- 1Department of Hematology, Guangzhou Women and Children’s Medical Center, Guangzhou Medical University, Guangzhou, China
- 2Pediatrics Department, The First Affiliated Hospital, Sun Yat-Sen University, Guangzhou, China
- 3Department of Pediatrics, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, Guangzhou, China
- 4Pediatric Center of Zhujiang Hospital, Southern Medical University, Guangzhou, China
- 5Department of Pediatrics, Nanfang Hospital, Southern Medical University, Guangzhou, China
- 6Division of Birth Cohort Study, Guangzhou Women and Children’s Medical Center, Guangzhou Medical University, Guangzhou, China
- 7Department of Pediatrics, The Third Affiliated Hospital, Sun Yat-sen University, Guangzhou, China
Objective: To reveal the contributing role of METTL3 gene SNPs in pediatric ALL risk.
Patients and Methods: A total of 808 pediatric ALL cases and 1,340 cancer-free controls from five hospitals in South China were recruited. A case-control study by genotyping three SNPs in the METTL3 gene was conducted. Genomic DNA was abstracted from peripheral blood. Three SNPs (rs1263801 C>G, rs1139130 A>G, and rs1061027 A>C) in the METTL3 gene were chosen to be detected by taqman real-time polymerase chain reaction assay.
Results: That rs1263801 C>G, rs1139130 A>G, and rs1061027 A>C polymorphisms were significantly associated with increased pediatric ALL risk was identified. In stratification analyses, it was discovered that rs1263801 CC, rs1061027 AA, and rs1139130 GG carriers were more likely to develop ALL in subgroups of common B-ALL, MLL gene fusion. Rs1263801 CC and rs10610257 AA carriers were more possible to increase the risk of ALL in subgroups of low hyperdiploid, and all of these three SNPs exhibited a trend toward the risk of ALL. All of these three polymorphisms were associated with the primitive/naïve lymphocytes and MRD in marrow after chemotherapy in ALL children. Rs1263801 CC and rs1139130 AA alleles provided a protective effect on MRD ≥0.01% among CCCG-treated children. As for rs1139130, AA alleles provided a protective effect on MRD in marrow ≥0.01% on 33 days and 12 weeks among CCCG-treated children, but provided a risk effect on MRD in the marrow ≥0.01% among SCCLG-treated children. As for rs1263801 CC and rs1139130 AA, these two alleles provided a protective effect on MRD in the marrow ≥0.01% among CCCG-treated children.
Conclusion: In this study, we revealed that METTL3 gene polymorphisms were associated with increased pediatric ALL risk and indicated that METTL3 gene polymorphisms might be a potential biomarker for choosing ALL chemotherapeutics.
Introduction
Acute lymphoblastic leukemia (ALL) is the most common type of pediatric cancer in the world; in China, it accounts for 70–80% of pediatric leukemia (1). ALL can be classified by immune cell phenotype as B-cell ALL and T-cell ALL. B-cell ALL is the most common ALL; T-cell ALL is typically more aggressive (2). As traditional chemotherapy combined with novel therapies makes great progress, higher survival rates and reduced morbidities have been achieved in ALL. Recently, the 5-year overall survival rate of ALL children younger than 14 years has been achieved >90% (3). However, recurrence occurs in 15–20% of ALL children, and 15% pediatric ALL patients were therapeutic failures, which resulted in early age mortality (4). ALL is characterized by multiple genetic alterations (5).
Heritable variations in genes are risk factors for ALL and play a strong role in the development of pediatric ALL (6). Populations with different races are well distinguished by genetic polymorphisms. Genome-wide association studies (GWAS) have identified a number of loci, and single nucleotide polymorphism (SNP) associations in several genes are associated with the risk of ALL. Genetic alterations in pediatric ALL are found to be very different from those in adult ALL (7).
N6-methyladenosine (m6A) is the most abundant internal modification of messenger RNAs (mRNAs) in eukaryotic organisms. Methylation at the sixth N atom on adenine base is m6A. M6A regulates mRNA expression posttranscriptionally in a dynamic and reversible manner (8). M6A modification is regulated by several key regulators, including writers [RNA methyltransferase complex methyltransferase-like 3 (METTL3)/methyltransferase-like 14 (METTL14)/Wilms’ tumor 1-associating protein (WTAP)], erasers [demethylases fat mass and obesity-associated protein (FTO) and AlkB homolog 5 (ALKBH5)], and readers (YTHD family proteins) (9). It was reported that dysregulation of m6A is associated with multiple tumors including acute myeloid leukemia (AML) (10). M6A methylation writer METTL3 was discovered playing an oncogenic role in carcinogenesis, such as colorectal carcinoma (11), bladder cancer (12), breast cancer (13), etc. METTL3 mRNA and protein are expressed abundantly in AML cells, and their depletion induces cell differentiation and apoptosis and delays leukemia progression (14). Some genetic variations in m6A-related gene regions may affect m6A methylation, subsequently regulating mRNA expression (15). Studies identified that m6A-associated SNPs were potential functional variants for periodontitis (16) and coronary artery disease (17). Genetic alterations in the m6A demethyltransferase FTO gene were shown to be associated with ALL and AML risk, and there is evidence that indicates dysregulation of m6A methyltransferase METTL3 in AML (18, 19). However the relationship between genetic variations of the METTL3 gene and ALL is still unclear.
In the present study, a total of three SNPs were selected to assess the relationship between METTL3 polymorphisms and pediatric ALL. The current study was a case-control study that was performed using samples from five hospitals in South China.
Materials and Methods
Study Subjects
A Southern Chinese population that included 808 pediatric ALL patients and 1,340 age-matched, gender-matched, and ethnicity-matched healthy controls is summarized in Table S1. ALL cases were collected from Guangzhou Women and Children’s Medical Center (GWCMC), Guangzhou Medical University (n=582), The First Affiliated Hospital, Sun Yat-sen University (n=74), Sun Yat-sen Memorial Hospital, Sun Yat-sen University (n=26), Nanfang Hospital, Southern Medical University (SMU) (n=100), and Zhujiang Hospital, Southern Medical University (n=26), from January 2017 to May 2019. All children were diagnosed with ALL by at least two hematologists. The control subjects were free from any type of hematological diseases or any other malignancy or autoimmune disorder and were recruited from the same hospital.
The major clinical and biological characteristics of the ALL children, including age, gender, immunophenotype, gene fusion type, risk level, karyotype, clinical manifestations, rate of primitive/naive lymphocytes in the marrow, and minimal residual disease on 19 days, 33 days, and 12 weeks after chemotherapy and chemotherapy regimen were collected. The information is summarized in Table S1.
The study was approved by the institutional ethics committee of every participating hospital, and written informed consent was acquired from all participants in accordance with the Declaration of Helsinki.
METTL3 SNPs Selection and Genotyping
The included potentially functional candidate SNPs were selected as follows: located in the 5’ untranslated region, 3’ untranslated region, 5’ flanking region, and exon of the METTL3 gene. The NCBI dbSNP database (http://www.ncbi.nlm.nih.gov/projects/SNP) and the SNPinfo (https://snpinfo.niehs.nih.gov/snpinfo/snpfunc.html) online software were used to perform the above selection. Three SNPs (rs1263801 C>G, rs1139130 G>A, and rs1061027 A>C) in the METTL3 gene were chosen. Genomic DNA was extracted from peripheral blood. The reaction system and condition of the Taqman RT-PCR assay was according to the published reference (20, 21). To ensure the accuracy of these genotyping results, 10% of the samples were randomly selected to be genotyped by a DNA sequencing method. A concordance rate of 100% for the quality control samples was obtained (21).
Statistical Analysis
The goodness-of-fit χ2 test was performed to assess if the selected METTL3 SNPs deviated from Hardy–Weinberg equilibrium among controls. The two-sided χ2 test was used to compare demographic variables and genotype frequencies of the cases and controls. ORs and their corresponding 95% CIs were computed by unconditional logistic regression analyses with or without adjustment for age and gender. The SAS statistical package (version 9.1; SAS Institute, Cary, NC) was used to perform all statistical analyses. All reported p values were two sided, and a p value < 0.05 was considered statistically significant.
Results
Population Characteristics
The demographic and clinical characteristics data of ALL cases and cancer-free controls are summarized in Table S1. No significant differences were observed between cases and controls for the Southern Chinese children regarding age (p=0.082) and gender (p=0.059). Among ALL cases, 28.22% (228 cases) were pro B cell ALL, 35.27% (285 cases) were common B cell ALL, 20.67% (167 cases) were pre-B cell ALL, 0.67% (3 cases) were mature B ALL, 8.54% (69 cases) were T cell ALL, and 6.93% (56 cases) were undefined immunophenotype. Regarding the gene fusion type, 3.34% (27 cases) had BCR-ABL gene fusion, 16.83% (136 cases) had TEL-AML, 2.97% (24 cases) had E2A-PBX, 0.99% (8 cases) had SIL-TAL, 1.98% (16 cases) had MLL, 3.09% (25 cases) had other gene fusions, 68.19% (551 cases) were normal, and 21 were undefined. A total of 258 patients (33.73%) were with low risk, 360 cases (47.06%) were with medium risk, 77 cases (10.07%) were with high risk, and 70 cases (9.15%) were undefined. Regarding the karyotype, 64.40% (517 cases) were normal diploid, 5.25% (45 cases) were abnormal diploid, 2.69% (22 cases) were hypodiploid, 3.46% (27 cases) were low hyperdiploid, and 7.81% (61 cases) were high hyperdiploid.
Correlation of METTL3 Gene Polymorphisms With ALL Risk
The genotype frequencies of METTL3 associated with ALL risk are shown in Table 1. In the single-locus analysis, carriers of rs1263801 (CC vs. GG: adjusted OR= 4.18, 95% CI=3.21–5.43, p<0.001) and rs1061027 (CA vs. CC: adjusted OR=2.42, 95% CI=2.00–2.94, p<0.001; AA vs. CC: adjusted OR=6.21, 95% CI=4.38–8.81, p<0.001) variant alleles showed significant enhanced risk of pediatric ALL. On the contrary, rs1139130 (GA vs. GG: adjusted OR=1.41, 95% CI=1.15–1.73, p=0.001; AA vs. GG: adjusted OR=1.52, 95% CI=1.81–3.06, p<0.001) variant alleles contribute to decreased risk of pediatric ALL.
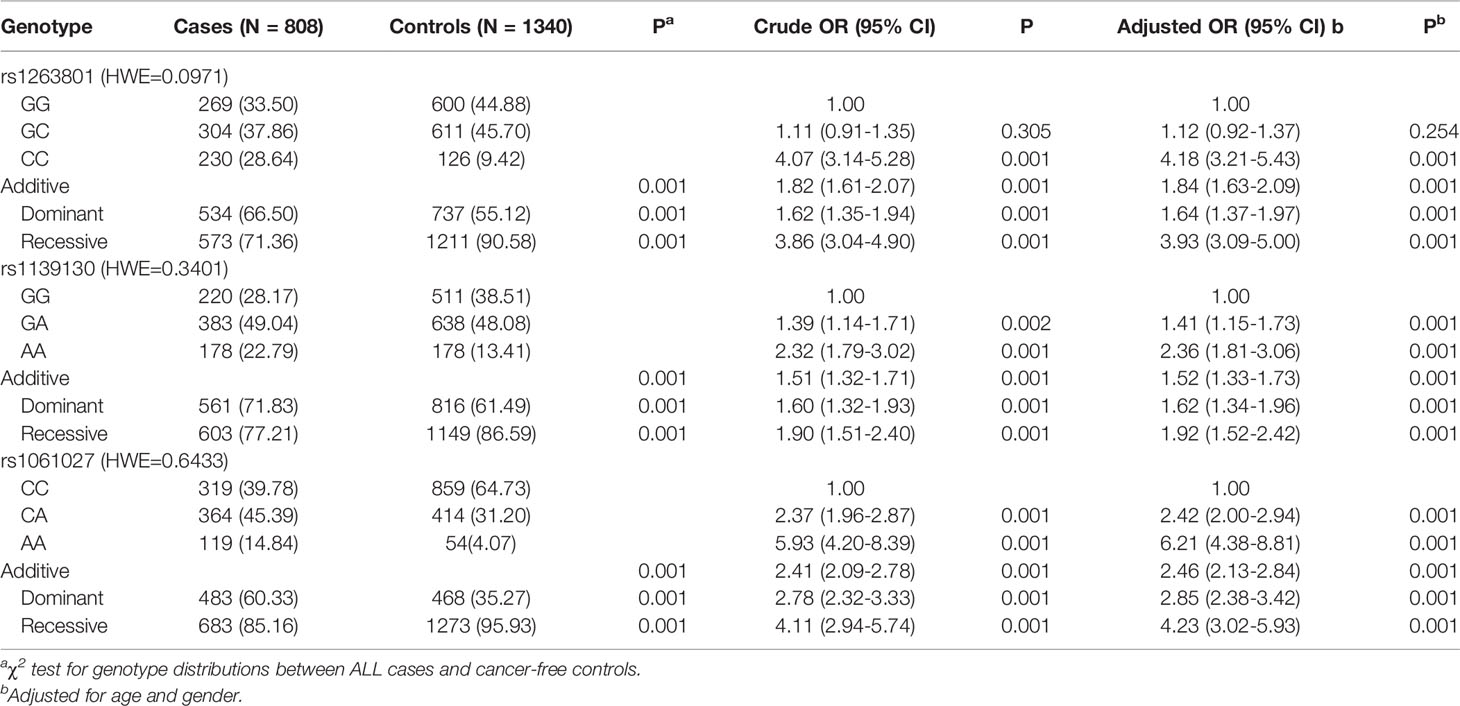
Table 1 Logistic regression analysis of associations between METTL3 polymorphisms and ALL susceptibility.
Stratification Analysis of Identified SNPs
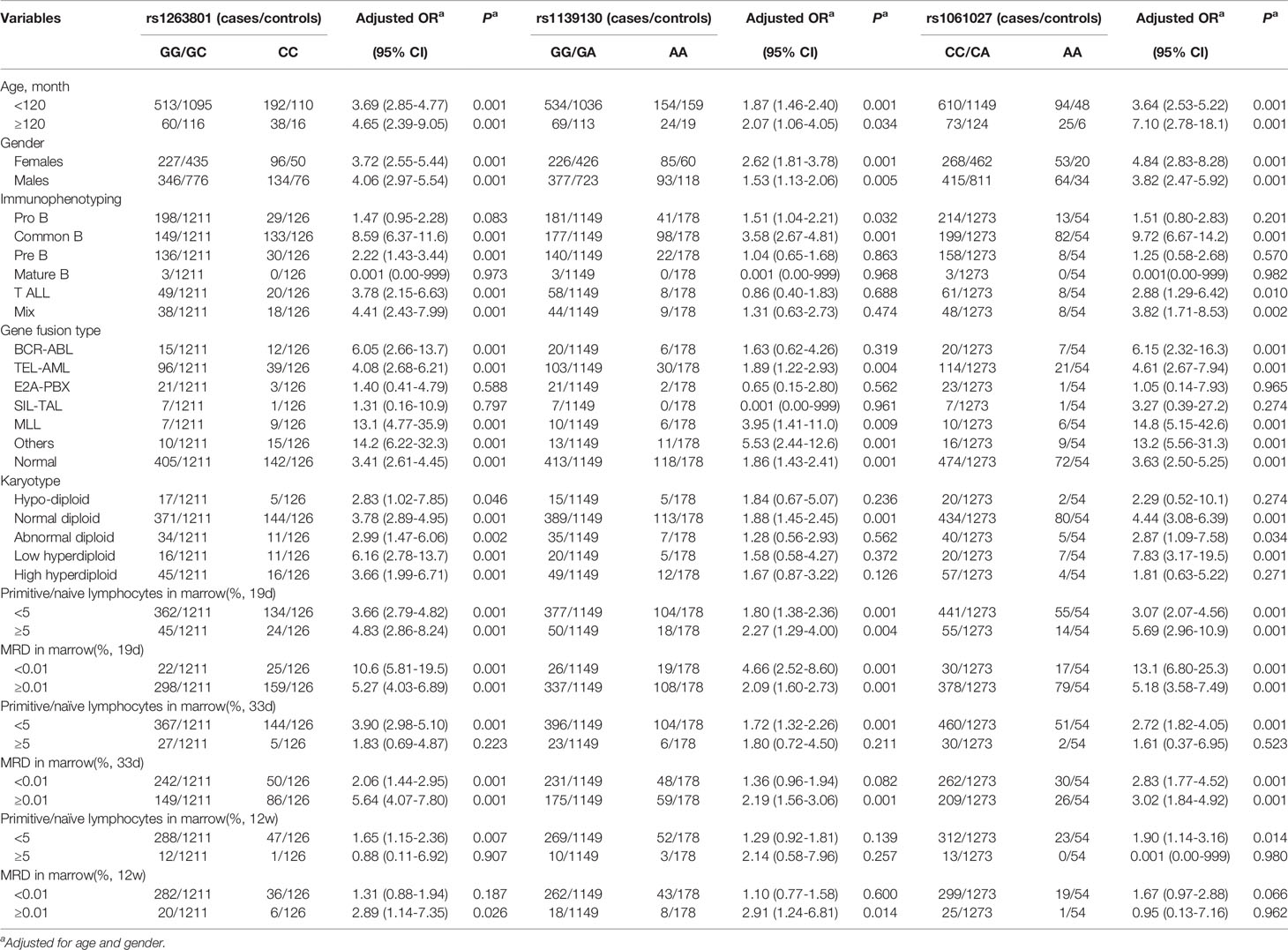
We further analyzed whether the selected METTL3 polymorphisms (rs1263801 C>G, rs1139130 A>G, and rs1061027 A>C) preferentially predispose to any specific subtype of ALL (Table 2). A stronger risk effect of rs1263801 was found among children older than 10 years (adjusted OR= 4.65, 95% CI=2.39–9.05, p<0.001), male (adjusted OR= 4.06, 95% CI=2.97–5.54, p=0.001), common B subtype ALL (adjusted OR= 8.59, 95% CI=6.37–11.6, p<0.001), MLL gene fusion type (adjusted OR= 13.1, 95% CI=4.77–35.9, p<0.001), low hyperdiploid (adjusted OR= 6.16, 95% CI=2.78–13.7, p<0.001), primitive/naive lymphocytes in marrow ≥ 5% on 19 days (adjusted OR=4.83, 95% CI=2.86–8.24, p<0.001) after chemotherapy, primitive/naive lymphocytes in marrow < 5% on 33 days (adjusted OR= 3.90, 95% CI=2.98–5.10, p<0.001) and 12 weeks (adjusted OR=1.65, 95% CI=1.15–2.36, p=0.007) after chemotherapy, MRD in marrow < 0.01% on 19 days (adjusted OR=10.6, 95% CI=5.81–19.5, p<0.001), MRD ≥ 0.01% on 33 days (adjusted OR=5.64, 95% CI=4.07–7.80–8.24, p<0.001), and ≥0.01% on 12 weeks (adjusted OR=2.89, 95% CI=1.14–7.35, p=0.026). As for the rs1139130 polymorphism, a more significant risk association was identified with those children age ≥10 years (adjusted OR=2.07, 95% CI= 1.06–4.05, p=0.034), female (adjusted OR=2.62, 95% CI= 18.1–3.78, p<0.001), common B subtype (adjusted OR=3.58, 95% CI= 2.67–4.81, p<0.001), MLL gene fusion type (adjusted OR=3.95, 95% CI=1.41–11.0, p=0.009), normal diploid (adjusted OR=1.88, 95% CI=1.45–2.45, p<0.001), primitive/naive lymphocytes in marrow ≥ 5% on 19 days (adjusted OR= 2.27, 95% CI=1.29–4.00, p<0.001) and <5% on 33 days (adjusted OR=1.72, 95% CI=1.32–2.26, p<0.001) after chemotherapy, MRD in marrow <0.01% on 19 days (adjusted OR= 4.66, 95% CI=2.52–8.60, p<0.001), MRD ≥ 0.01% on 33 days (adjusted OR= 2.19, 95% CI=1.56–3.06, p<0.001), and ≥0.01% on 12 weeks (adjusted OR= 2.91, 95% CI=1.24–6.81, p=0.014). As for the rs1061027 polymorphism, a stronger risk association was revealed with those children age ≥10 years (adjusted OR=7.10, 95% CI= 2.78–18.1, p<0.001), female (adjusted OR=4.84, 95% CI= 2.83–8.28, p<0.001), common B subtype (adjusted OR=9.72, 95% CI= 6.67–14.2, p<0.001), MLL gene fusion type (adjusted OR=14.8, 95% CI= 5.15–42.6, p<0.001), low hyperdiploid (adjusted OR=7.83, 95% CI= 3.17–19.5, p<0.001), primitive/naive lymphocytes in marrow ≥5% on 19 days (adjusted OR= 5.69, 95% CI=2.96–10.9, p<0.001) and <5% on 33 days (adjusted OR=2.72, 95% CI= 1.82–4.05, p<0.001) after chemotherapy, MRD in marrow <0.01% on 19 days (adjusted OR= 13.1, 95% CI=6.80–25.3, p<0.001), and MRD ≥0.01% on 33 days (adjusted OR= 3.02, 95% CI=1.84–4.92, p<0.001). No correlation was found between the rs1061027 polymorphism and MRD on 12 weeks.
Association of METTL3 Polymorphisms With Chemotherapeutics in Southern Chinese Pediatric ALL Patients
All patients were treated with Chinese Children Cancer Group chemotherapeutics (CCCG) or South China Children Leukemia Group chemotherapeutics (SCCLG). We compared the MRD in the marrow of patients with different alleles after being treated with CCCG and SCCLG (Table 3). As for rs1263801, CC alleles provided a protective effect on MRD in the marrow ≥0.01% on 33 days (adjusted OR= 0.17, 95% CI= 0.10–0.30, p<0.001) and 12 weeks (adjusted OR= 0.30, 95% CI= 0.10–0.90, p=0.030) among CCCG-treated children. As for rs1139130, AA alleles provided a protective effect on MRD in marrow ≥0.01% on 33 days (adjusted OR= 0.50, 95% CI= 0.29–0.83, p=0.008) and 12 weeks (adjusted OR= 0.32, 95% CI= 0.12–0.87, p=0.030) among CCCG-treated children but provided a risk effect on MRD in marrow ≥0.01% among SCCLG-treated children (adjusted OR=5.70, 95% CI=1.37–23.7, p=0.017). As for rs1061.27, AA alleles provided a risk effect on MRD in the marrow ≥0.01% among CCCG-treated children (adjusted OR=8.63, 95% CI=2.31–32.3, p=0.002). These results indicated that SCCLG chemotherapeutics is more suitable for rs1263801 CC and rs1139130 AA carriers; CCCG chemotherapeutics is more efficient for rs1061027 AA carriers.
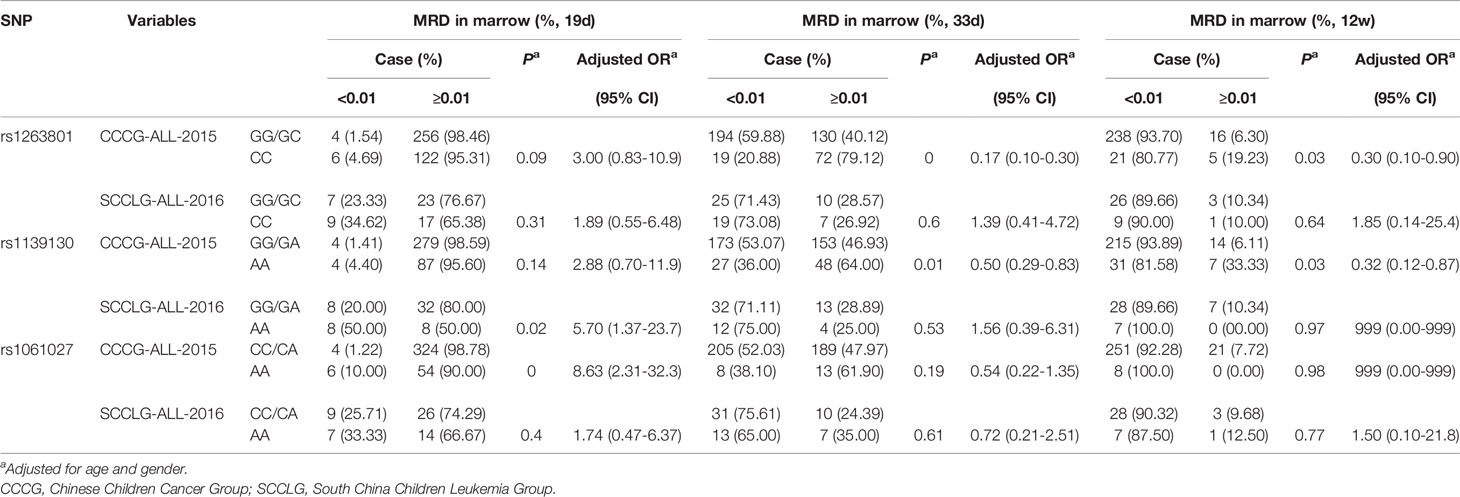
Table 3 The correlation between METTL3 polymorphisms and South China pediatric ALL patients’ response to different chemotherapeutics.
Discussion
In the current case-control study with 808 pediatric ALL case and 1,340 healthy controls from Southern Chinese populations, we explored the potential association between METTL3 gene polymorphisms and pediatric ALL risk. We certificated that three polymorphisms, namely rs1263801 C>G, rs1139130 A>G, and rs1061027 A>C, were associated with an increased susceptibility of pediatric ALL. To our knowledge, this study is the first to identify the association between METTL3 polymorphisms and pediatric ALL susceptibility.
Epigenetic alterations, including DNA methylation, histone modifications, and noncoding RNAs, have been reported to contribute to ALL progression (22). In recent years, another epigenetic modification, RNA methylation, is considered to play an important role in carcinogenesis (11). M6A is the most common modification of RNA on the posttranscriptional level, mainly in mRNA and long noncoding RNA (lncRNA) (23). The complex composed of METTL3, METTL14, and WTAP induces m6A-methylation of mRNA or lncRNA. METTL3 is the essential component of the complex. Dysregulation of METTL3 was identified to be a key role in the progression of multiple malignant tumors, such as endometrial cancer (24), bladder cancer (25), pancreatic cancer (26), etc. A number of articles infer that METTL3 can promote tumor progression through multiple mechanisms. METTL3 can promote growth, survival, and invasion by interacting with the translation initiation element to enhance mRNA translation in lung adenocarcinoma (27). Lin et al. (28) revealed that deletion of METTL3 could impair the epithelial-mesenchymal transition (EMT). In breast cancer, METTL3 is upregulated by HBXIP and promotes the cancer progression by suppressing let-7g (29). METTL3 promotes self-renewal of glioblastoma stem cells to induce tumorigenesis (30). METTL3 can directly interact with the eukaryotic translation initiation factor e subunit h (eIF3h). The interaction between METTL3 and eIF3h is essential for translation and oncogenic transformation in lung cancer (31). Promoter-bound METTL3 promotes m6A modification within the coding region of mRNA transcript and enhances translation by inhibiting ribosome stalling. METTL3 regulates mRNA expression in this way to facilitate the progression of acute myeloid leukemia (32). However, the function of METTL3 in ALL is still unknown.
Herein, we investigated whether METTL3 gene polymorphisms could influence the susceptibility of ALL in South China children for the first time. With regard to the remaining three METTL3 gene polymorphisms (rs1263801 C>G, rs1139130 A>G, and rs1061027 A>C), we identified the association between these three SNPs and pediatric ALL susceptibility. The location and predicted function was analyzed using the online software SNPinfo. The rs1263801 C>G polymorphism was located in intron 1 of the METTL3 gene and was predicted to be the transcriptional factor binding site. The rs1139130 A>G located in the exon 5 of the METTL3 gene was predicted to affect splicing and protein function. The rs1061027 A>C polymorphism located in intron 8 was predicted to be associated with miRNA function. In 2018, Bertero et al. reported that the interactome of transcriptional factors SMAD2/3 promoted another transcriptional factor TGFβ to control the METTL3/METTL14/WTAP complex mediated m6A mRNA methylation in human pluripotent stem cells (33). Xia et al. reported that Zmettl3 mutation disrupts gamete maturation and reduces fertility in zebrafish (34). Other studies identified that METTL3 mRNA could be targeted by miR-600 (35) and miR-33a (36). However, there was no evidence certifying that genetic variations of METTL3 could affect the transcriptional factor or miRNAs binding to METLL3 and the coding of METTL3 mRNA. Our results suggested that the rs1263801 CC phenotype, rs1139130 GG phenotypes, and rs1061027 CA/CC phenotypes are associated with an increased risk of pediatric ALL in South China. Lin et al. reported that the combination of rs1139130, rs1263801, rs1061026, and rs1061027 reduced the risk of Wilms tumor in Chinese children (37). Bian et al. (38) identified that these four polymorphisms were associated with an increased risk of neuroblastoma. It suggested that METTL3 polymorphisms function diversely in different tumors.
We next examined whether the METTL3 SNP genotype preferentially predisposes to any pediatric ALL subtype, including immunophenotype, gene fusion type, karyotype, primitive/naïve lymphocytes, and MRD in the marrow after chemotherapy. The METTL3 rs1263801 CC phenotype and the rs1061027 AA phenotype were considered to increase the risk of ALL in the B-ALL, mature B ALL, and T-ALL subtype. In BCR-ABL, TEL-AML, and MLL gene fusion types, rs1263801 CC phenotype and rs1061027 AA phenotype carriers showed a higher risk for ALL. The rs1139130 GG carriers were revealed to have a higher risk for ALL in B-ALL, mature B ALL subtype, and medium risk level subtype. We failed to identify the association between the FAB subtype and these three METTL3 polymorphisms.
In stratification analysis, we tried to reveal the relationship between clinical characteristic, response to different chemotherapeutics, and METTL3 polymorphisms. The results showed that rs1263801 C>G, rs1139130 A>G, and rs1061027 A>C could remarkably increase the risk of the common B type and MLL fusion type ALL in Southern Chinese children. All these three selected polymorphisms were more strongly associated with the primitive/naïve lymphocytes over 5% and MRD less than 0.01% on the 19th day, and also with the primitive/naïve lymphocytes less than 5% and MRD more than 0.01% on the 33rd day after chemotherapy. After chemotherapy treatment of 12 weeks, rs1263801 C>G and rs1061027 A>C were identified to increase susceptibility to primitive/naïve lymphocytes less than 5%; rs1263801 C>G and rs1139130 A>G may increase susceptibility to MRD more than 0.01% in ALL patients. And we also identified that SCCLG chemotherapeutics was more suitable for rs1263801 CC and rs1139130 AA carriers; CCCG chemotherapeutics was more efficient for rs1061027 AA carriers.
Several limitations should be noted in the current study. First, the sample size was not large enough. Second, this was a retrospective study; information bias and selection bias were inevitable. We have reduced these biases by frequency-matching of cases and controls by age and gender, and recruiting subjects from six hospitals in South China. Third, our study focused on the analysis of genetic factors in pediatric ALL risk. However, other important information such as environment and dietary intake was not available for analysis. Finally, the association between METTL3 gene polymorphisms and prognosis of pediatric ALL was not analyzed in the current study.
In summary, our results suggest that polymorphisms rs1263801 C>G, rs1139130 A>G, and rs1061027 A>C in the METTL3 gene were significantly associated with increased pediatric ALL risk, and SCCLG chemotherapeutics is more suitable for rs1263801 CC and rs1139130 AA carriers; CCCG chemotherapeutics is more efficient for rs1061027 AA carriers in the Southern Chinese ALL children. Further studies are necessary to elucidate the biological function of METTL3 gene risk SNPs in the etiology of pediatric ALL.
Conclusion
METTL3 gene polymorphisms were associated with increased pediatric ALL risk. These three polymorphisms (rs1263801 C>G, rs1139130 A>G, and rs1061027 A>C) were likely to contribute to the sensitivity of different chemotherapies in pediatric ALL. The results indicated that METTL3 gene polymorphisms might be a potential biomarker for ALL susceptibility and when choosing chemotherapeutics.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The study was approved by the institutional ethics committee of Guangzhou Women and Children’s Medical Center, Guangzhou Medical University; The First Affiliated Hospital, Sun Yat-sen University; Sun Yat-sen Memorial Hospital, Sun Yat-sen University; Nanfang Hospital, Southern Medical University; and Zhujiang Hospital, Southern Medical University. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Author Contributions
XpL and LH are equal to this work in writing the manuscript. KH, LY, XW, JW, and YC collected ALL blood samples. XY analyzed the data. AL, MC, XdL, and YY performed qPCR. LX and HJ supplied the idea and funding. All authors contributed to the article and approved the submitted version.
Funding
National Natural Science Foundation of China (81672496 and 81870115), Natural Science Foundation of Guangdong Province (2020A1515010188), and Guangzhou Municipal Science and Technology Project (201804010042).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the Clinical Biological Resource Bank of Guangzhou Women and Children’s Medical Center for providing part of the clinical samples.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.635251/full#supplementary-material
References
1. Wu C, Li W. Genomics and Pharmacogenomics of Pediatric Acute Lymphoblastic Leukemia. Crit Rev Oncol Hematol (2018) 126:100–11. doi: 10.1016/j.critrevonc.2018.04.002
2. Schiffman JD. Applying Molecular Epidemiology in Pediatric Leukemia. J Investig Med (2016) 64(2):355–60. doi: 10.1097/JIM.0000000000000204
3. Jabbour E, Pui CH, Kantarjian H. Progress and Innovations in the Management of Adult Acute Lymphoblastic Leukemia. JAMA Oncol (2018) 4(10):1413–20. doi: 10.1001/jamaoncol.2018.1915
4. Eryilmaz E, Canpolat C. Novel Agents for the Treatment of Childhood Leukemia: An Update. Onco Targets Ther (2017) 10:3299–306. doi: 10.2147/OTT.S126368
5. Horowitz NA, Akasha D, Rowe JM. Advances in the Genetics of Acute Lymphoblastic Leukemia in Adults and the Potential Clinical Implications. Expert Rev Hematol (2018) 11(10):781–91. doi: 10.1080/17474086.2018.1509702
6. Enciso-Mora V, Hosking FJ, Sheridan E, Kinsey SE, Lightfoot T, Roman E, et al. Common Genetic Variation Contributes Significantly to the Risk of Childhood B-Cell Precursor Acute Lymphoblastic Leukemia. Leukemia (2012) 26(10):2212–5. doi: 10.1038/leu.2012.89
7. de Smith AJ, Walsh KM, Francis SS, Zhang C, Hansen HM, Smirnov I, et al. BMI1 Enhancer Polymorphism Underlies Chromosome 10p12.31 Association With Childhood Acute Lymphoblastic Leukemia. Int J Cancer (2018) 143(11):2647–58. doi: 10.1002/ijc.31622
8. Deng X, Su R, Weng H, Huang H, Li Z, Chen J. RNA N(6)-Methyladenosine Modification in Cancers: Current Status and Perspectives. Cell Res (2018) 28(5):507–17. doi: 10.1038/s41422-018-0034-6
9. Weng H, Huang H, Chen J. RNA N (6)-Methyladenosine Modification in Normal and Malignant Hematopoiesis. Adv Exp Med Biol (2019) 1143:75–93. doi: 10.1007/978-981-13-7342-8_4
10. Huang Y, Su R, Sheng Y, Dong L, Dong Z, Xu H, et al. Small-Molecule Targeting of Oncogenic FTO Demethylase in Acute Myeloid Leukemia. Cancer Cell (2019) 35(4):677–91.e10. doi: 10.1016/j.ccell.2019.03.006
11. Li T, Hu PS, Zuo Z, Lin JF, Li X, Wu QN, et al. METTL3 Facilitates Tumor Progression via an M(6)A-IGF2BP2-Dependent Mechanism in Colorectal Carcinoma. Mol Cancer (2019) 18(1):112. doi: 10.1186/s12943-019-1038-7
12. Han J, Wang JZ, Yang X, Yu H, Zhou R, Lu HC, et al. METTL3 Promote Tumor Proliferation of Bladder Cancer by Accelerating Pri-Mir221/222 Maturation in M6a-Dependent Manner. Mol Cancer (2019) 18(1):110. doi: 10.1186/s12943-019-1036-9
13. Wu L, Wu D, Ning J, Liu W, Zhang D. Changes of N6-Methyladenosine Modulators Promote Breast Cancer Progression. BMC Cancer (2019) 19(1):326. doi: 10.1186/s12885-019-5538-z
14. Vu LP, Pickering BF, Cheng Y, Zaccara S, Nguyen D, Minuesa G, et al. The N(6)-Methyladenosine (M(6)A)-Forming Enzyme METTL3 Controls Myeloid Differentiation of Normal Hematopoietic and Leukemia Cells. Nat Med (2017) 23(11):1369–76. doi: 10.1038/nm.4416
15. Zheng Y, Nie P, Peng D, He Z, Liu M, Xie Y, et al. M6avar: A Database of Functional Variants Involved in M6a Modification. Nucleic Acids Res (2018) 46(D1):D139–45. doi: 10.1093/nar/gkx895
16. Lin W, Xu H, Wu Y, Wang J, Yuan Q. In Silico Genome-Wide Identification of M6a-Associated SNPs as Potential Functional Variants for Periodontitis. J Cell Physiol (2020) 235(2):900–8. doi: 10.1002/jcp.29005
17. Mo XB, Lei SF, Zhang YH, Zhang H. Detection of M(6)A-Associated SNPs as Potential Functional Variants for Coronary Artery Disease. Epigenomics (2018) 10(10):1279–87. doi: 10.2217/epi-2018-0007
18. Szymon S, Bik-Multanowski M, Balwierz W, Pietrzyk JJ, Surmiak M, Strojny W, et al. Homozygosity for the Rs9939609t Allele of the FTO Gene may Have Protective Effect on Becoming Overweight in Survivors of Childhood Acute Lymphoblastic Leukaemia. J Genet (2011) 90(2):365–8. doi: 10.1007/s12041-011-0089-3
19. Kwiecinska K, Strojny W, Pietrys D, Bik-Multanowski M, Siedlar M, Balwierz W, et al. Late Effects in Survivors of Childhood Acute Lymphoblastic Leukemia in the Context of Selected Gene Polymorphisms. Ital J Pediatr (2018) 44(1):92. doi: 10.1186/s13052-018-0526-5
20. He J, Wang F, Zhu J, Zhang R, Yang T, Zou Y, et al. Association of Potentially Functional Variants in the XPG Gene With Neuroblastoma Risk in a Chinese Population. J Cell Mol Med (2016) 20(8):1481–90. doi: 10.1111/jcmm.12836
21. He J, Zou Y, Liu X, Zhu J, Zhang J, Zhang R, et al. Association of Common Genetic Variants in Pre-microRNAs and Neuroblastoma Susceptibility: A Two-Center Study in Chinese Children. Mol Ther Nucleic Acids (2018) 11:1–8. doi: 10.1016/j.omtn.2018.01.003
22. Cruz-Rodriguez N, Combita AL, Zabaleta J. Epigenetics in Hematological Malignancies. Methods Mol Biol (2018) 1856:87–101. doi: 10.1007/978-1-4939-8751-1_5
23. Alarcon CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-Methyladenosine Marks Primary microRNAs for Processing. Nature (2015) 519(7544):482–5. doi: 10.1038/nature14281
24. Liu J, Eckert MA, Harada BT, Liu SM, Lu Z, Yu K, et al. M(6)A mRNA Methylation Regulates AKT Activity to Promote the Proliferation and Tumorigenicity of Endometrial Cancer. Nat Cell Biol (2018) 20(9):1074–83. doi: 10.1038/s41556-018-0174-4
25. Jin H, Ying X, Que B, Wang X, Chao Y, Zhang H, et al. N(6)-Methyladenosine Modification of ITGA6 mRNA Promotes the Development and Progression of Bladder Cancer. EBioMedicine (2019) 47:195–207. doi: 10.1016/j.ebiom.2019.07.068
26. Zhang J, Bai R, Li M, Ye H, Wu C, Wang C, et al. Excessive miR-25-3p Maturation via N(6)-Methyladenosine Stimulated by Cigarette Smoke Promotes Pancreatic Cancer Progression. Nat Commun (2019) 10(1):1858. doi: 10.1038/s41467-019-09712-x
27. Lin S, Choe J, Du P, Triboulet R, Gregory RI. The M(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol Cell (2016) 62(3):335–45. doi: 10.1016/j.molcel.2016.03.021
28. Lin X, Chai G, Wu Y, Li J, Chen F, Liu J, et al. RNA M(6)A Methylation Regulates the Epithelial Mesenchymal Transition of Cancer Cells and Translation of Snail. Nat Commun (2019) 10(1):2065. doi: 10.1038/s41467-019-09865-9
29. Cai X, Wang X, Cao C, Gao Y, Zhang S, Yang Z, et al. HBXIP-Elevated Methyltransferase METTL3 Promotes the Progression of Breast Cancer via Inhibiting Tumor Suppressor Let-7g. Cancer Lett (2018) 415:11–9. doi: 10.1016/j.canlet.2017.11.018
30. Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, et al. M(6)A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell Rep (2017) 18(11):2622–34. doi: 10.1016/j.celrep.2017.02.059
31. Choe J, Lin S, Zhang W, Liu Q, Wang L, Ramirez-Moya J, et al. mRNA Circularization by METTL3-Eif3h Enhances Translation and Promotes Oncogenesis. Nature (2018) 561(7724):556–60. doi: 10.1038/s41586-018-0538-8
32. Barbieri I, Tzelepis K, Pandolfini L, Shi J, Millan-Zambrano G, Robson SC, et al. Promoter-Bound METTL3 Maintains Myeloid Leukaemia by M(6)A-Dependent Translation Control. Nature (2017) 552(7683):126–31. doi: 10.1038/nature24678
33. Bertero A, Brown S, Madrigal P, Osnato A, Ortmann D, Yiangou L, et al. The SMAD2/3 Interactome Reveals That TGFbeta Controls M(6)A mRNA Methylation in Pluripotency. Nature (2018) 555(7695):256–9. doi: 10.1038/nature25784
34. Xia H, Zhong C, Wu X, Chen J, Tao B, Xia X, et al. Mettl3 Mutation Disrupts Gamete Maturation and Reduces Fertility in Zebrafish. Genetics (2018) 208(2):729–43. doi: 10.1534/genetics.117.300574
35. Wei W, Huo B, Shi X. miR-600 Inhibits Lung Cancer via Downregulating the Expression of METTL3. Cancer Manag Res (2019) 11:1177–87. doi: 10.2147/CMAR.S181058
36. Du M, Zhang Y, Mao Y, Mou J, Zhao J, Xue Q, et al. MiR-33a Suppresses Proliferation of NSCLC Cells via Targeting METTL3 mRNA. Biochem Biophys Res Commun (2017) 482(4):582–9. doi: 10.1016/j.bbrc.2016.11.077
37. Lin A, Zhou M, Hua RX, Zhang J, Zhou H, Li S, et al. METTL3 Polymorphisms and Wilms Tumor Susceptibility in Chinese Children: A Five-Center Case-Control Study. J Gene Med (2020) 22(11):e3255. doi: 10.1002/jgm.3255
Keywords: methyltransferase-like 3, acute lymphoid leukemia, polymorphism, pediatric, susceptibility
Citation: Liu X, Huang L, Huang K, Yang L, Yang X, Luo A, Cai M, Wu X, Liu X, Yan Y, Wen J, Cai Y, Xu L and Jiang H (2021) Novel Associations Between METTL3 Gene Polymorphisms and Pediatric Acute Lymphoblastic Leukemia: A Five-Center Case-Control Study. Front. Oncol. 11:635251. doi: 10.3389/fonc.2021.635251
Received: 30 November 2020; Accepted: 23 July 2021;
Published: 09 September 2021.
Edited by:
Hua Tan, University of Texas Health Science Center at Houston, United StatesReviewed by:
Matteo Chinello, Integrated University Hospital Verona, ItalyMeenakshi Devidas, St. Jude Children’s Research Hospital, United States
Copyright © 2021 Liu, Huang, Huang, Yang, Yang, Luo, Cai, Wu, Liu, Yan, Wen, Cai, Xu and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hua Jiang, amlhbmdfaHVhMThAc2luYS5jbg==; Ling Xu, bHVveHVsNjRAMTI2LmNvbQ==
†These authors have contributed equally to this work
 Xiaoping Liu1†
Xiaoping Liu1† Lihua Yang
Lihua Yang Xu Yang
Xu Yang Xuedong Wu
Xuedong Wu Ling Xu
Ling Xu Hua Jiang
Hua Jiang