- Department of Obstetrics and Gynecology, Affiliated Haikou Hospital, Xiangya Medical College of Central South University, Haikou, China
Metastasis remains to be a huge challenge in cancer therapy. The mechanism underlying cervical cancer metastasis is not well understood and needs to be elucidated. Recent studies have highlighted the diverse roles of non-coding RNAs in cancer progression and metastasis. Increasing numbers of miRNAs, lncRNAs and circRNAs are found to be dysregulated in cervical cancer, associated with metastasis. They have been shown to regulate metastasis through regulating metastasis-related genes, epithelial-mesenchymal transition, signaling pathways and interactions with tumor microenvironment. Moreover, miRNAs can interact with lncRNAs and circRNAs respectively during this complex process. Herein, we review literatures up to date involving non-coding RNAs in cervical cancer metastasis, mainly focus on the underlying mechanisms and highlight the interaction network between miRNAs and lncRNAs, as well as circRNAs. Finally, we discuss the therapeutic prospects.
Introduction
Cervical cancer is a common gynecological cancer which ranks fourth for both incidence and mortality in women worldwide (1). Over the last few decades, there has been a decline in incidence and mortality rates in many populations worldwide, owing to the screening techniques and application of HPV vaccines (2). However, in some developing countries, cervical cancer ranks second or remains the most frequently diagnosed cancer and the leading cause of cancer death among women (3). Furthermore, the prognosis is very poor for patients with advanced or metastatic cervical cancer, whereas current treatment is rather limited (4–6). Therefore, there is an urgent need to decipher the mechanism of metastasis and find new therapeutic targets. Clinically, cervical cancer cells usually invade adjacent tissues and metastasis through lymphatic or blood vessels to distant sites. Over the last decades, studies on mechanisms underlying cervical cancer metastasis focused mainly on oncogenes and protein-related signaling pathways (7, 8). However, the leading regulator of this process is still not well understood. Metastasis is a complex process including several steps and large amounts of molecular interactions (9). Cancer cells need to undergo phenotype changes during this complicated process. In recent years, our understanding of the non-coding transcripts has been largely advanced through high-throughput sequencing technology. Increasing evidence is pointing toward non-coding RNAs as important regulators in many aspects of cancer metastasis (10, 11).
Non-coding RNAs (NcRNAs) are transcripts that do not code for proteins, which can be roughly divided into small non-coding RNAs (smaller than 200 nt) and long non-coding RNAs (lncRNAs, longer than 200 nt) (12, 13). NcRNAs account for the majority of transcriptome, interestingly the amount of ncRNAs correlates with organismal complexity (14). Emerging evidence showed that ncRNAs have regulatory roles in diverse cellular processes both in biological and pathological conditions including cancer (15). Their roles in cancer progression and metastasis are being appreciated (16, 17). Among them, microRNAs (miRNAs), lncRNAs and circular RNAs (circRNAs) are actively studied in recent years. Research is accelerating to decipher the underlying mechanism of ncRNA-regulated cervical cancer metastasis (18, 19). In this review, we summarize the recently identified ncRNAs in cervical cancer metastasis, describe the mechanism of action and discuss their therapeutic perspectives. Particularly, we show the interaction networks between miRNAs and lncRNAs, as well as circRNAs. We hope to provide insights into the aspect of ncRNAs-regulated metastasis and their potential as therapeutic targets.
MicroRNAs in Cervical Cancer Metastasis
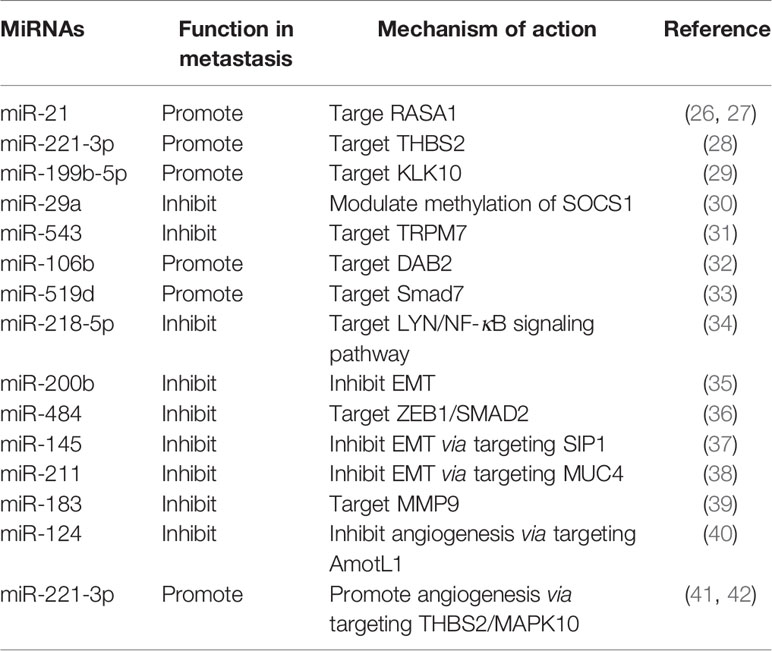
MiRNAs are a class of conserved small endogenous RNAs defined as single-stranded RNAs of ~22 nucleotides in length with no protein-coding potential. Thousands of miRNAs have been identified and annotated among different species (20). Since the discovery of miRNAs over two decades ago, the biology of miRNAs has been extensively reviewed. Most miRNA genes are transcribed by RNA polymerase II (Pol II) and processed in the nucleus, then cleaved in the cytoplasm and incorporated into Argonaute protein, formulating the RNA-induced silencing complex (miRISC) containing the mature miRNA strand (21). MiRNAs are critical regulators of gene expression, they can guide miRISCs to target mRNAs by base pairing with the 3′ untranslated regions (UTRs) of mRNAs, resulting in degradation or translational repression of the mRNA targets (22). Both the 5’ and 3’ regions of miRNA provide information for the specific target recognition (23). Computational approaches to predict miRNA targets revealed that a single miRNA can target several mRNAs and a single mRNA can be regulated by different miRNAs (20). Since the early finding in 2004 revealed that nearly one-half of miRNA genes are located in fragile sites or in caner-associated genomic regions (24), a great number of miRNAs have been reported to be dysregulated in cancer with pro- or anti-tumor potential (25). Over the past decade, a series of miRNAs have been found to be aberrantly expressed in cervical cancer and correlate with metastasis (Table 1).
MicroRNAs Regulate Metastasis-Related Genes
Detection of circulating miRNAs in serum of cervical cancer patients found that miR-21 was related to lymph node metastasis by inhibiting RASA1 (26). Subsequent in vivo study also found that miR-21 could promote lymph node metastasis in orthotopic xenograft mouse model of cervical cancer (27). Thrombospondin-2 (THBS2) is a matricellular protein with antiangiogenic activity, which can modulate extracellular matrix assembly (43), and correlates with cancer metastasis (44). In cervical cancer, miR-221-3p was found to be upregulated by the transcription factor twist2, and promote lymph node metastasis via targeting THBS2 (28). Another miRNA 199b-5p was reported to promote metastasis in cervical cancer by downregulating kallikrein-related peptidase 10 (KLK10) (29). Metastasis-inhibiting miRNAs have also been documented, such as miR-29a inhibits invasion and metastasis of cervical cancer through modulating methylation of suppressor of cytokine signaling protein 1 (SOCS1) (30), and miR-543 inhibits cervical cancer metastasis by targeting transient receptor potential melastatin 7 (TRPM7) (31).
MicroRNAs Regulate Metastasis-Related Signaling Pathways
TGF-β signaling pathway has been reported to correlate with lymph node metastasis in cervical cancer (45). Recently, miR-106b was found to be involved in TGF-β-induced cell migration by targeting disabled homolog 2 (DAB2) in cervical carcinoma (32). Smad 7 is a negative regulator in TGF-β signaling pathway. Study showed that miR-519d facilitates progression and metastasis of cervical cancer through targeting smad7 (33). A recent study based on bioinformatic analysis found that miR-218-5p could inhibit cervical cancer cell metastasis via targeting LYN/NF-κB signaling pathway (34).
MicroRNAs Regulate Epithelial-Mesenchymal Transition
Epithelial-mesenchymal transition (EMT) is a plastic and dynamic biological process orchestrating cell morphological changes, the reverse program of this process is called mesenchymal-epithelial transition (MET) (46). The roles of EMT plasticity in cancer metastasis have been actively studied and extensively reviewed (47, 48). EMT may contribute to the early stage of metastasis by conferring upon epithelium-derived cancer cells the capacity of migration and invasion, while MET is thought to be important for colonization of the disseminated cancer cells at distant sites (48). MiRNAs are emerging as crucial regulators of EMT by targeting multiple components of this program. The miR-200 family (miR-200a, -200b, -200c, -141 and -429) has been recognized as tumor suppressor miRNAs by inhibiting EMT (49, 50). MiR-200b has been reported to suppress cervical cancer cell invasion and metastasis through inhibiting EMT (35). Some other miRNAs have also been reported to modulate EMT and metastasis in cervical cancer. MiR-484 (36), miR-145 (37) and miR-211 (38) have recently been reported to inhibit EMT and invasion of cervical cancer cells via targeting ZEB1/SMAD2, SMAD-interacting protein 1 (SIP1) and mucin 4 (MUC4), respectively.
MicroRNAs Regulate Tumor Microenvironment
The tumor microenvironment includes numerous types of stroma cells and the extracellular matrix. The reciprocal interactions between tumor cells and tumor microenvironment during tumor initiation and progression have long been recognized (51). Moreover, miRNAs have been revealed to regulate tumor microenvironment through different aspects (52). Matrix metalloproteinases (MMPs) are prominent extracellular proteinases which can influence the primary tumor invasion and metastasis (53). Research in cervical cancer found that miR-183 can target MMP-9 and then inhibit cellular invasion and metastasis (39). The induction of angiogenesis is indispensable for growth and metastasis of solid tumors in the tumor microenvironment (54). Research has reported that miR-124 could target angiomotin-like protein AmotL1 and then represses vasculogenic mimicry and cell motility in cervical carcinoma cells (40). Recent data shows that cancer-derived exosomes can transport miRNAs to regulate angiogenesis and invasion in cervical cancer. Cervical squamous cell carcinoma-secreted exosomal miR-221-3p has been shown to promote angiogenesis by targeting THBS2 (41). Additionally, another report implicated that the exosomal miR-221-3p promotes invasion, migration and angiogenesis in cervical cancer by decreasing MAPK10 (42).
Long Non-Coding RNAs in Cervical Cancer Metastasis
LncRNAs are defined as transcripts longer than 200nt with no significant open reading frames and encode no proteins (55). The biogenesis of lncRNAs is much like mRNAs. Many lncRNAs are transcribed by RNA polymerase II, polyadenylated, spliced and 5′-capped, but tend to be shorter than mRNAs (56). Moreover, lncRNAs are expressed at relatively low levels compared with mRNAs, but show more cell-type and tissue-type specificity (56). LncRNAs can be roughly divided into five classes according to their location in the genome where they are transcribed, including long intergenic noncoding RNAs (lincRNAs), natural antisense transcripts, pseudogenes, long intronic ncRNAs and the ncRNAs produced from the transcription start sites, such as promoter-associated RNAs and enhancer RNAs (57).
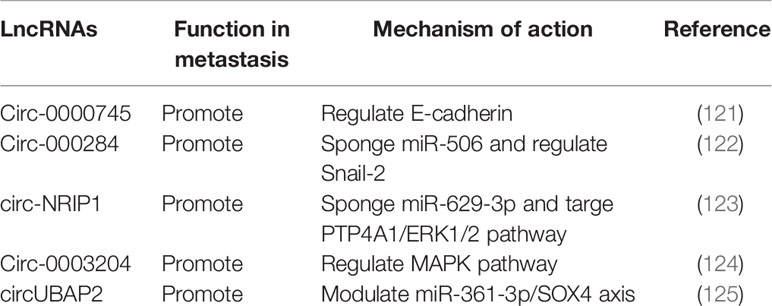
Increasing numbers of lncRNAs have been identified, but only a small fraction of them have been functionally characterized (58). Unlike miRNAs which function predominantly in the cytoplasmic compartment, lncRNAs are found both in the nucleus and the cytoplasm (59), indicating that lncRNAs may function through diverse mechanisms. LncRNAs have been shown to regulate gene expression at different levels, and they can regulate gene expression either in cis or in trans (14, 60). Cis-acting lncRNAs can regulate the expression or chromatin state of nearby genes through three common mechanisms: (1) sequence-dependent lncRNA regulation, the lncRNA transcript can recruit regulatory factors to specific gene loci (Figure 1A) (61); (2) transcription or splicing-dependent regulation, the act of lncRNA transcription rather than the transcript itself can affect gene expression (Figure 1B) (62); and (3) the cis-acting DNA elements within lncRNA loci can also regulate adjacent gene expression (Figure 1C) (63). While, the trans-acting lncRNAs can leave the site of transcription and regulate gene expression at independent sites (60). For example, they can regulate gene expression at distant sites by interacting with promoters, enhancers or proteins binding with these sites (Figure 2A) (64), or modulating chromatin states (65) and RNA polymerase activities (Figure 2B) (66). Moreover, some lncRNAs may affect nuclear architecture to influence gene expression (Figure 2B) (67). Additionally, some trans-acting lncRNAs can bind to and regulate the activity or abundance of proteins or RNAs by function as decoys or competing endogenous RNAs (ceRNAs) (Figure 2C) (68, 69). In consideration of the diverse functions of lncRNAs, recent studies have highlighted the significant roles of lncRNAs in cancer progression (70, 71). A series of lncRNAs have been revealed to play crucial roles in cervical cancer metastasis (Table 2).
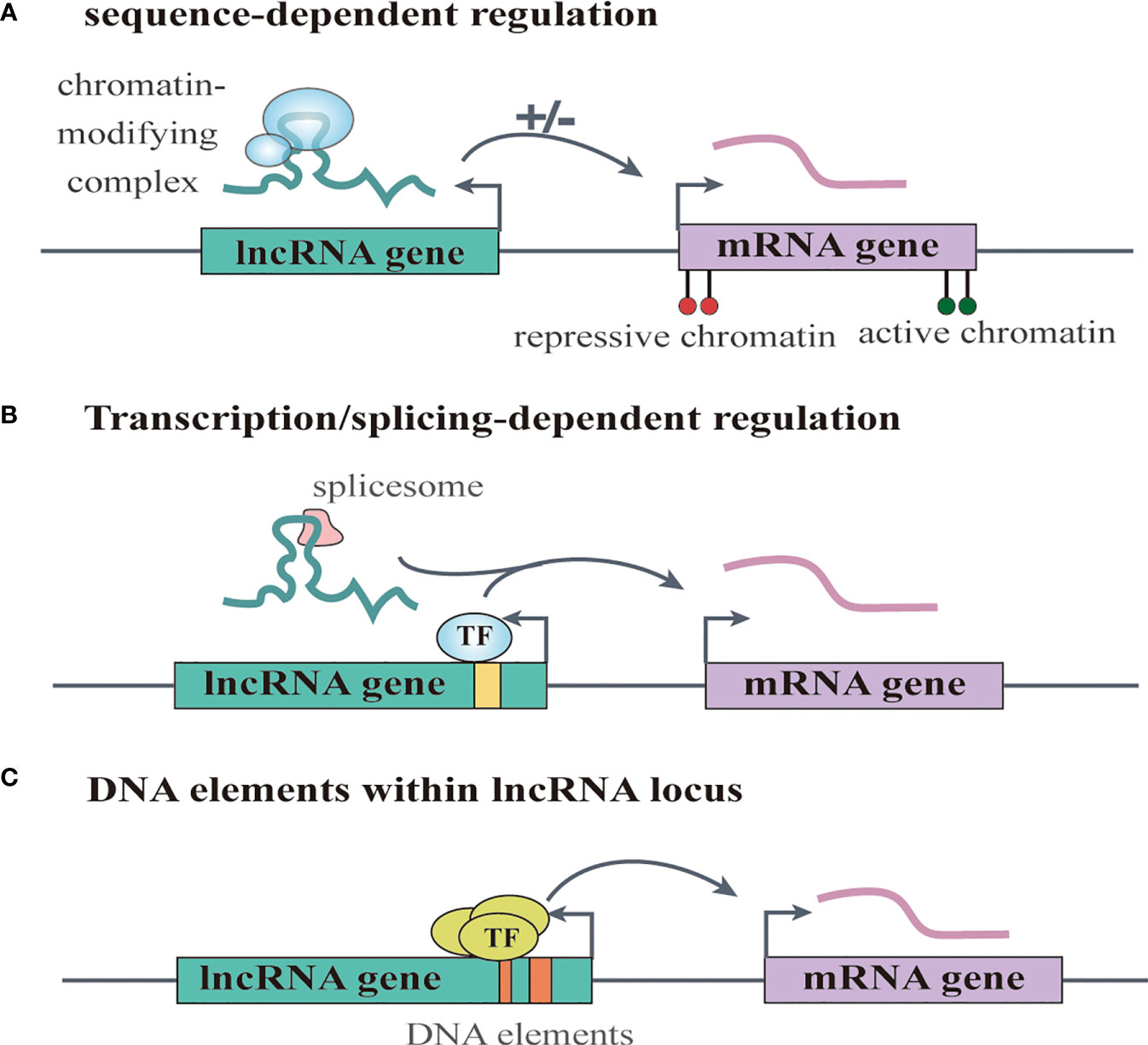
Figure 1 LncRNAs regulate gene expression in cis. (A) sequence-dependent regulation, the lncRNA transcript can recruit regulatory factors to specific gene loci. (B) transcription or splicing-dependent regulation, the act of lncRNA transcription rather than the transcript itself can affect gene expression. (C) the cis-acting DNA elements within lncRNA loci can also regulate adjacent gene expression.
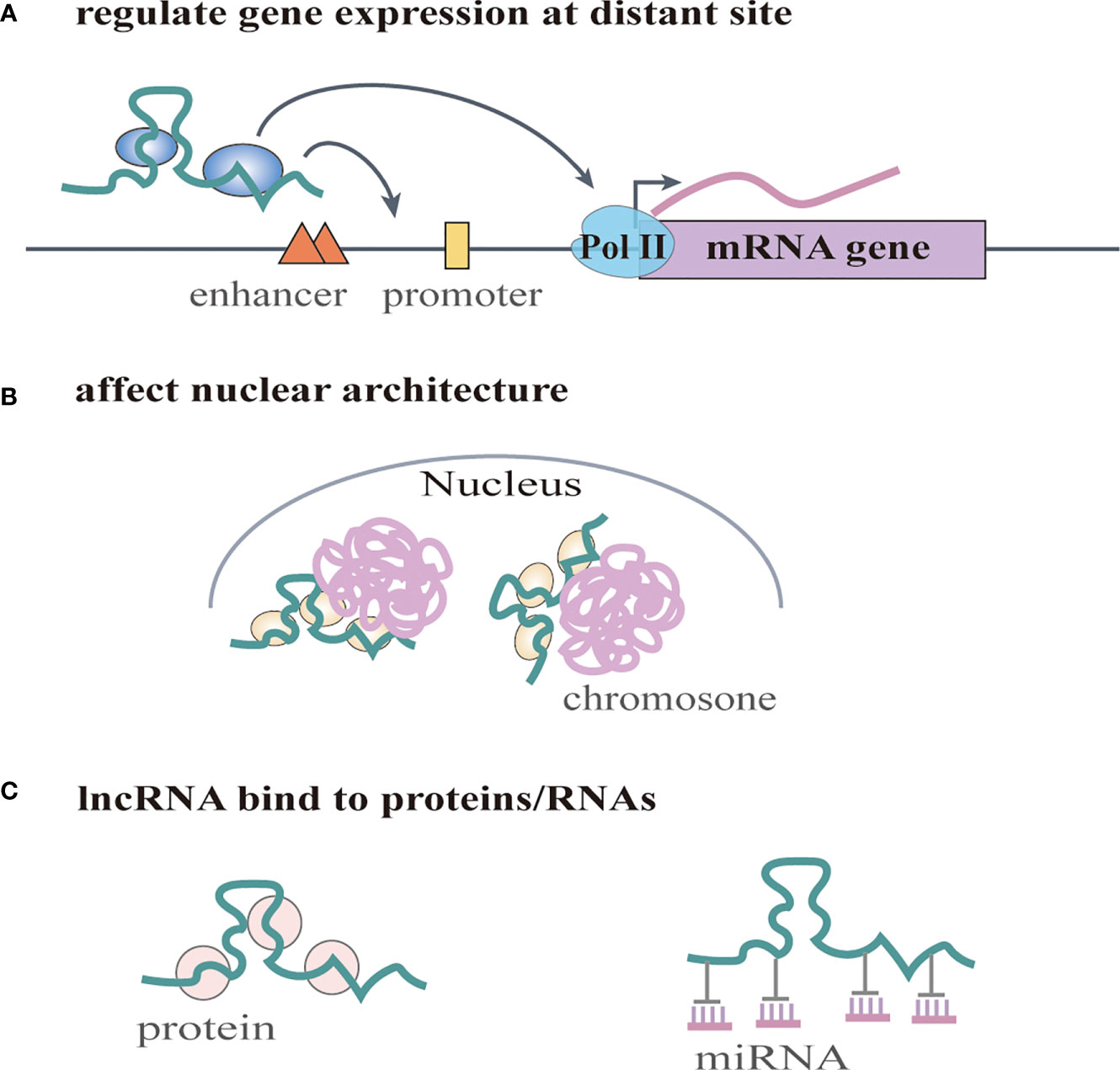
Figure 2 LncRNAs regulate gene expression in trans. (A) lncRNAs regulate gene expression at distant site by interacting with promoters, enhancers or proteins binding with these sites. They can also regulate RNA polymerase activities. (B) lncRNAs affect nuclear architecture to influence gene expression. (C) lncRNAs can bind to and regulate the activity or abundance of proteins or RNAs by function as decoys or competing endogenous RNAs (ceRNAs).
Long Non-Coding RNAs Regulate Epithelial-Mesenchymal Transition
Some lncRNAs have been found to regulate EMT-related genes and EMT-transcription factors. Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is a highly conserved and abundant lncRNA, which was originally described to play crucial roles in lung cancer metastasis (93). It has also been reported to associate with metastasis of many other tumors (94). In cervical cancer, study showed that MALAT1 promotes invasion and metastasis of cervical cancer cells via inducing EMT (72). Another lncRNA EBIC which can bind to enhancer of zeste homolog 2 (EZH2) in cervical cancer has been reported to promote cell invasion by repressing E-cadherin (75). Taurine-upregulated gene 1 (TUG1) is an oncogenic lncRNA in multiple human cancers (95). It has been found to regulate cervical cancer cells migration and invasion by promoting EMT (76). A novel lncRNA CTS identified from the lncRNA microarray database was found to promote metastasis and EMT of cervical cancer by regulating miR-505/ZEB2 axis (77).
Long Non-Coding RNAs Regulate Metastasis by Interacting With MicroRNAs
The vast majority of transcripts in the genome can interact with each other through different mechanisms (96, 97). For example, lncRNAs can act as competing endogenous RNAs (ceRNAs) to sequester miRNAs, regulating the abundance and activity of miRNAs, leading to derepression of genes targeted by corresponding miRNAs (98, 99). The ceRNA network is also implicated in cancer progression (100).
As mentioned previously, MALAT1 is involved in cervical cancer metastasis. Recent study showed that MALAT1 can sponge miR-429 to promote cervical cancer metastasis and progression both in vitro and in vivo (73). Another study also found that MALAT1 can promote invasion of cervical cancer cells through sponging miR-202-3p and upregulating expression of periostin (74). HOX transcript antisense intergenic RNA (HOTAIR) is a trans-acting lncRNA which was originally found to promote metastasis in breast cancer by reprograming chromatin state (101). In cervical cancer, HOTAIR was reported to enhance metastatic potential by sponging miR-23b and modulating the expression of MAPK1 (78). Xist is a well-known lncRNA derived from XIST gene which can regulate X-chromosome inactivation (102), and recognized as a tumor promoter in various malignant tumors (103). It has been revealed to promote cervical cancer cell invasion and migration via competitively binding miR-889-3p and upregulating SIX1 (81). LncRNA 799 is a lncRNA identified from microarray analysis, which has been shown to promote metastasis of SiHa cells via competing for miR-454-3p and upregulating transducing β-like protein1-related protein (TBL1XR1) (82). Another microarray-identified lncRNA XLOC_006390 was found to facilitate metastasis as a ceRNA against miR-331-3p and miR-338-3p in cervical cancer (83). A novel lncRNA TTN-AS1 was found to promote metastasis of cervical cancer cells via sponging miR-573 and regulating E2F3 (84). LncRNA Zinc finger protein 667-antisense RNA 1 (ZNF667-AS1) was revealed to suppress metastasis in cervical cancer by sponging miR-93-3p and upregulating PEG3 (85). Another cancer-related lncRNA DANCR was found to act as a ceRNA for miR-665 and promote metastasis of cervical cancer through the ERK/SMAD pathway (86).
Besides, lncRNAs can regulate expression of miRNAs through epigenetic regulation. PVT1 is an oncogenic lncRNA involved in a variety of cancer types, correlates with the copy number of the MYC oncogene (104). Increased PVT1 expression in cervical cancer contributes to cancer phenotype and associates with poor prognosis (105). Further studies showed that PVT1 could contribute to cervical cancer progression and metastasis through epigenetically silencing miR-200b (87) and miR-195 (88) respectively and modulating EMT. Growth arrest-specific transcript 5 (GAS5) is down-regulated in several cancers and recognized as a tumor suppressing lncRNA. The antisense transcript of GAS5 (GAS5-AS1) has been reported to suppress metastasis of cervical cancer by modulating GAS5 epigenetically and increasing its stability (89).
Long Non-Coding RNAs Regulate Metastasis-Related Signaling Pathways
LncRNAs have also been revealed to drive different cancer phenotypes through regulating the intracellular signaling networks (106). HOTAIR has been revealed to target the Notch signaling pathway (79) or synergize with STAT3 (80) to promote metastasis of cervical cancer cells. DGCR5 (Digeorge syndrome critical gene 5, also known as linc00037) is a lncRNA downregulated in Huntington’s disease neurodegeneration, which has also been implicated in cancer progression. It has been demonstrated that DGCR5 suppressed migration and invasion of cervical cancer cells by targeting WNT signaling (90). Additionally, ANRIL knockdown inhibits cell proliferation and metastasis in vitro, and its inhibition guides inactivation of the PI3K/Akt pathway in cervical cancer (91, 92).
Other Long Non-Coding RNAs Involved in Cervical Cancer Metastasis
Recent findings add to a growing list of lncNRAs associated with cervical cancer metastasis, pending further mechanistic investigation. For example, Colon cancer associated lncRNA (CCAT1) was found to be highly expressed in cervical cancer, and silencing of CCAT1 led to suppression of metastasis of Hela cells (107). LncRNA ATB is a lncRNA activated by TGF-β, originally identified in hepatocellular carcinoma with critical roles in invasion-metastasis cascade (108). Later, a series of studies revealed that ATB could promote metastasis in other cancers (109, 110). Study in cervical cancer showed that ATB is upregulated in cervical cancer tissues and cell lines, and correlates with lymph node metastasis and poor prognosis (111). LncRNA small nucleolar RNA host gene 1 (SNHG1) is enriched in nuclear and found to regulate gene transcription either in cis or in trans (112). Recently, SNHG1 was reported to be highly expressed in cervical cancer and knock-down of SNHG1 decreased migration and invasion of cancer cells (113). Another nuclear-enriched transcript lncRNA NEAT1 is an essential component of paraspeckle, correlates with p53 activation and chemosensitivity (114). Study in cervical cancer showed that high expression of NEAT1 predicted poor prognosis and promoted migration and invasion of cervical cancer cells (115). Upregulation of lncRNA CCHE1 in cervical cancer is correlated with advanced FIGO stage, larger tumor size, lymph node metastasis, invasion of the uterine corpus and poor prognosis (116). Further study is needed to identify the underlying mechanism of the lncRNAs discussed above.
Circular RNAs in Cervical Cancer Metastasis
Circular RNAs (circRNAs) are newly identified class of ncRNAs, different from linear RNAs, they are covalently closed and lack polyadenylation (117). CircRNAs can arise from exons or introns within precursor mRNAs (pre-mRNAs), formed by backsplicing or intronic lariats (118, 119). Interestingly, genome-wide analyses have shown that many circRNAs are abundant, highly stable and evolutionarily conserved in mammalian cells (120, 121). However, biological functions of circRNAs still need further investigation. It has been suggested that circRNAs can function as miRNA or protein sponges, regulate transcription of parental genes in cis, even under some circumstances circRNAs can be translated (119, 122). In recent years, roles of circRNAs in cancer are emerging (123). In cervical cancer, several circRNAs have been implicated in cancer metastasis (Table 3).
High-throughput sequencing technology has been employed to explore circRNA expression profile in cervical cancer tissues and cell lines. A couple of circRNAs have been reported to regulate metastasis through EMT and interaction with signaling pathways. Circ-0000745 was found to be upregulated in cervical cancer tissues, associated with poor differentiation and vascular/lymphatic invasion. Inhibition of circ-0000745 led to down regulation of E-cadherin and reduced migration and invasion (124). Circ-000284 was found to be upregulated in cervical cancer cells. Loss of function assay showed that circ-000284 can promote proliferation and invasion of cervical cancer via sponging miR-506 and regulating Snail-2 (125). Another upregulated circRNA circ-NRIP1 was reported to be relevant to lymphovascular space invasion. Mechanistic investigation showed that circ-NRIP1 can promote migration and invasion of cervical cancer by sponging miR-629-3p and targeting the PTP4A1/ERK1/2 pathway (126). Circ-0003204 was identified by RNA sequencing, which was also upregulated in cervical cancer, found to promote proliferation, migration and invasion of cervical cancer cells through regulating MAPK pathway (127). Recently, the roles of circUBAP2 have been implicated in different cancers. In cervical cancer, it was found to be upregulated and contribute to tumor growth and metastasis by modulating miR-361-3p/SOX4 axis (128). Roles of circRNAs in cervical cancer metastasis remain largely unknown and research is increasing.
Conclusions and Perspectives
The improvement in sequencing technology led to exploding increase in different types of ncRNAs. NcRNAs are emerging as active regulators of cellular process in cancer. MiRNAs are well studied over the past decade, lncRNAs are actively studied for their diverse roles in gene expression regulation, circRNAs are recently identified and their functions need further investigation. Besides, the ncRNAs themselves can interact with each other, such as lncRNAs and circRNAs can sponge miRNAs. The RNA molecules sharing miRNA response elements (MREs) can act as ceRNAs and crosstalk with each other, indicating an enormous and complex regulatory network orchestrating cellular process. As described above, a lot of ncRNAs are dysregulated in cervical cancer, exhibiting either metastasis-promoting or -inhibiting roles through different ways. These findings give insights into target therapy based on RNAs.
Given the diverse roles of ncRNAs in cervical cancer metastasis and their highly expression specificity, therapeutics targeting these regulatory ncRNAs may be appreciated. Overexpression and knock-down approaches used in experimental studies to modulate gene expression and metastasis have shown promising results. Indeed, therapeutic approaches targeting RNAs such as small interfering RNA (siRNA) and antisense oligonucleotide (ASO) have been exploited for many years, as reviewed elsewhere (129–131). Progress is gratifying for more than 100 RNA-targeted therapies have reached clinical development and some have been approved for commercial use in rare disease or chronic disease (132, 133). At present, some phase 1 clinical trials of ASO-based drugs in advanced metastatic cancer treatment are undergoing (e.g., NCT00466583, NCT01120288 and NCT00471432).
The field of ncRNAs in cancer metastasis is promising, which is progressing rapidly for the new classes of ncRNAs are emerging. Large amount of experimental work is still needed to decipher the mechanisms underlying metastasis and fully assess their therapeutic potential. With the development of nucleic acid therapeutics, we hope to identify ncRNAs which can be targeted in advanced and metastatic cervical cancer patients.
Author Contributions
TC performed the literature search and wrote the manuscript. SH reviewed and edited the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer J Clin (2018) 68(6):394–424. doi: 10.3322/caac.21492
2. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA: Cancer J Clin (2021) 71(1):7–33. doi: 10.3322/caac.21654
3. Vaccarella S, Laversanne M, Ferlay J, Bray F. Cervical cancer in Africa, Latin America and the Caribbean and Asia: Regional inequalities and changing trends. Int J Cancer (2017) 141(10):1997–2001. doi: 10.1002/ijc.30901
4. Wright JD, Chen L, Tergas AI, Burke WM, Hou JY, Neugut AI, et al. Population-level trends in relative survival for cervical cancer. Am J Obstet Gynecol (2015) 213(5):670 e1–7. doi: 10.1016/j.ajog.2015.07.012
5. Pfaendler KS, Tewari KS. Changing paradigms in the systemic treatment of advanced cervical cancer. Am J Obstet Gynecol (2016) 214(1):22–30. doi: 10.1016/j.ajog.2015.07.022
6. Buda A, Cesarin J, Mueller M, Fanfani F, Zapardiel I, Mereu L, et al. The impact of low-volume metastasis on disease-free survival of women with early-stage cervical cancer. J Cancer Res Clin Oncol (2020). doi: 10.1007/s00432-020-03435-z
7. Qureshi R, Arora H, Rizvi MA. EMT in cervical cancer: its role in tumour progression and response to therapy. Cancer Lett (2015) 356(2 Pt B):321–31. doi: 10.1016/j.canlet.2014.09.021
8. Zeng YT, Liu XF, Yang WT, Zheng PS. REX1 promotes EMT-induced cell metastasis by activating the JAK2/STAT3-signaling pathway by targeting SOCS1 in cervical cancer. Oncogene (2019) 38(43):6940–57. doi: 10.1038/s41388-019-0906-3
9. Lambert AW, Pattabiraman DR, Weinberg RA. Emerging Biological Principles of Metastasis. Cell (2017) 168(4):670–91. doi: 10.1016/j.cell.2016.11.037
10. Crea F, Clermont PL, Parolia A, Wang Y, Helgason CD. The non-coding transcriptome as a dynamic regulator of cancer metastasis. Cancer Metastasis Rev (2014) 33(1):1–16. doi: 10.1007/s10555-013-9455-3
11. Cao MX, Tang YL, Zhang WL, Tang YJ, Liang XH. Non-coding RNAs as Regulators of Lymphangiogenesis in Lymphatic Development, Inflammation, and Cancer Metastasis. Front Oncol (2019) 9:916:916. doi: 10.3389/fonc.2019.00916
12. Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature (2012) 489(7414):101–8. doi: 10.1038/nature11233
13. Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet (2015) 47(3):199–208. doi: 10.1038/ng.3192
14. Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol (2013) 14(11):699–712. doi: 10.1038/nrm3679
15. Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol Rev (2016) 96(4):1297–325. doi: 10.1152/physrev.00041.2015
16. Markopoulos GS, Roupakia E, Tokamani M, Chavdoula E, Hatziapostolou M, Polytarchou C, et al. A step-by-step microRNA guide to cancer development and metastasis. Cell Oncol (Dordrecht) (2017) 40(4):303–39. doi: 10.1007/s13402-017-0341-9
17. Dhamija S, Diederichs S. From junk to master regulators of invasion: lncRNA functions in migration, EMT and metastasis. Int J Cancer (2016) 139(2):269–80. doi: 10.1002/ijc.30039
18. Wang JY, Chen LJ. The role of miRNAs in the invasion and metastasis of cervical cancer. Biosci Rep (2019) 39(3):1–16. doi: 10.1042/bsr20181377
19. Shang C, Zhu W, Liu T, Wang W, Huang G, Huang J, et al. Characterization of long non-coding RNA expression profiles in lymph node metastasis of early-stage cervical cancer. Oncol Rep (2016) 35(6):3185–97. doi: 10.3892/or.2016.4715
20. Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res (2019) 47(D1):D155–d62. doi: 10.1093/nar/gky1141
21. Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol (2014) 15(8):509–24. doi: 10.1038/nrm3838
22. Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet (2015) 16(7):421–33. doi: 10.1038/nrg3965
23. Broughton JP, Lovci MT, Huang JL, Yeo GW, Pasquinelli AE. Pairing beyond the Seed Supports MicroRNA Targeting Specificity. Mol Cell (2016) 64(2):320–33. doi: 10.1016/j.molcel.2016.09.004
24. Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci United States America (2004) 101(9):2999–3004. doi: 10.1073/pnas.0307323101
25. Bracken CP, Scott HS, Goodall GJ. A network-biology perspective of microRNA function and dysfunction in cancer. Nat Rev Genet (2016) 17(12):719–32. doi: 10.1038/nrg.2016.134
26. Zhang L, Zhan X, Yan D, Wang Z. Circulating MicroRNA-21 Is Involved in Lymph Node Metastasis in Cervical Cancer by Targeting RASA1. Int J Gynecol Cancer Off J Int Gynecol Cancer Soc (2016) 26(5):810–6. doi: 10.1097/igc.0000000000000694
27. Wei WF, Han LF, Liu D, Wu LF, Chen XJ, Yi HY, et al. Orthotopic Xenograft Mouse Model of Cervical Cancer for Studying the Role of MicroRNA-21 in Promoting Lymph Node Metastasis. Int J Gynecol Cancer Off J Int Gynecol Cancer Soc (2017) 27(8):1587–95. doi: 10.1097/igc.0000000000001059
28. Wei WF, Zhou CF, Wu XG, He LN, Wu LF, Chen XJ, et al. MicroRNA-221-3p, a TWIST2 target, promotes cervical cancer metastasis by directly targeting THBS2. Cell Death Dis (2017) 8(12):3220. doi: 10.1038/s41419-017-0077-5
29. Xu LJ, Duan Y, Wang P, Yin HQ. MiR-199b-5p promotes tumor growth and metastasis in cervical cancer by down-regulating KLK10. Biochem Biophys Res Commun (2018) 503(2):556–63. doi: 10.1016/j.bbrc.2018.05.165
30. Gong Y, Wan JH, Zou W, Lian GY, Qin JL, Wang QM. MiR-29a inhibits invasion and metastasis of cervical cancer via modulating methylation of tumor suppressor SOCS1. Future Oncol (London England) (2019) 15(15):1729–44. doi: 10.2217/fon-2018-0497
31. Liu X, Gan L, Zhang J. miR-543 inhibites cervical cancer growth and metastasis by targeting TRPM7. Chem Biol Interact (2019) 302:83–92. doi: 10.1016/j.cbi.2019.01.036
32. Cheng Y, Guo Y, Zhang Y, You K, Li Z, Geng L. MicroRNA-106b is involved in transforming growth factor beta1-induced cell migration by targeting disabled homolog 2 in cervical carcinoma. J Exp Clin Cancer Res CR (2016) 35:11. doi: 10.1186/s13046-016-0290-6
33. Zhou JY, Zheng SR, Liu J, Shi R, Yu HL, Wei M. MiR-519d facilitates the progression and metastasis of cervical cancer through direct targeting Smad7. Cancer Cell Int (2016) 16:21. doi: 10.1186/s12935-016-0298-1
34. Xu Y, He Q, Lu Y, Tao F, Zhao L, Ou R. MicroRNA-218-5p inhibits cell growth and metastasis in cervical cancer via LYN/NF-κB signaling pathway. Cancer Cell Int (2018) 18:198. doi: 10.1186/s12935-018-0673-1
35. Cheng YX, Zhang QF, Hong L, Pan F, Huang JL, Li BS, et al. MicroRNA-200b suppresses cell invasion and metastasis by inhibiting the epithelial-mesenchymal transition in cervical carcinoma. Mol Med Rep (2016) 13(4):3155–60. doi: 10.3892/mmr.2016.4911
36. Hu Y, Xie H, Liu Y, Liu W, Liu M, Tang H. miR-484 suppresses proliferation and epithelial-mesenchymal transition by targeting ZEB1 and SMAD2 in cervical cancer cells. Cancer Cell Int (2017) 17:36. doi: 10.1186/s12935-017-0407-9
37. Sathyanarayanan A, Chandrasekaran KS, Karunagaran D. microRNA-145 modulates epithelial-mesenchymal transition and suppresses proliferation, migration and invasion by targeting SIP1 in human cervical cancer cells. Cell Oncol (Dordrecht) (2017) 40(2):119–31. doi: 10.1007/s13402-016-0307-3
38. Xu D, Liu S, Zhang L, Song L. MiR-211 inhibits invasion and epithelial-to-mesenchymal transition (EMT) of cervical cancer cells via targeting MUC4. Biochem Biophys Res Commun (2017) 485(2):556–62. doi: 10.1016/j.bbrc.2016.12.020
39. Fan D, Wang Y, Qi P, Chen Y, Xu P, Yang X, et al. MicroRNA-183 functions as the tumor suppressor via inhibiting cellular invasion and metastasis by targeting MMP-9 in cervical cancer. Gynecol Oncol (2016) 141(1):166–74. doi: 10.1016/j.ygyno.2016.02.006
40. Wan HY, Li QQ, Zhang Y, Tian W, Li YN, Liu M, et al. MiR-124 represses vasculogenic mimicry and cell motility by targeting amotL1 in cervical cancer cells. Cancer Lett (2014) 355(1):148–58. doi: 10.1016/j.canlet.2014.09.005
41. Wu XG, Zhou CF, Zhang YM, Yan RM, Wei WF, Chen XJ, et al. Cancer-derived exosomal miR-221-3p promotes angiogenesis by targeting THBS2 in cervical squamous cell carcinoma. Angiogenesis (2019) 22(3):397–410. doi: 10.1007/s10456-019-09665-1
42. Zhang L, Li H, Yuan M, Li M, Zhang S. Cervical Cancer Cells-Secreted Exosomal microRNA-221-3p Promotes Invasion, Migration and Angiogenesis of Microvascular Endothelial Cells in Cervical Cancer by Down-Regulating MAPK10 Expression. Cancer Manage Res (2019) 11:10307–19. doi: 10.2147/cmar.S221527
43. Calabro NE, Kristofik NJ, Kyriakides TR. Thrombospondin-2 and extracellular matrix assembly. Biochim Biophys Acta (2014) 1840(8):2396–402. doi: 10.1016/j.bbagen.2014.01.013
44. Liu JF, Lee CW, Tsai MH, Tang CH, Chen PC, Lin LW, et al. Thrombospondin 2 promotes tumor metastasis by inducing matrix metalloproteinase-13 production in lung cancer cells. Biochem Pharmacol (2018) 155:537–46. doi: 10.1016/j.bcp.2018.07.024
45. Noordhuis MG, Fehrmann RS, Wisman GB, Nijhuis ER, van Zanden JJ, Moerland PD, et al. Involvement of the TGF-beta and beta-catenin pathways in pelvic lymph node metastasis in early-stage cervical cancer. Clin Cancer Res Off J Am Assoc Cancer Res (2011) 17(6):1317–30. doi: 10.1158/1078-0432.ccr-10-2320
46. Nieto MA, Huang RY, Jackson RA, Thiery JP. EMT: 2016. Cell (2016) 166(1):21–45. doi: 10.1016/j.cell.2016.06.028
47. Mittal V. Epithelial Mesenchymal Transition in Tumor Metastasis. Annu Rev Pathol (2018) 13:395–412. doi: 10.1146/annurev-pathol-020117-043854
48. Lu W, Kang Y. Epithelial-Mesenchymal Plasticity in Cancer Progression and Metastasis. Dev Cell (2019) 49(3):361–74. doi: 10.1016/j.devcel.2019.04.010
49. Feng X, Wang Z, Fillmore R, Xi Y. MiR-200, a new star miRNA in human cancer. Cancer Lett (2014) 344(2):166–73. doi: 10.1016/j.canlet.2013.11.004
50. Title AC, Hong SJ, Pires ND, Hasenöhrl L, Godbersen S, Stokar-Regenscheit N, et al. Genetic dissection of the miR-200-Zeb1 axis reveals its importance in tumor differentiation and invasion. Nat Commun (2018) 9(1):4671. doi: 10.1038/s41467-018-07130-z
51. Friedl P, Alexander S. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell (2011) 147(5):992–1009. doi: 10.1016/j.cell.2011.11.016
52. Drak Alsibai K, Meseure D. Tumor microenvironment and noncoding RNAs as co-drivers of epithelial-mesenchymal transition and cancer metastasis. Dev Dyn Off Publ Am Assoc Anat (2018) 247(3):405–31. doi: 10.1002/dvdy.24548
53. Deryugina EI, Quigley JP. Tumor angiogenesis: MMP-mediated induction of intravasation- and metastasis-sustaining neovasculature. Matrix Biol J Int Soc Matrix Biol (2015) 44-46:94–112. doi: 10.1016/j.matbio.2015.04.004
54. Paduch R. The role of lymphangiogenesis and angiogenesis in tumor metastasis. Cell Oncol (Dordrecht) (2016) 39(5):397–410. doi: 10.1007/s13402-016-0281-9
55. Kashi K, Henderson L, Bonetti A, Carninci P. Discovery and functional analysis of lncRNAs: Methodologies to investigate an uncharacterized transcriptome. Biochim Biophys Acta (2016) 1859(1):3–15. doi: 10.1016/j.bbagrm.2015.10.010
56. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet (2016) 17(1):47–62. doi: 10.1038/nrg.2015.10
57. Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics (2013) 193(3):651–69. doi: 10.1534/genetics.112.146704
58. Zhao Y, Li H, Fang S, Kang Y, Wu W, Hao Y, et al. NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic Acids Res (2016) 44(D1):D203–8. doi: 10.1093/nar/gkv1252
59. Cabili MN, Dunagin MC, McClanahan PD, Biaesch A, Padovan-Merhar O, Regev A, et al. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol (2015) 16:20. doi: 10.1186/s13059-015-0586-4
60. Kopp F, Mendell JT. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell (2018) 172(3):393–407. doi: 10.1016/j.cell.2018.01.011
61. da Rocha ST, Heard E. Novel players in X inactivation: insights into Xist-mediated gene silencing and chromosome conformation. Nat Struct Mol Biol (2017) 24(3):197–204. doi: 10.1038/nsmb.3370
62. Anderson KM, Anderson DM, McAnally JR, Shelton JM, Bassel-Duby R, Olson EN. Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature (2016) 539(7629):433–6. doi: 10.1038/nature20128
63. Groff AF, Sanchez-Gomez DB, Soruco MML, Gerhardinger C, Barutcu AR, Li E, et al. In Vivo Characterization of Linc-p21 Reveals Functional cis-Regulatory DNA Elements. Cell Rep (2016) 16(8):2178–86. doi: 10.1016/j.celrep.2016.07.050
64. Negishi M, Wongpalee SP, Sarkar S, Park J, Lee KY, Shibata Y, et al. A new lncRNA, APTR, associates with and represses the CDKN1A/p21 promoter by recruiting polycomb proteins. PloS One (2014) 9(4):e95216. doi: 10.1371/journal.pone.0095216
65. Portoso M, Ragazzini R, Brenčič Ž, Moiani A, Michaud A, Vassilev I, et al. PRC2 is dispensable for HOTAIR-mediated transcriptional repression. EMBO J (2017) 36(8):981–94. doi: 10.15252/embj.201695335
66. Xing YH, Yao RW, Zhang Y, Guo CJ, Jiang S, Xu G, et al. SLERT Regulates DDX21 Rings Associated with Pol I Transcription. Cell (2017) 169(4):664–78.e16. doi: 10.1016/j.cell.2017.04.011
67. Lin Y, Schmidt BF, Bruchez MP, McManus CJ. Structural analyses of NEAT1 lncRNAs suggest long-range RNA interactions that may contribute to paraspeckle architecture. Nucleic Acids Res (2018) 46(7):3742–52. doi: 10.1093/nar/gky046
68. Lee S, Kopp F, Chang TC, Sataluri A, Chen B, Sivakumar S, et al. Noncoding RNA NORAD Regulates Genomic Stability by Sequestering PUMILIO Proteins. Cell (2016) 164(1-2):69–80. doi: 10.1016/j.cell.2015.12.017
69. Du Z, Sun T, Hacisuleyman E, Fei T, Wang X, Brown M, et al. Integrative analyses reveal a long noncoding RNA-mediated sponge regulatory network in prostate cancer. Nat Commun (2016) 7:10982. doi: 10.1038/ncomms10982
70. Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res (2017) 77(15):3965–81. doi: 10.1158/0008-5472.Can-16-2634
71. Huang Q, Yan J, Agami R. Long non-coding RNAs in metastasis. Cancer Metastasis Rev (2018) 37(1):75–81. doi: 10.1007/s10555-017-9713-x
72. Sun R, Qin C, Jiang B, Fang S, Pan X, Peng L, et al. Down-regulation of MALAT1 inhibits cervical cancer cell invasion and metastasis by inhibition of epithelial-mesenchymal transition. Mol Biosyst (2016) 12(3):952–62. doi: 10.1039/c5mb00685f
73. Shen F, Zheng H, Zhou L, Li W, Xu X. Overexpression of MALAT1 contributes to cervical cancer progression by acting as a sponge of miR-429. J Cell Physiol (2019) 234(7):11219–26. doi: 10.1002/jcp.27772
74. Han X, Wang Q, Wang Y, Hu B, Dong X, Zhang H, et al. Long non-coding RNA metastasis-associated lung adenocarcinoma transcript 1/microRNA-202-3p/periostin axis modulates invasion and epithelial-mesenchymal transition in human cervical cancer. J Cell Physiol (2019) 234(8):14170–80. doi: 10.1002/jcp.28113
75. Sun NX, Ye C, Zhao Q, Zhang Q, Xu C, Wang SB, et al. Long noncoding RNA-EBIC promotes tumor cell invasion by binding to EZH2 and repressing E-cadherin in cervical cancer. PloS One (2014) 9(7):e100340. doi: 10.1371/journal.pone.0100340
76. Hu Y, Sun X, Mao C, Guo G, Ye S, Xu J, et al. Upregulation of long noncoding RNA TUG1 promotes cervical cancer cell proliferation and migration. Cancer Med (2017) 6(2):471–82. doi: 10.1002/cam4.994
77. Feng S, Liu W, Bai X, Pan W, Jia Z, Zhang S, et al. LncRNA-CTS promotes metastasis and epithelial-to-mesenchymal transition through regulating miR-505/ZEB2 axis in cervical cancer. Cancer Lett (2019) 465:105–17. doi: 10.1016/j.canlet.2019.09.002
78. Li Q, Feng Y, Chao X, Shi S, Liang M, Qiao Y, et al. HOTAIR contributes to cell proliferation and metastasis of cervical cancer via targetting miR-23b/MAPK1 axis. Biosci Rep (2018) 38(1):1–13. doi: 10.1042/bsr20171563
79. Lee M, Kim HJ, Kim SW, Park SA, Chun KH, Cho NH, et al. The long non-coding RNA HOTAIR increases tumour growth and invasion in cervical cancer by targeting the Notch pathway. Oncotarget (2016) 7(28):44558–71. doi: 10.18632/oncotarget.10065
80. Zhang Y, Cheng X, Liang H, Jin Z. Long non-coding RNA HOTAIR and STAT3 synergistically regulate the cervical cancer cell migration and invasion. Chem Biol Interact (2018) 286:106–10. doi: 10.1016/j.cbi.2018.03.010
81. Liu X, Xie S, Zhang J, Kang Y. Long Noncoding RNA XIST Contributes to Cervical Cancer Development Through Targeting miR-889-3p/SIX1 Axis. Cancer Biother Radiopharmaceuticals (2020) 35(9):640-49. doi: 10.1089/cbr.2019.3318
82. Liao LM, Zhang FH, Yao GJ, Ai SF, Zheng M, Huang L. Role of Long Noncoding RNA 799 in the Metastasis of Cervical Cancer through Upregulation of TBL1XR1 Expression. Mol Ther Nucleic Acids (2018) 13:580–9. doi: 10.1016/j.omtn.2018.10.007
83. Luan X, Wang Y. LncRNA XLOC_006390 facilitates cervical cancer tumorigenesis and metastasis as a ceRNA against miR-331-3p and miR-338-3p. J Gynecol Oncol (2018) 29(6):e95. doi: 10.3802/jgo.2018.29.e95
84. Chen P, Wang R, Yue Q, Hao M. Long non-coding RNA TTN-AS1 promotes cell growth and metastasis in cervical cancer via miR-573/E2F3. Biochem Biophys Res Commun (2018) 503(4):2956–62. doi: 10.1016/j.bbrc.2018.08.077
85. Li YJ, Yang Z, Wang YY, Wang Y. Long noncoding RNA ZNF667-AS1 reduces tumor invasion and metastasis in cervical cancer by counteracting microRNA-93-3p-dependent PEG3 downregulation. Mol Oncol (2019) 13(11):2375–92. doi: 10.1002/1878-0261.12565
86. Cao L, Jin H, Zheng Y, Mao Y, Fu Z, Li X, et al. DANCR-mediated microRNA-665 regulates proliferation and metastasis of cervical cancer through the ERK/SMAD pathway. Cancer Sci (2019) 110(3):913–25. doi: 10.1111/cas.13921
87. Zhang S, Zhang G, Liu J. Long noncoding RNA PVT1 promotes cervical cancer progression through epigenetically silencing miR-200b. APMIS Acta Pathol Microbiol Immunol Scand (2016) 124(8):649–58. doi: 10.1111/apm.12555
88. Shen CJ, Cheng YM, Wang CL. LncRNA PVT1 epigenetically silences miR-195 and modulates EMT and chemoresistance in cervical cancer cells. J Drug Targeting (2017) 25(7):637–44. doi: 10.1080/1061186x.2017.1307379
89. Wang X, Zhang J, Wang Y. Long noncoding RNA GAS5-AS1 suppresses growth and metastasis of cervical cancer by increasing GAS5 stability. Am J Trans Res (2019) 11(8):4909–21.
90. Liu Y, Chang Y, Lu S, Xiang YY. Downregulation of long noncoding RNA DGCR5 contributes to the proliferation, migration, and invasion of cervical cancer by activating Wnt signaling pathway. J Cell Physiol (2019) 234(7):11662–9. doi: 10.1002/jcp.27825
91. Zhang D, Sun G, Zhang H, Tian J, Li Y. Long non-coding RNA ANRIL indicates a poor prognosis of cervical cancer and promotes carcinogenesis via PI3K/Akt pathways. Biomed Pharmacother = Biomed Pharmacother (2017) 85:511–6. doi: 10.1016/j.biopha.2016.11.058
92. Zhang WY, Liu YJ, He Y, Chen P. Down-regulation of long non-coding RNA ANRIL inhibits the proliferation, migration and invasion of cervical cancer cells. Cancer Biomarkers Section A Dis Markers (2018) 23(2):243–53. doi: 10.3233/cbm-181467
93. Gutschner T, Hammerle M, Eissmann M, Hsu J, Kim Y, Hung G, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res (2013) 73(3):1180–9. doi: 10.1158/0008-5472.can-12-2850
94. Sun Y, Ma L. New Insights into Long Non-Coding RNA MALAT1 in Cancer and Metastasis. Cancers (2019) 11(2):216. doi: 10.3390/cancers11020216
95. Li Z, Shen J, Chan MT, Wu WK. TUG1: a pivotal oncogenic long non-coding RNA of human cancers. Cell Proliferation (2016) 49(4):471–5. doi: 10.1111/cpr.12269
96. Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell (2011) 147(2):344–57. doi: 10.1016/j.cell.2011.09.029
97. Ulitsky I. Interactions between short and long noncoding RNAs. FEBS Lett (2018), 2874–83, 592(17):2874–83. doi: 10.1002/1873-3468.13085
98. Yoon JH, Abdelmohsen K, Gorospe M. Functional interactions among microRNAs and long noncoding RNAs. Semin Cell Dev Biol (2014) 34:9–14. doi: 10.1016/j.semcdb.2014.05.015
99. Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet (2016) 17(5):272–83. doi: 10.1038/nrg.2016.20
100. Chan JJ, Tay Y. Noncoding RNA:RNA Regulatory Networks in Cancer. Int J Mol Sci (2018) 19(5):1310. doi: 10.3390/ijms19051310
101. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature (2010) 464(7291):1071–6. doi: 10.1038/nature08975
102. Brockdorff N, Bowness JS, Wei G. Progress toward understanding chromosome silencing by Xist RNA. Genes Dev (2020) 34(11-12):733–44. doi: 10.1101/gad.337196.120
103. Xing F, Liu Y, Wu SY, Wu K, Sharma S, Mo YY, et al. Loss of XIST in Breast Cancer Activates MSN-c-Met and Reprograms Microglia via Exosomal miRNA to Promote Brain Metastasis. Cancer Res (2018) 78(15):4316–30. doi: 10.1158/0008-5472.Can-18-1102
104. Cui M, You L, Ren X, Zhao W, Liao Q, Zhao Y. Long non-coding RNA PVT1 and cancer. Biochem Biophys Res Commun (2016) 471(1):10–4. doi: 10.1016/j.bbrc.2015.12.101
105. Iden M, Fye S, Li K, Chowdhury T, Ramchandran R, Rader JS. The lncRNA PVT1 Contributes to the Cervical Cancer Phenotype and Associates with Poor Patient Prognosis. PloS One (2016) 11(5):e0156274. doi: 10.1371/journal.pone.0156274
106. Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell (2016) 29(4):452–63. doi: 10.1016/j.ccell.2016.03.010
107. Jia L, Zhang Y, Tian F, Chu Z, Xin H. Long noncoding RNA colon cancer associated transcript1 promotes the proliferation, migration and invasion of cervical cancer. Mol Med Rep (2017) 16(4):5587–91. doi: 10.3892/mmr.2017.7302
108. Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell (2014) 25(5):666–81. doi: 10.1016/j.ccr.2014.03.010
109. Yue B, Qiu S, Zhao S, Liu C, Zhang D, Yu F, et al. LncRNA-ATB mediated E-cadherin repression promotes the progression of colon cancer and predicts poor prognosis. J Gastroenterol Hepatol (2016) 31(3):595–603. doi: 10.1111/jgh.13206
110. Lu G, Zhang Y. Long non-coding RNA ATB promotes human non-small cell lung cancer proliferation and metastasis by suppressing miR-141-3p. PloS One (2020) 15(2):e0229118. doi: 10.1371/journal.pone.0229118
111. Cao W, Peng T, Zhou Y. Long noncoding RNA activated by transforming growth factor-beta promotes cancer development and is a prognostic marker in cervical cancer. J Cancer Res Ther (2017) 13(5):801–6. doi: 10.4103/jcrt.JCRT_256_17
112. Sun Y, Wei G, Luo H, Wu W, Skogerbø G, Luo J, et al. The long noncoding RNA SNHG1 promotes tumor growth through regulating transcription of both local and distal genes. Oncogene (2017) 36(49):6774–83. doi: 10.1038/onc.2017.286
113. Liu Y, Yang Y, Li L, Liu Y, Geng P, Li G, et al. LncRNA SNHG1 enhances cell proliferation, migration, and invasion in cervical cancer. Biochem Cell Biol = Biochim Biol Cell (2018) 96(1):38–43. doi: 10.1139/bcb-2017-0188
114. Adriaens C, Standaert L, Barra J, Latil M, Verfaillie A, Kalev P, et al. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat Med (2016) 22(8):861–8. doi: 10.1038/nm.4135
115. Wang HL, Hou SY, Li HB, Qiu JP, Bo L, Mao CP. Biological Function and Mechanism of Long Noncoding RNAs Nuclear-Enriched Abundant Transcript 1 in Development of Cervical Cancer. Chin Med J (2018) 131(17):2063–70. doi: 10.4103/0366-6999.239308
116. Chen Y, Wang CX, Sun XX, Wang C, Liu TF, Wang DJ. Long non-coding RNA CCHE1 overexpression predicts a poor prognosis for cervical cancer. Eur Rev Med Pharmacol Sci (2017) 21(3):479–83.
117. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet (2019) 20(11):675–91. doi: 10.1038/s41576-019-0158-7
118. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature (2013) 495(7441):333–8. doi: 10.1038/nature11928
119. Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular intronic long noncoding RNAs. Mol Cell (2013) 51(6):792–806. doi: 10.1016/j.molcel.2013.08.017
120. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol (2014) 32(5):453–61. doi: 10.1038/nbt.2890
121. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA (New York NY) (2013) 19(2):141–57. doi: 10.1261/rna.035667.112
122. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol (2015) 22(3):256–64. doi: 10.1038/nsmb.2959
123. Bach DH, Lee SK, Sood AK. Circular RNAs in Cancer. Mol Ther Nucleic Acids (2019) 16:118–29. doi: 10.1016/j.omtn.2019.02.005
124. Jiao J, Zhang T, Jiao X, Huang T, Zhao L, Ma D, et al. hsa_circ_0000745 promotes cervical cancer by increasing cell proliferation, migration, and invasion. J Cell Physiol (2020) 235(2):1287–95. doi: 10.1002/jcp.29045
125. Ma HB, Yao YN, Yu JJ, Chen XX, Li HF. Extensive profiling of circular RNAs and the potential regulatory role of circRNA-000284 in cell proliferation and invasion of cervical cancer via sponging miR-506. Am J Trans Res (2018) 10(2):592–604.
126. Li X, Ma N, Zhang Y, Wei H, Zhang H, Pang X, et al. Circular RNA circNRIP1 promotes migration and invasion in cervical cancer by sponging miR-629-3p and regulating the PTP4A1/ERK1/2 pathway. Cell Death Dis (2020) 11(5):399. doi: 10.1038/s41419-020-2607-9
127. Huang XB, Song KJ, Chen GB, Liu R, Jiang ZF, He YL. Circular RNA hsa_circ_0003204 promotes cervical cancer cell proliferation, migration, and invasion by regulating MAPK pathway. Cancer Biol Ther (2020) 21(10):972–82. doi: 10.1080/15384047.2020.1824513
128. Meng L, Jia X, Yu W, Wang C, Chen J, Liu F. Circular RNA UBAP2 contributes to tumor growth and metastasis of cervical cancer via modulating miR-361-3p/SOX4 axis. Cancer Cell Int (2020) 20:357. doi: 10.1186/s12935-020-01436-z
129. Wahlestedt C. Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat Rev Drug Discovery (2013) 12(6):433–46. doi: 10.1038/nrd4018
130. Crooke ST, Witztum JL, Bennett CF, Baker BF. RNA-Targeted Therapeutics. Cell Metab (2018) 27(4):714–39. doi: 10.1016/j.cmet.2018.03.004
131. Adams BD, Parsons C, Walker L, Zhang WC, Slack FJ. Targeting noncoding RNAs in disease. J Clin Invest (2017) 127(3):761–71. doi: 10.1172/jci84424
132. Bennett CF. Therapeutic Antisense Oligonucleotides Are Coming of Age. Annu Rev Med (2019) 70:307–21. doi: 10.1146/annurev-med-041217-010829
Keywords: non-coding RNAs, long non-coding RNAs (lncRNAs), circular RNAs (circRNAs), cervical cancer, metastasis
Citation: Cheng T and Huang S (2021) Roles of Non-Coding RNAs in Cervical Cancer Metastasis. Front. Oncol. 11:646192. doi: 10.3389/fonc.2021.646192
Received: 25 December 2020; Accepted: 03 February 2021;
Published: 11 March 2021.
Edited by:
Alberto Farolfi, Romagnolo Scientific Institute for the Study and Treatment of Tumors (IRCCS), ItalyCopyright © 2021 Cheng and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shouguo Huang, aHNnMTExQGNzdS5lZHUuY24=
 Tanchun Cheng
Tanchun Cheng Shouguo Huang
Shouguo Huang