
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol. , 29 April 2021
Sec. Head and Neck Cancer
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.616202
This article is part of the Research Topic Non-coding RNAs in Head and Neck Squamous Cell Carcinoma: Functional and Clinical Implications. View all 26 articles
 Panchun Li1
Panchun Li1 Kunjie Zhu2
Kunjie Zhu2 Yongzhen Mo3
Yongzhen Mo3 Xiangying Deng3
Xiangying Deng3 Xianjie Jiang3
Xianjie Jiang3 Lei Shi4
Lei Shi4 Can Guo3
Can Guo3 Wenling Zhang3
Wenling Zhang3 Zhaoyang Zeng3
Zhaoyang Zeng3 Guiyuan Li3
Guiyuan Li3 Wei Xiong3
Wei Xiong3 Shanshan Zhang5
Shanshan Zhang5 Zhaojian Gong1*
Zhaojian Gong1*Circular RNAs (circRNAs) are a novel type of non-coding RNAs. Because of their characteristics of a closed loop structure, disease- and tissue-specificity, and high conservation and stability, circRNAs have the potential to be biomarkers for disease diagnosis. Head and neck cancers are one of the most common malignant tumors with high incidence rates globally. Affected patients are often diagnosed at the advanced stage with poor prognosis, owing to the concealment of anatomic sites. The characteristics, functions, and specific mechanisms of circRNAs in head and neck cancers are increasingly being discovered, and they have important clinical significance for the early diagnosis, treatment, and prognosis evaluation of patients with cancer. In this study, the generation, characteristics, and functions of circRNAs, along with their regulatory mechanisms in head and neck cancers have been summarized. We report that circRNAs interact with molecules such as transcription and growth factors to influence specific pathways involved in tumorigenesis. We conclude that circRNAs have an important role to play in the proliferation, invasion, metastasis, energy and substance metabolism, and treatment resistance in cancers.
Head and neck cancers (HNC) are malignant tumors occurring in the oral cavity, hypopharynx, nasopharynx, and larynx. Depending on the location, HNC can be divided into hypopharyngeal squamous cell carcinoma (HSCC), oral squamous cell carcinoma (OSCC), laryngeal squamous cell carcinoma (LSCC), and nasopharyngeal carcinoma (NPC). With about 600,000 new patients per year, HNC ranks seventh in the incidence rate of malignant tumors worldwide (1). There are many ways to treat HNC, such as surgical treatment, radiotherapy, and chemotherapy. However, due to the lack of early diagnosis methods, patients with HNC are often diagnosed in the late stage. The 5-year overall survival rate of patients is 54.7%–65.9% and has not significantly improved over the years (2, 3). Therefore, it is necessary to elucidate the potential molecular regulation mechanism of HNC and to find possible molecular markers and targets. Circular RNAs (circRNAs) have the potential to be HNC biomarkers (4). This review summarizes the research progress of circRNAs in HNC and seeks to understand the occurrence, development, and metastasis of HNC, and the possible molecular markers and therapeutic targets.
According to the composition of the source, circRNA can be divided into 4 categories: exonic circRNAs (ecircRNA), intronic circRNAs (ciRNAs), exon-intron circRNAs (EIciRNA), and tRNA intronic circRNA(tricRNA) (5). Although the formation mechanism of circRNAs is not completely clear, the current research results reveal two different formation modes (4).
Lariat-driven circularization: Lasso-driven cyclization is also known as the exon-skipping model (4), whereby non-adjacent exons of pre-mRNA are close to each other during transcription, causing part of exons to be skipped and eventually not appear on mature mRNA. By exon skipping, the exons and introns that are skipped form a lasso structure, and after the introns are removed, the ecircRNA is formed.
Intron-pairing-driven circularization model: There are reverse complementary sequences in flanking introns of pre-mRNA, which can assist pre-mRNA to form lasso structures through complementary base pairing. For example, inverted repeated Alu sequences can make introns move close to each other in space and form a hairpin structure, and finally form a lasso structure containing exons and introns. After the introns are removed, ecircRNA containing only the exons is formed (6). During this process, introns are sometimes retained in the circRNA resulting in formation of EIciRNA (7). However, the specific mechanisms of EIciRNA formation are not clear. In addition, when the two ends of an intron have complementary sequences, the cyclization of a single intron can occur and form ciRNA (8). The formation of ciRNA (mainly occurring in the nucleus) depends on a consensus motif containing a seven-nucleotide (nt) GU-rich element near the 5′ splice site and an 11 nt C-rich element close to the branchpoint site (8).
RNA-binding proteins (RBP) are also involved in the circRNA cyclization; they can bind to introns, assist the interaction of upstream and downstream introns, and promote circRNA cyclization (8). Quaking is an RBP, which can form dimers and bind to the binding site of flanking introns, and promote the production of circRNA (9).
Up to now, more than one million circRNAs have been found in human tissues by high-throughput sequencing technology. Because of their characteristics of a closed loop structure, disease- and tissue-specificity, and high conservation and stability, circRNAs have the potential to be biomarkers for disease diagnosis.
The highly stable structure of circRNAs is a closed loop, without 3′ and 5′ ends and a poly (A) tail. Therefore, compared with linear RNAs, circRNAs are not easily hydrolyzed by RNase. It is reported that the half-life of circRNAs can reach more than 48 h, while that of linear RNAs is only 10 h (10).
Types and expression abundance: circRNAs are numerous and widely available in human cells (11). Over 1,000,000 circRNAs have been found in human tissues using high-throughput sequencing technology (4). The expression abundance of different circRNAs varies greatly. The expression of some circRNAs is much higher than that of linear mRNAs, and even more than 10 times that of their linear isomers (4). However, most circRNAs have low expression abundance.
Conservation: Because of the stability of circRNAs, most of them are conserved in the evolution process of different species (12, 13). For example, 5¬–30% of circRNAs from human and mouse homologous genes are completely conserved (14). The conservation of circRNAs proves that they are not by-products of transcription but play an important role in life activities.
Expression specificity: The expression of circRNAs is tissue- and disease-specific (12). The expression of circRNAs in embryo, brain, heart, stomach, lung, liver, and other organs is very different. For example, in the brain, the amount of circRNAs is higher than that of other tissues (15, 16). Expression specificity indicates that circRNAs may play a specific function in disease development and may serve as a biomarker for disease diagnosis.
miRNA sponging: CircRNAs are rich in miRNA binding sites, which can act as competitive endogenous RNA (ceRNA) to adsorb miRNA, thereby regulating the activity of downstream target genes of miRNA (11). For example, CDR1as contains more than 70 miR-7 binding sites, which act as ceRNA regulatory target genes in many biological processes (16–19).
Combination with protein: CircRNAs can directly bind to proteins and affect the function of downstream proteins. For example, circ-CCND1 can inhibit the binding of HuR to CCND1 mRNA by binding to HUR (20). In addition, circRNAs can act as a scaffold to make multiple proteins form complex and accelerate the reaction.
Translation of proteins: CircRNAs without 5’cap and 3’poly(A) tail lack internal ribosome entry site, so circRNAs are defined as non-coding RNA. However, studies have shown that circRNAs with open reading frames can translate functional proteins through internal ribosome entry site (IRES) or m6A-mediated translation initiation mechanisms (21). IRES can fold into a tRNA-like structure and mediate the binding of ribosomes to RNA. For example, circAKT3 in brain tumors can encode functional protein AKT3-174aa through IRES, and regulate PI3K/AKT signaling pathway (22). In addition, m6A modified circRNAs can directly bind to eukaryotic initiation factor 3, realizing a cap-independent translation initiation mode (23).
With the development of high-throughput sequencing, microarray technology, and bioinformatics, it has been increasingly found that circRNAs have abnormal expression and potential functions in HNC (24–27). For example, in HSCC, Feng et al. analyzed the expression profiles of four paired HSCC tissues and found 173 differentially expressed circRNAs. The authors performed enrichment analysis on 40 circRNAs with the most significant differences and found that these circRNAs are most likely to play a role in the ErbB and Hippo signaling pathways (28). In addition, Cheng et al. constructed a circRNA expression profile of LSCC by performing second generation sequencing on five paired LSCC tissues. They detected 21,444 circRNAs, and further analysis using the Kyoto Encyclopedia of Genes and Genomes showed that peroxisome proliferator-activated receptor-, axon guidance, and Wnt signaling pathways are the key pathways of circRNAs in the occurrence of LSCC (26).
CircRNAs are not only widely expressed in HNC but are also correlated to the prognosis of patients. They have the potential as biomarkers for tumor diagnosis and prognosis (29–37). For example, the expression of hsa_circ_0092125 is related to the tumor size, tumor node metastasis stage, and lymph node metastasis in OSCC patients. Survival analysis shows that patients with a higher expression of hsa_circ_0092125 survive longer (29).
The receiver operating characteristic (ROC) curve and the area under curve (AUC) can be used as indices to evaluate the accuracy of biomarkers. The AUC values of hsa_circ_0072387, hsa_circ_001242, hsa_circ_009755, hsa_circ_0004491, hsa_circ_0036722, hsa_circ_0008309 and hsa_circ_0086414 are 0.746, 0.784, 0.782, 0.751, 0.838, 0.764 and 0.749, indicating the reliability of these molecules as biomarkers (30–37). Body fluids also contain a large amount of circRNAs. For example, Zhao et al. reported that the expression of hsa_circ_0001874 and hsa_circ_0001971 was upregulated in OSCC saliva, and the AUC value of the combination of these two molecules was 0.922 (33). Serum exosomes can reflect the pathological conditions of the body and are considered as a potential auxiliary diagnostic marker. CircRASSF2 is upregulated in the serum exosomes of patients with LSCC and can affect the progression of LSCC (38). Those circRNAs in body fluids can be easily detected and have a great clinical value and significance.
In addition, circRNAs have the potential to be therapeutic targets. Fucoidan is a component of the seaweed cell wall, which can inhibit OSCC growth, migration, and invasion. Zhang et al. reported that circFLNA is a fucoidan target. They found that fucoidan increased the expression of circFLNA in OSCC cell lines and affected the expression of key proteins related to growth, apoptosis, migration, and invasion (39).
The uncontrolled proliferation of tumor cells is the basis of tumorigenesis. Multiple circRNAs have been demonstrated to be involved in HNC development through the regulation of the proliferation process (Figure 1). CircATRNL1 (40), hsa_circ_0007059 (41), circRNA_002178 (42), hsa_circ_0046263 (43), hsa_circ_0005379 (44), circUHRF1 (45), circPVT1 (46, 47), circHIPK3 (48, 49), circRNA_0000140 (50), circRNA_100290 (51), circDOCK1 (52), hsa_circ_0055538 (53), circITCH (54), hsa_circRNA_101036 (55), circCCND1 (20), circMYLK (56), CDR1as (17, 57, 58), circPKD2 (59), circLARP4 (60), hsa_circ_0042666 (61), hsa_circRNA_100533 (62), circMDM2 (63), circ_0067934 (64), circRPMS1 (65), circ-ZNF609 (66, 67), hsa_circ_0072387 (68), circRASSF2 (38), hsa_circ_0023028 (69), circCTDP1 (70), circ_0008450 (71), circMATR3 (72), hsa_circ_0057481 (73), circ_0001742 (74), circ_0000218 (75), circ_0001971 (76), circCORO1C (77), hsa_circ_0109291 (78), hsa_circ_0036722 (34), hsa_circ_0002203 (79) and hsa_circ_0066755 (80) are associated with tumor proliferation (Table 1).
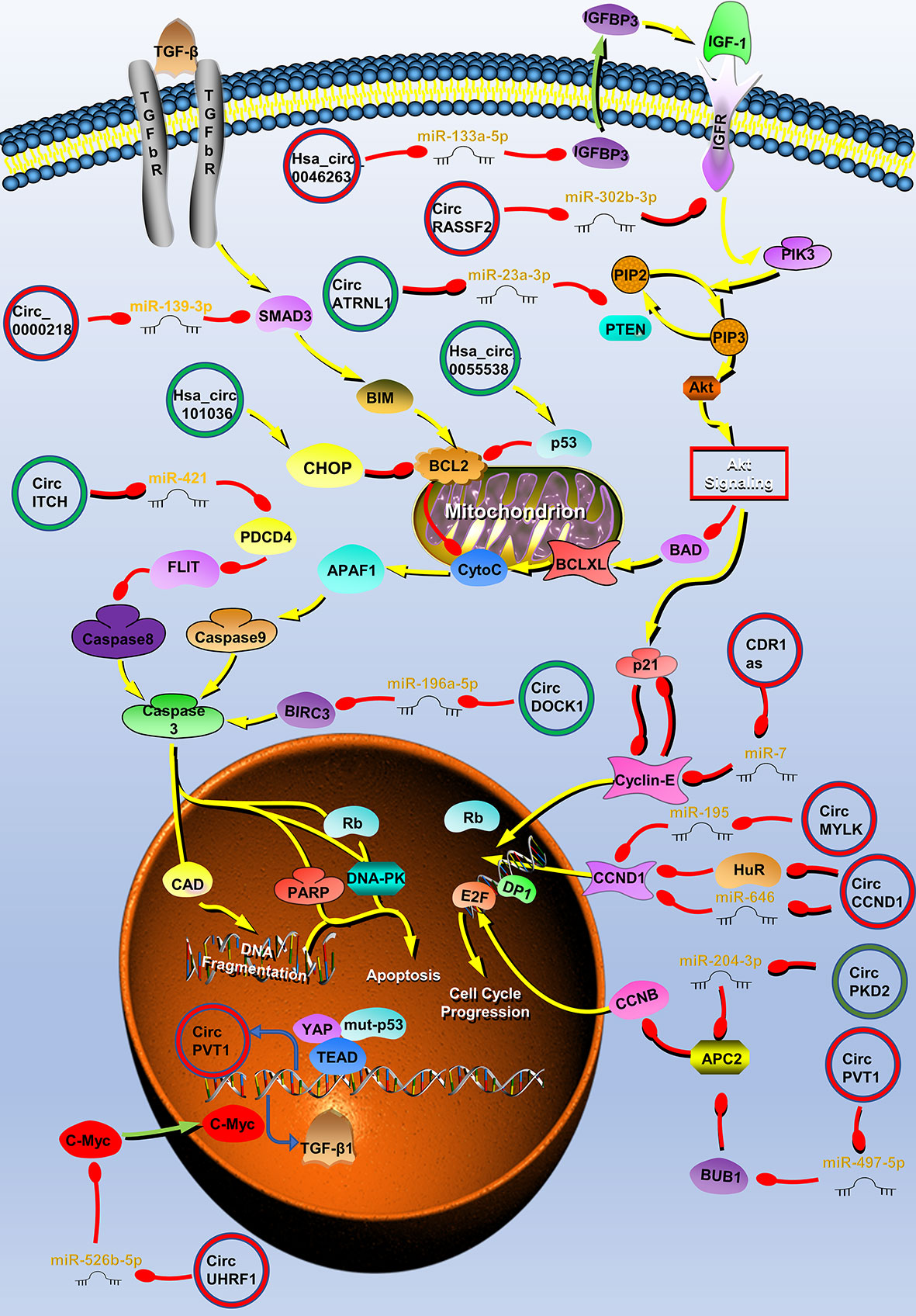
Figure 1 Circular RNAs (circRNAs) associated with proliferation in head and neck cancers (HNC). CircRNAs that are known to regulate proliferation processes and their validated targets in HNC. CircRNAs regulate the growth of HNC by regulating PI3K/Akt/mTOR, insulin-like growth factor (IGFR), transforming growth factor (TGF) β, p53 signaling pathways, apoptosis, and cell cycle-related molecules. Red circle, upregulated circRNAs in HNC; green circle, downregulated circRNAs in HNC; yellow arrow, positive regulation; red arrow, negative regulation.
Tumor proliferation is regulated by multiple cellular pathways, such as the PI3K/Akt/mTOR signaling pathway, and some circRNAs are involved in the regulation of tumor proliferation through these pathways. In OSCC, circATRNL1 can induce apoptosis and cell cycle arrest through the PI3K/Akt/mTOR signaling pathway (40). CircATRNL1 is downregulated in OSCC and can target miR-23a-3p to eliminate the inhibition of phosphatase and tensin homolog (PTEN) by miR-23a-3p. PTEN is one of the most common tumor suppressor genes, which regulates cell apoptosis and cell cycle through PI3K/Akt/mTOR signaling pathway (82). PTEN can enhance BAD activity, inhibit caspase-3 activation, and induce apoptosis by antagonizing the PI3K/Akt/mTOR signaling pathway (83, 84). In addition, PTEN induces the expression of the cell cycle inhibitor p21, resulting in cell G0/G1 arrest (85). Therefore, circATRNL1 affects the progression of OSCC through the PTEN/PI3K/Akt/mTOR signaling pathway (40). Similarly, hsa_circ_0007059 and circRNA_002178 also affect OSCC growth through AKT signaling pathway (41, 42). The transforming growth factor β (TGF-β) signaling pathway can also regulate cell proliferation (86). Zhao et al. reported that circUHRF1 positively regulates the c-Myc oncogene by competitively binding miR-526b-5. In addition to activating the TGF-β1 pathway, c-Myc can also promote the expression of ESRP1. ESRP1 promotes the generation of circUHRF1 by combining with the flanking introns of circUHRF1, and finally forms a positive feedback regulation loop. CircUHRF1 eventually activates the TGF-β pathway to promote tumor growth (45).
Various growth factors such as insulin-like growth factors (IGFs) and their receptors are also involved in the growth of HNC (87, 88). When IGF-1R is combined with its ligand, it can activate the PI3K/Akt/mTOR signaling pathway, and play an important role in the proliferation and apoptosis of tumor cells (88, 89). CircRASSF2 can adsorb miR-302b-3p as a competitive endogenous RNA (ceRNA), increase the expression of IGF-1R, and promote the progression of LSCC (38). IGFBP3 is the transporter of IGF-1, and it can extend the half-life of IGF-1 and promote the binding of IGF-1 and IGF-1R. IGFBP3 is the downstream target of miR-133a-5p and hsa_circ_0046263 acts as a sponge of miR-133a-5p in NPC and promotes IGFBP3 expression. Therefore, hsa_circ_0046263 can also activate the IGFR signaling pathway to promote LSCC (43).
P53 is the most commonly mutated gene in HNC; the mutation and deletion of this gene leads to the disorder of intracellular signal pathways and the uncontrolled growth of tumor cells (90). The expression of circPVT1 is upregulated in patients with HNC with a p53 mutation. Further research found that the complex formed by mutant p53, yes-associated protein, and TEA domain combined with the circPVT1 promoter to regulate circPVT1 expression at the transcriptional level. CircPVT1 acts as an oncogene and can regulate the expression of miR-497-5p and its downstream genes, which are involved in cell proliferation (46). In addition to the above mechanisms, circRNAs can regulate the proliferation of HNC through different mechanisms such as affecting apoptosis and cell cycle.
Resistance to cell death is one of the ten characteristics of tumors. Apoptosis is the main way to clear cells that are potentially harmful to the body, and almost all cancer cells have anti-apoptotic mechanisms (91). CircRNAs can directly regulate molecules related to the apoptosis pathway. For example, Wang et al. found that circDOCK1 can act as the ceRNA of miR−196a−5p to regulate the expression of Baculoviral IAP repeat-containing protein 3 (BIRC3). BIRC3 is an endogenous inhibitor of a cysteine proteolytic enzyme (caspase), which can prevent the conversion of the p19 subunit of caspase-3 to the p17 subunit and inhibit the activity of caspase 3 (92, 93). Wang et al. used tumor necrosis factor α to stimulate the OSCC cell line CAL-27 to establish an OSCC apoptosis model, establish an apoptosis-related circRNA profile through microarray sequencing, and screen the apoptosis-related molecule circDOCK1 (52). Finally, the authors found that circDOCK1 regulates the expression of BIRC3 by acting as a ceRNA and participates in the process of OSCC cell apoptosis (52). The p53 molecule is the central mediating switch of apoptosis and can downregulate the expression of Bcl-2 to promote cell apoptosis (94). Su et al. found that the overexpression of hsa_circ_0055538 in OSCC increased the expression levels of p53, Bax, and caspase 3, and decreased the expression of Bcl-2. These indicate that hsa_circ_0055538 may affect OSCC apoptosis through the p53/Bcl-2/caspase signaling pathway (53). Programmed cell death 4 (PDCD4) is a tumor suppressor gene related to apoptosis. Studies have found that PDCD4 inhibits FLIP-mediated tumor cell apoptosis. In addition, circ-ITCH can directly bind to miR-421 and block its inhibition of PDCD4 in OSCC and induce cell apoptosis (54). Endoplasmic reticulum stress is another way to induce apoptosis. The main signs of endoplasmic reticulum stress are the upregulation of related proteins CHOP and Bcl-2 (95). After the overexpression of hsa_circRNA_101036 in OSCC, the levels of these proteins levels of CHOP and Bcl-2 can be found to increase, and hsa_circRNA_101036 may exert a tumor suppressor effect by regulating the endoplasmic reticulum stress in cancer cells (55). Autophagy is also a way of cell death, but under severe conditions, tumor cells can promote cell survival through autophagy. According to reports, CDR1as can promote autophagy and growth of OSCC by adsorbing miR-671-5p and promoting the AKT/ERK/mTOR signaling pathway (58).
The cell cycle is closely related to the growth of tumors (96). Abnormal expression of cyclin will result in abnormal cell growth. Cyclin D (CCND1) plays an important positive regulatory role in G1/S phase transition. Its high expression can shorten the G1 phase and accelerate the cell cycle. CircCCND1 is a circRNA derived from the CCND1 gene (97). CircCCND1 can block the inhibition of CCND1 by adsorbing miR-646. In addition, circ-CCND1 also inhibits the binding of the human antigen R to CCND1 mRNA by binding to HUR, thereby enhancing the stability of CCND1 mRNA (20). Briefly, circ-CCND1 regulates CCND1 in these two ways and regulates the transformation of cells from G1 to S phase. circMYLK also promotes CCND1 expression in LSCC by binding to miR-195 (56). Cyclin E1 also plays an important role in the G1/S phase transition (98). As the first reported circRNA molecule, CDR1as can inhibit miR-7, upregulate the expression of cyclin E1, and promote LSCC progression (17). Gao et al. found that the expression of circPKD2 was downregulated in the OSCC circRNA expression profile and experimentally proved that circPKD2 can induce cell cycle arrest. CircPKD2 also acts as a sponge for miR-204-3p, inhibiting miR-204-3p from acting on APC2 (59). APC2 can work with DNA-PKcs to inhibit the stability of CCND1 (81). Therefore, circ-PKD2 plays a tumor suppressor effect by promoting APC2.
Invasion and metastasis are the most essential biological features of malignant tumors and are also the direct cause of treatment failure in tumor patients. CircRNAs in HNC have important roles in HNC metastasis and invasion (Figure 2). Hsa_circ_0007059 (41), circUHRF1 (45), circHIPK3 (48), circRNA_0000140 (50), circRNA_100290 (51), hsa_circ_0055538 (53), CDR1as (17, 57), circPKD2 (59), circLARP4 (60), hsa_circ_0042666 (61), hsa_circRNA_100533 (62), circ_0067934 (64), circRPMS1 (65), circ-ZNF609 (66), hsa_circ_0072387 (68), circRASSF2 (38), hsa_circ_0023028 (69), circCTDP1 (70), circ_0008450 (71), circMATR3 (72), hsa_circ_0057481 (73), circ_0001742 (74), circCORO1C (77), hsa_circ_0004491 (37), circ_0001971 (33), circFLNA (99), hsa_circ_0001162 (100), circARHGAP12 (101), circCRIM1 (102), circEPSTI1 (103), hsa_circ_0109291 (78), circRNA_002178 (42), hsa_circ_0002203 (79) and hsa_circ_0066755 (80) are involved in the regulation of HNC invasion and metastasis (Table 2).
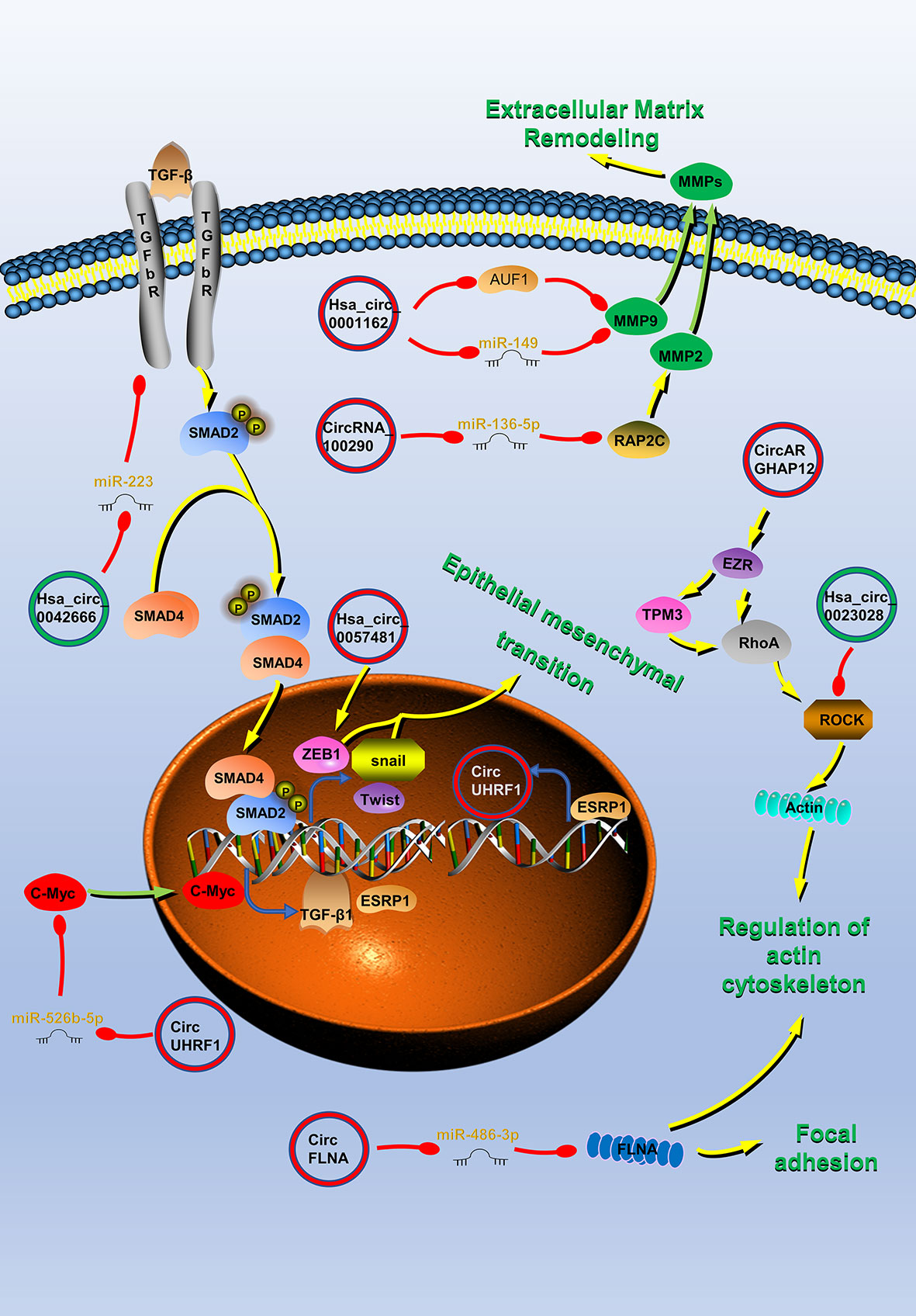
Figure 2 Circular RNAs (circRNAs) associated with invasion and metastasis in head and neck cancers (HNC). CircRNAs that are known to regulate invasion and metastasis processes and their validated targets in HNC. CircRNAs regulate the cytoskeleton remodeling, local adhesion, extracellular matrix remodeling, and the epithelial–mesenchymal transition (EMT) of HNC by regulating invasion and metastasis-related molecules to promote tumor invasion and metastasis. Red circle, upregulated circRNAs in HNC; green circle, downregulated circRNAs in HNC; yellow arrow, positive regulation; red arrow, negative regulation.
The PI3K/Akt/mTOR, IGF-1/IGF-1R, Hippo, P53, and TGFβ signaling pathways mentioned above also play a role in the regulation of HNC invasion and metastasis by circRNA (38, 40–42, 45, 50, 75, 104). For example, TGF-β can act on the transcription factors Snail, Twist, etc., and is a key factor in inducing epithelial mesenchymal transition (EMT) (105). EMT is an important biological process for malignant tumor cells derived from epithelial cells to acquire the ability of migration and invasion (105). As mentioned above, circUHRF1 competitively binds miR-526b-5 and positively regulates c-Myc, which activates the TGF-β1 pathway (45). Therefore, circUHRF1 can promote the EMT and invasion and metastasis of HNC by activating the TGF-β1 pathway.
ZEB1 is also an important factor which promotes tumor EMT (106). It can work with snail and slug to inhibit downstream adhesion molecules and promoter molecules, and ultimately induce EMT. Hsa_circ_0057481 can directly capture miR-200c, promote the expression of the ZEB1, and promote the invasion and metastasis of LSCC (73).
Matrix metalloproteinases (MMPs) can promote tumor invasion and metastasis by promoting the degradation of the extracellular matrix (107). Hsa_circ_0001162 is a circRNA derived from MMP9, which blocks the inhibitory effect of miR-149 on the 3′ end of MMP9 by adsorbing miR-149, and promotes the migration and invasion of OSCC (100). In addition, there is also an AU-rich element RBP 1 binding motif on MMP9 mRNA 3′-UTR, which does not overlap with the binding site of miR-149 on MMP9 3′-UTR. Therefore, hsa_circ_0001162 can also enhance the stability of MMP9 mRNA by antagonizing the attenuation of MMP9 mRNA induced by the AU-rich element RBP 1 in OSCC (100). Rap2c is a member of the RAS oncogene family and it is reported that it can increase the activity of MMP2. Highly-expressed circRNA_100290 promotes the migration of LSCC by blocking miR-136-5p inhibiting this protein (51).
The realization of tumor migration and invasion requires the rearrangement of intracellular cytoskeletal proteins. The protein encoded by the EZR gene is a component of the microvilli, acting as an intermediate product between the plasma membrane and the actin cytoskeleton, and can regulate cell adhesion and skeletal reorganization (108). Studies have found that circARHGAP12 can bind to the 3′-UTR of EZR mRNA and promote its stability (101). EZR can interact with TPM3 and RhoA to regulate cytoskeleton remodeling and promote NPC invasion and metastasis (101). Rock1 protein plays a key role in cell surface structure adhesion and cytoskeleton recombination (109). Pan et al. proved that ROCK1 is the direct target of circLARP4, which inhibits cell migration and invasion of NPC by regulating the kinase (60). FLNA can limit tumor migration by regulating the activation of integrins and the morphology and degradation of focal adhesions (110). CircFLNA can act as a miRNA sponge for miR-486-3p, thereby protecting FLNA from miR-486-3p-mediated silencing, and inhibiting the migration of LSCC cells (99).
The various biological characteristics of tumors are closely related to the metabolism of substances and energy. Tumor metabolism provides energy for the biological behavior of tumors, promotes tumor growth, metastasis, and other activities (111). CircRNAs such as circPVT1 (112), circ_0000140 (113), circRNA_100290 (114), CDR1as (57), circMDM2 (63), hsa_circ_0072387 (68) and circGDI2 (115) (Table 3) can also affect tumor growth through glucose metabolism (Figure 3).
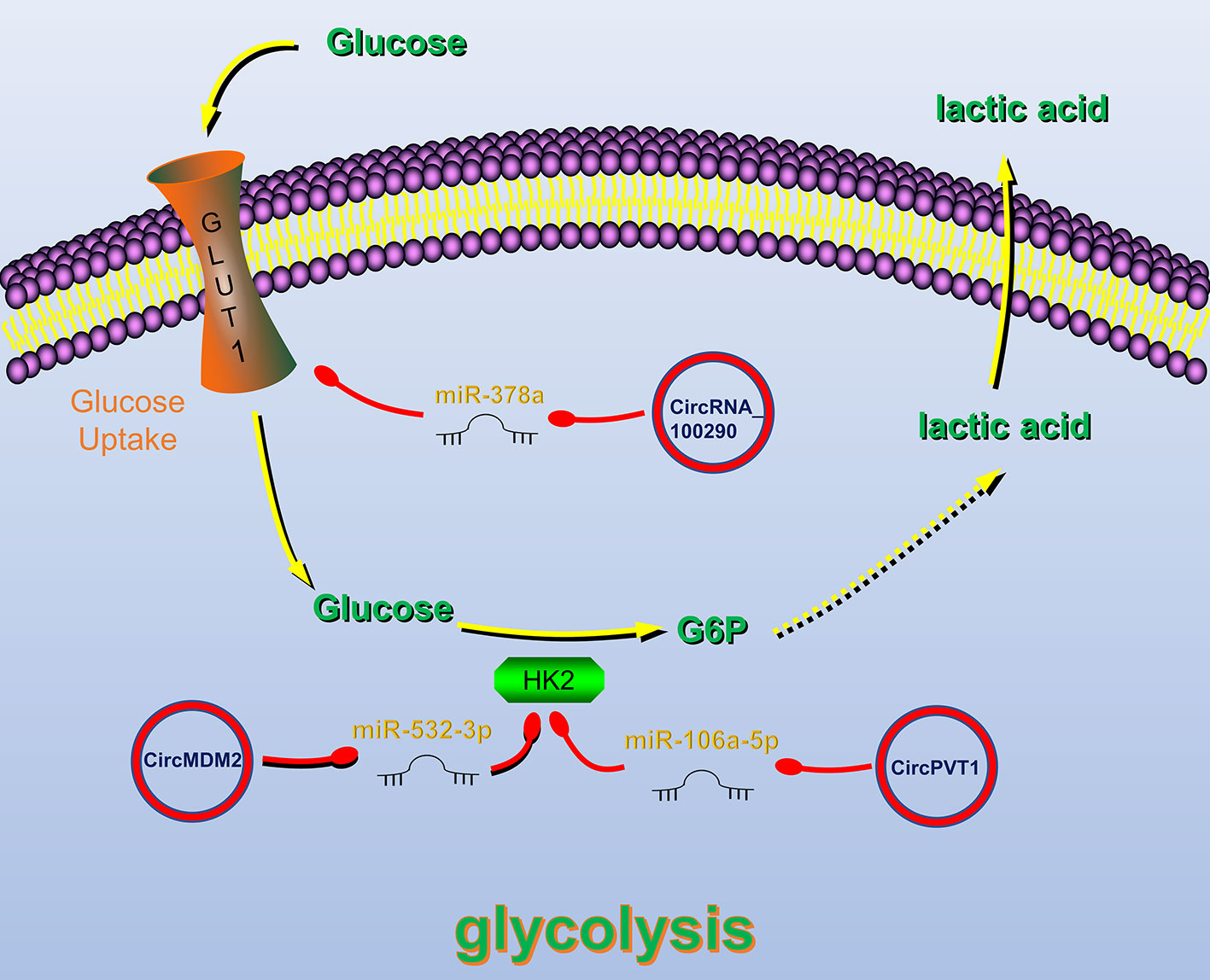
Figure 3 Circular RNAs (circRNAs) related to regulation of tumor glucose metabolism in head and neck cancers (HNC). CircRNAs that are known to regulate glucose metabolism processes and their validated targets in HNC. CircRNAs regulate HNC glucose metabolism by regulating Glucose transporter 1 (GLUT1) and Hexokinase 2, key enzymes of glycolysis. Red circle, upregulated circRNAs in HNC; green circle, downregulated circRNAs in HNC; yellow arrow, positive regulation; red arrow, negative regulation.
Glucose transporter 1 (GLUT1) plays an important role in the glycolysis process by regulating glucose uptake (116). CircRNA_100290 can alleviate the inhibitory effect of miR-378a on GLUT1. Therefore, overexpression of circRNA_100290 can increase the glucose uptake rate of tumor cells and meet their high metabolic need (114). Hexokinase 2 (HK2) is also a key enzyme in the glycolysis pathway, and its expression in HNC cells is significantly increased (117). Studies have shown that circRNAs can regulate the HK2 gene, thereby affecting the glycolysis of HNC cells. For example, in OSCC, circPVT1 and circMDM2 upregulate HK2 expression by inhibiting miR-497-5p and miR-532-3p, respectively, thereby promoting glycolysis (63, 112).
Surgery combined with radiotherapy and chemotherapy is the main treatment for HNC, but many patients develop radiochemotherapy resistance. Therefore, exploring the molecular mechanisms of HNC radiochemotherapy resistance has important practical significance for further improving the therapeutic effect. Recently, several lines of evidence have suggested that circRNAs, such as circRNA_000543 (118), circRNA_0000285 (119), hsa_circRNA_001387 (120), circATRNL1 (40) circ_0001971 (33), hsa_circ_0109291 (78), circCRIM1 (102), hsa_circ_0028007 (121), hg_circ_0005033 (122) and hsa_circ_0005379 (44) (Table 4) are likely to play vital roles in radiochemotherapy resistance in HNC (Figure 4).
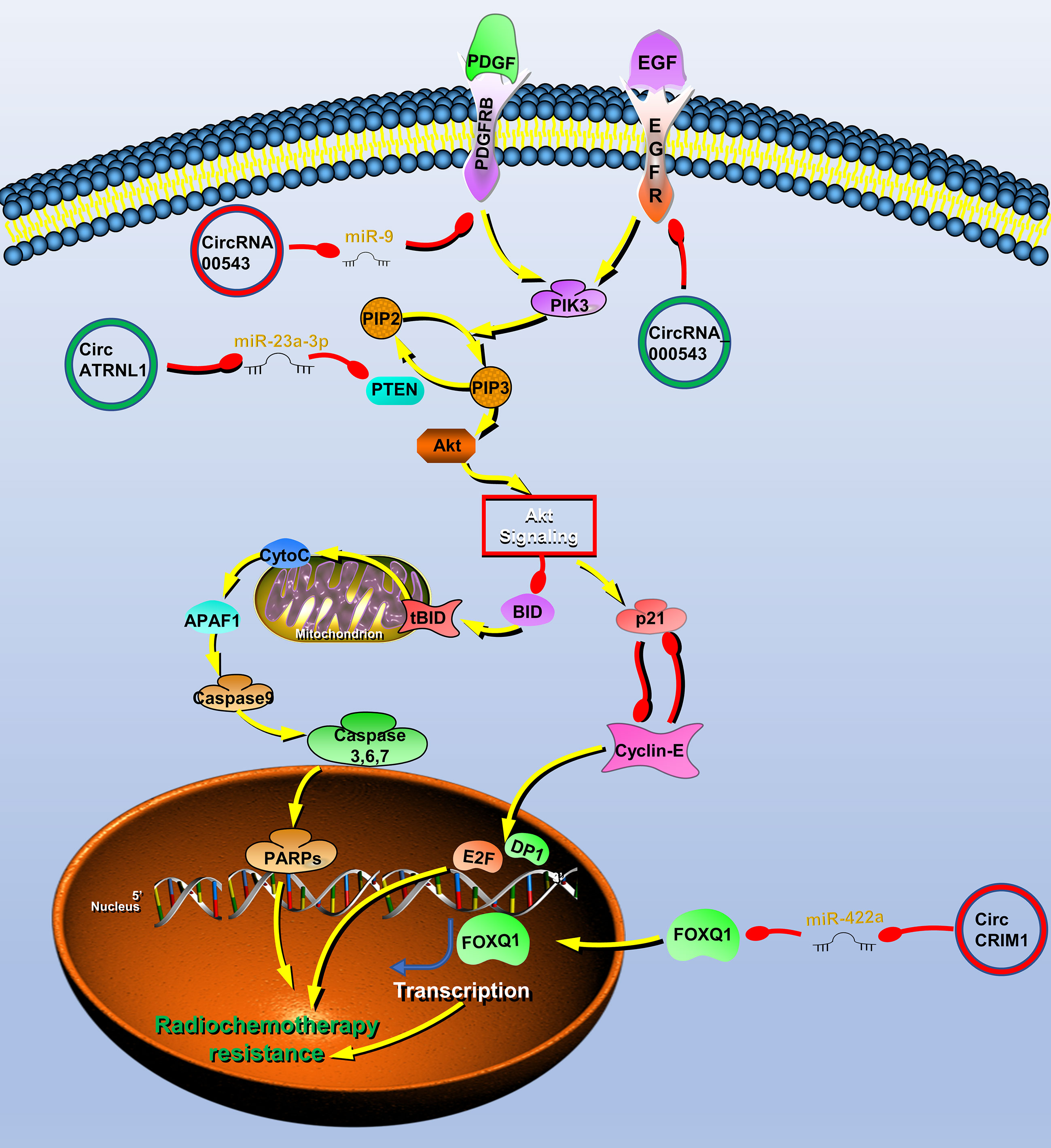
Figure 4 Circular RNAs (circRNAs) associated with radiochemotherapy resistance in head and neck cancers (HNC). CircRNAs that are known to regulate radiochemotherapy resistance processes and their validated targets in HNC. CircRNAs regulate the radiochemotherapy resistance of HNC through phosphatase and tensin homolog (PTEN)/PI3K/Akt/mTOR, epidermal growth factor receptor (EGFR), platelet-derived growth factor receptor A (PDGFR) signaling pathway and transcription factor forkhead box Q1 (FOXQ1). Red circle, upregulated circRNAs in HNC; green circle, downregulated circRNAs in HNC; yellow arrow, positive regulation; red arrow, negative regulation.
To find radiotherapy resistance-related circRNAs, Chen et al. performed high-throughput sequencing on radiotherapy-sensitive and -insensitive NPC tissues and found that the expression of circRNA_000543 was upregulated in radiotherapy-insensitive NPC, which may be a carcinogenic circRNA involved in radiotherapy resistance. Further studies have shown that miR-9 can bind to circRNA_000543, and a bioinformatics software predicts that miR-9 can bind to the 3′ UTR of the PDFGRB. Therefore, the author finally proved that circRNA_000543 is involved in radiation resistance of NPC by blocking inhibition of miR-9 to PDFGRB (77).
Radiotherapy is also one of the main treatment methods of OSCC. The resistance to radiation often leads to poor prognosis and tumor recurrence in patients with OSCC. As mentioned above, circATRNL1 in OSCC induces tumor apoptosis and cell cycle arrest through PTEN/PI3K/AKT. Therefore, circATRNL1 also improves the radiosensitivity of OSCC cells through this mechanism (40).
Systemic induction chemotherapy is also an important auxiliary method. Recent studies reported that circCRIM1 can promote NPC metastasis and docetaxel chemoresistance. CircRNA sequencing was performed on high- and low-metastatic ability cell lines and obtained metastasis-related circRNA expression profiles, among which circCRIM1 was the most upregulated molecule. Further studies have shown that circCRIM1 upregulates FOXQ1 expression by acting as a miR-422a sponge, thereby promoting the process of NPC. The author performs enrichment analysis on the expression profile of NPC with high and low expression of FOXQ1 and found that chemoresistance was significantly rich. Furthermore, in vitro and in vivo experiments both showed that circCRIM1 promotes chemoresistance of NPC docetaxel (102). Cetuximab is an IgG1 monoclonal antibody with high affinity for the epidermal growth factor receptor (EGFR) (44). It can inhibit cell cycle progression and induce tumor cell apoptosis by specifically binding to the extracellular EGFR domain. Su et al. found that hsa_circ_0005379 can act on EGFR to significantly enhance the sensitivity of OSCC to cetuximab drugs (123). Functional experiments show that hsa_circ_0005379 can inhibit the growth and metastasis of OSCC, and increase the sensitivity to cetuximab drugs (123).
EBV and HPV are closely related to the pathogenesis of HNC. It is reported that about 98% of NPC patients are EBV positive (124), and about 70% of oral and pharyngeal squamous cell carcinoma are associated with HPV infection (125). Therefore, it is very important to understand the carcinogenic mechanism of the virus. Studies have shown that DNA viruses can produce circRNA in host cells, and these circRNAs may also be a potential mechanism of virus carcinogenesis (126, 127).
EBV is the first virus reported to produce circRNAs in the host genome. Toptan et al. used circRNA Seq to sequence the EBV genome and found that the BART, LMP2, and BHLF1 genes in the EBV genome can produce circRNA (126). In another study, the authors developed a tool called circleVis and found more than 30 circRNAs produced by EBV (127). Among them, circRPMS1 encoded by BART has been verified by other studies, and it has been found that circRPMS1 can promote the proliferation and metastasis of NPC by acting as a miRNA sponge (65, 128). circLMP2A was also verified to exist in gastric cancer, but there is no research of circLMP2A in HNC (129). Similarly, HPV can also produce circRNAs. Wang et al. found that the E7 gene in HPV16 virus can produce circular RNA-CircE7 in cervical cancer, and CircE7 can be translated into protein and promote tumor growth (130). However, the study of circRNAs produced by HPV in HNC has not been reported yet. Whether the circRNA produced by these tumor viruses also functions in HNC is worthy of further investigation.
CircRNAs have a closed-loop structure, lack free ends, and are not easily hydrolyzed by RNase. In addition, because of their high conservation, stability, expression abundance, and tissue and disease expression-specificity, circRNAs have the potential to become diagnostic or predictive biomarkers for disease. Notably, circRNAs can be detected in body fluids including peripheral blood, saliva, and urine. For example, there are a large number of circRNAs in human saliva (131), and hsa_circ_0001874 and hsa_circ_0001971 in saliva may be used as biomarkers for OSCC (33). In addition, circRNAs are enriched in exosomes (132). CircRASSF2 has been reported to be upregulated in serum exosomes in patients with LSCC and affects the progression of LSCC (38). These characteristics make the detection of circRNAs in body fluids of patients with HNC possible and have great clinical value and significance.
In HNC, circRNAs can affect tumor development through various mechanisms such as cell cycle, apoptosis, EMT, metabolism, and radiochemotherapy resistance. However, circRNAs also affect tumor development in other ways, such as immune escape, cancer stem cells. Immune checkpoint refers to a group of factors involved in negative regulation in the immune system and is closely related to the immune escape mechanism (133). Recently, inhibitors of immune checkpoint PD1 or PDL1 have been approved for the treatment of advanced HNC (134), and have become a new trend in the clinical treatment of HNC. Studies have shown that circRNA can also play a regulatory role in immune checkpoints, and may be used as a target for immune checkpoint therapy. For example, Circ-CPA4 acts as miRNA sponge for let-7 to down-regulate the expression of PD-L1, thereby promoting lung cancer cell death (135), while circFGFR1 can induce lung cancer cell resistance to PD-1 drugs by inhibiting miR-381-3p (136). However, the study of circRNA related to PD1 or PD-L1 in HNC has not been reported yet, and further research is needed. In addition, anti-tumor research for innate immunity is also very necessary. It has recently been reported that exogenous circRNA can activate innate immunity and is an effective adjuvant for inducing anti-tumor immunity in vivo (137). Exogenous viruses associated with HNC can also produce circRNA, such as EBV and HPV (65, 138–140). Therefore, the role of exogenous circRNA encoded by EBV or HPV genome in innate immunity deserves further study. In short, more efforts are needed to promote the research on the immune regulation of circRNA in HNC.
Cancer stem cells have the ability to induce tumor self-renewal and high chemoradiotherapy resistance in tumors, so cancer stem cells are one of the main reasons for drug resistance and high recurrence in tumor treatment (141). In HNC, circRNA participates in chemoradiotherapy resistance by regulating cancer stem cells. For example, curcumin mediates the regulation of cancer stem cell-like cells through the circRNA network and enhances the radiosensitization effect of NPC (142). In addition, Hg19_circ_0005033 affects LSCC stem cells, promotes tumor occurrence and chemotherapy resistance (122). However, the research on cancer stem cells in HNC just emerges, and there may be other regulatory mechanisms for circRNA. The main pathways involved in cancer stem cells are Hedgehog, Wnt, PI3K/PTEN, Notch, and Hippo signaling pathways (143), and many circRNAs in HNC are related to some of these signaling pathways (28, 82). Therefore, circRNAs may also participate in the regulation of cancer stem cells through these signaling pathways. Studying the role of circRNA in cancer stem cells will contribute to the improvement of tumor diagnosis and treatment methods, and further research is needed.
As with some studies, this manuscript has also limitations. In HNC, circRNAs affect tumor progression mainly by acting as miRNA sponges or binding proteins. But a few circRNAs with open reading frames can translate functional proteins under the mediation of IRES or m6A. Since there is no report on circRNA encoded protein in HNC, this article does not cover it. However, it cannot be ruled out that the circRNA in HNC can also play a role through the mechanism of encoding protein. In addition, the current research on circRNA has just emerged, and more clinical research is needed before clinical application. However, with the development of research, circRNAs will play a more important role. The emergence of circRNAs has greatly enriched the regulatory network of non-coding RNAs in human diseases and which may be used as new disease biomarkers and therapeutic targets with great potential.
PL, KZ, YM, XD, XJ, LS, CG, WZ, ZZ, GL WX, SZ, and ZG conceived and wrote the manuscript. PL and ZG did the figure. All authors contributed to the article and approved the submitted version.
The present study was supported by the National Natural Science Foundation of China (grant nos. 81803025 and 81772901).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer YW declared a shared affiliation, with no collaboration, with the authors to the handling editor at the time of the review.
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2018) 68(6):394–424. doi: 10.3322/caac.21492
2. Siegel R, Ward E, Brawley O, Jemal A. Cancer Statistics, 2011: The Impact of Eliminating Socioeconomic and Racial Disparities on Premature Cancer Deaths. CA Cancer J Clin (2011) 61(4):212–36. doi: 10.3322/caac.20121
3. Pulte D, Brenner H. Changes in Survival in Head and Neck Cancers in the Late 20th and Early 21st Century: A Period Analysis. Oncolog (2010) 15(9):994–1001. doi: 10.1634/theoncologist.2009-0289
4. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are Abundant, Conserved, and Associated With ALU Repeats. RNA (New York NY) (2013) 19(2):141–57. doi: 10.1261/rna.035667.112
5. Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, et al. Circular RNA: A New Star of Noncoding Rnas. Cancer Lett (2015) 365(2):141–8. doi: 10.1016/j.canlet.2015.06.003
6. Jeck WR, Sharpless NE. Detecting and Characterizing Circular Rnas. Nat Biotechnol (2014) 32(5):453–61. doi: 10.1038/nbt.2890
7. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-Intron Circular RNAs Regulate Transcription in the Nucleus. Nat Struct Mol Biol (2015) 22(3):256–64. doi: 10.1038/nsmb.2959
8. Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular Intronic Long Noncoding Rnas. Mol Cell (2013) 51(6):792–806. doi: 10.1016/j.molcel.2013.08.017
9. Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, et al. The RNA Binding Protein Quaking Regulates Formation of Circrnas. Cell (2015) 160(6):1125–34. doi: 10.1016/j.cell.2015.02.014
10. Suzuki H, Zuo Y, Wang J, Zhang MQ, Malhotra A, Mayeda A. Characterization of RNase R-Digested Cellular RNA Source That Consists of Lariat and Circular RNAs From pre-mRNA Splicing. Nucleic Acids Res (2006) 34(8):e63. doi: 10.1093/nar/gkl151
11. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a Large Class of Animal RNAs With Regulatory Potency. Nature (2013) 495(7441):333–8. doi: 10.1038/nature11928
12. Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-Type Specific Features of Circular RNA Expression. PloS Genet (2013) 9(9):e1003777. doi: 10.1371/journal.pgen.1003777
13. Wang PL, Bao Y, Yee MC, Barrett SP, Hogan GJ, Olsen MN, et al. Circular RNA is Expressed Across the Eukaryotic Tree of Life. PloS One (2014) 9(6):e90859. doi: 10.1371/journal.pone.0090859
14. Guo JU, Agarwal V, Guo H, Bartel DP. Expanded Identification and Characterization of Mammalian Circular Rnas. Genome Biol (2014) 15(7):409. doi: 10.1186/s13059-014-0409-z
15. Xia S, Feng J, Lei L, Hu J, Xia L, Wang J, et al. Comprehensive Characterization of Tissue-Specific Circular RNAs in the Human and Mouse Genomes. Brief Bioinform (2017) 18(6):984–92. doi: 10.1093/bib/bbw081
16. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA Circles Function as Efficient microRNA Sponges. Nature (2013) 495(7441):384–8. doi: 10.1038/nature11993
17. Zhang J, Hu H, Zhao Y, Zhao Y. CDR1as is Overexpressed in Laryngeal Squamous Cell Carcinoma to Promote the Tumour’s Progression Via miR-7 Signals. Cell Prolif (2018) 51(6):e12521. doi: 10.1111/cpr.12521
18. Huang H, Wei L, Qin T, Yang N, Li Z, Xu Z, et al. Circular RNA ciRS-7 Triggers the Migration and Invasion of Esophageal Squamous Cell Carcinoma Via miR-7/KLF4 and NF-κb Signals. Cancer Biol Ther (2019) 20(1):73–80. doi: 10.1080/15384047.2018.1507254
19. Liu L, Liu F-B, Huang M, Xie K, Xie Q-S, Liu C-H, et al. Circular RNA ciRS-7 Promotes the Proliferation and Metastasis of Pancreatic Cancer by Regulating Mir-7-Mediated EGFR/STAT3 Signaling Pathway. Hepatobil Pancreat Dis Int (2019) 18(6):580–6. doi: 10.1016/j.hbpd.2019.03.003
20. Zang Y, Li J, Wan B, Tai Y. circRNA circ-CCND1 Promotes the Proliferation of Laryngeal Squamous Cell Carcinoma Through Elevating CCND1 Expression Via Interacting With HuR and Mir-646. J Cell Mol Med (2020) 24(4):2423–33. doi: 10.1111/jcmm.14925
21. Wu P, Mo Y, Peng M, Tang T, Zhong Y, Deng X, et al. Emerging Role of Tumor-Related Functional Peptides Encoded by lncRNA and Circrna. Mol Cancer (2020) 19(1):22. doi: 10.1186/s12943-020-1147-3
22. Xia X, Li X, Li F, Wu X, Zhang M, Zhou H, et al. A Novel Tumor Suppressor Protein Encoded by Circular AKT3 RNA Inhibits Glioblastoma Tumorigenicity by Competing With Active Phosphoinositide-Dependent Kinase-1. Mol Cancer (2019) 18(1):131. doi: 10.1186/s12943-019-1056-5
23. Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, et al. Extensive Translation of Circular RNAs Driven by N-Methyladenosine. Cell Res (2017) 27(5):626–41. doi: 10.1038/cr.2017.31
24. Qiu X, Ke X, Ma H, Han L, Chen Q, Zhang S, et al. Profiling and Bioinformatics Analyses Reveal Differential Expression of Circular RNA in Tongue Cancer Revealed by High-Throughput Sequencing. J Cell Biochem (2019) 120(3):4102–12. doi: 10.1002/jcb.27695
25. Yang J, Gong Y, Jiang Q, Liu L, Li S, Zhou Q, et al. Circular RNA Expression Profiles in Nasopharyngeal Carcinoma by Sequence Analysis. Front Oncol (2020) 10:601. doi: 10.3389/fonc.2020.00601
26. Lu C, Shi X, Wang AY, Tao Y, Wang Z, Huang C, et al. Rna-Seq Profiling of Circular RNAs in Human Laryngeal Squamous Cell Carcinomas. Mol Cancer (2018) 17(1):86. doi: 10.1186/s12943-018-0833-x
27. Cao S, Wei D, Li X, Zhou J, Li W, Qian Y, et al. Novel Circular RNA Expression Profiles Reflect Progression of Patients With Hypopharyngeal Squamous Cell Carcinoma. Oncotarget (2017) 8(28):45367–79. doi: 10.18632/oncotarget.17488
28. Feng C, Li Y, Lin Y, Cao X, Li D, Zhang H, et al. CircRNA-associated ceRNA Network Reveals ErbB and Hippo Signaling Pathways in Hypopharyngeal Cancer. Int J Mol Med (2019) 43(1):127–42. doi: 10.3892/ijmm.2018.3942
29. Gao L, Wang QB, Zhi Y, Ren WH, Li SM, Zhao CY, et al. Down-Regulation of Hsa_Circ_0092125 is Related to the Occurrence and Development of Oral Squamous Cell Carcinoma. Int J Oral Maxillofac Surg (2020) 49(3):292–7. doi: 10.1016/j.ijom.2019.07.014
30. Dou Z, Li S, Ren W, Wang Q, Liu J, Kong X, et al. Decreased Expression of Hsa_Circ_0072387 as a Valuable Predictor for Oral Squamous Cell Carcinoma. Oral Dis (2019) 25(5):1302–8. doi: 10.1111/odi.13094
31. Sun S, Li B, Wang Y, Li X, Wang P, Wang F, et al. Clinical Significance of the Decreased Expression of Hsa_Circ_001242 in Oral Squamous Cell Carcinoma. Dis Markers (2018) 2018:6514795. doi: 10.1155/2018/6514795
32. Wang Z, Tang J, Wang Y, Sun S, Chen Y, Shen Y, et al. Circular RNA Hsa_Circ_009755 Downregulation Correlates With Clinicopathology in Oral Squamous Cell Carcinoma. OncoTargets Ther (2019) 12:4025–31. doi: 10.2147/ott.S196618
33. Zhao SY, Wang J, Ouyang SB, Huang ZK, Liao L. Salivary Circular Rnas Hsa_Circ_0001874 and Hsa_Circ_0001971 as Novel Biomarkers for the Diagnosis of Oral Squamous Cell Carcinoma. Cell Physiol Biochem (2018) 47(6):2511–21. doi: 10.1159/000491624
34. Guo Y, Huang Q, Zheng J, Hsueh CY, Yuan X, Heng Y, et al. Diagnostic Role of Dysregulated Circular RNA Hsa_Circ_0036722 in Laryngeal Squamous Cell Carcinoma. Onco Targets Ther (2020) 13:5709–19. doi: 10.2147/ott.S231076
35. Li B, Wang F, Li X, Sun S, Shen Y, Yang H. Hsa_Circ_0008309 May be a Potential Biomarker for Oral Squamous Cell Carcinoma. Dis Markers (2018) 2018:7496890. doi: 10.1155/2018/7496890
36. Li L, Zhang ZT. Hsa_Circ_0086414 Might be a Diagnostic Biomarker of Oral Squamous Cell Carcinoma. Med Sci Monit (2020) 26:e919383. doi: 10.12659/msm.919383
37. Li X, Zhang H, Wang Y, Sun S, Shen Y, Yang H. Silencing Circular RNA Hsa_Circ_0004491 Promotes Metastasis of Oral Squamous Cell Carcinoma. Life Sci (2019) 239:116883. doi: 10.1016/j.lfs.2019.116883
38. Tian L, Cao J, Jiao H, Zhang J, Ren X, Liu X, et al. CircRASSF2 Promotes Laryngeal Squamous Cell Carcinoma Progression by Regulating the miR-302b-3p/IGF-1R Axis. Clin Sci (Lond) (2019) 133(9):1053–66. doi: 10.1042/cs20190110
39. Zhang N, Gao L, Ren W, Li S, Zhang D, Song X, et al. Fucoidan Affects Oral Squamous Cell Carcinoma Cell Functions In Vitro by Regulating FLNA-derived Circular RNA. Ann New York Acad Sci (2020) 1462(1):65–78. doi: 10.1111/nyas.14190
40. Chen G, Li Y, He Y, Zeng B, Yi C, Wang C, et al. Upregulation of Circular Rna circATRNL1 to Sensitize Oral Squamous Cell Carcinoma to Irradiation. Mol Ther Nucleic Acids (2020) 19:961–73. doi: 10.1016/j.omtn.2019.12.031
41. Su W, Wang Y, Wang F, Zhang B, Zhang H, Shen Y, et al. Circular RNA Hsa_Circ_0007059 Indicates Prognosis and Influences Malignant Behavior Via AKT/mTOR in Oral Squamous Cell Carcinoma. J Cell Physiol (2019) 234(9):15156–66. doi: 10.1002/jcp.28156
42. Yang Y, Ci HS, Mao YL, Li JW, Zuo JH. CircRNA_002178 Promotes the Proliferation and Migration of Oral Squamous Cell Carcinoma Cells by Activating the Akt/mTOR Pathway. Eur Rev Med Pharmacol Sci (2020) 24(11):6122–30. doi: 10.26355/eurrev_202006_21507
43. Yin L, Chen J, Ma C, Pei S, Du M, Zhang Y, et al. Hsa_Circ_0046263 Functions as a ceRNA to Promote Nasopharyngeal Carcinoma Progression by Upregulating IGFBP3. Cell Death Dis (2020) 11(7):562–. doi: 10.1038/s41419-020-02785-3
44. London M, Gallo E. Epidermal Growth Factor Receptor (EGFR) Involvement in Epithelial-Derived Cancers and its Current Antibody-Based Immunotherapies. Cell Biol Int (2020) 44:1267–82. doi: 10.1002/cbin.11340
45. Zhao W, Cui Y, Liu L, Qi X, Liu J, Ma S, et al. Splicing Factor Derived Circular RNA circUHRF1 Accelerates Oral Squamous Cell Carcinoma Tumorigenesis Via Feedback Loop. Cell Death Differ (2020) 27(3):919–33. doi: 10.1038/s41418-019-0423-5
46. Verduci L, Ferraiuolo M, Sacconi A, Ganci F, Vitale J, Colombo T, et al. The Oncogenic Role of circPVT1 in Head and Neck Squamous Cell Carcinoma is Mediated Through the Mutant p53/YAP/TEAD Transcription-Competent Complex. Genome Biol (2017) 18(1):237. doi: 10.1186/s13059-017-1368-y
47. He T, Li X, Xie D, Tian L. Overexpressed circPVT1 in Oral Squamous Cell Carcinoma Promotes Proliferation by Serving as a miRNA Sponge. Mol Med Rep (2019) 20(4):3509–18. doi: 10.3892/mmr.2019.10615
48. Ke Z, Xie F, Zheng C, Chen D. CircHIPK3 Promotes Proliferation and Invasion in Nasopharyngeal Carcinoma by Abrogating Mir-4288-Induced ELF3 Inhibition. J Cell Physiol (2019) 234(2):1699–706. doi: 10.1002/jcp.27041
49. Wang J, Zhao SY, Ouyang SS, Huang ZK, Luo Q, Liao L. [Circular RNA circHIPK3 Acts as the Sponge of microRNA-124 to Promote Human Oral Squamous Cell Carcinoma Cells Proliferation]. Zhonghua Kou Qiang Yi Xue Za Zhi (2018) 53(8):546–51. doi: 10.3760/cma.j.issn.1002-0098.2018.08.009
50. Peng Q-S, Cheng Y-N, Zhang W-B, Fan H, Mao Q-H, Xu P. circRNA_0000140 Suppresses Oral Squamous Cell Carcinoma Growth and Metastasis by Targeting miR-31 to Inhibit Hippo Signaling Pathway. Cell Death Dis (2020) 11(2):112. doi: 10.1038/s41419-020-2273-y
51. Wang Z, Huang C, Zhang A, Lu C, Liu L. Overexpression of circRNA_100290 Promotes the Progression of Laryngeal Squamous Cell Carcinoma Through the miR-136-5p/RAP2C Axis. Biomed Pharmacother Biomed Pharmacother (2020) 125:109874. doi: 10.1016/j.biopha.2020.109874
52. Wang L, Wei Y, Yan Y, Wang H, Yang J, Zheng Z, et al. CircDOCK1 Suppresses Cell Apoptosis Via Inhibition of miR−196a−5p by Targeting BIRC3 in OSCC. Oncol Rep (2018) 39(3):951–66. doi: 10.3892/or.2017.6174
53. Su W, Sun S, Wang F, Shen Y, Yang H. Circular RNA Hsa_Circ_0055538 Regulates the Malignant Biological Behavior of Oral Squamous Cell Carcinoma Through the p53/Bcl-2/caspase Signaling Pathway. J Trans Med (2019) 17(1):76. doi: 10.1186/s12967-019-1830-6
54. Hao C, Wangzhou K, Liang Z, Liu C, Wang L, Gong L, et al. Circular RNA Itch Suppresses Cell Proliferation But Induces Apoptosis in Oral Squamous Cell Carcinoma by Regulating miR-421/PDCD4 Axis. Cancer Manag Res (2020) 12:5651–8. doi: 10.2147/cmar.S258887
55. Deng W, Fu J, Wang T, Chen JX, Fu LB, Peng W. Hsa_circRNA_101036 Acts as Tumor-Suppressor in Oral Squamous Cell Carcinoma Cells Via Inducing Endoplasmic Reticulum Stress. Eur Rev Med Pharmacol Sci (2020) 24(11):6111–21. doi: 10.26355/eurrev_202006_21506
56. Duan X, Shen N, Chen J, Wang J, Zhu Q, Zhai Z. Circular RNA MYLK Serves as an Oncogene to Promote Cancer Progression Via Microrna-195/Cyclin D1 Axis in Laryngeal Squamous Cell Carcinoma. Biosci Rep (2019) 39(9):BSR20190227 doi: 10.1042/bsr20190227
57. Zhong Q, Huang J, Wei J, Wu R. Circular RNA CDR1as Sponges miR-7-5p to Enhance E2F3 Stability and Promote the Growth of Nasopharyngeal Carcinoma. Cancer Cell Int (2019) 19:252. doi: 10.1186/s12935-019-0959-y
58. Gao L, Dou Z-C, Ren W-H, Li S-M, Liang X, Zhi K-Q. CircCDR1as Upregulates Autophagy Under Hypoxia to Promote Tumor Cell Survival Via AKT/ERK(½)/mTOR Signaling Pathways in Oral Squamous Cell Carcinomas. Cell Death Dis (2019) 10(10):745–. doi: 10.1038/s41419-019-1971-9
59. Gao L, Zhao C, Li S, Dou Z, Wang Q, Liu J, et al. circ-PKD2 Inhibits Carcinogenesis Via the miR-204-3p/APC2 Axis in Oral Squamous Cell Carcinoma. Mol Carcinogenesis (2019) 58(10):1783–94. doi: 10.1002/mc.23065
60. Pan JY, Bao XL, Zhu K. Circular RNA_LARP4 Inhibits Cell Proliferation and Invasion of Nasopharyngeal Carcinoma by Repressing ROCK1. Eur Rev Med Pharmacol Sci (2019) 23(22):9915–22. doi: 10.26355/eurrev_201911_19557
61. Wei Z, Chang K, Fan C. Hsa_Circ_0042666 Inhibits Proliferation and Invasion Via Regulating miR-223/TGFBR3 Axis in Laryngeal Squamous Cell Carcinoma. Biomed Pharmacother (2019) 119:109365. doi: 10.1016/j.biopha.2019.109365
62. Zhu X, Shao P, Tang Y, Shu M, Hu WW, Zhang Y. Hsa_circRNA_100533 Regulates GNAS by Sponging Hsa_miR_933 to Prevent Oral Squamous Cell Carcinoma. J Cell Biochem (2019) 120(11):19159–71. doi: 10.1002/jcb.29245
63. Zheng Z, Ma X, Li H. Circular RNA circMDM2 Accelerates the Glycolysis of Oral Squamous Cell Carcinoma by Targeting Mir-532-3p/HK2. J Cell Mol Med (2020) 24(13):7531–7. doi: 10.1111/jcmm.15380
64. Chu YL. Circ_0067934 Correlates With Poor Prognosis and Promotes Laryngeal Squamous Cell Cancer Progression by Sponging Mir-1324. Eur Rev Med Pharmacol Sci (2020) 24(8):4320–7. doi: 10.26355/eurrev_202004_21013
65. Liu Q, Shuai M, Xia Y. Knockdown of EBV-encoded circRNA circRPMS1 Suppresses Nasopharyngeal Carcinoma Cell Proliferation and Metastasis Through Sponging Multiple Mirnas. Cancer Manag Res (2019) 11:8023–31. doi: 10.2147/CMAR.S218967
66. Zhu L, Liu Y, Yang Y, Mao XM, Yin ZD. Circrna ZNF609 Promotes Growth and Metastasis of Nasopharyngeal Carcinoma by Competing With Microrna-150-5p. Eur Rev Med Pharmacol Sci (2019) 23(7):2817–26. doi: 10.26355/eurrev_201904_17558
67. Li M, Li Y, Yu M. Circrna ZNF609 Knockdown Suppresses Cell Growth Via Modulating Mir-188/ELF2 Axis in Nasopharyngeal Carcinoma. Onco Targets Ther (2020) 13:2399–409. doi: 10.2147/ott.S234230
68. Han L, Cheng J, Li A. Hsa_Circ_0072387 Suppresses Proliferation, Metastasis, and Glycolysis of Oral Squamous Cell Carcinoma Cells by Downregulating Mir-503-5p. Cancer Biother Radiopharmaceut (2020) 36(1):84–94. doi: 10.1089/cbr.2019.3371
69. Chen X, Su X, Zhu C, Zhou J. Knockdown of Hsa_Circ_0023028 Inhibits Cell Proliferation, Migration, and Invasion in Laryngeal Cancer by Sponging Mir-194-5p. Biosci Rep (2019) 39(6):BSR20190177. doi: 10.1042/bsr20190177
70. Li H, You J, Xue H, Tan X, Chao C. CircCTDP1 Promotes Nasopharyngeal Carcinoma Progression Via a Microrna−320b/HOXA10/Tgfβ2 Pathway. Int J Mol Med (2020) 45(3):836–46. doi: 10.3892/ijmm.2020.4467
71. Wei H, Liu D, Sun J, Mao Y, Zhao L, Zhu W, et al. Circular RNA Circ_0008450 Upregulates CXCL9 Expression by Targeting miR-577 to Regulate Cell Proliferation and Invasion in Nasopharyngeal Carcinoma. Exp Mol Pathol (2019) 110:104288. doi: 10.1016/j.yexmp.2019.104288
72. Wang Z, Wei P, Wei D, Cao S, Liu H, Chen L, et al. Effect of Up-Regulation of circMATR3 on the Proliferation, Metastasis, Progression and Survival of Hypopharyngeal Carcinoma. J Cell Mol Med (2020) 24(8):4687–97. doi: 10.1111/jcmm.15134
73. Fu D, Huang Y, Gao M. Hsa_Circ_0057481 Promotes Laryngeal Cancer Proliferation and Migration by Modulating the miR-200c/ZEB1 Axis. Int J Clin Exp Pathol (2019) 12(11):4066–76.
74. Hu YT, Li XX, Zeng LW. Circ_0001742 Promotes Tongue Squamous Cell Carcinoma Progression Via miR-431-5p/ATF3 Axis. Eur Rev Med Pharmacol Sci (2019) 23(23):10300–12. doi: 10.26355/eurrev_201912_19668
75. Bai Y, Hou J, Wang X, Geng L, Jia X, Xiang L, et al. Circ_0000218 Plays a Carcinogenic Role in Laryngeal Cancer Through Regulating microRNA-139-3p/Smad3 Axis. Pathol Res Pract (2020) 216(9):153103. doi: 10.1016/j.prp.2020.153103
76. Tan X, Zhou C, Liang Y, Lai YF, Liang Y. Circ_0001971 Regulates Oral Squamous Cell Carcinoma Progression and Chemosensitivity by Targeting miR-194/miR-204 In Vitro and In Vivo. Eur Rev Med Pharmacol Sci (2020) 24(5):2470–81. doi: 10.26355/eurrev_202003_20515
77. Wu Y, Zhang Y, Zheng X, Dai F, Lu Y, Dai L, et al. Circular RNA circCORO1C Promotes Laryngeal Squamous Cell Carcinoma Progression by Modulating the let-7c-5p/PBX3 Axis. Mol Cancer (2020) 19(1):99. doi: 10.1186/s12943-020-01215-4
78. Ouyang SB, Wang J, Zhao SY, Zhang XH, Liao L. CircRNA_0109291 Regulates Cell Growth and Migration in Oral Squamous Cell Carcinoma and its Clinical Significance. Iran J Basic Med Sci (2018) 21(11):1186–91. doi: 10.22038/ijbms.2018.30347.7313
79. Su W, Wang YF, Wang F, Yang HJ, Yang HY. [Effect of Circular RNA Hsa_Circ_0002203 on the Proliferation, Migration, Invasion, and Apoptosis of Oral Squamous Cell Carcinoma Cells]. Hua Xi Kou Qiang Yi Xue Za Zhi (2019) 37(5):509–15. doi: 10.7518/hxkq.2019.05.011
80. Wang J, Kong J, Nie Z, Chen D, Qiang J, Gao W, et al. Circular RNA Hsa_Circ_0066755 as an Oncogene Via Sponging miR-651 and as a Promising Diagnostic Biomarker for Nasopharyngeal Carcinoma. Int J Med Sci (2020) 17(11):1499–507. doi: 10.7150/ijms.47024
81. Shang ZF, Tan W, Liu XD, Yu L, Li B, Li M, et al. DNA-Pkcs Negatively Regulates Cyclin B1 Protein Stability Through Facilitating its Ubiquitination Mediated by Cdh1-APC/C Pathway. Int J Biol Sci (2015) 11(9):1026–35. doi: 10.7150/ijbs.12443
82. Haddadi N, Lin Y, Travis G, Simpson AM, Nassif NT, McGowan EM. Pten/Ptenp1: ‘Regulating the Regulator of RTK-dependent PI3K/Akt Signalling’, New Targets for Cancer Therapy. Mol Cancer (2018) 17(1):37. doi: 10.1186/s12943-018-0803-3
83. Zhu Y, Tang H, Zhang L, Gong L, Wu G, Ni J, et al. Suppression of miR-21-3p Enhances TRAIL-mediated Apoptosis in Liver Cancer Stem Cells by Suppressing the PI3K/Akt/Bad Cascade Via Regulating PTEN. Cancer Manag Res (2019) 11:955–68. doi: 10.2147/cmar.S183328
84. She Q-B, Solit DB, Ye Q, O’Reilly KE, Lobo J, Rosen N. The BAD Protein Integrates Survival Signaling by EGFR/MAPK and PI3K/Akt Kinase Pathways in PTEN-deficient Tumor Cells. Cancer Cell (2005) 8(4):287–97. doi: 10.1016/j.ccr.2005.09.006
85. Choi HJ, Chung TW, Kang SK, Lee YC, Ko JH, Kim JG, et al. Ganglioside GM3 Modulates Tumor Suppressor PTEN-mediated Cell Cycle Progression–Transcriptional Induction of p21(WAF1) and p27(kip1) by Inhibition of PI-3K/AKT Pathway. Glycobiology (2006) 16(7):573–83. doi: 10.1093/glycob/cwj105
86. Zhang L, Zhou F, ten Dijke P. Signaling Interplay Between Transforming Growth Factor-β Receptor and PI3K/AKT Pathways in Cancer. Trends Biochem Sci (2013) 38(12):612–20. doi: 10.1016/j.tibs.2013.10.001
87. Vlasova-St Louis I, Bohjanen PR. Post-Transcriptional Regulation of Cytokine and Growth Factor Signaling in Cancer. Cytokine Growth Factor Rev (2017) 33:83–93. doi: 10.1016/j.cytogfr.2016.11.004
88. Kasprzak A, Kwasniewski W, Adamek A, Gozdzicka-Jozefiak A. Insulin-Like Growth Factor (IGF) Axis in Cancerogenesis. Mutat Res Rev Mutat Res (2017) 772:78–104. doi: 10.1016/j.mrrev.2016.08.007
89. Galvan V, Logvinova A, Sperandio S, Ichijo H, Bredesen DE. Type 1 Insulin-Like Growth Factor Receptor (IGF-IR) Signaling Inhibits Apoptosis Signal-Regulating Kinase 1 (ASK1). J Biol Chem (2003) 278(15):13325–32. doi: 10.1074/jbc.M211398200
90. Sarig R, Rivlin N, Brosh R, Bornstein C, Kamer I, Ezra O, et al. Mutant p53 Facilitates Somatic Cell Reprogramming and Augments the Malignant Potential of Reprogrammed Cells. J Exp Med (2010) 207(10):2127–40. doi: 10.1084/jem.20100797
91. Gudipaty SA, Conner CM, Rosenblatt J, Montell DJ. Unconventional Ways to Live and Die: Cell Death and Survival in Development, Homeostasis, and Disease. Annu Rev Cell Dev Biol (2018) 34:311–32. doi: 10.1146/annurev-cellbio-100616-060748
92. Pocsfalvi G, Stanly C, Fiume I, Vékey K. Chromatography and its Hyphenation to Mass Spectrometry for Extracellular Vesicle Analysis. J Chromatogr A (2016) 1439:26–41. doi: 10.1016/j.chroma.2016.01.017
93. Kavanagh E, Rodhe J, Burguillos MA, Venero JL, Joseph B. Regulation of Caspase-3 Processing by cIAP2 Controls the Switch Between Pro-Inflammatory Activation and Cell Death in Microglia. Cell Death Dis (2014) 5(12):e1565. doi: 10.1038/cddis.2014.514
94. Aubrey BJ, Kelly GL, Janic A, Herold MJ, Strasser A. How Does p53 Induce Apoptosis and How Does This Relate to p53-mediated Tumour Suppression? Cell Death Differ (2018) 25(1):104–13. doi: 10.1038/cdd.2017.169
95. Iurlaro R, Muñoz-Pinedo C. Cell Death Induced by Endoplasmic Reticulum Stress. FEBS J (2016) 283(14):2640–52. doi: 10.1111/febs.13598
96. Diaz-Moralli S, Tarrado-Castellarnau M, Miranda A, Cascante M. Targeting Cell Cycle Regulation in Cancer Therapy. Pharmacol Ther (2013) 138(2):255–71. doi: 10.1016/j.pharmthera.2013.01.011
97. Gao X, Leone GW, Wang H. Cyclin D-CDK4/6 Functions in Cancer. Adv Cancer Res (2020) 148:147–69. doi: 10.1016/bs.acr.2020.02.002
98. Caruso JA, Duong MT, Carey JPW, Hunt KK, Keyomarsi K. Low-Molecular-Weight Cyclin E in Human Cancer: Cellular Consequences and Opportunities for Targeted Therapies. Cancer Res (2018) 78(19):5481–91. doi: 10.1158/0008-5472.Can-18-1235
99. Wang JX, Liu Y, Jia XJ, Liu SX, Dong JH, Ren XM, et al. Upregulation of circFLNA Contributes to Laryngeal Squamous Cell Carcinoma Migration by circFLNA-miR-486-3p-FLNA Axis. Cancer Cell Int (2019) 19:196. doi: 10.1186/s12935-019-0924-9
100. Xia B, Hong T, He X, Hu X, Gao Y. A Circular RNA Derived From MMP9 Facilitates Oral Squamous Cell Carcinoma Metastasis Through Regulation of MMP9 mRNA Stability. Cell Transplant (2019) 28(12):1614–23. doi: 10.1177/0963689719875409
101. Fan C, Qu H, Xiong F, Tang Y, Tang T, Zhang L, et al. CircARHGAP12 Promotes Nasopharyngeal Carcinoma Migration and Invasion Via Ezrin-Mediated Cytoskeletal Remodeling. Cancer Lett (2021) 496:41–56. doi: 10.1016/j.canlet.2020.09.006
102. Hong X, Liu N, Liang Y, He Q, Yang X, Lei Y, et al. Circular RNA CRIM1 Functions as a ceRNA to Promote Nasopharyngeal Carcinoma Metastasis and Docetaxel Chemoresistance Through Upregulating FOXQ1. Mol Cancer (2020) 19(1):33. doi: 10.1186/s12943-020-01149-x
103. Wang J, Jiang C, Li N, Wang F, Xu Y, Shen Z, et al. The circEPSTI1/mir-942-5p/LTBP2 Axis Regulates the Progression of OSCC in the Background of OSF Via EMT and the PI3K/Akt/mTOR Pathway. Cell Death Dis (2020) 11(8):682. doi: 10.1038/s41419-020-02851-w
104. Qin JB, Chang W, Yuan GH, Huang L, Qiu ZF. Circular RNA Hsa_Circ_0000285 Acts as an Oncogene in Laryngocarcinoma by Inducing Wnt/β-Catenin Signaling Pathway. Eur Rev Med Pharmacol Sci (2019) 23(24):10803–9. doi: 10.26355/eurrev_201912_19783
105. Inan S, Hayran M. Cell Signaling Pathways Related to Epithelial Mesenchymal Transition in Cancer Metastasis. Crit Rev Oncog (2019) 24(1):47–54. doi: 10.1615/CritRevOncog.2018029509
106. Wu HT, Zhong HT, Li GW, Shen JX, Ye QQ, Zhang ML, et al. Oncogenic Functions of the EMT-related Transcription Factor ZEB1 in Breast Cancer. J Transl Med (2020) 18(1):51. doi: 10.1186/s12967-020-02240-z
107. Conlon GA, Murray GI. Recent Advances in Understanding the Roles of Matrix Metalloproteinases in Tumour Invasion and Metastasis. J Pathol (2019) 247(5):629–40. doi: 10.1002/path.5225
108. Zhan XH, Jiao JW, Zhang HF, Xu XE, He JZ, Li RL, et al. Loxl2 Upregulates Phosphorylation of Ezrin to Promote Cytoskeletal Reorganization and Tumor Cell Invasion. Cancer Res (2019) 79(19):4951–64. doi: 10.1158/0008-5472.Can-19-0860
109. Yuan J, Chen L, Xiao J, Qi XK, Zhang J, Li X, et al. SHROOM2 Inhibits Tumor Metastasis Through RhoA-ROCK Pathway-Dependent and -Independent Mechanisms in Nasopharyngeal Carcinoma. Cell Death Dis (2019) 10(2):58. doi: 10.1038/s41419-019-1325-7
110. Xu Y, Bismar TA, Su J, Xu B, Kristiansen G, Varga Z, et al. Filamin A Regulates Focal Adhesion Disassembly and Suppresses Breast Cancer Cell Migration and Invasion. J Exp Med (2010) 207(11):2421–37. doi: 10.1084/jem.20100433
111. Boroughs LK, DeBerardinis RJ. Metabolic Pathways Promoting Cancer Cell Survival and Growth. Nat Cell Biol (2015) 17(4):351–9. doi: 10.1038/ncb3124
112. Zhu X, Du J, Gu Z. Circ-PVT1/miR-106a-5p/HK2 Axis Regulates Cell Growth, Metastasis and Glycolytic Metabolism of Oral Squamous Cell Carcinoma. Mol Cell Biochem (2020) 474(1–2):147–58. doi: 10.1007/s11010-020-03840-5
113. Guo J, Su Y, Zhang M. Circ_0000140 Restrains the Proliferation, Metastasis and Glycolysis Metabolism of Oral Squamous Cell Carcinoma Through Upregulating CDC73 Via Sponging Mir-182-5p. Cancer Cell Int (2020) 20:407. doi: 10.1186/s12935-020-01501-7
114. Chen X, Yu J, Tian H, Shan Z, Liu W, Pan Z, et al. Circle RNA Hsa_circRNA_100290 Serves as a ceRNA for miR-378a to Regulate Oral Squamous Cell Carcinoma Cells Growth Via Glucose Transporter-1 (GLUT1) and Glycolysis. J Cell Physiol (2019) 234(11):19130–40. doi: 10.1002/jcp.28692
115. Zhang Y, Tang K, Chen L, Du M, Qu Z. Exosomal CircGDI2 Suppresses Oral Squamous Cell Carcinoma Progression Through the Regulation of MiR-424-5p/SCAI Axis. Cancer Manag Res (2020) 12:7501–14. doi: 10.2147/cmar.S255687
116. Ma L, Zong X. Metabolic Symbiosis in Chemoresistance: Refocusing the Role of Aerobic Glycolysis. Front Oncol (2020) 10:5. doi: 10.3389/fonc.2020.00005
117. Li WC, Huang CH, Hsieh YT, Chen TY, Cheng LH, Chen CY, et al. Regulatory Role of Hexokinase 2 in Modulating Head and Neck Tumorigenesis. Front Oncol (2020) 10:176. doi: 10.3389/fonc.2020.00176
118. Chen L, Zhou H, Guan Z. CircRNA_000543 Knockdown Sensitizes Nasopharyngeal Carcinoma to Irradiation by Targeting miR-9/platelet-derived Growth Factor Receptor B Axis. Biochem Biophys Res Commun (2019) 512(4):786–92. doi: 10.1016/j.bbrc.2019.03.126
119. Shuai M, Hong J, Huang D, Zhang X, Tian Y. Upregulation of circRNA_0000285 Serves as a Prognostic Biomarker for Nasopharyngeal Carcinoma and is Involved in Radiosensitivity. Oncol Lett (2018) 16(5):6495–501. doi: 10.3892/ol.2018.9471
120. Shuai M, Huang L. High Expression of Hsa_circRNA_001387 in Nasopharyngeal Carcinoma and the Effect on Efficacy of Radiotherapy. Onco Targets Ther (2020) 13:3965–73. doi: 10.2147/ott.S249202
121. Qiongna D, Jiafeng Z, Yalin H, Ping H, Chuan Z, Xiaojie J, et al. Implication of Hsa_Circ_0028007 in Reinforcing Migration, Invasion, and Chemo-Tolerance of Nasopharyngeal Carcinoma Cells. J Clin Lab Anal (2020) 34(9):e23409. doi: 10.1002/jcla.23409
122. Wu Y, Zhang Y, Niu M, Shi Y, Liu H, Yang D, et al. Whole-Transcriptome Analysis of CD133+CD144+ Cancer Stem Cells Derived From Human Laryngeal Squamous Cell Carcinoma Cells. Cell Physiol Biochem (2018) 47(4):1696–710. doi: 10.1159/000490992
123. Su W, Wang Y, Wang F, Sun S, Li M, Shen Y, et al. Hsa_Circ_0005379 Regulates Malignant Behavior of Oral Squamous Cell Carcinoma Through the EGFR Pathway. BMC Cancer (2019) 19(1):400. doi: 10.1186/s12885-019-5593-5
124. Tsao SW, Tsang CM, To KF, Lo KW. The Role of Epstein-Barr Virus in Epithelial Malignancies. J Pathol (2015) 235(2):323–33. doi: 10.1002/path.4448
125. Westra WH. The Changing Face of Head and Neck Cancer in the 21st Century: The Impact of HPV on the Epidemiology and Pathology of Oral Cancer. Head Neck Pathol (2009) 3(1):78–81. doi: 10.1007/s12105-009-0100-y
126. Toptan T, Abere B, Nalesnik MA, Swerdlow SH, Ranganathan S, Lee N, et al. Circular DNA Tumor Viruses Make Circular Rnas. Proc Natl Acad Sci U S A (2018) 115(37):E8737–45. doi: 10.1073/pnas.1811728115
127. Ungerleider N, Concha M, Lin Z, Roberts C, Wang X, Cao S, et al. The Epstein Barr Virus Circrnaome. PloS Pathog (2018) 14(8):e1007206. doi: 10.1371/journal.ppat.1007206
128. Huang JT, Chen JN, Gong LP, Bi YH, Liang J, Zhou L, et al. Identification of Virus-Encoded Circular RNA. Virology (2019) 529:144–51. doi: 10.1016/j.virol.2019.01.014
129. Gong LP, Chen JN, Dong M, Xiao ZD, Feng ZY, Pan YH, et al. Epstein-Barr Virus-Derived Circular RNA LMP2A Induces Stemness in EBV-associated Gastric Cancer. EMBO Rep (2020) 21(10):e49689. doi: 10.15252/embr.201949689
130. Zhao J, Lee EE, Kim J, Yang R, Chamseddin B, Ni C, et al. Transforming Activity of an Oncoprotein-Encoding Circular RNA From Human Papillomavirus. Nat Commun (2019) 10(1):2300. doi: 10.1038/s41467-019-10246-5
131. Bahn JH, Zhang Q, Li F, Chan TM, Lin X, Kim Y, et al. And Circular RNA in Human Saliva. Clin Chem (2015) 61(1):221–30. doi: 10.1373/clinchem.2014.230433
132. Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. Circular RNA is Enriched and Stable in Exosomes: A Promising Biomarker for Cancer Diagnosis. Cell Res (2015) 25(8):981–4. doi: 10.1038/cr.2015.82
133. Goleva E, Lyubchenko T, Kraehenbuehl L, LaCouture ME, Leung DYM, Kern JA. Our Current Understanding of Checkpoint Inhibitor Therapy in Cancer Immunotherapy. Ann Allergy Asthma Immunol (2021). doi: 10.1016/j.anai.2021.03.003
134. Ye J, Ji X, Dennis PA, Abdullah H, Mukhopadhyay P. Relationship Between Progression-Free Survival, Objective Response Rate, and Overall Survival in Clinical Trials of PD-1/PD-L1 Immune Checkpoint Blockade: A Meta-Analysis. Clin Pharmacol Ther (2020) 108(6):1274–88. doi: 10.1002/cpt.1956
135. Hong W, Xue M, Jiang J, Zhang Y, Gao X. Circular RNA circ-CPA4/ Let-7 miRNA/PD-L1 Axis Regulates Cell Growth, Stemness, Drug Resistance and Immune Evasion in non-Small Cell Lung Cancer (NSCLC). J Exp Clin Cancer Res (2020) 39(1):149. doi: 10.1186/s13046-020-01648-1
136. Zhang PF, Pei X, Li KS, Jin LN, Wang F, Wu J, et al. Circular RNA circFGFR1 Promotes Progression and anti-PD-1 Resistance by Sponging miR-381-3p in non-Small Cell Lung Cancer Cells. Mol Cancer (2019) 18(1):179. doi: 10.1186/s12943-019-1111-2
137. Chen YG, Chen R, Ahmad S, Verma R, Kasturi SP, Amaya L, et al. N6-Methyladenosine Modification Controls Circular Rna Immunity. Mol Cell (2019) 76(1):96–109.e9. doi: 10.1016/j.molcel.2019.07.016
138. Shannon-Lowe C, Rickinson A. The Global Landscape of EBV-Associated Tumors. Front Oncol (2019) 9:713. doi: 10.3389/fonc.2019.00713
139. Sun Y, Zhang Y, Liu L, Song X, Li G. Genetic Polymorphisms and HPV Infection in Oral Squamous Cell Carcinomas. Curr Opin Virol (2015) 14:1–6. doi: 10.1016/j.coviro.2015.05.004
140. Tsang CM, Lui VWY, Bruce JP, Pugh TJ, Lo KW. Translational Genomics of Nasopharyngeal Cancer. Semin Cancer Biol (2020) 61:84–100. doi: 10.1016/j.semcancer.2019.09.006
141. Peiris-Pages M, Martinez-Outschoorn UE, Pestell RG, Sotgia F, Lisanti MP. Cancer Stem Cell Metabolism. Breast Cancer Res (2016) 18(1):55. doi: 10.1186/s13058-016-0712-6
142. Zhu D, Shao M, Yang J, Fang M, Liu S, Lou D, et al. Curcumin Enhances Radiosensitization of Nasopharyngeal Carcinoma Via Mediating Regulation of Tumor Stem-Like Cells by a CircRNA Network. J Cancer (2020) 11(8):2360–70. doi: 10.7150/jca.39511
Keywords: circRNA, non-coding RNA, biomarker, head and neck cancer, miRNA sponge
Citation: Li P, Zhu K, Mo Y, Deng X, Jiang X, Shi L, Guo C, Zhang W, Zeng Z, Li G, Xiong W, Zhang S and Gong Z (2021) Research Progress of circRNAs in Head and Neck Cancers. Front. Oncol. 11:616202. doi: 10.3389/fonc.2021.616202
Received: 11 October 2020; Accepted: 07 April 2021;
Published: 29 April 2021.
Edited by:
Ming Yann Lim, Tan Tock Seng Hospital, SingaporeReviewed by:
Yumin Wang, Central South University, ChinaCopyright © 2021 Li, Zhu, Mo, Deng, Jiang, Shi, Guo, Zhang, Zeng, Li, Xiong, Zhang and Gong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhaojian Gong, Z29uZ3poYW9qaWFuNDQ1OEBjc3UuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.