- 1School of Public Health, North China University of Science and Technology, Tangshan, China
- 2College of Life Science, North China University of Science and Technology, Tangshan, China
- 3School of Basic Medical Sciences, North China University of Science and Technology, Tangshan, China
- 4Affiliated Tangshan Gongren Hospital, North China University of Science and Technology, Tangshan, China
This study aims to analyze the methylation regulation of TLR3 in lung adenocarcinoma (LUAD) and to explore the association of TLR3 expression with immune microenvironment. TLR3 has a decreased expression in LUAD tissues and low expression of TLR3 is not only associated with poor prognosis in patients with LUAD, but also can be used as a diagnostic marker. Bisulfite sequencing PCR (BSP) results showed that the methylation level in the promoter of TLR3 was negatively correlated with the level of TLR3 mRNA in LUAD tissues. TIMER analysis showed that TLR3 was negatively correlated with the tumor purity of LUAD and positively with immune cell infiltration to some extent. ESTIMATE analysis also suggested that TLR3 expression and its methylation had significant correlation with immune score. The lower immune scores were associated with the late stage of LUAD and poor prognosis. The high expression of TLR3 might inhibit the development of LUAD by activating apoptosis pathway. The proteins interacted with TLR3 were mainly involved in the apoptosis pathway and positively correlated with the key genes (MYD88, Caspase 8, BIRC3, PIK3R1) in this pathway. Therefore, TLR3 as a key biomarker for prognosis and diagnosis in LUAD, might be considered as a potential epigenetic and immunotherapeutic target.
Introduction
Lung adenocarcinoma (LUAD) is the major histological subtype of non-small cell lung cancer which accounts for about 80–85% of all lung cancer (1). Due to the lack of clinical symptoms and effective screening techniques, most patients are diagnosed at late-stage and have a poor prognosis. Therefore, the effective LUAD biomarkers are still needed to be developed.
Toll-like receptors (TLRs) are one of important pattern recognition receptors which have been demonstrated to be associated with the tumor progress. Several of TLR agonists have been investigated in clinical studies for their potential function in antitumor immunity (2–4). Toll-like receptor 3 (TLR3), as one of important TLR members, is participated in the development of many tumors by recognizing and binding to its corresponding ligands (5–9). TLR3 has been reported to express on normal epithelial cells, such as lung tissue. The abnormal expression of TLR3 has been reported on various of cancer tissues (10, 11). TLR3 has also been recognized as a favorable prognosis biomarker of lung cancer by activating apoptosis or promoting autophagy (12, 13). TLR3 agonist enhanced T cell infiltration in lung tissue, which is essential for tumor immunity within the tumor microenvironment (14–16). A clinical trial showed that Poly-ICLC (Hiltinol), an advanced form of TLR3 agonist, could kill cancer cells and inhibit the metastasis of non-small cell lung cancer by activating several cancer suppressors and rebuilding an immunosuppressive tumor microenvironment (17).
Tumor microenvironments (TMEs) are largely populated with stromal and immune cells, which play the crucial role in the initiation and progression of lung cancer (18). Studies revealed that the proportion and character of T lymphocytes within TMEs had important effects on the progression and prognosis of lung cancer (16, 19, 20). Studies also revealed that the innate immune cells, such as macrophages and dendritic cells, when presenting in TMEs, contributed to the lung cancer initiation and played a key role in cancer immunotherapy (21–23). The crosstalk between cancer cells and the proximal immune cells ultimately is favorable to foster tumor growth and metastasis (24).
DNA methylation, one of Epigenetic mechanisms, plays a crucial role in controlling gene expression. The aberrant alteration of methylation has been demonstrated to be associated with the development of lung cancer (25, 26). The alterations of DNA methylation promote tumorigenesis by silencing tumor suppressor genes (27). Several candidate methylation biomarkers have been identified to serve as lung cancer detection and prognostic evaluation (28–30).
The anti-cancer activity induced by TLRs is partly due to the functional regulation of immune cells infiltrating in TME (31, 32). In order to investigate the interaction between tumor cells and tumors microenvironment, we used estimation of stromal and immune cells in malignant tumor tissues using expression data (ESTIMATE) algorithm to estimate the stromal and immune cells scores in LUAD and further evaluated the association of TLR3 expression with immune microenvironment. We also evaluated the effects of TLR3 on LUAD development and prognosis and to further shed light on the methylation regulatory of TLR3.
Materials and Methods
Patients and Tissue Samples
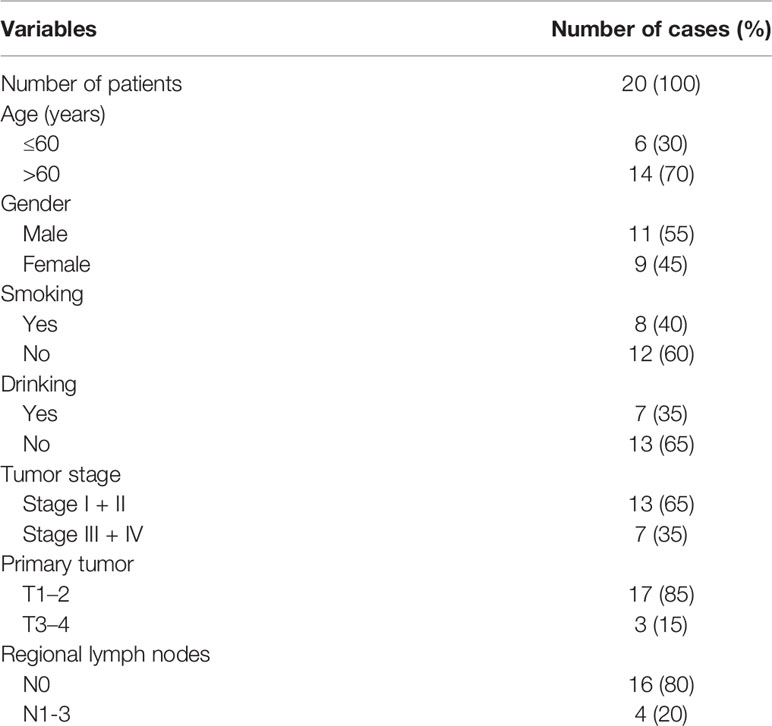
Tumor and its normal adjacent lung specimen from 20 patients with primary lung adenocarcinoma were collected from Affiliated Tangshan Gongren Hospital of North China University of Science and Technology. The detailed clinicopathological characteristics of these cases are summarized in Table 1. This study was approved by the Institutional Review Board of North China University of Science and Technology and informed consent was signed by all participants.
Prognosis Analysis
To measure the separability by TLR3 mRNA, receiver operating characteristics (ROC) curve was drawn and the area under curve (AUC) was calculated by “pROC” package. Based on the best separation cut-off value, overall survival (OS) and progression-free survival (PFS) was analyzed by KM Plotter Online Tool.
Cell Culture and Treatment
The lung cancer cell lines (A549 and NCI-H460) and human normal bronchial epithelial cell BEAS-2B were all bought from American Type Culture Collection (ATCC). A549 and NCI-H460 cells were cultured in RPMI-1640 medium and BEAS-2B cells was cultured in DMEM medium. All mediums were mixed with 10% fetal bovine serum (FBS; Life Technologies, NY, USA) and antibiotics (100 U/ml penicillin and 100 μg/ml streptomycin) in a humidified incubator with 5% CO2 at 37°C. For TLR3 agonist treatment, 10 and 50 µg/ml Poly (I:C) (ThermoFisher, USA) were used to treat A549 cells for 48 h.
Bisulfite Sequencing and DNA Demethylation Treatment
Genomic DNA was extracted from lung cancer cells and subjected to sodium bisulfite modification using EpiTect Bisulfite Kits (TIANGEN, Beijing, China) following the manufacturer’s protocols. Sodium bisulfite-treated DNA was amplified using Bisulfite sequencing PCR (BSP) primers (5’-AGT ATT TTG GGA GGT TAA GGT GG-3’ and 5’-AAT AAC AAA CCA AAT ATA ATT AAC AAA T-3’). PCR products were subcloned into pGM-T vector according to the manufacturer’s instruction. Ten colonies from each sample were then picked up for sequencing. To test the effect of methylation on the expression of TLR3, the methylation inhibitor 5-Aza-2-deoxycytidine (Sigma, CA, USA) was used to treat lung cancer cells (A549 and NCI-H460) in different concentration (0, 5, 10 μmol/L) for 72 h.
RNA Extraction and qRT-PCR
Total RNA was extracted from 20 pairs of LUAD and matched para-cancer tissues by using Trizol reagent (Invitrogen, CA, USA) and were then reversely transcribed into cDNA with RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, NY, USA). TLR3 mRNA was detected using Power SYBR Green PCR Master Mix (Thermo Fisher Scientific, NY, USA) in ABIPRISM® 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, USA). The amplification procedure was: 50°C for 2 min, 95°C for 2 min, followed by 40 cycles of 95°C for 15 s and 60°C for 2 min. GAPDH was used as the reference gene. The relative mRNA expression was analyzed using the 2−△△Ct method. The experiment was repeated three times. The qRT-PCR primers of TLR3 are as follows: 5’-CAA ACA CAA GCA TTC GGA ATC TG-3’ and 5’-AAG GAA TCG TTA CCA ACC ACA TT-3’.
Stromal/Immune Scores and Tumor-Infiltrating Immune Cells Infiltration
ESTIMATE is a newly developed algorithm that takes advantage of the unique properties of the transcriptional profiles of cancer tissues to infer tumor cellularity as well as the different infiltrating normal cells (33). We used this algorithm to impute stromal and immune scores and then to predict the level of infiltrating stromal and immune cells based on the gene expression profile. Furthermore, the abundance of infiltrating immune cells was evaluated by Tumor Immune Estimation Resource (TIMER) algorithm. For survival analysis, LUAD patients were classified by stromal/immune scores after getting the optimal cutoff value by X-tile Software.
Western Blot Analysis
Lung cancer cells were lysed in a lysis buffer and protein samples were run on 8% SDS polyacrylamide gel and then transferred to Nitrocellulose (NC) membrane. After incubated with the specified primary antibody, and then corresponding horseradish peroxidase (HRP) conjugated secondary antibody, specific protein was developed by enhanced chemiluminescence (ECL) luminescence reagents (Amersham, UK). β-actin was applied as reference control. All antibodies (anti-PI3 kinase p85 alpha, anti-caspase 8, anti-MyD88 and anti-BIRC3 antibodies) were purchased from Abcam (Eugene, USA).
Gene Enrichment and GSVA Analysis
We used the “ggplot2” R package and the DAVID 6.8 database (https://david.ncifcrf.gov/) to conduct Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis. Using TCGA-LUAD RNA-seq data, we performed the Gene Set Variation Analysis (GSVA) using GSVA R package to determine the pathways which most related to TLR3. The gene set “2.cp.kegg.v6.2.symbols.gmt” from the Molecular Signature Database was selected as the reference.
Statistical Analyses
All analyses were performed with R version 3.6.1 and its appropriate packages. Statistical significance of Student’s t-test was used for two-group comparisons. Survival analysis was estimated by the Kaplan–Meier method and the log-rank test. P <0.05 was considered as significant.
Results
The Expression and Clinical Significance of TLR3 in Human LUAD
To determine the differential expression of TLR3, we analyzed the data from the cancer genome atlas (TCGA) by TIMER and found that TLR3 was significantly decreased in LUAD tissues than in adjacent lung tissues (Figure 1A). To verify the result from TCGA, we measured the mRNA level of TLR3 in 20 paired LUAD tissues and adjacent normal tissues by qRT-PCR which turned out to be consistent with the finding of database (Figure 1B).
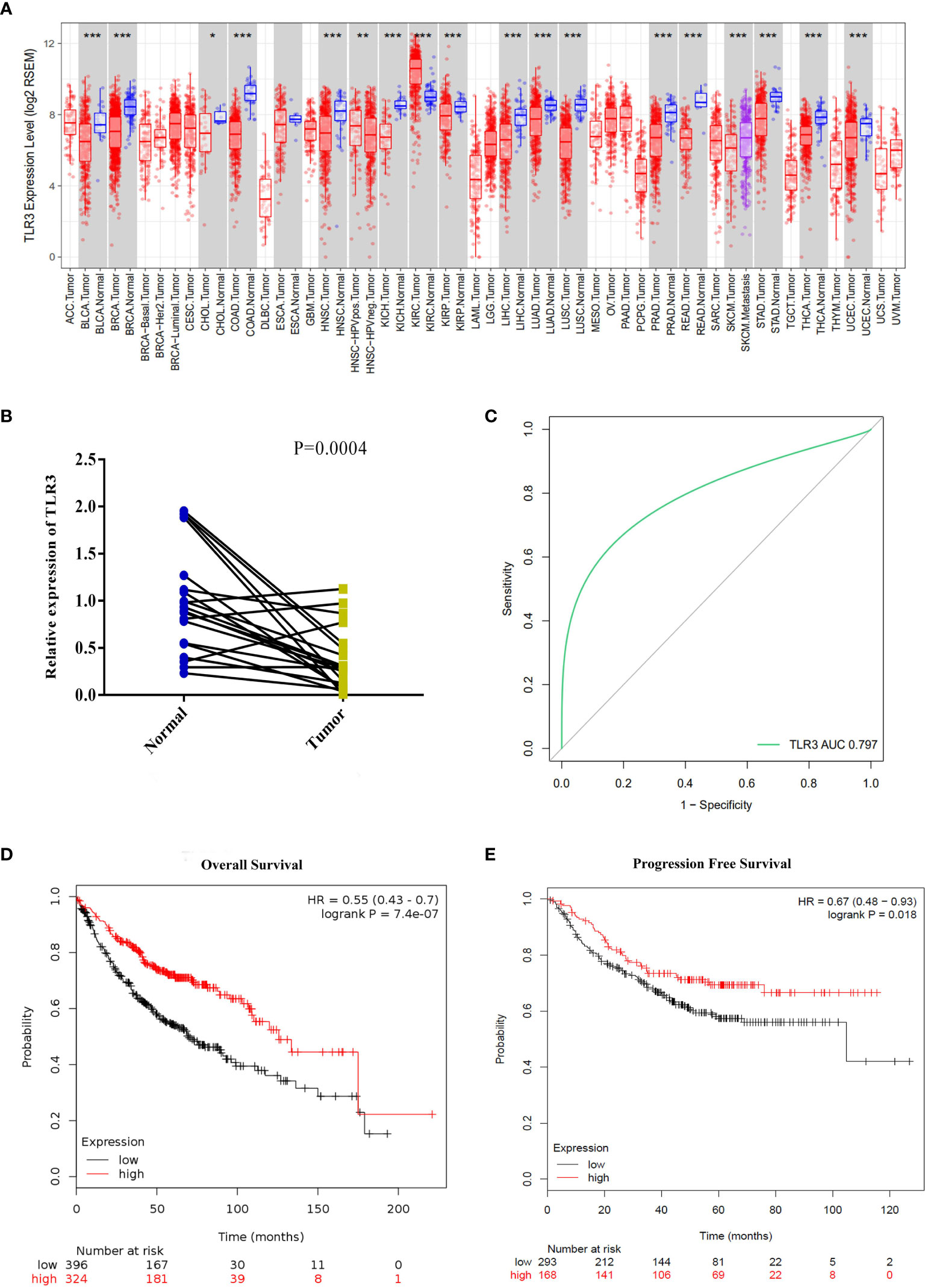
Figure 1 TLR3 expression levels in lung adenocarcinoma and its relationship with prognosis. (A) TLR3 expression levels in different tumor types from TCGA database were determined by TIMER (*P < 0.05, **P < 0.01, ***P < 0.001). (B) Quantification of TLR3 expression levels in lung adenocarcinoma and paired adjacent normal tissues by qRT-PCR. P-values were obtained by Student’s t-test. (C) ROC curve from qRT-PCR shows TLR3 is a marker to distinguish lung adenocarcinoma tissues from normal tissues. (D, E) Overall survival and disease-free survival analysis of lung adenocarcinoma patients based on TLR3 expression. Data was analyzed using Kaplan–Meier Plotter.
To measure the separability by the expression of TLR3, ROC curve was plotted (Figure 1C). The area under the curve for TLR3 expression was 0.806, which means a good measure of separability by TLR3. Using the Kaplan–Meier plotter data, we found that the expression of TLR3 was positively correlated with the overall survival (HR = 0.55, 95%CI = 0.43–0.7, P = 7.4e−07) and progression-free survival (HR = 0.67, 95%CI = 0.48–0.93, P = 0.018) in LUAD cancer patients (Figures 1D, E).
TLR3 Is Regulated by DNA Methylation
Using the MethPrimer program, we found that there was a CpG island with fourteen CpG sites in the proximal promoter region (3,000 bp upstream the TSS site) of TLR3 (Figure 2A). Upon analyzing TCGA methylation data of LUAD, we found that TLR3 methylation was negatively correlated with the level of TLR3 mRNA (Figure 2B). The sequencing of bisulfite converted DNA presented a higher methylation level in A549 and NCI-H460 cancer cells than that in BEAS-2B cells (Figure 2C).
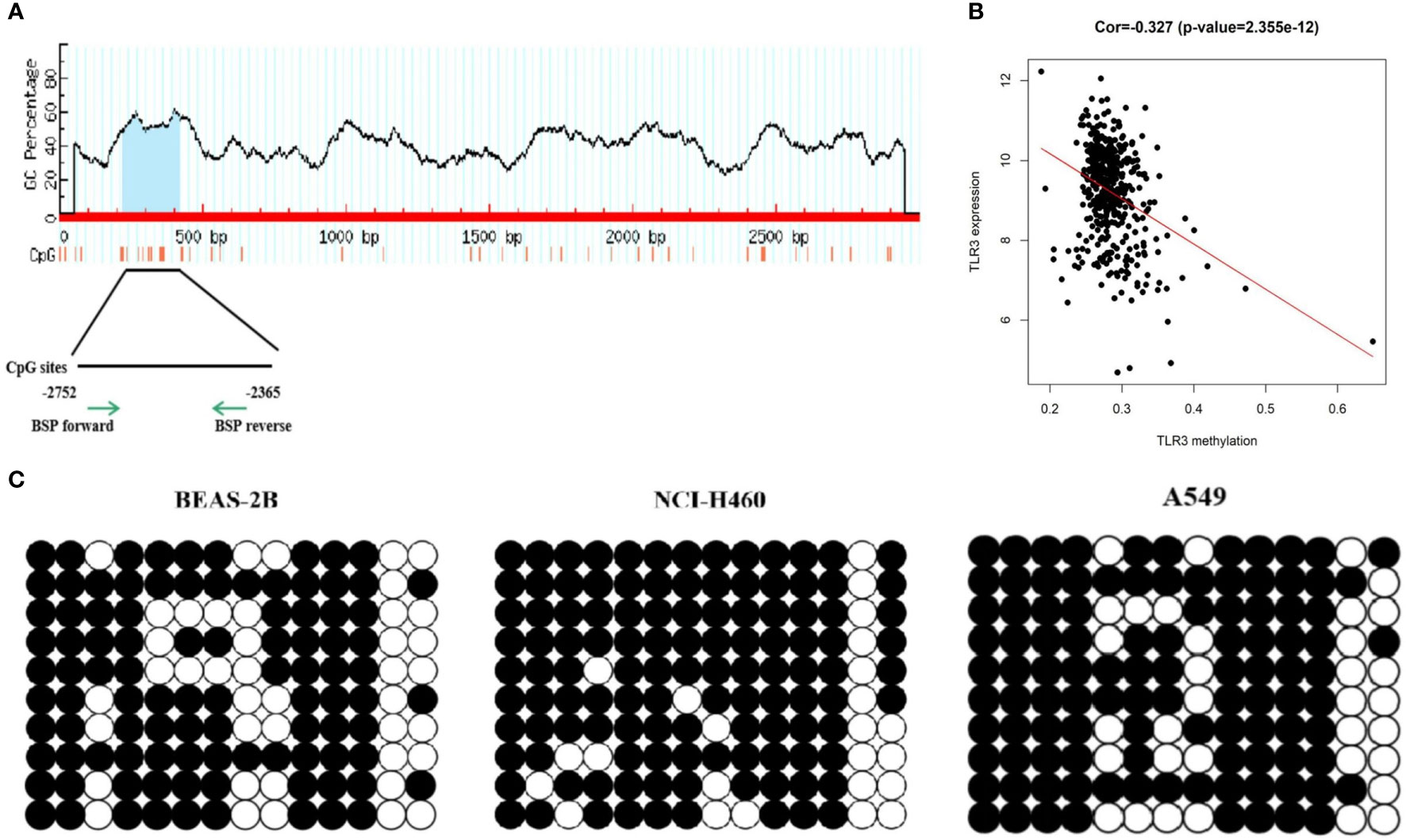
Figure 2 DNA methylation detection of TLR3 promoter. (A) MethPrimer database predicts CpG islands in the TLR3 promoter region. (B) Pearson correlation analysis of TLR3 expression and methylation degree. (C) BSP analysis of methylation status in the TLR3 promoter region.
After detecting the mRNA and protein level of TLR3 in lung cancer cells (A549, NCI-H460, NCI-H1975 and SPC-A1) (Figures 3A, B), we investigated the effect of methylation on the expression of TLR3. We treated lung cancer cells with methyltransferase inhibitor 5-aza-dc and found that TLR3 mRNA was significantly increased in dose-dependent manner (Figure 3C).
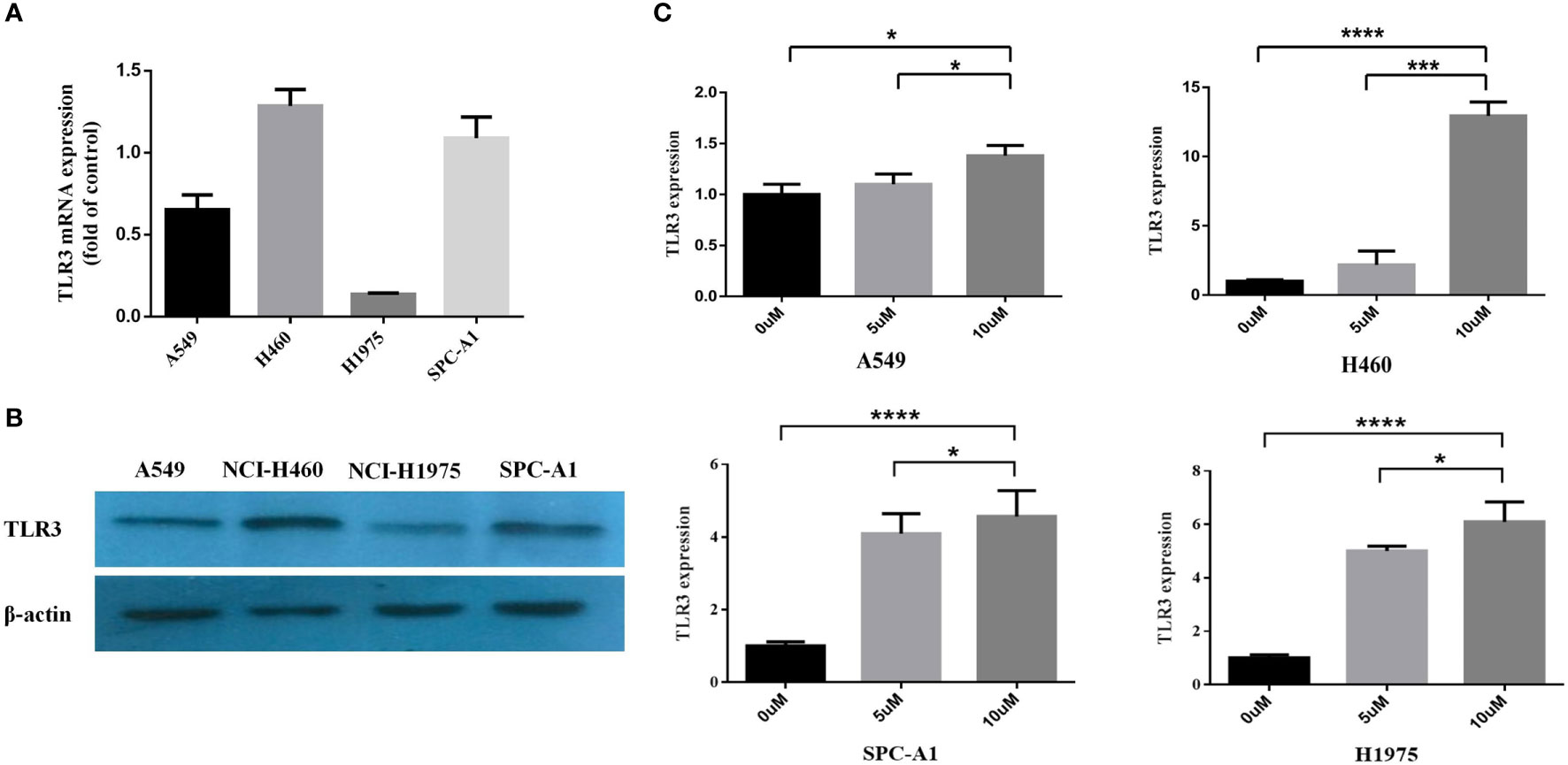
Figure 3 TLR3 expression is regulated by DNA methylation of its promoter. (A, B) The expression levels of TLR3 mRNA and protein were measured by qRT-PCR and western blot analysis in A549, NCI-H460, NCI-H1975 and SPC-A1 cells, respectively. (C) The expression of TLR3 restored in A549, NCI-H460, NCI-H1975 and SPC-A1 cell lines after treatment with 5-aza-2′-deoxycytidine for 72 h. Data are from at least three independent experiments. *P < 0.05, ***P < 0.001, ****P < 0.0001.
Estimation of Infiltrating Cells and Tumor Purity and Their Impact on LUAD Progress and Prognosis
In tumor microenvironment, infiltrating stromal and immune cells play an important role in the development, progression and prognosis of lung cancer (15, 34). ESTIMATE algorithm was performed for each sample to predict the abundance of infiltrating stromal and immune cells using immune and stromal scores and to infer tumor purity using ESTIMATE score (33). Samples with higher stromal or immune scores were represented for larger amount of the immune or stromal components in tumor microenvironment. After estimating the stromal, immune and ESTIMATE scores, we evaluated the impact of these scores on the progression and prognosis of LUAD. As shown in Figure 4A, immune scores and ESTIMATE scores roughly decreased with increasing tumor stage (P = 0.027; P = 0.043). After separated into two groups by immune/stromal/ESTIMAT score, patients with higher these scores had significantly longer overall survival time (P = 4.83e−03, 5.76e−02 and 3.74e−02) (Figure 4B).
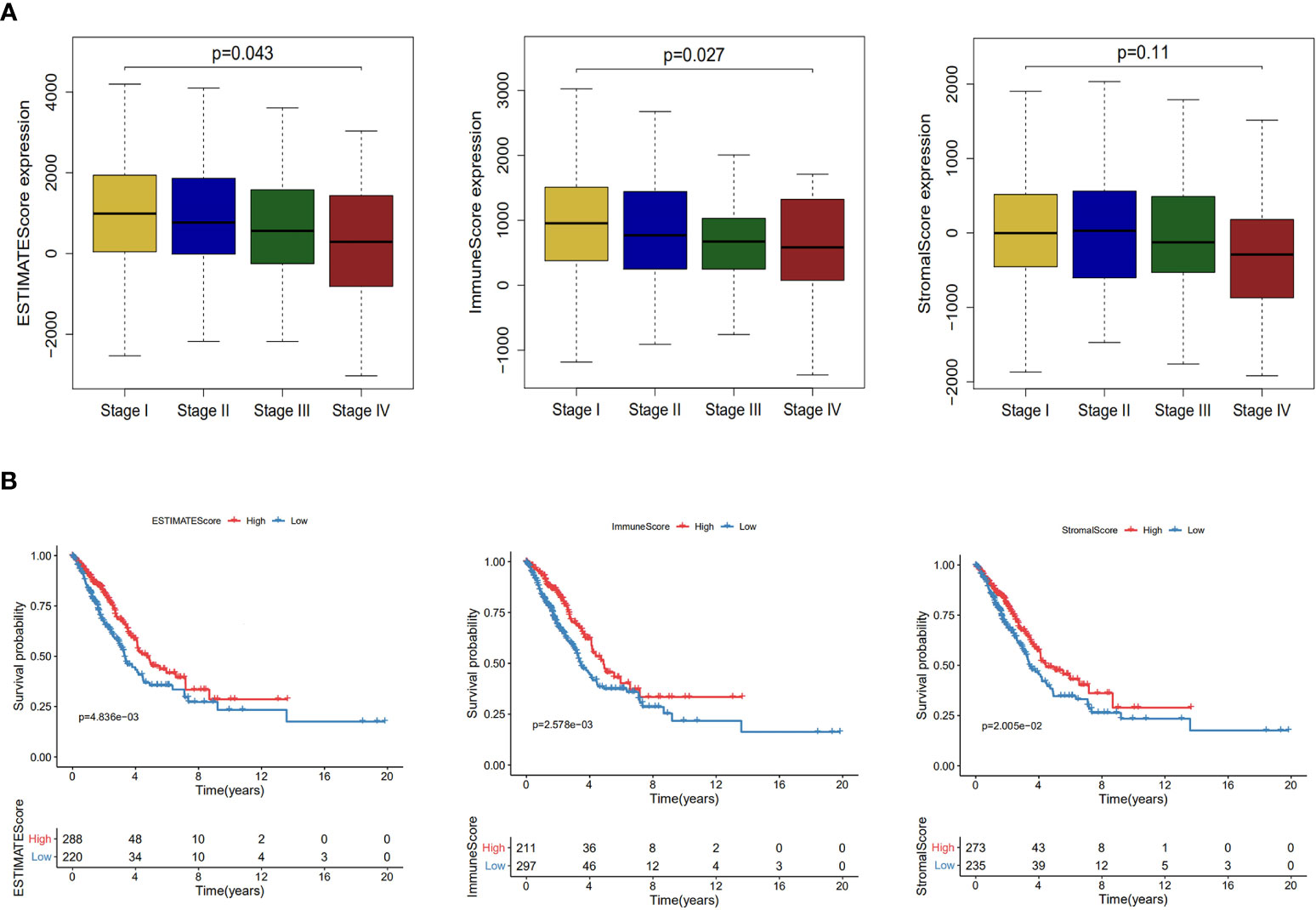
Figure 4 Association of stromal and immune scores with lung adenocarcinoma pathology and prognosis. (A) Distributions and comparisons of ESTIMATE score, immune score and stromal score among different tumor stages. (B) Kaplan–Meier plot of overall survival for patients with ESTIMATE score, immune score and stromal score.
TLR3 Expression Is Correlated With Immune Cells Infiltrating and Tumor Purity in LUAD
Due to the important role of toll-like receptors in innate immunity, we analyzed the relationship between TLR3 expression and immune cell infiltration by TIMER (Figure 5A) and found that TLR3 expression was related to infiltrating levels of B cells (r = 0.151, P = 8.82e−04), CD8+ T cells (r = 0.287, P = 1.06e−10), CD4+ T cells (r = 0.209, P = 3.44e−06), Macrophages (r = 0.193, P = 1.97e−05), Neutrophils (r = 0.286, P = 1.55e−10), and Dendritic cells (r = 0.383, P = 1.86e−18) in LUAD. We also found that TLR3 copy number variation (CNV) had effect on the infiltration level of several immune cells (B cell, CD4+ T cells, Macrophages, Neutrophils and Dendritic cells) (Figure 5B).
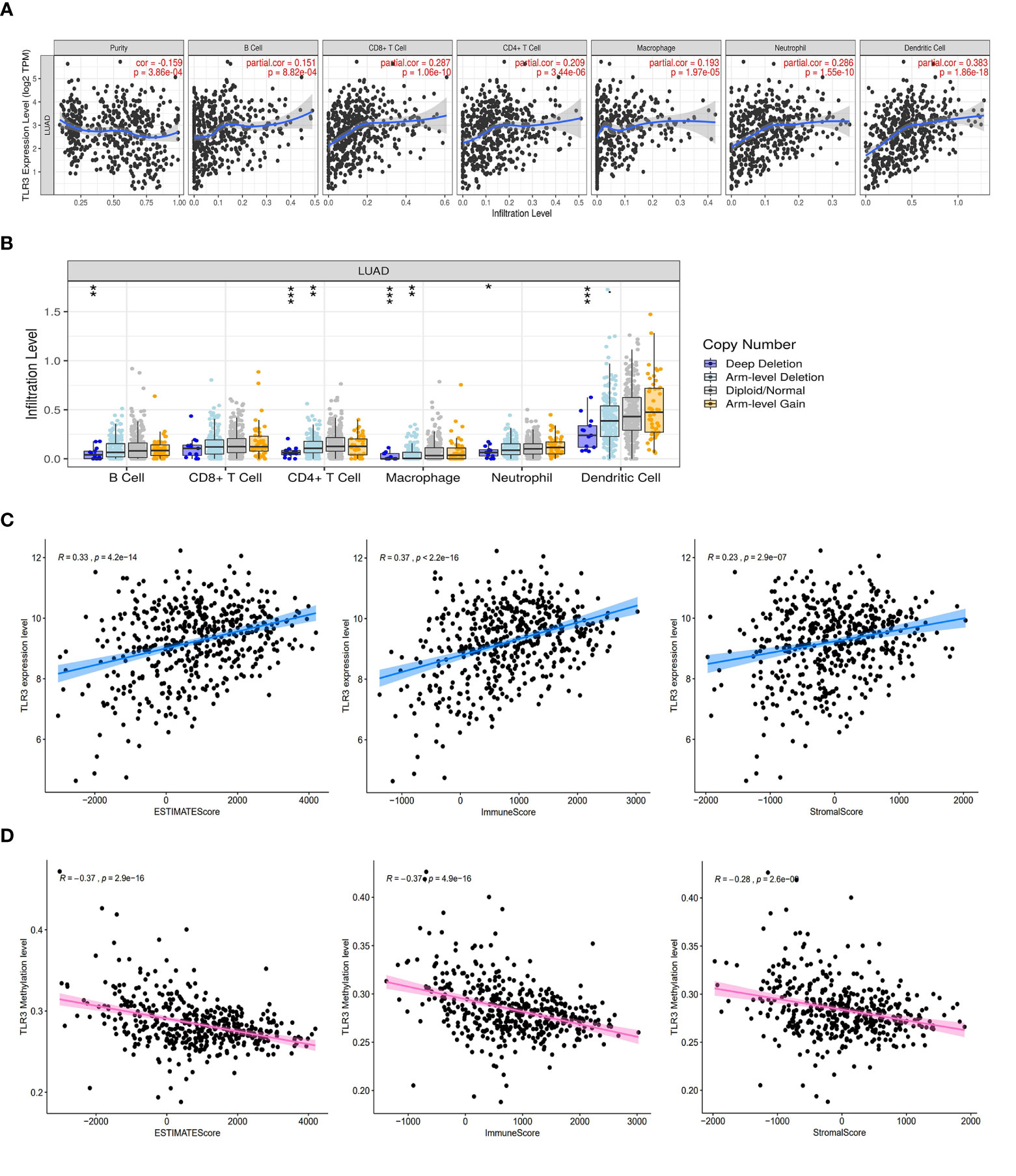
Figure 5 TLR3 expression is correlated with immune cells infiltrating and tumor purity in LUAD. (A) Correlation analysis of TLR3 expression and immune cells. (B) Correlation analysis of TLR3 copy number variation and immune cells. (C) TLR3 expression was positively correlated with ESTIMATE score, immune score and stromal score in lung adenocarcinoma. (D) TLR3 methylation was negative correlated with ESTIMATE score, immune score and stromal score in lung adenocarcinoma. *P < 0.05, **P < 0.01,***P < 0.001.
To further evaluate the impact of TLR3 on the level of immune cells infiltrating and tumor purity, we analyzed the correlation between TLR3 and the immune/stromal/ESTIMAT score. Our results showed that stromal score, immune score and ESTIMATE score in LUAD were positively associated with TLR3 expression, but negatively associated with TLR3 methylation (Figures 5C, D).
The Potential Mechanism of TLR3 in LUAD Development
To discover the potential mechanism of TLR3 in the development of LUAD, we used the compartmentalized protein-protein interaction database (ComPPI) to construct the protein–protein interaction network of TLR3. As a result, a total of 38 central interactive proteins were filtered out with an average Interaction Score of 0.749 (Figure 6A). Based on these interactive proteins, we performed GO and KEGG enrichment analysis. The enrichment analysis showed that these proteins were involved in apoptosis pathway (Figures 6B, C) which contained nine genes, including receptor-interacting protein kinase 1(RIPK1), inhibitor of nuclear factor kappa B kinase regulatory subunit gamma (IKBKG), myeloiddifferentiationfactor88 (MYD88), Caspase 8, baculoviral iap repeat containing 3 (BIRC3), phosphoinositide-3-kinase regulatory subunit 1 (PIK3R1), baculoviral iap repeat containing 2 (BIRC2), interleukin 1 receptor associated kinase 2 (IRAK2) and fas associated via death domain (FADD). LUAD samples were divided into two groups by the median of TLR3 mRNA level. GSVA confirmed that most samples were enriched in pathways closely correlated with tumorigenesis, including apoptosis (Figure 6D). We then evaluated the relationship between TRL3 and these apoptosis-related genes in RNA level using TCGA dataset. The results turned out a significant correlations betweenTLR3 and the expression of MYD88, Caspase 8, BIRC3 and PIK3R1 (r >0.3; P <0.001) (Figure 7A). To further verify the above results, we used TLR3 agonist Poly(I:C) to treat lung cancer cells (A549) and found that the expressions of MYD88, Caspase 8, BIRC3 and PIK3R1 were in line with the expression of TLR3 (Figures 7B, C).
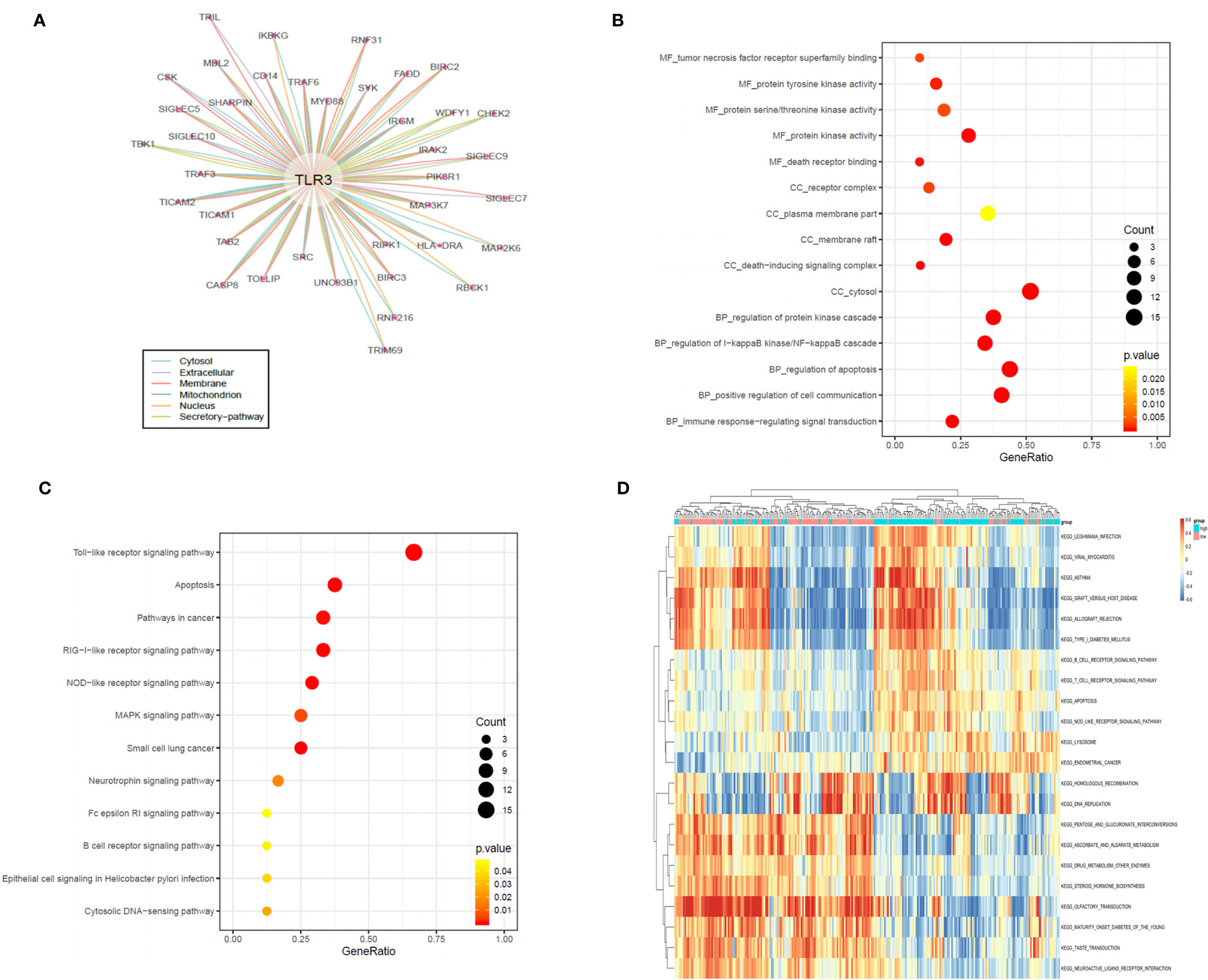
Figure 6 TLR3 regulate LUAD development by apoptotic pathways (A). Protein–protein interaction (PPI) analysis of the network linking TLR3 by the ComPPI database. (B, C). TLR3 interacting proteins performing GO function enrichment and KEGG pathway analysis. (D) GSVA-derived clustering heatmaps of differentially expressed pathways for TLR3. Only signaling pathways with log(foldchange) > 0.2 are shown.
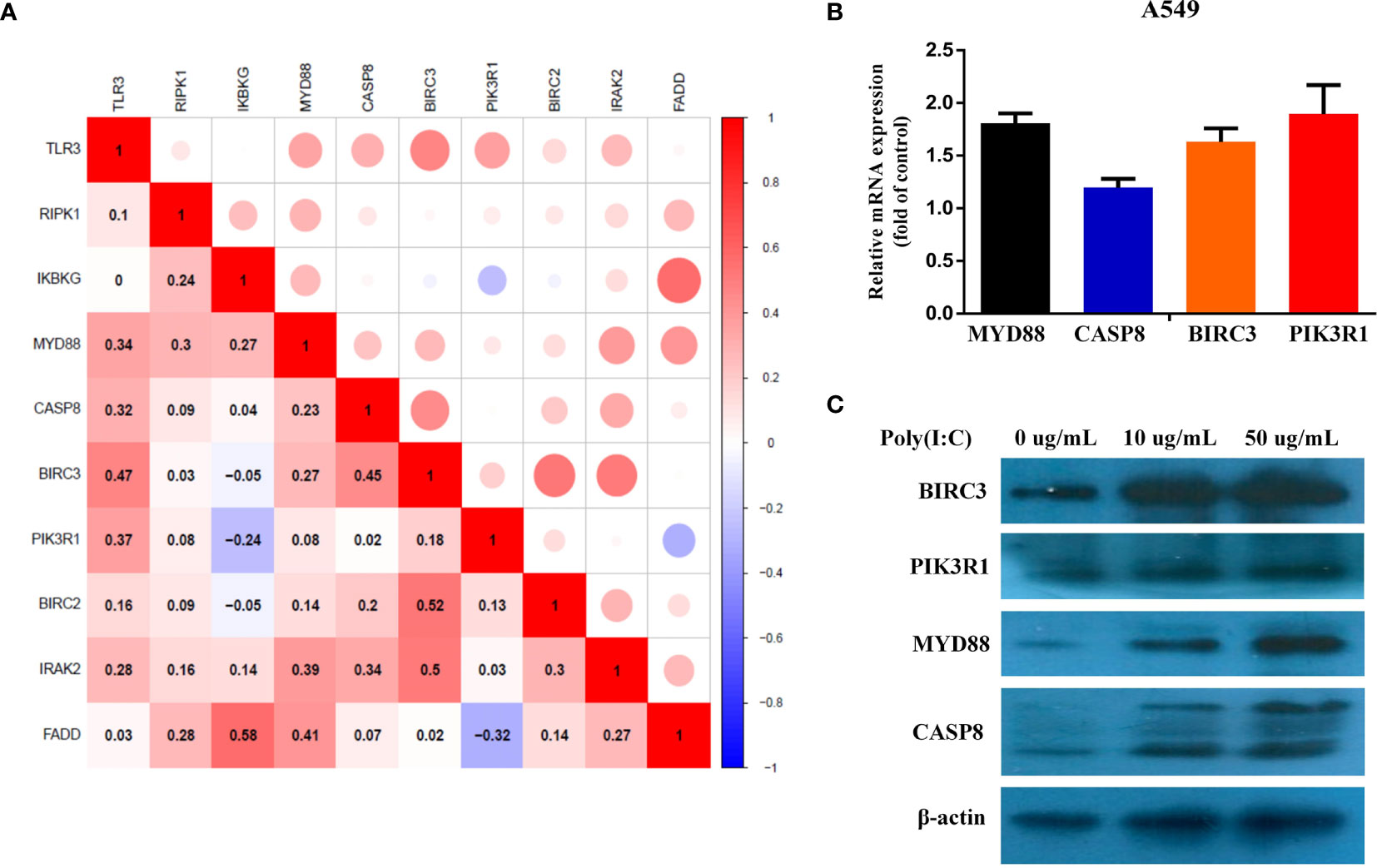
Figure 7 Evaluate the correlation between different TLR3 expression and apoptosis genes. (A). Pearson Correlation analysis of TLR3 and apoptosis genes. (B, C). The expression levels of MYD88, CASP8, BIRC3 and PIK3R1 mRNA and protein were measured by qRT-PCR and western blot analysis after 24 h Poly(I:C) treatment in the A549 cells, respectively.
Discussion
Toll-like receptors are one of the most well-known pattern recognition receptor family members. TLR3 is the only one member of TLRs which signals through TRIF instead of MyD88. In recent years, TLR3 has been reported to be aberrantly expressed in several cancer tissues and is involved in cancer therapy and prognosis of certain cancers. For example, TLR3 resisted the DNA damage induced by cisplatin, which leads to the resistance of head and neck cancer cells to cisplatin (5). TLR3 expression in neuroblastoma was associated with the improving prognosis (35). The suppression of TLR3 was related to tumor proliferation, angiogenesis, and the inhibition of apoptosis in liver cancer (6, 10). However, several reports also demonstrated that TLR3 protein level on tumor cells was associated with improving overall survival in a stage-I cohort study of non-small cell lung cancer (36). Similarly, in our current study, we presented that TLR3 was not only downregulated, but also served as an independent prognostic marker for LUAD patients. The low expression of TLR3 is considered to be related to a poor outcome of LUAD. Go and KEGG enrichment analyses presented that TLR3 was correlated with several key proteins in the apoptotic pathway. GSVA analysis showed that highly expressed TLR3 could also activate the apoptotic pathway.
As one of important epigenetic mechanisms, DNA methylation has received more and more attention in recent decades. Hypermethylation in gene promoter regions adjacent to transcription start sites (TSSs) often leads to gene silencing, and then result in the alterations of gene function in several cancer types (37, 38). Some studies have shown that DNA methylation was widely used for early diagnosis and treatment in breast cancer, gastric cancer, and liver cancer (39). In this study, we systematically evaluated the relationship between DNA methylation and the expression of TLR3 in LUAD and found that the hypermethylation of the TLR3 inhibited the activity of the gene. The expression of TLR3 increased when LUAD cells were treated with 5-Aza-dc. These results provided new insights into the role of DNA methylation in the regulation of LUAD.
Tumor progression is also affected by its extrinsic tumor microenvironment. Studies have shown that TIME played key roles in regulating the process of carcinogenesis, tumor invasion, and metastasis (40). The immune suppressive microenvironment induced by tumor infiltrating regulatory T lymphocytes prevents effective anti-tumor immunity and has become the key factor of immunotherapy against cancer (41–43). Studies also showed that LUAD had higher levels of immune infiltration than other histopathological types, therefore LUAD patients were more sensitive to immunotherapy (44, 45). TLRs are expressed not only on human tumor cells, but also on immune cells (46–48). As one of the important TLR family members, TLR3 have been confirmed to promote the activation of CD8+ T lymphocytes and IFN production in dendritic cells by binding to specific ligands (49, 50). Noteworthy, our results indicated a substantial explicit connection between TLR3 expression and infiltration levels of immune cells in LUAD.
TME is a complicated system, in which immune and stromal cells are two major non-tumor components which are associated with tumor growth and prognosis (51, 52). ESTIMATE algorithm has been used to predict the purity of tumors and the levels of infiltrating matrix and immune cells in tumor tissues (33, 53–57). A recent meta-analysis with 29 studies indicated that tumor-infiltrating immune cells had a superior prognostic impact on lung cancer (58). In current study, we demonstrated that LUAD patients with late clinical pathological stages or poor prognosis had low-stromal and immune scores. Our data also presented a positive correlation between stromal/immune scores and TLR3 expression. These results suggested that increased TLR3 level might inhibit cancer cell proliferation by activating immune cells infiltrating.
Taken together, our findings serve for a better understanding of the role of TLR3 in the development of lung cancer. Furthermore, if confirmed in further in-deep study, TLR3 could be considered as a promising lung cancer biomarker for tumor aggressiveness and prognosis.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary materials. Further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by Institutional Review Board of North China University of Science and Technology. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
AL drafted the manuscript. AL and YZ performed the bioinformatic analysis. AL and QT conducted methylation analysis. ZZ and HW performed statistical analyses. ZZ collected tissue samples. XZ designed and supervised the research. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by Key Project of Natural Science Foundation of Hebei province of China (grant number H2017209233).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-Small Cell Lung Cancer: Epidemiology, Risk Factors, Treatment, and Survivorship. Mayo Clin Proc (2008) 83(5):584–94. doi: 10.4065/83.5.584
2. Le Noci V, Tortoreto M, Gulino A, Storti C, Bianchi F, Zaffaroni N, et al. Poly(I:C) and CpG-ODN Combined Aerosolization to Treat Lung Metastases and Counter the Immunosuppressive Microenvironment. Oncoimmunology (2015) 4(10):e1040214. doi: 10.1080/2162402x.2015.1040214
3. Le Noci V, Sommariva M, Tortoreto M, Zaffaroni N, Campiglio M, Tagliabue E, et al. Reprogramming the Lung Microenvironment by Inhaled Immunotherapy Fosters Immune Destruction of Tumor. Oncoimmunology (2016) 5(11):e1234571. doi: 10.1080/2162402x.2016.1234571
4. De Cesare M, Sfondrini L, Pennati M, De Marco C, Motta V, Tagliabue E, et al. CpG-oligodeoxynucleotides Exert Remarkable Antitumor Activity Against Diffuse Malignant Peritoneal Mesothelioma Orthotopic Xenografts. J Transl Med (2016) 14:25. doi: 10.1186/s12967-016-0781-4
5. Chuang HC, Chou MH, Chien CY, Chuang JH, Liu YL. Triggering TLR3 Pathway Promotes Tumor Growth and Cisplatin Resistance in Head and Neck Cancer Cells. Oral Oncol (2018) 86:141–9. doi: 10.1016/j.oraloncology.2018.09.015
6. Bonnin M, Fares N, Testoni B, Estornes Y, Weber K, Vanbervliet B, et al. Toll-Like Receptor 3 Downregulation is an Escape Mechanism From Apoptosis During Hepatocarcinogenesis. J Hepatol (2019) 71(4):763–72. doi: 10.1016/j.jhep.2019.05.031
7. Sato Y, Motoyama S, Wakita A, Kawakita Y, Liu J, Nagaki Y, et al. TLR3 Expression Status Predicts Prognosis in Patients With Advanced Thoracic Esophageal Squamous Cell Carcinoma After Esophagectomy. Am J Surg (2018) 216(2):319–25. doi: 10.1016/j.amjsurg.2018.01.038
8. Fan L, Zhou P, Hong Q, Chen AX, Liu GY, Yu KD, et al. Toll-Like Receptor 3 Acts as a Suppressor Gene in Breast Cancer Initiation and Progression: A Two-Stage Association Study and Functional Investigation. Oncoimmunology (2019) 8(6):e1593801. doi: 10.1080/2162402x.2019.1593801
9. Zeljic K, Supic G, Jovic N, Kozomara R, Brankovic-Magic M, Obrenovic M, et al. Association of TLR2, Tlr3, TLR4 and CD14 Genes Polymorphisms With Oral Cancer Risk and Survival. Oral Dis (2014) 20(4):416–24. doi: 10.1111/odi.12144
10. Yuan MM, Xu YY, Chen L, Li XY, Qin J, Shen Y. TLR3 Expression Correlates With Apoptosis, Proliferation and Angiogenesis in Hepatocellular Carcinoma and Predicts Prognosis. BMC Cancer (2015) 15:245. doi: 10.1186/s12885-015-1262-5
11. Shi S, Xu C, Fang X, Zhang Y, Li H, Wen W, et al. Expression Profile of Toll−like Receptors in Human Breast Cancer. Mol Med Rep (2020) 21(2):786–94. doi: 10.3892/mmr.2019.10853
12. Bianchi F, Alexiadis S, Camisaschi C, Truini M, Centonze G, Milione M, et al. Tlr3 Expression Induces Apoptosis in Human non-Small-Cell Lung Cancer. Int J Mol Sci (2020) 21(4):1440. doi: 10.3390/ijms21041440
13. Jakobsen CJ. Is Knowledge of Hemodynamics Really Dangerous? J Cardiothorac Vasc Anesth (2016) 30(3):575–8. doi: 10.1053/j.jvca.2016.02.017
14. Jin JO, Yu Q. Systemic Administration of TLR3 Agonist Induces IL-7 Expression and IL-7-dependent CXCR3 Ligand Production in the Lung. J Leukoc Biol (2013) 93(3):413–25. doi: 10.1189/jlb.0712360
15. Zheng X, Hu Y, Yao C. The Paradoxical Role of Tumor-Infiltrating Immune Cells in Lung Cancer. Intractable Rare Dis Res (2017) 6(4):234–41. doi: 10.5582/irdr.2017.01059
16. Schulze AB, Evers G, Görlich D, Mohr M, Marra A, Hillejan L, et al. Tumor Infiltrating T Cells Influence Prognosis in Stage I-III non-Small Cell Lung Cancer. J Thorac Dis (2020) 12(5):1824–42. doi: 10.21037/jtd-19-3414a
17. Chang SC, Zhang BX, Su EC, Wu WC, Hsieh TH, Salazar AM, et al. Hiltonol Cocktail Kills Lung Cancer Cells by Activating Cancer-Suppressors, PKR/OAS, and Restraining the Tumor Microenvironment. Int J Mol Sci (2021) 22(4):1626. doi: 10.3390/ijms22041626
18. Quail DF, Joyce JA. Microenvironmental Regulation of Tumor Progression and Metastasis. Nat Med (2013) 19(11):1423–37. doi: 10.1038/nm.3394
19. Stankovic B, Bjørhovde HAK, Skarshaug R, Aamodt H, Frafjord A, Müller E, et al. Immune Cell Composition in Human non-Small Cell Lung Cancer. Front Immunol (2018) 9:3101. doi: 10.3389/fimmu.2018.03101
20. Yu Y, Zeng D, Ou Q, Liu S, Li A, Chen Y, et al. Association of Survival and Immune-Related Biomarkers With Immunotherapy in Patients With Non-Small Cell Lung Cancer: A Meta-Analysis and Individual Patient-Level Analysis. JAMA Netw Open (2019) 2(7):e196879. doi: 10.1001/jamanetworkopen.2019.6879
21. Lavin Y, Kobayashi S, Leader A, Amir ED, Elefant N, Bigenwald C, et al. Innate Immune Landscape in Early Lung Adenocarcinoma by Paired Single-Cell Analyses. Cell (2017) 169(4):750–65.e17. doi: 10.1016/j.cell.2017.04.014
22. Milette S, Fiset PO, Walsh LA, Spicer JD, Quail DF. The Innate Immune Architecture of Lung Tumors and its Implication in Disease Progression. J Pathol (2019) 247(5):589–605. doi: 10.1002/path.5241
23. Corrales L, Matson V, Flood B, Spranger S, Gajewski TF. Innate Immune Signaling and Regulation in Cancer Immunotherapy. Cell Res (2017) 27(1):96–108. doi: 10.1038/cr.2016.149
24. Hinshaw DC, Shevde LA. The Tumor Microenvironment Innately Modulates Cancer Progression. Cancer Res (2019) 79(18):4557–66. doi: 10.1158/0008-5472.Can-18-3962
25. Battram T, Richmond RC, Baglietto L, Haycock PC, Perduca V, Bojesen SE, et al. Appraising the Causal Relevance of DNA Methylation for Risk of Lung Cancer. Int J Epidemiol (2019) 48(5):1493–504. doi: 10.1093/ije/dyz190
26. Duruisseaux M, Esteller M. Lung Cancer Epigenetics: From Knowledge to Applications. Semin Cancer Biol (2018) 51:116–28. doi: 10.1016/j.semcancer.2017.09.005
27. Sharma S, Kelly TK, Jones PA. Epigenetics in Cancer. Carcinogenesis (2010) 31(1):27–36. doi: 10.1093/carcin/bgp220
28. Zhang Y, Wang R, Song H, Huang G, Yi J, Zheng Y, et al. Methylation of Multiple Genes as a Candidate Biomarker in non-Small Cell Lung Cancer. Cancer Lett (2011) 303(1):21–8. doi: 10.1016/j.canlet.2010.12.011
29. Heller G, Babinsky VN, Ziegler B, Weinzierl M, Noll C, Altenberger C, et al. Genome-Wide CpG Island Methylation Analyses in non-Small Cell Lung Cancer Patients. Carcinogenesis (2013) 34(3):513–21. doi: 10.1093/carcin/bgs363
30. Dmitriev AA, Kashuba VI, Haraldson K, Senchenko VN, Pavlova TV, Kudryavtseva AV, et al. Genetic and Epigenetic Analysis of non-Small Cell Lung Cancer With Noti-Microarrays. Epigenetics (2012) 7(5):502–13. doi: 10.4161/epi.19801
31. Schoenhals JE, Seyedin SN, Anderson C, Brooks ED, Li YR, Younes AI, et al. Uncovering the Immune Tumor Microenvironment in non-Small Cell Lung Cancer to Understand Response Rates to Checkpoint Blockade and Radiation. Transl Lung Cancer Res (2017) 6(2):148–58. doi: 10.21037/tlcr.2017.03.06
32. Yu X, Wang X. Tumor Immunity Landscape in non-Small Cell Lung Cancer. PeerJ (2018) 6:e4546. doi: 10.7717/peerj.4546
33. Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring Tumour Purity and Stromal and Immune Cell Admixture From Expression Data. Nat Commun (2013) 4:2612. doi: 10.1038/ncomms3612
34. Bremnes RM, Al-Shibli K, Donnem T, Sirera R, Al-Saad S, Andersen S, et al. The Role of Tumor-Infiltrating Immune Cells and Chronic Inflammation at the Tumor Site on Cancer Development, Progression, and Prognosis: Emphasis on non-Small Cell Lung Cancer. J Thorac Oncol (2011) 6(4):824–33. doi: 10.1097/JTO.0b013e3182037b76
35. Hsu WM, Huang CC, Wu PY, Lee H, Huang MC, Tai MH, et al. Toll-Like Receptor 3 Expression Inhibits Cell Invasion and Migration and Predicts a Favorable Prognosis in Neuroblastoma. Cancer Lett (2013) 336(2):338–46. doi: 10.1016/j.canlet.2013.03.024
36. Bianchi F, Milione M, Casalini P, Centonze G, Le Noci VM, Storti C, et al. Toll-Like Receptor 3 as a New Marker to Detect High Risk Early Stage Non-Small-Cell Lung Cancer Patients. Sci Rep (2019) 9(1):14288. doi: 10.1038/s41598-019-50756-2
37. Ehrlich M. DNA Methylation in Cancer: Too Much, But Also Too Little. Oncogene (2002) 21(35):5400–13. doi: 10.1038/sj.onc.1205651
38. Yang B, Guo M, Herman JG, Clark DP. Aberrant Promoter Methylation Profiles of Tumor Suppressor Genes in Hepatocellular Carcinoma. Am J Pathol (2003) 163(3):1101–7. doi: 10.1016/s0002-9440(10)63469-4
39. Pan Y, Liu G, Zhou F, Su B, Li Y. DNA Methylation Profiles in Cancer Diagnosis and Therapeutics. Clin Exp Med (2018) 18(1):1–14. doi: 10.1007/s10238-017-0467-0
40. Solinas G, Germano G, Mantovani A, Allavena P. Tumor-Associated Macrophages (TAM) as Major Players of the Cancer-Related Inflammation. J Leukoc Biol (2009) 86(5):1065–73. doi: 10.1189/jlb.0609385
41. Bense RD, Sotiriou C, Piccart-Gebhart MJ, Haanen J, van Vugt M, de Vries EGE, et al. Relevance of Tumor-Infiltrating Immune Cell Composition and Functionality for Disease Outcome in Breast Cancer. J Natl Cancer Inst (2017) 109(1):djw192. doi: 10.1093/jnci/djw192
42. Kurebayashi Y, Emoto K, Hayashi Y, Kamiyama I, Ohtsuka T, Asamura H, et al. Comprehensive Immune Profiling of Lung Adenocarcinomas Reveals Four Immunosubtypes With Plasma Cell Subtype a Negative Indicator. Cancer Immunol Res (2016) 4(3):234–47. doi: 10.1158/2326-6066.Cir-15-0214
43. Campa MJ, Moody MA, Zhang R, Liao HX, Gottlin EB, Patz EF Jr. Interrogation of Individual Intratumoral B Lymphocytes From Lung Cancer Patients for Molecular Target Discovery. Cancer Immunol Immunother (2016) 65(2):171–80. doi: 10.1007/s00262-015-1787-0
44. Varn FS, Wang Y, Mullins DW, Fiering S, Cheng C. Systematic Pan-Cancer Analysis Reveals Immune Cell Interactions in the Tumor Microenvironment. Cancer Res (2017) 77(6):1271–82. doi: 10.1158/0008-5472.Can-16-2490
45. Li B, Severson E, Pignon JC, Zhao H, Li T, Novak J, et al. Comprehensive Analyses of Tumor Immunity: Implications for Cancer Immunotherapy. Genome Biol (2016) 17(1):174. doi: 10.1186/s13059-016-1028-7
46. Furrie E, Macfarlane S, Thomson G, Macfarlane GT. Toll-Like receptors-2, -3 and -4 Expression Patterns on Human Colon and Their Regulation by Mucosal-Associated Bacteria. Immunology (2005) 115(4):565–74. doi: 10.1111/j.1365-2567.2005.02200.x
47. Schmausser B, Andrulis M, Endrich S, Lee SK, Josenhans C, Müller-Hermelink HK, et al. Expression and Subcellular Distribution of Toll-Like Receptors TLR4, TLR5 and TLR9 on the Gastric Epithelium in Helicobacter Pylori Infection. Clin Exp Immunol (2004) 136(3):521–6. doi: 10.1111/j.1365-2249.2004.02464.x
48. Huhta H, Helminen O, Kauppila JH, Salo T, Porvari K, Saarnio J, et al. The Expression of Toll-like Receptors in Normal Human and Murine Gastrointestinal Organs and the Effect of Microbiome and Cancer. J Histochem Cytochem (2016) 64(8):470–82. doi: 10.1369/0022155416656154
49. Seya T, Matsumoto M. The Extrinsic RNA-sensing Pathway for Adjuvant Immunotherapy of Cancer. Cancer Immunol Immunother (2009) 58(8):1175–84. doi: 10.1007/s00262-008-0652-9
50. Matsumoto M, Seya T. TLR3: Interferon Induction by Double-Stranded RNA Including Poly(I:C). Adv Drug Delivery Rev (2008) 60(7):805–12. doi: 10.1016/j.addr.2007.11.005
51. Xiong Y, Wang K, Zhou H, Peng L, You W, Fu Z. Profiles of Immune Infiltration in Colorectal Cancer and Their Clinical Significant: A Gene Expression-Based Study. Cancer Med (2018) 7(9):4496–508. doi: 10.1002/cam4.1745
52. Liu Z, Zhu Y, Xu L, Zhang J, Xie H, Fu H, et al. Tumor Stroma-Infiltrating Mast Cells Predict Prognosis and Adjuvant Chemotherapeutic Benefits in Patients With Muscle Invasive Bladder Cancer. Oncoimmunology (2018) 7(9):e1474317. doi: 10.1080/2162402x.2018.1474317
53. Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, et al. Genomic Analyses Identify Molecular Subtypes of Pancreatic Cancer. Nature (2016) 531(7592):47–52. doi: 10.1038/nature16965
54. Mertins P, Mani DR, Ruggles KV, Gillette MA, Clauser KR, Wang P, et al. Proteogenomics Connects Somatic Mutations to Signalling in Breast Cancer. Nature (2016) 534(7605):55–62. doi: 10.1038/nature18003
55. Guinney J, Dienstmann R, Wang X, de Reyniès A, Schlicker A, Soneson C, et al. The Consensus Molecular Subtypes of Colorectal Cancer. Nat Med (2015) 21(11):1350–6. doi: 10.1038/nm.3967
56. Shah N, Wang P, Wongvipat J, Karthaus WR, Abida W, Armenia J, et al. Regulation of the Glucocorticoid Receptor Via a BET-dependent Enhancer Drives Antiandrogen Resistance in Prostate Cancer. Elife (2017) 6:e27861. doi: 10.7554/eLife.27861
57. Klemm F, Joyce JA. Microenvironmental Regulation of Therapeutic Response in Cancer. Trends Cell Biol (2015) 25(4):198–213. doi: 10.1016/j.tcb.2014.11.006
Keywords: TLR3, methylation, lung adenocarcinoma, immune infiltration, apoptosis
Citation: Li A, Wu H, Tian Q, Zhang Y, Zhang Z and Zhang X (2021) Methylation Regulation of TLR3 on Immune Parameters in Lung Adenocarcinoma. Front. Oncol. 11:620200. doi: 10.3389/fonc.2021.620200
Received: 30 October 2020; Accepted: 26 April 2021;
Published: 20 May 2021.
Edited by:
Umberto Malapelle, University of Naples Federico II, ItalyReviewed by:
Pasquale Pisapia, University of Naples Federico II, ItalyGiuseppina Roscigno, University of Naples Federico II, Italy
Copyright © 2021 Li, Wu, Tian, Zhang, Zhang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuemei Zhang, anl4dWVtZWlAZ21haWwuY29t
 Ang Li
Ang Li Hongjiao Wu1
Hongjiao Wu1 Xuemei Zhang
Xuemei Zhang