
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol., 10 June 2021
Sec. Cancer Immunity and Immunotherapy
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.592854
This article is part of the Research TopicImmunotherapy for NSCLC with Oncogenic Driver VariantsView all 19 articles
 Yi Zheng1
Yi Zheng1 Shiying Hao2
Shiying Hao2 Cheng Xiang1
Cheng Xiang1 Yaguang Han1
Yaguang Han1 Yanhong Shang3
Yanhong Shang3 Qiang Zhen1
Qiang Zhen1 Yiyi Zhao1
Yiyi Zhao1 Miao Zhang1
Miao Zhang1 Yan Zhang1*
Yan Zhang1*Background: Immune checkpoint inhibitors have achieved breakthrough efficacy in treating lung adenocarcinoma (LUAD) with wild-type epidermal growth factor receptor (EGFR), leading to the revision of the treatment guidelines. However, most patients with EGFR mutation are resistant to immunotherapy. It is particularly important to study the differences in tumor microenvironment (TME) between patients with and without EGFR mutation. However, relevant research has not been reported. Our previous study showed that secreted phosphoprotein 1 (SPP1) promotes macrophage M2 polarization and PD-L1 expression in LUAD, which may influence response to immunotherapy. Here, we assessed the role of SPP1 in different populations and its effects on the TME.
Methods: We compared the expression of SPP1 in LUAD tumor and normal tissues, and in samples with wild-type and mutant EGFR. We also evaluated the influence of SPP1 on survival. The LUAD data sets were downloaded from TCGA and CPTAC databases. Clinicopathologic characteristics associated with overall survival in TCGA were assessed using Cox regression analysis. GSEA revealed that several fundamental signaling pathways were enriched in the high SPP1 expression group. We applied CIBERSORT and xCell to calculate the proportion and abundance of tumor-infiltrating immune cells (TICs) in LUAD, and compared the differences in patients with high or low SPP1 expression and wild-type or mutant EGFR. In addition, we explored the correlation between SPP1 and CD276 for different groups.
Results: SPP1 expression was higher in LUAD tumor tissues and in people with EGFR mutation. High SPP1 expression was associated with poor prognosis. Univariate and multivariate cox analysis revealed that up-regulated SPP1 expression was independent indicator of poor prognosis. GSEA showed that the SPP1 high expression group was mainly enriched in immunosuppressed pathways. In the SPP1 high expression group, the infiltration of CD8+ T cells was lower and M2-type macrophages was higher. These results were also observed in patients with EGFR mutation. Furthermore, we found that the SPP1 expression was positively correlated with CD276, especially in patients with EGFR mutation.
Conclusion: SPP1 levels might be a useful marker of immunosuppression in patients with EGFR mutation, and could offer insight for therapeutics.
Lung cancer has become one of the most serious threats to human health, and its global morbidity and mortality rank first among all cancer types (1). Approximately 85% of lung cancers are non-small cell lung cancer (NSCLC), and lung adenocarcinoma (LUAD) accounts for 40%–50% of NSCLC. In China, 50%–60% of patients with LUAD also have epidermal growth factor receptor (EGFR) mutation. Epidermal growth factor receptor-tyrosine kinase inhibitor (EGFR-TKI) targeted therapy has been recommended for treating patients with EGFR sensitive mutations and such therapy has significantly improved survival in advanced NSCLC (2, 3). However, EGFR-TKI resistance has been observed in patients with NSCLC, which is challenging the prognosis of the disease (4).
Recently, immune checkpoint inhibitors (ICIs), represented by programmed cell death-1 (PD-1) and programmed cell death-ligand 1(PD-L1) monoclonal antibodies, have presented a new approach for NSCLC treatment (5, 6). ICIs achieve long-term disease control in patients who have developed an anti-tumor response, by activating the body’s immune system for tumor cell recognition and removal (7). However, ICIs had poor efficacy and adverse effects in patients with EGFR mutation or secondary T790M mutation (8–10).
The IMpower 150 study found that a combination of atezolizumab, bevacizumab, and chemotherapy improved overall survival (OS) in patients with EGFR mutation (11). This study was the first randomized phase III trial of immunotherapy that showed a benefit in patients with EGFR mutation, suggesting that “primary drug resistance” could be reversed. In-vitro studies showed that the non-inflammatory tumor microenvironment (TME) changed in EGFR-mutated NSCLC after partial drug intervention, improving the efficacy of immunotherapy (12). Hence, there might be a connection between the EGFR-mutated NSCLC immune microenvironment and the mechanism of primary resistance to ICIs. Exploring the microenvironment characteristics may improve our knowledge of drug resistance mechanism and offer clues to reversing resistance.
Osteopontin (OPN, encoded by SPP1) is a secreted phosphorylated glycoprotein, which is produced by T, NK and other immune cells, myeloid cells, osteoblasts, bone cells, epithelial cells, etc. (13, 14). It is also a kind of multifunctional cytokine. Previously, we found that lung adenocarcinoma cells induced M2 polarization of macrophages through SPP1, and activation of T cells were observed after SPP1 silencing (15). However, the role of SPP1 mediated immunosuppression in patients with EGFR mutations remains unclear. What is more, the B7 family is important in regulating T cell immune response. PD-1 and B7-H3 (CD276) are both members of B7/CD28 family and had similar effects in TME (16). The up-regulated of CD276 expression can promote immune escaping of tumor cells, including inhibit the proliferation of T cells, reduce the secretion of IFN-γ, tumor necrosis factor-alpha (TNF-α), and other cytokines (16). As a co-inhibitory molecule of T cell, CD276 is an attractive target for cancer immunotherapy (17, 18).
In this study, we explored the TME of patients with EGFR mutation, which has been highlighted for its potential impact on the resistance of ICIs. We analyzed SPP1 expression in LUAD with or without EGFR mutation, and explored its association with clinicopathologic characteristics and patient outcomes. We further evaluated the differences in the tumor-infiltrating immune cells (TICs) in the immune microenvironment between groups with different levels of SPP1 expression, and those with or without EGFR mutation. In addition, the correlation between SPP1 and CD276 for different groups was compared. A study workflow is presented in Figure 1.
LUAD patient datasets were downloaded from The Cancer Genome Atlas (TCGA, https://portal.gdc.cancer.gov/) (19, 20), including transcriptome RNA-seq gene expression profiles (Level 3), and clinical information. A total of 497 tumor tissues and 54 adjacent tissues were included. We compared SPP1 protein expression in 102 normal and 109 cancer tissues, using The Clinical Proteomic Tumor Analysis Consortium (CPTAC, https://proteomics.cancer.gov/programs/cptac) (21, 22). We collected the mutation information of TCGA cohort from UCSC Xena database (https://xenabrowser.net/datapages/) (23), in which there are 472 cases with mutation information available including 409 EGFR wild-type and 63 EGFR mutant cases. Additionally, survival of patients with LUAD was analyzed in relation to SPP1 expression in TCGA and CPTAC databases.
Univariate and multivariate cox regression analysis was performed in 435 patients (missing clinical information were excluded) to screen factors significantly associated with OS in TCGA. Gene Set Enrichment Analysis (GSEA) (http://software.broadinstitute.org/gsea/) was performed to determine the biological differences and pathways affected by differential expression of SPP1 (one-fourth cutoff) in Gene Set c2(c2.cp. Kegg.v7.0.symbols) and c5(c5.all.v7.0.symbols). The number of random sample permutations was set at 1000. The significance threshold was P < 0.05 and false discovery rate (FDR) < 0.25.
CIBERSORT(http://cibersort.stanford.edu/) is an immune cell infiltrating assessment analysis tool (24, 25). We assessed the influence of SPP1 expression and EGFR mutation to 22 types of immune cells by CIBERSORT. The filter criteria of each sample is set as the P < 0.05, which indicating that the inferred proportion of each TICs subtype are accurate and suitable for further analysis. We calculated the correlation between different immune cells, and marked those with P < 0.05. xCell is a gene signatures-based method, which performs cell type enrichment analysis from gene expression data for 64 immune and stroma cell types (26). To verify the results of CIBERSORT, we downloaded the results of immune cell abundance in TCGA LUAD by xCell algorithm from TIMER database (http://timer.cistrome.org/) (27). Besides, the correlation between SPP1 and CD276 for different groups was calculated using Spearman correlation coefficients.
Statistical analysis of data from TCGA and CPTAC were performed using R-3.6.1. The independent samples t-test or Wilcoxon’s rank sum-tests were used to compare continuous variables between two groups. Kruskal-Wallis one-way analysis of variance followed by a posthoc Kruskal-Dunn test with BH’s method for adjusting for multiple comparisons. Prism8 software was used to plot the survival curves using the Kaplan-Meier method, and the log-rank test was used to compare the survival curves. Uni- and multi-variate analyses were performed using Cox proportional hazard models, where P < 0.05 was considered statistically significant. The correlation between SPP1 and CD276 was calculated using Spearman’s correlation coefficient (R), with P < 0.01 was considered statistically significant.
LUAD cohorts consisted of a total of 477 patients in TCGA and 109 patients in CPTAC. The clinical characteristics between low and high SPP1 expression groups were listed in Table 1 and Supplementary Table 1. SPP1 expression was significantly higher in tumor tissues than in adjacent tissues, regardless of RNA or protein level (Figures 2A, B, P < 0.001). SPP1 expression was higher in EGFR-mutated tumor samples than in wild-type samples (Figure 2C, P = 0.017). The median level of SPP1 expression was used to dichotomize patients into high- or low- expression groups, and Kaplan-Meier survival analysis was performed separately for TCGA and CPTAC. Increased SPP1 expression was significantly correlated with poor OS, and the median OS of the TCGA cohort was 4.73 vs. 3.37 (Figure 2D; HR:1.48 95%CI 1.10-2.00; P= 0.009). Median OS was not achieved in CPTAC cohort, but differences were observed (Figure 2E; HR: 3.40 95%CI 1.15-10.08; P = 0.047). In addition, subgroup analysis of TCGA LUAD showed that patients with EGFR mutation in SPP1 high-expression group had a poor prognosis (Supplementary Figure 1A; HR: 1.62 95%CI 0.90-2.93; P = 0.055). There was no difference in survival between wild and mutant EGFR patients in SPP1 low-expression group (Supplementary Figure 1B; HR: 1.17 95%CI 0.56-2.44; P = 0.662).
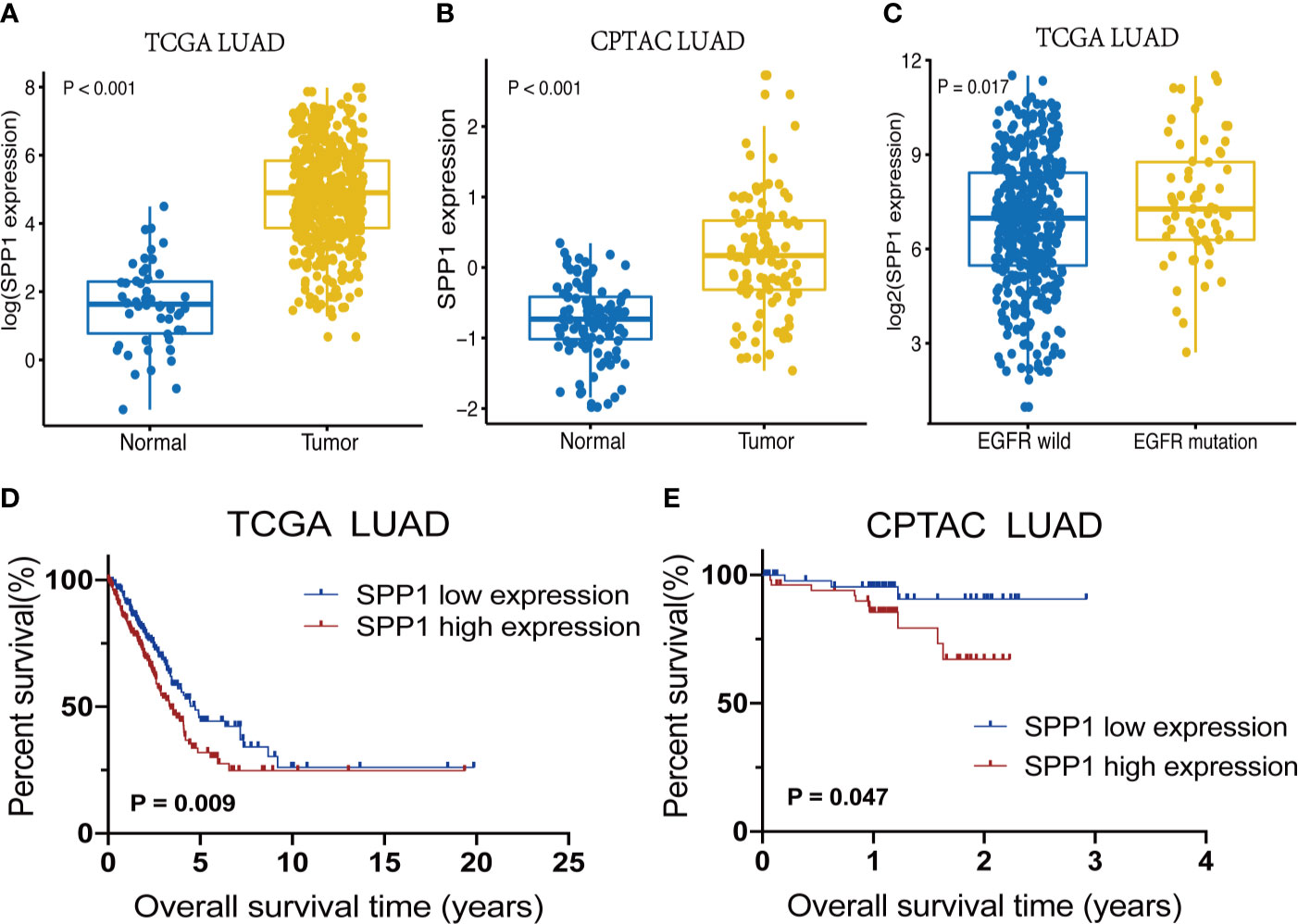
Figure 2 SPP1 expression differences and survival outcomes in LUAD. (A) SPP1 RNA expression levels in Normal vs. Tumor samples. (B) Expression of SPP1 protein in Normal vs. Tumor samples. (C) SPP1 RNA expression in EGFR wild-type vs. EGFR mutant samples. (D) Kaplan–Meier survival curves for high and low SPP1 expression groups in TCGA. (E) Kaplan–Meier survival curves for high and low SPP1 expression groups in CPTAC.
A Cox proportional hazards model including differentiation age, gender, stage, T stage, N stage, EGFR status and SPP1 expression was used. The resulted of univariate and multivariate analysis revealed that different SPP1 expression in patients was significantly associated with OS. Moreover, early tumor stage, and T stage are also the independent factors of favorable prognosis (Figures 3A, B).
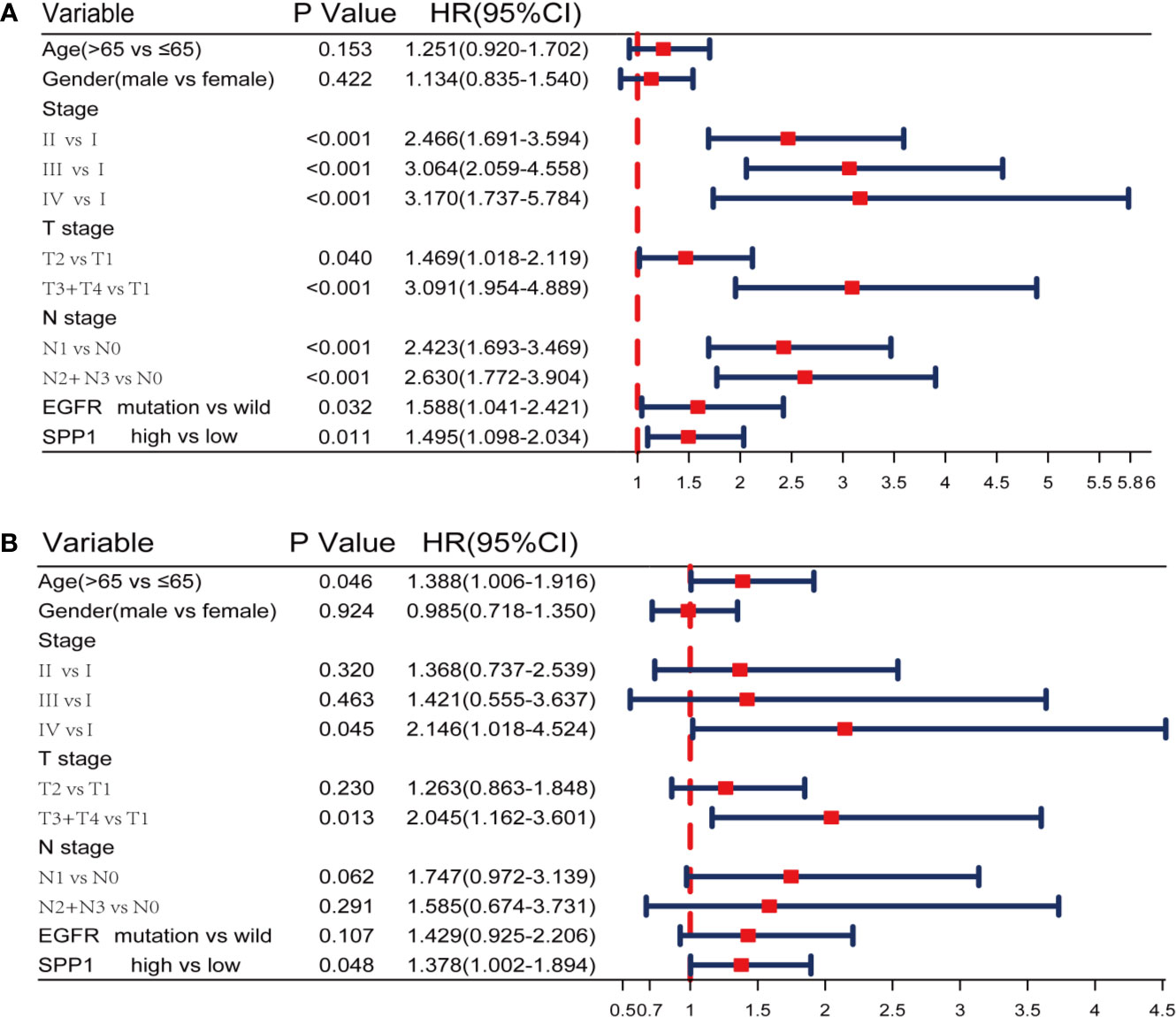
Figure 3 SPP1 was an independent prognostic biomarker in TCGA. (A, B) Univariate and Multivariate Cox analysis of SPP1 expression and other clinicopathological factors.
To interrogate potential signaling pathways related to SPP1 gene in LUAD, we used GSEA analysis (Figure 4). We found that the high SPP1 expression group was significantly associated with extracellular matrix (ECM) receptor interaction (NES = 1.737, P = 0.028), Fc gamma r mediated phagocytosis (NES = 1.813, P = 0.010), glycolysis and gluconeogenesis (NES = 1.770, P = 0.006), and the Toll like receptor (TLR) signaling pathway (NES = 2.025, P < 0.001). Meanwhile, GO analysis showed that the high SPP1 expression group was positively associated with integrin-mediated cell adhesion (NES = 2.008, P < 0.001), interleukin 6 production (NES = 1.960, P < 0.001), Nik Nf-Kappa B (NF-κB) signaling (NES = 1.961, P = 0.044), and phagocytosis (NES = 2.043, P < 0.001).
Previous analyses suggested that TICs could be markers of response to ICIs in several cancers (28). In this study, we examined how EGFR mutation and SPP1 expression are related to immune infiltration in LUAD. TCGA LUAD tumor samples (n = 477) were analyzed by CIBERSORT and 368 cases in the wild-type group and 63 cases in the mutant group met the CIBERSORT screening criteria. The results showed that EGFR mutations contribute to reducing the infiltration of CD4+ T cells, CD8+ T cells, M1 macrophages and other immune effector cells in the TME (Figure 5A). Taken together, these results indicate that EGFR mutations confer immunosuppressive effects. Additionally, 207 cases in the SPP1 low-expression group and 233 cases in the SPP1 high-expression group met the screening criteria. The results show that high SPP1 expression may play a role in regulating macrophage polarization to the M2 phenotype, reducing TICs such as CD8+ T cells, B cells, follicular helper T cells, NK cells, and activated dendritic cells (Figure 5B). These results support the contentions that SPP1 promotes host tumor immune tolerance and immune escape. The correlation heat map (Figure 5C) reveals that the different TIC subpopulations are weakly or moderately correlated. Subgroup analysis showed that the proportion of CD8+ T cells infiltration was the highest in patients with wild-type EGFR in SPP1 low-expression group, and the lowest in patients with EGFR mutation in SPP1 high-expression group (Figure 5D, P < 0.001). The infiltration of M2 macrophages was the most in patients with EGFR mutation in SPP1 high-expression group, and the least in patients with wild type EGFR in SPP1 low-expression group (Figure 5E, P = 0.071).
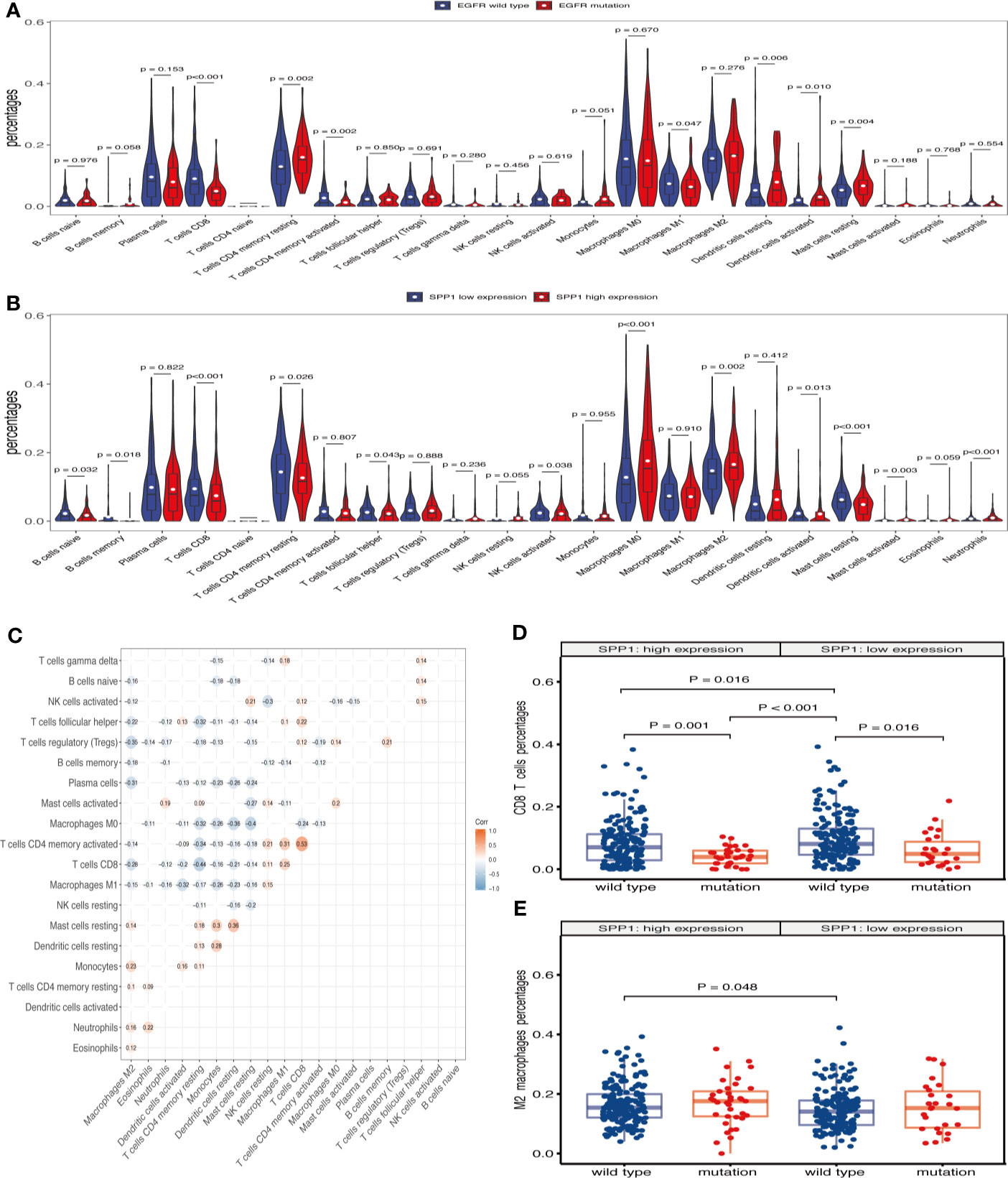
Figure 5 SPP1-related immune infiltration alteration. (A) Violin plot showing the ratio differentiation of 22 kinds of TICs in EGFR wild-type and mutant samples. Wilcoxon rank sum was used for the significance test. (B) Violin plot showing the ratio differentiation of 22 kinds of TICs in low and high SPP1 expression groups. (C) The correlation between different TICs subpopulations. (D) Differences in CD8+ T cells infiltration between EGFR mutation and wild-type patients with SPP1 high- or low-expression group. (E) Differences in M2 macrophages infiltration between EGFR mutation and wild-type patients with SPP1 high- or low-expression group.
xCell results showed that the abundance of CD8+ T cells was less (P = 0.009) and M2 macrophages was more (P = 0.073) in patients with EGFR mutation (Figure 6A). There were more CD8+ T cells (P = 0.036) and less M2 macrophages (P = 0.018) in SPP1 low-expression group (Figure 6B). The abundance of CD8+ T cells was the highest in EGFR wild-type patients with SPP1 low-expression group (Figure 6C). M2-type macrophages in SPP1 high-expression group with EGFR mutation was higher than those of the group with SPP1 low-expression and EGFR wild type (Figure 6D, P = 0.073). And we compared CD8 and M2 between the group (SPP1 high and EGFR mutation) and the group (SPP1 low and EGFR wild) in supplement Figure 2. Moreover, we found that the SPP1 expression was positively correlated with CD276, especially in patients with EGFR mutation (Figures 6E–G).
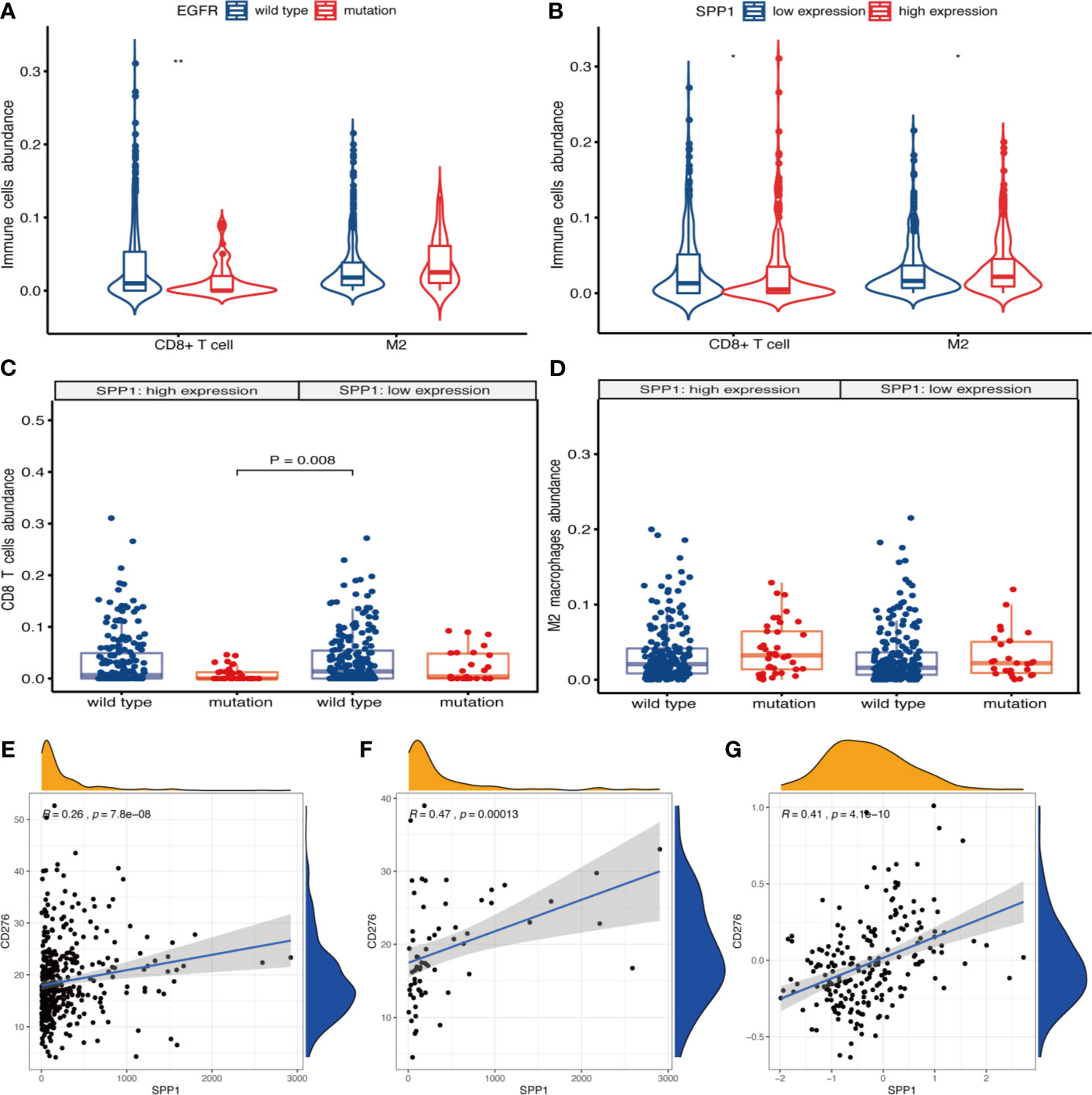
Figure 6 SPP1 promotes immunosuppressive microenvironment in patients with EGFR mutation. (A) xCell calculated the abundance of CD8+ T cells and M2 macrophages in EGFR wild-type and mutant samples. (B) The abundance of CD8+ T cells and M2 macrophages in different SPP1 expression groups were evaluated by xCell. (C) Differences in CD8+ T cells abundance between EGFR mutation and wild-type patients with SPP1 high- or low-expression group. (D) Differences in M2 macrophages abundance between EGFR mutation and wild-type patients with SPP1 high- or low-expression group. (E) The expression of SPP1 was correlated with CD276 in EGFR wild-type patients at mRNA level. (F) The expression of SPP1 was correlated with CD276 in EGFR mutation patients at mRNA level. (G) The expression of SPP1 was correlated with CD276 at the protein level. *P < 0.05, **P < 0.01.
Immunotherapy can eliminate tumor cells through the body’s immune system and bring long-term survival benefits to patients with NSCLC, significantly ushering in a new era of antitumor therapy. EGFR mutation is a predictor of the therapeutic effects of EGFR-TKIs in patients with LUAD (2, 3). However, it was once considered as a marker of immune resistance (9, 29–31). Here, we recognized the characteristics of TICs in LUAD using bioinformatics analysis. The immune-tolerant TME is more likely to correlate with patients harboring EGFR mutation. Additionally, the observed outcomes indicate that SPP1 may be a potential indicator for patients nonresponsive to ICIs.
In this study, we showed that the SPP1 expression is significantly higher in LUAD tumor tissues and in patients with EGFR mutation. Several previous studies showed SPP1 expression is directly related to CD8+ T cell activation (32, 33), and M2 macrophage polarization (15). Consistently, we found that SPP1 expression was negatively correlated with CD8+ T cell numbers and positively correlated with M2 macrophage numbers. By applying two methods for immunocyte enrichment or proportion analysis, we can find that M2 macrophages enriched more (CD8+ T cells enriched less) in SPP1 high group or EGFR mutation group, and the phenomenon seems more obvious in the group with both SPP1 high and EGFR mutation, which indicated that tumors with both SPP1 high and EGFR mutation tend to show immune evasion phenotype. Therefore, we speculate that SPP1 might cause immune resistance in NSCLC with EGFR mutation.
OPN involved in many physiological and pathological processes, including inflammatory, angiogenesis, tumor metastasis, immune suppression in TME (34, 35). GSEA analysis results indicate the involvement of extracellular matrix (ECM) receptor interaction, Fc gamma r mediated phagocytosis, TLR signaling pathway, integrin- mediated cell adhesion, interleukin 6 production, NF-κB signaling, and phagocytosis in the high SPP1 expression group. OPN is an important component of ECM, regulating matrix interactions and cell adhesion (13). It plays a key role in tumor cell migration by interacting with integrins and CD44 (36). The interaction between OPN and CD44 transmembrane glycoprotein suppressed the CD8+ T cell activation and IFN-γ production. OPN regulation by TLR and NF-κB signaling can reshape the immune inflammatory environment (37, 38). IL-6 binds to IL-6R and activates the Janus kinase (JAK)-STAT3 pathway and the JAK-SHP-2-mitogen-activated protein (MAP) kinase pathway via gp130 (39). Previously, it was shown that activation of the JAK/STAT3 pathway can suppress the immune response (40) and that this pathway is activated in patients with EGFR mutation (41). However, it is not clear whether anti-OPN agents, combined with ICIs, can reverse primary resistance in EGFR mutated NSCLC. Therefore, increased SPP1 expression is consistent with a role in immunosuppression, indicating a possible mechanism through which ICIs is ineffective in EGFR-mutated NSCLC.
Pre-clinical studies, involving co-culture of tumor cells and peripheral blood mononuclear cells, show that combined EGFR-TKI and anti-PD-1 antibody therapies do not produce synergistic tumor cell killing effects (42). Multiple clinical studies of EGFR-TKIs combined with ICIs were terminated due to poor efficacy or severe toxicity (43). As a target antibody of vascular endothelial growth factor (VEGF), bevacizumab not only has the effect of anti-angiogenesis, but promotes T cell activation and invasion in tumor tissues (44). Combined with other immunoregulatory drugs, ICIs could be more efficient for the treatment of EGFR-TKI resistance in patients with EGFR mutation. OPN act as an important chemokine and contributes to immune suppression in human colon cancer and other cancers (14, 30). CIBERSORT analysis indicated a significant relationship between SPP1 expression and increased levels of M2 macrophages infiltration, and reduced CD8 + T cell, activated NK cell and activated dendritic cell infiltration. Furthermore, the similarities between CD276 and other immune checkpoints (PD-1/PD-L1, CTLA4) have led to the targeting of CD276 in novel immunotherapy strategy (16, 17). We propose that a combination regimen using anti-OPN and ICIs may be a promising treatment option for LUAD with EGFR mutation, especially with high expression of SPP1. However, further studies should be conducted.
In summary, differences in TICs in patients with EGFR mutation and those with wild-type LUAD may affect the efficacy of ICIs. We observed that LUAD with EGFR mutated have less infiltration of anti-tumor immune cells, including CD8+ T cells, activated CD4+ T cells and M1 macrophages, and increased M2 macrophages infiltration. However, SPP1 likely has an essential influence on TICs, though combined with anti-OPN therapy, has the potential to reverse immune resistance in LUAD with EGFR mutation.
Publicly available datasets were analyzed in this study. This data can be found here: https://portal.gdc.cancer.gov/, https://proteomics.cancer.gov/programs/cptac.
YaZ and SH found the clinical problem and proposed the research direction. YiZ made use of the public databases for bioinformatics analysis. CX and YH searched literatures and analyzed data results. YS and QZ contributed to the original writing of the manuscript and data consolidation. YYZ and MZ undertook the task of modifying the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by grants from the Natural Science Foundation of Hebei Province, China (H2019106033).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.592854/full#supplementary-material
Supplementary Figure 1 | Effects of EGFR mutation and wild-type on prognosis of LUAD in different SPP1 expression groups in TCGA database. (A) Prognosis comparison of EGFR wild-type and mutant patients in SPP1 high expression group. (B) Prognosis comparison of EGFR wild-type and mutant patients in SPP1 low expression group.
Supplementary Figure 2 | CD8+ T cells and M2 macrophage differed between groups with high SPP1 expression and EGFR mutation and those with low SPP1 expression and wild-type EGFR. (A) Differences in CD8+ T cells infiltration calculated by CIBERSORT between SPP1 high-expression with EGFR mutation and SPP1 low-expression with EGFR wild type. (B) Differences in M2 macrophages infiltration calculated by CIBERSORT between SPP1 high-expression with EGFR mutation and SPP1 low-expression with EGFR wild type. (C) The difference in CD8+ T cells abundance assessed by xCell was compared between the group of SPP1 high-expression with EGFR mutation and SPP1 low-expression with EGFR wild type samples. (D) The difference in M2 macrophage abundance assessed by xCell was compared between the group of SPP1 high-expression with EGFR mutation and SPP1 low-expression with EGFR wild type samples.
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2018) 68:394–424. doi: 10.3322/caac.21492
2. Han JY, Park K, Kim SW, Lee DH, Kim HY, Kim HT, et al. First-SIGNAL: First-Line Single-Agent Iressa Versus Gemcitabine and Cisplatin Trial in Never-Smokers With Adenocarcinoma of the Lung. J Clin Oncol (2012) 30:1122–8. doi: 10.1200/JCO.2011.36.8456
3. Shi YK, Wang L, Han BH, Li W, Yu P, Liu YP, et al. First-Line Icotinib Versus Cisplatin/Pemetrexed Plus Pemetrexed Maintenance Therapy for Patients With Advanced EGFR Mutation-Positive Lung Adenocarcinoma (CONVINCE): A Phase 3, Open-Label, Randomized Study. Ann Oncol (2017) 28:2443–50. doi: 10.1093/annonc/mdx359
4. Lu X, Yu L, Zhang Z, Ren X, Smaill JB, Ding K. Targeting EGFR(L858R/T790M) and EGFR(L858R/T790M/C797S) Resistance Mutations in NSCLC: Current Developments in Medicinal Chemistry. Med Res Rev (2018) 38:1550–81. doi: 10.1002/med.21488
5. Reck M, Rodriguez-Abreu D, Robinson AG, Hui R, Csoszi T, Fulop A, et al. Pembrolizumab Versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl J Med (2016) 375:1823–33. doi: 10.1056/NEJMoa1606774
6. Tomasini P, Greillier L, Boyer A, Jeanson A, Barlesi F. Durvalumab After Chemoradiotherapy in Stage III Non-Small Cell Lung Cancer. J Thorac Dis (2018) 10:S1032–S36. doi: 10.21037/jtd.2018.04.61
7. Ribas A, Wolchok JD. Cancer Immunotherapy Using Checkpoint Blockade. Science (2018) 359:1350–55. doi: 10.1126/science.aar4060
8. Garassino MC, Cho BC, Kim JH, Mazieres J, Vansteenkiste J, Lena H, et al. Durvalumab as Third-Line or Later Treatment for Advanced non-Small-Cell Lung Cancer (ATLANTIC): An Open-Label, Single-Arm, Phase 2 Study. Lancet Oncol (2018) 19:521–36. doi: 10.1016/S1470-2045(18)30144-X
9. Gainor JF, Shaw AT, Sequist LV, Fu X, Azzoli CG, Piotrowska Z, et al. Egfr Mutations and ALK Rearrangements Are Associated With Low Response Rates to PD-1 Pathway Blockade in Non-Small Cell Lung Cancer: A Retrospective Analysis. Clin Cancer Res (2016) 22:4585–93. doi: 10.1158/1078-0432.CCR-15-3101
10. Haratani K, Hayashi H, Tanaka T, Kaneda H, Togashi Y, Sakai K, et al. Tumor Immune Microenvironment and Nivolumab Efficacy in EGFR Mutation-Positive Non-Small-Cell Lung Cancer Based on T790M Status After Disease Progression During EGFR-TKI Treatment. Ann Oncol (2017) 28:1532–39. doi: 10.1093/annonc/mdx183
11. Socinski MA, Jotte RM, Cappuzzo F, Orlandi F, Stroyakovskiy D, Nogami N, et al. Atezolizumab for First-Line Treatment of Metastatic Nonsquamous Nsclc. N Engl J Med (2018) 378:2288–301. doi: 10.1056/NEJMoa1716948
12. Sugiyama E, Togashi Y, Takeuchi Y, Shinya S, Tada Y, Kataoka K, et al. Blockade of EGFR Improves Responsiveness to PD-1 Blockade in EGFR-mutated Non-Small Cell Lung Cancer. Sci Immunol (2020) 5:eaav3937. doi: 10.1126/sciimmunol.aav3937
13. Lamort AS, Giopanou I, Psallidas I, Stathopoulos GT. Osteopontin as a Link Between Inflammation and Cancer: The Thorax in the Spotlight. Cells (2019) 8:815. doi: 10.3390/cells8080815
14. Anborgh PH, Mutrie JC, Tuck AB, Chambers AF. Role of the Metastasis-Promoting Protein Osteopontin in the Tumour Microenvironment. J Cell Mol Med (2010) 14:2037–44. doi: 10.1111/j.1582-4934.2010.01115.x
15. Zhang Y, Du W, Chen Z, Xiang C. Upregulation of PD-L1 by SPP1 Mediates Macrophage Polarization and Facilitates Immune Escape in Lung Adenocarcinoma. Exp Cell Res (2017) 359:449–57. doi: 10.1016/j.yexcr.2017.08.028
16. Castellanos JR, Purvis IJ, Labak CM, Guda MR, Tsung AJ, Velpula KK, et al. B7-H3 Role in the Immune Landscape of Cancer. Am J Clin Exp Immunol (2017) 6:66–75.
17. Picarda E, Ohaegbulam KC, Zang X. Molecular Pathways: Targeting B7-H3 (CD276) for Human Cancer Immunotherapy. Clin Cancer Res (2016) 22:3425–31. doi: 10.1158/1078-0432.CCR-15-2428
18. Kontos F, Michelakos T, Kurokawa T, Sadagopan A, Schwab JH, Ferrone CR, et al. B7-H3: An Attractive Target for Antibody-Based Immunotherapy. Clin Cancer Res (2021) 27:1227–35. doi: 10.1158/1078-0432.CCR-20-2584
19. Tomczak K, Czerwinska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): An Immeasurable Source of Knowledge. Contemp Oncol (Pozn) (2015) 19:A68–77. doi: 10.5114/wo.2014.47136
20. Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, et al. An Integrated Tcga Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell (2018) 173:400–16.e11. doi: 10.1016/j.cell.2018.02.052
21. Rudnick PA, Markey SP, Roth J, Mirokhin Y, Yan X, Tchekhovskoi DV, et al. A Description of the Clinical Proteomic Tumor Analysis Consortium (CPTAC) Common Data Analysis Pipeline. J Proteome Res (2016) 15:1023–32. doi: 10.1021/acs.jproteome.5b01091
22. Whiteaker JR, Halusa GN, Hoofnagle AN, Sharma V, MacLean B, Yan P, et al. Cptac Assay Portal: A Repository of Targeted Proteomic Assays. Nat Methods (2014) 11:703–4. doi: 10.1038/nmeth.3002
23. Goldman M, Craft B, Swatloski T, Cline M, Morozova O, Diekhans M, et al. The UCSC Cancer Genomics Browser: Update 2015. Nucleic Acids Res (2015) 43:D812–7. doi: 10.1093/nar/gku1073
24. Petitprez F, Vano YA, Becht E, Giraldo NA, de Reynies A, Sautes-Fridman C, et al. Transcriptomic Analysis of the Tumor Microenvironment to Guide Prognosis and Immunotherapies. Cancer Immunol Immunother (2018) 67:981–88. doi: 10.1007/s00262-017-2058-z
25. Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA. Profiling Tumor Infiltrating Immune Cells With CIBERSORT. Methods Mol Biol (2018) 1711:243–59. doi: 10.1007/978-1-4939-7493-1_12
26. Aran D, Hu Z, Butte AJ. xCell: Digitally Portraying the Tissue Cellular Heterogeneity Landscape. Genome Biol (2017) 18:220. doi: 10.1186/s13059-017-1349-1
27. Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, et al. Timer: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res (2017) 77:e108–e10. doi: 10.1158/0008-5472.CAN-17-0307
28. Klauschen F, Muller KR, Binder A, Bockmayr M, Hagele M, Seegerer P, et al. Scoring of Tumor-Infiltrating Lymphocytes: From Visual Estimation to Machine Learning. Semin Cancer Biol (2018) 52:151–57. doi: 10.1016/j.semcancer.2018.07.001
29. Dong ZY, Zhang JT, Liu SY, Su J, Zhang C, Xie Z, et al. EGFR Mutation Correlates With Uninflamed Phenotype and Weak Immunogenicity, Causing Impaired Response to PD-1 Blockade in Non-Small Cell Lung Cancer. Oncoimmunology (2017) 6:e1356145. doi: 10.1080/2162402X.2017.1356145
30. Li HY, McSharry M, Bullock B, Nguyen TT, Kwak J, Poczobutt JM, et al. The Tumor Microenvironment Regulates Sensitivity of Murine Lung Tumors to PD-1/PD-L1 Antibody Blockade. Cancer Immunol Res (2017) 5:767–77. doi: 10.1158/2326-6066.CIR-16-0365
31. Jia Y, Li X, Jiang T, Zhao S, Zhao C, Zhang L, et al. EGFR-Targeted Therapy Alters the Tumor Microenvironment in EGFR-Driven Lung Tumors: Implications for Combination Therapies. Int J Cancer (2019) 145:1432–44. doi: 10.1002/ijc.32191
32. Klement JD, Paschall AV, Redd PS, Ibrahim ML, Lu C, Yang D, et al. An Osteopontin/CD44 Immune Checkpoint Controls CD8+ T Cell Activation and Tumor Immune Evasion. J Clin Invest (2018) 128:5549–60. doi: 10.1172/JCI123360
33. Wei J, Marisetty A, Schrand B, Gabrusiewicz K, Hashimoto Y, Ott M, et al. Osteopontin Mediates Glioblastoma-Associated Macrophage Infiltration and is a Potential Therapeutic Target. J Clin Invest (2019) 129:137–49. doi: 10.1172/JCI121266
34. Denhardt DT, Noda M, O’Regan AW, Pavlin D, Berman JS. Osteopontin as a Means to Cope With Environmental Insults: Regulation of Inflammation, Tissue Remodeling, and Cell Survival. J Clin Invest (2001) 107(9):1055–61. doi: 10.1172/JCI12980
35. Pang X, Gong K, Zhang X, Wu S, Cui Y, Qian BZ. Osteopontin as a Multifaceted Driver of Bone Metastasis and Drug Resistance. Pharmacol Res (2019) 144:235–44. doi: 10.1016/j.phrs.2019.04.030
36. Maeda N, Maenaka K. The Roles of Matricellular Proteins in Oncogenic Virus-Induced Cancers and Their Potential Utilities as Therapeutic Targets. Int J Mol Sci (2017) 18:2198. doi: 10.3390/ijms18102198
37. Salvi V, Scutera S, Rossi S, Zucca M, Alessandria M, Greco D, et al. Dual Regulation of Osteopontin Production by TLR Stimulation in Dendritic Cells. J Leukoc Biol (2013) 94:147–58. doi: 10.1189/jlb.0412194
38. Friedmann-Morvinski D, Narasimamurthy R, Xia Y, Myskiw C, Soda Y, Verma IM. Targeting NF-kappaB in Glioblastoma: A Therapeutic Approach. Sci Adv (2016) 2:e1501292. doi: 10.1126/sciadv.1501292
39. Tanaka T, Narazaki M, Kishimoto T. IL-6 in Inflammation, Immunity, and Disease. Cold Spring Harb Perspect Biol (2014) 6:a016295. doi: 10.1101/cshperspect.a016295
40. Yu H, Pardoll D, Jove R. Stats in Cancer Inflammation and Immunity: A Leading Role for STAT3. Nat Rev Cancer (2009) 9:798–809. doi: 10.1038/nrc2734
41. Gao SP, Mark KG, Leslie K, Pao W, Motoi N, Gerald WL, et al. Mutations in the EGFR Kinase Domain Mediate STAT3 Activation Via IL-6 Production in Human Lung Adenocarcinomas. J Clin Invest (2007) 117:3846–56. doi: 10.1172/JCI31871
42. Chen N, Fang W, Zhan J, Hong S, Tang Y, Kang S, et al. Upregulation of PD-L1 by EGFR Activation Mediates the Immune Escape in EGFR-Driven NSCLC: Implication for Optional Immune Targeted Therapy for NSCLC Patients With EGFR Mutation. J Thorac Oncol (2015) 10:910–23. doi: 10.1097/JTO.0000000000000500
43. Ahn MJ, Sun JM, Lee SH, Ahn JS, Park K. Egfr TKI Combination With Immunotherapy in Non-Small Cell Lung Cancer. Expert Opin Drug Saf (2017) 16:465–69. doi: 10.1080/14740338.2017.1300656
Keywords: immune checkpoint inhibitors, lung adenocarcinoma, epidermal growth factor receptor, tumor microenvironment, secreted phosphoprotein 1, tumor-infiltrating immune cells
Citation: Zheng Y, Hao S, Xiang C, Han Y, Shang Y, Zhen Q, Zhao Y, Zhang M and Zhang Y (2021) The Correlation Between SPP1 and Immune Escape of EGFR Mutant Lung Adenocarcinoma Was Explored by Bioinformatics Analysis. Front. Oncol. 11:592854. doi: 10.3389/fonc.2021.592854
Received: 08 August 2020; Accepted: 17 May 2021;
Published: 10 June 2021.
Edited by:
Qian Chu, Huazhong University of Science and Technology, ChinaReviewed by:
Yuanning Guo, Clinical Research Institute, IsraelCopyright © 2021 Zheng, Hao, Xiang, Han, Shang, Zhen, Zhao, Zhang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Zhang, emhhbmd5YW4yMDE4MTBAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.