- 1Department of Radiation Oncology, Fudan University Shanghai Cancer Center, Shanghai, China
- 2Department of Oncology, Shanghai Medical College, Fudan University, Shanghai, China
- 3Department of Oncology Intervention, The Affiliated Municipal Hospital of Taizhou University, Taizhou, China
- 4Department of Infectious Disease, The Affiliated Municipal Hospital of Taizhou University, Taizhou, China
- 5Department of Radiology, The Affiliated Municipal Hospital of Taizhou University, Taizhou, China
- 6Department of Hepatobiliary Surgery, The Affiliated Municipal Hospital of Taizhou University, Taizhou, China
Purpose/Objectives(s): Salivary gland tumors are a rare, histologically heterogeneous group of tumors. The distinction between malignant and benign tumors of the parotid gland is clinically important. This study aims to develop and evaluate a deep-learning network for diagnosing parotid gland tumors via the deep learning of MR images.
Materials/Methods: Two hundred thirty-three patients with parotid gland tumors were enrolled in this study. Histology results were available for all tumors. All patients underwent MRI scans, including T1-weighted, CE-T1-weighted and T2-weighted imaging series. The parotid glands and tumors were segmented on all three MR image series by a radiologist with 10 years of clinical experience. A total of 3791 parotid gland region images were cropped from the MR images. A label (pleomorphic adenoma and Warthin tumor, malignant tumor or free of tumor), which was based on histology results, was assigned to each image. To train the deep-learning model, these data were randomly divided into a training dataset (90%, comprising 3035 MR images from 212 patients: 714 pleomorphic adenoma images, 558 Warthin tumor images, 861 malignant tumor images, and 902 images free of tumor) and a validation dataset (10%, comprising 275 images from 21 patients: 57 pleomorphic adenoma images, 36 Warthin tumor images, 93 malignant tumor images, and 89 images free of tumor). A modified ResNet model was developed to classify these images. The input images were resized to 224x224 pixels, including four channels (T1-weighted tumor images only, T2-weighted tumor images only, CE-T1-weighted tumor images only and parotid gland images). Random image flipping and contrast adjustment were used for data enhancement. The model was trained for 1200 epochs with a learning rate of 1e-6, and the Adam optimizer was implemented. It took approximately 2 hours to complete the whole training procedure. The whole program was developed with PyTorch (version 1.2).
Results: The model accuracy with the training dataset was 92.94% (95% CI [0.91, 0.93]). The micro-AUC was 0.98. The experimental results showed that the accuracy of the final algorithm in the diagnosis and staging of parotid cancer was 82.18% (95% CI [0.77, 0.86]). The micro-AUC was 0.93.
Conclusion: The proposed model may be used to assist clinicians in the diagnosis of parotid tumors. However, future larger-scale multicenter studies are required for full validation.
Introduction
Parotid gland tumors are rare tumors, accounting for approximately 5% of head and neck tumors, and approximately 75% of them are benign. The most common types of parotid gland benign tumors are pleomorphic adenomas and Warthin tumors (1, 2).
The preoperative diagnosis of benign and malignant tumors of the parotid gland is of great clinical significance and can have an important impact on surgical planning. The choice of surgical procedure depends on the histological type of the tumor. Approximately 5% to 10% of pleomorphic adenomas have a risk of malignant transformation and a high risk of recurrence, and radical surgery is usually used to treat them. The malignant transformation of Warthin tumors is extremely rare, occurring for only 0.3% of patients. Tumor removal or conservative observation is recommended in clinical practice to avoid the risk of facial nerve injury due to surgery (3). Malignant tumors of the parotid gland request extensive resection (4).
It is difficult to diagnose malignant tumors of the parotid gland via clinical manifestations (5). Fine-needle aspiration cytology (FNAC) is often used for the preoperative diagnosis of parotid gland tumors, in which the accuracy in discriminating benign and malignant diseases is 87.8%-97% (6, 7). However, due to the difficulty of sampling and the heterogeneity of the tumor, fine-needle aspiration cytology is sometimes uncertain and not representative of the true nature of the tumor. It may also lead to the spread of tumor cells, increasing the possibility of local recurrence and sometimes increasing the risk of infection (8). Therefore, preoperative imaging plays an important role in evaluating the location and nature of the tumor for the surgical plan (9–11). Ultrasound and CT are common imaging methods for diagnosing parotid gland tumor (12). However, an inflammatory lump is not easily distinguishable from a tumor on the ultrasound image, and when the difference between the density of the tumor and the density of the parotid tissue was small, the clear boundary could not be obtained by general CT (12–14). MRI is also an important method for the diagnosis of benign and malignant tumors of the parotid gland due to the high resolution of soft tissues. The sensitivity of MRI for parotid gland tumors is 86%, and the specificity is 90% (15). However, because parotid gland tumors are relatively rare and the tumors are heterogeneous, there are obvious differences in the judgment of benign and malignant parotid gland tumors.
In recent years, with the development of artificial intelligence, the application of deep learning in the medical field has made rapid progress. Deep learning uses simple neurons to form a complex neural network (15). For medical image diagnostic assistance, deep-learning methods can outperform many traditional machine learning methods. Antropova et al. (16) extracted mammograms, ultrasound and MRI features and used them for deep-learning training combined with traditional computer-aid diagnosis (CAD) methods to develop systems that are superior to the traditional CAD analysis of single images. Wang et al. (17) used pretraining and transfer learning methods to fine-tune the network model for other classification tasks in predicting benign and malignant prostate cancer (PCa) on MR images. Yang et al. used multiparameter magnetic resonance imaging (mp-MRI) to diagnose and detect prostate cancer through a CNN and SVM cotrained model (18). These studies demonstrate that deep learning, especially of convolutional neural networks (CNNs), is superior to non–deep-learning methods.
In terms of practical application, many studies have shown that the performance of artificial intelligence or deep learning can reach or exceed that of physicians (19–23). Several studies have shown that deep-learning techniques are comparable to radiologists’ detection and segmentation tasks in MRI examinations (24). In a recent report, Zhao et al. (25) developed a deep-learning autoLNDS (lymph node detection and segmentation) model based on mp-MRI. The model can detect and segment LNs (lymph nodes) quickly, yield good clinical efficiency and reduce the difference among physicians with different levels of experience. However, the application of deep learning in medical image diagnosis is limited (26). Wang et al.’s (27) research shows that incorporating diagnostic features into neural networks is a promising direction for future study. Ma et al. (28) used complementary patch-based CNNs to extract low-level and high-level features and fused the feature maps to performing classification. This kind of operation may yield information from important feature domains, but networks based on single imaging series still have limitations. Therefore, this study aims to propose a multiseries-input CNN to boost the performance of MR image classification tasks.
At present, there is no relevant research based on neural networks to predict the type of parotid gland tumor (15). This is mainly due to the rarity of parotid gland tumors, as the incidence of common cancers is higher than that of parotid gland cancer (29), which therefore lack sufficient data. On the other hand, due to the rarity of and limited clinical experience with parotid gland tumors, the significance of their auxiliary diagnosis will be substantial. The distinction between malignant and benign tumors of the parotid gland is clinically important.
This study aims to develop a system to act as an intelligent assistant in medical image diagnosis based on deep-learning technology and to design a model for predicting parotid gland tumors.
Methods and Materials
Workflow Introduction
Figure 1 shows the whole workflow of our research. The MR images of 233 patients were collected. The tumor and parotid gland were segmented manually by physicians. A modified ResNet18 model was used to discriminate different parotid lesions.
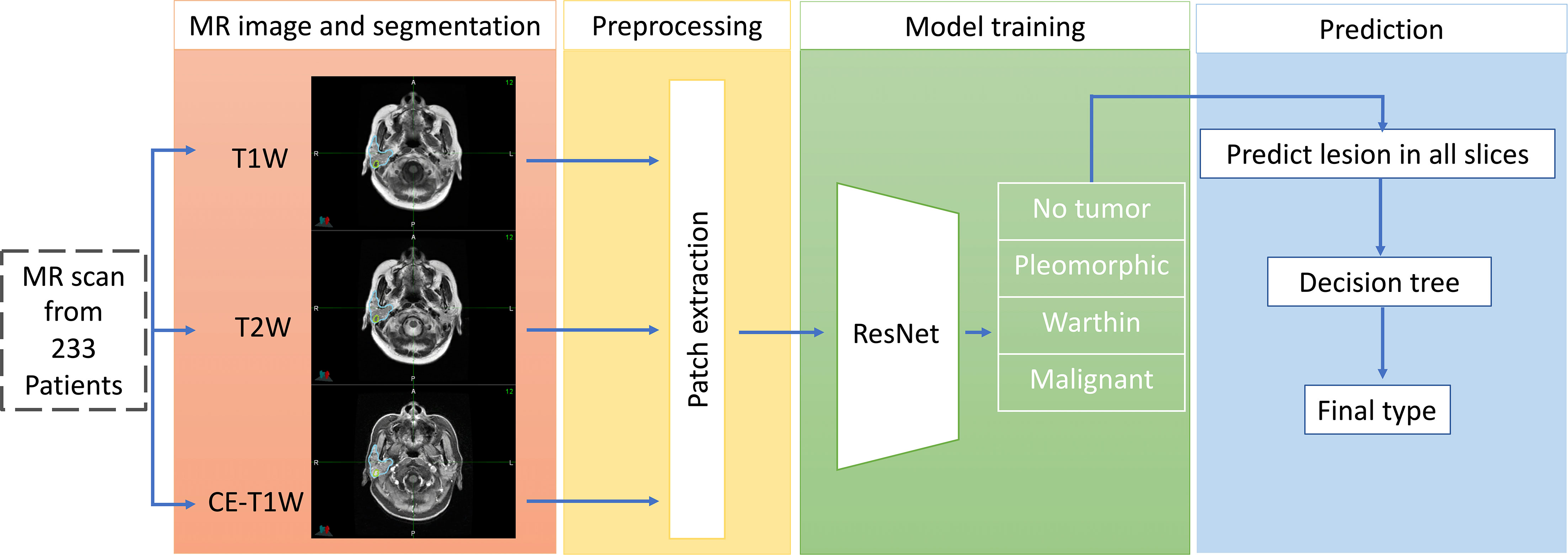
Figure 1 The workflow of our proposed deep-learning framework for the differentiation of benign from malignant parotid lesions. The first part shows multimodal MR images and tumor segmentation. The second part shows the preprocessing stage for the MR images. The third part shows the training network prediction model and tumor type classification. The final part comprehensively shows that predictions are made for all slices to determine the tumor type.
Patients and MR Image Acquisition
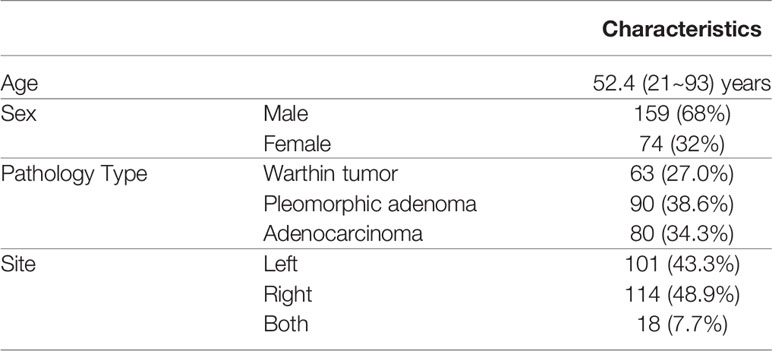
Two hundred thirty-three patients were enrolled in our study (Table 1). All patients were treated from 2014-2018 at Fudan University Shanghai Cancer Center. There were 159 males and 74 females with an average age of 52.4 (range, 21-93 years). The histopathology results were acquired by the operation, and each tumor have only one histology result from patient’s pathology report. Patient pathology information was collected from the EMR system.
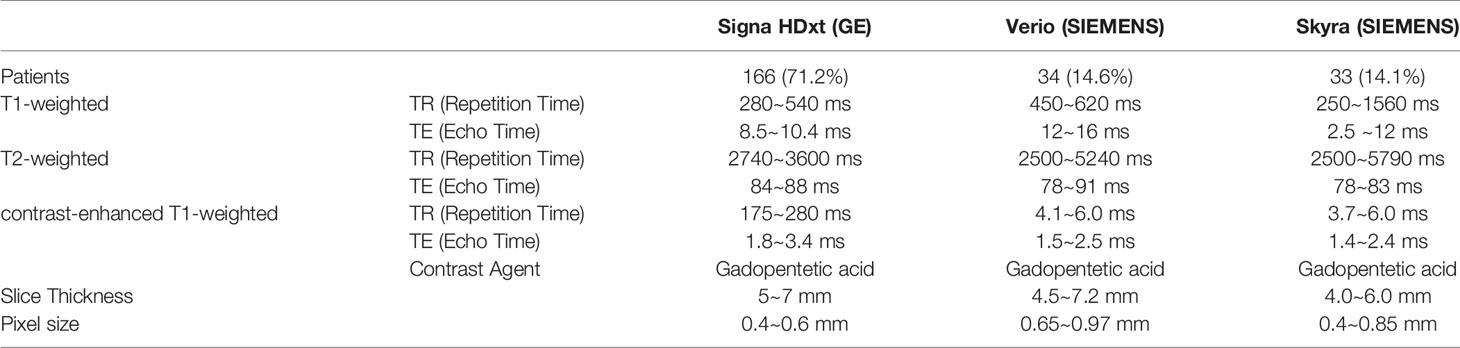
This study focuses on two types of benign tumors (pleomorphic adenoma and Warthin tumor) and one type of malignant tumors (adenocarcinoma). The MRI scan parameters were based on our parotid gland MR scanning protocol and fine-tuned during scanning by the MRI operator. The details of the imaging parameters are shown in Table 2. All patients were scanned with at least three series (T1-weighted, T2-weighted and contrast-enhanced T1-weighted).
As a reference, 215 patients’ contralateral normal parotid glands (991 slice images in total) Were selected as negative samples.
Parotid Gland and ROI Delineation
The parotid gland tumors were segmented by a radiologist with 10 years of clinical experience based on the MR series. The delineation tasks were performed on MIM (version 6.8.10, Cleveland, US). These contours were double-checked by a physicist. To improve the performance of the deep-learning model, the entire parotid gland was also segmented. Figure 2 shows an example of this delineation.
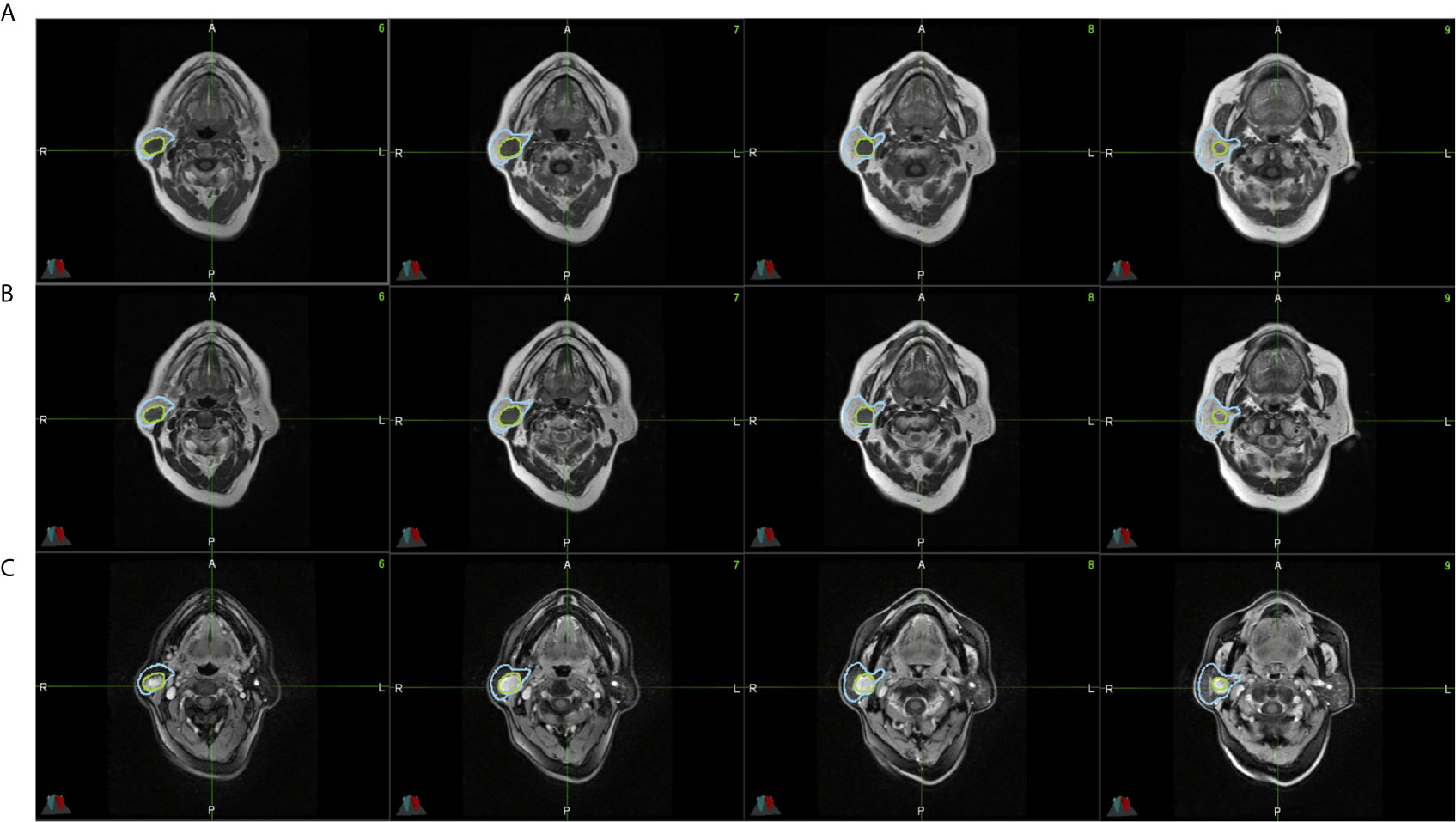
Figure 2 All parotid gland and tumor ROIs from a single patient’s lesion on MR images. (A) shows T1-weighted MR images. (B) shows CE-T1-weighted MR images. (C) shows T2-weighted MR images. The blue region is the parotid gland, and the green region is the tumor.
Dataset and MR Image Preprocessing
After tumor and parotid gland segmentation, 3791 parotid gland region images were cropped from the MR images. A label (pleomorphic adenoma, Warthin tumor, malignant tumor or free of tumor), which was based on histological results, was assigned to each image.
To train the deep-learning model, these data were randomly divided into training and test sets at a ratio of 9:1, with 212 patients in the training set and 21 patients in the test set. The training set included 73 adenocarcinoma, 83 pleomorphic adenoma and 58 Warthin tumor patients, 2133 total slices with lesions (861 adenocarcinoma slices, 714 pleomorphic adenoma slices, and 558 Warthin tumor slices), and 902 total slices without lesions. The test set included 8 adenocarcinoma, 8 pleomorphic adenoma and 5 Warthin tumor patients, 186 total slices with lesions (93 adenocarcinoma, 57 pleomorphic adenoma, and 36 Warthin tumor slices), and 89 slices without lesions. Details on the processing workflow for the MR images are shown in Figure 3. A four-channel image was generated as the model input; the first three channels consisted of T1-weighted, CE-T1-weighted, and T2-weighted MR images, and the fourth channel included images of the parotid glands, which were contoured by a radiotherapist. A total of 991 parotid gland images without lesions were used as negative samples.
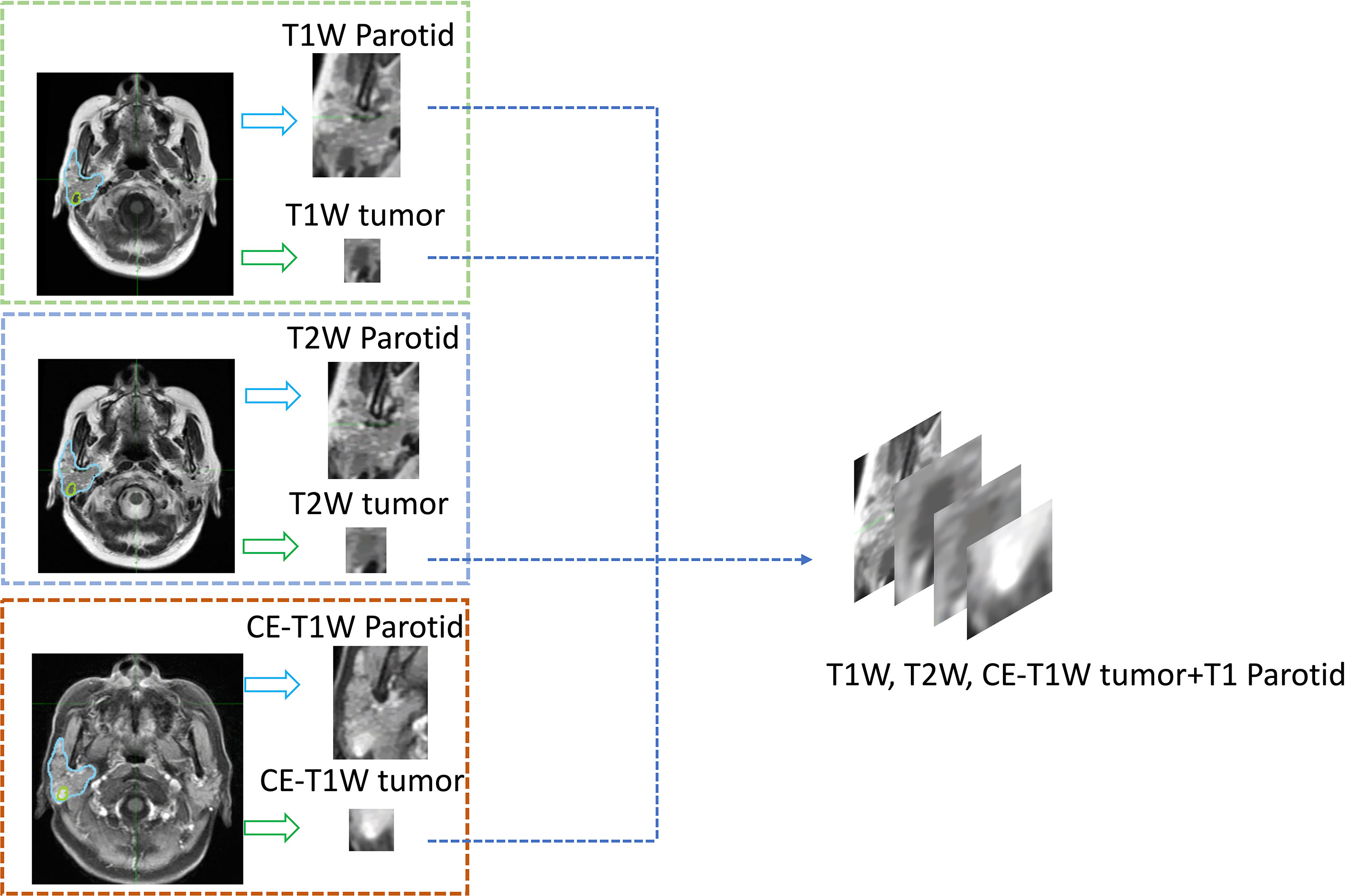
Figure 3 An example of the four-channel input. All parotid glands and tumors were cropped from segmented MR images, and then the three series of tumor images and the T1-weighted parotid gland image were input into different channels of one image.
Deep-Learning Network Structure
The detailed prediction network structure is shown in Figure 4. The network architecture is based on ResNet18. The input images were resized to 224*224 pixels. Random image flipping, contrast adjustment, color jitter, and affine transform were used for data augmentation. All image pixel values were normalized to [0, 1]. The batch size of training set and test set was 32. To avoid overfitting, we reduced the number of network layers (30) (i.e., the number of residual blocks were reduced to 2) and adjusted the number of network layers appropriately. The cross-entropy loss function (31) and the Adam optimizer (32) were used. The model was trained for 1200 epochs; the learning rate was 1e-6 for the first 600 epochs and was then multiplied by a factor of 0.8 every 100 epochs. An Intel I7-8700K CPU and Nvidia GeForce 1080 Ti GPU were used for model training. It took approximately 2 hours to complete the whole training procedure. The program was developed with PyTorch (version 1.2).
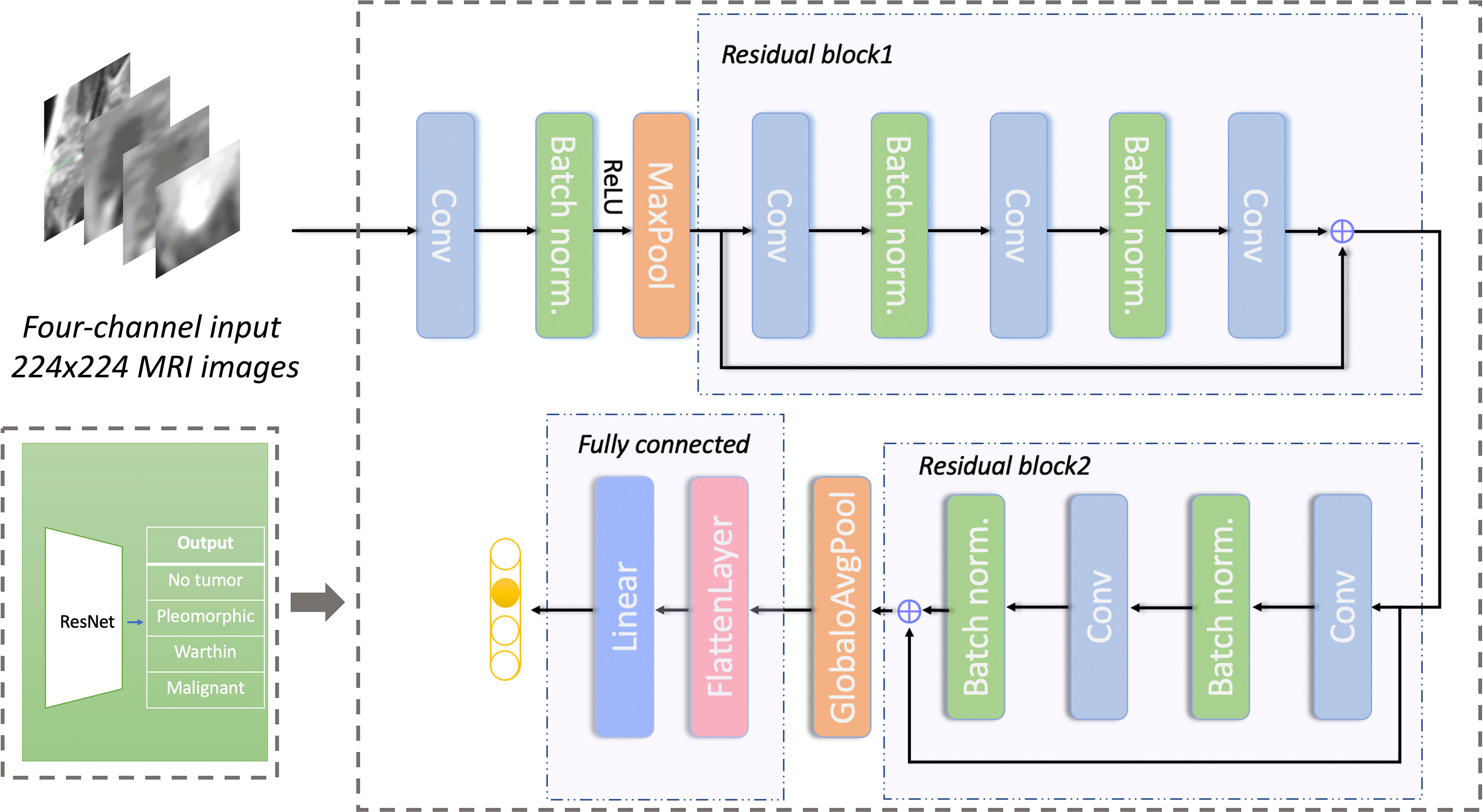
Figure 4 Network structure for predicting different types of parotid gland tumors based on ResNet. The network has 2 residual blocks. Conv, convolutional layer; Batch norm, batch normalization; Maxpool, max-pooling layer; GlobaloAvgpool, global average pooling layer; Linear, linear layer.
Performance Evaluation
The model performance was reported as its accuracy, which for binary classification was computed by:
where TP denotes the number of true positives, TN denotes the number of true negatives, and Total represents the number of total samples. For multiclassification, the calculation of accuracy was computed by:
where FPi represents the number of false negatives of the ith-class negative sample.
The Impact of Different Image Series
To investigate the impact of different image series on the prediction accuracy, we filtered the input to the model using only a specific sequence of input models. Each dataset was trained separately until the accuracy of the model converged.
Automatic Patient Diagnosis
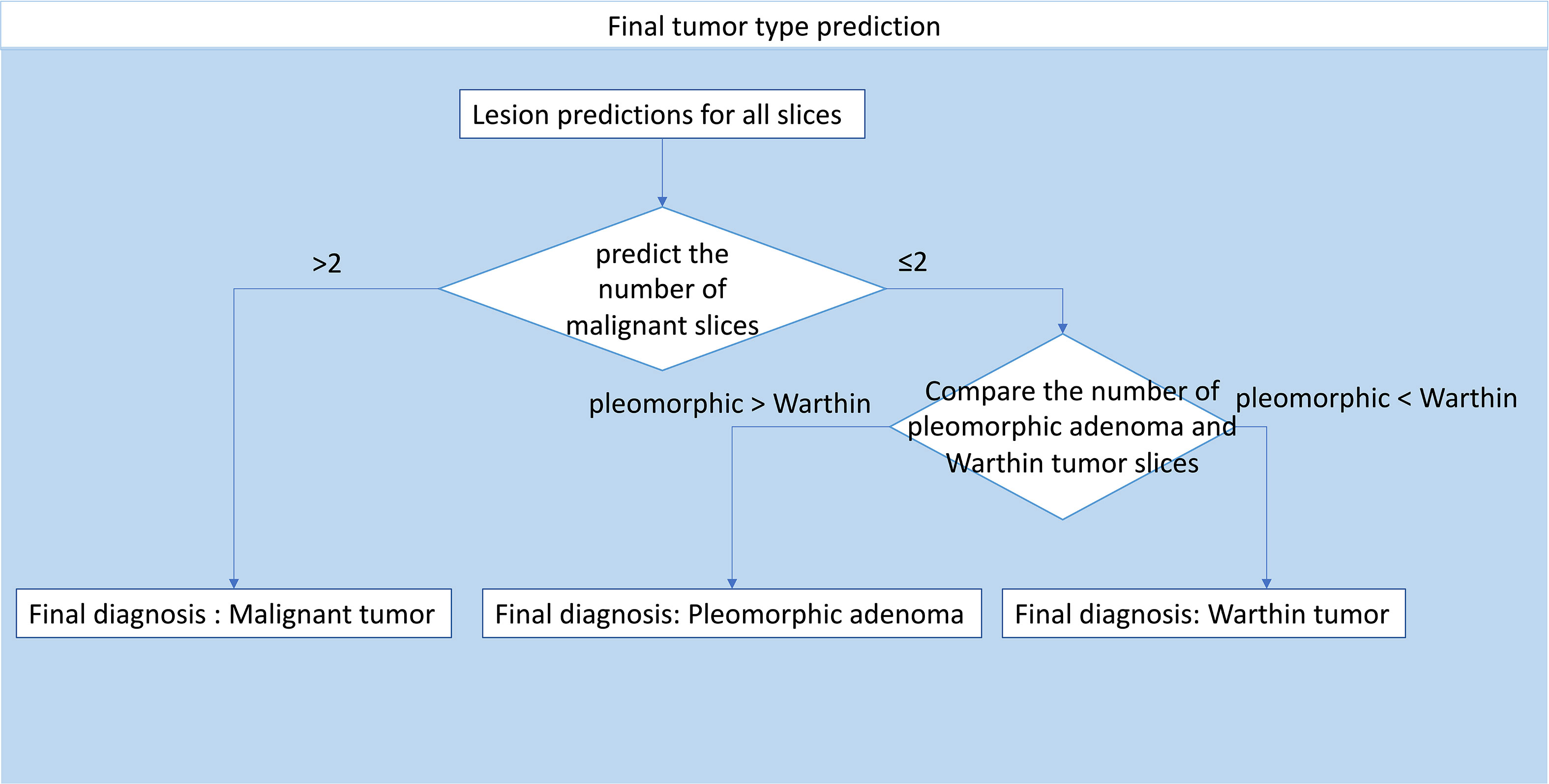
The slice probability was acquired by model prediction. To obtain patient diagnoses, a decision-making process was developed (Figure 5). The decision-making process was as follows: 1. Patients with more than two malignant slices were diagnosed as having a malignant tumor, and patients with no more than two malignant slices were diagnosed as having a benign tumor; 2. For patients with benign tumors, the tumor type was decided by slice number comparison. If the number of pleomorphic adenoma slices was greater than the number of Warthin tumor slices, the patient was diagnosed with pleomorphic adenoma, and vice versa.
Results
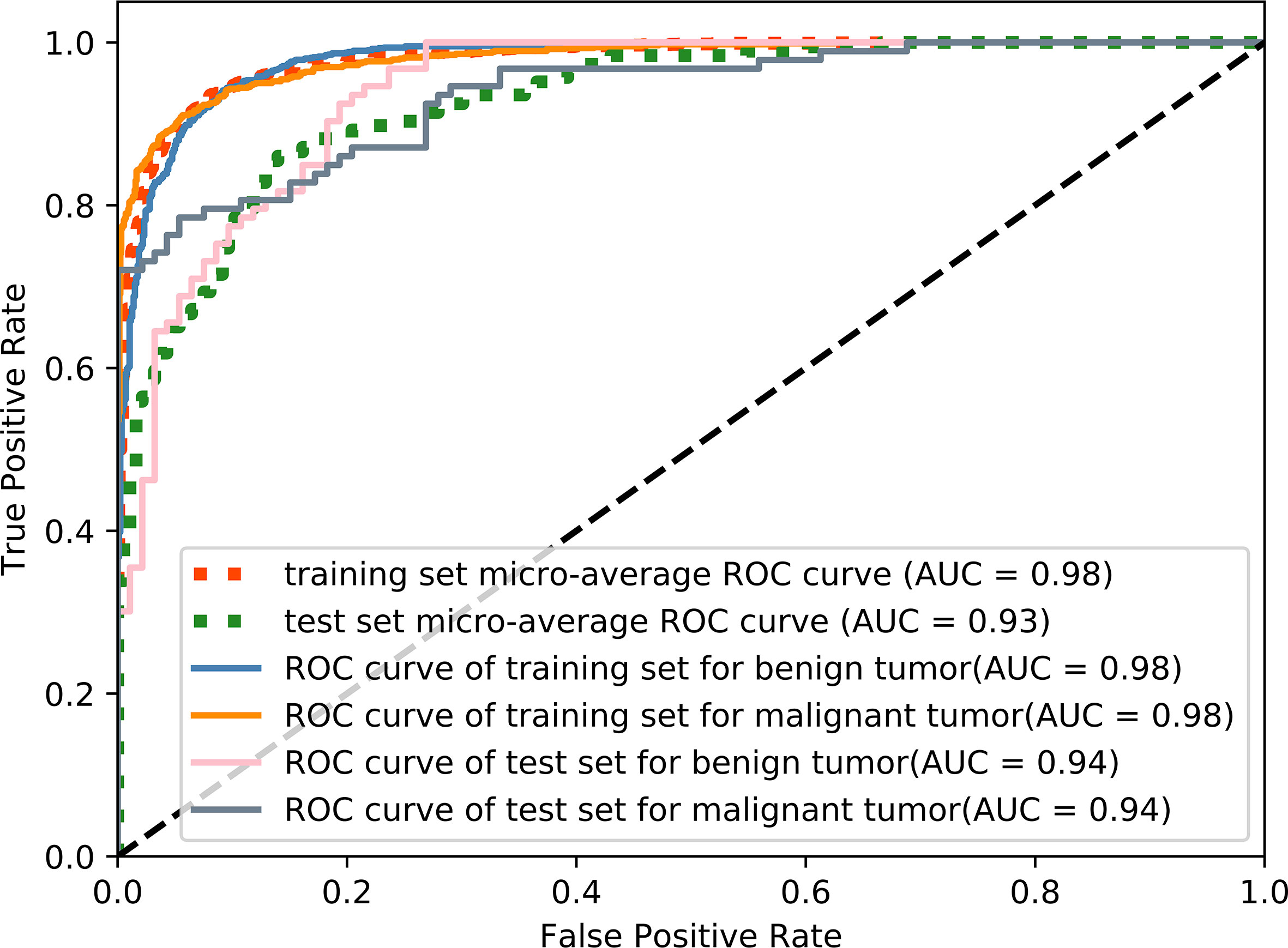
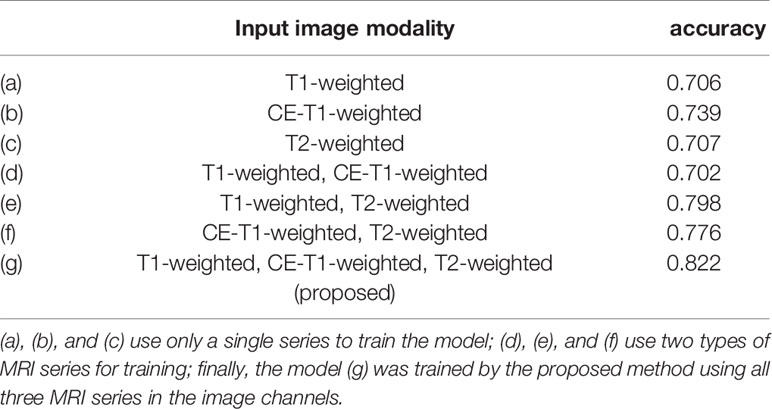
The accuracy of slice classification was 92.94% for the training set and 82.18% for the test set. Figure 6 shows that the area under the micro-average ROC curve (micro-AUC) was 0.98 for the training set and 0.93 for the test set. Table 3 shows results of different training strategy for the single MR slice. For (a-c), the model was trained with single-modality image series. Pairs of MR series were integrated into the different image channels for training the model in (d-f), and all three MR series were used for the image channels in (g). The accuracy of the model when trained using single-modality image series in the channels was 0.706 (T1-weighted), 0.739 (CE-T1-weighted), and 0.707 (T2-weighted). Using pairs of image series boost the accuracy (Table 3). The use of all three image series (T1-weighted, CE-T1-weighted, T2-weighted) and the parotid gland contour images in the channels yields the best performance.
Details of the results for the single MR slice prediction are shown in Table 4. Our model has good performance in distinguishing images that did and did not contain lesions and achieves an accuracy of 0.882 in identifying benign from malignant tumors. The accuracy in distinguishing pleomorphic adenoma and Warthin tumors among benign tumors was 63.4%.

Table 4 Parotid gland tumor classification results for different types of tumors in a single MR slice.
Figure 7 shows the confusion matrix of our proposed prediction model for individual slices. Each element (x, y) in the confusion matrix represents the number of samples with true class x that was classified as being in class y. The overall accuracy was 81.45%. The accuracy for the first class was perfect, as all 89 cases were correctly classified. In identifying benign tumor cases, sixteen cases (28%) and eighteen cases (50%) of pleomorphic adenomas and Warthin tumors, respectively, were misclassified. The accuracy in predicting malignant tumors was 81.72%.
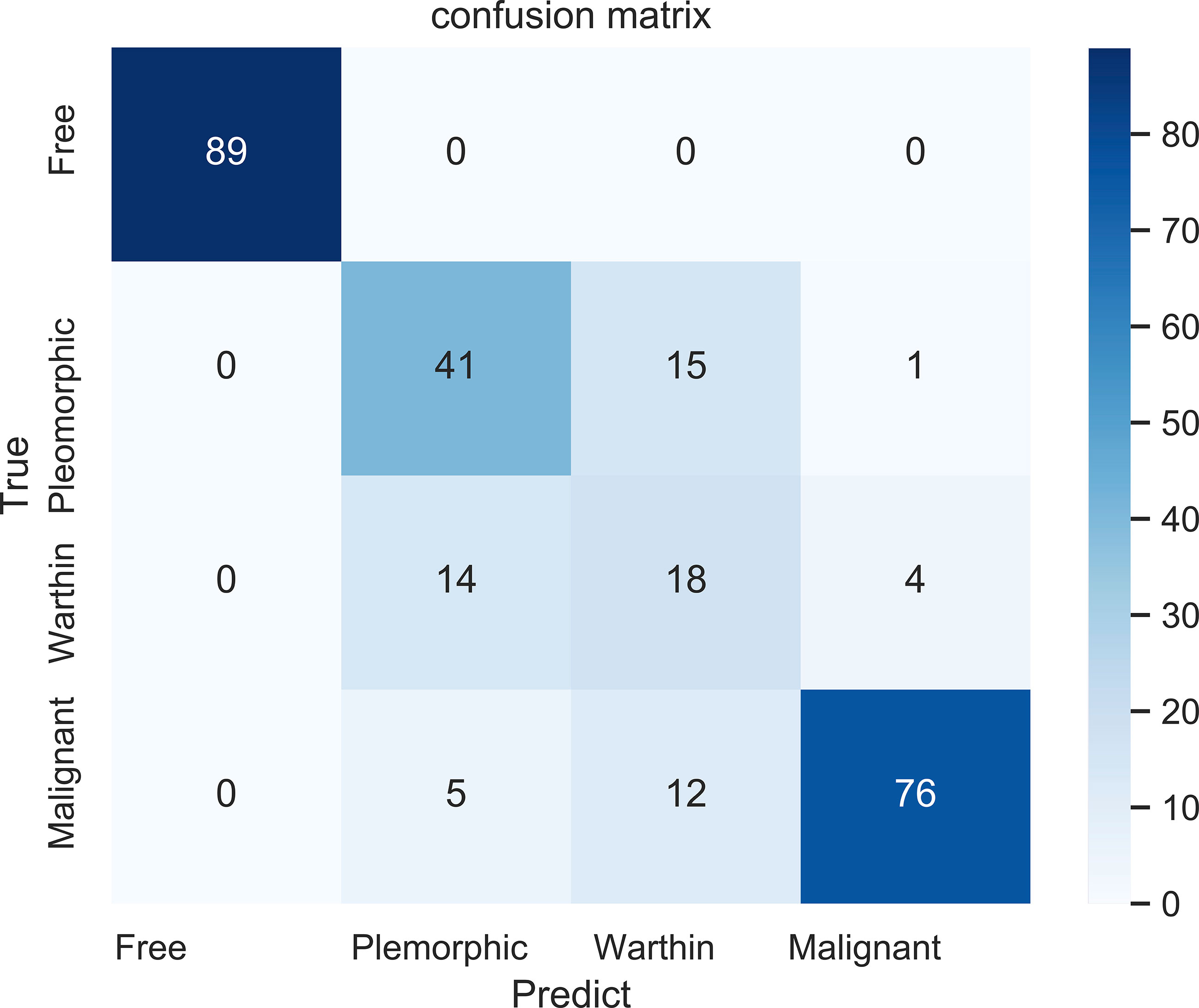
Figure 7 The confusion matrix for the four classifications: free (no tumor), pleomorphic adenoma, Warthin tumor, and malignant tumor.
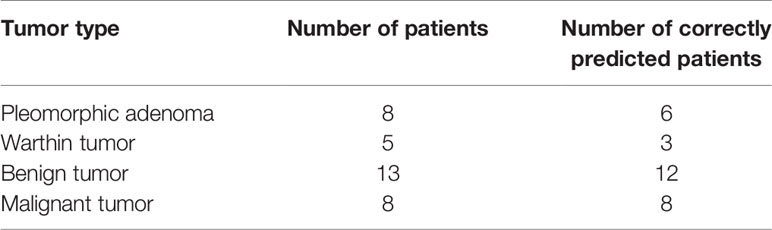
Table 5 shows the results from diagnosing twenty-one patients in the test set (8 with pleomorphic adenomas, 5 with Warthin tumors, and 8 with malignant tumors) by using the decision tree method (Figure 5). Six (75%) and three patients (60%) were correctly classified as pleomorphic adenoma and Warthin tumor, respectively. Only one patient in the benign tumor class was misidentified, and all eight patients with malignant tumors were correctly predicted.
Discussion
In this study, we developed a model to predict the type of parotid gland tumor. The results show that our model can identify the type of parotid gland tumor at the slice level. Experiments show that using multichannel images as the input can improve the model’s ability to identify tumor features. The model can thus assist doctors in quickly determining tumor classifications in clinical practice. Using artificial intelligence modeling methods, the accuracy in predicting benign and malignant parotid tumors can be further improved, the prognosis can be evaluated, and a reasonable diagnosis and treatment plan can be formulated for patients.
Pathological analysis takes time and is expensive, resulting in a heavy financial and psychological burden for the patient (33). The main advantage of adopting deep learning into the prediction of the type of parotid gland tumor is that the deep-learning method can inform the physicist which patient may have a tumor or cancer faster than pathological analysis (34). With the use of deep-learning models, the patient’s condition could be narrowed and locked into benign or malignant type. This method may be useful for improving the efficiency of routine clinical practice and saving time in patient treatment.
In MR image preprocessing, due to the limited size of the image itself, we compared the performance between the multichannel images and single-channel images as the network input. Each dataset was trained with the same strategy, and the final average accuracy was 71.7% for single-channel input and 75.8% for double-channel input, which were 10.5% and 6.4% lower, respectively, than our proposed method. The neural network can extract features from different MR series, so we hypothesize that the use of multiple channels may boost model performance in diagnosing the type of parotid gland tumor; the obtained experimental results show that the performance of the model is effective. From Table 3, as the number of channels and integrated MR series increases, the accuracy of the model also gradually increases.
In training the model, we chose the current prevalent ResNet18 network as the backbone. The residual blocks prevent disappearance of the gradient to minimize the effects of the degradation problem after many iterations (35, 36). In our practice, we made some modifications to ResNet18 to adapt it to our classification task.
It should be noted that the segmentation of the tumor and parotid gland had to be performed manually by a physician, and the last prediction step involved a simple decision tree. In the future, these steps could also be performed by the deep learning-based model (autosegmentation of the tumor and parotid gland ROIs and integration of all patient slices to predict the tumor type by using a neural network).
The final model showed good performance in predicting pleomorphic adenoma and Warthin tumors. The prediction result for Warthin tumors seemed to be worse than that of the other classes, as fourteen cases (77.8%) of Warthin tumors were misidentified as being pleomorphic adenomas. We consider two reasons for this. First, our dataset was uneven, as the number of Warthin tumors was too low; therefore, the model performance in distinguishing pleomorphic adenomas and Warthin tumors was worse than that in identifying benign from malignant tumors. Second, benign tumors may share certain features that makes it difficult to distinguish the two types. In the future, a specific model for predicting different benign tumor types will be generated that may outperform the current model. Consequently, here we proposed a script that could can accurately distinguish between benign and malignant tumors.
There were some limitations in this study. First, we did not validate our model with an external dataset, which could have been valuable in demonstrating the reliable performance of the model. Second, we combined three types of MRI series (T1W, T2W, CE-T1W). During routine diagnostics, some series may not be acquired. Third, we proved the feasibility of using multiple channels to predict the type of tumor; however, the performance between benign tumors was not sufficiently precise. Furthermore, our data did not include examples of three other classes of parotid gland tumors. Given the lack of data on these other three classes of tumor, this study merely explored the feasibility of the above methods for the three classes with sufficient data.
In the future, larger-scale, multicenter studies are required for full validation of the model. We will enroll more patients in our study to train the model for diagnosing all six classes of parotid gland cancer.
Conclusion
In this study, we proposed a novel method combining clinical experience and a deep-learning method to diagnose parotid gland tumors. We proved the feasibility of the method, trained the model for predicting tumor types, and developed a script to analyze the final prediction. We propose that the results of this study will help physicians diagnose tumor types in patients faster. It can improve the effectiveness of routine clinical practice for these tumors. In the future, this model could be used to assist young doctors in preventing misdiagnoses and other mistakes that could be made from working long hours.
Data Availability Statement
The datasets presented in this article are not readily available because the data are not publicly available due to privacy restrictions. Requests to access the datasets should be directed to BF, NDExNTkzMTUwQHFxLmNvbQ==.
Ethics Statement
The studies involving human participants were reviewed and approved by ethics committee, Fudan University Shanghai Cancer Center.
Author Contributions
BF and XX: drafting the manuscript. BF: processing the data, developing the deep learning model and script. XX, QH, YY, and YM: acquisition of data and generated research ideas. JW and WH: provided guidance on methodology and overall project, and reviewed manuscript. LS: provided lab and technical support. All authors contributed to the article and approved the submitted version.
Funding
The work was supported by the Zhejiang Provincial Health Science and Technology Project. Grant number: 2021KY396.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
CAD, computer-aided diagnosis; FNAC, Fine-needle aspiration cytology; T1W, T1-weighted; CE-T1W, contrast-enhanced T1-weighted; T2W, T2-weighted; TR, Repetition Time; TE, Echo Time.
References
1. Guzzo M, Locati LD, Prott FJ, Gatta G, McGurk M, Licitra L. Major and Minor Salivary Gland Tumors. Crit Rev Oncol Hematol (2010) 74:134–48. doi: 10.1016/j.critrevonc.2009.10.004
2. Sentani K, Ogawa I, Ozasa K, Sadakane A, Utada M, Tsuya T, et al. Characteristics of 5015 Salivary Gland Neoplasms Registered in the Hiroshima Tumor Tissue Registry Over a Period of 39 Years. J Clin Med (2019) 8:566. doi: 10.3390/jcm8050566
3. Kanatas A, Ho MWS, Mücke T. Current Thinking About the Management of Recurrent Pleomorphic Adenoma of the Parotid: A Structured Review. Br J Oral Maxillofac Surg (2018) 56:243–8. doi: 10.1016/j.bjoms.2018.01.021
4. Lim YC, Lee SY, Kim K, Lee JS, Koo BS, Shin HA, et al. Conservative Parotidectomy for the Treatment of Parotid Cancers. Oral Oncol (2005) 41:1021–7. doi: 10.1016/j.oraloncology.2005.06.004
5. Correia-Sá I, Correia-Sá M, Costa-Ferreira P, Silva A, Marques M. Fine-Needle Aspiration Cytology (FNAC): Is it Useful in Preoperative Diagnosis of Parotid Gland Lesions? Acta Chir Belg (2017) 117(2):110–4. doi: 10.1080/00015458.2016.1262491
6. Eytan DF, Yin LX, Maleki Z, Koch WM, Tufano RP, Eisele DW, et al. Utility of Preoperative Fine Needle Aspiration in Parotid Lesions. Laryngoscope (2018) 128:398–402. doi: 10.1002/lary.26776
7. Liu CC, Jethwa AR, Khariwala SS, Johnson J, Shin JJ. Sensitivity, Specificity, and Posttest Probability of Parotid Fine-Needle Aspiration: A Systematic Review and Meta-Analysis. Otolaryngol Head Neck Surg (2016) 154:9–23. doi: 10.1177/0194599815607841
8. Mezei T, Mocan S, Ormenisan A, Baróti B, Iacob A. The Value of Fine Needle Aspiration Cytology in the Clinical Management of Rare Salivary Gland Tumors. J Appl Oral Sci (2018) 26:e20170267. doi: 10.1590/1678-7757-2017-0267
9. Elmokadem AH, Abdel Khalek AM, Abdel Wahab RM, Tharwat N, Gaballa GM, Elata MA, et al. Diagnostic Accuracy of Multiparametric Magnetic Resonance Imaging for Differentiation Between Parotid Neoplasms. Can Assoc Radiol J (2019) 70:264–72. doi: 10.1016/j.carj.2018.10.010
10. Xu Z, Zheng S, Pan A, Cheng X, Gao M. A Multiparametric Analysis Based on DCE-MRI to Improve the Accuracy of Parotid Tumor Discrimination. Eur J Nucl Med Mol Imaging (2019) 46:2228–34. doi: 10.1007/s00259-019-04447-9
11. Liang YY, Xu F, Guo Y, Wang J. Diagnostic Accuracy of Magnetic Resonance Imaging Techniques for Parotid Tumors, a Systematic Review and Meta-Analysis. Clin Imaging (2018) 52:36–43. doi: 10.1016/j.clinimag.2018.05.026
12. Choi DS, Na DG, Byun HS, Ko YH, Kim CK, Cho JM, et al. Salivary Gland Tumors: Evaluation With Two-Phase Helical CT. Radiology (2000) 214(1):231–6. doi: 10.1148/radiology.214.1.r00ja05231
13. Yerli H, Aydin E, Coskun M, Geyik E, Ozluoglu LN, Haberal N, et al. Dynamic Multislice Computed Tomography Findings for Parotid Gland Tumors. J Comput Assist Tomogr (2007) 31(2):309–16. doi: 10.1097/01.rct.0000236418.82395.b3
14. Jin GQ, Su DK, Xie D, Zhao W, Liu LD, Zhu XN. Distinguishing Benign From Malignant Parotid Gland Tumours: Low-Dose Multi-Phasic CT Protocol With 5-Minute Delay. Eur Radiol (2011) 21:1692–8. doi: 10.1007/s00330-011-2101-y
15. Mazurowski MA, Buda M, Saha A, Bashir MR. Deep Learning in Radiology: An Overview of the Concepts and a Survey of the State of the Art With Focus on MRI. J Magn Reson Imaging (2019) 49:939–54. doi: 10.1002/jmri.26534
16. Antropova N, Huynh BQ, Giger ML. A Deep Feature Fusion Methodology for Breast Cancer Diagnosis Demonstrated on Three Imaging Modality Datasets. Med Phys (2017) 44:5162–71. doi: 10.1002/mp.12453
17. Wang X, Yang W, Weinreb J, Han J, Li Q, Kong X, et al. Searching for Prostate Cancer by Fully Automated Magnetic Resonance Imaging Classification: Deep Learning Versus non-Deep Learning. Sci Rep (2017) 7:15415. doi: 10.1038/s41598-017-15720-y
18. Yang X, Liu C, Wang Z, Yang J, Min HL, Wang L, et al. Co-Trained Convolutional Neural Networks for Automated Detection of Prostate Cancer in Multi-Parametric MRI. Med Image Anal (2017) 42:212–27. doi: 10.1016/j.media.2017.08.006
19. Thrall JH, Li X, Li Q, Cruz C, Do S, Dreyer K, et al. Artificial Intelligence and Machine Learning in Radiology: Opportunities, Challenges, Pitfalls, and Criteria for Success. J Am Coll Radiol (2018) 15:504–8. doi: 10.1016/j.jacr.2017.12.026
20. Lian C, Liu M, Zhang J, Shen D. Hierarchical Fully Convolutional Network for Joint Atrophy Localization and Alzheimer’s Disease Diagnosis Using Structural Mri. IEEE Trans Pattern Anal Mach Intell (2020) 42:880–93. doi: 10.1109/TPAMI.2018.2889096
21. Jean N, Burke M, Xie M, Davis WM, Lobell DB, Ermon S. Combining Satellite Imagery and Machine Learning to Predict Poverty. Science (2016) 353:790–4. doi: 10.1126/science.aaf7894
22. Gao L, Liu R, Jiang Y, Song W, Wang Y, Liu J, et al. Computer-Aided System for Diagnosing Thyroid Nodules on Ultrasound: A Comparison With Radiologist-Based Clinical Assessments. Head Neck (2018) 40:778–83. doi: 10.1002/hed.25049
23. Gao Y, Maraci MA, Noble JA. “Describing Ultrasound Video Content Using Deep Convolutional Neural Networks”. In: 2016 IEEE 13th International Symposium on Biomedical Imaging (Isbi). Prague: IEEE (The Institute of Electrical and Electronics Engineers) (2016). p. 787–90. doi: 10.1109/ISBI.2016.7493384
24. Ghafoorian M, Karssemeijer N, Heskes T, van Uden IWM, Sanchez CI, Litjens G, et al. Location Sensitive Deep Convolutional Neural Networks for Segmentation of White Matter Hyperintensities. Sci Rep (2017) 7:5110. doi: 10.1038/s41598-017-05300-5
25. Zhao X, Xie P, Wang M, Li W, Pickhardt PJ, Xia W, et al. Deep Learning-Based Fully Automated Detection and Segmentation of Lymph Nodes on Multiparametric-Mri for Rectal Cancer: A Multicentre Study. EBioMedicine (2020) 56:102780. doi: 10.1016/j.ebiom.2020.102780
26. Cheng JZ, Ni D, Chou YH, Qin J, Tiu CM, Chang YC, et al. Computer-Aided Diagnosis With Deep Learning Architecture: Applications to Breast Lesions in US Images and Pulmonary Nodules in CT Scans. Sci Rep (2016) 6:24454. doi: 10.1038/srep24454
27. Wang H, Zhou Z, Li Y, Chen Z, Lu P, Wang W, et al. Comparison of Machine Learning Methods for Classifying Mediastinal Lymph Node Metastasis of non-Small Cell Lung Cancer From 18F-FDG PET/CT Images. EJNMMI Res (2017) 7:11. doi: 10.1186/s13550-017-0260-9
28. Ma S, Li YT, Wu JB, Geng TW, Wu Z. Performance Analyses of Subcarrier BPSK Modulation Over M Turbulence Channels With Pointing Errors. Optoelectron Lett (2016) 12:221–5. doi: 10.1007/s11801-016-6054-x
29. Torre LA, Siegel RL, Ward EM, Jemal A. Global Cancer Incidence and Mortality Rates and Trends–An Update. Cancer Epidemiol Biomarkers Prev (2016) 25:16–27. doi: 10.1158/1055-9965.EPI-15-0578
30. Ahmed KB, Hall LO, Goldgof DB, Liu R, Gatenby RA. Fine-Tuning Convolutional Deep Features for MRI Based Brain Tumor Classification. Proc SPIE 10134 Med Imaging 2017: Computer-Aided Diagn (2017) 10134:2E. doi: 10.1117/12.2253982
31. Heaton J, Goodfellow I, Bengio Y, Courville A. Deep Learning. Massachusetts: The MIT Press (2016).
32. Bock S, Weiß M. “A Proof of Local Convergence for the Adam Optimizer”. In: 2019 International Joint Conference on Neural Networks (Ijcnn). Budapest, Hungary: IEEE (The Institute of Electrical and Electronics Engineers) (2019). p. 1–8. doi: 10.1109/IJCNN.2019.8852239
33. Madabhushi A, Lee G. Image Analysis and Machine Learning in Digital Pathology: Challenges and Opportunities. Med Image Anal (2016) 33:170–5. doi: 10.1016/j.media.2016.06.037
34. Komura D, Ishikawa S. Machine Learning Approaches for Pathologic Diagnosis. Virchows Arch (2019) 475:131–8. doi: 10.1007/s00428-019-02594-w
35. He K, Zhang X, Ren S, Sun J. Deep Residual Learning for Image Recognition. IEEE (2016) 770–8. doi: 10.1109/CVPR.2016.90
Keywords: MR image, parotid gland tumor, deep learning, classification, image processing
Citation: Xia X, Feng B, Wang J, Hua Q, Yang Y, Sheng L, Mou Y and Hu W (2021) Deep Learning for Differentiating Benign From Malignant Parotid Lesions on MR Images. Front. Oncol. 11:632104. doi: 10.3389/fonc.2021.632104
Received: 22 November 2020; Accepted: 07 June 2021;
Published: 23 June 2021.
Edited by:
Jiuquan Zhang, Chongqing University, ChinaReviewed by:
Xiance Jin, Wenzhou Medical University, ChinaYidong Yang, University of Science and Technology of China, China
Copyright © 2021 Xia, Feng, Wang, Hua, Yang, Sheng, Mou and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liang Sheng, c2w5ODAxOTVAc2luYS5jb20=; Yonghua Mou, bW91eWgyMDA4QDE2My5jb20=; Weigang Hu, amFja2h1d2dAZ21haWwuY29t
†These authors have contributed equally to this work
 Xianwu Xia
Xianwu Xia Bin Feng
Bin Feng Jiazhou Wang
Jiazhou Wang Qianjin Hua3
Qianjin Hua3 Yonghua Mou
Yonghua Mou