
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol. , 15 March 2021
Sec. Cancer Immunity and Immunotherapy
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.638357
Extracellular vesicles are small membrane particles derived from various cell types. EVs are broadly classified as ectosomes or small extracellular vesicles, depending on their biogenesis and cargoes. Numerous studies have shown that EVs regulate multiple physiological and pathophysiological processes. The roles of small extracellular vesicles in cancer growth and metastasis remain to be fully elucidated. As endogenous products, small extracellular vesicles are an ideal drug delivery platform for anticancer agents. However, several aspects of small extracellular vesicle biology remain unclear, hindering the clinical implementation of small extracellular vesicles as biomarkers or anticancer agents. In this review, we summarize the utility of cancer-related small extracellular vesicles as biomarkers to detect early-stage cancers and predict treatment outcomes. We also review findings from preclinical and clinical studies of small extracellular vesicle-based cancer therapies and summarize interventional clinical trials registered in the United States Food and Drug Administration and the Chinese Clinical Trials Registry. Finally, we discuss the main challenges limiting the clinical implementation of small extracellular vesicles and recommend possible approaches to address these challenges.
Extracellular vesicles (EVs) are small lipid bilayer-bound vesicles released from living cells into the extracellular environment. These vesicles lack functional nuclei and cannot replicate (1). Traditionally, EVs can be roughly classified into two main subtypes regarding their characteristics and biogenesis pathway: ectosomes and small extracellular vesicles (sEVs). Ectosomes have a diameter between 100 and 1000 nm and are generated by cytoplasmic membrane budding (2). sEVs, also referred to as “exosomes” by some researchers, are smaller in diameter (30–150 nm) and released by the fusion of the multivesicular bodies (MVBs) with the plasma membrane. According to the updated guidelines of the International Society for Extracellular Vesicles of 2018 (MISEV2018), when naming a new EV subtype, the term “EVs” is recommended, followed by a description of vesicle features, such as size, density, cell of origin, and experimental conditions, making EV names more descriptive and informative (2, 3). Some experts have a greater attachment to the traditional term “exosomes” and we consider it quite a personal preference to call them either “exosomes” or “small extracellular vesicles (sEVs)” (3). In congruence with the latest suggestion, we refer to “exosomes” as “sEVs” throughout this review.
In 2018, Zhang et al. discovered another group of non-membranous secretory extracellular nanoparticles (<50 nm) termed “exomeres”, which were initially identified by asymmetric flow field-flow fractionation (AF4) and consisted of various biological molecules (4). Later on, a simple ultracentrifugation-based method was employed to isolate exomeres from sEVs (5). Although the research on exomeres is limited by technical barriers, such as the rarity of AF4 and the lack of the universally recognized nomenclature system, researchers have reported some biological functions of exomeres. For example, the β-galactoside α2,6-sialyltransferase 1 (ST6Gal-I), an exomere protein from human colorectal cancer cells, could sialylate the β1-integrin of recipient cells. Another exomere protein amphiregulin (AREG) could serve as the EGFR ligand, regulate the EGFR signaling pathway in normal intestinal organoids, and significantly promote the growth of colonic cancer organoids (5). Although exomeres and sEVs have comparable protein profiles, the stability in the circulatory system regarding the absence of bilayered membrane, and the biological safety of exomeres in vivo are still unclear. However, similar issues related to sEVs have been well addressed with extensive investigation, leaving sEVs much more accessible therapeutic tools.
EVs play a critical role in intracellular hemostasis and intercellular communication. In 1946, researchers discovered that ultracentrifugation pellets of plasma could activate platelets and blood clotting factors; these effects were attributed to the EVs in the pellets (6, 7). Subsequently, reticulocyte-derived EVs were identified by electronic microscopy as “discarded waste” that maintains intracellular homeostasis during erythrocyte maturation (8, 9). As the knowledge of sEVs has grown exponentially since 2000, some research groups have unveiled more biological functions and established novel clinical applications of sEVs, especially sEVs as messengers in various pathological processes and an efficient and targeted drug delivery system (10–13). In this review article, we focus on the recent development of sEVs as biomarkers for early cancer detection and follow-up care, as well as therapeutic particles for cancer treatment.
sEV biogenesis is complex and involves the formation of specialized intracellular compartments, known as MVBs/late endosomes, a key sorting point in the endocytic pathway (14). The inward budding of MVB membrane produces numerous intraluminal vesicles (ILVs), a group of small (~25–30 nm in diameter) spheroids containing cytosolic components and certain proteins from the invaginated MVB membrane (15). ILVs are expelled into the extracellular environment when MVBs fuse with the plasma membrane, referred to as sEVs. sEVs are spheroids in solution but sometimes they display a sauce-like shape under transmission electron microscopy as an artifact of drying during preparation (16). Typically, sEVs collected by fraction collection have a density range from 1.1 to 1.2 g/ml (17). Alternatively, MVBs with ILVs can fuse with lysosomes for hydrolysis of endocytosed macromolecules (18). However, some ILVs can fuse with MVB membrane and the proteins of ILVs membrane can be incorporated into endosome or lysosome membrane, a process called “backfusion”. As a result, some ILV membrane proteins, such as tetraspanin proteins and mannose-6-phosphate receptors, can escape the lysosomal degradation and recycle to the trans-Golgi network or plasma membrane (15, 19) (Figure 1).
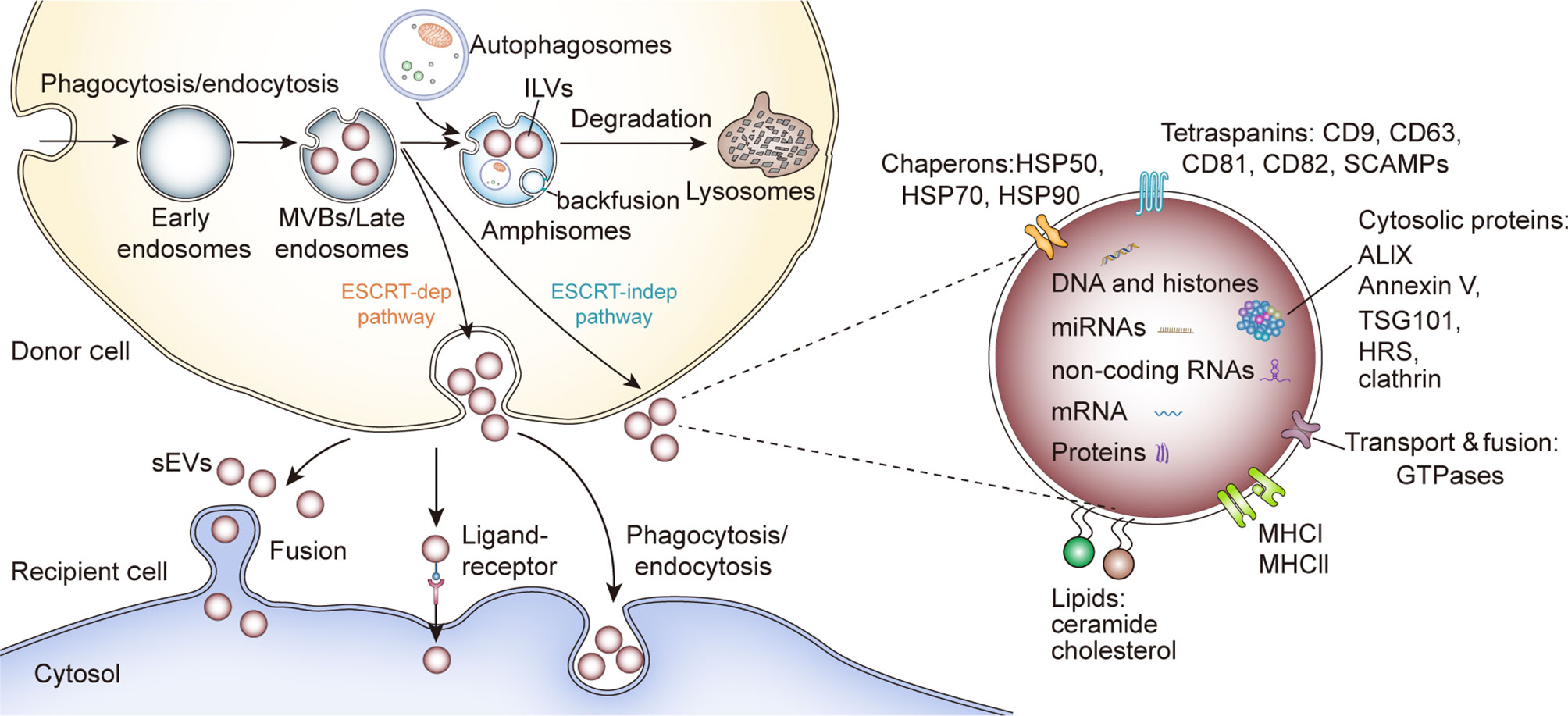
Figure 1 sEV biogenesis, cargo contents and uptake. HSP, Heat shock proteins; SCAMPs, Secretory carrier membrane proteins; ALIX, Apoptosis-linked gene 2–interacting protein X; HRS, Hepatocyte growth factor-regulated tyrosine kinase substrate; TSG101, Tumor susceptibility gene 101.
Although the formation of ILVs and the protein cargo sorting system have not been fully elucidated, researchers showed that the endosomal sorting complex required for transport (ESCRT) functions in this process (20). As complicated protein machinery, ESCRT consists of four main complexes (ESCRT-0, -I, -II, and -III) and works cooperatively with associated proteins (VPS4, VTA1, ALIX) to promote MVB and ILV biogenesis. The ESCRT-0 complex recognizes and sequesters ubiquitinated domains of endosomal membrane proteins via its ubiquitin-binding subunits, such as hepatocyte growth factor-regulated tyrosine kinase substrate protein (HRS); whereas ESCRT-I and-II facilitate the formation of the buds with sequestered proteins. Then the whole complex interacts with ESCRT-III/Vps4 complex, which stabilizes tubular endosomes and cleaves the bud to form ILVs (20, 21). Finally, the disassembly and recycling of ESCRT machinery requires the energy supplied by AAA-ATPase Vps4 (20). Besides, several ESCRT components (TSG101, ALIX) and ubiquitinated proteins have been identified in purified sEVs from various cell types. Typically, the sEV marker protein ALIX was reported to collaborate with other ESCRT proteins (TSG101 and CHMP4) and facilitate cargo selection, endosomal membrane budding and vesicle detachment in the form of syntenin-syndecan-ALIX complex (22). These results support the theory that ESCRT is critical in sEV biogenesis. However, some research indicated ESCRT-independent mechanisms for ILVs formation and release (Figure 1). Trajkovic et al. reported that the depletion of sphingomyelinases, instead of ESCRT inhibition, in oligodendroglial cells could significantly reduce the formation of MVBs and associated proteolipid protein (PLP)-enriched sEVs, an inspirational phenomenon referred to as “ceramide-dependent sEV biogenesis” (23). Further investigation showed that some lipid raft-based microdomains of the endosomal membrane contain numerous sphingomyelinases, enzymes hydrolyzing sphingomyelins to ceramides (24). Ceramides could induce negative curvatures of the endosomal membrane and lateral phase separation, thus functioning as coalescers in the process of ILV abscission (25, 26). Nonetheless, whether this ceramide-dependent machinery could be generalizable to other cell types and intraluminal contents is still unclear. Interestingly, some tetraspanins, such as CD63, CD82 and Tspan8, have been regarded as key points in cargo sequestration and sEVs formation, in a ceramide-independent and ESCRT-independent manner. Niel et al. reported that a CD63-dependent mechanism facilitated the formation and release of melanosomes-enriched sEVs, while downregulating CD63 led these melanosomes into ESCRT-dependent degradation in human melanoma cells (27). Similarly, the elevated expression of CD9 and CD82 could increase the secretion of β-catenin-enriched sEVs from human embryonic kidney 293 cells (HEK293), and a study utilizing the rat adenocarcinoma model showed that Tspan8 participated in the selective recruitment of proteins into sEVs (28, 29). Additionally, thousands of molecules have been reported to contribute to selective cargo sorting into ILVs and the release of sEVs, such as Rab GTPase (30), glycosphingolipids and flotillins (31), chaperone HSC70 (32, 33) and small integral membrane proteins of the lysosome/late endosome (SIMPLE) (34).
Similar to the specific recruitment of protein contents, the sorting of particular nucleic acids is dedicatedly monitored by multiple mechanisms. Generally, smaller size, higher abundance, and greater accessibility to membranous intracellular organelles and cytoplasmic locations promote the accumulation of RNAs in sEVs. Since most RNAs are relocated to specific cellular compartments in association with RNA-binding proteins (RBPs) and 25% of sEV proteins have been identified as RBPs, it is rational to assume that RBPs are important in transferring RNAs into sEVs (35, 36). For example, Beltri et al. reported that a four nucleotide motif (GGAG) could bind to sumoylated heterogeneous ribonucleoprotein A2B1 (hnRNPA2B1), thus delivering miRNAs into sEVs derived from human T cells (37). HnRNPA2B1 could also deliver unspliced HIV genomic RNA into sEVs via the interaction with several sequences (A2RE and nucleotides 557-663) (38, 39). Besides, miRNAs with 3′-end uridylation appeared to be directed into sEVs but 3′-end adenylated isoforms were more likely to be confined within the cells, indicating that post-transcriptional modification plays a role in the RNA sorting system (40, 41). Subsequently, RNA secondary configuration and unique motifs (42, 43), the interaction with membrane lipids (43) and innumerable RBPs, such as AG02 (44), major vault protein (MVP) (45), YBX1 (46, 47) and lupus La protein (48), have also been implicated in RNA packaging into sEVs. In summary, there are multiple mechanisms of cargo sorting and ILVs formation and it is still obscure whether the sequestering and sorting of specific molecules requires different pathways or how these pathways regulate various ILV subpopulations even within one MVB.
The components of sEVs indicate their cellular origin and potential biological functions. As oncologists, we have a particular interest in identifying sEV-related oncogenic factors (49–52). Significant efforts have been made to characterize the oncogenic alterations in sEVs and inhibit tumor-specific sEV pathways as a new class of anticancer therapies. Here, we briefly discuss how the malignant transformation of parent cells affects sEV components to promote cancer progression and described sEV as cancer biomarkers. For instance, upregulation of oncogenes can increase the levels of sEV oncoproteins, such as EGFR variant III and HRS (53, 54). Additionally, researchers have reported specific collections of sEV proteins associated with different donor cells. For example, Hurwitz et al. reported that nine different cancer types had only 213 sEV proteins in common through proteomic analysis of sEVs from 60 cancer cell lines provided by the National Cancer Institute (NCI-60). Most of the 6,071 proteins identified were unique for each cancer type, suggesting that sEV protein enrichment analysis could provide insight into the cells of origin (55). In line with these findings, p53 loss and oncogenic aberrations in EGFR in human bronchial epithelial cells (HBECs) leads to sEV protein patterns that are distinct from those in non-malignant HBECs (52).
Nucleic acids and lipid components can also be altered by oncogenic mutations (56, 57). In patients with non-small-cell lung cancer (NSCLC), the levels of let-7f and miR-30e-3p in circulating sEVs are positively correlated with disease severity (58). Llorente et al. showed that, compared with parental cells, sEVs from prostate cancer cells had significant enrichment of distinct lipids and a higher lipid-to-protein ratio, indicating that the lipid profile of sEVs could provide further information about sEV biogenesis (59).
Numerous randomized clinical trials have revealed different clinical outcomes in patients with the same cancer after treatment with different therapeutic regimens, raising the possibility that different treatments or stress factors in tumor cells can alter sEV cargo patterns and downstream signaling pathways. Harmati et al. obtained B16F1 cell-derived sEVs released under heat, oxidative, or cytostatic stress and observed unique miRNA and protein patterns in each sEV group (60). Besides, each sEV group demonstrated distinct oncogenic functions; for instance, doxorubicin-elicited sEVs enhanced melanoma cell migration, and oxidative stress-elicited sEVs promoted Ki-67 upregulation in mesenchymal stem cells (60). These results suggest that we may be able to predict therapeutic outcomes and design more effective personalized cancer treatment plans based on the analysis of sEVs from the patients’ tumor tissue, organoid models, or xenograft models. Using these models, we could precisely mimic the complex tumor microenvironments and obtain reliable information about the disease.
Recently, the importance of sEVs in intercellular communication has attracted increasing attention, especially in the context of tumorigenesis and metastasis. The bilayer lipid membranes of sEVs protect their cargo, enabling the transfer of signaling molecules from sEVs to the nearby cells or the distant sites via circulatory and lymphatic systems (61–64). It has become evident that sEVs from cancer cells can promote tumor progression and metastasis through multiple pathways (65–68). For instance, breast cancer-derived sEVs promoted oncogenic transformation in non-malignant epithelial cells (65, 69, 70), neoangiogenesis (71–73), cancer cell invasion, metastasis (74), chemoresistance (75, 76), and immune suppression (77, 78). Recently, Gao et al. reported that cancer-associated fibroblasts (CAFs) promoted tamoxifen resistance in breast cancer by sending miR-22 to silence ERα and PTEN expression in recipient cancer cells, suggesting the engagement of tumor stroma-derived sEVs in oncogenesis (79).
Additionally, sEVs may serve as “waste disposals” to eliminate unwanted cellular components (8, 9). Recent studies have identified the crosstalk between lysosomal degradation, autophagy and sEV biogenesis (80). As a traditional proton pump V-ATPase inhibitor, bafilomycin A1 could increase the pH of lysosomes and interfere with the trafficking between lysosomes and other intracellular compartments, especially MVBs and autophagosomes, thus enhancing sEV secretion in various cell types (81, 82). Some researchers reported that bafilomycin A1 could remarkably reduce the tumor load in hepatocellular carcinoma(HCC) xenograft mice and confirmed its potential as antitumor medicine (83). However, this conclusion is challenged by other experiments. Guo et al. discovered that the autophagy-related protein 5 (ATG5) could improve sEV secretion by dissociating V1V0-ATPase that acidifies MVBs and lysosomes, similar to the mechanism that bafilomycin A1 promoted sEV secretion; the increased sEV secretion could accelerate tumor migration in breast cancer mice models (84). These conflicting results may be due to the relatively small sample size of animal experiments, different cell types, and more importantly, the intricate relationship between sEV secretion and autophagy, warranting extensive investigation to illuminate the underlying mechanism.
Another interesting study showed that bafilomycin A1 could rescue the sEV secretion from human T cells, which was impaired by the IFN-1 induced TSG101(a component of ESCRT complex) ISGylation (85). In a ubiquitin-like fashion, interferon-stimulated gene 15 (ISG15) is covalently linked to target proteins, a process called ISGylation. Some researchers reported that downregulation of ISG15 increased the camptothecin resistance in breast cancer, while another study showed that high ISG15 expression in breast cancer indicated radiation resistance and poor prognosis (86, 87). Therefore, bafilomycin A1 might serve as a powerful tool to help us understand the double-edged sword role ISGylation plays in oncogenesis with a mechanism involving membrane vesicle trafficking.
ExoCarta is a manually curated database of sEVs proteins, RNAs and lipids, providing imperative information for sEV research. As of January 21, 2021, the number of recorded protein (9,769) has far exceeded that of RNAs (3,408) and lipids (1,116), indicating extensive studies on sEV proteins (88). Therefore, it is safe to suppose that sEV proteins could function as biomarkers for sEVs-related biological events. Several proteins, such as CD9, CD63, and CD81, are enriched on most sEVs and recognized as markers to verify the presence of sEVs in heterogeneous populations of EVs (89). Further research showed that some unique sEV proteins were associated with the existence of tumor cells, thus facilitating the early cancer detection and prognosis prediction (Table 1).
The application of sEVs as liquid biopsy samples has gained substantial attention (Figure 2). Although blood is the primary sample used for liquid biopsy, saliva is a promising alternative approach to detect early-stage oral cancer since obtaining saliva is less invasive and closer to malignant lesions than phlebotomy. For instance, increased expression of alpha-2-macroglobulin, haptoglobin, and mucin-5B on oral cancer-derived sEVs could serve as biomarkers for oral cancer diagnosis (103). Saliva biopsies are also becoming increasingly common for head and neck cancer diagnosis, in addition to the detection of other solid tumors, such as breast cancer (104) and pancreatic cancer (105, 106). Urinary sEV protein analysis also identified several markers associated with pancreatic cancer and has emerged as another noninvasive method to identify biomarkers for cancer diagnosis (107).
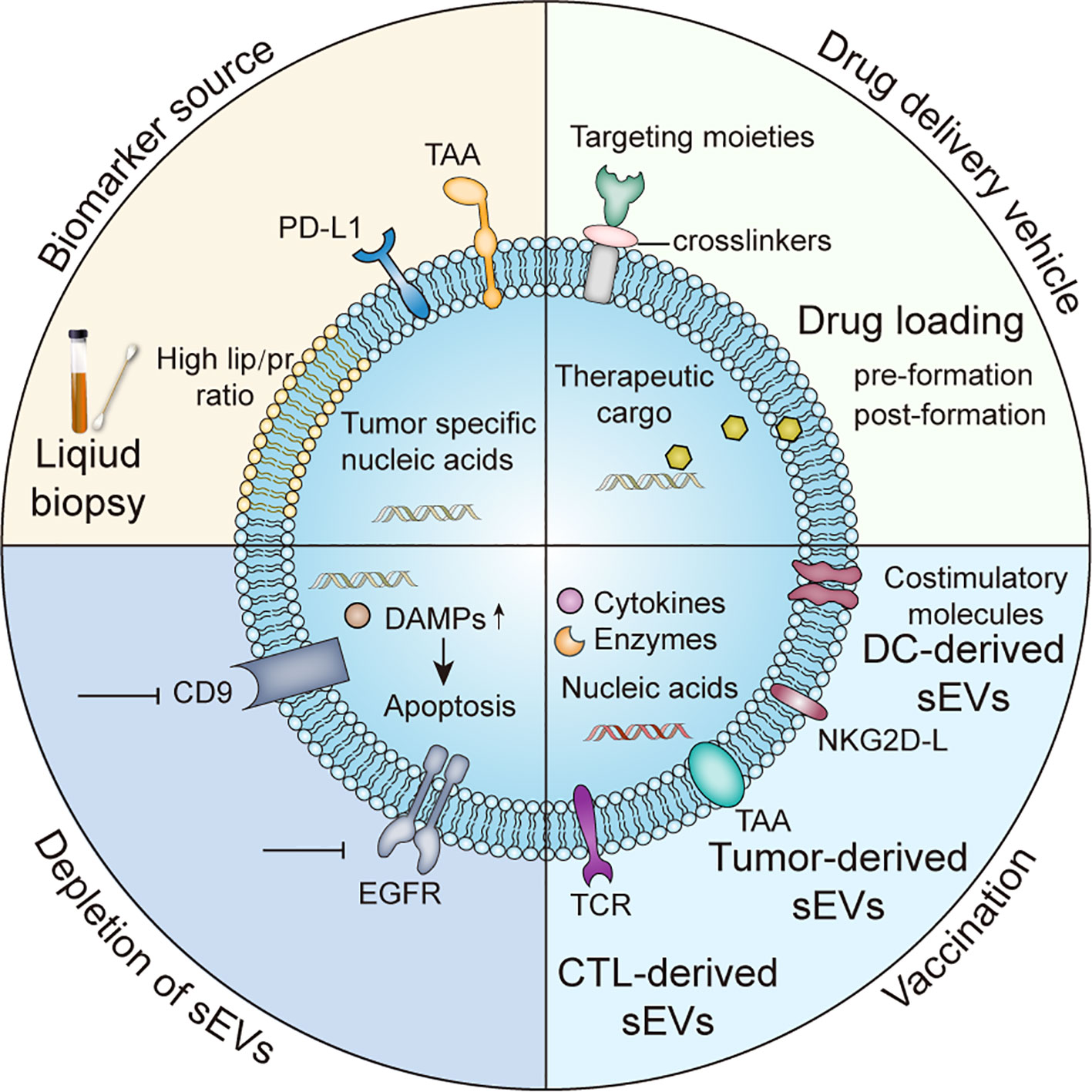
Figure 2 Clinical applications of sEVs TAA, tumor-associated antigens; DAMPs, damage-associated molecular patterns; NKG2D, Natural Killer Group 2D receptor ligands.
Some surface proteins can guide the cellular tropism of sEVs, leading to organotypic metastasis. For instance, breast cancer-derived sEVs coated with integrin α6β4 and α6β1 were more likely to be uptaken by pulmonary fibroblasts and epithelium, establishing a premetastatic niche in the lungs (74). Several surface proteins from melanoma-derived sEVs, such as very late antigen 4 (VLA-4) and tyrosinase-related protein-2 (TYRP2), were also shown to recruit bone marrow progenitor cells to premetastatic niches via the MET signaling pathway, promoting bone metastasis and outgrowth (67, 108). Compared with without liver metastasis and healthy control subjects, stage 1 pancreatic ductal adenocarcinoma (PDAC) patients who later developed liver metastasis had remarkably higher levels of migration inhibitory factor (MIF) in their circulating sEVs (66). Depletion of MIF in PDAC-derived sEVs strongly inhibited the formation of fibrotic microenvironment created by Kuffer cells, thereby reducing premetastatic niche in preclinical mice models (66). These unique surface proteins could potentially be used to predict metastatic sites.
However, to which extend a single sEV protein marker can serve as a diagnostic and predictive biomarker in cancer is still under debate. Glypican 1 (GPC1) levels on pancreatic cancer-derived sEVs indicated cancer burden in patients before and after surgery, exhibiting significant diagnostic and predictive potentials (96, 109). In contrast, other studies have failed to confirm the diagnostic value of GPC1 alone in pancreatic cancer (110), highlighting the need for the combination of GPC1 with other biomarkers to increase diagnosis accuracy (111, 112). These studies emphasize that we could not sensitively detect early-stage cancerous lesions with a single protein biomarker, probably owing to the high heterogeneity of tumors.
Certain sEV proteins have been shown to modulate the tumor immune microenvironment through various mechanisms. Notably, the melanoma-derived sEV cargo proteins S100A8 and S100A9 downregulated the expression of the costimulatory marker CD83 in dendritic cells (DCs), thereby inhibiting DC maturation and cytokine production, as well as establishing a premetastatic niche (113). In melanoma mouse models, sEVs containing high PD-L1 levels have been shown to bind PD-1 receptors on T cells, exacerbating T cell exhaustion; pembrolizumab administration diminished this immunosuppression (77, 78). Therefore, further investigations of circulating PD-L1+ EVs are required to identify the mechanisms underlying anti-PD-L1 therapy failure and determine the ability of PD-L1+ sEVs to predict clinical outcomes of anti-PD-L1 treatment (Figure 2).
Intriguingly, blockade of PD-L1+ sEVs reinforced systemic antitumor responses in mouse models of prostate cancer (78), underpinning these cancer-promoting sEVs as a promising therapeutic target. Several studies have demonstrated that PD-L1 on sEVs acts as a decoy inhibiting immune responses in draining lymph nodes (78, 114, 115). Furthermore, increased levels of serum PD-L1+ sEVs predicted poor prognosis in patients with head and neck cancer (116). Although these data suggest serum PD-L1+ as a promising predictor of immunotherapy response and that targeting tumor-associated sEVs may augment the antitumor effects of immunotherapy, the underlying mechanisms remain unclear. In addition to immune checkpoints, tumor-infiltrating lymphocytes and innate immune cells are also crucial for the success of immunotherapy (117, 118). However, future studies are required to confirm the ability of circulating PD-L1+ sEVs to predict response to anti-PD-L1 therapy.
sEVs also play a critical role in innate immune responses. Pancreatic cancer-derived sEVs were found to express tumor-associated antigens (TAA)(Fig.2) and bind to circulating autoantibodies, thereby inhibiting complement-mediated cancer cell lysis (119). Furthermore, gastric cancer cell-derived sEVs were enriched in high mobility group box-1 (HMGB1) and activated tumor-promoting neutrophils via the TLR4/NF-κB signaling pathway (120). However, a recent study has shown that sEVs released by breast cancer cells with higher expressing of NFAT3 could inhibit cancer cell invasion; the underlying mechanisms are under extensive investigation (121). It is widely accepted that tumor-associated sEVs are key modulators of immune responses, exerting anti-inflammatory or pro-inflammatory effects. Therefore, sEVs can be used as robust biomarkers to predict cancer prognosis and response to immunotherapy (122).
In addition to protein sEV cargoes, nucleic acids contained in sEVs have also emerged as promising biomarkers. Nucleic acid components include DNA, coding mRNA, and non-coding RNA (ncRNA) molecules (13, 123). Double-stranded (ds) mitochondrial and genomic DNA molecules have been detected in serum sEVs from cancer patients (124, 125). Importantly, sEV dsDNA reflected driver mutations in patients with pancreatic cancer (50, 126, 127). In addition to shared genetic mutation patterns, sEV DNA and genomic DNA have similar nucleic acid modifications. Thakur et al. have identified shared methylation patterns between sEVs DNA and chromosomal DNA from parental cells, suggesting that sEV DNA methylation analysis might have strong clinical implications (126).
Ten HCC-related mRNAs in sEVs were identified by EV Click Chip analysis and these mRNAs could be used to accurately detect early-stage HCC (128). A scoring system based on these mRNAs was superior in distinguishing HCC patients from liver cirrhosis patients. Compared with traditional serum AFP tests, this system provided a higher area under the curve (AUC) of 0.69 versus 0.93, 0.68 versus 0.91, and 0.70 versus 0.92, according to BCLC stage 0-A, Milan criteria, and UNOS DS criteria respectively (128). These data provide strong evidence of sEV mRNA utilization for early-stage cancer detection.
ncRNAs, such as microRNAs (miRNAs), circular RNAs, and long ncRNAs (lncRNAs), are also abundant in sEVs (36, 129). The levels of certain miRNAs have been associated with cancer development and progression (130–132). Several studies have shown that, in HCC patients, high levels of miR-92a-3p in circulating HCC-derived sEVs were associated with HCC cell proliferation and poor overall survival (130, 133, 134). In multiple myeloma-derived sEVs, let-7b, and miR-18a levels accurately stratified patients according to survival outcomes (132). Although one or two specific miRNAs could strongly predict cancer prognosis, a recent study has shown that using a miRNA signature improves the sensitivity and specificity of sEVs as biomarkers (135–137). A miRNA cluster from breast cancer-derived circulating sEVs has been confirmed as a predictor for bone metastasis and is currently under investigation as a new therapeutic target (138).
ncRNAs from sEVs are also promising clinical biomarkers. In cholangiocarcinoma, several lncRNAs (139) and circular RNAs (140, 141) from circulating sEVs have been correlated with metastasis and have been proposed as cancer biomarkers (142). An ongoing clinical trial is investigating the ability of a group of serum lncRNAs to detect early-stage lung tumors to improve clinical outcomes (143).
An obstacle in the discovery of liquid cancer biomarkers is the low yield of sEV isolation methods, which are not yet suitable for clinical practice. Genetic engineering of donor cells and manipulating the cell culture medium have been employed to scale up the isolation of sEVs (144). Furthermore, many techniques have been established to purify sEVs based on their unique physicochemical and biochemical properties (145, 146). Particularly, sEV isolation by size-exclusion separation chromatography can significantly reduce the sEV damage and protect sEV nucleic acids and other cargoes (145).
Compared with sEV proteins and nucleic acids, sEV lipids have been studied less as cancer biomarkers, possibly due to the limited diversity in sEV lipids and the limitations of lipid analysis techniques. In 2003, distinct lipid patterns were identified in sEVs from human B cells. The accumulation of certain lipids and a relatively higher lipid-to-protein ratio were also validated in sEVs secreted by colorectal cancer cells and prostate cancer cells (Figure 2) (59, 147). Recent studies evaluated the lipidomic profiles of cancer-derived sEVs. Cheng et al. have demonstrated that sEVs from human ovarian cancer cells SKOV-3 had higher amounts of cholesterol esters and zymosterol species than sEVs derived from a different human ovarian epithelial cancer cell line (HOSEPiC), indicating that the lipid profile of sEVs could indicate the origin of sEVs (148). However, considering that these cholesterol esters assemble dynamically into lipid rafts and are rapidly exchanged between neighboring cells, future studies are warranted to determine the feasibility and sensitivity of using cholesterol esters as cancer biomarkers.
In the past decade, we have witnessed the exponential growth of sEVs modification strategies. These strategies could be classified into two types according to the source of parental cells: altering sEVs from unmodified parental cells and harvesting sEVs from well-modified parental cells (Figure 3).
Numerous efforts have been made to increase the targeting ability of modified sEVs. Aminoethylanisamide-polyethylene glycol (AA-PEG) vector moiety targeting the sigma receptor on cancer cells was integrated into sEVs from macrophages via crosslinkers (149) (Figure 2). Compared with unmodified parental sEVs, modified sEVs accumulated in cancer cells and prolonged overall survival in mice (149). Tian et al. established an efficient and rapid method to add integrin αvβ3 targeting peptides [c(RGDyK)] onto the sEVs via crosslinkers, considering the high expression of integrin αvβ3 on the surface of reactive vascular endothelial cells (150) (Figure 3). Although this technology was initially employed in cerebral ischemia models, it can be adapted for anti-angiogenesis therapy in cancer models, given the role of integrin αvβ3 in promoting tumor angiogenesis (151). Modified nucleic acids could also be used for targeted delivery. Pi et al. developed an innovative method to anchor arrow-shaped RNA nanoparticles, known as aptamers, on the surface of sEVs, which carried therapeutic siRNA and miRNA (152). These aptamers could be stably attached to the external surface of the membrane due to their unique configuration and the cholesterols added to their tails (152) (Figure 3). The aptamers specifically recognized surface cancer markers, such as EGFR in breast cancer and PSMA in prostate cancer, thus guiding the sEVs to cancer cells (152).
Compared with EVs from genetically engineered donor cells, surface-modified purified sEVs provides more options in terms of conjugated ligands. However, how to protect sEV structure and cargoes from acidic environments and high-speed centrifugation in the modification process are critical challenges that need to be addressed.
Compared with liposomes, carbon nanoparticles, and other artificial drug delivery systems, autologous sEVs are less toxic and immunogenic (153, 154). sEVs can also target tumor cells and immune cells with a higher specificity than liposomes and are ideal drug delivery vehicles (155, 156). Numerous efforts have been made to engineer isolated sEVs and donor cells (Figure 3). Some commercial kits have been available for loading nucleic acids into sEVs (64). The techniques used to load sEVs with various cargo include free-thaw cycles to fuse sEVs and liposomes, sonication, extrusion, permeabilization with saponin, and electroporation (157–159). Electroporation is commonly used to transfer small interfering RNAs (siRNAs) and other cytotoxic agents into sEVs (160–164). Particularly, mesenchymal stromal cell-derived sEVs loaded with siRNA or other oligonucleotides specific for oncogenic KrasG12D strongly inhibited KrasG12D expression in pancreatic ductal cancer cells and inhibited orthotopic tumor growth (162). Importantly, sEVs loaded with KrasG12D-targeting siRNA significantly suppressed pancreatic cancer growth in mice (162). The strong antitumor effects of these modified sEVs were attributed to their high CD47 levels, which protected sEVs from macrophage-mediated elimination (162). Recently, Faruqu et al. used electroporation to transfer siRNAs into sEVs from human embryonic kidney cells (HEK-293 cells); these modified sEVs were internalized by pancreatic cancer cells with high efficiency, providing a standard procedure for making siRNA-containing sEVs via electroporation (164) (Figure 3). Although sEV loading by electroporation may cause less damage than the genetic modification of donor cells, RNA precipitation and altered biochemical and physiochemical features of the membrane during electroporation are critical challenges that need to be addressed (165).
Although the ability of sEVs to target different cell types has been well demonstrated, their targeting ability as a drug delivery system can be further enhanced by introducing targeting molecules into the donor cells, which release the targeting ligand-enriched sEVs. The neuron-targeting peptide RVG has been successfully conjugated with Lamp2b, a transmembrane protein abundantly found on the surface of sEVs (161, 166). This reconstructed protein enabled the delivery of therapeutic siRNA-packaged sEVs to neurons, maximizing their safety and efficacy (161). Although this method was primarily tested in mouse models of neurodegenerative diseases, it provided an optimized method to improve the targeting ability of sEVs. Recently, HEK293T cell-derived sEVs with Lamp2b-anti-HER2 affibody fusion protein were shown to selectively bind HER2-expressing colon cancer cells, delivering 5-fluorouracil and miRNA inhibitors to tumor cells and suppressing tumor growth (Figure 3) (167).
Significant progress has also been made in combining various nanoparticles and sEVs via chemical modification of donor cell membranes, providing a novel method to generate therapeutic sEVs. For instance, nanobodies attached to the sEV surface via a glycosylphosphatidylinositol (GPI) linker can recognize different therapeutic targets, such as EGFR expressed on tumor cells (Figure 3) (168, 169).
Modification of donor cells is another strategy to load anticancer agents into sEVs and improve the targeting ability of sEVs. An early study showed that paclitaxel-treated donor cells secreted paclitaxel-containing sEVs, which significantly inhibited the proliferation of pancreatic cancer cells in vitro (170). Guo et al. designed a novel system to generate methotrexate (MTX)-loaded EVs from UVB-irritated donor cells treated with MTX previously (Figure 3) (171). These EVs (30–930 nm in diameter) remarkably increased the overall survival of mice with malignant pleural effusion and decreased pleural effusion in a small group of patients with late-stage lung cancer (171). These preliminary findings open a new avenue for the production of therapeutic EVs by manipulating donor cells. However, the clinical implementation of this method is hindered by several factors. Importantly, a balance between achieving an ideal loading efficacy and minimizing cytotoxicity caused by co-incubation with chemotherapy drugs is required. The stability of the pharmaceutics in the cytoplasm is another key concern, given that some commonly used drugs (e.g., cisplatin and paclitaxel) can be inactivated by intracellular enzymes (170, 172, 173). Indeed, isolated sEVs, rather than intact donor cells, have been regarded as more suitable carriers to transport chemotherapeutics because of the limitations of current techniques and platforms.
Transfecting donor cells with vectors carrying genes of interest is a possible method to generate sEVs enriched in specific proteins. Previous studies have reported the successful generation of sEVs carrying 5-fluorocytosine-activating enzymes by transfecting 293T cells; co-administration of these sEVs with 5- fluorocytosine significantly inhibited the proliferation of glioma cells (Figure 3) (174, 175). The feasibility of this method has been validated by a study showing that caspase-1-containing sEVs released from transfected 293T cells inhibited the growth of schwannoma cells without causing significant neurotoxicity (176). However, this sEV generation approach has been used less than the production of siRNA/miRNA-enriched sEVs. One of the challenges in producing soluble protein-enriched sEVs is the cytotoxicity to parental cells caused by the overexpression of these proteins, which significantly reduces sEVs production yield and alters sEV composition (174). Another obstacle is the complexity of the protein sorting and trafficking system involved in sEV biogenesis. This system randomly delivers target proteins into sEVs, hindering the reproducibility of the method (174, 177). Therefore, other approaches have been established to specifically transport target proteins into sEVs, including conjugation of the target protein with sEVs-associated proteins and ubiquitination of target proteins.
Enhancing the expression of target proteins and exogenous leader peptide complexes in donor cells has emerged as a promising method to generate sEVs. Viral envelope proteins are an ideal source of leader peptides. The mechanisms underlying viral recognition of target cells and viral spread via cellular vesicles have been extensively studied for decades, and several links between viral proteins and sEV biogenesis have been identified (178, 179). Viral proteins can be delivered to sEVs by using endosomal sorting complexes required for transport (ESCRT). CD8 ectodomain-Env chimeric protein was loaded to K562 cell-derived sEVs by the interaction of the Env protein of the bovine leukemia virus with ESCRT (180). A mutated HIV-Nef protein, also known as NefMuf, was transported to sEVs without causing significant damage, indicating its potential to lead target molecules to sEVs (181–183). Manfredi et al. designed a NefMuf/E7 DNA carrier to incorporate NefMuf-HPV E7 fusion protein into 293T cell-derived sEVs (184). The HPV E7 protein conjugated to the C-terminal of NefMuf accumulated in the sEVs; these sEVs remained intact and activated anti-E7 CD8+ T cells, inhibiting TC-1 tumor cell proliferation in mice (Figure 3) (184). Vesicular stomatitis virus glycoprotein (VSVG) among other viral proteins, has also been used as leader peptides to target fusion proteins into sEVs (185). Therefore, viral leader peptides may provide a novel avenue to produce therapeutic sEVs.
Interestingly, some intracellular signal tags have been demonstrated to facilitate the targeted delivery of modified molecules to sEVs. Cheng et al. reported that recombinant proteins with C-terminal ubiquitination were ten times more likely to be loaded into HEK 293 cell-derived sEVs than unmodified isoforms (186); sEVs carrying ubiquitin (Ub)-tagged proteins boosted T cell responses (Figure 3) (186). Additionally, monoubiquitinated syntaxin 3 (Stx3) was rapidly internalized from the basolateral plasma membrane and was secreted apically into sEVs along with Stx3-binding partners (187). However, mutated forms of Stx3 that cannot be ubiquitinated could not be loaded into sEVs and reduced the shedding of human cytomegalovirus (HCMV) from infected cells, highlighting this to be a promising approach to produce customized sEVs (187). Besides, some plasma membrane anchors, such as N-terminal myristoylation and palmitoylation, selectively guided fusion proteins to sEVs (188). Sterzenbach et al. labeled Cre recombinase with WW tag, a signal peptide recognized by NDFIP1 involved in ESCRT system assembly. The modified Cre recombinase was ubiquitinated and loaded into sEVs (189). Interestingly, the recipient cells underwent Cre-mediated genomic editing after sEV internalization, opening a new avenue for reversing oncogenic mutations in tumor cells (Figure 3) (189).
However, one problem of using reconstructed proteins is the fragility of signal peptides during sEV biogenesis (166). Besides, whether these cargo modifications work independently of the ESCRT machinery is still under debate. Studies have shown that ubiquitinated MHCII β-chain levels were withdrawn from the cell surface and accumulated into secretory sEVs (190, 191). In contrast, Gauvreau et al. argued that MHCII molecules are loaded into sEVs via a ubiquitin-independent pathway (192). Additionally, it remains unclear whether these tags, especially ubiquitin, induce the degradation of fusion proteins by proteasomes and alter the therapeutic effects of sEVs.
Another possible method to generate therapeutic ligand-enriched sEVs is the conjugation of target proteins with constitutive sEV proteins, especially transmembrane proteins and peptides. These hybrid proteins have successfully been delivered to sEVs and the antitumor effects of these sEVs are under investigation (193) (Table 2). For example, CD9 is abundantly found on the sEV surface. Yim et al. developed an optically controlled delivery system by using the interaction between cargo-photoreceptor cryptochrome 2 (CRY2) soluble protein and CIBN-CD9 membrane complex. Under blue light, the interaction between CIBN and CRY2 subunits attaches the cargo protein to the intraluminal site of the sEV membrane, and the cargo can be released into the intraluminal space in the absence of blue light (Figure 3) (205). Moreover, sEVs from genetically engineered immune cells, a widely used immunotherapy, can deliver specific proteins to desired organs and cells. Compared with chimeric antigen receptor (CAR)-T cells, sEVs from CAR-T cells caused less severe cytokine release syndrome and comparable antitumor effects in mouse models (206). Moreover, CAR-T cell-derived sEVs with minimal PD-1 expression are less immunosuppressive than CAR-T cells, which express higher levels of PD-1 (206). Therefore, more research is needed to validate these promising preclinical findings and confirm the therapeutic effects of CAR sEVs in patients. Additionally, the ability of CAR sEVs to improve the safety and antitumor efficacy of CAR-T cell therapy merit further investigation.
Due to their natural characteristics, such as low homing in the liver and better penetration across the blood-brain barrier, sEVs have been utilized as reliable bio-shuttles for gene therapy, especially delivering brittle nucleic acid agents to target cells (207). Since miRNA and siRNA are the most favorable tools to regulate genomic expression, many efforts have been made to produce sEVs containing miRNAs as therapeutic agents by transfecting donor cells with DNA vectors (208). sEVs from mesenchymal stem cells carrying miR-122 expression plasmids delivered enormous amounts of miRNAs to liver cancer cells, sensitizing xenograft tumors to sorafenib (209). Abels et al. isolated miR-21-enriched sEVs from transfected glioma cells; these sEVs were internalized by microglia and suppressed the expression of specific mRNA targets (210). Although cell transfection is a simple and feasible approach to generate siRNA-enriched sEVs, cytoplasmic siRNA instability and miRNA-mediated cytotoxicity in donor cells are challenges that remain to be addressed.
Given that cancer cell-derived sEVs promote cancer progression and metastasis, the depletion of circulating oncogenic sEVs has been proposed as a method to suppress cancer progression (172, 211). In xenograft mouse models of human breast cancer, administration of anti-CD9 and anti-CD63 antibodies suppressed the spread of cancer cells to lymph nodes, lungs, and the thoracic cavity by targeting cancer-derived sEVs carrying CD9 and CD63 (212). Subsequent studies have demonstrated the preferential internalization of antibody-tagged sEVs by macrophages, suggesting that blocking specific surface protein biomarkers on sEVs could be a promising anticancer strategy (212).
Aptamer-modified nanoparticles were employed to transport circulating lung cancer-derived sEVs from the circulation to the intestinal tract; these agents inhibited sEV-induced metastasis in mice. Notably, EGFR-targeting aptamers were attached to positively charged mesoporous silica nanoparticles, and when they were injected intravenously, they recognized circulating EGFR+ sEVs. sEVs-nanoparticle complexes were uptaken by Kuffer cells and secreted into the bile; their elimination from the circulation suppressed cancer cell metastasis (211). Hence, selective depletion of circulating oncogenic sEVs can improve therapeutic outcomes in cancer patients and imply various potential clinical applications.
Chemotherapy and radiotherapy can cause the accumulation of DNA fragments in the cytoplasm of cancer cells and some DNA pieces are disposed of into sEVs (213–215). As sEVs could remove damage-associated molecular patterns (DAMPs) from source cells to maintain intracellular homeostasis, inhibiting sEVs secretion can increase DAMPs levels in the cytoplasm, triggering cancer cell apoptosis (214). Cannabidiol, ketotifen, and simvastatin can block sEV biogenesis and release from cancer cells and monocytes through various mechanisms, which amongst others are linked to mitochondrial dysfunction and calcium dysregulation, thereby reversing chemotherapy resistance and magnifying antitumor immune responses (216–219). Interestingly, these inhibitors target different steps of sEV biogenesis and secretion; thus, combination therapies could synergistically inhibit sEV secretion.
Numerous research has focused on developing cancer vaccines to augment antitumor immunity. In April 2001, the US Food and Drug Administration (FDA) approved Provenge® (sipuleucel-T) as the first cancer vaccine to treat metastatic, hormone-resistant prostate cancer (220, 221). Unlike traditional prophylactic vaccines against infectious diseases, this anticancer vaccine primes DCs with prostate cancer-associated antigens in vitro, and the activated DCs are then re-infused into patients (222). In a phase III trial, sipuleucel-T significantly prolonged the overall survival of 127 patients with metastatic prostate cancer survival without causing significant toxicity (223). However, not all clinical trials of sipuleucel-T showed encouraging results (224), raising the need for further investigation of the clinical efficacy of sipuleucel-T. The inconsistent clinical outcomes may be due to the variable sensitivity of DCs to immunosuppressive molecules, and the cost of priming and storing autologous DCs is very high. Therefore, we wonder whether cell-free vaccines, such as sEV-based vaccines, could facilitate antitumor immunity. Cancer cell-derived sEVs could carry numerous tumor-associated neoantigens, MHC class I molecules and HSP70, which are recognized as the stimulus for the immune response against cancer (225, 226). Rao et al. showed that HCC-derived sEVs could carry several HCC-associated antigens and trigger a stronger tumor suppression than cell lysates in murine HCC models (227). Biopsy showed improved HCC tumor microenvironments by sEVs, with increased T lymphocyte infiltration, elevated IFN-γ and decreased interleukin-10 expression, indicating sEV cancer vaccines as competent immune modulators (227).
DAMPs involved pathways have also been reported in the application of sEV cancer vaccines. Due to the hypoxia, malnutrition and cytotoxic agents in tumor microenvironments, misfolded proteins accumulate in cancer cells and trigger endoplasmic reticulum (ER) stress (228). ER stress could promote MVB formation and the release of the DAMPs-rich sEVs, which could inhibit tumor progression via an inflammatory response (229, 230). DNA fragments resulted from chemotherapy are a well-known example of DAMPs. For instance, chromosomal DNA-rich sEVs from cancer cells treated with topotecan activated DCs via the cGAS-STING signaling pathway, thereby inducing cancer cell apoptosis and inhibiting tumor progression (213, 231, 232). Given that these sEVs may promote antitumor immunity, careful selection of DNA-containing sEVs subpopulations is critical.
Furthermore, sEVs from irradiated cancer cells could inhibit the growth of nearby nonirradiated cells, a phenomenon known as the radiation-induced bystander effect (RIBE) (233). Numerous sEV contents have been identified to mediate the RIBE, including cytokines, antigens, free radicals, and immune modulators (234, 235). For instance, high levels of miR-1246 have been detected in sEVs from irradiated BEP2D cells, and miR-1246 has been shown to inhibit non-homologous end joining (NHEJ) and the proliferation of unirradiated recipient cells (236). Besides, several studies have shown that adjusting wavelengths and doses of electromagnetic radiation could remarkably alter the number of sEVs produced by mesenchymal tissues and tumor cells via several mechanisms, such as the activation of Wnt signaling and enhanced p53 expression (228, 235, 237, 238). These results indicate that not only the contents but the number of sEVs might account for RIBE, which varies with the adaptation of radiotherapy modalities. In a recent study, Wan et al. reported that injecting irradiated lung cancer cell-derived EVs with a mean diameter of approximately 400 nm into the pleural space activated macrophages and significantly prolonged overall survival in a pleural malignancy mouse model (239). Therefore, we wonder if the administration of larger EVs in addition to sEV vaccines could significantly augment the antitumor immunity. These inspiring results have deepened our understanding of RIBE, indicating advanced clinical strategies when managing patients with inoperable cancerous lesions.
Although tumor cell-derived sEVs could perform as potential cell-free vaccines, the culture and expansion of patients’ tumor cells in vitro is time-consuming in clinical practice. Malignant effusion, such as ascites from colorectal cancer (CRC) patients, provides an approachable source of adequate tumor cell-derived sEVs. Dai et al. conducted a phase I clinical trial of treating CRC patients with either ascites-derived sEVs alone or the sEVs plus GM-CSF. The combination regimen (sEVs plus GM-CSF) showed remarkable tumor inhibition and confirmed biosafety while administering sEVs alone did not achieve a significant therapeutic effect (240). This study offered a convenient and safe method to harvest autologous sEVs, but the researchers did not compare the clinical outcomes of using GM-CSF alone with the combinatory therapy, requiring more investigation to demonstrate how ascites-derived sEVs enhance antitumor immunity in conjugation with immune-modulatory cytokines (240).
Some immune cells-derived sEVs could also regulate antitumor immunity. DC-derived sEVs (DEVs) carry peptides/MHC complexes, costimulatory molecules, such as CD80 and CD86, and tumor-associated antigens to prime T cells directly in vitro (241). However, in vivo experiments showed that DEVs were less able to prime naïve T cells via direct sEVs-to-T cells interaction (240). Instead, DEVs could be internalized into or fuse with nearby APCs and tumor cells, followed by the antigen presentation on the surface of these recipient cells indirectly (242). Zitvogel et al. reported that cancer cells cocultured with DEVs could more efficiently reactivate previously primed T cells, showing the higher expression of interferon-γ (IFN-γ) (243) Besides, DEVs possess Natural Killer Group 2D receptor ligands (NKG2D-L), which bind to NKG2D on NK cell surface and activate NK cells; the tumor necrosis factor (TNF) in these sEVs could enhance the INF-γ release from NK cells (244). A completed phase II clinical trial conducted in advanced lung cancer patients confirmed the biosafety and activation of NK cells when using DEVs as the maintenance immunotherapy (245). Therefore, DEVs in combination with NK cells for cancer immunotherapy should be considerable.
Cytotoxic T cells (CTLs) are key regulators in antitumor immunity and many methods have been adopted to reactive CTLs in tumor microenvironments. One of the problems when applying these methods is that CTLs can hardly penetrate the dense stromal barriers, which shield cancer cells from the CTL-mediated cytotoxicity. As nano-sized particles with the better penetration across biological barriers, CTL-derived sEVs could efficiently approach the lesions, carry TCR and costimulatory molecules to bind to peptides/MHC complexes and contain cytotoxic enzymes to trigger targeted cell death (246). A recent study reported that with the stimulation of IL-12, high-affinity CTLs could secrete sEVs that activated low-affinity CTLs to produce more granzyme B and IFN-γ (247). However, Xie et al. reported that sEVs from exhausted CD+8 T cells could impair the activity of functional CD+8 T cells and a set of lncRNAs involved in the immunosuppression was identified (246). These paradoxical discoveries allow us to better understand the multiple functions of CTL-derived sEVs under different circumstances and intravital imaging could provide a powerful tool to directly observe the interaction between sEVs vaccines and recipient cells (248). Other source cells for producing therapeutic sEVs have been disclosed, such as NK cells (249), myeloid-derived suppressor cells (MDSCs) (250), tumor-associated macrophages (159), mast cells (251) and neutrophils (252), which shed light on the systemic study of disorganized antitumor immunity.
As of October 4, 2020, nine clinical trials testing sEVs as potential therapeutics have been registered on ClinicalTrials.gov and Chictr.org.cn (Table 3). No sEV-derived therapeutics have been approved by the US FDA or the Chinese National Medical Products Administration (NMPA). The evaluation of the safety and therapeutic efficacy of sEVs remains a significant clinical challenge caused by the lack of understanding of sEV biogenesis and functions. Besides, the pharmacokinetic characteristics of sEVs, such as the rapid clearance through the liver and kidneys, are factors limiting their therapeutic efficacy (201, 253). Notably, nanoparticle drug delivery systems have been approved by the FDA as anticancer agents because of their excellent performance in maximizing the efficacy and minimizing the side effects of chemotherapy (254). Therefore, advancing sEV therapeutics using similar techniques as those used for nanoparticles, such as fusing liposomes with sEV membrane (159) to increase the cellular uptake, improve the performance of sEVs as drug delivery vehicles. Recently, some biotechnology companies have announced plans to start sEV-related clinical trials as early as 2020, providing hope for the clinical application of sEVs (251).
In this review, we discuss the role of sEVs in cancer development and the recent advances in using sEVs to diagnose and treat various cancers. Despite significant progress in this field, several important issues remain to be solved. For instance, how to obtain homogenous sEVs subpopulations and distinguish between oncogenic sEVs and antitumor sEVs are significant obstacles in translating these scientific findings into clinical practice. Here, we propose several possible solutions. First, future studies are required to deepen our understanding of sEV biology, such as intraluminal components in different experimental conditions and various downstream events when sEVs are internalized by recipient cells. Second, advanced techniques for large-scale sEV production and purification are in demand. A combinatory approach consisted of immune-magnetic isolation, size-exclusion chromatography, and centrifugation could increase the homogeneity of final products (65, 144, 252, 255, 256). However, this approach could reduce the yield of sEVs due to its multiple steps. Therefore, a balance between obtaining homogenous sEV subpopulations and maximizing sEV yield is required. Establishing a standardized nomenclature system and elucidating the mechanisms underlying sEV biogenesis, release, and interaction with surrounding cells are also required.
Although there are multiple ongoing clinical trials evaluating sEVs as biomarkers for cancer detection and monitoring, only a few clinical trials are investigating the therapeutic effects of modified sEVs; thus, the long-term safety and clinical efficacy of sEVs as therapeutic targets or drug carriers remain unclear. In contrast to using sEVs as drug carriers, inhibiting sEVs release and depleting circulating sEVs are far less studied in preclinical models since sEVs clearance remains a challenge and the available sEV inhibitors cannot selectively inhibit specific sEV subtypes (257). Besides, the biodistribution and safety of sEVs absorption agents is still a major clinical concern (258). Hence, collective efforts are required to address the remaining technical challenges in the development of sEV-based biomarkers and therapies.
YG and YQ contributed to the article writing. CW, YS, JM, YH, and JH contributed to the data collection. HJ and KY supervised and revised the article. All authors contributed to the article and approved the submitted version.
This study was conducted with the support by the National Natural Science Foundation of China (Grant Nos. 2020BHB021, 81874233, 81874222, and 81672978) and the Key International Cooperation Project of Hubei Province (82022040).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
AF4, asymmetric flow field-flow fractionation; ATG5, autophagy-related protein 5; AREG, amphiregulin; CAR-T cells, chimeric antigen receptor T cells; ChiCTR, Chinese Clinical Trials Registry; CRY2, cryptochrome 2; DAMPs, damage-associated molecular patterns; DCs, dendritic cells; DEVs, DC-derived sEVs; EGFR, epidermal growth factor receptor; ESCRT, endosomal sorting complexes required for transport; EVs, extracellular vesicles; GPC1, glypican 1; GPI, glycosylphosphatidylinositol; HBECs, human bronchial epithelial cells; HCC, hepatocellular carcinoma; HCMV, human cytomegalovirus; HEK-293 cells, human embryonic kidney cells; HMGB1, high mobility group box-1; hnRNPA2B1, sumoylated heterogeneous ribonucleoprotein A2B1; HRS, hepatocyte growth factor-regulated tyrosine kinase substrate protein; ISG15, interferon-stimulated gene 15; MDSCs, myeloid-derived suppressor cells; MIF, migration inhibitory factor; MISEV2018, International Society for Extracellular Vesicles in 2018; MTX, methotrexate; MVB, multivesicular body; MVP, major vault protein; NFAT3, nuclear factor of activated T cells 3; NHEJ, non-homologous end-joining; NSCLC, non-small-cell lung cancer; OVA, ovalbumin; PDAC, pancreatic ductal adenocarcinoma; PLP, proteolipid protein; RIBE, radiation-induced bystander effect; sEVs, small extracellular vesicles; SIMPLE, small integral membrane protein of the lysosome/late endosome; Stx3, syntaxin 3; ST6Gal-I, β-galactoside α2,6-sialyltransferase 1; TYRP2, tyrosinase-related protein-2; Ub, ubiquitin; VLA-4, very late antigen 4; VSVG, vesicular stomatitis virus glycoprotein.
1. Woith E, Fuhrmann G, Melzig MF. Extracellular Vesicles-Connecting Kingdoms. Int J Mol Sci (2019) 20(22):5695. doi: 10.3390/ijms20225695
2. Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles (2018) 7(1):1535750. doi: 10.1080/20013078.2018.1535750
3. Witwer KW, Théry C. Extracellular vesicles or exosomes? On primacy, precision, and popularity influencing a choice of nomenclature. J Extracell Vesicles (2019) 8(1):1648167. doi: 10.1080/20013078.2019.1648167
4. Zhang H, Freitas D, Kim HS, Fabijanic K, Li Z, Chen H, et al. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat Cell Biol (2018) 20(3):332–43. doi: 10.1038/s41556-018-0040-4
5. Zhang Q, Higginbotham JN, Jeppesen DK, Yang Y-P, Li W, McKinley ET, et al. Transfer of Functional Cargo in Exomeres. Cell Rep (2019) 27(3):940–54.e6. doi: 10.1016/j.celrep.2019.01.009
6. Chargaff E, West R. The biological significance of the thromboplastic protein of blood. J Biol Chem (1946) 166(1):189–97.
7. Nielsen T, Kristensen AF, Pedersen S, Christiansen G, Kristensen SR. Investigation of procoagulant activity in extracellular vesicles isolated by differential ultracentrifugation. J Extracell Vesicles (2018) 7(1):1454777–. doi: 10.1080/20013078.2018.1454777
8. Harding C, Heuser J, Stahl P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J Cell Biol (1983) 97(2):329–39. doi: 10.1083/jcb.97.2.329
9. Pan BT, Teng K, Wu C, Adam M, Johnstone RM. Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J Cell Biol (1985) 101(3):942–8. doi: 10.1083/jcb.101.3.942
10. Baj-Krzyworzeka M, Szatanek R, Weglarczyk K, Baran J, Urbanowicz B, Brański P, et al. Tumour-derived microvesicles carry several surface determinants and mRNA of tumour cells and transfer some of these determinants to monocytes. Cancer Immunol Immunother (2006) 55(7):808–18. doi: 10.1007/s00262-005-0075-9
11. Ratajczak J, Wysoczynski M, Hayek F, Janowska-Wieczorek A, Ratajczak MZ. Membrane-derived microvesicles: important and underappreciated mediators of cell-to-cell communication. Leukemia (2006) 20(9):1487–95. doi: 10.1038/sj.leu.2404296
12. Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol (2007) 9(6):654–9. doi: 10.1038/ncb1596
13. Rahbarghazi R, Jabbari N, Sani NA, Asghari R, Salimi L, Kalashani SA, et al. Tumor-derived extracellular vesicles: reliable tools for Cancer diagnosis and clinical applications. Cell Commun Signal (2019) 17(1):73–. doi: 10.1186/s12964-019-0390-y
14. Zhang Y, Liu Y, Liu H, Tang WH. Exosomes: biogenesis, biologic function and clinical potential. Cell Biosci (2019) 9(1):19. doi: 10.1186/s13578-019-0282-2
15. Huotari J, Helenius A. Endosome maturation. EMBO J (2011) 30(17):3481–500. doi: 10.1038/emboj.2011.286
16. Yellon DM, Davidson SM. Exosomes: nanoparticles involved in cardioprotection? Circ Res (2014) 114(2):325–32. doi: 10.1161/circresaha.113.300636
17. Doyle LM, Wang MZ. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells (2019) 8(7):727. doi: 10.3390/cells8070727
18. Luzio JP, Mullock BM, Pryor PR, Lindsay MR, James DE, Piper RC. Relationship between endosomes and lysosomes. Biochem Soc Trans (2001) 29(Pt 4):476–80. doi: 10.1042/bst0290476
19. Trombetta ES, Mellman I. Cell biology of antigen processing in vitro and in vivo. Annu Rev Immunol (2005) 23:975–1028. doi: 10.1146/annurev.immunol.22.012703.104538
20. Colombo M, Moita C, van Niel G, Kowal J, Vigneron J, Benaroch P, et al. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J Cell Sci (2013) 126(24):5553–65. doi: 10.1242/jcs.128868
21. McCullough J, Frost A, Sundquist WI. Structures, Functions, and Dynamics of ESCRT-III/Vps4 Membrane Remodeling and Fission Complexes. Annu Rev Cell Dev Biol (2018) 34:85–109. doi: 10.1146/annurev-cellbio-100616-060600
22. Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol (2012) 14(7):677–85. doi: 10.1038/ncb2502
23. Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, et al. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science (New York NY) (2008) 319(5867):1244–7. doi: 10.1126/science.1153124
24. Airola MV, Hannun YA. Sphingolipid metabolism and neutral sphingomyelinases. Handb Exp Pharmacol (2013) 215):57–76. doi: 10.1007/978-3-7091-1368-4_3
25. López-Montero I, Monroy F, Vélez M, Devaux PF. Ceramide: From lateral segregation to mechanical stress. Biochim Biophys Acta (BBA) - Biomembr (2010) 1798(7):1348–56. doi: 10.1016/j.bbamem.2009.12.007
26. Elsherbini A, Bieberich E. Ceramide and Exosomes: A Novel Target in Cancer Biology and Therapy. Adv Cancer Res (2018) 140:121–54. doi: 10.1016/bs.acr.2018.05.004
27. van Niel G, Charrin S, Simoes S, Romao M, Rochin L, Saftig P, et al. The tetraspanin CD63 regulates ESCRT-independent and -dependent endosomal sorting during melanogenesis. Dev Cell (2011) 21(4):708–21. doi: 10.1016/j.devcel.2011.08.019
28. Nazarenko I, Rana S, Baumann A, McAlear J, Hellwig A, Trendelenburg M, et al. Cell surface tetraspanin Tspan8 contributes to molecular pathways of exosome-induced endothelial cell activation. Cancer Res (2010) 70(4):1668–78. doi: 10.1158/0008-5472.can-09-2470
29. Chairoungdua A, Smith DL, Pochard P, Hull M, Caplan MJ. Exosome release of β-catenin: a novel mechanism that antagonizes Wnt signaling. J Cell Biol (2010) 190(6):1079–91. doi: 10.1083/jcb.201002049
30. Blanc L, Vidal M. New insights into the function of Rab GTPases in the context of exosomal secretion. Small GTPases (2018) 9(1-2):95–106. doi: 10.1080/21541248.2016.1264352
31. Phuyal S, Hessvik NP, Skotland T, Sandvig K, Llorente A. Regulation of exosome release by glycosphingolipids and flotillins. FEBS J (2014) 281(9):2214–27. doi: 10.1111/febs.12775
32. Sahu R, Kaushik S, Clement CC, Cannizzo ES, Scharf B, Follenzi A, et al. Microautophagy of cytosolic proteins by late endosomes. Dev Cell (2011) 20(1):131–9. doi: 10.1016/j.devcel.2010.12.003
33. Takeuchi T, Suzuki M, Fujikake N, Popiel HA, Kikuchi H, Futaki S, et al. Intercellular chaperone transmission via exosomes contributes to maintenance of protein homeostasis at the organismal level. Proc Natl Acad Sci (2015) 112(19):E2497–E506. doi: 10.1073/pnas.1412651112
34. Juan T, Fürthauer M. Biogenesis and function of ESCRT-dependent extracellular vesicles. Semin Cell Dev Biol (2018) 74:66–77. doi: 10.1016/j.semcdb.2017.08.022
35. Sork H, Corso G, Krjutskov K, Johansson HJ, Nordin JZ, Wiklander OPB, et al. Heterogeneity and interplay of the extracellular vesicle small RNA transcriptome and proteome. Sci Rep (2018) 8(1):10813. doi: 10.1038/s41598-018-28485-9
36. O’Brien K, Breyne K, Ughetto S, Laurent LC, Breakefield XO. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat Rev Mol Cell Biol (2020) 21(10):585–606. doi: 10.1038/s41580-020-0251-y
37. Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, Pérez-Hernández D, Vázquez J, Martin-Cofreces N, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun (2013) 4:2980–. doi: 10.1038/ncomms3980
38. Columba Cabezas S, Federico M. Sequences within RNA coding for HIV-1 Gag p17 are efficiently targeted to exosomes. Cell Microbiol (2013) 15(3):412–298. doi: 10.1111/cmi.12046
39. Lévesque K, Halvorsen M, Abrahamyan L, Chatel-Chaix L, Poupon V, Gordon H, et al. Trafficking of HIV-1 RNA is mediated by heterogeneous nuclear ribonucleoprotein A2 expression and impacts on viral assembly. Traffic (Copenhagen Denmark) (2006) 7(9):1177–93. doi: 10.1111/j.1600-0854.2006.00461.x
40. Xu P, Bailey-Bucktrout S, Xi Y, Xu D, Du D, Zhang Q, et al. Innate antiviral host defense attenuates TGF-β function through IRF3-mediated suppression of Smad signaling. Mol Cell (2014) 56(6):723–37. doi: 10.1016/j.molcel.2014.11.027
41. Koppers-Lalic D, Hackenberg M, Bijnsdorp IV, van Eijndhoven MAJ, Sadek P, Sie D, et al. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep (2014) 8(6):1649–58. doi: 10.1016/j.celrep.2014.08.027
42. Mateescu B, Kowal EJ, van Balkom BW, Bartel S, Bhattacharyya SN, Buzás EI, et al. Obstacles and opportunities in the functional analysis of extracellular vesicle RNA - an ISEV position paper. J Extracell Vesicles (2017) 6(1):1286095. doi: 10.1080/20013078.2017.1286095
43. Ragusa M, Barbagallo C, Cirnigliaro M, Battaglia R, Brex D, Caponnetto A, et al. Asymmetric RNA Distribution among Cells and Their Secreted Exosomes: Biomedical Meaning and Considerations on Diagnostic Applications. Front Mol Biosci (2017) 4:66. doi: 10.3389/fmolb.2017.00066
44. Tan J, McKenzie C, Vuillermin PJ, Goverse G, Vinuesa CG, Mebius RE, et al. Dietary Fiber and Bacterial SCFA Enhance Oral Tolerance and Protect against Food Allergy through Diverse Cellular Pathways. Cell Rep (2016) 15(12):2809–24. doi: 10.1016/j.celrep.2016.05.047
45. Wang L, Guan P, Teng J, Liu P, Chen D, Xie W, et al. New twinning route in face-centered cubic nanocrystalline metals. Nat Commun (2017) 8(1):2142. doi: 10.1038/s41467-017-02393-4
46. Kossinova OA, Gopanenko AV, Tamkovich SN, Krasheninina OA, Tupikin AE, Kiseleva E, et al. Cytosolic YB-1 and NSUN2 are the only proteins recognizing specific motifs present in mRNAs enriched in exosomes. Biochim Biophys Acta (BBA) - Proteins Proteomics (2017) 1865(6):664–73. doi: 10.1016/j.bbapap.2017.03.010
47. Yanshina DD, Kossinova OA, Gopanenko AV, Krasheninina OA, Malygin AA, Venyaminova AG, et al. Structural features of the interaction of the 3’-untranslated region of mRNA containing exosomal RNA-specific motifs with YB-1, a potential mediator of mRNA sorting. Biochimie (2018) 144:134–43. doi: 10.1016/j.biochi.2017.11.007
48. Temoche-Diaz MM, Shurtleff MJ, Nottingham RM, Yao J, Fadadu RP, Lambowitz AM, et al. Distinct mechanisms of microRNA sorting into cancer cell-derived extracellular vesicle subtypes. eLife (2019) 8. doi: 10.7554/eLife.47544
49. Wen SW, Lima LG, Lobb RJ, Norris EL, Hastie ML, Krumeich S, et al. Breast Cancer-Derived Exosomes Reflect the Cell-of-Origin Phenotype. Proteomics (2019) 19(8):e1800180. doi: 10.1002/pmic.201800180
50. Kahlert C, Melo SA, Protopopov A, Tang J, Seth S, Koch M, et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J Biol Chem (2014) 289(7):3869–75. doi: 10.1074/jbc.C113.532267
51. Novo D, Heath N, Mitchell L, Caligiuri G, MacFarlane A, Reijmer D, et al. Mutant p53s generate pro-invasive niches by influencing exosome podocalyxin levels. Nat Commun (2018) 9(1):5069. doi: 10.1038/s41467-018-07339-y
52. Lobb RJ, Hastie ML, Norris EL, van Amerongen R, Gorman JJ, Möller A. Oncogenic transformation of lung cells results in distinct exosome protein profile similar to the cell of origin. Proteomics (2017) 17(23-24):1600432. doi: 10.1002/pmic.201600432
53. Xiao Y, Zhong J, Zhong B, Huang J, Jiang L, Jiang Y, et al. Exosomes as potential sources of biomarkers in colorectal cancer. Cancer Lett (2020) 476:13–22. doi: 10.1016/j.canlet.2020.01.033
54. Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol (2008) 10(5):619–24. doi: 10.1038/ncb1725
55. Hurwitz SN, Rider MA, Bundy JL, Liu X, Singh RK, Meckes DG Jr. Proteomic profiling of NCI-60 extracellular vesicles uncovers common protein cargo and cancer type-specific biomarkers. Oncotarget (2016) 7(52):86999–7015. doi: 10.18632/oncotarget.13569
56. Osaki M, Okada F. Exosomes and Their Role in Cancer Progression. Yonago Acta Med (2019) 62(2):182–90. doi: 10.33160/yam.2019.06.002
57. Toda Y, Takata K, Nakagawa Y, Kawakami H, Fujioka S, Kobayashi K, et al. Effective internalization of U251-MG-secreted exosomes into cancer cells and characterization of their lipid components. Biochem Biophys Res Commun (2015) 456(3):768–73. doi: 10.1016/j.bbrc.2014.12.015
58. Silva J, García V, Zaballos Á, Provencio M, Lombardía L, Almonacid L, et al. Vesicle-related microRNAs in plasma of nonsmall cell lung cancer patients and correlation with survival. Eur Respir J (2011) 37(3):617–23. doi: 10.1183/09031936.00029610
59. Llorente A, Skotland T, Sylvänne T, Kauhanen D, Róg T, Orłowski A, et al. Molecular lipidomics of exosomes released by PC-3 prostate cancer cells. Biochim Biophys Acta (BBA) - Mol Cell Biol Lipids (2013) 1831(7):1302–9. doi: 10.1016/j.bbalip.2013.04.011
60. Harmati M, Gyukity-Sebestyen E, Dobra G, Janovak L, Dekany I, Saydam O, et al. Small extracellular vesicles convey the stress-induced adaptive responses of melanoma cells. Sci Rep (2019) 9(1):15329. doi: 10.1038/s41598-019-51778-6
61. Yellon DM, Davidson SM. Exosomes. Circ Res (2014) 114(2):325–32. doi: 10.1161/CIRCRESAHA.113.300636
62. Srinivasan S, Vannberg FO, Dixon JB. Lymphatic transport of exosomes as a rapid route of information dissemination to the lymph node. Sci Rep (2016) 6(1):24436. doi: 10.1038/srep24436
63. Yáñez-Mó M, Siljander PRM, Andreu Z, Bedina Zavec A, Borràs FE, Buzas EI, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles (2015) 4(1):27066. doi: 10.3402/jev.v4.27066
64. Shokrollahi E, Nourazarian A, Rahbarghazi R, Salimi L, Karbasforush S, Khaksar M, et al. Treatment of human neuroblastoma cell line SH-SY5Y with HSP27 siRNA tagged-exosomes decreased differentiation rate into mature neurons. J Cell Physiol (2019) 234(11):21005–13. doi: 10.1002/jcp.28704
65. Melo SA, Sugimoto H, O’Connell JT, Kato N, Villanueva A, Vidal A, et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell (2014) 26(5):707–21. doi: 10.1016/j.ccell.2014.09.005
66. Costa-Silva B, Aiello NM, Ocean AJ, Singh S, Zhang H, Thakur BK, et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol (2015) 17(6):816–26. doi: 10.1038/ncb3169
67. Peinado H, Alečković M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med (2012) 18(6):883–91. doi: 10.1038/nm.2753
68. Xavier CPR, Caires HR, Barbosa MAG, Bergantim R, Guimarães JE, Vasconcelos MH. The Role of Extracellular Vesicles in the Hallmarks of Cancer and Drug Resistance. Cells (2020) 9(5). doi: 10.3390/cells9051141
69. Antonyak MA, Li B, Boroughs LK, Johnson JL, Druso JE, Bryant KL, et al. Cancer cell-derived microvesicles induce transformation by transferring tissue transglutaminase and fibronectin to recipient cells. Proc Natl Acad Sci U S A (2011) 108(12):4852–7. doi: 10.1073/pnas.1017667108
70. Jabbari N, Akbariazar E, Feqhhi M, Rahbarghazi R, Rezaie J. Breast cancer-derived exosomes: Tumor progression and therapeutic agents. J Cell Physiol (2020) 235(10):6345–56. doi: 10.1002/jcp.29668
71. Sato S, Vasaikar S, Eskaros A, Kim Y, Lewis JS, Zhang B, et al. EPHB2 carried on small extracellular vesicles induces tumor angiogenesis via activation of ephrin reverse signaling. JCI Insight (2019) 4(23). doi: 10.1172/jci.insight.132447
72. Mao Y, Wang Y, Dong L, Zhang Y, Zhang Y, Wang C, et al. Hypoxic exosomes facilitate angiogenesis and metastasis in esophageal squamous cell carcinoma through altering the phenotype and transcriptome of endothelial cells. J Exp Clin Cancer Res (2019) 38(1):389. doi: 10.1186/s13046-019-1384-8
73. Ahmadi M, Rezaie J. Tumor cells derived-exosomes as angiogenenic agents: possible therapeutic implications. J Trans Med (2020) 18(1):249. doi: 10.1186/s12967-020-02426-5
74. Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, et al. Tumour exosome integrins determine organotropic metastasis. Nature (2015) 527(7578):329–35. doi: 10.1038/nature15756
75. Santos JC, Lima NDS, Sarian LO, Matheu A, Ribeiro ML, Derchain SFM. Exosome-mediated breast cancer chemoresistance via miR-155 transfer. Sci Rep (2018) 8(1):829. doi: 10.1038/s41598-018-19339-5
76. Li J, Yang X, Guan H, Mizokami A, Keller ET, Xu X, et al. Exosome-derived microRNAs contribute to prostate cancer chemoresistance. Int J Oncol (2016) 49(2):838–46. doi: 10.3892/ijo.2016.3560
77. Chen G, Huang AC, Zhang W, Zhang G, Wu M, Xu W, et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature (2018) 560(7718):382–6. doi: 10.1038/s41586-018-0392-8
78. Poggio M, Hu T, Pai CC, Chu B, Belair CD, Chang A, et al. Suppression of Exosomal PD-L1 Induces Systemic Anti-tumor Immunity and Memory. Cell (2019) 177(2):414–27.e13. doi: 10.1016/j.cell.2019.02.016
79. Gao Y, Li X, Zeng C, Liu C, Hao Q, Li W, et al. CD63+ Cancer-Associated Fibroblasts Confer Tamoxifen Resistance to Breast Cancer Cells through Exosomal miR-22. Adv Sci (2020) 7(21):2002518. doi: 10.1002/advs.202002518
80. Xu J, Camfield R, Gorski SM. The interplay between exosomes and autophagy - partners in crime. J Cell Sci (2018) 131(15). doi: 10.1242/jcs.215210
81. Vinod V, Padmakrishnan CJ, Vijayan B, Gopala S. ‘How can I halt thee?’ The puzzles involved in autophagic inhibition. Pharmacol Res (2014) 82:1–8. doi: 10.1016/j.phrs.2014.03.005
82. Hikita T, Miyata M, Watanabe R, Oneyama C. Sensitive and rapid quantification of exosomes by fusing luciferase to exosome marker proteins. Sci Rep (2018) 8(1):14035–. doi: 10.1038/s41598-018-32535-7
83. Graham RM, Thompson JW, Webster KA. Inhibition of the vacuolar ATPase induces Bnip3-dependent death of cancer cells and a reduction in tumor burden and metastasis. Oncotarget (2014) 5(5):1162–73. doi: 10.18632/oncotarget.1699
84. Guo H, Chitiprolu M, Roncevic L, Javalet C, Hemming FJ, Trung MT, et al. Atg5 Disassociates the V(1)V(0)-ATPase to Promote Exosome Production and Tumor Metastasis Independent of Canonical Macroautophagy. Dev Cell (2017) 43(6):716–30. doi: 10.1016/j.devcel.2017.11.018
85. Villarroya-Beltri C, Baixauli F, Mittelbrunn M, Fernández-Delgado I, Torralba D, Moreno-Gonzalo O, et al. ISGylation controls exosome secretion by promoting lysosomal degradation of MVB proteins. Nat Commun (2016) 7:13588–. doi: 10.1038/ncomms13588
86. Han HG, Moon HW, Jeon YJ. ISG15 in cancer: Beyond ubiquitin-like protein. Cancer Lett (2018) 438:52–62. doi: 10.1016/j.canlet.2018.09.007
87. Wood LM, Pan Z-K, Seavey MM, Muthukumaran G, Paterson Y. The ubiquitin-like protein, ISG15, is a novel tumor-associated antigen for cancer immunotherapy. Cancer Immunol Immunother (2012) 61(5):689–700. doi: 10.1007/s00262-011-1129-9
88. exocarta. http://exocarta.org/index.html(2021). [cited 2021].
89. Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci U S A (2016) 113(8):E968–77. doi: 10.1073/pnas.1521230113
90. Arbelaiz A, Azkargorta M, Krawczyk M, Santos-Laso A, Lapitz A, Perugorria MJ, et al. Serum extracellular vesicles contain protein biomarkers for primary sclerosing cholangitis and cholangiocarcinoma. Hepatology (2017) 66(4):1125–43. doi: 10.1002/hep.29291
91. Sun Y, Huo C, Qiao Z, Shang Z, Uzzaman A, Liu S, et al. Comparative Proteomic Analysis of Exosomes and Microvesicles in Human Saliva for Lung Cancer. J Proteome Res (2018) 17(3):1101–7. doi: 10.1021/acs.jproteome.7b00770
92. Li Y, Zhang Y, Qiu F, Qiu Z. Proteomic identification of exosomal LRG1: a potential urinary biomarker for detecting NSCLC. Electrophoresis (2011) 32(15):1976–83. doi: 10.1002/elps.201000598
93. Chen I-H, Xue L, Hsu C-C, Paez JSP, Pan L, Andaluz H, et al. Phosphoproteins in extracellular vesicles as candidate markers for breast cancer. Proc Natl Acad Sci (2017) 114(12):3175–80. doi: 10.1073/pnas.1618088114
94. Khan S, Bennit HF, Turay D, Perez M, Mirshahidi S, Yuan Y, et al. Early diagnostic value of survivin and its alternative splice variants in breast cancer. BMC Cancer (2014) 14(1):176. doi: 10.1186/1471-2407-14-176
95. Galindo-Hernandez O, Villegas-Comonfort S, Candanedo F, González-Vázquez MC, Chavez-Ocaña S, Jimenez-Villanueva X, et al. Elevated concentration of microvesicles isolated from peripheral blood in breast cancer patients. Arch Med Res (2013) 44(3):208–14. doi: 10.1016/j.arcmed.2013.03.002
96. Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature (2015) 523(7559):177–82. doi: 10.1038/nature14581
97. Li J, Chen Y, Guo X, Zhou L, Jia Z, Peng Z, et al. GPC1 exosome and its regulatory miRNAs are specific markers for the detection and target therapy of colorectal cancer. J Cell Mol Med (2017) 21(5):838–47. doi: 10.1111/jcmm.12941
98. Tian Y, Ma L, Gong M, Su G, Zhu S, Zhang W, et al. Protein Profiling and Sizing of Extracellular Vesicles from Colorectal Cancer Patients via Flow Cytometry. ACS Nano (2018) 12(1):671–80. doi: 10.1021/acsnano.7b07782
99. Shender VO, Pavlyukov MS, Ziganshin RH, Arapidi GP, Kovalchuk SI, Anikanov NA, et al. Proteome-metabolome profiling of ovarian cancer ascites reveals novel components involved in intercellular communication. Mol Cell Proteomics (2014) 13(12):3558–71. doi: 10.1074/mcp.M114.041194
100. Madhankumar AB, Mrowczynski OD, Patel SR, Weston CL, Zacharia BE, Glantz MJ, et al. Interleukin-13 conjugated quantum dots for identification of glioma initiating cells and their extracellular vesicles. Acta Biomater (2017) 58:205–13. doi: 10.1016/j.actbio.2017.06.002
101. Alegre E, Zubiri L, Perez-Gracia JL, González-Cao M, Soria L, Martín-Algarra S, et al. Circulating melanoma exosomes as diagnostic and prognosis biomarkers. Clin Chim Acta Int J Clin Chem (2016) 454:28–32. doi: 10.1016/j.cca.2015.12.031
102. Nilsson J, Skog J, Nordstrand A, Baranov V, Mincheva-Nilsson L, Breakefield XO, et al. Prostate cancer-derived urine exosomes: a novel approach to biomarkers for prostate cancer. Br J Cancer (2009) 100(10):1603–7. doi: 10.1038/sj.bjc.6605058
103. Winck FV, Prado Ribeiro AC, Ramos Domingues R, Ling LY, Riaño-Pachón DM, Rivera C, et al. Insights into immune responses in oral cancer through proteomic analysis of saliva and salivary extracellular vesicles. Sci Rep (2015) 5(1):16305. doi: 10.1038/srep16305
104. Lau CS, Wong DT. Breast cancer exosome-like microvesicles and salivary gland cells interplay alters salivary gland cell-derived exosome-like microvesicles in vitro. PLoS One (2012) 7(3):e33037. doi: 10.1371/journal.pone.0033037
105. Katsiougiannis S, Chia D, Kim Y, Singh RP, Wong DT. Saliva exosomes from pancreatic tumor-bearing mice modulate NK cell phenotype and antitumor cytotoxicity. FASEB J (2017) 31(3):998–1010. doi: 10.1096/fj.201600984R
106. Xie C, Ji N, Tang Z, Li J, Chen Q. The role of extracellular vesicles from different origin in the microenvironment of head and neck cancers. Mol Cancer (2019) 18(1):83. doi: 10.1186/s12943-019-0985-3
107. Hildonen S, Skarpen E, Halvorsen TG, Reubsaet L. Isolation and mass spectrometry analysis of urinary extraexosomal proteins. Sci Rep (2016) 6:36331. doi: 10.1038/srep36331
108. Sceneay J, Smyth MJ, Möller A. The pre-metastatic niche: finding common ground. Cancer Metastasis Rev (2013) 32(3-4):449–64. doi: 10.1007/s10555-013-9420-1
109. Frampton AE, Prado MM, López-Jiménez E, Fajardo-Puerta AB, Jawad ZAR, Lawton P, et al. Glypican-1 is enriched in circulating-exosomes in pancreatic cancer and correlates with tumor burden. Oncotarget (2018) 9(27):19006–13. doi: 10.18632/oncotarget.24873
110. Lucien F, Lac V, Billadeau DD, Borgida A, Gallinger S, Leong HS. Glypican-1 and glycoprotein 2 bearing extracellular vesicles do not discern pancreatic cancer from benign pancreatic diseases. Oncotarget (2019) 10(10):1045–55. doi: 10.18632/oncotarget.26620
111. Buscail E, Chauvet A, Quincy P, Degrandi O, Buscail C, Lamrissi I, et al. CD63-GPC1-Positive Exosomes Coupled with CA19-9 Offer Good Diagnostic Potential for Resectable Pancreatic Ductal Adenocarcinoma. Transl Oncol (2019) 12(11):1395–403. doi: 10.1016/j.tranon.2019.07.009
112. Yang KS, Im H, Hong S, Pergolini I, Del Castillo AF, Wang R, et al. Multiparametric plasma EV profiling facilitates diagnosis of pancreatic malignancy. Sci Trans Med (2017) 9(391). doi: 10.1126/scitranslmed.aal3226
113. Maus RLG, Jakub JW, Nevala WK, Christensen TA, Noble-Orcutt K, Sachs Z, et al. Human Melanoma-Derived Extracellular Vesicles Regulate Dendritic Cell Maturation. Front Immunol (2017) 8:358. doi: 10.3389/fimmu.2017.00358
114. Tang Y, Zhang P, Wang Y, Wang J, Su M, Wang Y, et al. The Biogenesis, Biology, and Clinical Significance of Exosomal PD-L1 in Cancer. Front Immunol (2020) 11:604. doi: 10.3389/fimmu.2020.00604
115. Morrissey SM, Yan J. Exosomal PD-L1: Roles in Tumor Progression and Immunotherapy. Trends Cancer (2020) 6(7):550–8. doi: 10.1016/j.trecan.2020.03.002
116. Theodoraki MN, Yerneni SS, Hoffmann TK, Gooding WE, Whiteside TL. Clinical Significance of PD-L1(+) Exosomes in Plasma of Head and Neck Cancer Patients. Clin Cancer Res (2018) 24(4):896–905. doi: 10.1158/1078-0432.ccr-17-2664
117. Teng MW, Ngiow SF, Ribas A, Smyth MJ. Classifying Cancers Based on T-cell Infiltration and PD-L1. Cancer Res (2015) 75(11):2139–45. doi: 10.1158/0008-5472.can-15-0255
118. Moynihan KD, Irvine DJ. Roles for Innate Immunity in Combination Immunotherapies. Cancer Res (2017) 77(19):5215–21. doi: 10.1158/0008-5472.can-17-1340
119. Capello M, Vykoukal JV, Katayama H, Bantis LE, Wang H, Kundnani DL, et al. Exosomes harbor B cell targets in pancreatic adenocarcinoma and exert decoy function against complement-mediated cytotoxicity. Nat Commun (2019) 10(1):254. doi: 10.1038/s41467-018-08109-6
120. Zhang X, Shi H, Yuan X, Jiang P, Qian H, Xu W. Tumor-derived exosomes induce N2 polarization of neutrophils to promote gastric cancer cell migration. Mol Cancer (2018) 17(1):146. doi: 10.1186/s12943-018-0898-6
121. de Camargo LCB, Guaddachi F, Bergerat D, Ourari N, Coillard L, Parietti V, et al. Extracellular vesicles produced by NFAT3-expressing cells hinder tumor growth and metastatic dissemination. Sci Rep (2020) 10(1):8964. doi: 10.1038/s41598-020-65844-x
122. US National Library of Medicine. Exosomes and Immunotherapy in Non-Hodgkin B-cell Lymphomas (ExoReBLy). (2019).
123. Kalluri R. The biology and function of exosomes in cancer. J Clin Invest (2016) 126(4):1208–15. doi: 10.1172/jci81135
124. Guescini M, Genedani S, Stocchi V, Agnati LF. Astrocytes and Glioblastoma cells release exosomes carrying mtDNA. J Neural Transm (Vienna Austria 1996) (2010) 117(1):1–4. doi: 10.1007/s00702-009-0288-8
125. Balaj L, Lessard R, Dai L, Cho YJ, Pomeroy SL, Breakefield XO, et al. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun (2011) 2:180. doi: 10.1038/ncomms1180
126. Thakur BK, Zhang H, Becker A, Matei I, Huang Y, Costa-Silva B, et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res (2014) 24(6):766–9. doi: 10.1038/cr.2014.44
127. Lee TH, Chennakrishnaiah S, Audemard E, Montermini L, Meehan B, Rak J. Oncogenic ras-driven cancer cell vesiculation leads to emission of double-stranded DNA capable of interacting with target cells. Biochem Biophys Res Commun (2014) 451(2):295–301. doi: 10.1016/j.bbrc.2014.07.109
128. Sun N, Lee YT, Zhang RY, Kao R, Teng PC, Yang Y, et al. Purification of HCC-specific extracellular vesicles on nanosubstrates for early HCC detection by digital scoring. Nat Commun (2020) 11(1):4489. doi: 10.1038/s41467-020-18311-0
129. Zhou R, Chen KK, Zhang J, Xiao B, Huang Z, Ju C, et al. The decade of exosomal long RNA species: an emerging cancer antagonist. Mol Cancer (2018) 17(1):75. doi: 10.1186/s12943-018-0823-z
130. Yang B, Feng X, Liu H, Tong R, Wu J, Li C, et al. High-metastatic cancer cells derived exosomal miR92a-3p promotes epithelial-mesenchymal transition and metastasis of low-metastatic cancer cells by regulating PTEN/Akt pathway in hepatocellular carcinoma. Oncogene (2020). doi: 10.1038/s41388-020-01450-5
131. Hannafon BN, Trigoso YD, Calloway CL, Zhao YD, Lum DH, Welm AL, et al. Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res (2016) 18(1):90. doi: 10.1186/s13058-016-0753-x
132. Manier S, Liu CJ, Avet-Loiseau H, Park J, Shi J, Campigotto F, et al. Prognostic role of circulating exosomal miRNAs in multiple myeloma. Blood (2017) 129(17):2429–36. doi: 10.1182/blood-2016-09-742296
133. Wang L, Wu J, Xie C. miR-92a promotes hepatocellular carcinoma cells proliferation and invasion by FOXA2 targeting. Iran J Basic Med Sci (2017) 20(7):783–90. doi: 10.22038/ijbms.2017.9010
134. Balacescu O, Sur D, Cainap C, Visan S, Cruceriu D, Manzat-Saplacan R, et al. The Impact of miRNA in Colorectal Cancer Progression and Its Liver Metastases. Int J Mol Sci (2018) 19(12). doi: 10.3390/ijms19123711
135. Asano N, Matsuzaki J, Ichikawa M, Kawauchi J, Takizawa S, Aoki Y, et al. A serum microRNA classifier for the diagnosis of sarcomas of various histological subtypes. Nat Commun (2019) 10(1):1299. doi: 10.1038/s41467-019-09143-8
136. Tang S, Cheng J, Yao Y, Lou C, Wang L, Huang X, et al. Combination of Four Serum Exosomal MiRNAs as Novel Diagnostic Biomarkers for Early-Stage Gastric Cancer. Front Genet (2020) 11:237(237). doi: 10.3389/fgene.2020.00237
137. Jin G, Liu Y, Zhang J, Bian Z, Yao S, Fei B, et al. A panel of serum exosomal microRNAs as predictive markers for chemoresistance in advanced colorectal cancer. Cancer Chemother Pharmacol (2019) 84(2):315–25. doi: 10.1007/s00280-019-03867-6
138. US National Library of Medicine. Identification and Characterization of Predictive Factors of Onset of Bone Metastases in Cancer Patients (PreMetOn). (2020).
139. Ge X, Wang Y, Nie J, Li Q, Tang L, Deng X, et al. The diagnostic/prognostic potential and molecular functions of long non-coding RNAs in the exosomes derived from the bile of human cholangiocarcinoma. Oncotarget (2017) 8(41):69995–70005. doi: 10.18632/oncotarget.19547
140. Louis C, Desoteux M, Coulouarn C. Exosomal circRNAs: new players in the field of cholangiocarcinoma. Clin Sci (2019) 133(21):2239–44. doi: 10.1042/cs20190940
141. Bai M, Fu W, Su G, Cao J, Gao L, Huang C, et al. The role of extracellular vesicles in cholangiocarcinoma. Cancer Cell Int (2020) 20(1):435. doi: 10.1186/s12935-020-01526-y
142. Chen S, Zhu X, Huang S. Clinical applications of extracellular vesicle long RNAs. Crit Rev Clin Lab Sci (2020) 1–14. doi: 10.1080/10408363.2020.1751584
143. US National Library of Medicine. Serum Exosomal Long Noncoding RNAs as Potential Biomarkers for Lung Cancer Diagnosis. (2019).
144. Jafari D, Malih S, Eini M, Jafari R, Gholipourmalekabadi M, Sadeghizadeh M, et al. Improvement, scaling-up, and downstream analysis of exosome production. Crit Rev Biotechnol (2020) 1–15. doi: 10.1080/07388551.2020.1805406
145. Yang D, Zhang W, Zhang H, Zhang F, Chen L, Ma L, et al. Progress, opportunity, and perspective on exosome isolation - efforts for efficient exosome-based theranostics. Theranostics (2020) 10(8):3684–707. doi: 10.7150/thno.41580
146. Li P, Kaslan M, Lee SH, Yao J, Gao Z. Progress in Exosome Isolation Techniques. Theranostics (2017) 7(3):789–804. doi: 10.7150/thno.18133
147. Lydic TA, Townsend S, Adda CG, Collins C, Mathivanan S, Reid GE. Rapid and comprehensive ‘shotgun’ lipidome profiling of colorectal cancer cell derived exosomes. Methods (San Diego Calif) (2015) 87:83–95. doi: 10.1016/j.ymeth.2015.04.014
148. Cheng L, Zhang K, Qing Y, Li D, Cui M, Jin P, et al. Proteomic and lipidomic analysis of exosomes derived from ovarian cancer cells and ovarian surface epithelial cells. J Ovarian Res (2020) 13(1):9–. doi: 10.1186/s13048-020-0609-y
149. Kim MS, Haney MJ, Zhao Y, Yuan D, Deygen I, Klyachko NL, et al. Engineering macrophage-derived exosomes for targeted paclitaxel delivery to pulmonary metastases: in vitro and in vivo evaluations. Nanomedicine (2018) 14(1):195–204. doi: 10.1016/j.nano.2017.09.011
150. Tian T, Zhang H-X, He C-P, Fan S, Zhu Y-L, Qi C, et al. Surface functionalized exosomes as targeted drug delivery vehicles for cerebral ischemia therapy. Biomaterials (2018) 150:137–49. doi: 10.1016/j.biomaterials.2017.10.012
151. Liu Z, Wang F, Chen X. Integrin alpha(v)beta(3)-Targeted Cancer Therapy. Drug Dev Res (2008) 69(6):329–39. doi: 10.1002/ddr.20265
152. Pi F, Binzel DW, Lee TJ, Li Z, Sun M, Rychahou P, et al. Nanoparticle orientation to control RNA loading and ligand display on extracellular vesicles for cancer regression. Nat Nanotechnol (2018) 13(1):82–9. doi: 10.1038/s41565-017-0012-z
153. Bastos N, Ruivo CF, da Silva S, Melo SA. Exosomes in cancer: Use them or target them? Semin Cell Dev Biol (2018) 78:13–21. doi: 10.1016/j.semcdb.2017.08.009
154. Ohno S, Drummen GP, Kuroda M. Focus on Extracellular Vesicles: Development of Extracellular Vesicle-Based Therapeutic Systems. Int J Mol Sci (2016) 17(2):172. doi: 10.3390/ijms17020172
155. Smyth TJ, Redzic JS, Graner MW, Anchordoquy TJ. Examination of the specificity of tumor cell derived exosomes with tumor cells in vitro. Biochim Biophys Acta (2014) 1838(11):2954–65. doi: 10.1016/j.bbamem.2014.07.026
156. Markov O, Oshchepkova A, Mironova N. Immunotherapy Based on Dendritic Cell-Targeted/-Derived Extracellular Vesicles-A Novel Strategy for Enhancement of the Anti-tumor Immune Response. Front Pharmacol (2019) 10:1152. doi: 10.3389/fphar.2019.01152
157. Haney MJ, Klyachko NL, Zhao Y, Gupta R, Plotnikova EG, He Z, et al. Exosomes as drug delivery vehicles for Parkinson’s disease therapy. J Control Release (2015) 207:18–30. doi: 10.1016/j.jconrel.2015.03.033
158. Lamichhane TN, Raiker RS, Jay SM. Exogenous DNA Loading into Extracellular Vesicles via Electroporation is Size-Dependent and Enables Limited Gene Delivery. Mol Pharm (2015) 12(10):3650–7. doi: 10.1021/acs.molpharmaceut.5b00364
159. Sato YT, Umezaki K, Sawada S, Mukai S-A, Sasaki Y, Harada N, et al. Engineering hybrid exosomes by membrane fusion with liposomes. Sci Rep (2016) 6(1):21933. doi: 10.1038/srep21933
160. Lu M, Xing H, Xun Z, Yang T, Ding P, Cai C, et al. Exosome-based small RNA delivery: Progress and prospects. Asian J Pharm Sci (2018) 13(1):1–11. doi: 10.1016/j.ajps.2017.07.008
161. Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol (2011) 29(4):341–5. doi: 10.1038/nbt.1807
162. Kamerkar S, LeBleu VS, Sugimoto H, Yang S, Ruivo CF, Melo SA, et al. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature (2017) 546(7659):498–503. doi: 10.1038/nature22341
163. Zhou Y, Yuan Y, Liu M, Hu X, Quan Y, Chen X. Tumor-specific delivery of KRAS siRNA with iRGD-exosomes efficiently inhibits tumor growth. ExRNA (2019) 1(1):28. doi: 10.1186/s41544-019-0034-9
164. Faruqu FN, Xu L, Al-Jamal KT. Preparation of Exosomes for siRNA Delivery to Cancer Cells. J Vis Exp (2018) 142):10.3791/58814. doi: 10.3791/58814
165. Kooijmans SAA, Stremersch S, Braeckmans K, de Smedt SC, Hendrix A, Wood MJA, et al. Electroporation-induced siRNA precipitation obscures the efficiency of siRNA loading into extracellular vesicles. J Control Release (2013) 172(1):229–38. doi: 10.1016/j.jconrel.2013.08.014
166. Hung ME, Leonard JN. Stabilization of exosome-targeting peptides via engineered glycosylation. J Biol Chem (2015) 290(13):8166–72. doi: 10.1074/jbc.M114.621383
167. Liang G, Zhu Y, Ali DJ, Tian T, Xu H, Si K, et al. Engineered exosomes for targeted co-delivery of miR-21 inhibitor and chemotherapeutics to reverse drug resistance in colon cancer. J Nanobiotechnology (2020) 18(1):10. doi: 10.1186/s12951-019-0563-2
168. Kooijmans SAA, Aleza CG, Roffler SR, van Solinge WW, Vader P, Schiffelers RM. Display of GPI-anchored anti-EGFR nanobodies on extracellular vesicles promotes tumour cell targeting. J Extracell Vesicles (2016) 5(1):31053. doi: 10.3402/jev.v5.31053
169. Barile L, Vassalli G. Exosomes: Therapy delivery tools and biomarkers of diseases. Pharmacol Ther (2017) 174:63–78. doi: 10.1016/j.pharmthera.2017.02.020
170. Pascucci L, Coccè V, Bonomi A, Ami D, Ceccarelli P, Ciusani E, et al. Paclitaxel is incorporated by mesenchymal stromal cells and released in exosomes that inhibit in vitro tumor growth: a new approach for drug delivery. J Control Release (2014) 192:262–70. doi: 10.1016/j.jconrel.2014.07.042
171. Guo M, Wu F, Hu G, Chen L, Xu J, Xu P, et al. Autologous tumor cell–derived microparticle-based targeted chemotherapy in lung cancer patients with malignant pleural effusion. Sci Trans Med (2019) 11(474):eaat5690. doi: 10.1126/scitranslmed.aat5690
172. Sun H, Burrola S, Wu J, Ding W-Q. Extracellular Vesicles in the Development of Cancer Therapeutics. Int J Mol Sci (2020) 21(17):6097. doi: 10.3390/ijms21176097
173. Zhang X, Liu L, Tang M, Li H, Guo X, Yang X. The effects of umbilical cord-derived macrophage exosomes loaded with cisplatin on the growth and drug resistance of ovarian cancer cells. Drug Dev Ind Pharm (2020) 46(7):1150–62. doi: 10.1080/03639045.2020.1776320
174. Hall J, Prabhakar S, Balaj L, Lai CP, Cerione RA, Breakefield XO. Delivery of Therapeutic Proteins via Extracellular Vesicles: Review and Potential Treatments for Parkinson’s Disease, Glioma, and Schwannoma. Cell Mol Neurobiol (2016) 36(3):417–27. doi: 10.1007/s10571-015-0309-0
175. Mizrak A, Bolukbasi MF, Ozdener GB, Brenner GJ, Madlener S, Erkan EP, et al. Genetically engineered microvesicles carrying suicide mRNA/protein inhibit schwannoma tumor growth. Mol Ther J Am Soc Gene Ther (2013) 21(1):101–8. doi: 10.1038/mt.2012.161
176. Prabhakar S, Taherian M, Gianni D, Conlon TJ, Fulci G, Brockmann J, et al. Regression of schwannomas induced by adeno-associated virus-mediated delivery of caspase-1. Hum Gene Ther (2013) 24(2):152–62. doi: 10.1089/hum.2012.094
177. Paggetti J, Haderk F, Seiffert M, Janji B, Distler U, Ammerlaan W, et al. Exosomes released by chronic lymphocytic leukemia cells induce the transition of stromal cells into cancer-associated fibroblasts. Blood (2015) 126(9):1106–17. doi: 10.1182/blood-2014-12-618025
178. Crenshaw BJ, Gu L, Sims B, Matthews QL. Exosome Biogenesis and Biological Function in Response to Viral Infections. Open Virol J (2018) 12:134–48. doi: 10.2174/1874357901812010134
179. Pegtel DM, Gould SJ. Exosomes. Annu Rev Biochem (2019) 88(1):487–514. doi: 10.1146/annurev-biochem-013118-111902
180. De Gassart A, Trentin B, Martin M, Hocquellet A, Bette-Bobillo P, Mamoun R, et al. Exosomal sorting of the cytoplasmic domain of bovine leukemia virus TM Env protein. Cell Biol Int (2009) 33(1):36–48. doi: 10.1016/j.cellbi.2008.10.001
181. Manfredi F, Di Bonito P, Arenaccio C, Anticoli S, Federico M. Incorporation of Heterologous Proteins in Engineered Exosomes. Methods Mol Biol (Clifton NJ) (2016) 1448:249–60. doi: 10.1007/978-1-4939-3753-0_18
182. Simons K, Sampaio JL. Membrane organization and lipid rafts. Cold Spring Harb Perspect Biol (2011) 3(10):a004697. doi: 10.1101/cshperspect.a004697
183. Foster JL, Denial SJ, Temple BRS, Garcia JV. Mechanisms of HIV-1 Nef function and intracellular signaling. J Neuroimmune Pharmacol (2011) 6(2):230–46. doi: 10.1007/s11481-011-9262-y
184. Di Bonito P, Chiozzini C, Arenaccio C, Anticoli S, Manfredi F, Olivetta E, et al. Antitumor HPV E7-specific CTL activity elicited by in vivo engineered exosomes produced through DNA inoculation. Int J Nanomedicine (2017) 12:4579–91. doi: 10.2147/IJN.S131309
185. Meyer C, Losacco J, Stickney Z, Li L, Marriott G, Lu B. Pseudotyping exosomes for enhanced protein delivery in mammalian cells. Int J Nanomed (2017) 12:3153–70. doi: 10.2147/ijn.s133430
186. Cheng Y, Schorey JS. Targeting soluble proteins to exosomes using a ubiquitin tag. Biotechnol Bioeng (2016) 113(6):1315–24. doi: 10.1002/bit.25884
187. Giovannone AJ, Reales E, Bhattaram P, Fraile-Ramos A, Weimbs T. Monoubiquitination of syntaxin 3 leads to retrieval from the basolateral plasma membrane and facilitates cargo recruitment to exosomes. Mol Biol Cell (2017) 28(21):2843–53. doi: 10.1091/mbc.E17-07-0461
188. Shen B, Wu N, Yang JM, Gould SJ. Protein targeting to exosomes/microvesicles by plasma membrane anchors. J Biol Chem (2011) 286(16):14383–95. doi: 10.1074/jbc.M110.208660
189. Sterzenbach U, Putz U, Low LH, Silke J, Tan SS, Howitt J. Engineered Exosomes as Vehicles for Biologically Active Proteins. Mol Ther J Am Soc Gene Ther (2017) 25(6):1269–78. doi: 10.1016/j.ymthe.2017.03.030
190. Ohmura-Hoshino M, Goto E, Matsuki Y, Aoki M, Mito M, Uematsu M, et al. A novel family of membrane-bound E3 ubiquitin ligases. J Biochem (2006) 140(2):147–54. doi: 10.1093/jb/mvj160
191. Shin JS, Ebersold M, Pypaert M, Delamarre L, Hartley A, Mellman I. Surface expression of MHC class II in dendritic cells is controlled by regulated ubiquitination. Nature (2006) 444(7115):115–8. doi: 10.1038/nature05261
192. Gauvreau ME, Côté MH, Bourgeois-Daigneault MC, Rivard LD, Xiu F, Brunet A, et al. Sorting of MHC class II molecules into exosomes through a ubiquitin-independent pathway. Traffic (Copenhagen Denmark) (2009) 10(10):1518–27. doi: 10.1111/j.1600-0854.2009.00948.x
193. Liu C, Su C. Design strategies and application progress of therapeutic exosomes. Theranostics (2019) 9(4):1015–28. doi: 10.7150/thno.30853
194. Amano T, Furuno T, Hirashima N, Ohyama N, Nakanishi M. Dynamics of intracellular granules with CD63-GFP in rat basophilic leukemia cells. J Biochem (2001) 129(5):739–44. doi: 10.1093/oxfordjournals.jbchem.a002914
195. Andreu Z, Yáñez-Mó M. Tetraspanins in extracellular vesicle formation and function. Front Immunol (2014) 5:442. doi: 10.3389/fimmu.2014.00442
196. Blanchard N, Lankar D, Faure F, Regnault A, Dumont C, Raposo G, et al. TCR activation of human T cells induces the production of exosomes bearing the TCR/CD3/zeta complex. J Immunol (Baltimore Md 1950) (2002) 168(7):3235–41. doi: 10.4049/jimmunol.168.7.3235
197. André F, Chaput N, Schartz NEC, Flament C, Aubert N, Bernard J, et al. Exosomes as Potent Cell-Free Peptide-Based Vaccine. I. Dendritic Cell-Derived Exosomes Transfer Functional MHC Class I/Peptide Complexes to Dendritic Cells. J Immunol (2004) 172(4):2126–36. doi: 10.4049/jimmunol.172.4.2126
198. Zeelenberg IS, Ostrowski M, Krumeich S, Bobrie A, Jancic C, Boissonnas A, et al. Targeting tumor antigens to secreted membrane vesicles in vivo induces efficient antitumor immune responses. Cancer Res (2008) 68(4):1228–35. doi: 10.1158/0008-5472.can-07-3163
199. Simpson RJ, Jensen SS, Lim JW. Proteomic profiling of exosomes: current perspectives. Proteomics (2008) 8(19):4083–99. doi: 10.1002/pmic.200800109
200. Khan S, Jutzy JMS, Aspe JR, McGregor DW, Neidigh JW, Wall NR. Survivin is released from cancer cells via exosomes. Apoptosis (2011) 16(1):1–12. doi: 10.1007/s10495-010-0534-4
201. Lai CP, Mardini O, Ericsson M, Prabhakar S, Maguire C, Chen JW, et al. Dynamic biodistribution of extracellular vesicles in vivo using a multimodal imaging reporter. ACS Nano (2014) 8(1):483–94. doi: 10.1021/nn404945r
202. Gastpar R, Gehrmann M, Bausero MA, Asea A, Gross C, Schroeder JA, et al. Heat shock protein 70 surface-positive tumor exosomes stimulate migratory and cytolytic activity of natural killer cells. Cancer Res (2005) 65(12):5238–47. doi: 10.1158/0008-5472.CAN-04-3804
203. Wyciszkiewicz A, Kalinowska-Łyszczarz A, Nowakowski B, Kaźmierczak K, Osztynowicz K, Michalak S. Expression of small heat shock proteins in exosomes from patients with gynecologic cancers. Sci Rep (2019) 9(1):9817. doi: 10.1038/s41598-019-46221-9
204. Radetskaya O, Lane RK, Friedman T, Garrett A, Borror MB, Russell J, et al. The PMK-3 (p38) Mitochondrial Retrograde Response Functions in Intestinal Cells to Extend Life via the ESCRT Machinery. bioRxiv (2019), 797308. doi: 10.1101/797308
205. Yim N, Ryu S-W, Choi K, Lee KR, Lee S, Choi H, et al. Exosome engineering for efficient intracellular delivery of soluble proteins using optically reversible protein–protein interaction module. Nat Commun (2016) 7(1):12277. doi: 10.1038/ncomms12277
206. Fu W, Lei C, Liu S, Cui Y, Wang C, Qian K, et al. CAR exosomes derived from effector CAR-T cells have potent antitumour effects and low toxicity. Nat Commun (2019) 10(1):4355. doi: 10.1038/s41467-019-12321-3
207. Rezaie J, Ajezi S, Avci ÇB, Karimipour M, Geranmayeh MH, Nourazarian A, et al. Exosomes and their Application in Biomedical Field: Difficulties and Advantages. Mol Neurobiol (2018) 55(4):3372–93. doi: 10.1007/s12035-017-0582-7
208. Marcus ME, Leonard JN. FedExosomes: Engineering Therapeutic Biological Nanoparticles that Truly Deliver. Pharmaceuticals (Basel Switzerland) (2013) 6(5):659–80. doi: 10.3390/ph6050659
209. Lou G, Song X, Yang F, Wu S, Wang J, Chen Z, et al. Exosomes derived from miR-122-modified adipose tissue-derived MSCs increase chemosensitivity of hepatocellular carcinoma. J Hematol Oncol (2015) 8:122. doi: 10.1186/s13045-015-0220-7
210. Abels ER, Maas SLN, Nieland L, Wei Z, Cheah PS, Tai E, et al. Glioblastoma-Associated Microglia Reprogramming Is Mediated by Functional Transfer of Extracellular miR-21. Cell Rep (2019) 28(12):3105–19.e7. doi: 10.1016/j.celrep.2019.08.036
211. Xie X, Nie H, Zhou Y, Lian S, Mei H, Lu Y, et al. Eliminating blood oncogenic exosomes into the small intestine with aptamer-functionalized nanoparticles. Nat Commun (2019) 10(1):5476. doi: 10.1038/s41467-019-13316-w
212. Nishida-Aoki N, Tominaga N, Takeshita F, Sonoda H, Yoshioka Y, Ochiya T. Disruption of Circulating Extracellular Vesicles as a Novel Therapeutic Strategy against Cancer Metastasis. Mol Ther J Am Soc Gene Ther (2017) 25(1):181–91. doi: 10.1016/j.ymthe.2016.10.009
213. Kitai Y, Kawasaki T, Sueyoshi T, Kobiyama K, Ishii KJ, Zou J, et al. DNA-Containing Exosomes Derived from Cancer Cells Treated with Topotecan Activate a STING-Dependent Pathway and Reinforce Antitumor Immunity. J Immunol (Baltimore Md 1950) (2017) 198(4):1649–59. doi: 10.4049/jimmunol.1601694
214. Takahashi A, Okada R, Nagao K, Kawamata Y, Hanyu A, Yoshimoto S, et al. Exosomes maintain cellular homeostasis by excreting harmful DNA from cells. Nat Commun (2017) 8:15287. doi: 10.1038/ncomms15287
215. Diamond JM, Vanpouille-Box C, Spada S, Rudqvist NP, Chapman JR, Ueberheide BM, et al. Exosomes Shuttle TREX1-Sensitive IFN-Stimulatory dsDNA from Irradiated Cancer Cells to DCs. Cancer Immunol Res (2018) 6(8):910–20. doi: 10.1158/2326-6066.cir-17-0581
216. Kosgodage US, Mould R, Henley AB, Nunn AV, Guy GW, Thomas EL, et al. Cannabidiol (CBD) Is a Novel Inhibitor for Exosome and Microvesicle (EMV) Release in Cancer. Front Pharmacol (2018) 9:889. doi: 10.3389/fphar.2018.00889
217. Khan FM, Saleh E, Alawadhi H, Harati R, Zimmermann WH, El-Awady R. Inhibition of exosome release by ketotifen enhances sensitivity of cancer cells to doxorubicin. Cancer Biol Ther (2018) 19(1):25–33. doi: 10.1080/15384047.2017.1394544
218. Kulshreshtha A, Singh S, Ahmad M, Khanna K, Ahmad T, Agrawal A, et al. Simvastatin mediates inhibition of exosome synthesis, localization and secretion via multicomponent interventions. Sci Rep (2019) 9(1):16373. doi: 10.1038/s41598-019-52765-7
219. Zhang H, Lu J, Liu J, Zhang G, Lu A. Advances in the discovery of exosome inhibitors in cancer. J Enzyme Inhib Med Chem (2020) 35(1):1322–30. doi: 10.1080/14756366.2020.1754814
221. Tan A, De La Peña H, Seifalian AM. The application of exosomes as a nanoscale cancer vaccine. Int J Nanomedicine (2010) 5:889–900. doi: 10.2147/ijn.s13402
222. Harzstark AL, Small EJ. Immunotherapy for prostate cancer using antigen-loaded antigen-presenting cells: APC8015 (Provenge). Expert Opin Biol Ther (2007) 7(8):1275–80. doi: 10.1517/14712598.7.8.1275
223. Small EJ, Schellhammer PF, Higano CS, Redfern CH, Nemunaitis JJ, Valone FH, et al. Placebo-controlled phase III trial of immunologic therapy with sipuleucel-T (APC8015) in patients with metastatic, asymptomatic hormone refractory prostate cancer. J Clin Oncol (2006) 24(19):3089–94. doi: 10.1200/jco.2005.04.5252
224. US National Library of Medicine. Provenge® (Sipuleucel-T) Active Cellular Immunotherapy Treatment of Metastatic Prostate Cancer After Failing Hormone Therapy. (2010).
225. André F, Schartz NE, Chaput N, Flament C, Raposo G, Amigorena S, et al. Tumor-derived exosomes: a new source of tumor rejection antigens. Vaccine (2002) 20(Suppl 4):A28–31. doi: 10.1016/s0264-410x(02)00384-5
226. Möller A, Lobb RJ. The evolving translational potential of small extracellular vesicles in cancer. Nat Rev Cancer (2020). doi: 10.1038/s41568-020-00299-w
227. Rao Q, Zuo B, Lu Z, Gao X, You A, Wu C, et al. Tumor-derived exosomes elicit tumor suppression in murine hepatocellular carcinoma models and humans in vitro. Hepatology (2016) 64(2):456–72. doi: 10.1002/hep.28549
228. Jabbari N, Nawaz M, Rezaie J. Ionizing Radiation Increases the Activity of Exosomal Secretory Pathway in MCF-7 Human Breast Cancer Cells: A Possible Way to Communicate Resistance against Radiotherapy. Int J Mol Sci (2019) 20(15). doi: 10.3390/ijms20153649
229. McAndrews KM, Kalluri R. Mechanisms associated with biogenesis of exosomes in cancer. Mol Cancer (2019) 18(1):52. doi: 10.1186/s12943-019-0963-9
230. Sun Y, Feng X, Wan C, Lovell JF, Jin H, Ding J. Role of nanoparticle-mediated immunogenic cell death in cancer immunotherapy. Asian J Pharm Sci (2020). doi: 10.1016/j.ajps.2020.05.004
231. Kwon J, Bakhoum SF. The Cytosolic DNA-Sensing cGAS-STING Pathway in Cancer. Cancer Discov (2020) 10(1):26–39. doi: 10.1158/2159-8290.cd-19-0761
232. Hoong BYD, Gan YH, Liu H, Chen ES. cGAS-STING pathway in oncogenesis and cancer therapeutics. Oncotarget (2020) 11(30). doi: 10.18632/oncotarget.27673
233. Kirolikar S, Prasannan P, Raghuram GV, Pancholi N, Saha T, Tidke P, et al. Prevention of radiation-induced bystander effects by agents that inactivate cell-free chromatin released from irradiated dying cells. Cell Death Dis (2018) 9(12):1142. doi: 10.1038/s41419-018-1181-x
234. Ni J, Bucci J, Malouf D, Knox M, Graham P, Li Y. Exosomes in Cancer Radioresistance. Front Oncol (2019) 9:869. doi: 10.3389/fonc.2019.00869
235. Jabbari N, Karimipour M, Khaksar M, Akbariazar E, Heidarzadeh M, Mojarad B, et al. Tumor-derived extracellular vesicles: insights into bystander effects of exosomes after irradiation. Lasers Med Sci (2020) 35(3):531–45. doi: 10.1007/s10103-019-02880-8
236. Mo L-J, Song M, Huang Q-H, Guan H, Liu X-D, Xie D-F, et al. Exosome-packaged miR-1246 contributes to bystander DNA damage by targeting LIG4. Br J Cancer (2018) 119(4):492–502. doi: 10.1038/s41416-018-0192-9
237. Bagheri HS, Mousavi M, Rezabakhsh A, Rezaie J, Rasta SH, Nourazarian A, et al. Low-level laser irradiation at a high power intensity increased human endothelial cell exosome secretion via Wnt signaling. Lasers Med Sci (2018) 33(5):1131–45. doi: 10.1007/s10103-018-2495-8
238. Lehmann BD, Paine MS, Brooks AM, McCubrey JA, Renegar RH, Wang R, et al. Senescence-associated exosome release from human prostate cancer cells. Cancer Res (2008) 68(19):7864–71. doi: 10.1158/0008-5472.can-07-6538
239. Wan C, Sun Y, Tian Y, Lu L, Dai X, Meng J, et al. Irradiated tumor cell-derived microparticles mediate tumor eradication via cell killing and immune reprogramming. Sci Adv (2020) 6(13):eaay9789. doi: 10.1126/sciadv.aay9789
240. Dai S, Wei D, Wu Z, Zhou X, Wei X, Huang H, et al. Phase I clinical trial of autologous ascites-derived exosomes combined with GM-CSF for colorectal cancer. Mol Ther J Am Soc Gene Ther (2008) 16(4):782–90. doi: 10.1038/mt.2008.1
241. Admyre C, Johansson SM, Paulie S, Gabrielsson S. Direct exosome stimulation of peripheral human T cells detected by ELISPOT. Eur J Immunol (2006) 36(7):1772–81. doi: 10.1002/eji.200535615
242. Gao D, Jiang L. Exosomes in cancer therapy: a novel experimental strategy. Am J Cancer Res (2018) 8(11):2165–75.
243. Zitvogel L, Regnault A, Lozier A, Wolfers J, Flament C, Tenza D, et al. Eradication of established murine tumors using a novel cell-free vaccine: dendritic cell-derived exosomes. Nat Med (1998) 4(5):594–600. doi: 10.1038/nm0598-594
244. Montecalvo A, Shufesky WJ, Stolz DB, Sullivan MG, Wang Z, Divito SJ, et al. Exosomes as a short-range mechanism to spread alloantigen between dendritic cells during T cell allorecognition. J Immunol (Baltimore Md 1950) (2008) 180(5):3081–90. doi: 10.4049/jimmunol.180.5.3081
245. Besse B, Charrier M, Lapierre V, Dansin E, Lantz O, Planchard D, et al. Dendritic cell-derived exosomes as maintenance immunotherapy after first line chemotherapy in NSCLC. Oncoimmunology (2016) 5(4):e1071008. doi: 10.1080/2162402x.2015.1071008
246. Xu Z, Zeng S, Gong Z, Yan Y. Exosome-based immunotherapy: a promising approach for cancer treatment. Mol Cancer (2020) 19(1):160. doi: 10.1186/s12943-020-01278-3
247. Wu SW, Li L, Wang Y, Xiao Z. CTL-Derived Exosomes Enhance the Activation of CTLs Stimulated by Low-Affinity Peptides. Front Immunol (2019) 10:1274. doi: 10.3389/fimmu.2019.01274
248. Lu L, Sun Y, Wan C, Hu Y, Lo P-C, Lovell JF, et al. Role of intravital imaging in nanomedicine-assisted anti-cancer therapy. Curr Opin Biotechnol (2021) 69:153–61. doi: 10.1016/j.copbio.2020.12.024
249. Jong AY, Wu CH, Li J, Sun J, Fabbri M, Wayne AS, et al. Large-scale isolation and cytotoxicity of extracellular vesicles derived from activated human natural killer cells. J Extracell Vesicles (2017) 6(1):1294368. doi: 10.1080/20013078.2017.1294368
250. Zöller M. Janus-Faced Myeloid-Derived Suppressor Cell Exosomes for the Good and the Bad in Cancer and Autoimmune Disease. Front Immunol (2018) 9:137. doi: 10.3389/fimmu.2018.00137
252. Taylor DD, Shah S. Methods of isolating extracellular vesicles impact down-stream analyses of their cargoes. Methods (San Diego Calif) (2015) 87:3–10. doi: 10.1016/j.ymeth.2015.02.019
253. Wiklander OP, Nordin JZ, O’Loughlin A, Gustafsson Y, Corso G, Mäger I, et al. Extracellular vesicle in vivo biodistribution is determined by cell source, route of administration and targeting. J Extracell Vesicles (2015) 4:26316. doi: 10.3402/jev.v4.26316
254. van der Meel R, Sulheim E, Shi Y, Kiessling F, Mulder WJM, Lammers T. Smart cancer nanomedicine. Nat Nanotechnol (2019) 14(11):1007–17. doi: 10.1038/s41565-019-0567-y
255. Kanada M, Bachmann MH, Hardy JW, Frimannson DO, Bronsart L, Wang A, et al. Differential fates of biomolecules delivered to target cells via extracellular vesicles. Proc Natl Acad Sci U S A (2015) 112(12):E1433–42. doi: 10.1073/pnas.1418401112
256. Clayton A, Court J, Navabi H, Adams M, Mason MD, Hobot JA, et al. Analysis of antigen presenting cell derived exosomes, based on immuno-magnetic isolation and flow cytometry. J Immunol Methods (2001) 247(1-2):163–74. doi: 10.1016/s0022-1759(00)00321-5
257. Catalano M, O’Driscoll L. Inhibiting extracellular vesicles formation and release: a review of EV inhibitors. J Extracell Vesicles (2019) 9(1):1703244–. doi: 10.1080/20013078.2019.1703244
Keywords: small extracellular vesicles, exosomes, early cancer detection, drug delivery systems, therapeutic strategy
Citation: Gao Y, Qin Y, Wan C, Sun Y, Meng J, Huang J, Hu Y, Jin H and Yang K (2021) Small Extracellular Vesicles: A Novel Avenue for Cancer Management. Front. Oncol. 11:638357. doi: 10.3389/fonc.2021.638357
Received: 06 December 2020; Accepted: 01 February 2021;
Published: 15 March 2021.
Edited by:
Jonathan Pol, Institut National de la Santé et de la Recherche Médicale (INSERM), FranceReviewed by:
Jafar Rezaie, Urmia University of Medical Sciences, IranCopyright © 2021 Gao, Qin, Wan, Sun, Meng, Huang, Hu, Jin and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Honglin Jin, amluQGh1c3QuZWR1LmNu; Kunyu Yang, eWFuZ2t1bnl1QGh1c3QuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.