some lncRNAs regulate EMT to promote BCa progression. LncRNA DLX6-AS1 (distal-less homeobox 6 antisense 1) is regulatory of members in the DLX gene family, which is localized on chromosome 7q21.3. DLX6-AS1 was up-regulated in BCa, which was related to LNM. Overexpression of DLX6-AS1 promoted the proliferation, invasion, and migration of BCa cells by regulating EMT and Wnt/β-catenin signaling pathway activity (41). DLX6-AS1-mediated miR-223 silencing can promote the growth and invasion of BCa through the up-regulation of HSP90B1 (40). LINC01296 is a novel intergenic lncRNA located at 14q11.2. The expression of LINC01296 was positively correlated with lymph node-positive BCa, and its up-regulated expression can promote BCa cells metastasis by activating the EMT pathway (42).
Another lncRNA, DANCR (differentiation antagonizing non-protein coding RNA), is located on chromosome 4q12.5, which is mainly distributed in the cytoplasm. Chen et al. (43) found that DANCR was significantly up-regulated in BCa tissues and positively correlated with LNM. DANCR promoted the LNM and BCa cells’ proliferation via DANCR guided LRPPRC (leucine-rich pentatricopeptide repeat containing) to stabilize its mRNA, then to activate IL-11-STAT3 signaling and increase CCND1 and PLAU expression. Zhan et al. (44) found that DANCR positively regulated the expression of MSI2 (musashi RNA binding protein 2) through sponging miR-149 to promote the malignant phenotype of BCa cells. Zhao et al. (100) found that the expression level of SPRY4-IT1 in BCa tissue was also higher than that of adjacent non-tumor tissues and was associated with LNM. SPRY4-IT1 is derived from the intron region of the SPRY4 gene and may contain several long hairpin secondary structures, which are located in 5q31.3. SPRY4-IT1 can promote proliferation and metastasis of BCa cells by sponging miR-101-3p to actively regulate the expression of EZH2 (45). Wu et al. (46) found that lncRNA NNT-AS1 was up-regulated in BCa, which was significantly associated with LNM. NNT-AS1 (nicotinamide nucleotide transhydrogenase antisense RNA 1) is located on chromosome 5p12 with 3 exons. NNT-AS1 promoted the proliferation, migration, and invasion of BCa cells by acting as a competing endogenous RNA for miR-496 to enhance the expression level of HMGB1. NNT-AS1 also targeted the miR-1301-3p/PODXL axis and activated the Wnt pathway, thereby enhancing BCa cells’ growth (47). LncRNA LNMAT2 (lymph node metastasis-associated transcript 2) was overexpressed in urinary-EXO and serum-EXO of patients with BCa, which was related to LNM. LNMAT2 was found to bind to the prospero homeobox 1 (PROX1) promoter by inducing H3K4 trimethylation, which enhanced PROX1 transcription, thus promoting lymphangiogenesis and lymph node metastasis in bladder cancer (48).
Additionally, several lncRNAs positively correlated with LNM, including: (1) HOXA-AS2, which inhibited the expression of miR-125b to promote the expression of Smad2, thus promoting the migration and invasion of BCa cells (49); (2)HNF1A-AS1, which positively regulated the expression of Bcl-2 by sponging miR-30b-5 to promote the proliferation of bladder cancer and inhibited its apoptosis (50, 101); (3) ROR1-AS1, which promoted the growth and migration of bladder cancer by regulating miR-504 (102); (4) RMRP, which promoted the proliferation, migration, and invasion of bladder cancer cells by regulating miR-206 as a sponge (103).
CircRNA is a type of non-coding RNA that forms a circular structure by covalent bonds but does not have a 5’-end cap and a 3’-end poly(A) tail. It is closely related to the occurrence and development of cancer. It can act as an mRNA ‘sponge’, regulate transcription and splicing, and interact with RNA-binding proteins (104). Most circRNA negatively regulates lymph node metastasis of bladder cancer, and some molecules positively regulate this process (Figure 5).
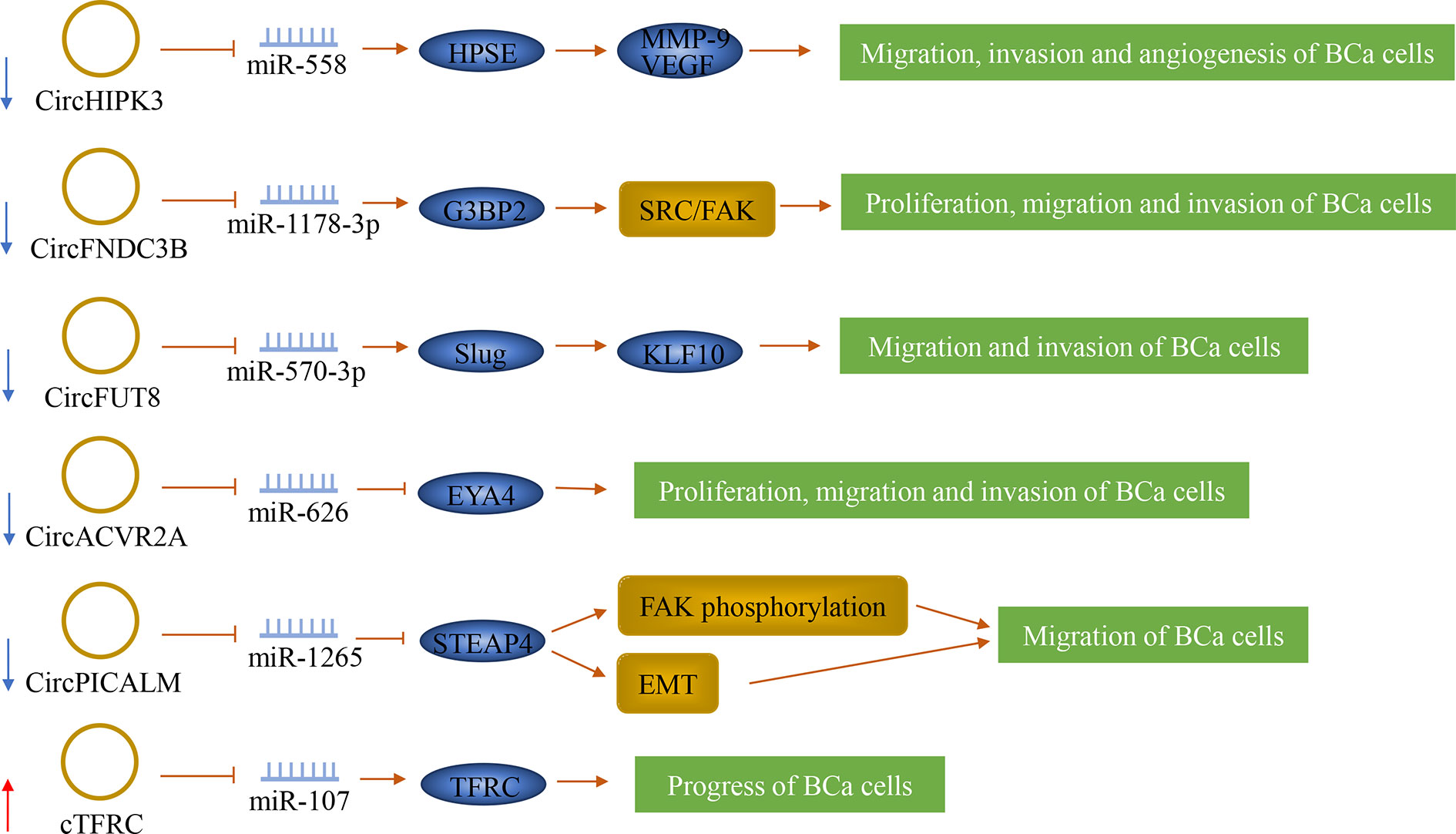
Figure 5 The role of circRNAs in bladder cancer lymph node metastasis. CircRNAs can play a role in bladder cancer as oncogenes and tumor suppressor genes. They can also predict lymph node metastasis.
CircHIPK3 (circRNA ID: hsa_circ_0000284), also known as bladder cancer-related circular RNA-2 (BCRC-2), was significantly down-regulated in BCa and was negatively correlated with LNM. It originates from the second exon of the Homeodomain-interacting protein kinase 3 (HIPK3) gene. CircHIPK3 sponged miR-558 and prevented miR-558 from being transported into the nucleus to bind the promoter of heparanase (HPSE) gene in BCa cells, thereby down-regulating the expression of HPSE and its downstream targets such as MMP-9, and VEGF, thus weakening the migration, invasion and angiogenesis of BCa cells (51). Additionally, Liu et al. (52) confirmed that circFNDC3B was significantly down-regulated in BCa tissue, and its low expression was significantly correlated with LNM. It is originated from exons 5 and 6 of the FNDC3B gene. CircFNDC3B acted as a sponge of miR-1178-3p to inhibit G3BP2 and further inhibit the downstream SRC/FAK signaling pathway, thereby inhibiting the proliferation, migration, and invasion of BCa cells.
By screening RNA sequencing data generated from human BCa tissues and matched adjacent normal bladder tissues, two novel tumor suppressors were separately identified, which are circFUT8 and circACVR2A. CircFUT8 (circBase: hsa_circ_0003028) was originated from exon 3 of the FUT8 gene. CircACVR2A was derived from exons 3, 4, and 5 of the ACVR2A gene. These two tumor suppressors were down-regulated in BCa tissues and were related to LNM (53, 54). CircFUT8 regulated the expression of Slug by sponging miR-570-3p to promote the expression of Krüpple-like-factor 10 (KLF10), thus inhibiting the metastasis and invasion of BCa cells (54). CircACVR2A can inhibit the proliferation, migration, and invasion of BCa cells by directly interacting with miR-626 and acting as a miRNA sponge to regulate EYA4 expression (53). In addition, circPICALM was found to suppress cancer progression. It is generated from exons 9-12 of PICALM. It was down-regulated in BCa tissues and associated with LNM. CircPICALM acted as a miR-1265 sponge to regulate STEAP4 and further affect FAK phosphorylation and EMT, thereby inhibiting the metastasis of BCa (55).
Serval other circRNAs were found to be possibly promoting cancer progression by inducing the malignant proliferation or migration and invasion of cancer cells. Su et al. identified a novel circular RNA called cTFRC. His study has shown that cTFRC was up-regulated in BCa tissues and was associated with LNM. The study also revealed that cTFRC might act as a sponge for miR-107 to up-regulate the expression of TFRC (transferrin receptor), further promoting the transitional phenotype of BCa cells from epithelial to mesenchymal, thereby promoting the progress of BCa. (56) Another circRNA, circPTK2, was significantly increased in BCa, and its expression level is closely related to LNM. CircPTK2 can promote the proliferation and migration of BCa cells, but its specific mechanisms are still unclear (105).
In addition to the molecules described above, studies on the tumor microenvironment and genetic modification can also help predict the lymphatic metastasis of bladder cancer.
Tumors often form a microenvironment that allows inflammatory cells to proliferate and produce large amounts of mediators. D’Andrea et al. (106) found that LMR (lymphocyte-to-monocyte ratio) and NLR (neutrophil-to-lymphocyte ratio) can be used as independent factors to predict the preoperative LNM and postoperative recurrence rate of BCa patients. Zhou et al. (107) found that lymphatic vessel density (LVD) within and around the tumor increases, and lymph node metastasis of bladder cancer also increase significantly. LVD is also related to the patient’s prognosis.
m6A (N6-methyladenosine) refers to methylation of the N6 position of adenosine bases. m6A RNA modification is a reversible posttranscriptional modification process maintained by a multicomponent methyltransferase ‘writer’ complex (KIAA1429, METTL3, METTL14, RBM15, WTAP, and ZC3H13) and removed by demethylases ‘erasers’ (FTO and ALKBH5). The function of m6A in mRNA metabolism primarily depends on reader proteins, which include HNRNPC, YTHDC1, YTHDC2, YTHDF1, and YTHDF2. These regulators were differentially associated with different clinicopathological variables of BCa patients. The expression of WTAP was significantly correlated with LNM (108). Han et al. (109) found that METTL3 was significantly increased in bladder cancer and correlated with high histological grade and poor prognosis. METTL3 interacted with the microprocessor protein DGCR8 and positively modulated the pri-miR221/222 processes, resulting in the reduction of PTEN, which ultimately leads to the progression of bladder cancer.
The prediction model includes many aspects, such as molecules, imaging, and pathology. With the advent of models, the predictive results of bladder cancer lymphatic metastasis have become more and more reliable.
Smith et al. (110) developed a 20-GEM (gene expression model) for predicting pathological node status, which is evaluable on primary tumor tissue from clinically node-negative (cN0) patients. The predictive efficacy of the model is modest. Seiler et al. (111) invented a KNN51 (K-nearest neighbor classifier 51) to predict pathological lymph node metastases, but the lack of external validation limited its application. Lu et al. (112) presented a preoperative nomogram incorporating the LNM signature and a genomic mutation of MLL2. The LNM signature consists of 48 selected features. The model demonstrated good discrimination and good calibration. KNN51 included 24 non-coding features from the 51 gene signature, but the LN20 signature was based only on coding genes. Clinical factors were not incorporated into the predictive models for evaluation.
Wu et al. (113, 114) developed and validated two types of radiomics nomograms incorporating the radiomics signature and CT/MRI-reported LN status for the preoperative prediction of LNM in patients with BCa, which was a non-invasive preoperative prediction tool. It shows favorable predictive accuracy, especially for cN0 patients. Multicenter validation should be performed to acquire high-level evidence for its clinical application.
Wu et al. (115) constructed an inclusive nomogram that incorporated the five-mRNA-based classifier, image-based LN status, transurethral resection (TUR) T stage, and TUR lymphovascular invasion (LVI) to predict LNM in BCa patients. Five LN-status-related mRNAs include ADRA1D, COL10A1, DKK2, HIST2H3D, and MMP11. It shows favorable discriminatory ability and may aid in clinical decision-making, especially for cN-patients. However, it requires multicenter prospective clinical trials to provide high-level evidence for clinical application.
Chen et al. (116) validated a genomic-radiomics nomogram incorporating CCR7 and CT to predict LNM in patients with BCa. The combined evaluation of CCR7 and CT appeared to be a more reliable marker for lymph node metastasis in BCa than the diagnosis by CT or CCR7 alone. However, these results require further confirmation by large sample and multi-center prospective studies.
In recent years, studies have found that some systemic diseases were closely related to tumor occurrence and development. Metabolic syndrome (MetS) was defined as the presence of three of the following: hypertension, hyperlipidemia, diabetes, or body mass index >30. Previous studies have proved that MetS cannot predict higher pathological stages and the risks of LVI and LNM, but a single component of metabolic syndrome was related to them. Body mass index, waist circumference, and hypertension were positively correlated with the risk of higher pathological stages. And higher BMI value was related to lymphatic invasion and lymph node metastasis (117, 118). Obesity was significantly related to recurrence-free survival, cancer-specific survival, and overall mortality. Adipose tissue can produce a variety of inflammatory factors, including leptin, adiponectin, and cytokines. Leptin played an anti-tumor effect by promoting the proliferation and activation of natural killer cells (119, 120). Nonalcoholic fatty liver was positively correlated with BCa, and it was a poor prognostic factor for BCa. Patients with nonalcoholic fatty liver disease had elevated vascular endothelial growth factor, interleukin 6, TNF-α, and IGF-1. These factors may increase the risk of BCa recurrence and lead to a poor prognosis (121). Studies have shown that patients with BCa had higher insulin resistance than those without cancer but with bladder disease (122). DM was associated with elevated BCa or cancer mortality risk, especially in men (123). Metformin is the most commonly used drug for patients with t2DM. Our team’s study found that the intake of metformin was positively associated with RFS, which improved PFS and cancer-specific survival (124). Metformin targeted a YAP1-TEAD4 complex via AMPKα to regulate CCNE1/2 in BCa cells (125). It can suppress cyclin D1, cyclin-dependent kinase 4 (CDK4), E2F1, and mammalian target of rapamycin (mTOR) (126). The use of insulin can increase the risk of BCa progression (127). High-dose human insulin and insulin glargine similarly promoted T24 BCa cell proliferation via PI3K-independent activation of Akt (128).
Environmental toxins are closely related to cancer occurrence and development, and arsenic is the most reported in BCa. Dimethylarsinic acid (DMAV) is a methylated metabolite of arsenicals found in most mammals, and long-term exposure to DMAV can lead to BCa. Previous studies have found that recurrent BCa with high arsenic levels in tissues was more aggressive and had a higher stage and grade, and recured earlier than people with low levels of arsenic (129). Zhou et al. found that chronic arsenic exposure can upregulate HER2 in human and rat bladder epithelial cells and promote the proliferation, migration, epithelial-mesenchymal transition, and angiogenesis of cancer cells by activating the MAPK, PI3K/AKT, and STAT3 pathways (130). Moreover, sodium arsenite can reduce the human urothelial WIF1 gene expression, increase its DNA methylation level, and promote cancer cells’ migration. The WIF1 gene expression and its DNA methylation can be considered as potential biomarkers for the diagnosis of human BCa (131).
For the LNM in BCa, three mechanisms are mainly involved: tumor cell proliferation, tumor cell migration and invasion, and chemosensitivity. Most biomarkers are related to the proliferation, migration, and invasion of BCa cells. Several biomarkers are involved in chemosensitivity. MiR-143, miR-101, miR-133b, MALAT1, CXCL5, and VEGF-C are related to all three of the above mechanisms. These biomarkers are more likely to be prognostic factors for BCa with LNM, but a large number of retrospective studies are still needed for further verification. Previous studies have shown that most biomarkers have a clear relationship with the prognosis of BCa patients (Table 2). However, the relationship between these eight biomarkers: ISYNA1, miR-539, miR-3658, OXCT1-AS1, DLX6-AS1, HOXA-AS2, circHIPK3, and circPTK2 and prognosis is still unclear; therefore, further research is needed to tap into their potential for the prognosis of BCa patients. Many biological assessment methods are economical and accurate. For example, peripheral blood can detect MMP, LMR, and NLR. Urine can detect the methylation status of GATA6 promoter, CXCL5, and MMP. Genetic testing for LNM is more sensitive and specific than traditional pathological examinations and is particularly suitable for micrometastasis diagnosis. Those test samples are easy to obtain before surgery, with strong reproducibility and high clinical feasibility. Recently, the research on SNP and m6A is also a hot spot. The relationship between them and bladder cancer with lymph node metastasis is not yet clear, and further investigation is needed, but it provides new directions for our future research. As for imaging, pathology, and molecular composition models, they are more accurate in terms of predicting lymphatic metastasis for bladder cancer, which should be studied in-depth and applied to clinical practice.
CZ contributed to reading the literature, preparing figures and the table, and writing the manuscript. JH, HL, HM, BO, WR, ZY, DQ, ZO, JC, and XZ assisted with writing and revised the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (81873626, 81902592), Hunan Natural Science Foundation (2020JJ5884), Hunan Province Key R&D Program (2019SK2202), and Xiangya Hospital Youth Fund (2018Q09).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We sincerely thank You-e He for editing the language of this article.
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2018) 68:394–424. doi: 10.3322/caac.21492
2. Zhang ZL, Dong P, Li YH, Liu ZW, Yao K, Han H, et al. Radical cystectomy for bladder cancer: oncologic outcome in 271 Chinese patients. Chin J Cancer (2014) 33:165–71. doi: 10.5732/cjc.012.10312
3. Li Y, Diao F, Shi S, Li K, Zhu W, Wu S, et al. Computed tomography and magnetic resonance imaging evaluation of pelvic lymph node metastasis in bladder cancer. Chin J Cancer (2018) 37:3. doi: 10.1186/s40880-018-0269-0
4. Hu Z, Lin Y, Chen H, Mao Y, Wu J, Zhu Y, et al. MicroRNA-101 suppresses motility of bladder cancer cells by targeting c-Met. Biochem Biophys Res Commun (2013) 435:82–7. doi: 10.1016/j.bbrc.2013.04.042
5. Long Y, Wu Z, Yang X, Chen L, Han Z, Zhang Y, et al. MicroRNA-101 inhibits the proliferation and invasion of bladder cancer cells via targeting c-FOS. Mol Med Rep (2016) 14:2651–6. doi: 10.3892/mmr.2016.5534
6. Chen L, Long Y, Han Z, Yuan Z, Liu W, Yang F, et al. MicroRNA-101 inhibits cell migration and invasion in bladder cancer via targeting FZD4. Exp Ther Med (2019a) 17:1476–85. doi: 10.3892/etm.2018.7084
7. Bu Q, Fang Y, Cao Y, Chen Q, Liu Y. Enforced expression of miR-101 enhances cisplatin sensitivity in human bladder cancer cells by modulating the cyclooxygenase-2 pathway. Mol Med Rep (2014) 10:2203–9. doi: 10.3892/mmr.2014.2455
8. Lei Y, Li B, Tong S, Qi L, Hu X, Cui Y, et al. miR-101 suppresses vascular endothelial growth factor C that inhibits migration and invasion and enhances cisplatin chemosensitivity of bladder cancer cells. PloS One (2015) 10:e0117809. doi: 10.1371/journal.pone.0117809
9. Song T, Zhang X, Wang C, Wu Y, Dong J, Gao J, et al. Expression of miR-143 reduces growth and migration of human bladder carcinoma cells by targeting cyclooxygenase-2. Asian Pac J Cancer Prev (2011) 12:929–33.
10. Tsujino T, Sugito N, Taniguchi K, Honda R, Komura K, Yoshikawa Y, et al. MicroRNA-143/Musashi-2/KRAS cascade contributes positively to carcinogenesis in human bladder cancer. Cancer Sci (2019) 110:2189–99. doi: 10.1111/cas.14035
11. Cai X, Qu L, Yang J, Xu J, Sun L, Wei X, et al. Exosome-transmitted microRNA-133b inhibited bladder cancer proliferation by upregulating dual-specificity protein phosphatase 1. Cancer Med (2020) 9:6009–19. doi: 10.1002/cam4.3263
12. Zhou Y, Wu D, Tao J, Qu P, Zhou Z, Hou J. MicroRNA-133 inhibits cell proliferation, migration and invasion by targeting epidermal growth factor receptor and its downstream effector proteins in bladder cancer. Scand J Urol (2013) 47:423–32. doi: 10.3109/00365599.2012.748821
13. Chen XN, Wang KF, Xu ZQ, Li SJ, Liu Q, Fu DH, et al. MiR-133b regulates bladder cancer cell proliferation and apoptosis by targeting Bcl-w and Akt1. Cancer Cell Int (2014) 14:70. doi: 10.1186/s12935-014-0070-3
14. Zhao F, Zhou LH, Ge YZ, Ping WW, Wu X, Xu ZL, et al. MicroRNA-133b suppresses bladder cancer malignancy by targeting TAGLN2-mediated cell cycle. J Cell Physiol (2019) 234:4910–23. doi: 10.1002/jcp.27288
15. Liao G, Chen F, Zhong J, Jiang X. MicroRNA−539 inhibits the proliferation and invasion of bladder cancer cells by regulating IGF−1R. Mol Med Rep (2018) 17:4917–24. doi: 10.3892/mmr.2018.8497
16. Wei Z, Hu X, Liu J, Zhu W, Zhan X, Sun S. MicroRNA-497 upregulation inhibits cell invasion and metastasis in T24 and BIU-87 bladder cancer cells. Mol Med Rep (2017) 16:2055–60. doi: 10.3892/mmr.2017.6805
17. Itesako T, Seki N, Yoshino H, Chiyomaru T, Yamasaki T, Hidaka H, et al. The microRNA expression signature of bladder cancer by deep sequencing: the functional significance of the miR-195/497 cluster. PloS One (2014) 9:e84311. doi: 10.1371/journal.pone.0084311
18. Zhang Y, Zhang Z, Li Z, Gong D, Zhan B, Man X, et al. MicroRNA-497 inhibits the proliferation, migration and invasion of human bladder transitional cell carcinoma cells by targeting E2F3. Oncol Rep (2016) 36:1293–300. doi: 10.3892/or.2016.4923
19. Zhao X, Ji Z, Xie Y, Liu G, Li H. MicroRNA-154 as a prognostic factor in bladder cancer inhibits cellular malignancy by targeting RSF1 and RUNX2. Oncol Rep (2017) 38:2727–34. doi: 10.3892/or.2017.5992
20. Zhang J, Mao S, Wang L, Zhang W, Zhang Z, Guo Y, et al. MicroRNA−154 functions as a tumor suppressor in bladder cancer by directly targeting ATG7. Oncol Rep (2019) 41:819–28. doi: 10.3892/or.2018.6879
21. Sugita S, Yoshino H, Yonemori M, Miyamoto K, Matsushita R, Sakaguchi T, et al. Tumor−suppressive microRNA−223 targets WDR62 directly in bladder cancer. Int J Oncol (2019) 54:2222–36. doi: 10.3892/ijo.2019.4762
22. Guo J, Cao R, Yu X, Xiao Z, Chen Z. MicroRNA-223-3p inhibits human bladder cancer cell migration and invasion. Tumour Biol (2017) 39:1010428317691678. doi: 10.1177/1010428317691678
23. Sugawara S, Yamada Y, Arai T, Okato A, Idichi T, Kato M, et al. Dual strands of the miR-223 duplex (miR-223-5p and miR-223-3p) inhibit cancer cell aggressiveness: targeted genes are involved in bladder cancer pathogenesis. J Hum Genet (2018) 63:657–68. doi: 10.1038/s10038-018-0437-8
24. Lombard AP, Mooso BA, Libertini SJ, Lim RM, Nakagawa RM, Vidallo KD, et al. miR-148a dependent apoptosis of bladder cancer cells is mediated in part by the epigenetic modifier DNMT1. Mol Carcinog (2016) 55:757–67. doi: 10.1002/mc.22319
25. Luan T, Zou R, Huang L, Li N, Fu S, Huang Y, et al. Hsa-miR-3658 Promotes Cell Proliferation, Migration and Invasion by Effecting LASS2 in Bladder Cancer. Clin Lab (2018) 64:515–25. doi: 10.7754/Clin.Lab.2017.171026
26. Ying L, Chen Q, Wang Y, Zhou Z, Huang Y, Qiu F. Upregulated MALAT-1 contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition. Mol Biosyst (2012) 8:2289–94. doi: 10.1039/c2mb25070e
27. Liu P, Li X, Cui Y, Chen J, Li C, Li Q, et al. LncRNA-MALAT1 mediates cisplatin resistance via miR-101-3p/VEGF-C pathway in bladder cancer. Acta Biochim Biophys Sin (Shanghai) (2019b) 51:1148–57. doi: 10.1093/abbs/gmz112
28. Xie H, Liao X, Chen Z, Fang Y, He A, Zhong Y, et al. LncRNA MALAT1 Inhibits Apoptosis and Promotes Invasion by Antagonizing miR-125b in Bladder Cancer Cells. J Cancer (2017) 8:3803–11. doi: 10.7150/jca.21228
29. Jiao D, Li Z, Zhu M, Wang Y, Wu G, Han X. LncRNA MALAT1 promotes tumor growth and metastasis by targeting miR-124/foxq1 in bladder transitional cell carcinoma (BTCC). Am J Cancer Res (2018) 8:748–60.
30. Liu Y, Gao S, Du Q, Zhao Q. Knockdown of long non-coding RNA metastasis associated lung adenocarcinoma transcript 1 inhibits the proliferation and migration of bladder cancer cells by modulating the microRNA-34a/cyclin D1 axis. Int J Mol Med (2019c) 43:547–56. doi: 10.3892/ijmm.2018.3959
31. Yu C, Longfei L, Long W, Feng Z, Chen J, Chao L, et al. LncRNA PVT1 regulates VEGFC through inhibiting miR-128 in bladder cancer cells. J Cell Physiol (2019) 234:1346–53. doi: 10.1002/jcp.26929
32. Tian Z, Cao S, Li C, Xu M, Wei H, Yang H, et al. LncRNA PVT1 regulates growth, migration, and invasion of bladder cancer by miR-31/CDK1. J Cell Physiol (2019) 234:4799–811. doi: 10.1002/jcp.27279
33. Chen JB, Zhu YW, Guo X, Yu C, Liu PH, Li C, et al. Microarray expression profiles analysis revealed lncRNA OXCT1-AS1 promoted bladder cancer cell aggressiveness via miR-455-5p/JAK1 signaling. J Cell Physiol (2019b) 234:13592–601. doi: 10.1002/jcp.28037
34. He W, Zhong G, Jiang N, Wang B, Fan X, Chen C, et al. Long noncoding RNA BLACAT2 promotes bladder cancer-associated lymphangiogenesis and lymphatic metastasis. J Clin Invest (2018) 128:861–75. doi: 10.1172/JCI96218
35. Chen C, He W, Huang J, Wang B, Li H, Cai Q, et al. LNMAT1 promotes lymphatic metastasis of bladder cancer via CCL2 dependent macrophage recruitment. Nat Commun (2018) 9:3826. doi: 10.1038/s41467-018-06152-x
36. Feng F, Chen A, Huang J, Xia Q, Chen Y, Jin X. Long noncoding RNA SNHG16 contributes to the development of bladder cancer via regulating miR-98/STAT3/Wnt/β-catenin pathway axis. J Cell Biochem (2018) 119:9408–18. doi: 10.1002/jcb.27257
37. Peng H, Li H. The encouraging role of long noncoding RNA small nuclear RNA host gene 16 in epithelial-mesenchymal transition of bladder cancer via directly acting on miR-17-5p/metalloproteinases 3 axis. Mol Carcinog (2019) 58:1465–80. doi: 10.1002/mc.23028
38. Chen W, Jiang T, Mao H, Gao R, Zhang H, He Y, et al. SNHG16 regulates invasion and migration of bladder cancer through induction of epithelial-to-mesenchymal transition. Hum Cell (2020b) 33:737–49. doi: 10.1007/s13577-020-00343-9
39. Yang H, Li G, Cheng B, Jiang R. ZFAS1 functions as an oncogenic long non-coding RNA in bladder cancer. Biosci Rep (2018) 38(3):BSR20180475. doi: 10.1042/BSR20180475
40. Fang C, Xu L, He W, Dai J, Sun F. Long noncoding RNA DLX6-AS1 promotes cell growth and invasiveness in bladder cancer via modulating the miR-223-HSP90B1 axis. Cell Cycle (2019) 18:3288–99. doi: 10.1080/15384101.2019.1673633
41. Guo J, Chen Z, Jiang H, Yu Z, Peng J, Xie J, et al. The lncRNA DLX6-AS1 promoted cell proliferation, invasion, migration and epithelial-to-mesenchymal transition in bladder cancer via modulating Wnt/β-catenin signaling pathway. Cancer Cell Int (2019b) 19:312. doi: 10.1186/s12935-019-1010-z
42. Wang X, Wang L, Gong Y, Liu Z, Qin Y, Chen J, et al. Long noncoding RNA LINC01296 promotes cancer-cell proliferation and metastasis in urothelial carcinoma of the bladder. Onco Targets Ther (2019a) 12:75–85. doi: 10.2147/OTT.S192809
43. Chen Z, Chen X, Xie R, Huang M, Dong W, Han J, et al. DANCR Promotes Metastasis and Proliferation in Bladder Cancer Cells by Enhancing IL-11-STAT3 Signaling and CCND1 Expression. Mol Ther (2019c) 27:326–41. doi: 10.1016/j.ymthe.2018.12.015
44. Zhan Y, Chen Z, Li Y, He A, He S, Gong Y, et al. Long non-coding RNA DANCR promotes malignant phenotypes of bladder cancer cells by modulating the miR-149/MSI2 axis as a ceRNA. J Exp Clin Cancer Res (2018) 37:273. doi: 10.1186/s13046-018-0921-1
45. Liu D, Li Y, Luo G, Xiao X, Tao D, Wu X, et al. LncRNA SPRY4-IT1 sponges miR-101-3p to promote proliferation and metastasis of bladder cancer cells through up-regulating EZH2. Cancer Lett (2017) 388:281–91. doi: 10.1016/j.canlet.2016.12.005
46. Wu D, Zhang T, Wang J, Zhou J, Pan H, Qu P. Long noncoding RNA NNT-AS1 enhances the malignant phenotype of bladder cancer by acting as a competing endogenous RNA on microRNA-496 thereby increasing HMGB1 expression. Aging (Albany NY) (2019a) 11:12624–40. doi: 10.18632/aging.102591
47. Liu Y, Wu G. NNT-AS1 enhances bladder cancer cell growth by targeting miR-1301-3p/PODXL axis and activating Wnt pathway. Neurourol Urodyn (2020) 39:547–57. doi: 10.1002/nau.24238
48. Chen C, Luo Y, He W, Zhao Y, Kong Y, Liu H, et al. Exosomal long noncoding RNA LNMAT2 promotes lymphatic metastasis in bladder cancer. J Clin Invest (2020c) 130:404–21. doi: 10.1172/JCI130892
49. Wang F, Wu D, Chen J, Chen S, He F, Fu H, et al. Long non-coding RNA HOXA-AS2 promotes the migration, invasion and stemness of bladder cancer via regulating miR-125b/Smad2 axis. Exp Cell Res (2019b) 375:1–10. doi: 10.1016/j.yexcr.2018.11.005
50. Wang YH, Liu YH, Ji YJ, Wei Q, Gao TB. Upregulation of long non-coding RNA HNF1A-AS1 is associated with poor prognosis in urothelial carcinoma of the bladder. Eur Rev Med Pharmacol Sci (2018c) 22:2261–5.
51. Li Y, Zheng F, Xiao X, Xie F, Tao D, Huang C, et al. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep (2017b) 18:1646–59. doi: 10.15252/embr.201643581
52. Liu H, Bi J, Dong W, Yang M, Shi J, Jiang N, et al. Invasion-related circular RNA circFNDC3B inhibits bladder cancer progression through the miR-1178-3p/G3BP2/SRC/FAK axis. Mol Cancer (2018) 17:161. doi: 10.1186/s12943-018-0908-8
53. Dong W, Bi J, Liu H, Yan D, He Q, Zhou Q, et al. Circular RNA ACVR2A suppresses bladder cancer cells proliferation and metastasis through miR-626/EYA4 axis. Mol Cancer (2019) 18:95. doi: 10.1186/s12943-019-1025-z
54. He Q, Yan D, Dong W, Bi J, Huang L, Yang M, et al. circRNA circFUT8 Upregulates Krüpple-like Factor 10 to Inhibit the Metastasis of Bladder Cancer via Sponging miR-570-3p. Mol Ther Oncolytics (2020) 16:172–87. doi: 10.1016/j.omto.2019.12.014
55. Yan D, Dong W, He Q, Yang M, Huang L, Kong J, et al. Circular RNA circPICALM sponges miR-1265 to inhibit bladder cancer metastasis and influence FAK phosphorylation. EBioMedicine (2019) 48:316–31. doi: 10.1016/j.ebiom.2019.08.074
56. Su H, Tao T, Yang Z, Kang X, Zhang X, Kang D, et al. Circular RNA cTFRC acts as the sponge of MicroRNA-107 to promote bladder carcinoma progression. Mol Cancer (2019) 18:27. doi: 10.1186/s12943-019-0951-0
57. Zu X, Tang Z, Li Y, Gao N, Ding J, Qi L. Vascular endothelial growth factor-C expression in bladder transitional cell cancer and its relationship to lymph node metastasis. BJU Int (2006) 98:1090–3. doi: 10.1111/j.1464-410X.2006.06446.x
58. Zhang HH, Qi F, Shi YR, Miao JG, Zhou M, He W, et al. RNA interference-mediated vascular endothelial growth factor-C reduction suppresses malignant progression and enhances mitomycin C sensitivity of bladder cancer T24 cells. Cancer Biother Radiopharm (2012a) 27:291–8. doi: 10.1089/cbr.2010.0919
59. Zhang HH, Qi F, Zu XB, Cao YH, Miao JG, Xu L, et al. A proteomic study of potential VEGF-C-associated proteins in bladder cancer T24 cells. Med Sci Monit (2012b) 18:Br441–9. doi: 10.12659/MSM.883537
60. Mccoll BK, Baldwin ME, Roufail S, Freeman C, Moritz RL, Simpson RJ, et al. Plasmin activates the lymphangiogenic growth factors VEGF-C and VEGF-D. J Exp Med (2003) 198:863–8. doi: 10.1084/jem.20030361
61. Pepper MS, Mandriota SJ, Jeltsch M, Kumar V, Alitalo K. Vascular endothelial growth factor (VEGF)-C synergizes with basic fibroblast growth factor and VEGF in the induction of angiogenesis in vitro and alters endothelial cell extracellular proteolytic activity. J Cell Physiol (1998) 177:439–52. doi: 10.1002/(SICI)1097-4652(199812)177:3<439::AID-JCP7>3.0.CO;2-2
62. Zhu H, Yun F, Shi X, Wang D. VEGF-C inhibition reverses resistance of bladder cancer cells to cisplatin via upregulating maspin. Mol Med Rep (2015) 12:3163–9. doi: 10.3892/mmr.2015.3684
63. Liu J, Yu HG, Yu JP, Wang XL, Zhou XD, Luo HS. Overexpression of cyclooxygenase-2 in gastric cancer correlates with the high abundance of vascular endothelial growth factor-C and lymphatic metastasis. Med Oncol (2005) 22:389–97. doi: 10.1385/MO:22:4:389
64. Al-Maghrabi B, Gomaa W, Abdelwahed M, Al-Maghrabi J. Increased COX-2 Immunostaining in Urothelial Carcinoma of the Urinary Bladder Is Associated with Invasiveness and Poor Prognosis. Anal Cell Pathol (Amst) (2019) 2019:5026939. doi: 10.1155/2019/5026939
65. Dong L, Li Y, Xue D, Liu Y. PCMT1 is an unfavorable predictor and functions as an oncogene in bladder cancer. IUBMB Life (2018) 70:291–9. doi: 10.1002/iub.1717
66. Islam SS, Mokhtari RB, Noman AS, Uddin M, Rahman MZ, Azadi MA, et al. Sonic hedgehog (Shh) signaling promotes tumorigenicity and stemness via activation of epithelial-to-mesenchymal transition (EMT) in bladder cancer. Mol Carcinog (2016) 55:537–51. doi: 10.1002/mc.22300
67. Nedjadi T, Salem N, Khayyat D, Al-Sayyad A, Al-Ammari A, Al-Maghrabi J. Sonic Hedgehog Expression is Associated with Lymph Node Invasion in Urothelial Bladder Cancer. Pathol Oncol Res (2019) 25:1067–73. doi: 10.1007/s12253-018-0477-6
68. Syed IS, Pedram A, Farhat WA. Role of Sonic Hedgehog (Shh) Signaling in Bladder Cancer Stemness and Tumorigenesis. Curr Urol Rep (2016) 17:11. doi: 10.1007/s11934-015-0568-9
69. Zhu X, Qiao Y, Liu W, Wang W, Shen H, Lu Y, et al. CXCL5 is a potential diagnostic and prognostic marker for bladder cancer patients. Tumour Biol (2016) 37:4569–77. doi: 10.1007/s13277-015-4275-4
70. Wang C, Li A, Yang S, Qiao R, Zhu X, Zhang J. CXCL5 promotes mitomycin C resistance in non-muscle invasive bladder cancer by activating EMT and NF-κB pathway. Biochem Biophys Res Commun (2018a) 498:862–8. doi: 10.1016/j.bbrc.2018.03.071
71. Zheng J, Zhu X, Zhang J. CXCL5 knockdown expression inhibits human bladder cancer T24 cells proliferation and migration. Biochem Biophys Res Commun (2014) 446:18–24. doi: 10.1016/j.bbrc.2014.01.172
72. Gao Y, Guan Z, Chen J, Xie H, Yang Z, Fan J, et al. CXCL5/CXCR2 axis promotes bladder cancer cell migration and invasion by activating PI3K/AKT-induced upregulation of MMP2/MMP9. Int J Oncol (2015) 47:690–700. doi: 10.3892/ijo.2015.3041
73. Mohammad MA, Ismael NR, Shaarawy SM, El-Merzabani MM. Prognostic value of membrane type 1 and 2 matrix metalloproteinase expression and gelatinase A activity in bladder cancer. Int J Biol Markers (2010) 25:69–74. doi: 10.1177/172460081002500202
74. Szarvas T, Becker M, Vom Dorp F, Gethmann C, Tötsch M, Bánkfalvi A, et al. Matrix metalloproteinase-7 as a marker of metastasis and predictor of poor survival in bladder cancer. Cancer Sci (2010) 101:1300–8. doi: 10.1111/j.1349-7006.2010.01506.x
75. Vasala K, Pääkko P, Turpeenniemi-Hujanen T. Matrix metalloproteinase-9 (MMP-9) immunoreactive protein in urinary bladder cancer: a marker of favorable prognosis. Anticancer Res (2008) 28:1757–61.
76. Zhao J, Xu W, He M, Zhang Z, Zeng S, Ma C, et al. Whole-exome sequencing of muscle-invasive bladder cancer identifies recurrent copy number variation in IPO11 and prognostic significance of importin-11 overexpression on poor survival. Oncotarget (2016) 7:75648–58. doi: 10.18632/oncotarget.12315
77. Zhao J, Shi L, Zeng S, Ma C, Xu W, Zhang Z, et al. Importin-11 overexpression promotes the migration, invasion, and progression of bladder cancer associated with the deregulation of CDKN1A and THBS1. Urol Oncol (2018) 36:311.e311–13. doi: 10.1016/j.urolonc.2018.03.001
78. Presler M, Wojtczyk-Miaskowska A, Schlichtholz B, Kaluzny A, Matuszewski M, Mika A, et al. Increased expression of the gene encoding stearoyl-CoA desaturase 1 in human bladder cancer. Mol Cell Biochem (2018) 447:217–24. doi: 10.1007/s11010-018-3306-z
79. Piao C, Cui X, Zhan B, Li J, Li Z, Li Z, et al. Inhibition of stearoyl CoA desaturase-1 activity suppresses tumour progression and improves prognosis in human bladder cancer. J Cell Mol Med (2019) 23:2064–76. doi: 10.1111/jcmm.14114
80. Du X, Wang QR, Chan E, Merchant M, Liu J, French D, et al. FGFR3 stimulates stearoyl CoA desaturase 1 activity to promote bladder tumor growth. Cancer Res (2012) 72:5843–55. doi: 10.1158/0008-5472.CAN-12-1329
81. Guo X, Li HH, Hu J, Duan YX, Ren WG, Guo Q, et al. ISYNA1 is overexpressed in bladder carcinoma and regulates cell proliferation and apoptosis. Biochem Biophys Res Commun (2019a) 519:246–52. doi: 10.1016/j.bbrc.2019.08.129
82. Chen J, Chou F, Yeh S, Ou Z, Shyr C, Huang C, et al. Androgen dihydrotestosterone (DHT) promotes the bladder cancer nuclear AR-negative cell invasion via a newly identified membrane androgen receptor (mAR-SLC39A9)-mediated Gαi protein/MAPK/MMP9 intracellular signaling. Oncogene (2020a) 39:574–86. doi: 10.1038/s41388-019-0964-6
83. Tang Y, Zu X, Xiong Y, Zhang X. [Expression of Maspin in bladder carcinoma and the relationship between Maspin and lymph node metastasis]. Zhong Nan Da Xue Xue Bao Yi Xue Ban (2015) 40:1306–12. doi:10.11817/j.issn.1672-7347.2015.12.004
84. Zhang HH, Qi F, Cao YH, Zu XB, Chen MF. Expression and clinical significance of microRNA-21, maspin and vascular endothelial growth factor-C in bladder cancer. Oncol Lett (2015) 10:2610–6. doi: 10.3892/ol.2015.3540
85. Juengel E, Beecken WD, Mundiyanapurath S, Engl T, Jonas D, Blaheta RA. Maspin modulates adhesion of bladder carcinoma cells to vascular endothelium. World J Urol (2010) 28:465–71. doi: 10.1007/s00345-010-0539-1
86. Sugimoto S, Maass N, Takimoto Y, Sato K, Minei S, Zhang M, et al. Expression and regulation of tumor suppressor gene maspin in human bladder cancer. Cancer Lett (2004) 203:209–15. doi: 10.1016/j.canlet.2003.09.010
87. Lin YH, Tsui KH, Chang KS, Hou CP, Feng TH, Juang HH. Maspin is a PTEN-Upregulated and p53-Upregulated Tumor Suppressor Gene and Acts as an HDAC1 Inhibitor in Human Bladder Cancer. Cancers (Basel) (2019) 12(1):10. doi: 10.3390/cancers12010010
88. Chen J, Wang L, Tang Y, Gong G, Liu L, Chen M, et al. Maspin enhances cisplatin chemosensitivity in bladder cancer T24 and 5637 cells and correlates with prognosis of muscle-invasive bladder cancer patients receiving cisplatin based neoadjuvant chemotherapy. J Exp Clin Cancer Res (2016a) 35:2. doi: 10.1186/s13046-015-0282-y
89. Wang C, Liu Q, Huang M, Zhou Q, Zhang X, Zhang J, et al. Loss of GATA6 expression promotes lymphatic metastasis in bladder cancer. FASEB J (2020) 34:5754–66. doi: 10.1096/fj.201903176R
90. Zhang Y, Jia L, Zhang Y, Ji W, Li H. Higher expression of FOXOs correlates to better prognosis of bladder cancer. Oncotarget (2017) 8:96313–22. doi: 10.18632/oncotarget.22029
91. Shiota M, Song Y, Yokomizo A, Kiyoshima K, Tada Y, Uchino H, et al. Foxo3a suppression of urothelial cancer invasiveness through Twist1, Y-box-binding protein 1, and E-cadherin regulation. Clin Cancer Res (2010) 16:5654–63. doi: 10.1158/1078-0432.CCR-10-0376
92. Chen X. MiR-101 acts as a novel bio-marker in the diagnosis of bladder carcinoma. Med (Baltimore) (2019) 98:e16051. doi: 10.1097/MD.0000000000016051
93. Liu X, Zhao W, Wang X, Zhu Y, Zhou Z, Shi B. Expression of mir-143 in serum of bladder cancer patients and its correlation with clinical features and prognosis. J Buon (2019a) 24:791–6.
94. Chen X, Wu B, Xu Z, Li S, Tan S, Liu X, et al. Downregulation of miR-133b predict progression and poor prognosis in patients with urothelial carcinoma of bladder. Cancer Med (2016b) 5:1856–62. doi: 10.1002/cam4.777
95. Ma L, Xu Z, Xu C, Jiang X. MicroRNA-148a represents an independent prognostic marker in bladder cancer. Tumour Biol (2016) 37:7915–20. doi: 10.1007/s13277-015-4688-0
96. Chen YJ, Wang HF, Liang M, Zou RC, Tang ZR, Wang JS. Upregulation of miR-3658 in bladder cancer and tumor progression. Genet Mol Res (2016c) 15(4). doi: 10.4238/gmr15049048
97. Li C, Cui Y, Liu LF, Ren WB, Li QQ, Zhou X, et al. High Expression of Long Noncoding RNA MALAT1 Indicates a Poor Prognosis and Promotes Clinical Progression and Metastasis in Bladder Cancer. Clin Genitourin Cancer (2017a) 15:570–6. doi: 10.1016/j.clgc.2017.05.001
98. Li B, Guo LH, Ban ZQ, Liu L, Luo L, Cui TY. Upregulation of lncRNA plasmacytoma variant translocation 1 predicts poor prognosis in patients with muscle-invasive bladder cancer. Med (Baltimore) (2020) 99:e21059. doi: 10.1097/MD.0000000000021059
99. Wang JS, Liu QH, Cheng XH, Zhang WY, Jin YC. The long noncoding RNA ZFAS1 facilitates bladder cancer tumorigenesis by sponging miR-329. BioMed Pharmacother (2018b) 103:174–81. doi: 10.1016/j.biopha.2018.04.031
100. Zhao XL, Zhao ZH, Xu WC, Hou JQ, Du XY. Increased expression of SPRY4-IT1 predicts poor prognosis and promotes tumor growth and metastasis in bladder cancer. Int J Clin Exp Pathol (2015) 8:1954–60.
101. Zhan Y, Li Y, Guan B, Wang Z, Peng D, Chen Z, et al. Long non-coding RNA HNF1A-AS1 promotes proliferation and suppresses apoptosis of bladder cancer cells through upregulating Bcl-2. Oncotarget (2017) 8:76656–65. doi: 10.18632/oncotarget.20795
102. Chen Q, Fu L. Upregulation of long non-coding RNA ROR1-AS1 promotes cell growth and migration in bladder cancer by regulation of miR-504. PloS One (2020) 15:e0227568. doi: 10.1371/journal.pone.0227568
103. Cao HL, Liu ZJ, Huang PL, Yue YL, Xi JN. lncRNA-RMRP promotes proliferation, migration and invasion of bladder cancer via miR-206. Eur Rev Med Pharmacol Sci (2019) 23:1012–21.
104. Liang Z, Guo W, Fang S, Zhang Y, Lu L, Xu W, et al. CircRNAs: Emerging Bladder Cancer Biomarkers and Targets. Front Oncol (2020) 10:606485. doi: 10.3389/fonc.2020.606485
105. Xu ZQ, Yang MG, Liu HJ, Su CQ. Circular RNA hsa_circ_0003221 (circPTK2) promotes the proliferation and migration of bladder cancer cells. J Cell Biochem (2018) 119:3317–25. doi: 10.1002/jcb.26492
106. D’andrea D, Moschini M, Gust KM, Abufaraj M, Özsoy M, Mathieu R, et al. Lymphocyte-to-monocyte ratio and neutrophil-to-lymphocyte ratio as biomarkers for predicting lymph node metastasis and survival in patients treated with radical cystectomy. J Surg Oncol (2017) 115:455–61. doi: 10.1002/jso.24521
107. Zhou M, He L, Zu X, Zhang H, Zeng H, Qi L. Lymphatic vessel density as a predictor of lymph node metastasis and its relationship with prognosis in urothelial carcinoma of the bladder. BJU Int (2011) 107:1930–5. doi: 10.1111/j.1464-410X.2010.09725.x
108. Chen M, Nie ZY, Wen XH, Gao YH, Cao H, Zhang SF. m6A RNA methylation regulators can contribute to malignant progression and impact the prognosis of bladder cancer. Biosci Rep (2019d) 39(12):BSR20192892. doi: 10.1042/BSR20192892
109. Han J, Wang JZ, Yang X, Yu H, Zhou R, Lu HC, et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol Cancer (2019) 18:110. doi: 10.1186/s12943-019-1036-9
110. Smith SC, Baras AS, Dancik G, Ru Y, Ding KF, Moskaluk CA, et al. A 20-gene model for molecular nodal staging of bladder cancer: development and prospective assessment. Lancet Oncol (2011) 12:137–43. doi: 10.1016/S1470-2045(10)70296-5
111. Seiler R, Lam LL, Erho N, Takhar M, Mitra AP, Buerki C, et al. Prediction of Lymph Node Metastasis in Patients with Bladder Cancer Using Whole Transcriptome Gene Expression Signatures. J Urol (2016) 196:1036–41. doi: 10.1016/j.juro.2016.04.061
112. Lu X, Wang Y, Jiang L, Gao J, Zhu Y, Hu W, et al. A Pre-operative Nomogram for Prediction of Lymph Node Metastasis in Bladder Urothelial Carcinoma. Front Oncol (2019) 9:488. doi: 10.3389/fonc.2019.00488
113. Wu S, Zheng J, Li Y, Yu H, Shi S, Xie W, et al. A Radiomics Nomogram for the Preoperative Prediction of Lymph Node Metastasis in Bladder Cancer. Clin Cancer Res (2017) 23:6904–11. doi: 10.1158/1078-0432.CCR-17-1510
114. Wu S, Zheng J, Li Y, Wu Z, Shi S, Huang M, et al. Development and Validation of an MRI-Based Radiomics Signature for the Preoperative Prediction of Lymph Node Metastasis in Bladder Cancer. EBioMedicine (2018a) 34:76–84. doi: 10.1016/j.ebiom.2018.07.029
115. Wu SX, Huang J, Liu ZW, Chen HG, Guo P, Cai QQ, et al. A Genomic-clinicopathologic Nomogram for the Preoperative Prediction of Lymph Node Metastasis in Bladder Cancer. EBioMedicine (2018b) 31:54–65. doi: 10.1016/j.ebiom.2018.03.034
116. Chen J, Cui YU, Liu L, Li C, Tang Y, Zhou XU, et al. CCR7 as a predictive biomarker associated with computed tomography for the diagnosis of lymph node metastasis in bladder carcinoma. Oncol Lett (2016d) 11:735–40. doi: 10.3892/ol.2015.3939
117. Cantiello F, Cicione A, Autorino R, Salonia A, Briganti A, Ferro M, et al. Visceral obesity predicts adverse pathological features in urothelial bladder cancer patients undergoing radical cystectomy: a retrospective cohort study. World J Urol (2014) 32:559–64. doi: 10.1007/s00345-013-1147-7
118. Garg T, Young AJ, O’keeffe-Rosetti M, Mcmullen CK, Nielsen ME, Murphy TE, et al. Association between metabolic syndrome and recurrence of nonmuscle-invasive bladder cancer in older adults. Urol Oncol (2020) 38:737.e717–23. doi: 10.1016/j.urolonc.2020.04.010
119. Chromecki TF, Cha EK, Fajkovic H, Rink M, Ehdaie B, Svatek RS, et al. Obesity is associated with worse oncological outcomes in patients treated with radical cystectomy. BJU Int (2013) 111:249–55. doi: 10.1111/j.1464-410X.2012.11322.x
120. Kwon T, Jeong IG, You D, Han KS, Hong S, Hong B, et al. Obesity and prognosis in muscle-invasive bladder cancer: the continuing controversy. Int J Urol (2014) 21:1106–12. doi: 10.1111/iju.12530
121. Chiang CL, Huang HH, Huang TY, Shih YL, Hsieh TY, Lin HH. Nonalcoholic Fatty Liver Disease Associated With Bladder Cancer. Am J Med Sci (2020) 360:161–5. doi: 10.1016/j.amjms.2020.04.031
122. Tarantino G, Crocetto F, Di Vito C, Creta M, Martino R, Pandolfo SD, et al. Association of NAFLD and Insulin Resistance with Non Metastatic Bladder Cancer Patients: A Cross-Sectional Retrospective Study. J Clin Med (2021) 10(2):346. doi: 10.3390/jcm10020346
123. Xu Y, Huo R, Chen X, Yu X. Diabetes mellitus and the risk of bladder cancer: A PRISMA-compliant meta-analysis of cohort studies. Med (Baltimore) (2017) 96:e8588. doi: 10.1097/MD.0000000000008588
124. Hu J, Chen JB, Cui Y, Zhu YW, Ren WB, Zhou X, et al. Association of metformin intake with bladder cancer risk and oncologic outcomes in type 2 diabetes mellitus patients: A systematic review and meta-analysis. Med (Baltimore) (2018) 97:e11596. doi: 10.1097/MD.0000000000011596
125. Wu Y, Zheng Q, Li Y, Wang G, Gao S, Zhang X, et al. Metformin targets a YAP1-TEAD4 complex via AMPKα to regulate CCNE1/2 in bladder cancer cells. J Exp Clin Cancer Res (2019b) 38:376. doi: 10.1186/s13046-019-1346-1
126. Zhang T, Guo P, Zhang Y, Xiong H, Yu X, Xu S, et al. The antidiabetic drug metformin inhibits the proliferation of bladder cancer cells in vitro and in vivo. Int J Mol Sci (2013) 14:24603–18. doi: 10.3390/ijms141224603
127. Newton CC, Gapstur SM, Campbell PT, Jacobs EJ. Type 2 diabetes mellitus, insulin-use and risk of bladder cancer in a large cohort study. Int J Cancer (2013) 132:2186–91. doi: 10.1002/ijc.27878
128. Liu S, Li Y, Lin T, Fan X, Liang Y, Heemann U. High dose human insulin and insulin glargine promote T24 bladder cancer cell proliferation via PI3K-independent activation of Akt. Diabetes Res Clin Pract (2011) 91:177–82. doi: 10.1016/j.diabres.2010.11.009
129. Pal DK, Agrawal A, Ghosh S, Ghosh A. Association of arsenic with recurrence of urinary bladder cancer. Trop Doct (2020) 50:325–30. doi: 10.1177/0049475520930155
130. Zhou Q, Jin P, Liu J, Li S, Liu W, Xi S. Arsenic-induced HER2 promotes proliferation, migration and angiogenesis of bladder epithelial cells via activation of multiple signaling pathways in vitro and in vivo. Sci Total Environ (2021) 753:141962. doi: 10.1016/j.scitotenv.2020.141962
Keywords: lymph node metastasis, bladder cancer, biomarkers, oncogenes, tumor suppressor genes
Citation: Zhang C, Hu J, Li H, Ma H, Othmane B, Ren W, Yi Z, Qiu D, Ou Z, Chen J and Zu X (2021) Emerging Biomarkers for Predicting Bladder Cancer Lymph Node Metastasis. Front. Oncol. 11:648968. doi: 10.3389/fonc.2021.648968
Received: 14 January 2021; Accepted: 05 March 2021;
Published: 19 March 2021.
Edited by:
Matteo Ferro, European Institute of Oncology (IEO), ItalyReviewed by:
Felice Crocetto, Federico II University Hospital, ItalyCopyright © 2021 Zhang, Hu, Li, Ma, Othmane, Ren, Yi, Qiu, Ou, Chen and Zu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiongbing Zu, enV4Ynh5eXlAMTI2LmNvbQ==; Jinbo Chen, Y2hlbmppbmJvMTk4OUB5YWhvby5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.