
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol. , 26 February 2021
Sec. Cancer Genetics
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.629390
With the aging of the population, the incidence of colorectal cancer in China is increasing. One of the epigenetic alterations: CpG island methylator phenotype (CIMP) plays an important role in the incidence of colorectal cancer. Recent studies have shown that CIMP is closely related to some specific clinicopathological phenotypes and multiple molecular phenotypes in colorectal cancer. In this paper, the newest progress of CIMP colorectal cancer in chemotherapeutic drugs, targeted agents and small molecular methylation inhibitors are going to be introduced. We hope to provide potential clinical treatment strategies for personalized and precise treatment of colorectal cancer patients.
Colorectal cancer (CRC) is one of the most common malignant gastrointestinal tumors that seriously threaten the human health. It has a high incidence and mortality worldwide, and the incidence of CRC in China is also increasing year by year (1). Tumorigenesis of colorectal cancer is multistep and complex process involving genetic and epigenetic alterations. Epigenetic alterations refer to changes in gene expression without changing in the DNA sequence, leading to silencing of transcriptional genes or inactivating DNA repair genes and tumor suppressor genes (2). Over past 30 years, more and more studies indicate that epigenetic changes including DNA methylation, histone modification, nucleosome localization and small non-coding RNAs etc. play a key role in the tumorigenesis of colorectal cancer (3).
In recent years, DNA methylation modification has been extensively studied. In humans, DNA methylation to form 5-methylcytosine occurs mainly at the cytosine residues’ fifth carbon position of CpG dinucleotides. Around 60–80% of CpG cytosines are methylated in human somatic cells, however CpG islands are regions with a high frequency of CpG sites mostly located near the transcription start site of promoter genes which are constitutively unmethylated. In 1999, Toyota et al. (4) first proposed a novel subset of CRCs positive for CpG island methylator phenotype (CIMP) that extensively displayed multiple cancer specific genes promoter DNA hypermethylation at some specific set of CpG islands in CRC tissues. CIMP is now considered as a distinct molecular subtype of sporadic CRC which is the initial event of the serrated neoplasia pathway in CRC’s tumorigenesis (5). CpG island methylator phenotype (CIMP) is mediated by DNA methyltransferases (DNMTs) which promotes hypermethylation in promoter associated CpG-rich regions of tumor suppressor genes which are inactivated by transcription, leading to development and progression of CRC (6). Although there is no consensus on the definition of CIMP and no methodology has been proven to be superior to another (7–9), CIMP has been still recognized as a hotspot research direction of colorectal cancer in these past 20 years.
CIMP in CRC was confirmed not only tightly associated with some specific clinicopathologic phenotypes, but also closely related to many molecular characteristics. In 2006, Weisenberger and colleagues (10) recommended a novel and sensitive panel of CIMP including five specific genes’ promoter DNA hypermethylation markers to identify CIMP-positive CRCs. The study demonstrated that CIMP-positive was significantly related to female gender, proximal location, MSI-H status, MLH1 methylation, BRAF mutation, and KRAS mutation. In their further large population-based sample analysis, older age, family history of CRC and NSAIDs using before diagnosis related to CIMP-positive were additionally observed. Furthermore, smoke and overweight statistically associated with only female CIMP-positive CRCs were reported (11). Ogino and colleagues (12) selected different promoter loci as CIMP panel markers to identify CIMP-High, CIMP-Low and non-CIMP phenotypes. Follow-up experiments showed that CIMP-Low tumors different from CIMP-High and non-CIMP tumors were tightly correlated to male gender and KRAS mutation (13). In further CIMP subgroup analysis, CIMP-positive tumors independent of MSI status were significantly associated with mucinous or signet ring cell morphology, tumor infiltrating lymphocytes (TIL), peritumoral lymphocytes (PLS), presence of Crohn-like infiltrates, tumor necrosis, tumor cell sheeting, and poor differentiation (14). Interestingly, low level intake of folate was proven to be associated with a trend towards an increased risk of non-CIMP-High colon tumors (15). Recent years, CIMP status was also reported to be positively correlated with F. nucleatum, the gut microbiome component in CRC (16, 17). Using more advanced methods or meta-analysis to evaluate the relationship between CIMP status and the clinicopathological and molecular characteristics in CRC, the conclusions obtained by Guinney et al. (18) and Advani et al. (19) were similar to those of Weisenberger and Ogino. In the past 20 years, CIMP-positive tumors were accepted as a consensus which were positively correlated with female, proximal location, MSI-H status, BRAF mutation and mucinous histology. These epidemiology associations help us explore the underlying cancer prevention and treatment strategies.
A growing number of studies suggest that CIMP might be a potential epigenetic predictor or prognostic biomarker contributed to individualized and precise treatment of colorectal cancer patients (20, 21).
As the most widely used chemotherapeutic drug for colorectal cancer, 5-fluorouracil (5-FU) principally acts as a thymidylate synthase (TS) inhibitor by interrupting DNA replication. In 2007, Shen et al. (22) investigated CIMP status in 188 advanced CRCs who received 5-FU based chemotherapy and found that the median survival in the CIMP-positive subset was 6 months versus 17 months in CIMP-negative subset (P < 0.001) and two-year survival rate was 8% in the CIMP-positive group versus 28% in the CIMP-negative group (Table 1). In multivariate analysis, CIMP-positive cases had a significantly shorter survival (hazard ratio, HR=2.9; P < 0.0001). Jover et al. (23) studied 196 stage II-III CRCs and found that CIMP-positive CRCs did not benefit from 5-FU based adjuvant treatment. The disease-free survival (DFS; log-rank=0.02) of CIMP-positive patients receiving adjuvant 5-FU based chemotherapy was lower than that of CIMP-negative patients. In CIMP-negative CRCs, adjuvant 5-FU based chemotherapy significantly prolonged DFS (log-rank=0.00001). However, it failed to improve DFS (log-rank=0.7) in CIMP-positive CRCs. Multivariate analysis showed adjuvant 5-FU based treatment was not an independent predictor of prognosis in CIMP-positive CRCs (HR=0.8; 95% confidence interval, CI=0.3–2.0). Min and colleagues (24) reached the opposite conclusion by an independent Asian population clinical trials. They performed 124 stage II–III CRCs and reported that CIMP-high CRCs (n=17; 3-year recurrence-free survival, RFS: 100%) who received 5-FU based regimen after surgery had significantly better RFS than those accepted surgery alone (n=7; 3-year RFS: 71.4%; P=0.022). Furthermore, Rijnsoever et al. (25) considered that CIMP-positive status was an independent significant predictor for the survival benefit treated with adjuvant 5-FU based chemotherapy in CRCs. They evaluated CIMP-positive status in 103 stage III CRCs treated with surgery alone and 103 cases treated with surgery plus adjuvant 5-FU based chemotherapy and provided evidence that CIMP positive CRCs could receive longer cumulative survival time from surgery plus 5-FU based treatment than surgery alone (P=0.002). However, CIMP-negative CRCs displayed no these association (P=0.6). For investigating the response of CIMP subtypes to 5-FU based chemotherapy, in the light of MSI, BRAF and KRAS molecular classification, Murcia et al. (26) classified 324 stage II-III CRCs into six study subgroups and reported that microsatellite stability (MSS), BRAF wild-type and CIMP-negative subtype CRCs had a longer time of DFS when they treated with 5-FU based chemotherapy (log rank P = 0.003 for KRAS mutation subgroup and P < 0.001 for KRAS wild-type subgroup). In a multivariate analysis, they found only MSS, BRAF wild-type, KRAS wild-type and CIMP-negative subgroup CRCs independently displayed significant benefit with 5-FU based chemotherapy (HR = 2.06, 95% CI 1.24–3.44, P = 0.005). In these independent clinical trials for stage II-III CRCs, the efficacy of 5-FU based regimen on CIMP or CIMP subgroup patients still seemed controversial. In my opinion, the main reason for the inconsistent results may be related to the different methylation markers and technologies used to define CIMP status, which may lead bias to distinguish the real CIMP-positive patients. For example, MethyLight based on real-time PCR with specific probe was considered much more reliable than MSP to define CIMP status. In Min and Murcia’s studies, the same method is used to define CIMP status. Furthermore, we should not merely consider the efficacy of 5-FU alone for adjuvant chemotherapy. the influence of oxaliplatin, irinotecan and radiotherapy should also be well evaluated. The application of 5-FU plus oxaliplatin- and irinotecan-based chemotherapy increased nearly double survival benefit of CRCs. FOLFIRI (5-FU, calcium folinate and irinotecan) and FOLFOX (5-FU, calcium folinate and oxaliplatin) were two important cytotoxic combined regimens treated with CRCs. Researchers confirmed that first-line FOLFIRI regimen and second-line FOLFOX regimen performed the same clinical efficacy comparing with reverse sequential regimen for metastatic CRCs (35). Zhang et al. (27) investigated 125 metastatic CRCs treated with combination chemotherapy and demonstrated that CIMP-positive metastatic CRCs showed significantly benefit from irinotecan-based regimen followed by FOLFOX rather than the reverse sequential regimen. Median progression free survival (mPFS) was prolonged 8.6 months (mPFS=15.2 vs 6.6 months, P = 0.043) and median overall survival (mOS) was extended 8 months (mOS=20.8 vs 12.8 months, P = 0.11). Shiovitz and colleagues (28) assessed the association between survival of 615 stage III colon cancer patients receiving 5-FU and leucovorin alone or with irinotecan (IFL) after surgery and CIMP status. The results showed that CIMP-positive patients receiving IFL versus FU/LV treatment were tended to increase OS (69 vs. 56%; 95% Cl: 0.37–1.05; P = 0.07), especially for the mismatch repair-intact (MMR-I) subgroup (P = 0.01), while CIMP-negative patients did not have this trend (HR = 1.38; 95% Cl: 1.00–1.89; P = 0.049). CIMP-positive patients with stage III CRC could obviously benefit from irinotecan-based regimen. This might be largely driven by MMR-I tumors which was associated with improved OS (28). For patients with metastatic colorectal cancer, irinotecan-based regimen also improved the survival of CIMP-positive patients. We speculated that stable DNA damage response (DDR) related genes (e.g. RECQ helicases) could enhance the hypersensitivity of CIMP-positive CRC to irinotecan. Of course, this need to be confirmed by further basic study. Previous studies have shown that demethylation treatment can activate multiple cancer cell signaling pathways not only allow the use of less toxic doses of irinotecan but also improve the efficacy of it (36, 37). It was seemed that CIMP-positive status was a potential biomarker to predict irinotecan-based chemotherapy regimen for CRCs.
Cohen et al. (29) studied CIMP status from 292 stage II-III CRCs who received adjuvant modified FOLFOX6 (mFOLFOX6) or XELOX (capecitabine and oxaliplatin). There was no significant difference in OS between CIMP-positive and CIMP-negative patients (HR=1.27; 95%CI: 0.58–2.80; P=0.55). Han et al. (30) analyzed CIMP status based on 322 stage II–III CRCs who received adjuvant FOLFOX chemotherapy and found that CIMP-high status CRCs had no significantly associated with 3-year DFS comparing with CIMP-Low or CIMP-negative CRCs (P=0.31). Although study illustrated that CIMP status could not be a significant prognostic biomarker for adjuvant oxaliplatin-based chemotherapy regimens in stage II-III CRCs, there seemed to be a tendency that the efficacy of oxaliplatin in CIMP-positive patients was worse than that in CIMP-negative patients. In a large-population clinical cohort research, Gallois et al. (31) investigated CIMP status of 1867 stage III CRCs who treated with adjuvant FOLFOX or FOLFOX plus cetuximab regimen and found that the OS (HR = 1.46; 95% CI: 1.02–1.94; P = 0.04) and survival after recurrence (SAR; HR = 1.76; 95% CI: 1.20–2.94; P < 0.0004) of CIMP-positive patients significantly shortened, but no significant difference of DFS (HR = 1.15; 95% CI: 0.86–1.54; P = 0.34) were observed. Cha et al. (32) divided 153 metastatic CRCs treated with systemic chemotherapy into three CIMP groups. The results were demonstrated that the OS were significantly different among the three CIMP groups with a median of 9.77, 22.2, and 35.7 months for the high, low and negative groups, respectively (P< 0.001). In 5-FU and oxaliplatin first-line chemotherapy (n=128), the median OS was 6.77, 23.8, and 37.9 months for the high, low and negative groups, respectively (P<0.001), while the median PFS was 1.83, 7.87 and 9.97 months, respectively (P=0.002). CIMP-high cases were significantly associated with worst efficacy of therapy. In 5-FU and irinotecan second-line chemotherapy (n=86), only the median OS was shown a significant difference according to the CIMP status with values of 2.90, 13.4, and 20.4 months for the high, low and negative groups, respectively (P<0.001). The CIMP-high status was considered as a negative prognostic factor for metastatic CRCs received with chemotherapy. Bae and colleagues (33) inspected 1,370 stage I–IV CRCs treated with surgery and/or chemotherapy. Compared with CIMP-P1 (CIMP-L), CIMP-negative CRCs showed better 5-year cancer-specific survival (CSS; HR=0.47; 95% CI: 0.28–0.78) and better 5-year RFS (HR=0.50; 95% CI: 0.29–0.88). The CIMP-H CRCs displayed best 5-year CSS from chemotherapy was observed, however no such trend was found in no chemotherapy analysis. Multiple clinical studies seemed to reach an agreement on CIMP-positive CRCs associated with poor survival, but failed to display a prognostic value of CIMP-positive CRCs who were treated with oxaliplatin-based adjuvant chemotherapy regimen. Albumin-bound paclitaxel is an anti-microtubule drug, which interferes with the rearrangement of microtubules, leading to the cessation of mitosis, thus inhibiting the growth of cancer cells. Overman et al. (34) reported a phase II clinical trial which enrolled 21 CIMP-high metastatic CRCs and no efficacy of nab-paclitaxel was observed. Oxaliplatin-based or paclitaxel-based chemotherapy might be indirectly affected by CRCs’ CIMP status.
The DNA repair gene O6-methylguanine-DNA methyltransferase (MGMT) promoter methylation is a frequent and early event in colorectal tumorigenesis which was considered benefit from alkylating agents such as temozolomide (TMZ) (38). In a phase II study, TMZ showed a modest activity and achieved an average 10% RR in heavily pretreated metastatic CRC patients with MGMT hypermethylation (39). A recent study was shown that irinotecan and TMZ (TEMIRI) combination regimen was reached the primary end point in irinotecan-sensitive, MGMT methylated and MSS pretreated metastatic CRC patients (40). Six out of 25 patients achieved PR (ORR=24%; 95% CI, 11–43%). The mPFS and mOS were 4.4 and 13.8 months, respectively. All patients with MGMT-positive IHC were non-responders. Consistently, patients with MGMT-negative/low tumors had a significantly longer mPFS than others (6.9 vs 2.0 months; HR = 0.29, 95% CI 0.02–0.41; P = 0.003). The reason of the efficacy of TEMIRI regimen for metastatic CRC patients with MGMT methylation and absent/low might be the inhibition of topoisomerase II enhances the cytotoxicity of alkylating agents.
Cetuximab, an epidermal growth factor receptor (EGFR) inhibitor, is an IgG1 monoclonal antibody specifically targeting EGFR overexpression and widely used in metastatic CRCs. In the 2 studies described above, Zhang et al. (27) demonstrated that the PFS of CIMP-positive and KRAS wild-type CRCs who treated with cetuximab was shorter than that of CIMP-negative CRCs with KRAS wild-type (mPFS, 2.1 vs. 5.1 months, P = 0.11) and objective response rate (ORR) was also decreased (20.0 vs. 24.4%, P = 0.90). Although this study did not show statistical significance, it seemed to suggest that CIMP-positive phenotype might be a biological negative predictor of efficacy of anti-EGFR antibody (Table 2). Also Gallois and colleagues (31) concluded that the application of cetuximab in CIMP-positive stage III CRCs bought a non-significant trend of negative efficacy. Ouchi et al. (41) analyzed 97 KRAS wild-type metastatic CRCs received anti-EGFR antibody by advanced genome-wide DNA methylation technique and divided patients into highly methylated epigenotype (HME), intermediate methylated epigenotype (IME), and low methylated epigenotype (LME). The results were shown that ORR (35.7 vs 6.3%, P = 0.03), disease control rate (DCR; 75 vs 31.3%, P = 0.005), PFS (HR = 0.22; 95% CI, 0.13–0.57; P < 0.001) and OS (HR = 0.19; 95% Cl, 0.06–0.54; P < 0.001) were significantly better in LME subgroup CRCs compared with HME subgroup CRCs. Although merely one study displayed statistical significance, we still believe that CIMP-positive phenotype is a negative efficacy indicator for KRAS wild-type metastatic CRCs who received anti-EGFR antibody. First, CIMP was significantly associated with the right side CRC which was considered as an independent negative prognostic factor to cetuximab therapy (42, 43). A clear mechanistic understanding to explain the worse outcome of CIMP-positive patients is currently lacking. EGFR promoter hypermethylation status may be responsible and has been reported to be more relevant than primary right colon in the prediction of negative response to anti-EGFR therapy in patients with metastatic CRCs (44). Secondly, we speculated that some hypermethylation genes increased the resistance of anti-EGFR antibody and lead to drug resistance. Demethylation agents were given to CIMP-positive CRCs which might reverse the resistance and improve patients’ survival. This is a potential research direction to change the therapeutic strategy of metastatic CRC in the future.
DNMT1, DNMT3A and DNMT3B are the canonical cytosine-5 DNMT enzymes. Their functions include not only the establishment and maintenance of DNA methylation patterns, but also the regulation of multiple gene functions, including transcriptional silencing, transcriptional activation and post-transcriptional regulation (45). Most widely studied DNA methylation inhibitors were 5-azacytidine (Azacitidine, 5-Aza-CR), 5-aza-2’-deoxycytidine (Decitabine, 5-Aza-CdR) and guadecitabine (SGI-110) formed irreversible complexes with DNMTs by substituting methylated cytosine targets during DNA replication, leading to the depletion of the enzyme and cytosine during cell division, passive DNA demethylation, tumor suppressor genes’ re-expression, proliferation control and carcinogenesis process inactivation (46, 47). It was suggested that CIMP-positive CRCs might potentially benefit from the treatment with DNA methylation inhibitors alone or combination (48, 49). Garrido-Laguna et al. (50) confirmed 20 patients with KRAS wild-type metastatic CRC receiving sequential decitabin and panitumumab were well tolerated in phase I/II clinical trials (Table 3). Two of patients previously received cetuximab had a partial response (PR). Ten patients had stable disease (SD). Although CIMP status was not inspected, the combination of DNA methylation inhibitor and panitumumab was shown activity in KRAS wild-type metastatic CRCs previously treated with cetuximab. Overman and colleagues (51) demonstrated that 26 metastatic CRCs treated with oxaliplatin-based regimen refractory receiving azacitidine and CAPOX (capecitibine and oxaliplatin) in phase I/II clinical trials. The results displayed 14 patients were CIMP-high. However, not correlated with SD and PFS. In this clinical study, CIMP status was failed in validating as a predictive factor for DNA methylation inhibitor. Azad et al. (52) enrolled 47 metastatic CRCs treated with azacitidine and histone deacetylase inhibitor entinostat in phase II clinical study. Although patients were tolerable with combination epigenetic therapy, no significant clinical activity was observed. Recently, Jansen and colleagues (53) investigated nine eligible patients with pretreated unresectable liver-predominant metastases from solid tumors and evaluated the safety and antitumor activity of administrating decitabine by hepatic arterial infusion (HAI) in phase I clinical trial. Four out of all patients suffered from CRC who were more heavily pretreated with chemotherapy. Results were shown decitabine could be safely administered by HAI. No objective response was observed, while after treatment, the upregulation of cancer testis antigens (CTAs) expression indicated decitabine combined with immunotherapy could be candidate treatment in the further study. From the above studies, it seemed that DNA methylation inhibitors combined with the traditional chemotherapy were shown no significant effective, while the efficacy of combination anti-EGFR antibody or immunotherapy seemed to be worth looking forward to. Although low-dose DNA methylation inhibitors show demethylation and promote apoptosis, inhibiting DNMT alone may not be sufficient to induce durable and robust transcriptional gene re-expression. DNA methylation inhibitors as immune modulators have been consequently considered inducing CTAs expression in CRC which stimulate cytotoxic T-cell responses and antitumor immunity (54). Furthermore, DNA hypermethylation of tumor-infiltrating immune cells or their ligands (e.g. PD-1, CTLA-4, TIM-3, TIGIT, PD-L1, and galectin-9) leading to tumor evasion from host immunosurveillance could be major contributors to the upregulation of immune checkpoints (55). Combining immunotherapy to evaluate the activity of DNA methylation inhibitors is the most promising research direction in the future. In addition, epigenetic therapies included not only DNA methylation inhibitors, but also HDAC inhibitor, BET inhibitor and EZH2 inhibitor. Different epigenetic alterations should be given appropriate interventions. Development of these therapies will provide exciting opportunities for novel and improved therapeutic interventions in CRC (56).
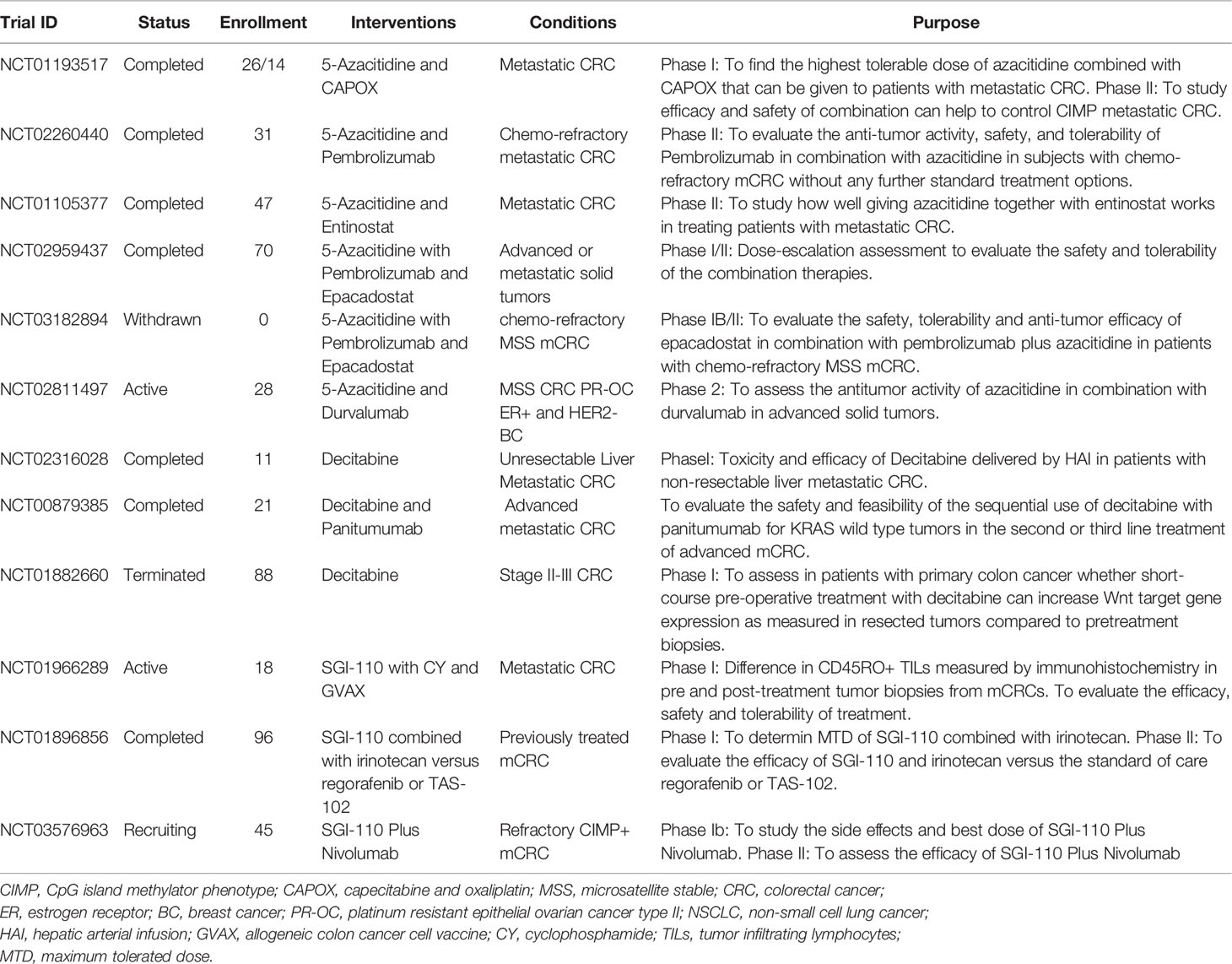
Table 3 Clinical trials involving DNA methylation inhibitors listed on.www.clinicaltrials.gov.
In summary, we reviewed the latest progress of CIMP CRCs’ characteristics and treatment. We clearly realize that CRCs with CIMP phenotype are tightly related to the pathological features of female, older age and right side colon, as well as molecular characteristics of BRAF mutation and MSI-H status. Certainly, with the wide application of next generation sequencing technology, more accurate method to distinguish CRCs’ CIMP status will be are constantly emerging. For chemotherapy, CIMP-positive CRCs were potentially benefit from irinotecan-based regimen rather than oxaliplatin-based regimen. For targeted therapy, negative efficacy from anti-EGFR antibodies seems to be associated with CIMP-positive CRCs. However, the mechanism of these phenomena needs to be further explored in the future. Clinicians are increasingly aware of the importance of CIMP phenotype in CRC. A various of DNA methylation inhibitors alone or especially combination with immunotherapy are undergoing clinical trials. These frontier studies provide potential individualized precise treatment strategies for patients with CRC.
All data collected, generated, or analyzed during this study are included in this published article.
The authors have contributed equally to this work. All authors contributed to the article and approved the submitted version.
This work was financially supported by the Guidance Plan of Natural Science Foundation of Liaoning Province (2019-ZD-0302), Dalian Youth Science and Technology Star Plan (2019RQ096), and Clinical plus Excellence Project (2020ZYA003) from Shanghai Nucleic Acid Chemistry and Nanomedicine Key Laboratory.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
5-Aza-CdR, 5-aza-2’-deoxycytidine; 5-Aza-CR, 5-azacytidine; CACNA1G, Calcium voltage-gated channel subunit alpha 1G; CI, Confidence interval; CIMP, CpG island methylation phenotype; CRABP1, Cellular retinoic acid binding protein1; CRC, Colorectal cancer; CSS, Cancer-specific survival; CTAs, Cancer testis antigens; DCR, Disease control rate; DFS, Disease-free survival; DNMT, DNA methyltransferase; EGFR, Epidermal growth factor receptor; FOLFIRI, 5-FU, calcium folinate and irinotecan; FOLFOX, 5-FU, calcium folinate and oxaliplatin; HAI, Hepatic arterial infusion; HME, Highly methylated epigenotype; HR, Hazard ratio; IEL, Intraepithelial lymphocytes; IFL, 5-FU and leucovorin alone or with irinotecan; IGF2, Insulin like growth factor 2; IME, Intermediate methylated epigenotype;
LME, Low methylated epigenotype; MDR1, Multi-drug resistance 1; mFOLFOX6, Modified FOLFOX6; MMR-I, Mismatch repair-intact; MSI, Microsatellite instability; MSP, Methylation-specific polymerase chain reaction; MSS, Microsatellite stability; MTHFR, Methylenetetrahydrofolate reductase; NEUROG, Neurogenin 1; ORR, Objective response rate; OS, Overall survival; PFS, Progression free survival; PLS, Peritumoral lymphocytes;
PR, Partial response; RFS, Recurrence-free survival; RUNX3, Runt-related transcription factor 3; SAM, S-adenosylmethionine; SAR, Survival after recurrence; SCNAs, Somatic copy number alterations; SD, Stable disease; SGI-110, Guadecitabine; SOCS1, Suppressor of cytokine signaling 1; TIL, Tumor infiltrating lymphocytes; TS, Thymidylate synthase; XELOX, Capecitabine and oxaliplatin.
1. Wei W, Zeng H, Zheng R, Zhang S, An L, Chen R, et al. Cancer registration in China and its role in cancer prevention and control. Lancet Oncol (2020) 21(7):e342–9. doi: 10.1016/S1470-2045(20)30073-5
2. Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science (2010) 330(6004):612–6. doi: 10.1126/science.1191078
3. Okugawa Y, Grady WM, Goel A. Epigenetic Alterations in Colorectal Cancer: Emerging Biomarkers. Gastroenterology (2015) 149(5):1204–25. doi: 10.1053/j.gastro.2015.07.011
4. Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA (1999) 96:8681–6. doi: 10.1073/pnas.96.15.8681
5. Advani SM, Advani PS, Brown DW, DeSantis SM, Korphaisarn K, VonVille HM, et al. Global differences in the prevalence of the CpG island methylator phenotype of colorectal cancer. BMC Cancer (2019) 19(1):964. doi: 10.1186/s12885-019-6144-9
6. Miranda Furtado CL, Dos Santos Luciano MC, Silva Santos RD, Furtado GP, Moraes MO, Pessoa C. Epidrugs: targeting epigenetic marks in cancer treatment. Epigenetics (2019) 14(12):1164–76. doi: 10.1080/15592294.2019.1640546
7. Hughes LA, Khalid-de Bakker CA, Smits KM, van den Brandt PA, Jonkers D, Ahuja N, et al. The CpG island methylator phenotype in colorectal cancer: progress and problems. Biochim Biophys Acta (2012) 1825(1):77–85. doi: 10.1016/j.bbcan.2011.10.005
8. Juo YY, Johnston FM, Zhang DY, Juo HH, Wang H, Pappou EP, et al. Prognostic value of CpG island methylator phenotype among colorectal cancer patients: a systematic review and meta-analysis. Ann Oncol (2014) 25(12):2314–27. doi: 10.1093/annonc/mdu149
9. Jia M, Gao X, Zhang Y, Hoffmeister M, Brenner H. Different definitions of CpG island methylator phenotype and outcomes of colorectal cancer: a systematic review. Clin Epigenet (2016) 8:25. doi: 10.1186/s13148-016-0191-8
10. Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet (2006) 38(7):787–93. doi: 10.1038/ng1834
11. Weisenberger DJ, Levine AJ, Long TI, Buchanan DD, Walters R, Clendenning M, et al. Colon Cancer Family Registry. Association of the colorectal CpG island methylator phenotype with molecular features, risk factors, and family history. Cancer Epidemiol Biomarkers Prev (2015) 24(3):512–9. doi: 10.1158/1055-9965.EPI-14-1161
12. Ogino S, Cantor M, Kawasaki T, Brahmandam M, Kirkner G, Weisenberger DJ, et al. CpG island methylator phenotype (CIMP) of colorectal cancer is best characterised by quantitative DNA methylation analysis and prospective cohort studies. Gut (2006) 55:1000–6. doi: 10.1136/gut.2005.082933
13. Ogino S, Kawasaki T, Kirkner GJ, Loda M, Fuchs CS. CpG island methylator phenotype-low (CIMP-low) in colorectal cancer: possible associations with male sex and KRAS mutations. J Mol Diagn (2006) 8(5):582–8. doi: 10.2353/jmoldx.2006.060082
14. Ogino S, Odze RD, Kawasaki T, Brahmandam M, Kirkner GJ, Laird PW, et al. Correlation of pathologic features with CpG island methylator phenotype (CIMP) by quantitative DNA methylation analysis in colorectal carcinoma. Am J Surg Pathol (2006) 30(9):1175–83. doi: 10.1097/01.pas.0000213266.84725.d0
15. Schernhammer ES, Giovannucci E, Baba Y, Fuchs CS, Ogino S. B vitamins, methionine and alcohol intake and risk of colon cancer in relation to BRAF mutation and CpG island methylator phenotype (CIMP). PloS One (2011) 6(6):e21102. doi: 10.1371/journal.pone.0021102
16. Tahara T, Yamamoto E, Suzuki H, Maruyama R, Chung W, Garriga J, et al. Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res (2014) 74(5):1311–8. doi: 10.1158/0008-5472.CAN-13-1865
17. Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut (2016) 65(12):1973–80. doi: 10.1136/gutjnl-2015-310101
18. Guinney J, Dienstmann R, Wang X, de Reyniès A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nat Med (2015) 21:1350–6. doi: 10.1038/nm.3967
19. Advani SM, Swartz MD, Loree J, Davis JS, Sarsashek AM, Lam M, et al. Epidemiology and Molecular-Pathologic Characteristics of CpG Island Methylator Phenotype (CIMP) in Colorectal Cancer. Clin Colorectal Cancer (2020) 12:S1533–0028(20)30136-5. doi: 10.1016/j.clcc.2020.09.007
20. Lange CP, Laird PW. Clinical applications of DNA methylation biomarkers in colorectal cancer. Epigenomics (2013) 5(2):105–8. doi: 10.2217/epi.13.4
21. Barault L, Amatu A, Siravegna G, Ponzetti A, Moran S, Cassingena A, et al. Discovery of methylated circulating DNA biomarkers for comprehensive non-invasive monitoring of treatment response in metastatic colorectal cancer. Gut (2018) 67(11):1995–2005. doi: 10.1136/gutjnl-2016-313372
22. Shen L, Catalano PJ, Benson AB3, O’Dwyer P, Hamilton SR, Issa JP. Association between DNA methylation and shortened survival in patients with advanced colorectal cancer treated with 5-fluorouracil based chemotherapy. Clin Cancer Res (2007) 13(20):6093–8. doi: 10.1158/1078-0432.CCR-07-1011
23. Jover R, Nguyen TP, Pérez-Carbonell L, Zapater P, Payá A, Alenda C, et al. 5-Fluorouracil adjuvant chemotherapy does not increase survival in patients with CpG island methylator phenotype colorectal cancer. Gastroenterology (2011) 140(4):1174–81. doi: 10.1053/j.gastro.2010.12.035
24. Min BH, Bae JM, Lee EJ, Yu HS, Kim YH, Chang DK, et al. The CpG island methylator phenotype may confer a survival benefit in patients with stage II or III colorectal carcinomas receiving fluoropyrimidine-based adjuvant chemotherapy. BMC Cancer (2011) 11:344. doi: 10.1186/1471-2407-11-344
25. Van Rijnsoever M, Elsaleh H, Joseph D, McCaul K, Iacopetta B. CpG island methylator phenotype is an independent predictor of survival benefit from 5-fluorouracil in stage III colorectal cancer. Clin Cancer Res (2003) 9(8):2898–903.
26. Murcia O, Juárez M, Rodríguez-Soler M, Hernández-Illán E, Giner-Calabuig M, Alustiza M, et al. Colorectal cancer molecular classification using BRAF, KRAS, microsatellite instability and CIMP status: Prognostic implications and response to chemotherapy. PloS One (2018) 13(9):e0203051. doi: 10.1371/journal.pone.0203051
27. Zhang X, Shimodaira H, Soeda H, Komine K, Takahashi H, Ouchi K, et al. CpG island methylator phenotype is associated with the efficacy of sequential oxaliplatin- and irinotecan-based chemotherapy and EGFR-related gene mutation in Japanese patients with metastatic colorectal cancer. Int J Clin Oncol (2016) 21(6):1091–101. doi: 10.1007/s10147-016-1017-6
28. Shiovitz S, Bertagnolli MM, Renfro LA, Nam E, Foster NR, Dzieciatkowski S, et al. CpG island methylator phenotype is associated with response to adjuvant irinotecan-based therapy for stage III colon cancer. Gastroenterology (2014) 147(3):637–45. doi: 10.1053/j.gastro.2014.05.009
29. Cohen SA, Wu C, Yu M, Gourgioti G, Wirtz R, Raptou G, et al. Evaluation of CpG Island Methylator Phenotype as a Biomarker in Colorectal Cancer Treated With Adjuvant Oxaliplatin. Clin Colorectal Cancer (2016) 15(2):164–9. doi: 10.1016/j.clcc.2015.10.005
30. Han SW, Lee HJ, Bae JM, Cho NY, Lee KH, Kim TY, et al. Methylation and microsatellite status and recurrence following adjuvant FOLFOX in colorectal cancer. Int J Cancer (2013) 132(9):2209–16. doi: 10.1002/ijc.27888
31. Gallois C, Taieb J, Le Corre D, Le Malicot K, Tabernero J, Mulot C, et al. Prognostic Value of Methylator Phenotype in Stage III Colon Cancer Treated with Oxaliplatin-based Adjuvant Chemotherapy. Clin Cancer Res (2018) 24(19):4745–53. doi: 10.1158/1078-0432.CCR-18-0866
32. Cha Y, Kim KJ, Han SW, Rhee YY, Bae JM, Wen X, et al. Adverse prognostic impact of the CpG island methylator phenotype in metastatic colorectal cancer. Br J Cancer (2016) 115(2):164–71. doi: 10.1038/bjc.2016.176
33. Bae JM, Kim JH, Kwak Y, Lee DW, Cha Y, Wen X, et al. Distinct clinical outcomes of two CIMP-positive colorectal cancer subtypes based on a revised CIMP classification system. Br J Cancer (2017) 116(8):1012–20. doi: 10.1038/bjc.2017.52
34. Overman MJ, Adam L, Raghav K, Wang J, Kee B, Fogelman D, et al. Phase II study of nab-paclitaxel in refractory small bowel adenocarcinoma and CpG island methylator phenotype (CIMP)-high colorectal cancer. Ann Oncol (2018) 29(1):139–44. doi: 10.1093/annonc/mdx688
35. Tournigand C, André T, Achille E, Lledo G, Flesh M, Mery-Mignard D, et al. FOLFIRI followed by FOLFOX6 or the reverse sequence in advanced colorectal cancer: a randomized GERCOR study. J Clin Oncol (2004) 22(2):229–37. doi: 10.1200/JCO.2004.05.113
36. Tsai HC, Li H, Van Neste L, Cai Y, Robert C, Rassool FV, et al. Transient low doses of DNA-demethylating agents exert durable antitumor effects on hematological and epithelial tumor cells. Cancer Cell (2012) 21(3):430–46. doi: 10.1016/j.ccr.2011.12.029
37. Sharma A, Vatapalli R, Abdelfatah E, Wyatt McMahon K, Kerner Z, A Guzzetta A, et al. Hypomethylating agents synergize with irinotecan to improve response to chemotherapy in colorectal cancer cells. PloS One (2017) 12(4):e0176139. doi: 10.1371/journal.pone.0176139
38. Pietrantonio F, Perrone F, de Braud F, Castano A, Maggi C, Bossi I, et al. Activity of temozolomide in patients with advanced chemorefractory colorectal cancer and MGMT promoter methylation. Ann Oncol (2014) 25(2):404–8. doi: 10.1093/annonc/mdt547
39. Calegari MA, Inno A, Monterisi S, Orlandi A, Santini D, Basso M, et al. A phase 2 study of temozolomide in pretreated metastatic colorectal cancer with MGMT promoter methylation. Br J Cancer (2017) 116(10):1279–86. doi: 10.1038/bjc.2017.109
40. Morano F, Corallo S, Niger M, Barault L, Milione M, Berenato R, et al. Temozolomide and irinotecan (TEMIRI regimen) as salvage treatment of irinotecan-sensitive advanced colorectal cancer patients bearing MGMT methylation. Ann Oncol (2018) 29(8):1800–6. doi: 10.1093/annonc/mdy197
41. Ouchi K, Takahashi S, Yamada Y, Tsuji S, Tatsuno K, Takahashi H, et al. DNA methylation status as a biomarker of anti-epidermal growth factor receptor treatment for metastatic colorectal cancer. Cancer Sci (2015) 106:1722–9. doi: 10.1111/cas.12827
42. Tejpar S, Stintzing S, Ciardiello F, Tabernero J, Van Cutsem E, Beier F, et al. Prognostic and Predictive Relevance of Primary Tumor Location in Patients With RAS Wild-Type Metastatic Colorectal Cancer: Retrospective Analyses of the CRYSTAL and FIRE-3 Trials. JAMA Oncol (2017) 3(2):194–201. doi: 10.1001/jamaoncol.2016.3797
43. Bahl A, Talwar V, Sirohi B, Mehta P, Arya D, Shrivastava G, et al. Primary Tumor Location as a Prognostic and Predictive Marker in Metastatic Colorectal Cancer (mCRC). Front Oncol (2020) 10:964. doi: 10.3389/fonc.2020.00964
44. Demurtas L, Puzzoni M, Giampieri R, Ziranu P, Pusceddu V, Mandolesi A, et al. The role of primary tumour sidedness, EGFR gene copy number and EGFR promoter methylation in RAS/BRAF wild-type colorectal cancer patients receiving irinotecan/cetuximab. Br J Cancer (2017) 117(3):315–21. doi: 10.1038/bjc.2017.178
45. Lyko F. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat Rev Genet (2018) 19(2):81–92. doi: 10.1038/nrg.2017.80
46. Stresemann C, Lyko F. Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine. Int J Cancer J Int Cancer (2008) 123:8–13. doi: 10.1002/ijc.23607
47. Chuang JC, Warner SL, Vollmer D, Vankayalapati H, Redkar S, Bearss DJ, et al. S110, a 5-Aza-2’-deoxycytidine-containing dinucleotide, is an effective DNA methylation inhibitor in vivo and can reduce tumor growth. Mol Cancer Ther (2010) 9(5):1443–50. doi: 10.1158/1535-7163.MCT-09-1048
48. Tse JWT, Jenkins LJ, Chionh F, Mariadason JM. Aberrant DNA Methylation in Colorectal Cancer: What Should We Target? Trends Cancer (2017) 3(10):698–712. doi: 10.1016/j.trecan.2017.08.003
49. Weisenberger DJ, Liang G, Lenz HJ. DNA methylation aberrancies delineate clinically distinct subsets of colorectal cancer and provide novel targets for epigenetic therapies. Oncogene (2018) 37(5):566–77. doi: 10.1038/onc.2017.374
50. Garrido-Laguna I, McGregor KA, Wade M, Weis J, Gilcrease W, Burr L, et al. A phase I/II study of decitabine in combination with panitumumab in patients with wild-type (wt) KRAS metastatic colorectal cancer. Invest New Drugs (2013) 31:1257–64. doi: 10.1007/s10637-013-9947-6
51. Overman MJ, Morris V, Moinova H, Manyam G, Ensor J, Lee MS, et al. Phase I/II study of azacitidine and capecitabine/oxaliplatin (CAPOX) in refractory CIMP-high metastatic colorectal cancer: evaluation of circulating methylated vimentin. Oncotarget (2016) 7(41):67495–506. doi: 10.18632/oncotarget.11317
52. Azad NS, El-Khoueiry A, Yin J, Oberg AL, Flynn P, Adkins D, et al. Combination epigenetic therapy in metastatic colorectal cancer (mCRC) with subcutaneous 5-azacitidine and entinostat: a phase 2 consortium/stand up 2 cancer study. Oncotarget (2017) 8(21):35326–38. doi: 10.18632/oncotarget.15108
53. Jansen YJL, Verset G, Schats K, Van Dam PJ, Seremet T, Kockx M, et al. Phase I clinical trial of decitabine (5-aza-2’-deoxycytidine) administered by hepatic arterial infusion in patients with unresectable liver-predominant metastases. ESMO Open (2019) 4(2):e000464. doi: 10.1136/esmoopen-2018-000464
54. Chiappinelli KB, Zahnow CA, Ahuja N, Baylin SB. Combining Epigenetic and Immunotherapy to Combat Cancer. Cancer Res (2016) 76(7):1683–9. doi: 10.1158/0008-5472.CAN-15-2125
55. Sasidharan Nair V, Toor SM, Taha RZ, Shaath H, Elkord E. DNA methylation and repressive histones in the promoters of PD-1, CTLA-4, TIM-3, LAG-3, TIGIT, PD-L1, and galectin-9 genes in human colorectal cancer. Clin Epigenet (2018) 10(1):104. doi: 10.1186/s13148-018-0539-3
Keywords: colorectal cancer, CpG island methylator phenotype, chemotherapy, targeted therapy, DNA methylation inhibitor
Citation: Zhang X, Zhang W and Cao P (2021) Advances in CpG Island Methylator Phenotype Colorectal Cancer Therapies. Front. Oncol. 11:629390. doi: 10.3389/fonc.2021.629390
Received: 18 November 2020; Accepted: 18 January 2021;
Published: 26 February 2021.
Edited by:
Shicheng Guo, University of Wisconsin-Madison, United StatesReviewed by:
Yang Zhang, Peking University Cancer Hospital, ChinaCopyright © 2021 Zhang, Zhang and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaofei Zhang, emhhbmd4aWFvZmVpQGhvdG1haWwuY28uanA=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.