
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol. , 22 July 2021
Sec. Gastrointestinal Cancers
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.628636
This article is part of the Research Topic Cholangiocarcinoma: A New Multimodality Approach View all 8 articles
Surgical treatment is the only possible cure for cholangiocarcinoma (CCA) at present. However, the high recurrence rate of postoperative CCA leads to a very poor prognosis for patients, effective postoperative chemotherapy is hence the key to preventing the recurrence of CCA. The sensitivity of CCA to cytotoxic chemotherapy drugs and targeted drugs varies from person to person, and therefore, the screening of sensitive drugs has become an important topic after CCA surgeries. Patient-Derived tumor Xenograft models (PDX) can stably retain the genetic and pathological characteristics of primary tumors, and better simulate the tumor microenvironment of CCA. The model is also of great significance in screening therapeutic targeted drugs after CCA, analyzing predictive biomarkers, and improving signal pathways in prognosis and basic research. This paper will review the current established methods and applications of the patient-derived tumor xenograft model of cholangiocarcinoma, aiming to provide new ideas for basic research and individualized treatment of cholangiocarcinoma after surgery.
Cholangiocarcinoma (CCA) is divided into intrahepatic cholangiocarcinoma (ICC), hilar cholangiocarcinoma (PCC), and Distal cholangiocarcinoma (DCC) based on the anatomical location (1). CCA accounts for about 15-20% of primary hepatobiliary malignancies (2). Its early clinical manifestations are not obvious and develop rapidly, characterized by a high mortality rate, strong invasiveness and poor prognosis. Its 5-year survival rate is only 10-20% (3–5). Usually, the occurrence of CCA is related to related risk factors such as cholestatic liver disease, cirrhosis and gallstones, chronic infection (hepatitis B, hepatitis C), inflammatory disease (inflammatory bowel disease), etc. (6). At present, surgical operation is the only possible treatment for CCA, but the recurrence rate is as high as 40%-80% (3–5). Therefore, postoperative adjuvant treatment has become an effective method to improve the CCA’s recurrence.
Current studies have shown that most CCA has multiple drug resistance (MDR), and there are complex drug resistance mechanisms (7). The mechanism of CCA chemoresistance is mostly related to the following aspects: ① The decrease in drug absorption and the increase of drug efflux leading to lower intracellular drug concentration (8); ② The reduced ability of tumor cells to activate prodrugs or the increased ability to detoxify (9); ③ Molecular target mutations (10, 11); ④ DNA repair mechanism: a part of chemotherapeutic drugs exert their function by damaging DNA, but the majority of cancer cells has impaired DNA damage response, such as base excision repair system, nucleotide excision repair system or mismatch repair system (8, 12); ⑤ Regulating the death-related signaling pathways: downregulating apoptosis pathways or necrosis pathways induced by chemotherapeutic drugs (13); ⑥ Tumor microenvironment changes (14), etc. Also, research on targeted therapy in CCA is still in its infancy, with fewer useful molecular targets, and the choice of therapeutic drugs.
The rapid development of Next Generation Sequencing(NGS) has had far reaching effects in the field of CCA research. Different types of gene mutations have been found in CCA, which affect cell cycle, cytokine signal transmission, genome stability and DNA repair, including CCA-related gene mutations, amplifications, deletions, fusions and methylation (15). In ICC, genetic changes in FGFR (15.8%), IDH1 (14.7%), IDH2 (5.3%), BAP1 (14.5%), PBRM1 (12.1%), MCL1 (21.4%), CDKN2A (21.1%) are more common. And in extrahepatic cholangiocarcinoma(ECC),genetic changes in TP53 (36.9%), KRAS (31.5%), CDKN2B (15.2%), Smad4 (14.3%) are more common (16, 17). NGS undoubtedly provides a unique opportunity to expand the current understanding of CCA’s molecular pathogenesis, and it can provide individualized therapeutic targets for clinical treatment. However, there are often many differentially expressed molecules screened by sequencing technology. Therefore, using effective animal models to verify the effectiveness of screening targets has become an obstacle that limits sequencing technology to individualized treatment.
Current research in CCA has explored mechanisms of tumor development, progress, and treatment. At present, there are many CCA animal models, including CCA animal models based on tumor carcinogens (18), mouse models of bile duct ligation that mimic the characteristics of cholestasis (19), and a genetically engineered mouse model of CCA (20). But none of them can accurately predict the efficacy of conventional drugs and new anti-tumor drugs. Patient-Derived tumor Xenograft (PDX) models bring hope that better treatment options will be developed for CCA patients.
In 1985, Hudd et al. (21) used patient-derived CCA cell lines to inject subcutaneously into the abdomen of mice and successfully established a CCA CDX model. Among them, 26 of 30 mice developed CCA tumors. The CDX model is easy to develop and operate. However, due to the long-term in vitro culture of tumor cell lines, tumor cell lines have specific mutations, gene silencing, and lack of interaction between the tumor microenvironment and the immune system. Biological behavior and tumor heterogeneity are quite different from the original tumor tissue, which leads to an unsatisfactory prediction of clinical efficacy (22–24). Therefore, in 1984, Braakhuis and others directly implanted the head and neck tumor tissue into athymic nude mice to establish a PDX model, which stably retained the primary tumor’s genetic characteristics and histopathological characteristics (20, 25, 26). Cavalloni et al. (27) used patient-derived CCA tissue to implant subcutaneously in mice directly and successfully established a CCA PDX model for the first time, then obtained the tumor tissue by surgery, implanted the second group of mice subcutaneously to form F2 generation, and so on. They observed that the F4 generation CCA PDX showed the same morphology, histology and immunohistochemistry as the primary tumor.The F4 CCA PDX and primary tumors had highly similar genetic and molecular profiles in gene expression, tissue arrangement, RNA expression and mutation.
The CCA PDX model based on the technology mentioned above has also been continuously used for CCA targeted drug screening, analysis of predictive markers, and CCA efficacy improvement. The PDX model of cholangiocarcinoma is of great significance and has good research value and application prospects.
Presently, the cholangiocarcinoma (CCA) PDX model establishment method is divided into the subcutaneous transplantation model and orthotopic transplantation model. The methods of establishing subcutaneous transplantation models (28) and the orthotopic transplantation models (29) have been described in detail in the literature. Usually, the F1-F3 generations are used for reproduction, and the F3 generations are used for drug testing. The F5–F8 generations can better retain the biological and genetic characteristics of primary tumor cells. Therefore, it is advantageous to conduct drug trials in F3–F8 to predict clinical efficacy (30, 31).
The advantage of subcutaneous transplantation is the high tumor formation rate. The PDX model constructed by Vaeteewoottacharn shows a rate as high as 75% (32), in which it is easy to generate models and monitor tumor size (33). However, the disadvantage is that it cannot provide a vital microenvironment for tumor development (34).
Moreover, the common construction method of the CCA PDX model is orthotopic transplantation. By implanting human intrahepatic CCA (ICC) tumor tissue into the liver of mice, an orthotopic CCA transplantation model is generated. Orthotopic transplantation can provide a better original tumor microenvironment than conventional subcutaneous transplantation, but it is relatively rare due to its difficulty and low tumor formation rate (20). Orthotopic transplantation of hilar and distal CCA is rare due to the difficulty of operation. There are several factors that affect the success of the model, such as transplantation within 60 min. If transplantation cannot be performed immediately or the first transplantation fails within 60 min, the loss can be reduced by cryopreservation technology. For the first time, Hernandez et al. established a PDX model of CCA within 60 min. Seventeen cases of CCA PDX models showed failed implantation. Then, 17 cases of CCA were immediately cryopreserved and implanted with resuscitation. In mice, 12 cases (70%) of PDX models of CCA were successfully constructed under the skin, which proved that long-term preservation of CCA could be achieved using cryopreservation technology (35).
Conversely, the success of PDX establishment varies with tumor origin and disease characteristics. More aggressive, recurrent, and highly metastatic tumors tend to show higher transplantation rates (36). According to reports, the KRAS mutation may be the key gene for its success (27).
In patients with advanced CCA, chemotherapy or targeted therapy is an important method to prolong overall survival. Presently, most guidelines recommend palliative treatment for patients with advanced CCA, such as chemotherapy regimens based on fluorouracil, gemcitabine, or platinum-based cytotoxic chemotherapeutics (37), but its effect is limited (38). Moreover, ICC is extremely resistant to chemotherapy drugs. Therefore, there is an urgent need for new systemic treatments to improve the prognosis of ICC. To date, a large number of studies have been conducted on CCA, and the mechanism of its development has been explored and understood, while various related targets have been discovered, for example, proto-oncogene c-Myc, fibroblast growth factor receptor, KRAS gene, Hippo-YAP signaling pathway, ubiquitin proteasome pathway, Notch signaling pathway, cyclin-dependent kinase family, and vascular endothelial growth factor (24, 39). However, none of the studies have shown targeted therapy that has advantages over systemic cytotoxic chemotherapy (38), and the reason is that there is no individualized screening of targeted drugs for CCA. Thus, exploration of CCA targets is still needed. The emergence of the PDX model links basic medical research to clinical trials. In contrast, the PDX model can be used for extensive sensitivity screening before targeted drugs. Moreover, the PDX model can be used to further explore the mechanism and development of new therapeutic targets (Figure 1).
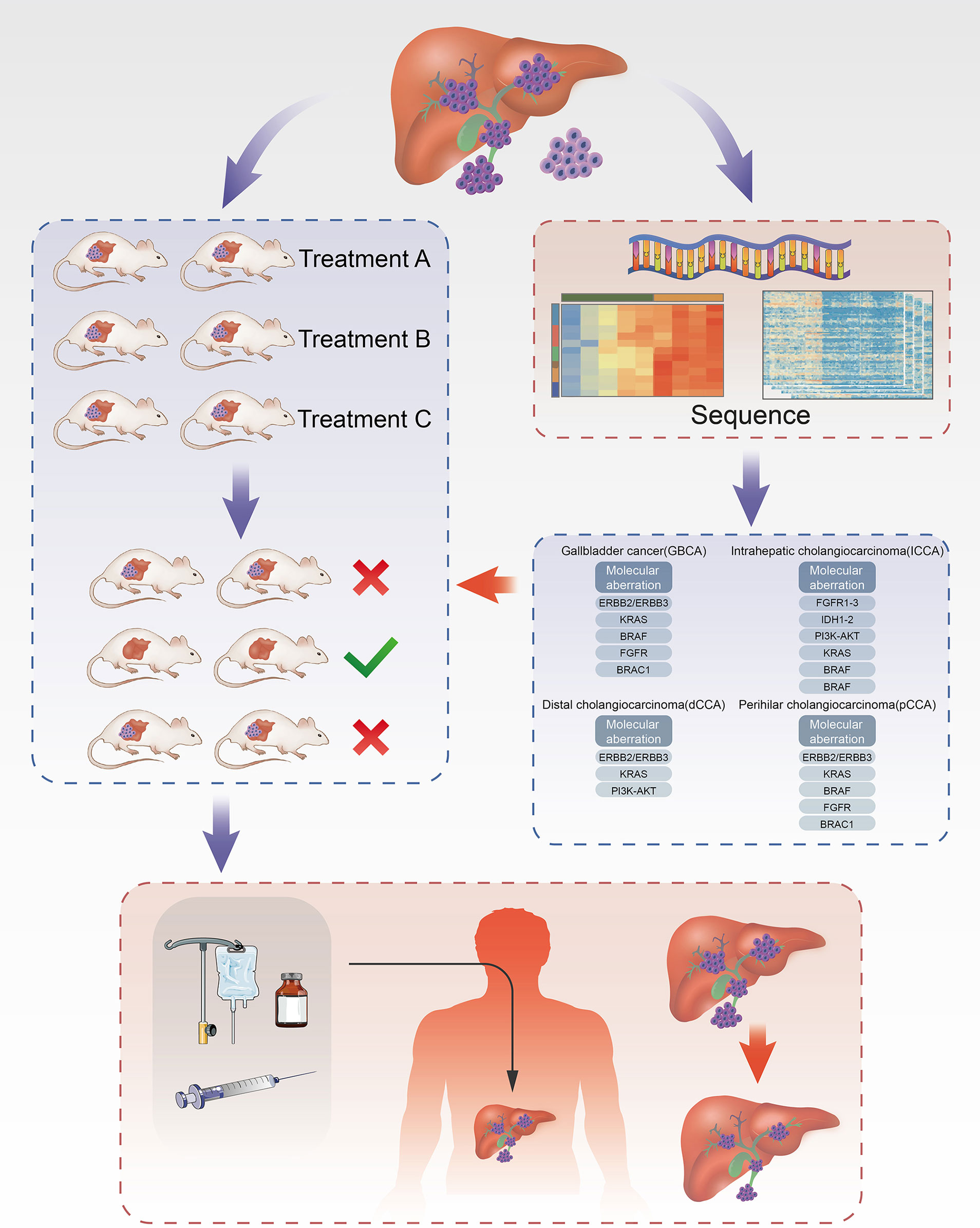
Figure 1 Establishment of ICC PDX models and application of drug screening. Part of the tissue of ICC after cholangiocarcinoma surgeries was transplanted into each group of mice correspondingly to cultivate cholangiocarcinoma. In the meanwhile, part of tissue was used for the analysis of targets for ICC using NGS technique to filter drugs with higher sensitivity and then administered on the treatment group. Through the analysis of drug efficacy against intrahepatic cholangiocarcinoma in the treatment group, the drugs with high sensitivity can be obtained and further acted on the human body.
The PDX model has been used to a certain extent in the screening of targeted drugs for cholangiocarcinoma (Table 1). The 26S proteasome is the main component of the ubiquitin-proteasome pathway responsible for regulating the protein degradation of more than 80% of mammalian cells (62). The ubiquitin-proteasome pathway degrades some apoptosis-related proteins through the 26S proteasome and regulates cell apoptosis. Studies have shown that actively proliferating malignant tumor cells are more sensitive to the ubiquitin-proteasome pathway inhibitors than normal cells (63). Using inhibitors against the ubiquitin-proteasome pathway is a new and powerful strategy for the treatment of malignant tumors.
The latest research shows that the ubiquitin receptor ADRM1 is overexpressed in liver cancer, gastric cancer, ovarian cancer and other malignant tumors. And gene knockout ADRM1 can inhibit the growth of malignant tumors. RA190 is a new small molecule ADRM1 inhibitor, confirmed to have a significant inhibitory effect on multiple myeloma (64–66). Yu et al. (44) have found through in vitro experiments that RA190 significantly inhibits the proliferation of primary ICC cells and ICC cell lines. To further explore the application value of ADRM1 inhibitor RA190 in ICC therapy, ICC PDX models were established by subcutaneous transplantation in mice.
Yu et al. (44) established the ICC PDX model and found that RA190 could significantly reduce the volume of ICC in vivo suggesting ADRM1 to be a promising anti-tumor target in ICC. In the study, ICC PDX mouse models were randomly divided into two groups (n=8), and they were given RA190 and buffer, respectively. After four weeks, the results showed that the tumor size and weight in the RA190 treatment group were significantly smaller than the buffer group. Also, the RA190 treatment group showed good tolerance. RA190 showed a significant therapeutic effect on ICC by inducing G2-M cell cycle arrest and apoptosis in vivo and in vitro.
Previous studies have shown that the Notch signaling pathway plays a central role in the occurrence of cholangiocarcinoma. The activation and expression of the NOTCH1/2 receptor and the typical ligand JAGGED1 lead to the overactive Notch signaling pathway and promote ICC’s occurrence and development (67).
Mancarella et al. (57) used the ICC PDX model to find that Notch signaling pathway inhibitor LY3039478 significantly inhibits Notch pathway and growth of tumor to the same extent as gemcitabine, and clarified the mechanism of LY3039478 inhibiting the growth of cholangiocarcinoma. The study used ICC tumor tissue implanted subcutaneously in the flanks of 4–5-week-old CD1 immunodeficient nude female mice to establish an ICC PDX model, divided into three groups and received buffer, LY3039478, and gemcitabine for eight weeks. Studies showed that the level of NOTCH1 in the LY3039478 treatment group was significantly reduced. The tumor size in the LY3039478 treatment group and the gemcitabine treatment group was considerably smaller than the control group (P<0.01), and the difference was little. The tumors in the LY3039478 treatment group began to resume growth 18 days after the end of treatment, and the gemcitabine group was 11 days later. Further analysis showed that LY3039478 inhibits the Notch signaling pathway in ICC while inhibiting the expression of HES1, DLL4, VEGFA, and MMP13, confirming the significance and value of Notch signaling pathway inhibitors in ICC targeted therapy. By constructing an ICC PDX model to evaluate the efficacy of LY3039478 in a highly restored human ICC tumor environment, inhibiting the Notch signaling pathway may become a new strategy for targeted treatment of ICC. In addition, the authors found that DLL4, VEGFA, and MMP13 genes are direct NOTCH targets in CCA, but the mechanism of action of LY3039478 on these target molecules still needs to be further studied.
CDK7 is a major member of the cyclin-dependent kinase (CDK) family, regulating the G2/M and G1/S cell cycle processes. Existing studies have shown that inhibiting CDK7 activity can inhibit transcription and cell cycle progression, thereby inhibiting tumor growth. CDK7 may be a new target for ICC targeted therapy (68). THZ1 is a highly specific CDK7 inhibitor that has shown effective anti-tumor activity in small cell lung cancer, ovarian cancer and triple-negative breast cancer. Chen et al. (58) studied the role of THZ1 in ICC. After subcutaneous implantation in BALB/c (nu/nu) mice for three generations, they established CDK7 overexpressing ICC PDX. Two groups (n=6) were randomly divided, and they were given intraperitoneal injections of PBS and THZ1 for 17 consecutive days. The results showed that the THZ1 treatment group’s tumor volume was significantly smaller than that of the PBS group, and THZ1 did not affect the weight and liver and kidney function of the mice. This study confirmed that CDK7 might be a feasible target for ICC targeted therapy.
Fibroblast growth factor receptor (FGFR) regulates cell proliferation and invasion, angiogenesis and tissue development in the body, and the fusion, rearrangement, translocation and amplification of the FGFR gene are closely related to the occurrence and development of tumors (69). In cholangiocarcinoma, FGFR2 mutations are as high as 20%, and up to 13% of ICCs have fibroblast growth factor receptor 2 (FGFR2) fusion genes (70).
Wang et al. (41) selected a fresh lung metastatic nodule tissue resected from a patient with stage IV ICC, and RNA sequencing showed that the tissue had FGFR2-CCDC6 fusion protein. They established the metastatic CCA PDX model and treated it with Ponatinib, dovitinib and BGJ398. The average tumor volume was 412.8 ± 53.82mm3, 269.7 ± 24.98mm3 and 204.2 ± 30.13mm3, respectively. BGJ398 had the best effect on inhibiting tumor growth. Rizvi et al. (42) also used the CCA PDX model to reach similar conclusions, confirming the therapeutic potential of BGJ398 for CCA patients with FGFR fusion protein again. Furthermore, they explored the relationship between the Hippo signaling pathway and FGFR fusion protein from the PDX model and proposed that the transcriptional regulator YAP in the Hippo signaling pathway may be a biomarker of FGFR in CCA targeted therapy. The expression of Mcl-1 in YAP-positive CCA tumor tissues is inhibited, and YAP can up-regulate the expression of FGFR1, FGFR2, and FGFR4 in CCA. The Hippo-YAP is an important pathway that can regulate cell proliferation and apoptosis (71). Changes in this pathway are related to the pathogenesis of CCA and other malignant tumors (72). YAP is its downstream effector molecule, which can be directly phosphorylated by LATS1/2, the signaling pathway’s core component. The phosphorylated YAP is eventually degraded, losing its growth-promoting and anti-apoptotic functions.
To explore the role of the Hippo-YAP signaling pathway in the mechanism of CCA occurrence and development, Sugihara et al. (45) used mouse CCA cells and CCA PDX models to conduct in vivo and in vitro experiments. The results showed that YAP is phosphorylated on tyrosine 357 (Y357). Compared with normal human bile duct cells, YAP tyrosine phosphorylation in CCA is enhanced. At the same time, the SFK inhibitor dasatinib blocks YAP tyrosine phosphorylation and induces YAP redistribution from the nucleus to the cytoplasm and down-regulates the expression of YAP target genes. siRNA targeted knockdown studies have shown that SFK member LCK plays a crucial role in mediating YAP Y357 phosphorylation and the YAP Y357 phosphorylation pathway in LCK-mediated CCA is a potential therapeutic target. Dasatinib showed tumor suppressor effects in both in vitro and in vivo experiments. The SFK inhibitor dasatinib provides theoretical support as a targeted therapy for CCA.
Kabashima et al. (46) also evaluated the efficacy of FGFR inhibitor LY2874455 on CCA through the PDX model and explored the FGFR inhibitor’s mechanism inducing CCA cell apoptosis. The results showed that FGFR inhibitors caused Mcl-1 expression inhibition in cholangiocarcinoma cell-matrix in the PDX model, leading to cell apoptosis. The research mentioned above is based on the construction of the CCA PDX model, which provides a strong basis for the study of FGFR inhibitors as individualized treatment options for CCA patients carrying the FGFR2 fusion gene, and promotes the process of individualized treatment.
In 23% to 50% of intrahepatic cholangiocarcinoma and 30% to 40% of extrahepatic cholangiocarcinoma, the KRAS gene often has mutations, which accelerate tumor progression (73, 74). Cavalloni et al. (27) implanted 17 cases of ICC tumors subcutaneously into NOD (non-obese diabetic)/Shi-SCID (severe immunodeficiency) mice. The results showed KRAS G12A mutation in the PDX model. After four months, the tumor volume reached 1000 mm3, and the tumor was removed and transplanted into the second generation of mice until the fourth generation. Further, using immunohistochemistry and genetic analysis techniques to evaluate biliary epithelial markers, tissue structure, genetic aberrations (including KRAS mutations), transcriptome, and microRNA profiles a high degree of gene consistency between the primary tumor and the 4th generation PDX model was confirmed. Previous studies have shown that KRAS mutations play an essential role in the metastasis of colorectal cancer (75), and speculated that KRAS mutations might be the driving factor for their successful implantation. KRAS mutation in colorectal cancer is one of the biological factors of resistance mechanism in EGFR targeted therapy (76). However, the role of KRAS mutations in the resistance mechanism of EGFR targeted therapy in CCA is still controversial. Therefore, this model may be suitable for evaluating the choice of targeted therapy drugs in CCA patients with KRAS mutations against EGFR. It improves the CCA PDX system and provides a promising platform for CCA targeted therapy to achieve precision medicine. However, the success rate of the PDX model is low (only 5.8%), which may be related to the KRAS mutation. Therefore, ways to improve the success rate of this mutant CCA is still an important question.
In summary, the targeted drugs screened by the PDX models can significantly delay the progress of CCA in mice and verify the mechanism of the development of CCA in vitro experiments. Although CCA PDX models are still in its infancy for drug screening and new drug targets mining at present, it has shown great potential.
Next-generation sequencing technology (NGS) has been increasingly applied to the field of tumor genomics research to promote individualized treatment in clinical oncology. It has become an effective method in tumor molecular diagnostics and targeted therapy research through efficient and accurate whole genome analysis and gene mutation detection of individual tumors. NGS technology is highly sensitive and can perform whole-genome sequencing, detect new mutations, small fragment insertion or small fragment deletion, copy number changes, and detection of gene fusion or rearrangement in small fragments (77). Javle et al. (78) used NGS technology to compare gene mutations between CCA subtypes in 412 ICC patients and 57 extrahepatic cholangiocarcinoma (EHC) patients. The analysis showed that the mutation rate in ICC of TP53 was 27%, KRAS 22%, IDH 16%, and in EHC the mutation rate of TP53 was 40%, KRAS 42%, and SMAD4 21%. NGS technology can better understand the genetic basis of the occurrence and development of CCA, and help to determine feasible treatment plans to guide the individualized treatment of CCA and accelerate the process of precision medicine.
Now in CCA research, the CCA PDX model and NGS technology are closely combined to provide new theoretical support for CCA targeted therapy, evaluate drug efficacy, and improve patient prognosis more comprehensively and accurately. The proto-oncogene c-Myc can regulate cell proliferation, apoptosis and the transcription of cell cycle-related genes. During the development of CCA, c-Myc expression is up-regulated. Down-regulating or knocking out c-Myc can reduce or inhibit the invasiveness of CCA. Bromodomain and extra terminal domain (BET) inhibitor JQ1 can down-regulate the expression of c-Myc (79–82). Although JQ1 has shown its potential in preclinical models of malignant tumors such as multiple myeloma, neuroblastoma, and pancreatic cancer (83), its usefulness in CCA requires further precise preclinical trials.
To deeply explore the significance and value of JQ1 in CCA treatment, Garcia et al. (24) used primary CCA tumors from 4 patients to establish a CCA (1–4) PDX model and evaluated the efficacy of JQ1 in CCA targeted therapy for the first time. The analysis showed that the CCA (2–4) PDX model retained the mutation type similar to the primary CCA tumor. Compared with other CCA models, JQ1 inhibited the growth of CCA2 and induced DNA damage and cell apoptosis. Immunohistochemical analysis showed that JQ1 inhibited the expression of c-Myc in the CCA2 PDX model, further indicating that for patients with CCA2 tumor origin, JQ1 may be a highly sensitive targeted therapy drug. It also proves that BET inhibitors (For example, JQ1) have a good application prospect in CCA personalized targeted therapy, worthy of further exploration.
The surgically resected CCA specimens combined with gene sequencing technology were used to screen mutated target genes. However, although clinical trials can verify whether inhibiting this target can improve patients’ prognosis and efficacy, researchers cannot effectively carry out individual therapies. Therefore, the CCA PDX model was established to highly simulate human CCA tumor environment and verify the existence of mutant targets with NGS technology. Then, the pharmacodynamic analysis was performed to promote personalized drugs and predict the prognosis of CCA patients, providing a platform with high accuracy and sensitivity for improving the efficacy of CCA and promoting the individualized treatment process. It paves the way for a deeper understanding of the occurrence and development of CCA. Based on precision medicine, CCA will gradually develop towards individualized treatment.
Comprehensive whole-exome and transcriptome sequencing has defined the genetic pattern of each CCA subtype. The technology that uses gene sequencing to screen the target of CCA drug has been relatively mature for individual patients. To date, the use of the CCA PDX model for targeted drug screening has been satisfactory in animal experiments, suggesting that the combined application of these two technologies will play an important role in the individualized clinical treatment of CCA. The ongoing registered clinical studies (ChiCTR1900024033, ChiCTR1900020978, and ChiCTR-ONC-17010678) are anticipated to provide strong backing for individualized treatment of CCA. However, it is worth considering that gene sequencing technology and PDX models for drug screening require a relatively long time in patients with advanced CCA. If the treatment target cannot be screened, treatment will be seriously delayed. Therefore, it is still recommended that patients with CCA receive palliative care based on the guidelines for CCA during the time of drug screening and testing (84).
The CCA PDX model can reflect the genetic characteristics, histopathology, and phenotypic characteristics of patients with CCA, but the CCA PDX model cannot simulate the interaction between tumor and immune cells in the original CCA tumor microenvironment. It is significant to ensure a high success rate of a model as soon as possible and complete drug screening in the best period widely using the PDX in CCA (22, 85).
It usually takes 4–8 months to establish a preclinical PDX model for patients with advanced CCA. This is such a substantial amount of time (86). Moreover, the success rate of orthotopic transplantation is low (40%) (29).
When the PDX model was used for drug screening after 3–5 sub generations, the tumor matrix was almost entirely replaced by the murine matrix, interfering with drug distribution (86). During the implantation process, the human matrix components quickly disappear and are replaced by the murine microenvironment. The tumor tissue of PDX model will have genomic instability after PDX continuous transplantation, and the expression of genes related to proliferation and angiogenesis will be up-regulated, while the expression of genes that inhibit apoptosis will be down-regulated. As a result, the sensitivity to chemotherapy and targeted therapy will be changed (87).
The clinical manifestations of CCA are unclear, and the diagnosis is often at an advanced stage, and the best opportunity for surgery is lost. The original CCA tumor specimens that are surgically removed are small, and sufficient transplantation volume cannot be obtained, making transplantation more difficult.
Orthotopic transplantation of hilar and distal CCA is rare due to the difficulty of operation. By implanting human ICC tumor tissue into the liver of mice, an orthotopic CCA transplantation model is generated. Orthotopic transplantation can have a better original tumor microenvironment than conventional subcutaneous transplantation, but it is relatively rare due to its difficult operation and low tumor formation rate (20). At the same time, the current transplantation method is relatively simple. Although orthotopic transplantation can provide a better original tumor microenvironment than subcutaneous transplantation, it is difficult, and the tumorigenic rate is low, so current studies on orthotopic transplantation are relatively rare.
The PDX model lacks a complete immune system and is not suitable for immunotherapy research.
The cost of the PDX model is relatively high, including the mice and various targeted drugs (36).
The PDX finder has been established globally, but there are still few CCA-related studies, with only 22 cases (www.pdxfinder.org), which is not conducive for global, multicenter collaboration research (88).
Current studies have confirmed that CCA’s tumor microenvironment includes the expression of immune cells (T cells, macrophages, dendritic cells and NK cells) and immune checkpoints. In recent years, Immune checkpoint inhibitor (ICI) therapy has become one of the key systemic treatments for many malignant tumors, including Programmed Cell Death-1 (PD-1) and Programmed Cell Death ligand 1 (PD-L1) and Programmed Death ligand 2 (PD-L2) checkpoint ICIs (89). However, there is no standard for immunotherapy in CCA treatment, and the PDX model lacks a complete immune system and is not suitable for immunotherapy research. At present, some scholars have established humanized mice with the immune system. Hematopoietic stem cells (HSC) derived from human was injected into the bone marrow cavity or tail vein of mice after radiation treatment of immunodeficient mice, thereby destroying the Hematopoietic function of bone marrow in the mice, making it a complete human immune system. The animal model established by implanting tumor tissue into humanized mice with the immune system is called Hu-PDX (humanized patient-derived xenograft) model (90–92). Hu-PDX model can reproduce the interaction between human tumors, the human immune system and tumor microenvironment, and has a good application prospect in preclinical research and immunotherapy drug screening. It has now highlighted its essential role and value in the immunotherapy research of malignant tumors such as liver cancer, colorectal cancer, and triple-negative breast cancer. Zhao et al. (90) established a Hu-HCC-PDX model of hepatocellular carcinoma (HCC). The results showed that the expressions of PD-1 and CTLA-4 in immune cells (especially Tc cells) were uniformly replicated in all Hu- HCCS -PDX models, which further improved the screening of immunotherapy drugs for hepatocellular carcinoma and deepened the research on the mechanism of drug resistance. Hence, it is essential to note that the role of Hu-PDX model in immunotherapy cannot be ignored. However, at present, the HU-PDX model has not been established in the immunotherapy research of CCA, which may become the research focus in the future. Promoting the research progress of the CCA PDX model in immunotherapy will provide a theoretical basis for CCA clinical trials to evaluate the efficacy and safety of anti-PD-1, PD-L1 therapy and CCA immune-microenvironment therapy, and a promising preclinical research platform for CCA immunotherapy. Simultaneously, efforts should be made to develop the next generation of humanized mice with the immune system, reflecting the characteristics of the human tumor microenvironment more accurately, and promote the acceleration of preclinical research using humanized mice models.
In addition, the next research focuses on how to shorten the time to build the PDX model. Improved transplantation methods may be used, such as ultrasound-guided fine needle aspiration. Ultrasound-guided fine needle aspiration technique can also be used for tissue acquisition (29).
The ultrasound monitor is used as a reference to push CCA cells into the liver with free hands. CCA cells from patients suspended in PBS were injected directly into the liver at a depth of 3 mm. The whole process from induction of anesthesia to recovery takes <5 min. Throughout the process, the physiological controller unit is used to monitor the animal’s heart and respiratory rate through the electrode pads on the ultrasound platform. This minimally invasive technique does not require postoperative analgesics.
Due to its minimally invasive nature, this method reduces the risk of complications in orthotopic transplantation and is fast and easy to perform. The tumor volume can be measured 1–3 weeks after injection. Compared with the traditional orthotopic transplantation model, it reduces the occurrence of inflammation and shortens the animal’s healing time. All mice survived after vaccination, which improves the survival rate of animals. Moreover, the tumor uptake rate is extremely high at 73%, and subsequent growth can be monitored longitudinally and noninvasively.
The current research is not entirely clear about the occurrence and development mechanism of CCA, and there is no standard for the construction of the CCA PDX model system. However, with the development of targeted therapy and immunotherapy based on precision medicine and other new treatments, the PDX model, NGS and other technologies have been applied to drug screening, efficacy evaluation, individual therapy, and basic research of CCA targeted therapy after surgery. The CCA PDX model can highly simulate the heterogeneity of human CCA tumors (Figure 2).
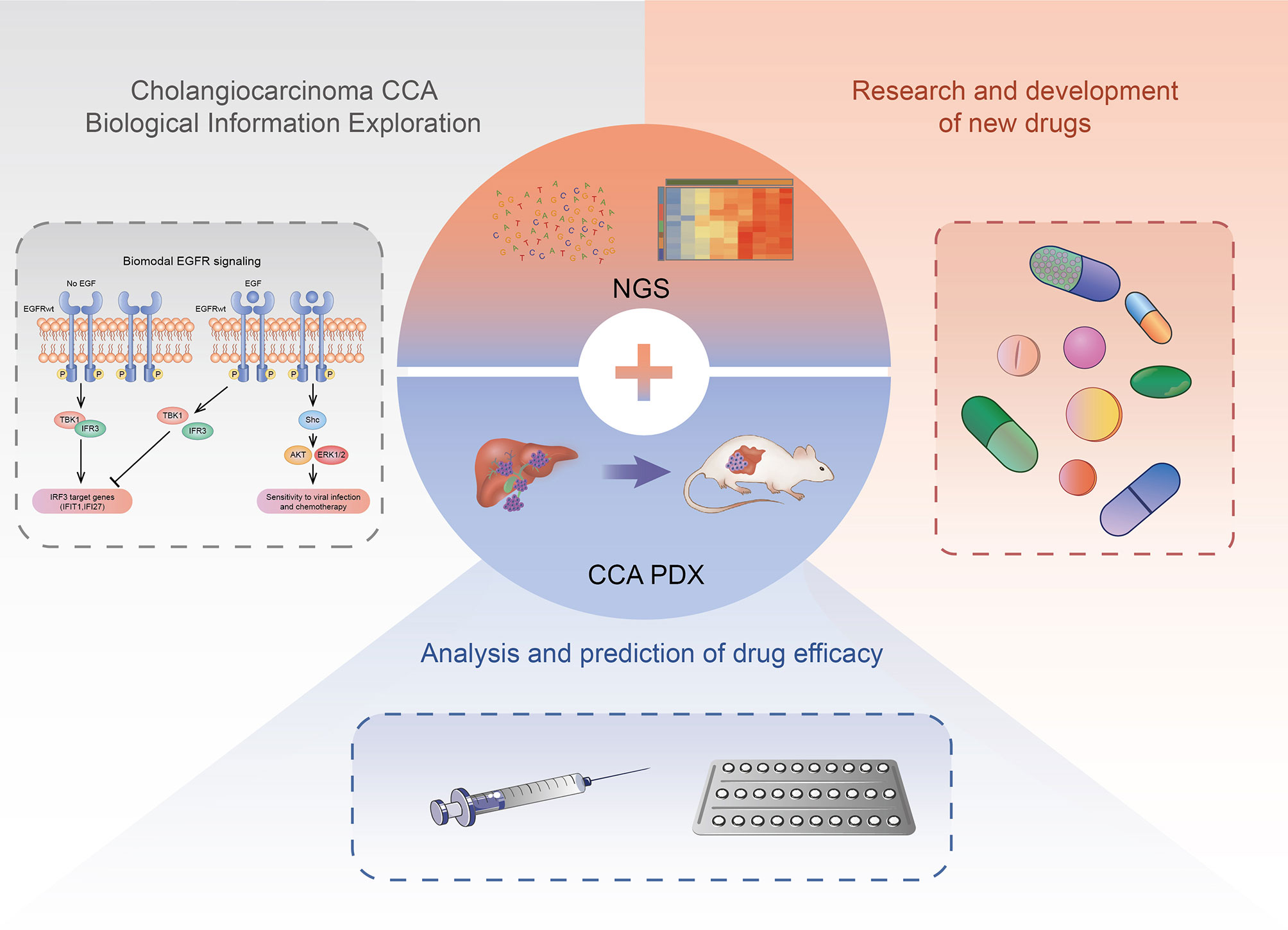
Figure 2 The application of CCA PDX models. The CCA PDX models have application potential in basic research, new drug development, and efficacy evaluation of the antineoplastic drug.
As the tumor-related animal model closest to clinical research at this stage, it has crucial translational significance for CCA preclinical research and evaluation of prognosis and good research value and application prospects. Through surgical resection and the construction of the CCA PDX model combined with targeted therapy and immunotherapy, CCA’s treatment will eventually move towards the era of individualized treatment.
JW and JS wrote the manuscript. HQ, MC, XZ and YY provided the critical revisions. All authors approved the final version of the manuscript for submission and approved it for publication.
This work was supported by grants from China Postdoctoral Science Foundation (2020M670864), Youth Support Project of Jilin Association for Science and Technology (202028), the Project of Hepatobiliary and Pancreatic Disease Translational Medicine Platform Construction (2017F009) and Medical and Health Talents Project of Jilin Province (2019SCZT003).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Razumilava N, Gores GJ. Cholangiocarcinoma. Lancet (2014) 383(9935):2168–79. doi: 10.1016/S0140-6736(13)61903-0
2. Blechacz B. Cholangiocarcinoma: Current Knowledge and New Developments. Gut Liver (2017) 11(1):13–26. doi: 10.5009/gnl15568
3. Banales JM, Cardinale V, Carpino G, Marzioni M, Andersen JB, Invernizzi P, et al. Expert Consensus Document: Cholangiocarcinoma: Current Knowledge and Future Perspectives Consensus Statement From the European Network for the Study of Cholangiocarcinoma (Ens-Cca). Nat Rev Gastroenterol Hepatol (2016) 13(5):261–80. doi: 10.1038/nrgastro.2016.51
4. Wirth TC, Vogel A. Surveillance in Cholangiocellular Carcinoma. Best Pract Res Clin Gastroenterol (2016) 30(6):987–99. doi: 10.1016/j.bpg.2016.11.001
5. Clements O, Eliahoo J, Kim JU, Taylor-Robinson SD, Khan SA. Risk Factors for Intrahepatic and Extrahepatic Cholangiocarcinoma: A Systematic Review and Meta-Analysis. J Hepatol (2020) 72(1):95–103. doi: 10.1016/j.jhep.2019.09.007
6. Labib PL, Goodchild G, Pereira SP. Molecular Pathogenesis of Cholangiocarcinoma. BMC Cancer (2019) 19(1):185. doi: 10.1186/s12885-019-5391-0
7. Marin JJG, Lozano E, Herraez E, Asensio M, Di Giacomo S, Romero MR, et al. Chemoresistance and Chemosensitization in Cholangiocarcinoma. Biochim Biophys Acta Mol Basis Dis (2018) 1864(4 Pt B):1444–53. doi: 10.1016/j.bbadis.2017.06.005
8. Marin JJG, Lozano E, Briz O, Al-Abdulla R, Serrano MA, Macias RIR. Molecular Bases of Chemoresistance in Cholangiocarcinoma. Curr Drug Targets (2017) 18(8):889–900. doi: 10.2174/1389450116666150223121508
9. Srijiwangsa P, Ponnikorn S, Na-Bangchang K. Effect of β-Eudesmol on NQO1 Suppression-Enhanced Sensitivity of Cholangiocarcinoma Cells to Chemotherapeutic Agents. BMC Pharmacol Toxicol (2018) 19(1):32. doi: 10.1186/s40360-018-0223-4
10. Peraldo-Neia C, Cavalloni G, Fenocchio E, Cagnazzo C, Gammaitoni L, Cereda S, et al. Prognostic and Predictive Role of EGFR Pathway Alterations in Biliary Cancer Patients Treated With Chemotherapy and Anti-EGFR. PloS One (2018) 13(1):e0191593. doi: 10.1371/journal.pone.0191593
11. Vaquero J, Lobe C, Tahraoui S, Clapéron A, Mergey M, Merabtene F, et al. The IGF2/IR/IGF1R Pathway in Tumor Cells and Myofibroblasts Mediates Resistance to EGFR Inhibition in Cholangiocarcinoma. Clin Cancer Res (2018) 24(17):4282–96. doi: 10.1158/1078-0432.Ccr-17-3725
12. Fisher SB, Fisher KE, Patel SH, Lim MG, Kooby DA, El-Rayes BF, et al. Excision Repair Cross-Complementing gene-1, Ribonucleotide Reductase Subunit M1, Ribonucleotide Reductase Subunit M2, and Human Equilibrative Nucleoside Transporter-1 Expression and Prognostic Value in Biliary Tract Malignancy. Cancer (2013) 119(2):454–62. doi: 10.1002/cncr.27739
13. Yokoi K, Kobayashi A, Motoyama H, Kitazawa M, Shimizu A, Notake T, et al. Survival Pathway of Cholangiocarcinoma Via AKT/mTOR Signaling to Escape RAF/MEK/ERK Pathway Inhibition by Sorafenib. Oncol Rep (2018) 39(2):843–50. doi: 10.3892/or.2017.6153
14. Marin JJG, Briz O, Herraez E, Lozano E, Asensio M, Di Giacomo S, et al. Molecular Bases of the Poor Response of Liver Cancer to Chemotherapy. Clin Res Hepatol Gastroenterol (2018) 42(3):182–92. doi: 10.1016/j.clinre.2017.12.006
15. Churi CR, Shroff R, Wang Y, Rashid A, Kang HC, Weatherly J, et al. Mutation Profiling in Cholangiocarcinoma: Prognostic and Therapeutic Implications. PloS One (2014) 9(12):e115383. doi: 10.1371/journal.pone.0115383
16. Lendvai G, Szekerczés T, Illyés I, Dóra R, Kontsek E, Gógl A, et al. Cholangiocarcinoma: Classification, Histopathology and Molecular Carcinogenesis. Pathol Oncol Res (2020) 26(1):3–15. doi: 10.1007/s12253-018-0491-8
17. Brandi G, Farioli A, Astolfi A, Biasco G, Tavolari S. Genetic Heterogeneity in Cholangiocarcinoma: A Major Challenge for Targeted Therapies. Oncotarget (2015) 6(17):14744–53. doi: 10.18632/oncotarget.4539
18. Urbanik T, Boger RJ, Longerich T, Becker K, Ehrenberg KR, Hövelmeyer N, et al. Liver Specific Deletion of CYLDexon7/8 Induces Severe Biliary Damage, Fibrosis and Increases Hepatocarcinogenesis in Mice. J Hepatol (2012) 57(5):995–1003. doi: 10.1016/j.jhep.2012.06.017
19. Thamavit W, Pairojkul C, Tiwawech D, Itoh M, Shirai T, Ito N. Promotion of Cholangiocarcinogenesis in the Hamster Liver by Bile Duct Ligation After Dimethylnitrosamine Initiation. Carcinogenesis (1993) 14(11):2415–7. doi: 10.1093/carcin/14.11.2415
20. Loeuillard E, Fischbach SR, Gores GJ, Rizvi S. Animal Models of Cholangiocarcinoma. Biochim Biophys Acta Mol Basis Dis (2019) 1865(5):982–92. doi: 10.1016/j.bbadis.2018.03.026
21. Hudd C, Euhus DM, LaRegina MC, Herbold DR, Palmer DC, Johnson FE. Effect of Cholecystokinin on Human Cholangiocarcinoma Xenografted Into Nude Mice. Cancer Res (1985) 45(3):1372–7.
22. Yoshida GJ. Applications of Patient-Derived Tumor Xenograft Models and Tumor Organoids. J Hematol Oncol (2020) 13(1):4. doi: 10.1186/s13045-019-0829-z
23. Murayama T, Gotoh N. Patient-Derived Xenograft Models of Breast Cancer and Their Application. Cells (2019) 8(6):621. doi: 10.3390/cells8060621
24. Garcia PL, Miller AL, Gamblin TL, Council LN, Christein JD, Arnoletti JP, et al. Jq1 Induces Dna Damage and Apoptosis, and Inhibits Tumor Growth in a Patient-Derived Xenograft Model of Cholangiocarcinoma. Mol Cancer Ther (2018) 17(1):107–18. doi: 10.1158/1535-7163.MCT-16-0922
25. Hu MH, Chen LJ, Chen YL, Tsai MS, Shiau CW, Chao TI, et al. Targeting SHP-1-STAT3 Signaling: A Promising Therapeutic Approach for the Treatment of Cholangiocarcinoma. Oncotarget (2017) 8(39):65077–89. doi: 10.18632/oncotarget.17779
26. Braakhuis BJ, Sneeuwloper G, Snow GB. The Potential of the Nude Mouse Xenograft Model for the Study of Head and Neck Cancer. Arch Otorhinolaryngol (1984) 239(1):69–79. doi: 10.1007/bf00454264
27. Cavalloni G, Peraldo-Neia C, Sassi F, Chiorino G, Sarotto I, Aglietta M, et al. Establishment of a Patient-Derived Intrahepatic Cholangiocarcinoma Xenograft Model With KRAS Mutation. BMC Cancer (2016) 16:90. doi: 10.1186/s12885-016-2136-1
28. Rudloff U, Bhanot U, Gerald W, Klimstra DS, Jarnagin WR, Brennan MF, et al. Biobanking of Human Pancreas Cancer Tissue: Impact of Ex-Vivo Procurement Times on RNA Quality. Ann Surg Oncol (2010) 17(8):2229–36. doi: 10.1245/s10434-010-0959-6
29. McVeigh LE, Wijetunga I, Ingram N, Marston G, Prasad R, Markham AF, et al. Development of Orthotopic Tumour Models Using Ultrasound-Guided Intrahepatic Injection. Sci Rep (2019) 9(1):9904. doi: 10.1038/s41598-019-46410-6
30. Xu W, Yang XW, Zhao ZY, Dong B, Guan XY, Tian XY, et al. Establishment of Pancreatic Cancer Patient-Derived Xenograft Models and Comparison of the Differences Among the Generations. Am J Transl Res (2019) 11(5):3128–39.
31. Lipner MB, Marayati R, Deng Y, Wang X, Raftery L, O’Neil BH, et al. Metformin Treatment Does Not Inhibit Growth of Pancreatic Cancer Patient-Derived Xenografts. PloS One (2016) 11(1):e0147113. doi: 10.1371/journal.pone.0147113
32. Vaeteewoottacharn K, Pairojkul C, Kariya R, Muisuk K, Imtawil K, Chamgramol Y, et al. Establishment of Highly Transplantable Cholangiocarcinoma Cell Lines From a Patient-Derived Xenograft Mouse Model. Cells (2019) 8(5):496. doi: 10.3390/cells8050496
33. Kim MP, Evans DB, Wang H, Abbruzzese JL, Fleming JB, Gallick GE. Generation of Orthotopic and Heterotopic Human Pancreatic Cancer Xenografts in Immunodeficient Mice. Nat Protoc (2009) 4(11):1670–80. doi: 10.1038/nprot.2009.171
34. Reiberger T, Chen Y, Ramjiawan RR, Hato T, Fan C, Samuel R, et al. An Orthotopic Mouse Model of Hepatocellular Carcinoma With Underlying Liver Cirrhosis. Nat Protoc (2015) 10(8):1264–74. doi: 10.1038/nprot.2015.080
35. Hernandez MC, Yang L, Leiting JL, Sugihara T, Bergquist JR, Ivanics T, et al. Successful Secondary Engraftment of Pancreatic Ductal Adenocarcinoma and Cholangiocarcinoma Patient-Derived Xenografts After Previous Failed Primary Engraftment. Transl Oncol (2019) 12(1):69–75. doi: 10.1016/j.tranon.2018.09.008
36. Collins AT, Lang SH. A Systematic Review of the Validity of Patient Derived Xenograft (PDX) Models: The Implications for Translational Research and Personalised Medicine. PeerJ (2018) 6:e5981. doi: 10.7717/peerj.5981
37. Lamarca A, Barriuso J, McNamara MG, Valle JW. Molecular Targeted Therapies: Ready for “Prime Time” in Biliary Tract Cancer. J Hepatol (2020) 73(1):170–85. doi: 10.1016/j.jhep.2020.03.007
38. Adeva J, Sangro B, Salati M, Edeline J, La Casta A, Bittoni A, et al. Medical Treatment for Cholangiocarcinoma. Liver Int (2019) 39 Suppl 1:123–42. doi: 10.1111/liv.14100
39. Valle JW, Lamarca A, Goyal L, Barriuso J, Zhu AX. New Horizons for Precision Medicine in Biliary Tract Cancers. Cancer Discovery (2017) 7(9):943–62. doi: 10.1158/2159-8290.CD-17-0245
40. Peraldo-Neia C, Cavalloni G, Soster M, Gammaitoni L, Marchiò S, Sassi F, et al. Anti-Cancer Effect and Gene Modulation of ET-743 in Human Biliary Tract Carcinoma Preclinical Models. BMC Cancer (2014) 14:918. doi: 10.1186/1471-2407-14-918
41. Wang Y, Ding X, Wang S, Moser CD, Shaleh HM, Mohamed EA, et al. Antitumor Effect of FGFR Inhibitors on a Novel Cholangiocarcinoma Patient Derived Xenograft Mouse Model Endogenously Expressing an FGFR2-CCDC6 Fusion Protein. Cancer Lett (2016) 380(1):163–73. doi: 10.1016/j.canlet.2016.05.017
42. Rizvi S, Yamada D, Hirsova P, Bronk SF, Werneburg NW, Krishnan A, et al. A Hippo and Fibroblast Growth Factor Receptor Autocrine Pathway in Cholangiocarcinoma. J Biol Chem (2016) 291(15):8031–47. doi: 10.1074/jbc.M115.698472
43. Peraldo Neia C, Cavalloni G, Chiorino G, Ostano P, Aglietta M, Leone F. Gene and microRNA Modulation Upon Trabectedin Treatment in a Human Intrahepatic Cholangiocarcinoma Paired Patient Derived Xenograft and Cell Line. Oncotarget (2016) 7(52):86766–80. doi: 10.18632/oncotarget.13575
44. Yu G-Y, Wang X, Zheng S-S, Gao X-M, Jia Q-A, Zhu W-W, et al. RA190, a Proteasome Subunit ADRM1 Inhibitor, Suppresses Intrahepatic Cholangiocarcinoma by Inducing Nf-Kb-Mediated Cell Apoptosis. Cell Physiol Biochem (2018) 47(3):1152–66. doi: 10.1159/000490210
45. Sugihara T, Werneburg NW, Hernandez MC, Yang L, Kabashima A, Hirsova P, et al. Yap Tyrosine Phosphorylation and Nuclear Localization in Cholangiocarcinoma Cells Are Regulated by LCK and Independent of LATS Activity. Mol Cancer Res (2018) 16(10):1556–67. doi: 10.1158/1541-7786.MCR-18-0158
46. Kabashima A, Hirsova P, Bronk SF, Hernandez MC, Truty MJ, Rizvi S, et al. Fibroblast Growth Factor Receptor Inhibition Induces Loss of Matrix MCL1 and Necrosis in Cholangiocarcinoma. J Hepatol (2018) 68(6):1228–38. doi: 10.1016/j.jhep.2018.01.026
47. Smoot RL, Werneburg NW, Sugihara T, Hernandez MC, Yang L, Mehner C, et al. Platelet-Derived Growth Factor Regulates YAP Transcriptional Activity Via Src Family Kinase Dependent Tyrosine Phosphorylation. J Cell Biochem (2018) 119(1):824–36. doi: 10.1002/jcb.26246
48. Sittithumcharee G, Suppramote O, Vaeteewoottacharn K, Sirisuksakun C, Jamnongsong S, Laphanuwat P, et al. Dependency of Cholangiocarcinoma on Cyclin D-Dependent Kinase Activity. Hepatology (2019) 70(5):1614–30. doi: 10.1002/hep.30704
49. Ursu S, Majid S, Garger C, de Semir D, Bezrookove V, Desprez PY, et al. Novel Tumor Suppressor Role of miRNA-876 in Cholangiocarcinoma. Oncogenesis (2019) 8(8):42. doi: 10.1038/s41389-019-0153-z
50. Hernandez MC, Bergquist JR, Leiting JL, Ivanics T, Yang L, Smoot RL, et al. Patient-Derived Xenografts Can Be Reliably Generated From Patient Clinical Biopsy Specimens. J Gastrointestinal Surg (2019) 23(4):818–24. doi: 10.1007/s11605-019-04109-z
51. Wu T, Jiang X, Xu B, Wang Y. [Ponatinib Inhibits Growth of Patient-Derived Xenograft of Cholangiocarcinoma Expressing FGFR2-CCDC6 Fusion Protein in Nude Mice]. Nan Fang Yi Ke Da Xue Xue Bao (2020) 40(10):1448–56. doi: 10.12122/j.issn.1673-4254.2020.10.10
52. Song F, Hu B, Cheng JW, Sun YF, Zhou KQ, Wang PX, et al. Anlotinib Suppresses Tumor Progression Via Blocking the VEGFR2/PI3K/AKT Cascade in Intrahepatic Cholangiocarcinoma. Cell Death Dis (2020) 11(7):14. doi: 10.1038/s41419-020-02749-7
53. Jiang TY, Pan YF, Wan ZH, Lin YK, Zhu B, Yuan ZG, et al. PTEN Status Determines Chemosensitivity to Proteasome Inhibition in Cholangiocarcinoma. Sci Transl Med (2020) 12(562):eaay0152. doi: 10.1126/scitranslmed.aay0152
54. Leiting JL, Murphy SJ, Bergquist JR, Hernandez MC, Ivanics T, Abdelrahman AM, et al. Biliary Tract Cancer Patient-Derived Xenografts: Surgeon Impact on Individualized Medicine. JHEP Rep (2020) 2(2):100068. doi: 10.1016/j.jhepr.2020.100068
55. Arora M, Bogenberger JM, Abdelrahman A, Leiting JL, Chen X, Egan JB, et al. Evaluation of NUC-1031: A First-in-Class ProTide in Biliary Tract Cancer. Cancer Chemother Pharmacol (2020) 85(6):1063–78. doi: 10.1007/s00280-020-04079-z
56. Saqub H, Proetsch-Gugerbauer H, Bezrookove V, Nosrati M, Vaquero EM, de Semir D, et al. Dinaciclib, a Cyclin-Dependent Kinase Inhibitor, Suppresses Cholangiocarcinoma Growth by Targeting CDK2/5/9. Sci Rep (2020) 10(1):18489. doi: 10.1038/s41598-020-75578-5
57. Mancarella S, Serino G, Dituri F, Cigliano A, Ribback S, Wang J, et al. Crenigacestat, a Selective NOTCH1 Inhibitor, Reduces Intrahepatic Cholangiocarcinoma Progression by Blocking VEGFA/DLL4/MMP13 Axis. Cell Death Differ (2020) 27:2330–43. doi: 10.1038/s41418-020-0505-4
58. Chen H-D, Huang C-S, Xu Q-C, Li F, Huang X-T, Wang J-Q, et al. Therapeutic Targeting of CDK7 Suppresses Tumor Progression in Intrahepatic Cholangiocarcinoma. Int J Biol Sci (2020) 16(7):1207–17. doi: 10.7150/ijbs.39779
59. Wang Y, Yu H, Xie X, Deng T, Ye L, Wu L, et al. Plasmalemma Vesicle-Associated Protein Promotes Angiogenesis in Cholangiocarcinoma Via the DKK1/CKAP4/PI3K Signaling Pathway. Oncogene (2021) 40:4324–37. doi: 10.1038/s41388-021-01844-z
60. Peraldo-Neia C, Massa A, Vita F, Basirico M, Raggi C, Bernabei P, et al. A Novel Multidrug-Resistant Cell Line From an Italian Intrahepatic Cholangiocarcinoma Patient. Cancers (2021) 13(9):21. doi: 10.3390/cancers13092051
61. Zhou ZJ, Wang PC, Sun RQ, Li J, Hu ZQ, Xin HY, et al. Tumor-Associated Neutrophils and Macrophages Interaction Contributes to Intrahepatic Cholangiocarcinoma Progression by Activating STAT3. J Immunother Cancer (2021) 9(3):12. doi: 10.1136/jitc-2020-001946.#
62. Collins GA, Goldberg AL. The Logic of the 26S Proteasome. Cell (2017) 169(5):792–806. doi: 10.1016/j.cell.2017.04.023
63. Bedford L, Lowe J, Dick LR, Mayer RJ, Brownell JE. Ubiquitin-Like Protein Conjugation and the Ubiquitin-Proteasome System as Drug Targets. Nat Rev Drug Discovery (2011) 10(1):29–46. doi: 10.1038/nrd3321
64. Yang X, Miao X, Wen Y, Hu J, Dai W, Yin B. A Possible Connection Between Adhesion Regulating Molecule 1 Overexpression and Nuclear Factor Kappa B Activity in Hepatocarcinogenesis. Oncol Rep (2012) 28(1):283–90. doi: 10.3892/or.2012.1767
65. Jang SH, Park JW, Kim HR, Seong JK, Kim HK. ADRM1 Gene Amplification Is a Candidate Driver for Metastatic Gastric Cancers. Clin Exp Metastasis (2014) 31(6):727–33. doi: 10.1007/s10585-014-9663-4
66. Fejzo MS, Anderson L, von Euw EM, Kalous O, Avliyakulov NK, Haykinson MJ, et al. Amplification Target Adrm1: Role as an Oncogene and Therapeutic Target for Ovarian Cancer. Int J Mol Sci (2013) 14(2):3094–109. doi: 10.3390/ijms14023094
67. Guest RV, Boulter L, Dwyer BJ, Kendall TJ, Man T-Y, Minnis-Lyons SE, et al. Notch3 Drives Development and Progression of Cholangiocarcinoma. Proc Natl Acad Sci USA (2016) 113(43):12250–5. doi: 10.1073/pnas.1600067113
68. Cao K, Shilatifard A. Inhibit Globally, Act Locally: CDK7 Inhibitors in Cancer Therapy. Cancer Cell (2014) 26(2):158–9. doi: 10.1016/j.ccr.2014.07.020
69. Yang T, Liang L, Wang M-D, Shen F. FGFR Inhibitors for Advanced Cholangiocarcinoma. Lancet Oncol (2020) 21(5):610–2. doi: 10.1016/S1470-2045(20)30152-2
70. Goyal L, Shi L, Liu LY, Fece de la Cruz F, Lennerz JK, Raghavan S, et al. Tas-120 Overcomes Resistance to ATP-Competitive Fgfr Inhibitors in Patients With FGFR2 Fusion-Positive Intrahepatic Cholangiocarcinoma. Cancer Discovery (2019) 9(8):1064–79. doi: 10.1158/2159-8290.CD-19-0182
71. Sugihara T, Isomoto H, Gores G, Smoot R. YAP and the Hippo Pathway in Cholangiocarcinoma. J Gastroenterol (2019) 54(6):485–91. doi: 10.1007/s00535-019-01563-z
72. Farshidfar F, Zheng S, Gingras M-C, Newton Y, Shih J, Robertson AG, et al. Integrative Genomic Analysis of Cholangiocarcinoma Identifies Distinct Idh-Mutant Molecular Profiles. Cell Rep (2017) 19(13):2878–80. doi: 10.1016/j.celrep.2017.06.008
73. O’Dell MR, Huang JL, Whitney-Miller CL, Deshpande V, Rothberg P, Grose V, et al. Kras(G12D) and p53 Mutation Cause Primary Intrahepatic Cholangiocarcinoma. Cancer Res (2012) 72(6):1557–67. doi: 10.1158/0008-5472.CAN-11-3596
74. Tian W, Hu W, Shi X, Liu P, Ma X, Zhao W, et al. Comprehensive Genomic Profile of Cholangiocarcinomas in China. Oncol Lett (2020) 19(4):3101–10. doi: 10.3892/ol.2020.11429
75. Huang D, Sun W, Zhou Y, Li P, Chen F, Chen H, et al. Mutations of Key Driver Genes in Colorectal Cancer Progression and Metastasis. Cancer Metastasis Rev (2018) 37(1):173–87. doi: 10.1007/s10555-017-9726-5
76. Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, et al. Emergence of KRAS Mutations and Acquired Resistance to anti-EGFR Therapy in Colorectal Cancer. Nature (2012) 486(7404):532–6. doi: 10.1038/nature11156
77. Atreya CE, Yaeger R, Chu E. Systemic Therapy for Metastatic Colorectal Cancer: From Current Standards to Future Molecular Targeted Approaches. Am Soc Clin Oncol Educ Book (2017) 37:246–56. doi: 10.14694/EDBK_175679
78. Javle M, Bekaii-Saab T, Jain A, Wang Y, Kelley RK, Wang K, et al. Biliary Cancer: Utility of Next-Generation Sequencing for Clinical Management. Cancer (2016) 122(24):3838–47. doi: 10.1002/cncr.30254
79. Nasi S, Ciarapica R, Jucker R, Rosati J, Soucek L. Making Decisions Through Myc. FEBS Lett (2001) 490(3):153–62. doi: 10.1016/S0014-5793(01)02118-4
80. Yang H, Li TWH, Peng J, Tang X, Ko KS, Xia M, et al. A Mouse Model of Cholestasis-Associated Cholangiocarcinoma and Transcription Factors Involved in Progression. Gastroenterology (2011) 141(1):378–88. doi: 10.1053/j.gastro.2011.03.044
81. Li Z-R, Wu Y-F, Ma C-Y, Nie S-D, Mao X-H, Shi Y-Z. Down-Regulation of c-Myc Expression Inhibits the Invasion of Bile Duct Carcinoma Cells. Cell Biol Int (2011) 35(8):799–802. doi: 10.1042/CBI20110099
82. Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective Inhibition of BET Bromodomains. Nature (2010) 468(7327):1067–73. doi: 10.1038/nature09504
83. Garcia PL, Miller AL, Kreitzburg KM, Council LN, Gamblin TL, Christein JD, et al. The BET Bromodomain Inhibitor JQ1 Suppresses Growth of Pancreatic Ductal Adenocarcinoma in Patient-Derived Xenograft Models. Oncogene (2016) 35(7):833–45. doi: 10.1038/onc.2015.126
84. Mancarella S, Serino G, Dituri F, Cigliano A, Ribback S, Wang J, et al. Crenigacestat, a Selective NOTCH1 Inhibitor, Reduces Intrahepatic Cholangiocarcinoma Progression by Blocking VEGFA/DLL4/MMP13 Axis. Cell Death Differ (2020) 27(8):2330–43. doi: 10.1038/s41418-020-0505-4
85. Okada S, Vaeteewoottacharn K, Kariya R. Application of Highly Immunocompromised Mice for the Establishment of Patient-Derived Xenograft (Pdx) Models. Cells (2019) 8(8):889. doi: 10.3390/cells8080889
86. Junttila MR, de Sauvage FJ. Influence of Tumour Micro-Environment Heterogeneity on Therapeutic Response. Nature (2013) 501(7467):346–54. doi: 10.1038/nature12626
87. Shi J, Li Y, Jia R, Fan X. The Fidelity of Cancer Cells in PDX Models: Characteristics, Mechanism and Clinical Significance. Int J Cancer (2020) 146(8):2078–88. doi: 10.1002/ijc.32662
88. Conte N, Mason JC, Halmagyi C, Neuhauser S, Mosaku A, Yordanova G, et al. Pdx Finder: A Portal for Patient-Derived Tumor Xenograft Model Discovery. Nucleic Acids Res (2019) 47(D1):D1073–D9. doi: 10.1093/nar/gky984
89. Jakubowski CD, Azad NS. Immune Checkpoint Inhibitor Therapy in Biliary Tract Cancer (Cholangiocarcinoma). Chin Clin Oncol (2020) 9(1):2. doi: 10.21037/cco.2019.12.10
90. Zhao Y, Shuen TWH, Toh TB, Chan XY, Liu M, Tan SY, et al. Development of a New Patient-Derived Xenograft Humanised Mouse Model to Study Human-Specific Tumour Microenvironment and Immunotherapy. Gut (2018) 67(10):1845–54. doi: 10.1136/gutjnl-2017-315201
91. Rosato RR, Dávila-González D, Choi DS, Qian W, Chen W, Kozielski AJ, et al. Evaluation of Anti-PD-1-based Therapy Against Triple-Negative Breast Cancer Patient-Derived Xenograft Tumors Engrafted in Humanized Mouse Models. Breast Cancer Res (2018) 20(1):108–. doi: 10.1186/s13058-018-1037-4
Keywords: patient-derived tumor xenograft models, cholangiocarcinoma, target treatment, individualized treatment, chemotherapy
Citation: Wu J, Sheng J, Qin H, Cui M, Yang Y and Zhang X (2021) The Application Progress of Patient-Derived Tumor Xenograft Models After Cholangiocarcinoma Surgeries. Front. Oncol. 11:628636. doi: 10.3389/fonc.2021.628636
Received: 12 November 2020; Accepted: 30 June 2021;
Published: 22 July 2021.
Edited by:
Fabrizio Bronte, Azienda Ospedaliera Ospedali Riuniti Villa Sofia Cervello, ItalyReviewed by:
Suraj Kadunganattil, Amala Cancer Research Centre, IndiaCopyright © 2021 Wu, Sheng, Qin, Cui, Yang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongsheng Yang, eXlzd3h0QDEyNi5jb20=; Xuewen Zhang, emhhbmd4d0BqbHUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.