
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 26 March 2021
Sec. Cancer Genetics
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.595675
 Xinhui Fu1†
Xinhui Fu1† Hanjie Lin1†
Hanjie Lin1† Xinjuan Fan1
Xinjuan Fan1 Yaxi Zhu1
Yaxi Zhu1 Chao Wang1
Chao Wang1 Zhiting Chen1
Zhiting Chen1 Xiaoli Tan1
Xiaoli Tan1 Jinglin Huang1
Jinglin Huang1 Yacheng Cai2,3
Yacheng Cai2,3 Yan Huang1*
Yan Huang1*Background: PIK3CA is a high-frequency mutation gene in colorectal cancer, while its prognostic value remains unclear. This study evaluated the mutation tendency, spectrum, prognosis power and predictive power in cetuximab treatment of PIK3CA in Chinese CRC cohort.
Methods: The PIK3CA exon 9 and 20 status of 5763 CRC patients was detected with Sanger sequencing and a high-resolution melting test. Clinicopathological characteristics of 5733 patients were analyzed. Kaplan-Meier method and nomogram were used to evaluate the overall survival curve and disease recurrence, respectively.
Results: Fifty-eight types of mutations in 13.4% (771/5733) of the patients were detected. From 2014 to 2018, the mutation rate of PIK3CA increased from 11.0% to 13.5%. At stage IV, exon 20 mutated patients suffered shorter overall survival time than wild-type patients (multivariate COX regression analysis, HR = 2.72, 95% CIs = 1.47-5.09; p-value = 0.012). At stage III, PIK3CA mutated patients were more likely to relapse (multivariate Logistic regression analysis, exon 9: OR = 2.54, 95% CI = 1.34-4.73, p = 0.003; exon 20: OR = 3.89, 95% CI = 1.66-9.10, p = 0.002). The concordance index of the nomogram for predicting the recurrence risk of stage III patients was 0.685. After cetuximab treatment, the median PFS of PIK3CA exon 9 wild-type patients (n = 9) and mutant patients (n = 5) did not reach a significant difference (3.6 months vs. 2.3 months, Log-rank test, p-value = 0.513).
Conclusions: We found that PIK3CA mutation was an adverse predictive marker for the overall survival of stage IV patients and recurrence of stage III patients, respectively. Further more, we suggested that PIK3CA exon 9 mutations are not negative predictors of cetuximab treatment in KRAS, NRAS, and BRAF wild-type mCRC patients.
Colorectal cancer (CRC) is the third most prevalent malignancy worldwide, which leads to more than 860,000 deaths every year (1). In China, CRC has become the third most common cancer and the fifth leading cause of cancer-related mortality (2), while the incidence is predicted to be growing (3).
To date, the most effective treatment of CRC is adjuvant chemotherapy after surgical resection. Recently, the significant predictive value of some genetic mutation status has been reported by various clinical studies. KRAS mutation status was proved to be a robust predictive biomarker for the efficacy of anti-epidermal growth factor receptor (anti-EGFR) therapies (4). BRAF mutation has been wildly recognized as a reliable indicator of poor prognosis (5, 6). PIK3CA is one of the most frequently mutated oncogenes in CRC. It was reported that about 15-20% of CRC patients carried PIK3CA mutation (7), 80% of which was found in exon 9 and exon 20 (8). Previous studies indicated that patients with PIK3CA mutation could benefit from regular Aspirin treatment (9, 10), while the prognostic impact of PIK3CA mutation has far been controversial (11–15). Besides, the relationship between PIK3CA mutant mCRC tumor and resistance of anti-EGFR agents, cetuximab, is lack of investigation and did not reach consistency (16–20).
Patients at stage III were recommended to receive chemotherapy treatments, and most of the chemotherapy regimens were 5-fluorouracil based, such as FOLFOX and XELOX (6). Some clinical studies reported that a large group of patients was found to show chemotherapy resistance (11–23). The predictive effect of PIK3CA mutations in chemotherapy regimens in CRC was rarely reported and remained unclear (24).
In this retrospective study, we analyzed PIK3CA mutations in 5763 CRC patients, using Sanger sequencing and high-resolution melting (HRM) test, and evaluated the associations between PIK3CA mutations and the clinicopathological characteristics. We also conducted a survival analysis to investigate the prognostic value of PIK3CA mutations, and evaluated the role of PIK3CA mutations in first-line chemotherapy. A nomogram was then constructed to predict the recurrence risk of CRC patients at stage III. Further, the predictive effect of PIK3CA exon 9 mutation in cetuximab treatment was investigated with wild-type KRAS, NRAS, BRAF mCRC patients.
This study was approved by the Ethics Committee of the Sixth Affiliated Hospital of Sun Yat-sen University (L2017ZSLYEC-003). All patients underwent an informed consent process approved by the Hospital Institutional Review Board.
Our study included 5763 CRC patients diagnosed at the Sixth Affiliated Hospital of Sun Yat-sen University from January 2014 to December 2018. In this study, colorectal tumor specimens were fixed in formalin, embedded in paraffin after surgery, and confirmed histologically. The clinicopathologic features of these patients were collected from their medical records. The definition of the rectum is 12 cm or less from the anal verge.
Among the 5763 CRC patients, a sub-cohort composed of 1946 patients with available follow-up records was used to evaluate the prognosis value of PIK3CA exon 9 and 20 mutations. Another sub-cohort composed of 377 stage III patients who had received at least six courses of 5-fluorouracil based chemotherapy treatment after surgical excision was used to evaluate the associations between PIK3CA mutations and CRC recurrence.
In this study, 5763 patients’ primary intestinal tumors were collected for genetic testing. Genomic DNA extraction and Sanger sequencing were followed by the procedure of our previous study (25).
Besides, high-resolution melting (HRM) test was also applied to detect the mutation status of PIK3CA. In brief, the PCR reagent used in the HRM test was LightCycler 480 High-Resolution Melting Master Reagent Kit (Cat no: 04909631001, Roche Diagnostics Indianapolis, USA). The reaction system of the HRM test was: 50-100ng genomic DNA, 10μl 2×Master mix, 2.8μl MgCl2, and 1μl 10µM of each primer for exon 9 test, or 0.8μl 10µM of each primer for exon 20 test, and the mixtures were added up to 15μl with H2O. The primers of the exon 9 HRM test were: 5’-GAGACAATGAATTAAGGGAAAATG-3’ and 5’-CACTTACCTGTGACTCCATAG-3’; The primers of the exon 20 HRM test were: 5’-ACCCTAGCCTTAGATAAAACTGAGC-3’ and 5’-TCCATTTTTGTTGTCCAGCCACCAT-3’. The reaction conditions of cycling and melting for exon 9 HRM test were: 95°C for 10 min; 45 cycles of 95°C for 20s, 60°C for 20s and 72°C for 20s; 95°C; for 30s and 37°C for 30s; followed by a high-resolution melt of 74°C to 87°C with 45 acquisitions/°C 40°C for 10s. The exon 20 HRM test’s reaction condition was the same as that of exon 9 except that the annealing temperature was 62°C.
The PIK3CA exon 9 and 20 HRM tests’ detection limits were evaluated with spiked plasmid samples containing wild-type copies and mutant-type copies at various percentages. In detail, the exon 9 mutation type was c.1633G>A, and the initial concentration of mutant and wild-type plasmids were both 9.2×105 copies/μl. The exon 20 mutation type was c.3140A>T, and the initial concentration of mutant and wild-type plasmids were both 4.9×105 copies/μl. PIK3CA mutant plasmid was mixed with its corresponding wild type plasmid at various dilutions, at ratio of 50%, 40%, 30%, 20%, 10%, 8%, 6%, 5%. The minimum detectable dilution serves as the detection limit in this study.
Sanger sequencing was wildly considered a golden standard method for mutation detection. According to the Sanger sequencing results, sensitivities and specificities were calculated for PIK3CA exon 9 and 20 by HRM tests. Only the samples that obtained consistent results from the two methods were included for this study’s subsequent analysis.
For association analysis, the mutation information of KRAS exon 2 and BRAF codon 600 in our cohort was also obtained by the previously described detection method (26).
Chi-square test, Mann-Whitney U test, and Fisher’s exact test were applied to analyze the association of PIK3CA exon 9 and 20 mutation status with clinical characteristics, including the mutation status of KRAS exon 2 and BRAFV600E. The analyses were initially evaluated with continuous variables, categories data analysis, and further accessed with logistic regression models to evaluate the association based on estimating the odds ratios (OR) and their 95% confidence intervals (CIs). The significance test was two-sided, and a p-value < 0.05 was considered as statistically significant. All the statistical analyses were performed with SPSS 20.0 packages (SPSS, Chicago, IL, USA).
The annual mutation rates of PIK3CA exon 9 and 20 from 2014 to 2018 were calculated separately, and the mutation rates tendency was analyzed with the joinpoint regression model (Joinpoint 4.6.0.0., Calverton, MD, USA).
Kaplan-Meier survival curves for overall survival (OS) of 1946 patients with available follow-up records were performed with GraphPad Prism 5 (Graph Pad Software Inc., San Diego, CA, USA). The Log-rank test was used to assess the significance, and the p-value less than 0.05 was considered statistically significant. Besides, the logistic regression was used to screen other clinical characteristics related to the recurrence of patients at stage III. These variables were then enrolled to create a nomogram with rms package (version 5.1-4, https://cran.r-project.org/web/packages/rms/) in R statistical software (version 3.4.3).
The study cohort involved 5733 patients (Figure 1), getting the general profiles of CRC patients in China. Some basic clinical features of the cohort were summarized in Table 1. Briefly, there were more male patients (60.9%) in this cohort. The age of the first diagnosis ranged from 17 to 96 years old. We assigned patients into four groups, younger than 45 years old, between 45 to 49 years old, between 50 to 75 years old, and older than 75 years old, with 13.8%, 9.4%, 67.2%, and 9.6% of the study cohort respectively. For all CRC patients’ tumor sites, 47.4% of tumors were located in the rectum, 30.9% of tumors were located in the left colon, and 21.6% of tumors were located in the right colon. The vast majority of patients included in this study were tubular adenocarcinoma, of which 17.6%, 74.6%, and 7.8% were well, moderately, and poorly differentiated, respectively. Among these 5733 patients, only 3153 patients’ TNM stages information was available, in which 10.9%, 37.6%, 34.9%, and 16.5% of patients were assigned to stage I, stage II, stage III, and stage IV.
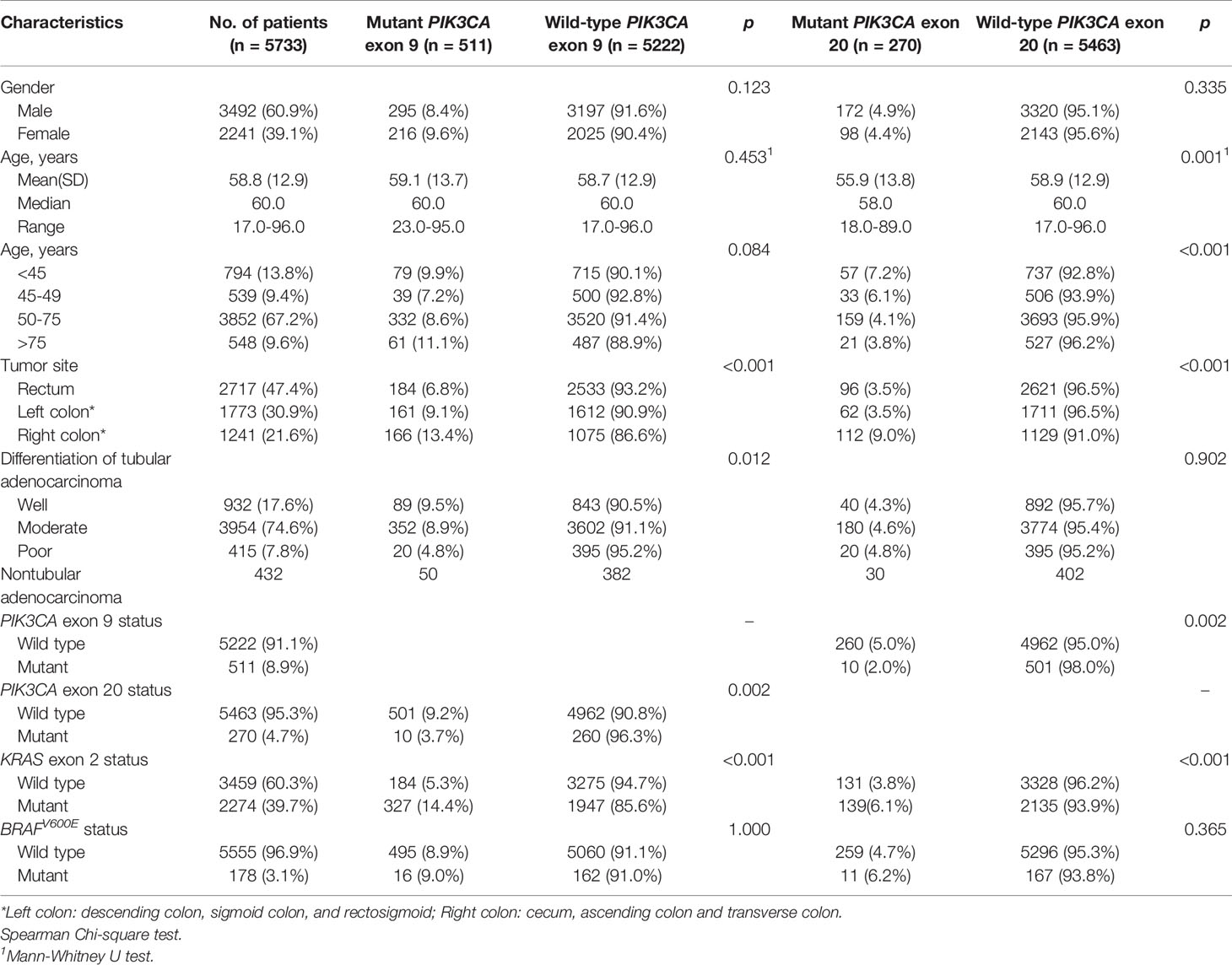
Table 1 Associations of PIK3CA exon 9 and 20 mutation status with clinicopathologic characteristics.
The associations of PIK3CA exon 9 and 20 mutations and clinicopathologic characteristics were shown in Table 1 and Table S1. In exon 9, mutations were more likely to occur at right colon (13.4%) than at other sides (6.8% at rectum, 9.1% at left side; p-value < 0.001), while less likely to present at poor differentiation tumors (4.8%) than at well and moderate differentiation tumors (9.5% and 8.9% respectively; p-value = 0.012). In TNM stage analysis, the exon 9 mutation frequency was higher in stage II (18.3%) than the other three stages (12.8% at stage I, 12.4% at stage III, and 15.5% at stage IV, p-value = 0.001, Table S1). In exon 20, mutations were more likely to occur in younger patients (7.2% in patients younger than 45 years old and 6.1% in patients between 45 to 49 years old) than those older than 50 years old (4.1% in patients between 50 to 75 years old and 3.8% at older than 75 years old, p-value < 0.001), as well as more likely to occur at the right colon (9.0%) than other sites (both 3.5% at the rectum and left colon, p-value < 0.001). In TNM stage analysis, exon 20 mutations were significantly more common at stage II (11.4%) than in other stages (5.5% at stage I, 5.9% at stage III, and 6.5% at stage IV; p-value < 0.001, Table S1).
Among all the 5733 patients, only ten patients mutated in both exons simultaneously, which indicated that exon 9 and exon 20 of PIK3CA are mutually exclusive in CRC (Table 1). KRAS exon 2 mutations were associated with PIK3CA exon 9 and exon 20 mutations (p-value < 0.001 for both exons), while BRAFV600E mutation did not show any association with both PIK3CA exons.
Features which were significantly associated with PIK3CA mutations in Table 1 or considered to have important effects in clinical practice were involved into logistic regression analysis, and the results were shown in Table S2. In both exons, mutations were more likely to present at right colon (in exon 9, OR = 1.86, 95% CIs = 1.53-2.26; in exon 20, OR = 2.72, 95% CIs = 2.12-3.50; p-value < 0.001 for both exons), as well as in stage II (in exon 9, OR= 1.46, 95% CIs = 1.20-1.78; in exon 20, OR = 2.01, 95% CIs = 1.55-2.60; p-value < 0.001 for both exons). In exon 9, mutations were less likely detected in poor differentiated tumor (OR = 0.51; 95% CIs = 0.32-0.81, p-value = 0.004). Higher PIK3CA exon 20 mutation frequency was associated with patients younger than 50 years old (OR = 1.70, 95% CIs = 1.31-2.21, p-value < 0.001).
The mutation rates of PIK3CA among Chinese CRC patients from 2014 to 2018 were shown in Figure S1 and Table S3. The mutation rate of PIK3CA exon 9 shows a slightly ascending tendency from 6.2% in 2014 to 8.9% in 2018 (Figure S1A. APC = 7.95, p-value > 0.05). The mutation rate of PIK3CA exon 20 remained nearly stable, as 5.00% in 2014 and 4.7% in 2018, (Figure S1B. APC = 0.30, p-value > 0.05). Combined both exons, the mutation rate of PIK3CA shows a gradually increasing tendency from 11.0% in 2014 to 13.5% in 2018 (Figure S1C. APC = 5.08, p-value > 0.05).
PIK3CA exon 9 and 20 mutations were tested in all 5763 samples with Sanger sequencing and the HRM test (Figure 1). Sanger sequencing failed to generate analyzable results from 16 samples, which can be detected by the HRM test. In the rest of 5747 samples, 14 had inconsistent results. The detection limits of PIK3CA exon 9 and exon 20 HRM tests were both 5% (Supplementary Figures S2 and S3), which is more sensitive than Sanger sequencing (20%). The specificities of PIK3CA exon 9 and exon 20 HRM tests were 99.77% and 99.96%, respectively, and the sensitivities of these two detecting methods were both 100% (Chinese patent, NO. 201610524420.9).
Among the final cohort with 5733 samples, 13.4% (771/5733) patients mutated in PIK3CA with 58 types of mutations (Table 2). Among these 771 mutated patients, 501 mutated at exon 9, 260 mutated at exon 20, and only 10 mutated at both exons simultaneously.
Among 58 mutation types, the 10 most frequent mutation types were: E545K (32.0%), H1047R (23.6%), E542K (17.6%), H1047L (3.9%), Q546K (3.5%), E545G (2.5%), Q546R (1.8%), E545A (1.8%), M1043I (1.7%) and H1047Y (1.0%). However, the other 48 types of mutations were only detected in 1.4% (81/5733) of CRC patients, which represented 10.5% (81/771) of PIK3CA mutated patients.
Among 58 mutation types, only 12 types of mutations could be detected by ARMS-based commercial PIK3CA mutation detection kits, which comprised 78.6% (606/771) of mutated patients.
In this study, we also annotated these somatic mutations following the instruction of ACMG guides on the interpretation of sequence variants (45). There were 4, 30, 21, and 3 mutation types in PIK3CA that were classified as Tier I, II, III, and IV variants in colorectal cancer, respectively (Table 2).
In order to evaluate the prognosis value of PIK3CA mutation in CRC patients, 1946 patients with available follow-up information were collected for survival analysis (clinicopathologic characteristics were shown in Table S4). The follow-up started from the day of surgery and ended on August 30, 2019. The median follow-up for the 1946 cohort is 16 months (0-64 months). Their overall survival rates were analyzed with the Kaplan-Meier method. In this follow-up cohort, 447 patients carried exon 9 mutations, 238 patients carried exon 20 mutations, and the remaining 1270 patients had wild-type PIK3CA gene. During the following-up, 178 patients died, among which 138 patients died from colorectal cancer or related diseases. No significant difference was detected between patients with and without PIK3CA mutation (Log-rank test, p-value = 0.289; Figure 2A), exon 9 mutation (Log-rank test, p-value = 0.241; Figure 2B), and exon 20 mutation (Log-rank test, p-value = 0.772; Figure 2C), respectively. For further analyses, patients were divided into four subgroups according to their TNM stages at the first diagnosis and conducted the survival analysis separately (Figures 2D–F). We found that only patients at stage IV who carried exon 20 mutations experienced significantly shorter OS than wild-type patients (median follow-up: 14 months; median survival for PIK3CA wild-typed and exon 20 mutated patients: 40 months vs. 23 months; Log-rank test, p-value = 0.002; Figure 2F). To assess the PIK3CA exon 20 mutations’ influence on the survival of stage IV patients, the COX regression model was applied. In comparison with patients carried PIK3CA exon 20 wild-type tumors, those with PIK3CA exon 20 mutated stage IV patients showed a decrease in OS (univariate HR = 2.52, 95% CIs = 1.38-4.59; p-value = 0.003, Table 3). In the multivariate COX regression model, PIK3CA exon 20 mutation was associated with a significant decrease in OS of stage IV CRC patients (HR = 2.72, 95% CIs = 1.47-5.09; p-value =0.012, Table 3).
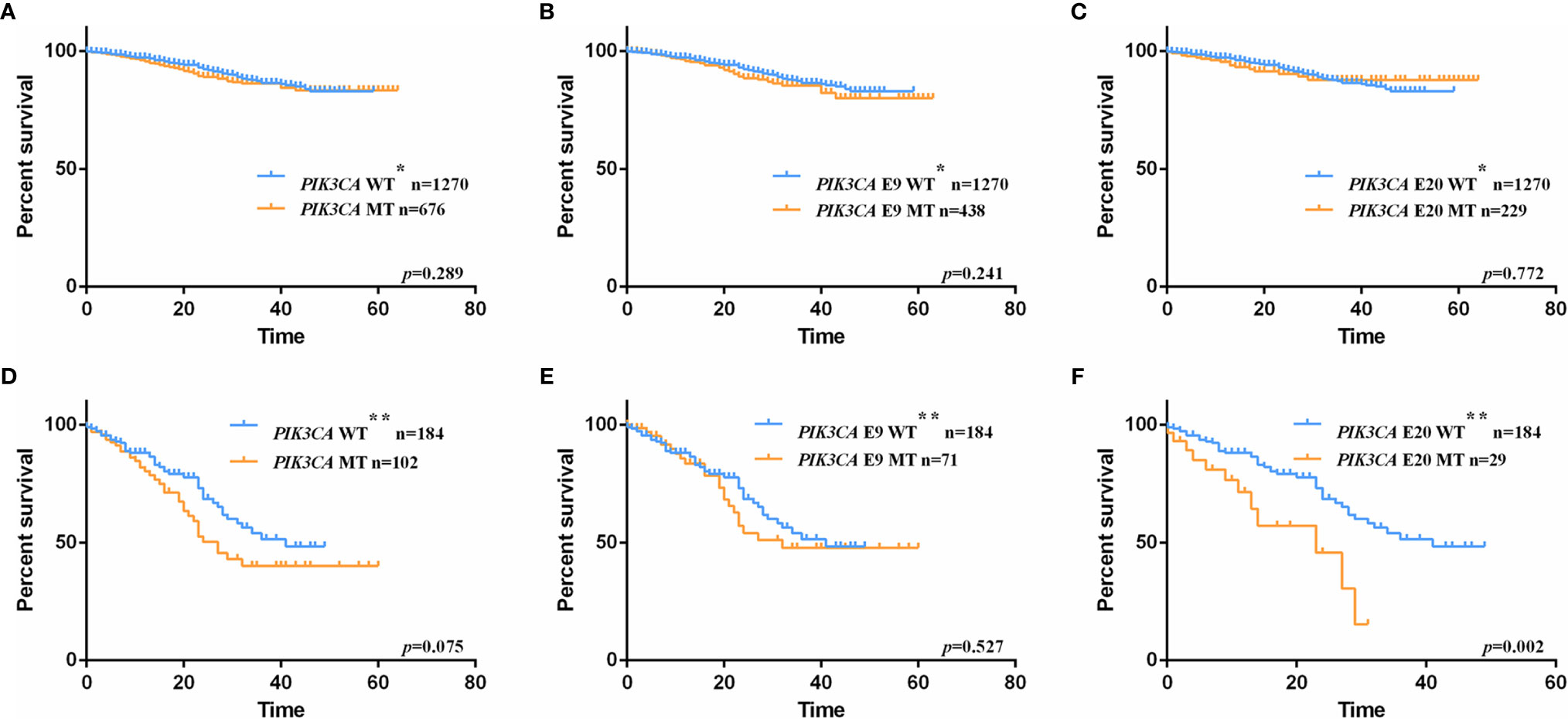
Figure 2 Kaplan-Meier plots of overall survival (OS) for CRC patients with and without PIK3CA mutation. (A) at all stage, patients with PIK3CA mutation vs. patients with wild-type allele, p-value = 0.289; (B) at all stage, patients with exon 9 mutation vs. patients with wild-type allele, p-value = 0.241; (C). at all stage, patients with exon 20 mutation vs. patients with wild-type allele, p-value = 0.772; (D) at stage IV, patients with PIK3CA mutation vs. patients with wild-type allele, p-value = 0.075; (E) at stage IV, patients with exon 9 mutation vs. patients with wild-type allele, p-value = 0.527; (F) at stage IV, patients with exon 20 mutation vs. patients with wild-type allele, p-value = 0.002. WT* represents stage I-IV wild-type patients with neither exon 9 nor exon 20 mutations. WT** represents stage IV wild-type patients with neither exon 9 nor exon 20 mutations.
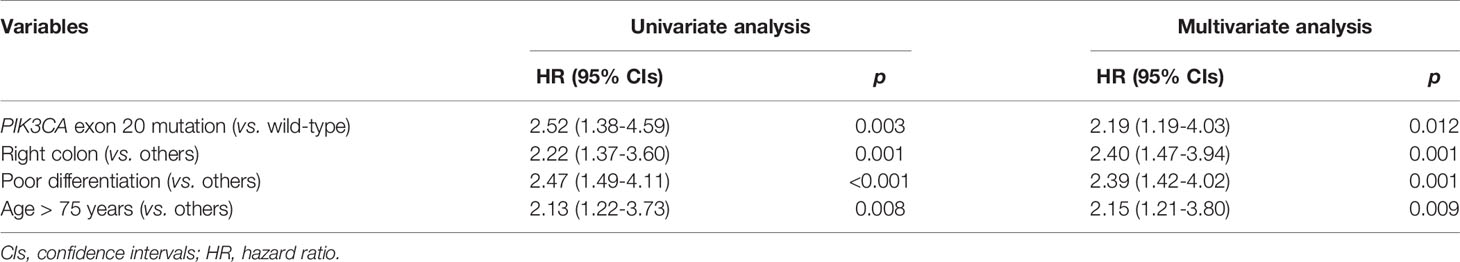
Table 3 Cox regression model associations between clinicopathologic characteristics and mortality in stage IV CRC patients.
In this study, the impact of PIK3CA mutations on 5-fluorouracil based chemotherapy treatment was evaluated in stage III patients who had received consistent chemotherapy in our hospital (clinicopathologic characteristics were shown in Table S5). In total, 377 patients were included in this sub-cohort, and 76 (20.2%) were observed to have disease progression (median follow-up: 13 months; median time-to relapse: 13 months). Univariate and multivariate logistic analyses were applied to screen independent factors relative to disease recurrence. In univariate and multivariate analysis, PIK3CA exon 9, and tumor sites were related to stage III patients’ disease relapse (p-value < 0.05 for all, Table 4), indicating that these variants were independent predictive markers in recurrence. Besides, PIK3CA exon 20 was found to be on the cusp of conventional statistical significance (OR = 2.15, 95% CIs = 0.99-4.65, p-value = 0.053) in univariate analysis. In multivariate analysis, it showed significantly associated with recurrence in stage III patients (OR = 3.89, 95%CIs = 1.66-9.10, p-value = 0.002). The same situation also occurred in patients’ gender and tumor differentiation (Table 4).
A nomogram incorporated these five variables of stage III patients’ disease recurrence was established (Figure 3A), and the concordance index of this nomogram was 0.685, which serves as a reasonable accuracy for prediction. The calibration curves also showed high coherence between the observed and predicted disease relapse in the nomogram (Figure 3B).
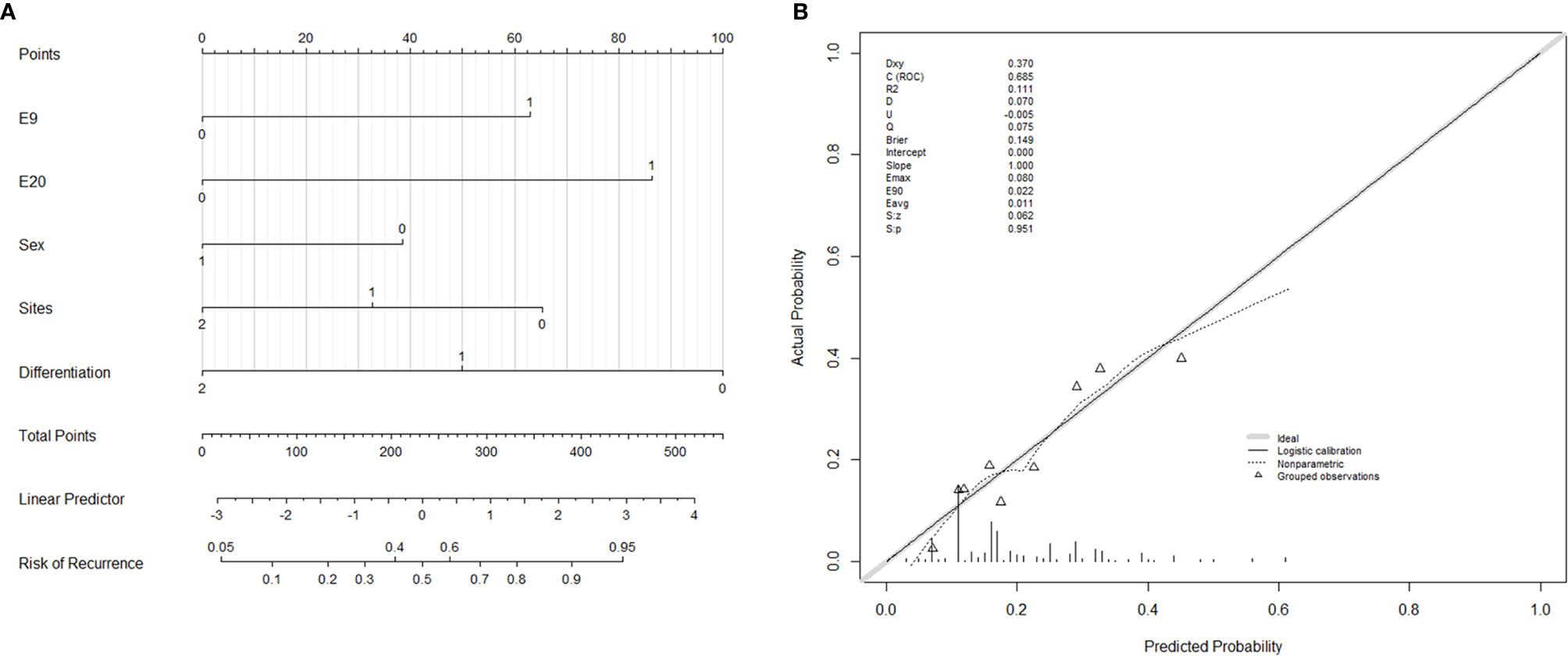
Figure 3 Nomogram and calibration curves for predicting the probability of disease recurrence in Stage III CRC patients. (A) E9: PIK3CA exon 9, 0 = wild-type, 1 = mutation; E20: PIK3CA exon 20, 0 = wild-type, 1 = mutation; Sex: 0 = female, 1 = male; Sites: tumor sites, 0 = rectum, 1 = left colon, 2 = right colon; Differentiation: tubular adenocarcinoma differentiation, 0 = poor differentiation, 1 = moderate differentiation, 2 = well differentiation. (B) Calibrate curve of nomogram. The C-index of this nomogram is 0.685.
We collected a cohort consist of 14 stage IV patients who received cetuximab treatment (Table 5) from the 5733 patients. All of these 14 patients carried wild-typed KRAS, NRAS, and BRAF. Five patients carried PIK3CA exon 9 mutations, and the rest nine patients carried wild-typed PIK3CA. For wild-typed PIK3CA patients, most of them (7/9) were evaluated as stable disease (SD), one patient was evaluated as partial response (PR), and one patient was evaluated as progressive disease (PD). For PIK3CA exon 9 mutant patients, four patients were evaluated as SD, and one was evaluated as PR. The disease control rate (DCR) was 88.9% (8/9) in wild-typed patients and 100% (5/5) in mutant patients. The progression-free survival (PFS) time ranged from 1.2 to 9.4 months in wild-type patients and from 1.3 to 12.2 months in mutant patients. The median PFS of the two groups did not reach a significant difference (3.6 months vs. 2.3 months, Log-rank test, p-value =0.513).
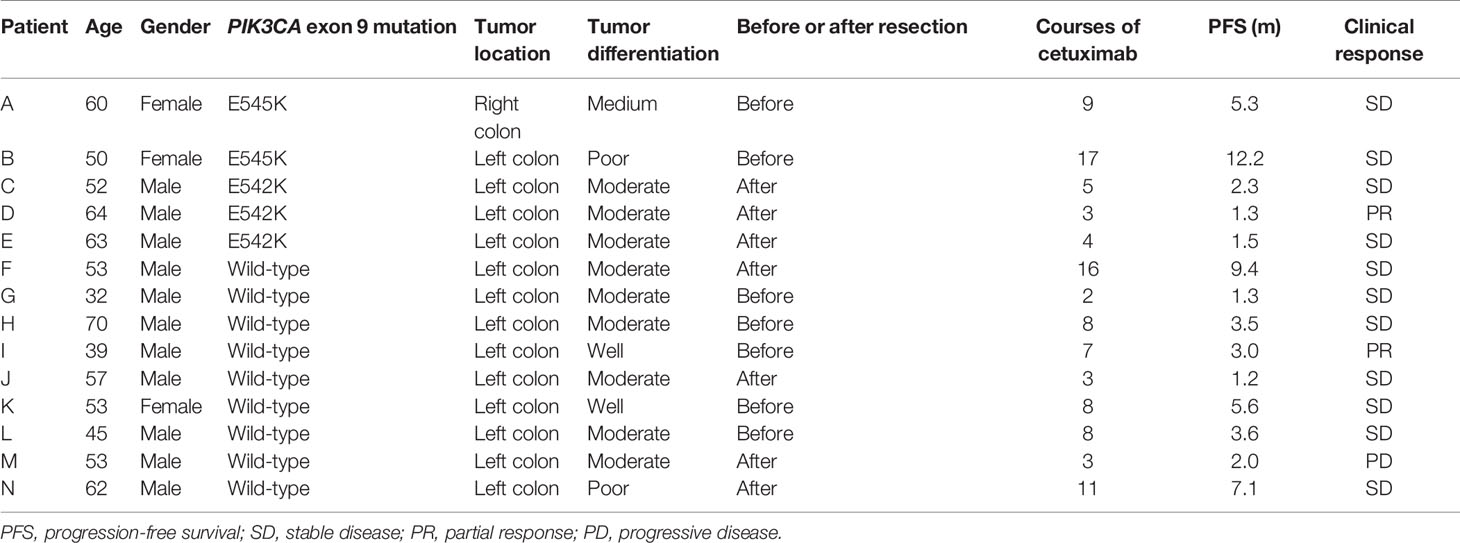
Table 5 Cetuximab response in 14 wild-type KRAS, NRAS and BRAF mCRC patients with/without PIK3CA exon 9 mutations.
In this study, we reported 58 types of PIK3CA mutation in Chinese CRC patients with two detecting methods. Most of the commercial kits for PIK3CA mutation detection were based on the ARMS-PCR technique, which can only detect five types of PIK3CA mutations (E542K, E545K, E545D, H1047L, and H1047R). In this case, there were only 12 mutation types in our real-world data that could be detected by such commercial kits, while 21.4% of mutated patients will be missed. Since we found that PIK3CA mutation was a valuable predictive biomarker in survival and disease recurrence, it is essential to achieve precise results in clinical testing. Here we proved that the HRM test was a more sensitive detection method than Sanger sequencing at a lower cost, which can also cover all the 58 mutation types we detected. For these reasons, it can be used as an auxiliary detection method.
Moreover, our study indicated that PIK3CA mutation was a neutral biomarker when considered patients at all stages (n = 1946). However, PIK3CA exon 20 was an adverse prognostic factor for CRC patients at stage IV (n = 213). We also found that PIK3CA mutation was a potential molecular biomarker for predicting resistance to 5-fluorouracil based chemotherapy regimens in stage III CRC patients. These findings may provide instructive information in clinical practice.
Mutations at PIK3CA could activate the PI3K/AKT signaling pathway, which is an essential factor that leads to the occurrence of various human malignancies (46–48). High mutation frequency was observed in multiple cancers, including breast cancer, bladder cancer, and colorectal cancer (8). Previous researches reported that the mutation rates of PIK3CA in CRC are 10-20% (7). In this study, we found that about 13.4% of Chinese CRC patients carried PIK3CA mutation, 8.7% were at exon 9, 4.5% were at exon 20, and 0.2% were found at both exons. The mutation rates of PIK3CA in Chinese CRC patients were varied from different studies, which might be due to the size of the cohort and detection technique (49, 50). Our research collected a large cohort of Chinese CRC patients and detected PIK3CA mutations with two different methods, which ensured that the mutation rates reported in this study are representative.
Though the mutation rate was relatively high, the prognostic effect of PIK3CA in CRC remains controversial. Some studies showed that PIK3CA mutation was associated with a shorter OS. For example, Ogino et al. found that PIK3CA mutations were an adverse prognostic factor in CRC patients at stage I-III, while this effect was only restricted to patients with wild-type KRAS (11). Besides, some studies indicated that mutations at different exons carried different prognostic effects. Research conducted by Farina et al. found that patients with PIK3CA exon 20 mutations conferred a more reduced disease-free survival (DFS) than patients with wild-type PIK3CA in stage III, and such an adverse effect was not observed in patients with PIK3CA exon 9 mutations (13). More studies, however, found that PIK3CA is a neutral prognostic factor that did not show any prognostic effects in CRC patients (14, 15). The reasons that lead to inconsistency may arise from different mutation sites, patient ethnicity, and the cohorts’ size. PIK3CA exon 9 and exon 20 were located in the helical and kinase domain separately. Mutations at these two domains show a gain of enzymatic function by activating the AKT signaling pathway to induce oncogenic transformation (51). A previous in vitro study suspected that the oncogenic mechanisms of these two domains were different. Briefly, mutations in the helical and kinase domain are both required to bind to p85 to induce the gain of function, while the former is required to interact with RAS-GTP simultaneously (52). The different mechanisms could be the reason that leads to the different prognostic effects of PIK3CA exon 9 and exon 20.
Chemotherapeutic treatment after surgical resection is a mainstream treatment in stage III CRC patients. 5-fluorouracil is the most widely used drug and is usually combined with other drugs as regimens, such as FOLFOX and XELOX. Recently, more and more CRC patients showed resistance to first-line chemotherapy (21–23). It is urgent to search for biomarkers that could predict chemotherapy resistance in CRC. A few studies suspected that PIK3CA mutation is a potential predictive marker in chemotherapy resistance, while the study cohorts were limited and the effect did not focus on stage III CRC patients (24), while patients at stage III with PIK3CA mutation were more likely to have disease recurrence or progression.
Nomogram is an effective method to visualize the regression model with various factors, which is valuable in clinical application. Most of the studies focus on constructing nomograms for survival prediction in CRC, including risk factors like TNM stage, age, and tumor location (53, 54). Our study is one of few that tried to construct a nomogram for predicting the recurrence risk of stage III CRC patients (55), and it is the first nomogram that involved PIK3CA mutation as a risk factor. The concordance index and the calibration curve indicated that the nomogram we constructed in this study served a well predictive function in clinical practice.
Previous studies indicated that mutation in PIK3CA exon 9 did not influence the outcome of anti-EGFR target therapy, while exon 20 mutation might suffer worse outcomes (16). Other studies, however, suspected that PIK3CA gene as a whole was a negative biomarker to cetuximab treatment in mCRC patients. The reason that leads to this inconsistency might be that these researches did not exclude KRAS, NRAS, and BRAF mutations during the analysis (17–20). In clinical treatment, bevacizumab treatment is more wildly used for patients with PIK3CA mutations. So in our study, only five mCRC patients with PIK3CA exon 9 mutations were treated with cetuximab. We found that the DCR in PIK3CA exon 9 mutation patients was higher than wild-type patients (100% vs. 88.9%), though the Kaplan-Meier analysis found that the PFS between these two subgroups did not differ significantly (Log-rank test, p-value =0.513). With these results, we suggested that PIK3CA exon 9 mutation did not affect cetuximab treatment among patients with wild-type KRAS, NRAS, and BRAF. For the low mutation rate, we could not obtain data of exon 20 mutation to cetuximab response. A larger cohort is required to clarify the role of PIK3CA in cetuximab resistance in the future.
There are some shortcomings in our study. First of all, though we collected a substantial cohort, patients involved in this study came from a single hospital, so there is a risk of bias in the cohort’s characteristics. Secondly, the concordance index in this nomogram was not very high (0.685). Additional researches were required to build a more reliable predictive model.
In this study, we uncover the PIK3CA mutation profile with a 5763 cohort. We found that 13.4% of Chinese CRC patients carried PIK3CA mutation with 58 types of mutants. And 21.4% of mutated patients could be missed if tested with commercial detection kits. We also found that the mutation trend of PIK3CA in our cohort remains steady from 2014 to 2018. In survival analysis, we found that PIK3CA exon 20 was an adverse biomarker in stage IV CRC patients, and Stage III patients with PIK3CA mutation were more likely to have disease recurrence than those with wild-type PIK3CA. We also found that PIK3CA exon 9 mutations is not a negative biomarker for wild-type KRAS, NRAS, and BRAF mCRC patients in cetuximab treatment.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The studies involving human participants were reviewed and approved by the ethics committee of the Sixth Affiliated Hospital of Sun Yat-sen University. The patients/participants provided their written informed consent to participate in this study.
XHF: Conceptualization, methodology, funding acquisition, writing-original draft. HL: methodology, investigation, data curation, writing-original draft. XJF: investigation, data curation, formal analysis. YZ: investigation, data curation; CW: data curation. ZC: investigation, data curation. XT: data curation, investigation. JH: data curation, investigation; YC: software, validation. YH: conceptualization, supervision, writing-review, and editing. All authors contributed to the article and approved the submitted version.
This work was supported by the National Key Research and Development Program of China (2017YFC1308800 to Ping Lan), National Natural Science Foundation of China (81971999 to Xin-hui Fu, 81201581 to Jian-ping Wang, 30872488 to Lei Wang), Science and Technology Achievements Transformation Project of Sun Yat-sen University (88000-18843232 to Xin-hui Fu), Young Teacher Training Program of Sun Yat-sen University (14YKPY31 to Xin-hui Fu), Science and Technology Planning Project of Guangdong Province (2012B031800355 to Xin-hui Fu), “985” Project of Sun Yat-sen University (4202037 to Jian-ping Wang), and China Scholarship Council (201706385049 to Xin-hui Fu) and National Key Clinical Discipline. The funder did not influence study design, data collection, and analysis, the decision to publish, or the manuscript preparation.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.595675/full#supplementary-material
Supplementary Figure 1 | The tendency of PIK3CA mutation rates from 2014 to 2018. (A) The mutation rate of PIK3CA exon 9 in CRC patients is rising, and the estimated value of 2019 is 9.6%. (B) The mutation rate of PIK3CA exon 20 in CRC patients is slightly increased, and the estimated value in 2019 is 4.7%. (C) The mutation rates of PIK3CA in CRC patients are increasing, and the estimated value of 2019 is 14.2%.
Supplementary Figure 2 | The detection limit of PIK3CA exon 9 mutations by the HRM test. The percentage of mutant-type spiked plasmid range from 50% to 0%, and in the 5% test, it still showed a clear bimodal image.
Supplementary Figure 3 | The detection limit of PIK3CA exon 20 mutations by the HRM test. The percentage of mutant-type spiked plasmid range from 50% to 0%, and in the 5% test, it showed a clear bimodal image.
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2018) 68(6):394–424. doi: 10.3322/caac.21492
2. Zheng RS, Sun KX, Zhang SW, Zeng HM, Zou XN, Chen R, et al. [Report of cancer epidemiology in China, 2015]. Zhonghua Zhong Liu Za Zhi (2019) 41(1):19–28. doi: 10.3760/cma.j.issn.0253-3766.2019.01.005
3. Zhang L, Cao F, Zhang G, Shi L, Chen S, Zhang Z, et al. Trends in and Predictions of Colorectal Cancer Incidence and Mortality in China From 1990 to 2025. Front Oncol (2019) 9:98. doi: 10.3389/fonc.2019.00098
4. Deng Y, Wang L, Tan S, Kim GP, Dou R, Chen D, et al. KRAS as a predictor of poor prognosis and benefit from postoperative FOLFOX chemotherapy in patients with stage II and III colorectal cancer. Mol Oncol (2015) 9(7):1341–7. doi: 10.1016/j.molonc.2015.03.006
5. Ahn TS, Jeong D, Son MW, Jung H, Park S, Kim H, et al. The BRAF mutation is associated with the prognosis in colorectal cancer. J Cancer Res Clin Oncol (2014) 140(11):1863–71. doi: 10.1007/s00432-014-1735-y
6. Benson AB, Venook AP, Al-Hawary MM, Cederquist L, Chen YJ, Ciombor KK, et al. NCCN Guidelines Insights: Colon Cancer, Version 2.2018. J Natl Compr Canc Netw (2018) 16(4):359–69. doi: 10.6004/jnccn.2018.0021
8. Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science (2004) 304(5670):554. doi: 10.1126/science.1096502
9. Liao X, Lochhead P, Nishihara R, Morikawa T, Kuchiba A, Yamauchi M, et al. Aspirin use, tumor PIK3CA mutation, and colorectal-cancer survival. N Engl J Med (2012) 367(17):1596–606. doi: 10.1056/NEJMoa1207756
10. Gray RT, Cantwell MM, Coleman HG, Loughrey MB, Bankhead P, McQuaid S, et al. Evaluation of PTGS2 Expression, PIK3CA Mutation, Aspirin Use and Colon Cancer Survival in a Population-Based Cohort Study. Clin And Trans Gastroenterology (2017) 8(4):e91. doi: 10.1038/ctg.2017.18
11. Ogino S, Nosho K, Kirkner GJ, Shima K, Irahara N, Chan SKT, et al. PIK3CA mutation is associated with poor prognosis among patients with curatively resected colon cancer. J Clin Oncol (2009) 27(9):1477–84. doi: 10.1200/JCO.2008.18.6544
12. Iida S, Kato S, Ishiguro M, Matsuyama T, Ishikawa T, Kobayashi H, et al. PIK3CA mutation and methylation influences the outcome of colorectal cancer. Oncol Lett (2012) 3(3):565–70. doi: 10.3892/ol.2011.544
13. Farina SA, Zeestraten EC, van Wezel T, van Lijnschoten G, van Eijk R, Dekker JW, et al. PIK3CA kinase domain mutation identifies a subgroup of stage III colon cancer patients with poor prognosis. Cell Oncol (Dordr) (2011) 34(6):523–31. doi: 10.1007/s13402-011-0054-4
14. Mouradov D, Domingo E, Gibbs P, Jorissen RN, Li S, Soo PY, et al. Survival in stage II/III colorectal cancer is independently predicted by chromosomal and microsatellite instability, but not by specific driver mutations. Am J Gastroenterol (2013) 108(11):1785–93. doi: 10.1038/ajg.2013.292
15. Ye ZL, Qiu MZ, Tang T, Wang F, Zhou YX, Lei MJ, et al. Gene mutation profiling in Chinese colorectal cancer patients and its association with clinicopathological characteristics and prognosis. Cancer Med (2019) 9(2):745–56. doi: 10.1002/cam4.2727
16. De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol (2010) 11(8):753–62. doi: 10.1016/S1470-2045(10)70130-3
17. Li QH, Wang YZ, Tu J, Liu CW, Yuan YJ, Lin R, et al. Anti-EGFR therapy in metastatic colorectal cancer: mechanisms and potential regimens of drug resistance. Gastroenterol Rep (Oxf) (2020) 8(3):179–91. doi: 10.1093/gastro/goaa026
18. Sartore-Bianchi A, Martini M, Molinari F, Veronese S, Nichelatti M, Artale S, et al. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res (2009) 69(5):1851–7. doi: 10.1158/0008-5472.CAN-08-2466
19. Perrone F, Lampis A, Orsenigo M, Di Bartolomeo M, Gevorgyan A, Losa M, et al. PI3KCA/PTEN deregulation contributes to impaired responses to cetuximab in metastatic colorectal cancer patients. Ann Oncol (2009) 20(1):84–90. doi: 10.1093/annonc/mdn541
20. Molinari F, Felicioni L, Buscarino M, De Dosso S, Buttitta F, Malatesta S, et al. Increased detection sensitivity for KRAS mutations enhances the prediction of anti-EGFR monoclonal antibody resistance in metastatic colorectal cancer. Clin Cancer Res (2011) 17(14):4901–14. doi: 10.1158/1078-0432.CCR-10-3137
21. Van der Jeught K, Xu HC, Li YJ, Lu XB, Ji G. Drug resistance and new therapies in colorectal cancer. World J Gastroenterol (2018) 24(34):3834–48. doi: 10.3748/wjg.v24.i34.3834
22. Longley DB, Allen WL, Johnston PG. Drug resistance, predictive markers and pharmacogenomics in colorectal cancer. Biochim Biophys Acta (2006) 1766(2):184–96. doi: 10.1016/j.bbcan.2006.08.001.502
23. Goldberg RM, Sargent DJ, Morton RF, Fuchs CS, Ramanathan RK, Williamson SK, et al. A randomized controlled trial of fluorouracil plus leucovorin, irinotecan, and oxaliplatin combinations in patients with previously untreated metastatic colorectal cancer. J Clin Oncol (2004) 22(1):23–30. doi: 10.1200/JCO.2004.09.046
24. Wang Q, Shi YL, Zhou K, Wang LL, Yan ZX, Liu YL, et al. PIK3CA mutations confer resistance to first-line chemotherapy in colorectal cancer. Cell Death Dis (2018) 9(7):739. doi: 10.1038/s41419-018-0776-6.510
25. Fu XH, Chen ZT, Wang WH, Fan XJ, Huang Y, Wu XB, et al. KRAS G12V Mutation is an Adverse Prognostic Factor of Chinese Gastric Cancer Patients. J Cancer (2019) 10(4):821–8. doi: 10.7150/jca.27899
26. Fu X, Huang Y, Fan X, Deng Y, Liu H, Zou H, et al. Demographic trends and KRAS/BRAF(V600E) mutations in colorectal cancer patients of South China: A single-site report. Int J Cancer (2019) 144(9):2109–17. doi: 10.1002/ijc.31973
27. Gradishar WJ, Anderson BO, Abraham J, Aft R, Agnese D, Allison KH, et al. Breast Cancer, Version 3.2020, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw (2020) 18(4):452–78. doi: 10.6004/jnccn.2020.0016
28. Andre F, Ciruelos E, Rubovszky G, Campone M, Loibl S, Rugo HS, et al. Alpelisib for PIK3CA-Mutated, Hormone Receptor-Positive Advanced Breast Cancer. N Engl J Med (2019) 380(20):1929–40. doi: 10.1056/NEJMoa1813904
29. Gymnopoulos M, Elsliger MA, Vogt PK. Rare cancer-specific mutations in PIK3CA show gain of function. Proc Natl Acad Sci U S A (2007) 104(13):5569–74. doi: 10.1073/pnas.0701005104
30. Blesinger H, Kaulfuss S, Aung T, Schwoch S, Prantl L, Rossler J, et al. PIK3CA mutations are specifically localized to lymphatic endothelial cells of lymphatic malformations. PloS One (2018) 13(7):e200343. doi: 10.1371/journal.pone.0200343
31. Ng PK, Li J, Jeong KJ, Shao S, Chen H, Tsang YH, et al. Systematic Functional Annotation of Somatic Mutations in Cancer. Cancer Cell (2018) 33(3):450–62. doi: 10.1016/j.ccell.2018.01.021
32. Dogruluk T, Tsang YH, Espitia M, Chen F, Chen T, Chong Z, et al. Identification of Variant-Specific Functions of PIK3CA by Rapid Phenotyping of Rare Mutations. Cancer Res (2015) 75(24):5341–54. doi: 10.1158/0008-5472.CAN-15-1654
33. Ikenoue T, Kanai F, Hikiba Y, Obata T, Tanaka Y, Imamura J, et al. Functional analysis of PIK3CA gene mutations in human colorectal cancer. Cancer Res (2005) 65(11):4562–7. doi: 10.1158/0008-5472.CAN-04-4114
34. Ross RL, Askham JM, Knowles MA. PIK3CA mutation spectrum in urothelial carcinoma reflects cell context-dependent signaling and phenotypic outputs. Oncogene (2013) 32(6):768–76. doi: 10.1038/onc.2012.87
35. Murugan AK, Hong NT, Fukui Y, Munirajan AK, Tsuchida N. Oncogenic mutations of the PIK3CA gene in head and neck squamous cell carcinomas. Int J Oncol (2008) 32(1):101–11. doi: 10.3892/ijo.32.1.101
36. Mankoo PK, Sukumar S, Karchin R. PIK3CA somatic mutations in breast cancer: Mechanistic insights from Langevin dynamics simulations. Proteins (2009) 75(2):499–508. doi: 10.1002/prot.22265
37. El-Habr EA, Levidou G, Trigka EA, Sakalidou J, Piperi C, Chatziandreou I, et al. Complex interactions between the components of the PI3K/AKT/mTOR pathway, and with components of MAPK, JAK/STAT and Notch-1 pathways, indicate their involvement in meningioma development. Virchows Arch (2014) 465(4):473–85. doi: 10.1007/s00428-014-1641-3
38. Vasan N, Razavi P, Johnson JL, Shao H, Shah H, Antoine A, et al. Double PIK3CA mutations in cis increase oncogenicity and sensitivity to PI3Kalpha inhibitors. Science (2019) 366(6466):714–23. doi: 10.1126/science.aaw9032
39. Vatte C, Al AA, Cyrus C, Chathoth S, Alsayyah A, Ahmad A, et al. Helical and kinase domain mutations of PIK3CA, and their association with hormone receptor expression in breast cancer. Oncol Lett (2019) 18(3):2427–33. doi: 10.3892/ol.2019.10565
40. Garcia-Solano J, Conesa-Zamora P, Carbonell P, Trujillo-Santos J, Torres-Moreno DD, Pagan-Gomez I, et al. Colorectal serrated adenocarcinoma shows a different profile of oncogene mutations, MSI status and DNA repair protein expression compared to conventional and sporadic MSI-H carcinomas. Int J Cancer (2012) 131(8):1790–9. doi: 10.1002/ijc.27454
41. Muller CI, Miller CW, Hofmann WK, Gross ME, Walsh CS, Kawamata N, et al. Rare mutations of the PIK3CA gene in malignancies of the hematopoietic system as well as endometrium, ovary, prostate and osteosarcomas, and discovery of a PIK3CA pseudogene. Leuk Res (2007) 31(1):27–32. doi: 10.1016/j.leukres.2006.04.011
42. Kachrilas S, Dellis A, Papatsoris A, Avgeris S, Anastasiou D, Gavriil A, et al. PI3K/AKT pathway genetic alterations and dysregulation of expression in bladder cancer. J BUON (2019) 24(1):329–37.
43. Cui W, Zheng S, Liu Z, Wang W, Cai Y, Bi R, et al. PIK3CA expression in diffuse large B cell lymphoma tissue and the effect of its knockdown in vitro. Onco Targets Ther (2017) 10:2239–47. doi: 10.2147/OTT.S129970
44. Zhou XK, Tang SS, Yi G, Hou M, Chen JH, Yang B, et al. RNAi knockdown of PIK3CA preferentially inhibits invasion of mutant PIK3CA cells. World J Gastroenterol (2011) 17(32):3700–8. doi: 10.3748/wjg.v17.i32.3700
45. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med (2015) 17(5):405–24. doi: 10.1038/gim.2015.30
46. Chalhoub N, Baker SJ. PTEN and the PI3-kinase pathway in cancer. Annu Rev Pathol (2009) 4:127–50. doi: 10.1146/annurev.pathol.4.110807.092311
47. Noorolyai S, Shajari N, Baghbani E, Sadreddini S, Baradaran B. The relation between PI3K/AKT signalling pathway and cancer. Gene (2019) 698:120–8. doi: 10.1016/j.gene.2019.02.076
48. Liu P, Cheng H, Roberts TM, Zhao JJ. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discovery (2009) 8(8):627–44. doi: 10.1038/nrd2926
49. Guo F, Gong H, Zhao H, Chen J, Zhang Y, Zhang L, et al. Mutation status and prognostic values of KRAS, NRAS, BRAF and PIK3CA in 353 Chinese colorectal cancer patients. Sci Rep (2018) 8(1):6076. doi: 10.1038/s41598-018-24306-1
50. Gao XH, Yu GY, Hong YG, Lian W, Chouhan H, Xu Y, et al. Clinical significance of multiple gene detection with a 22-gene panel in formalin-fixed paraffin-embedded specimens of 207 colorectal cancer patients. Int J Clin Oncol (2019) 24(2):141–52. doi: 10.1007/s10147-018-1377-1
51. Courtney KD, Corcoran RB, Engelman JA. The PI3K pathway as drug target in human cancer. J Clin Oncol (2010) 28(6):1075–83. doi: 10.1200/JCO.2009.25.3641
52. Zhao L, Vogt PK. Helical domain and kinase domain mutations in p110alpha of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proc Natl Acad Sci U S A (2008) 105(7):2652–7. doi: 10.1073/pnas.0712169105
53. Kim C, Kim WR, Kim KY, Chon HJ, Beom SH, Kim H, et al. Predictive Nomogram for Recurrence of Stage I Colorectal Cancer After Curative Resection. Clin Colorectal Cancer (2018) 17(3):e513–8. doi: 10.1016/j.clcc.2018.03.011
54. Li J, Gu J, Ma X, Li X, Liu X, Kang F, et al. Development and validation of a nomogram for predicting survival in Chinese han patients with resected colorectal cancer. J Surg Oncol (2018) 118(6):1034–41. doi: 10.1002/jso.25213
Keywords: colorectal cancer, mutation spectrum, nomogram, PIK3CA, the predictive value, HRM test, cetuximab
Citation: Fu X, Lin H, Fan X, Zhu Y, Wang C, Chen Z, Tan X, Huang J, Cai Y and Huang Y (2021) The Spectrum, Tendency and Predictive Value of PIK3CA Mutation in Chinese Colorectal Cancer Patients. Front. Oncol. 11:595675. doi: 10.3389/fonc.2021.595675
Received: 19 August 2020; Accepted: 04 March 2021;
Published: 26 March 2021.
Edited by:
Gong Zhang, Jinan University, ChinaReviewed by:
Jiayue Qin, Annoroad Gene Technology, ChinaCopyright © 2021 Fu, Lin, Fan, Zhu, Wang, Chen, Tan, Huang, Cai and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Huang, aHVhbmd5MjdAbWFpbC5zeXN1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.