
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 10 March 2021
Sec. Thoracic Oncology
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.630717
 Weize Lv1,2†
Weize Lv1,2† Hua Cheng2,3†
Hua Cheng2,3† Di Shao4†
Di Shao4† Yajun Wei1,2
Yajun Wei1,2 Weiping Zhu5
Weiping Zhu5 Kui Wu6
Kui Wu6 Wenxi Jiang4
Wenxi Jiang4 Liyang Hu2,7
Liyang Hu2,7 Zhou Sha2,7
Zhou Sha2,7 Beilong Zhong2,3*
Beilong Zhong2,3* Xiaofeng Pei2,7*
Xiaofeng Pei2,7*Although the National Comprehensive Cancer Network and the Chinese Society of Clinical Oncology guidelines recommend comprehensive genomic profiling of lung adenocarcinoma, it has not been widely applied in Chinese hospitals. This observational study aimed to determine real-world evidence of whether comprehensive genomic profiling can benefit the survival of patients with lung cancer. We investigated the frequency of genomic alterations, treatment strategies, and clinical outcomes in 233 patients with advanced non-small cell lung carcinoma who were routinely screened using a 508-gene panel. The most prevalent drivers were mutations of EGFR (51%), KRAS (9%), PIK3CA (7%), ALK (7%), MET (6%), and BRAF (5%). Mutations in tumor suppressor genes included TP53, KEAP1, RB1, PTEN, and APC. Median overall survival (OS) was significantly shorter among patients harboring KRAS (mutant, n = 17; WT, n = 154) and TP53 (mutant, n = 103; WT n =68) mutations (11.3 vs. 24.0 months; P = 0.16 and 18.7 vs. 28.7 months; P = 0.018, respectively). The OS was longer among patients with tumors harboring EGFR (P = 0.069) and ALK (P = 0.51) mutations. Most patients (65.4%) with the driver gene-positive (EGFR, ALK, and ROS1) tumors were received TKI treatment, whereas those with driver gene wild tumors (53.1%) chose platinum-based therapy. Univariate and multivariate analyses associated a shorter OS among patients with tumors harboring concomitant TP53 and EGFR mutations. These findings provide additional evidence from real-world on the potential importance of targeted therapies as a treatment option in NSCLC patients harboring clinically actionable mutation.
According to the guidelines of the National Comprehensive Cancer Network, molecular tests of epidermal growth factor receptor (EGFR), anaplastic lymphoma kinase (ALK), c-ros oncogene 1 (ROS1), B-Raf Proto-Oncogene (BRAF), mesenchymal-epithelial transition (MET), and rearranged during transfection (RET) are recommended for all non-small cell lung cancers (NSCLC) if specimens are sufficient for molecular analysis (1). The Chinese Society of Clinical Oncology recommends tests for EGFR mutations as well as ALK and ROS1 fusion (category 1) in non-squamous NSCLC and next-generation sequencing to detect all clinically meaningful mutations (category 2) (2). Resistance to targeted agents also requires changes in treatment strategies through repeated biopsies and molecular tests; thus, molecular analysis of advanced NSCLC has become routine (3). The French National Cancer Institute (INCa) launched a nationwide program for the routine molecular profiling of patients with advanced non-squamous NSCLC at 28 certified molecular genetics centers (4). The results suggested that routine nationwide molecular profiling provided a clinical benefit to patients with advanced NSCLC (5). In 2014, the Lung Cancer Mutation Consortium published their first study (LCMC1) reporting on mutation data of 1,007 late-stage NSCLC patients evaluated at 14 institutions in United States, this study demonstrated multiplexed testing aided physicians in selecting therapies and patients for targeted trials (6). However, a similar nationwide project does not exist in China, where clinicopathological and genomic data are retained in hospitals and third-party testing institutions, respectively. These data should be combined to generate real-world evidence (RWE) that supports comprehensive genomic profiling and promotes the application of routine molecular profiling in more hospitals.
The emergence of targeted therapy has redefined the treatment strategies of driver gene-positive advanced lung cancer (7–10). These targeted agents have been approved based on the results of randomized controlled trials (RCTs). Drugs are undoubtedly effective in ideal populations defined by RCTs, but the effectiveness of these drugs in actual clinical practice requires further verification. Real-world studies (RWSs) summarize the diagnosis and therapeutic data of clinical patients and provide generalizable evidence for precise medicine interventions, thus overcoming the inadequacy of RCT conclusions that are limited to specific patients (11). In addition, RCTs of patients with lung cancers harboring particularly rare mutations, such as EGFR exon 20 insertions and RET-NTRK fusions, are difficult to implement. RWSs are designed to meet clinical needs and can include most oncology patients, accelerating patient recruitment (12). This single-center, prospective, observational study aimed to determine the feasibility of routine molecular profiling and the clinical benefits for patients with advanced NSCLC.
The Institutional Review Board of Fifth Affiliated Hospital of Sun Yat-Sen University approved this study. Written informed consent was obtained from each of 233 patients with NSCLC or their legal representatives. We then collected epidemiological and clinicopathologic data (age, sex, smoking status, date of disease diagnosis, TNM stage, and dates of disease progression). Overall survival (OS) was calculated as the interval between diagnosis and death or final follow-up. The cutoff for outcomes was May 2020.
Genomic analysis was performed using Oseq, which is a clinical test designed to detect mutations, copy number alterations, and select gene fusions among 508 cancer-associated genes (13). Genomic DNA (50–200 ng) or cell-free DNA (10–50 ng) extracted from formalin-fixed paraffin-embedded (FFPE) or plasma samples were processed to generate bar-coded libraries according to manufacturer’s instructions (Integrated DNA Technologies, Coralville, IA, USA). Then exons were captured using IDT custom-designed probes (Integrated DNA Technologies, Coralville, IA, USA). A control library was constructed using DNA from white blood cells to filter germline mutations. DNA sequencing was performed on MGISEQ-2000 sequencing system (MGI, Shenzhen, China). Genomic alterations including base substitutions, short insertions and deletions, copy number alterations, and gene fusions were detected using a customized analysis pipeline which is modified from The Genome Analysis Toolkit (14). Data interpretation was focused on genomic alterations associated with clinically available targeted treatment options according to the standards and guidelines of the NCCN, the Association for Molecular Pathology (AMP), the American Society of Clinical Oncology (ASCO) and the College of American Pathologists (CAP) (15). The tumor mutation burden (TMB) was calculated as the number of non-synonymous mutations in non-driver genes per sample divided by the genomic coverage for that sample (1.7 Mb). The median TMB served as the cutoff to define TMB-high and TMB-low samples. Microsatellite instability (MSI) status was determined using MSIsensor and MANTIS (16, 17).
Categorical variables were compared using χ² tests or Fisher’s exact tests. The median age was compared between groups using Wilcoxon rank sum tests. The date of the initial diagnosis of lung cancer until the date of death or final follow-up was defined as OS, which was estimated from Kaplan-Meier curves. Differences in OS were analyzed using log-rank tests. Independent effects of concurrent mutations were assessed using multivariable Cox proportional hazards models adjusted for age, sex, and smoking history. Data were statistically analyzed and figures were prepared using R 3.6.3 (R Foundation for Statistical Computing, Vienna, Austria) with a significance level of 0.05.
We obtained 321 test reports of routine comprehensive genomic profiling between January 2016 and October 2019. After excluding 88 of other histologic types, 233 patients with NSCLC were included in the study. Table 1 summarizes their clinical characteristics. The median age was 61 (IQR, 51–67) years, and 43.1% were female. Formalin-fixed paraffin-embedded tissues and plasma samples were obtained from 177 (76.3%) and 55 (23.7%) patients, respectively, for genomic profiling. The most prevalent histological subtype was lung adenocarcinoma (198, 85.0%). Most patients had stage IV tumors (200, 85.8%). The TMB significantly differed between females and males, and between smokers and non-smokers (P = 0.001), but not according to age or tumor stage (Table S2).
The spectrum of significantly mutated genes was similar to that in our previous study (18). The most common driver mutations were in EGFR (51%) followed by KRAS (9%), PIK3CA (7%), ALK (7%), MET (6%), and BRAF (5%) (Figure 1). Mutations were present in tumor suppressor genes including TP53, KEAP1, RB1, PTEN, and APC. ALK fusions, HER2 insertions, MEK mutations, KIF5B-RET, and CD74-ROSI were mutually exclusive. Figure S1 compares mutation in significantly mutated genes in adenocarcinoma between this cohort and The Cancer Genome Atlas (TCGA) lung adenocarcinoma cohort (n = 515). Notable differences from TCGA data included EGFR (51.1 vs 13.0%, P < 0.001), KRAS (9.4 vs 30.0%; P < 0.001), CDKN2A (10.7 vs 4.0%, P < 0.001), KEAP1 (9.9 vs 19%, P < 0.001), NAV3 (8.6 vs 20%, P < 0.001), FAT3 (8.1 vs 21%, P < 0.001), PTPRD (6.9 vs 17%, P < 0.001), and NF1 (5.1 vs 12%, P = 0.005). High TMB was more common in young women and smokers (P = 0.001) (Table S2). Genomic alterations in TP53, EGFR, BRAF, ROSI, and RET were associated with significantly higher TMB than wild types cases (Figure S2).
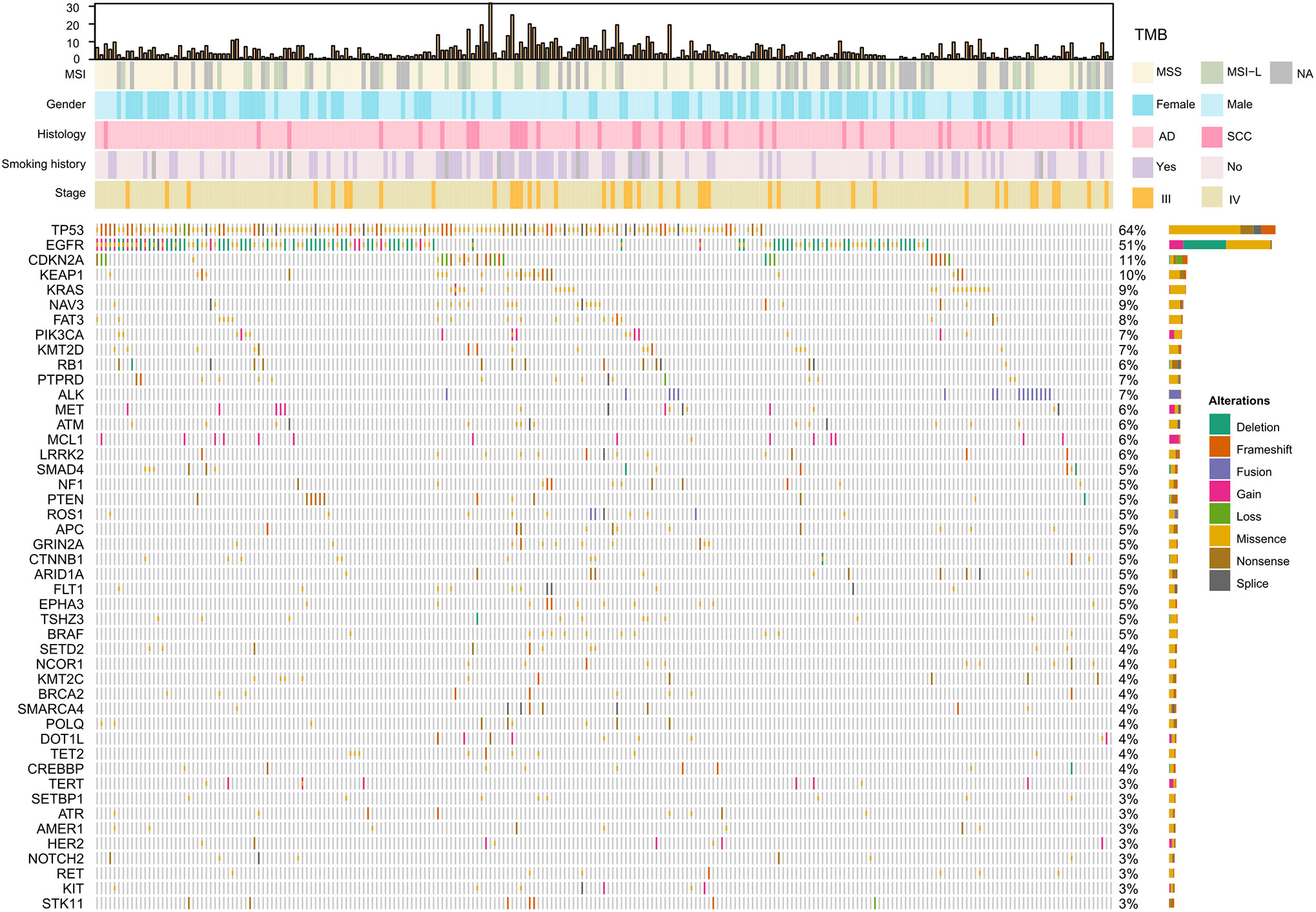
Figure 1 Significantly mutated genes and clinicopathological features of 233 patients with NSCLC. TMB, tumor mutation burden; MSS, microsatellite stability; MSI-L, Microsatellite instability—low; NA, not available; AD, adenocarcinoma; SCC, squamous cell carcinoma.
We also analyzed associations between driver alterations and clinical characteristics. EGFR mutations were more common in women(65.3 vs 34.7%; P <.001), and non-smokers (61.3 vs 35.1%; P <. 001) (Table 2). The mutation rate of KRAS was significantly higher in men than in women (12.9 vs 5.0%, P = 0.04) and in heavy smokers than in non-smokers (17.6 vs 6.0%, P = 0.006) (Table 2). Patients with ALK rearrangements were more often younger (mutant vs WT: 53.0 vs 61.0 years; P = 0.003) (Table 2).
The median follow-up of 171 patients with appropriate data and advanced disease was 14.4 (IQR 9.4–22.8) months. The mOS from initial diagnosis was 22.7 (95% CI, 17.7–28.7) months (Figure S3A). OS did not significantly differ between smokers and non-smokers (mOS: 19.6 vs 26.4 months; P = 0.54) (Figure S3C). Although the OS was longer for patients with adenocarcinoma (n = 146) than SCC (n = 25), the difference did not reach statistical significance (mOS: 24.0 vs 18.4 months; P = 0.57) (Figure S3D). We compared survival between patients with or without driver mutations. Patients harboring KRAS (mutant, n = 17; WT, n = 154) and TP53 (mutant, n = 103; WT, n = 68) mutations had a shorter OS (mOS: 11.3 vs 24.0 months; P = 0.16, and 18.7 vs 28.7 months, P = 0.018, respectively); whereas among patients harboring a driver mutation for which a targeted therapy was available, OS was longer for those whose tumors harbored EGFR (P = 0.069) and ALK (P = 0.51) mutations (Figures 2A–C and Figure S4A). Analysis of the potential prognostic implications of immune-related biomarkers associated a low TMB with longer OS (mOS: 18.4 vs 27.7 months; P = 0.061); OS did not significantly differ between low MSI (n = 27) and microsatellite stability (MSS) (n = 112) groups (P = 0.61) (Figure 2D and Figure S4D).
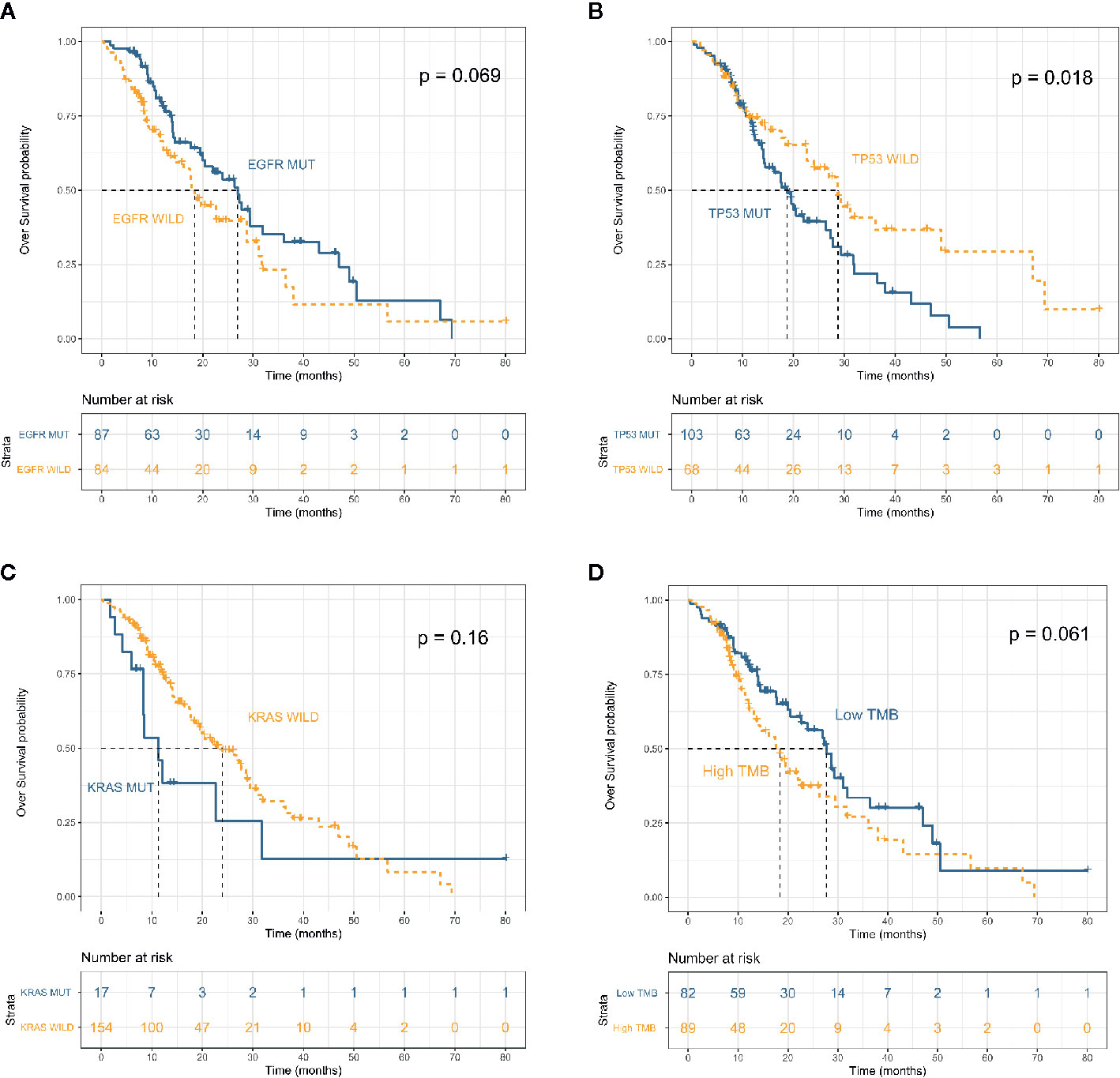
Figure 2 Overall survival in patients with advanced non–small cell lung cancer (n = 171) stratified by mutation status of EFGR (A), TP53 (B) and KRAS (C) and tumor mutation burden (D).
All advanced patients (n = 171) received first-line therapy. Most patients with driver gene-negative tumors (53.1%) were treated with platinum-based regimens, whereas most patients with mutated tumors (EGFR, ALK, and ROS1) (65.4%) were treated with TKIs. Table 3 shows the details of the treatment regimens. Most (66 [64.3%] of 87) patients with mutated EGFR were treated with the EGFR TKIs, gefitinib (38 [24.1%]), erlotinib (n = 3), icotinib (n = 4), and afatinib (n = 11). Similarly, most patients (8 [61.5%] of 13) with ALK fusions were treated with the ALK TKIs crizotinib, alectinib, and lorlatinib. OS was longer for patients treated with TKIs (EGFR, ALK, and ROS1) than with platinum-based chemotherapy (mOS: 27.7 vs 18.4 months; p = 0.038; Figure 3A). Survival outcomes in response to EGFR TKIs notably differed among exon 19 deletions, L858R mutations, and acquired resistance T790M in EGFR. The mOS was the longest and second longest among patients with an acquired T790M mutation and exon 19 del, respectively, and the shortest among patients with L858R point mutations (Figure 3B).
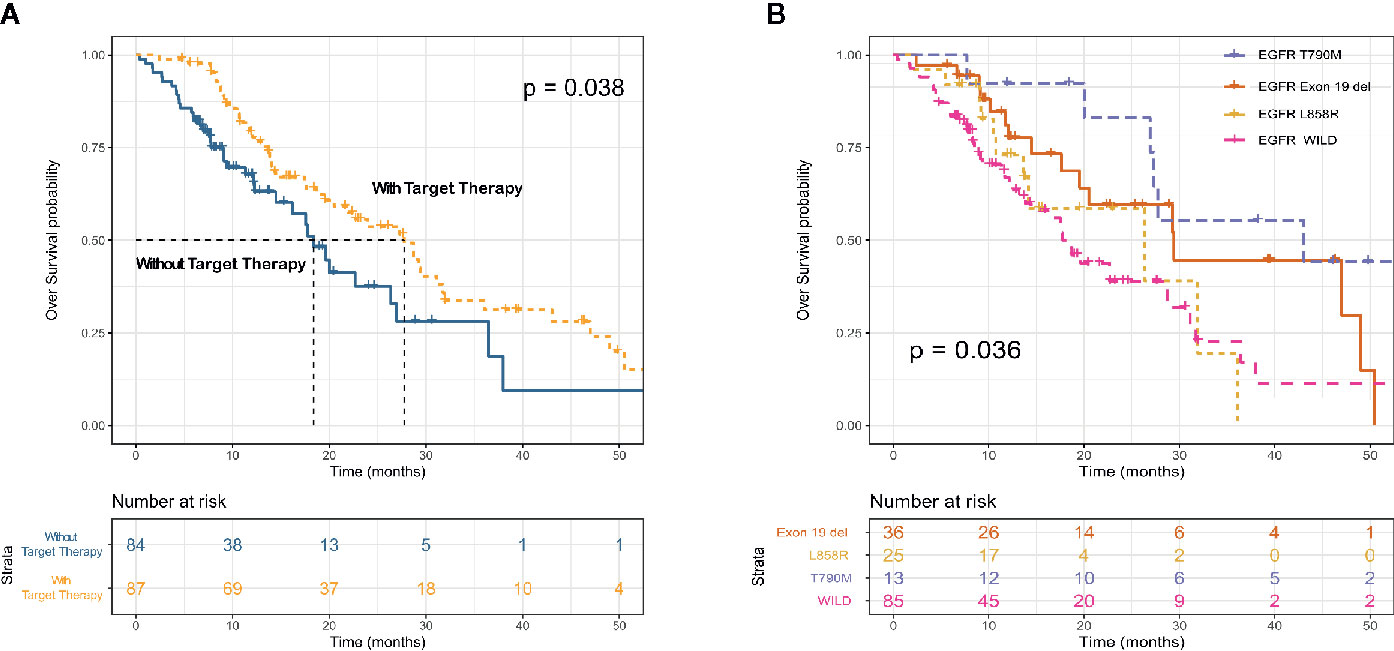
Figure 3 Kaplan–Meier survival curves for overall survival according to receipt of targeted therapy (EGFR TKI and ALK TKI) (A) and EGFR mutation subtype (B).
Comprehensive genomic profiling identified 86 EGFR mutated tumors among 171 patients with advanced cancers and complete follow-up information. Most of these were exon 19 deletions (n = 44, 51.1%) or L858R mutations (n = 29, 33.7%). The most prevalent concurrent mutations were TP53 in 64% and PIK3CA in 7.0% of tumors with EGFR mutations (Figure 4A). We further analyzed the clinical outcomes and possible resistance mechanisms among patients with concurrent mutations treated with EGFR TKIs. Univariate analysis associated a shorter OS with a concurrent mutation in TP53 (HR, 2.96; 95% CI, 1.07–8.22; P = 0.037). A PIK3CA mutation was also associated with a difference in OS, but the difference was statistically significant (HR, 3.79; 95% CI, 0.85–16.95; P = 0.08). Multivariable Cox proportional hazard models (adjusted for TP53, PIK3CA, age, sex, and stage) associated TP53 with a shorter treatment duration (HR, 0.37; 95% CI, 0.98–7.81; P = 0.05, Figure 4B).
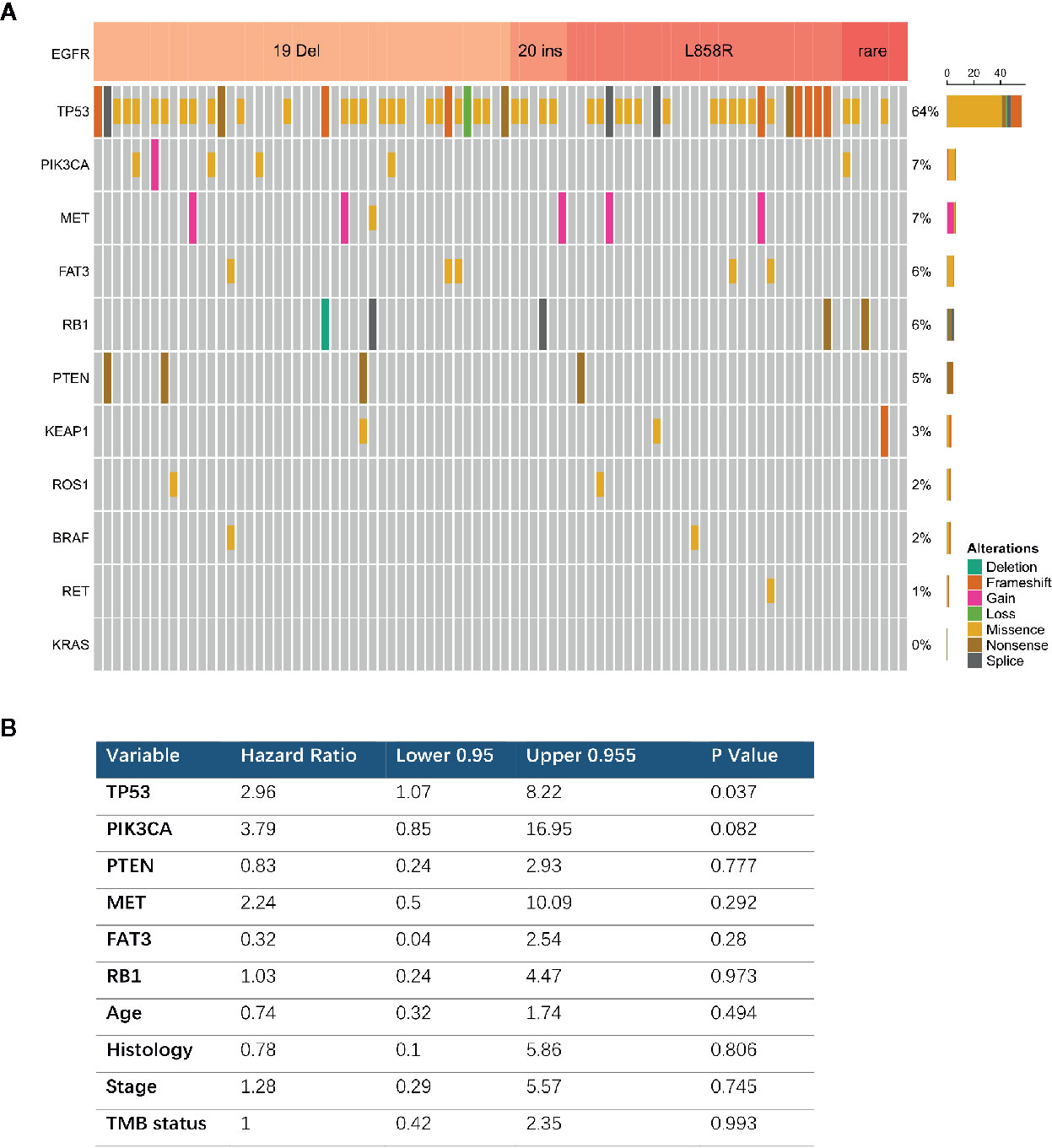
Figure 4 Concurrent genomic alteration with EGFR mutation and treatment efficacy of EGFR-TKIs. (A) Prevalence of concurrent genes in EGFR mutant patients (n = 86). (B) Cox Multivariable analysis of overall survival among patients with concurrent genomic alteration treaded with EGFR TKI.
The introduction of targeted therapy has changed the therapeutic landscape for lung cancers over the past decade (19). New targeted agents are continuously being approved by the FDA, and increasing numbers of patients are benefiting from precision medicines. The present study found that 62% of patients with NSCLC harbored actionable genetic alterations, meaning that tumor progression and metastasis could be managed with targeted drugs that also improved their quality of life. Precision medicine is the future of lung cancer management, and the primary prerequisite is a genomic map of tumors. Although the traditional molecular pathology detection methods (ARMS, IHC, and FISH) have been routinely applied, their limitations have become more obvious in the face of increasing numbers of targets. The emergence of high-throughput sequencing has provided a new tool for clinical molecular pathology detection. The simultaneous detection of all mutations related to therapy and prognosis is cost-effective and not only provides clinicians with a panoramic genetic map of cancer but also avoids the invasive collection of multiple clinical samples from patients. Based on the current evidence, ESMO recommends routine use of NGS on tumor samples in advanced NSCLC to accelerate cancer research and drug development through clinical trials, provide access to innovation to patients and to collect data (20). The turnaround time from specimen acquisition to clinical reports of assay findings at our molecular pathology laboratory is 6 days. Genomic reports provide clinicians with all information needed about possible sensitivity and resistance to targeted drugs. In addition to drug targets, our study proved that comprehensive genomic profiling also enabled the discovery of some mutations that can predict prognosis, such as TP53 mutation was associated with shorter survival, which is consistent with the findings of other studies (21, 22). The present findings provide RWE that comprehensive genomic profiling will benefit the survival of patients with lung cancer.
The EGFR 19 exon deletion (19del) and the 21 exon L858R mutation (21L858R) are classic sensitive mutations, but tumors with such mutations respond differently to TKIs. The ARCHER 1050 study of the second-generation EGFR TKI, dacomitinib, and the FLAURA study of the third-generation EGFR TKI, osimertinib, found that progression-free survival was longer for patients with 19del than with 21L858R (23, 24). Our subgroup analysis generated similar findings, indicating that better treatment options are needed for patients with EGFR 21L858R. The NEJ026 and CTONG 1509 study suggested that the anti-angiogenesis + EGFR TKI could confer a survival benefit upon patients with L858R-positive tumors (25, 26). Various TKIs have different effects on rare EGFR mutations such as E18, G719X, E21, L861Q, and E20 S768I. The objective response rate was better for the second-generation EGFR TKI agent afatinib than the first-generation agents gefitinib/erlotinib (27). The efficacy of both first- and second-generation EGFR-TKI is significantly lower than that of conventional chemotherapy for patients with E20 insertion mutations (28). Besides, it is founded that TP53 mutation and other oncogenic concurrent mutations were associated with worse OS in patients with EGFR mutant NSCLC (29–31). Therefore, patients with EGFR mutations require accurate molecular diagnosis before starting individualized therapy.
Acquired drug resistance in targeted therapy has always been an obstacle to improving the long-term survival of patients. Repeated biopsies after drug resistance are highly significant. Acquired drug resistance mutations can be monitored in real time, so physicians can make timely adjustments to treatment regimens. Replacing EGFR TKIs is the most direct strategy when the EGFR on-target resistance mechanism occurs. Osimertinib can be administered when a T790M mutation is acquired, and combined first- and third-generation TKIs can be administered when the osimertinib resistance mutation C797S is detected (32). Patients with EGFR TKI resistance caused by the activation of other driver genes can be treated with combined targeted therapies; for example, savolitinib+EGFR TKI can overcome EGFR TKI resistance caused by MET amplification (33). Patients in our cohort with acquired T790M mutations treated with osimertinib had the longest OS.
This study has some limitations. Although the rate of actionable genomic alterations was higher in our cohort than in the Caucasian population, the proportion of our patients treated with targeted drugs was lower than that in other studies. This is partly due to the fact that there were not many targeted drugs included in medical insurance when the patient received treatment. The CFDA has so far approved only the EGFR and ALK TKIs, gefitinib, erlotinib, afatinib, osimertinib, crizotinib, ceritinib, and alectinib, and these are covered by medical insurance. This issue will be resolved when more targeted drugs are approved such as capmatinib, selpercatinib, and lorlatinib to treat ROS1, MET exon 14 skipping, and RET fusion (34). Secondly, physicians will be able to recommend patients with more mutation-positive clinical and pathological features for comprehensive genomic testing. This selection bias will increase the proportions of specific mutations. Our comprehensive genomic testing also detected biomarkers that are related to the effects of immunotherapy such as TMB, MSI, and immune positively and negatively related gene mutations. However, because of the exorbitant cost of immunotherapy, only eight patients received PD-1/PD-L1 inhibitors. Therefore, we did not analyze correlations between these biomarkers and the effects of immune checkpoint inhibitors.
In summary, our findings presented a clear genomic landscape of the mutation frequencies of oncogenic drivers from 233 patients with NSCLC and their associations with sex, smoking status, and TMB. These findings provide additional RWE about the potential importance of targeted treatment options for patients with NSCLC harboring clinically actionable mutations.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: The data of this project has been submitted to CNGBdb (https://db.cngb.org/). The accession number of this project in CNGBdb is CNP0001436.
The studies involving human participants were reviewed and approved by the Institutional Review Board of Fifth Affiliated Hospital of Sun Yat-Sen University. The patients/participants provided their written informed consent to participate in this study.
WL, BZ, and DS participated in study conception and design. XP, HC, YW, WZ, and LH enrolled and managed patients. XP, HC, DS, WJ and ZS carried out collection and assembly of data. DS and WL were involved in data analysis and interpretation. DS prepared the manuscript and manuscript figures. WL and KW edited, critically read, and revised the manuscript. All authors contributed to the article and approved the submitted version.
DS and DJ is a current employee of BGI Genomics.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank all patients who participated in this study and their families. We also thank Huan Wang, and Hengliang Chen for providing research support.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.630717/full#supplementary-material
1. Ettinger DS, Wood DE, Aggarwal C, Aisner DL, Akerley W, Bauman JR, et al. NCCN Guidelines Insights: Non-Small Cell Lung Cancer, Version 1.2020. J Natl Compr Canc Netw (2019) 17(12):1464–72. doi: 10.6004/jnccn.2019.0059
2. Chinese Society of Clinical Oncology. Guidelines of Chinese society of clinical oncology: primary lung cancer. (2019), 48–9.
3. Recondo G, Facchinetti F, Olaussen KA, Besse B, Friboulet L. Making the first move in EGFR-driven or ALK-driven NSCLC: first-generation or next-generation TKI? Nat Rev Clin Oncol (2018) 15(11):694–708. doi: 10.1038/s41571-018-0081-4
4. Andre F, Nowak F, Arnedos M, Lacroix L, Viens P, Calvo F. Biomarker discovery, development, and implementation in France: a report from the French National Cancer Institute and cooperative groups. Clin Cancer Res (2012) 18(6):1555–60. doi: 10.1158/1078-0432.CCR-11-2201
5. Barlesi F, Mazieres J, Merlio JP, Debieuvre D, Mosser J, Lena H, et al. Routine molecular profiling of patients with advanced non-small-cell lung cancer: results of a 1-year nationwide programme of the French Cooperative Thoracic Intergroup (IFCT). Lancet (2016) 387(10026):1415–26. doi: 10.1016/S0140-6736(16)00004-0
6. Kris MG, Johnson BE, Berry LD, Kwiatkowski DJ, Iafrate AJ, Wistuba II, et al. USing multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. Jama (2014) 311(19):1998–2006. doi: 10.1001/jama.2014.3741
7. Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med (2004) 350(21):2129–39. doi: 10.1056/NEJMoa040938
8. Shaw AT, Kim DW, Nakagawa K, Seto T, Crino L, Ahn MJ, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med (2013) 368(25):2385–94. doi: 10.1056/NEJMoa1214886
9. Shaw AT, Ou SH, Bang YJ, Camidge DR, Solomon BJ, Salgia R, et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med (2014) 371(21):1963–71. doi: 10.1056/NEJMoa1406766
10. Wolf J ST, Han J, Reguart N, Garon EB, Groen HJM, et al. (2018). Results of the GEOMETRY mono-1 phase II study for evaluation of the MET inhibitor capmatinib (INC280) in patients (pts) with METΔex14 mutated advanced non-small cell lung cancer (NSCLC), in: Presented at: European Society for Medical Oncology, Munich, Germany, Oct19–23. doi: 10.1093/annonc/mdy424.090
11. Agarwala V, Khozin S, Singal G, O’Connell C, Kuk D, Li G, et al. Real-World Evidence In Support Of Precision Medicine: Clinico-Genomic Cancer Data As A Case Study. Health Aff (Millwood) (2018) 37(5):765–72. doi: 10.1377/hlthaff.2017.1579
12. Sherman RE, Anderson SA, Dal Pan GJ, Gray GW, Gross T, Hunter NL, et al. Real-World Evidence - What Is It and What Can It Tell Us? N Engl J Med (2016) 375(23):2293–7. doi: 10.1056/NEJMsb1609216
13. Liu L, Liu H, Shao D, Liu Z, Wang J, Deng Q, et al. Development and clinical validation of a circulating tumor DNA test for the identification of clinically actionable mutations in nonsmall cell lung cancer. Genes Chromosomes Cancer (2018) 57(4):211–20. doi: 10.1002/gcc.22522
14. DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet (2011) 43(5):491–8. doi: 10.1038/ng.806
15. Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S, et al. Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer: A Joint Consensus Recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn (2017) 19(1):4–23. doi: 10.1016/j.jmoldx.2016.10.002
16. Niu B, Ye K, Zhang Q, Lu C, Xie M, McLellan MD, et al. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics (2014) 30(7):1015–6. doi: 10.1093/bioinformatics/btt755
17. Kautto EA, Bonneville R, Miya J, Yu L, Krook MA, Reeser JW, et al. Performance evaluation for rapid detection of pan-cancer microsatellite instability with MANTIS. Oncotarget (2017) 8(5):7452–63. doi: 10.18632/oncotarget.13918
18. Liu L, Liu J, Shao D, Deng Q, Tang H, Liu Z, et al. Comprehensive genomic profiling of lung cancer using a validated panel to explore therapeutic targets in East Asian patients. Cancer Sci (2017) 108(12):2487–94. doi: 10.1111/cas.13410
19. Thomas A, Liu SV, Subramaniam DS, Giaccone G. Refining the treatment of NSCLC according to histological and molecular subtypes. Nat Rev Clin Oncol (2015) 12(9):511–26. doi: 10.1038/nrclinonc.2015.90
20. Mosele F, Remon J, Mateo J, Westphalen CB, Barlesi F, Lolkema MP, et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: a report from the ESMO Precision Medicine Working Group. Ann Oncol (2020) 31(11):1491–505. doi: 10.1016/j.annonc.2020.07.014
21. La Fleur L, Falk-Sorqvist E, Smeds P, Berglund A, Sundstrom M, Mattsson JS, et al. Mutation patterns in a population-based non-small cell lung cancer cohort and prognostic impact of concomitant mutations in KRAS and TP53 or STK11. Lung Cancer (2019) 130:50–8. doi: 10.1016/j.lungcan.2019.01.003
22. Singal G, Miller PG, Agarwala V, Li G, Kaushik G, Backenroth D, et al. Association of Patient Characteristics and Tumor Genomics with Clinical Outcomes Among Patients With Non-Small Cell Lung Cancer Using a Clinicogenomic Database. JAMA (2019) 321(14):1391–9. doi: 10.1001/jama.2019.3241
23. Wu YL, Cheng Y, Zhou X, Lee KH, Nakagawa K, Niho S, et al. Dacomitinib versus gefitinib as first-line treatment for patients with EGFR-mutation-positive non-small-cell lung cancer (ARCHER 1050): a randomised, open-label, phase 3 trial. Lancet Oncol (2017) 18(11):1454–66. doi: 10.1016/S1470-2045(17)30608-3
24. Soria JC, Ohe Y, Vansteenkiste J, Reungwetwattana T, Chewaskulyong B, Lee KH, et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med (2018) 378(2):113–25. doi: 10.1056/NEJMoa1713137
25. Saito H, Fukuhara T, Furuya N, Watanabe K, Sugawara S, Iwasawa S, et al. Erlotinib plus bevacizumab versus erlotinib alone in patients with EGFR-positive advanced non-squamous non-small-cell lung cancer (NEJ026): interim analysis of an open-label, randomised, multicentre, phase 3 trial. Lancet Oncol (2019) 20(5):625–35. doi: 10.1016/s1470-2045(19)30035-x
26. Zhou Y-LW Q, Cheng Y, Liu Y, Chen G, Cui J, Yang N, et al. CTONG 1509: Phase III study of bevacizumab with or without erlotinib in untreated Chinese patients with advanced EGFR-mutated NSCLC. Ann Oncol (2019) 30:v603. doi: 10.1093/annonc/mdz260.002
27. Yang JC, Sequist LV, Geater SL, Tsai CM, Mok TS, Schuler M, et al. Clinical activity of afatinib in patients with advanced non-small-cell lung cancer harbouring uncommon EGFR mutations: a combined post-hoc analysis of LUX-Lung 2, LUX-Lung 3, and LUX-Lung 6. Lancet Oncol (2015) 16(7):830–8. doi: 10.1016/S1470-2045(15)00026-1
28. Yasuda H, Kobayashi S, Costa DB. EGFR exon 20 insertion mutations in non-small-cell lung cancer: preclinical data and clinical implications. Lancet Oncol (2012) 13(1):e23–31. doi: 10.1016/S1470-2045(11)70129-2
29. Aggarwal C, Davis CW, Mick R, Thompson JC, Ahmed S, Jeffries S, et al. Influence of TP53 Mutation on Survival in Patients With Advanced EGFR-Mutant Non-Small-Cell Lung Cancer. JCO Precis Oncol (2018) 2:1–29. doi: 10.1200/PO.18.00107
30. Kim Y, Kim CH, Lee HY, Lee SH, Kim HS, Lee S, et al. Comprehensive Clinical and Genetic Characterization of Hyperprogression Based on Volumetry in Advanced Non-Small Cell Lung Cancer Treated with Immune Checkpoint Inhibitor. J Thorac Oncol (2019) 14(9):1608–18. doi: 10.1016/j.jtho.2019.05.033
31. Offin M, Chan JM, Tenet M, Rizvi HA, Shen R, Riely GJ, et al. Concurrent RB1 and TP53 Alterations Define a Subset of EGFR-Mutant Lung Cancers at risk for Histologic Transformation and Inferior Clinical Outcomes. J Thorac Oncol (2019) 14(10):1784–93. doi: 10.1016/j.jtho.2019.06.002
32. Arulananda S, Do H, Musafer A, Mitchell P, Dobrovic A, John T. Combination Osimertinib and Gefitinib in C797S and T790M EGFR-Mutated Non-Small Cell Lung Cancer. J Thorac Oncol (2017) 12(11):1728–32. doi: 10.1016/j.jtho.2017.08.006
33. Tang ZH, Lu JJ. Osimertinib resistance in non-small cell lung cancer: Mechanisms and therapeutic strategies. Cancer Lett (2018) 420:242–6. doi: 10.1016/j.canlet.2018.02.004
Keywords: non-small cell lung cancer, targeted therapy, real-world evidence, comprehensive genomic profiling, survival
Citation: Lv W, Cheng H, Shao D, Wei Y, Zhu W, Wu K, Jiang W, Hu L, Sha Z, Zhong B and Pei X (2021) Treatment Patterns and Survival of Patients With Advanced Non-Small Cell Lung Cancer Guided by Comprehensive Genomic Profiling: Real-World Single-Institute Study in China. Front. Oncol. 11:630717. doi: 10.3389/fonc.2021.630717
Received: 18 November 2020; Accepted: 04 January 2021;
Published: 10 March 2021.
Edited by:
Pasquale Pisapia, University of Naples Federico II, ItalyReviewed by:
Paola Ulivi, Istituto Scientifico Romagnolo per lo Studio e la Cura dei Tumori (IRCCS), ItalyCopyright © 2021 Lv, Cheng, Shao, Wei, Zhu, Wu, Jiang, Hu, Sha, Zhong and Pei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Beilong Zhong, YmVpbG9uZ3podWhhaUBob3RtYWlsLmNvbQ==; Xiaofeng Pei, cGVpeGYzQG1haWwuc3lzdS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.