
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 15 July 2021
Sec. Cancer Molecular Targets and Therapeutics
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.592393
 Lun Li1
Lun Li1 Min Chen1
Min Chen1 Shuyue Zheng1
Shuyue Zheng1 Hanlu Li1
Hanlu Li1 Weiru Chi1
Weiru Chi1 Bingqiu Xiu1
Bingqiu Xiu1 Qi Zhang1
Qi Zhang1 Jianjing Hou1
Jianjing Hou1 Jia Wang1
Jia Wang1 Jiong Wu1,2*
Jiong Wu1,2*Background: Trastuzumab shows excellent benefits for HER2+ breast cancer patients, although 20% treated remain unresponsive. We conducted a retrospective cohort study to optimize neoadjuvant chemotherapy and trastuzumab treatment in HER2+ breast cancer patients.
Methods: Six hundred patients were analyzed to identify clinical characteristics of those not achieving a pathological complete response (pCR) to develop a clinical predictive model. Available RNA sequence data was also reviewed to develop a genetic model for pCR.
Results: The pCR rate was 39.8% and pCR was associated with superior disease free survival and overall survival. ER negativity and PR negativity, higher HER2 IHC scores, higher Ki-67, and trastuzumab use were associated with improved pCR. Weekly paclitaxel and carboplatin had the highest pCR rate (46.70%) and the anthracycline+taxanes regimen had the lowest rate (11.11%). Four published GEO datasets were analyzed and a 10-gene model and immune signature for pCR were developed. Non-pCR patients were ER+PR+ and had a lower immune signature and gene model score. Hormone receptor status and immune signatures were independent predictive factors of pCR.
Conclusion: Hormone receptor status and a 10-gene model could predict pCR independently and may be applied for patient selection and drug effectiveness optimization.
Breast cancer is one of the most common cancers and the leading cause of cancer death among females all over the world (1). HER2 gene amplification or protein overexpression accounts for approximately 20% of invasive breast cancer cases (2). In the absence of HER2-targeted therapies, HER2 positivity is associated with a worse prognosis (3). Although targeted therapy is currently readily accessible, patients with large tumor size and clinically positive lymph nodes still exhibit the worst prognosis. Neoadjuvant chemotherapy and trastuzumab (NACT), an approach whereby patients receive preoperative treatments, has been a standard treatment strategy for locally advanced breast cancer. NACT could allow for treatment response monitoring by measuring changes in tumor size (4). Administering NACT to patients may increase the possibility of surgery with negative margins by downsizing the tumor stage and may further increase the probability of achieving a pathological complete response (pCR), which could be translated into better long-term survival (5). Several clinical trials have confirmed improved survival in HER2+ breast cancer patients who achieved pCR (4, 6–8). However, there was a high proportion of HER2+ breast cancer patients who failed to respond. Thus, better characterization of these patients might be helpful for improving their prognosis. To select patients most likely not to respond to NACT and to choose optimal treatment regimens, the characteristics of those who did not achieve pCR should be well described and models for the prediction of response to NACT are required.
Thus, in the present study, we aimed to summarize the clinical and genetic characteristics of patients who did not achieve pCR among HER2+ breast cancer patients in the neoadjuvant setting. In addition, we aimed to develop a predictive model for pCR in order to optimize the effectiveness for trastuzumab based chemotherapy.
This study was approved by Ethics Committee in Fudan University Shanghai Cancer Center (FUSCC). Written consent for study participation was waived due to the retrospective nature of the study.
Only those patients who were HER2-positive (HER2 3+ by immunohistochemistry (IHC), or fluorescence in situ hybridization (FISH) positive) breast cancer who received neoadjuvant chemotherapy with or without trastuzumab were included. Neoadjuvant chemotherapy was defined as chemotherapy before surgery (breast and axillary surgery). We excluded those patients in which all breast cancer tissue was resected before chemotherapy as for these patients, judging postoperative chemotherapy response is impossible, and those with distant metastasis (M1) confirmed by biology.
Patients were identified from the breast cancer database constructed by FUSCC. All information was retrieved from medical records, and double-checked by three authors (LL, MC, SY-Z). Prognostic information was retrieved from the breast cancer database. Our hospital has its own department to follow up the patients, and we also checked their recent visits.
The variables analyzed included the patent’s clinical stage (tumor size, nodal status), preoperative receptor (ER/PR) status, HER2 status, Ki-67 expression, neoadjuvant chemotherapy regimens, use of trastuzumab, surgery, postoperative response, pathological stage (tumor size, nodal status), and adjuvant treatments (chemotherapy, radiotherapy, and endocrine therapy). Clinical and pathological stages were classified according to the AJCC version 8 (9). Clinical stage before biology was assessed based on the data from Magnetic Resonance Imaging, ultrasonography, and physical examination. In most cases, these were consistent with each other. For those patients with inconsistent data, averaged values were used. The description of the breast tumor such as redness and swelling were also used in the clinical stage. ER and PR status were defined as positive when expression was ≥1%. Ki-67 was defined positivity if it was ≥20%. The pCR was defined as no invasive tumor or axillary lymph nodes (ypT0/is ypN0) (8). The prognostic outcomes assessed were overall survival (OS) and disease-free survival (DFS).
The RNA sequence data from GSE37946 (10), GSE50948 (11), GSE66305 (12), and GSE130788 (13) cohorts were analyzed. The characteristics of the published cohorts were presented in Supplemental Table 1. The expression of each gene was normalized relative to GAPDH (Δ=log2(x/g), x: the gene expression level; g: the expression level for GAPDH) and standardized from 0 to 10 [(x-min) × 10/(max-min)]. Differentially expressed genes (DEGs) were defined with a threshold of p-value < 0.05. The immune signatures for B cells, T cells, natural killer (NK) cells, chemokine, metabolic, and cell proliferation pathways were built based on DEGs between pCR and non-pCR patients.
The clinical model for pCR was developed based on the multivariate logistic regression analysis results. The training clinical cohort was based on the retrospective analysis of all HER2+ breast cancer patients that received neoadjuvant chemotherapy with or without trastuzumab from 2012 to 2016 at our center. The validation cohort was based on patient data from 2017 in our institution. In addition, published cohorts GSE22358 (14), GSE50948 (11), and GSE130788 (13) from GEO datasets were used for the validation.
For genetic models, the same DEGs across different GEO databases were used for model development. Three databases (GSE37946, GSE50948, GSE66305) were chosen as training cohorts, and GSE130788 was used to validate the model.
Data was analyzed using SPSS version 21.0 (SPSS Inc., Chicago, IL, USA, version 20) and R software. Categorical variables were expressed using frequency, and continuous data was expressed using mean and standard variance, as well as median and interquartile range (IQR). The chi-squared test or Fisher’s exact test, also by univariate logistic regression analysis was used to analyze the relationship between variables and pCR. The Odds ratio (OR) with its 95% CI was calculated. Those clinical variables with significant associations in univariate analyses were further analyzed by multivariate logistic regression analyses. Sensitivity analyses were conducted to check whether the results were stable across different kinds of population. The clinical model was developed based on multivariate logistic regression analyses.
The DEGs in each dataset were analyzed using R software “limma” packages. The lasso regression methods were used for the development of the genetic models. The predictive abilities for clinical and genetic models were assessed by the Area Under Curve (AUC) value based on the receiver operating characteristic curve (ROC) analysis.
Survival was estimated by the Kaplan-Meier method. Univariate and multivariate analyses were performed using Cox proportional hazards regression and the hazard ratio (HR) with its 95% confidence interval (95% CI) was calculated. All important factors that might influence DFS and OS were considered for multivariate analysis. p < 0.05 was considered statistically significant.
In the training cohort, 600 patients who received neoadjuvant trastuzumab from the FUSCC were included with a median follow-up 1484 days (IQR 1176.25-1939 days). Forty-nine patients were lost to follow-up, 60 (10%) patients died, and 112 patients (18.67%) experienced an event (relapse or metastasis). Most patients were stage II (347/600, 57.83%) and stage III (244/600, 40.67%). Specifically, most patients were staged cT2 (330/600, 55%) and cT3 (144/600, 24%), cN1 (346/600, 57.67%). There were 346 ER+ (346/600, 57.67%) and 404 PR+ (404/600, 67.33%) patients. Overall, 528 patients (88%) were scored 3+ by IHC, and 545 patients (90.83%) expressed ≥20% Ki-67. Most patients used PC (paclitaxel and carboplatin, 364/600, 60.67%) as the chemotherapy regimen and 318 (53%) patients who did not complete all predefined cycles of neoadjuvant chemotherapy. 517 patients (86.17%) received trastuzumab, 525 patients (87.5%) underwent mastectomy, and the most frequent axillary treatment was axillary lymph node dissection (ALND) (479/600, 79.83%). We developed a validation cohort using data from 2017 in our hospital, and 165 HER2+ breast cancer patients who received neoadjuvant trastuzumab were included. The characteristics of the validation cohort are listed in Supplemental Table 2. All patients did not receive neoadjuvant pertuzumab, lapatinib.
In the FUSCC training cohort, the 1, 3, 5, 7-year survival rates were 96.3%, 84.2%, 79.5%, and 77.4% for DFS and 99.3%, 93.8%, 87% and 85.3% for OS, respectively. Those patients with stages cT4 (DFS: HR 2.82, 95% CI 1.14-6.98, p=0.03; OS: HR 3.75, 95% CI 1.07-13.16, p=0.04) and cN3 (DFS: HR 2.25, 95% CI 1.22-4.16, p=0.01; OS: HR 3.98, 95% CI 1.67-9.49, p=0.002) had worse survival than cT1 and cN0 (Supplemental Tables 3, 4). Age, BMI, menopausal status, preoperative ER, PR, HER2 status, and higher Ki-67 were not associated with better DFS or OS.
Patients with pT0 and pTis had the best and similar survival (DFS: HR 1.21, 95% CI 0.49-3.00, p=0.68; OS: HR 0.52, 95% CI 0.07-4.19, p=0.54). Postoperative pathological T and N stages were negatively associated with OS and DFS. Higher pathological T and N stages had worse OS and DFS. However, postoperative invasive ductal cancer with or without DCIS were associated with the worst survival (DFS: HR 3.39, 95% CI 2.08-5.53, p<0.001; HR 3.47, 95% CI 1.83-6.57, p<0.001; OS: HR 5.02, 95% CI 2.35-10.71, p<0.001; HR 5.37, 95% CI 2.12-13.63, p<0.001). Patients achieving pCR were associated with longer survival (DFS: HR 0.23, 95% CI 0.13-0.38, p<0.001; OS: HR 0.08, 95% CI 0.02-0.24, p<0.001) than those who did not achieve (Figures 1A, B).
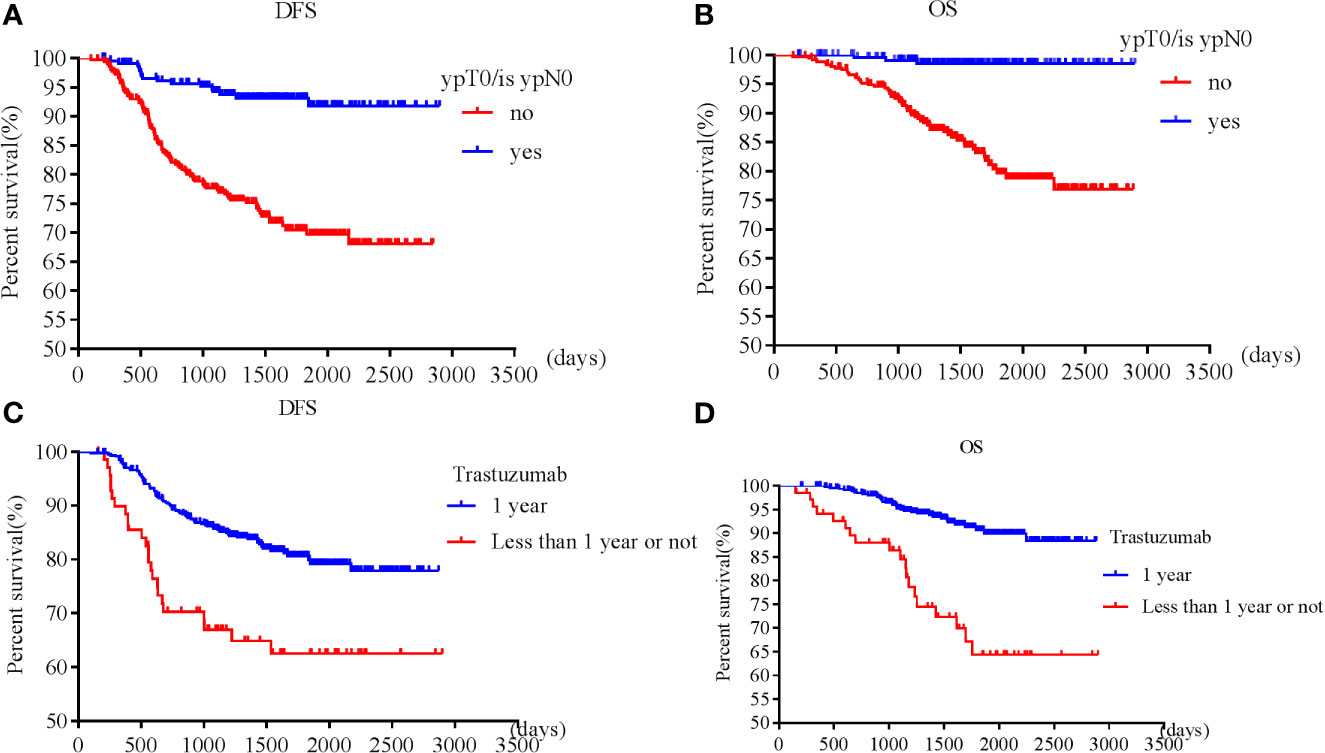
Figure 1 Survival according to pathological complete response [(A) disease free survival; (B) overall survival] and trastuzumab use [(C) disease free survival; (D) overall survival].
Patients who received adequate (12 months) trastuzumab therapy experienced longer survival than those with fewer than 12 months’ treatment or those without trastuzumab treatment (DFS: HR 0.49, 95% CI 0.29-0.85, p=0.01; OS: HR 0.30, 95% CI 0.15-0.60, p=0.001) (Figures 1C, D). pT, pN, postoperative residual invasive tumor, pCR and trastuzumab treatment for 1 year were all independent prognostic factors for DFS and OS, while cT, cN, preoperative ER, PR, and endocrine therapy were independent prognostic factors for OS.
When analyzing those who received neoadjuvant trastuzumab, pT, pN, postoperative residual invasive tumor, and pCR were all independent prognostic factors for DFS and OS. However, only pT2 and pT3 were independently associated with shorter DFS, while pT2 was independently associated with shorter OS (Supplemental Tables 5, 6). When analyzing those who received neoadjuvant wPC and trastuzumab, pN and pCR were all independent prognostic factors for DFS and OS (Supplemental Tables 7, 8).
The pCR rate was 39.8%, and did not differ across clinical T stages (χ2 = 3.16, p=0.37), N stages (χ2 = 0.19, p=0.98), age (χ2 = 0.66, p=0.72), BMI (χ2 = 0.58, p=0.45), or menopausal status (χ2 = 0.03, p=0.87) (Table 1). The pCR rates were lower in patients who were ER+ (χ2 = 39.37, p<0.001) and PR+ (χ2 = 32.52, p<0.001). IHC score of 1+ had the lowest pCR rate (χ2 = 11.31, p=0.003). Lower Ki67 (<20%) was associated with lower pCR possibility (χ2 = 3.99, p=0.046). Neoadjuvant trastuzumab was associated with higher pCR (χ2 = 19.03, p<0.001). Those patients who completed all predefined cycles of NACT (282/600, 47%) had a higher pCR rate than those who did not (46.45% vs. 33.96%, p=0.002). Among the neoadjuvant chemotherapy regimens, weekly paclitaxel and carboplatin (wPC) had the highest pCR rate (46.70%) and the anthracycline+taxanes regimen (TAC) regimens had the lowest pCR rate (11.11%). Paclitaxel and carboplatin administrated every three weeks (3wPC) and anthracycline followed taxanes (AC-T/P) had similar pCR rates (35.53% vs. 33.85%). Overall, negative ER and PR status was associated with higher pCR probability.
When analyzing those who received neoadjuvant trastuzumab, lower pCR rates were found in those who were ER+, PR+, HER2 1+, or those who did not complete all predefined cycles. Univariate and multivariate logistic analysis confirmed that ER, PR, HER2, and cycles of neoadjuvant chemotherapy were independent factors for pCR (Supplemental Tables 9, 10). When analyzing those who received neoadjuvant wPC and trastuzumab, only ER and PR statuses were associated with pCR by univariate logistic analysis. But only ER status was confirmed by multivariate logistic analysis (Supplemental Tables 11, 12).
Based on our multivariate logistic analysis, we developed a model that consisted of preoperative ER status, PR status, and HER2 status, chemotherapy regimen types and the number of cycles for those patients who received neoadjuvant trastuzumab and chemotherapy. The formula was:
y1=-0.695×ER-0.683×PR+0.688×HER2-0.210×chemotherapy+0.388×all-cycle-1.61 (Figure 2, supplemental materials).
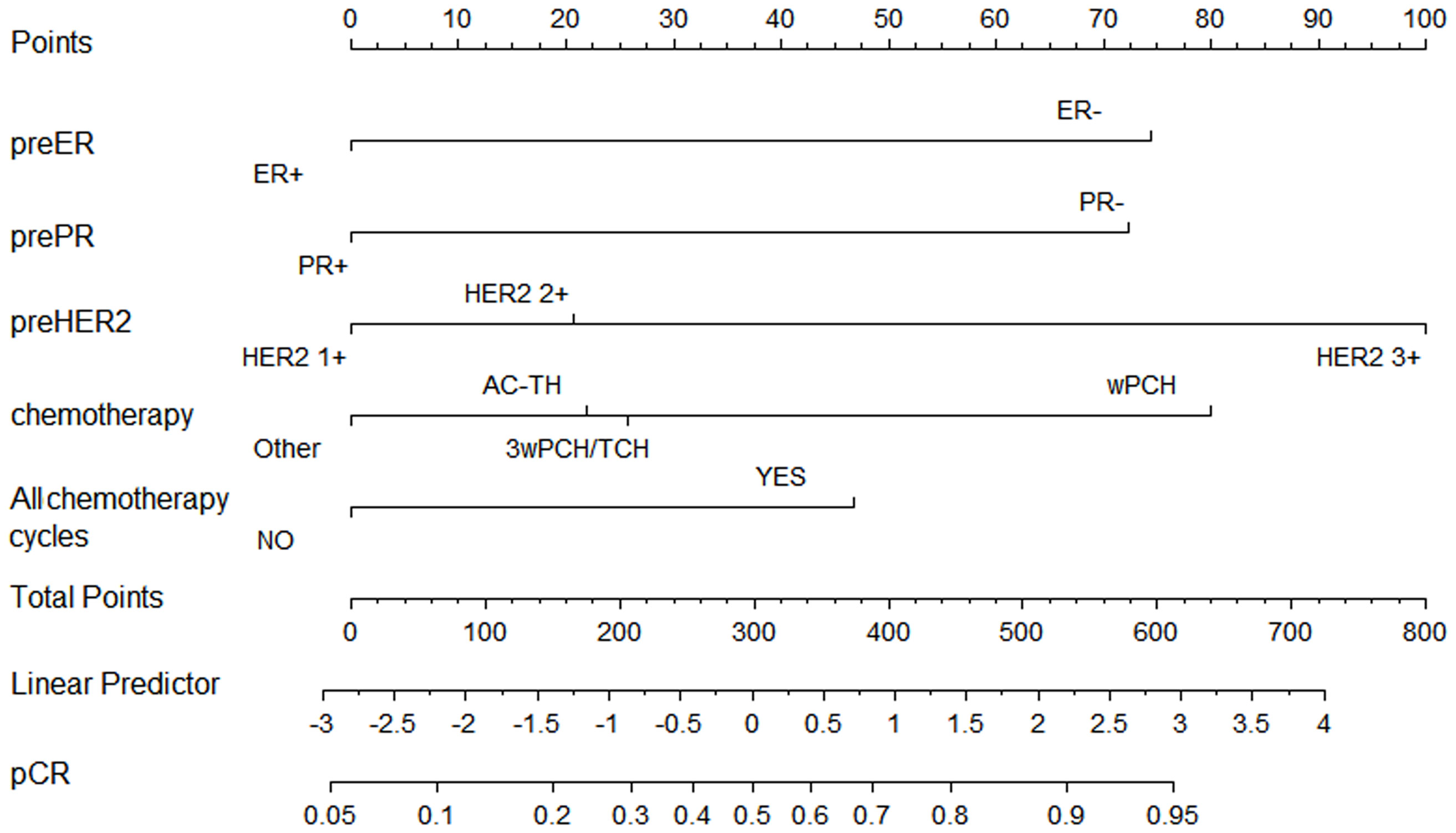
Figure 2 Nomogram to predict pathological complete response after preoperative chemotherapy and trastuzumab for HER2 positive breast cancer.
Although the AUC value was the highest for preoperative ER status (AUC = 0.63, p <0.001), the predictive value of this model (AUC =0.69, p <0.001) was higher than any of the single clinical factor (Supplemental Figure 1). Patients with a higher model score (>-0.5395) had a higher probability of achieving pCR in the training cohort (57.60% vs. 24.10%, p <0.001).
A validation cohort consisting of 165 patients in our institution was developed (Supplemental Table 2). In this validation cohort, ER negativity, PR negativity, HER2 positivity, higher Ki-67 expression, and trastuzumab use were associated with higher probability of pCR. However, in this cohort, the completion of all predefined cycles of chemotherapy was not associated with a higher pCR. The AUC was 0.65 (p=0.001) (Supplemental Figure 1B). Patients with higher clinical model scores (>-0.5395) were associated with higher pCR (58% vs. 29.6%, p = 0.001). This model was also validated using clinical data from GSE22358 and GSE130788 cohorts and the AUC were 0.75 (p=0.045) and 0.63 (p=0.045), respectively. All data from GSE22358, GSE37946, GSE50948, GSE76360 and GSE130788 were pooled together. A total of 268 patients were retrieved, and 116 patients (43.3%) achieved pCR. The predictive value of clinical factors was still stable (AUC = 0.61, p = 0.001).
Cluster analysis was used for the patients in the training cohort, and two clusters were found. In cluster 1, the pCR rate was 17.17% (34/198), among which most patients were ER+ (192/198) or PR+ (161/198). In cluster 2, the pCR rate was 53.43% (187/350), and most of patients were ER- (330/350) and PR- (344/350). Groups stratified according to the ER and PR status were: ER+PR+, ER+PR-, ER-PR+, ER-PR-. The pCR rates were 49.69% for ER-PR- (161/324), 66.67% for ER-PR+ (8/12), 36.84% for ER+PR-(21/57), and 20% for ER+PR+ (31/155). All ER+PR+ patients were in cluster 1, and all ER-PR- patients were in cluster 2 (Supplemental Figure 2). The ER-PR- subtype showed more sensitivity than the ER+PR+ subtype with wPC (57.64% vs. 25.69%), 3wPC (51.16% vs. 16.00%), AC-T/P (45.16% vs. 9.52%), TAC (25.00% vs. 0%). The pCR rates in ER-PR- were higher than that in ER+PR+ breast cancer based on data in the GSE22358 (15/19 vs. 0/10, p <0.001) and GSE50948 (26/43 vs. 1/7, p=0.023) cohorts.
However, the DFS and OS in ER-PR- patients did not differ from those with ER+PR+ HER2+ breast cancer patients. Among those who achieved pCR, no survival differences were observed between ER+PR+ and ER-PR- patients, but ER+PR+ patients showed significant survival benefits than ER-PR- patients among those who did not achieve pCR. Whether ER+PR+ or ER-PR- breast cancer patients, achieving pCR could improve survival (Figure 3).
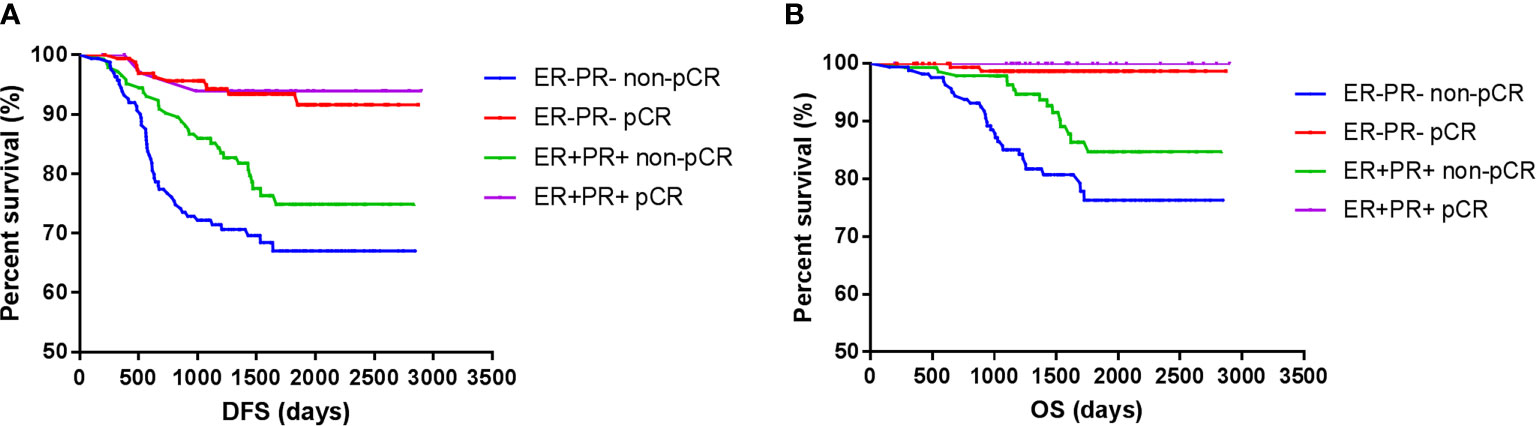
Figure 3 Survival according to pathological complete response and ER/PR. [(A) disease free survival; (B) over all survival].
Three databases (GSE37946, GSE50948, and GSE66305) were chosen as training cohorts, and 74 DEGs with the same trends in at least two databases were retrieved (Supplemental Table 13). These genes were T, B, and NK cell-associated genes, chemokines, genes in metabolic and cell proliferation pathways. T, B, NK cell, chemokine, genes in metabolic and cell proliferation pathways, and immune set 2 signatures were built (Supplemental Table 14). Low expressions of B cell, chemotaxis, immune set 2 and cell proliferation pathways were significantly correlated with lower probability of achieving pCR in all these datasets, while low expressions of T, NK, metabolic pathways were only correlated with lower probability of achieving pCR in GSE37946 and GSE50948 (Figure 4 and Supplemental Table 15). Using GSE130788 as a validation cohort, low expression levels of B, T, NK cell, chemotaxis, immune set 2, and metabolic pathways were associated with lower probability of achieving pCR (Supplemental Table 15).
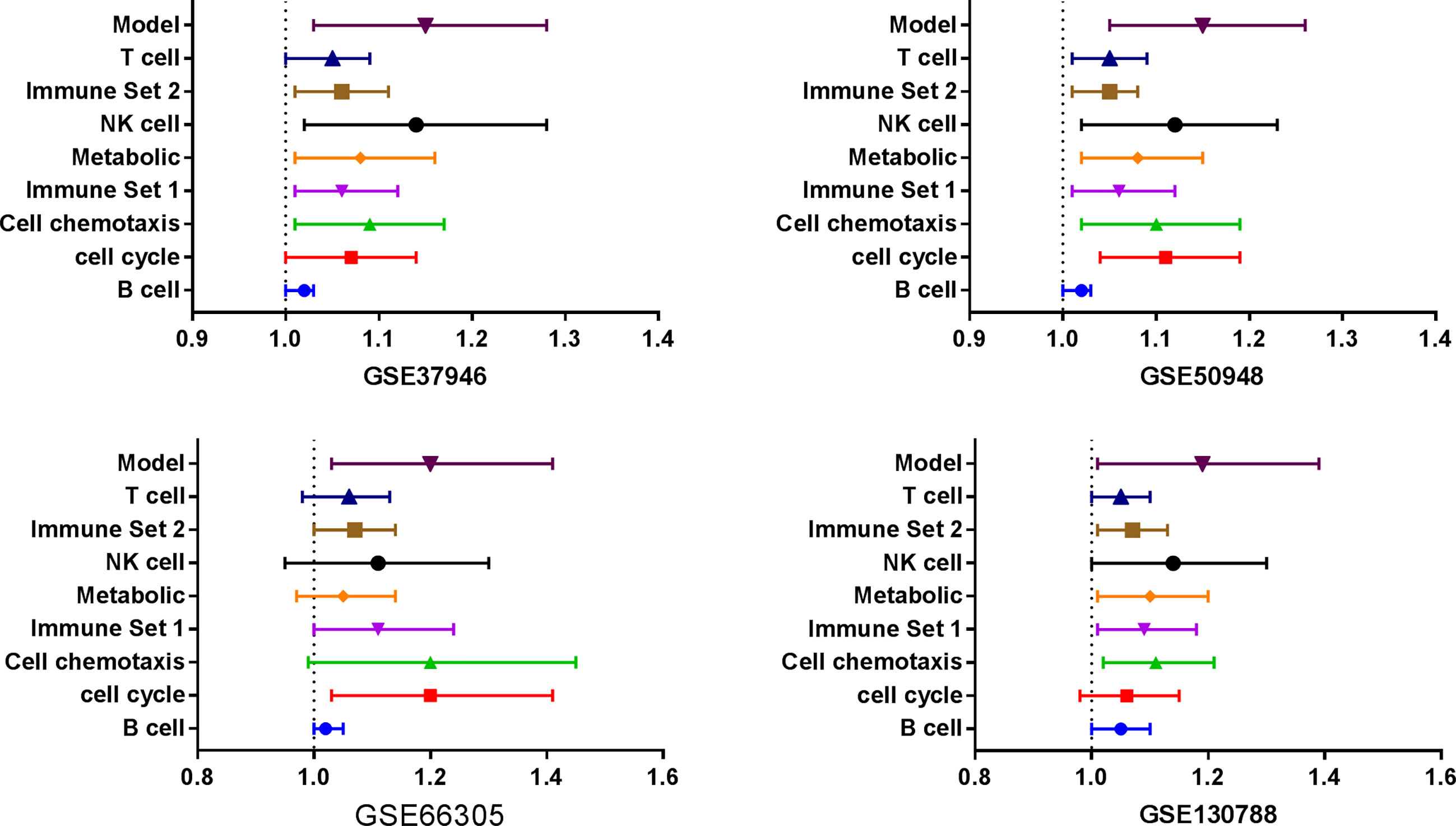
Figure 4 Immune signatures and model between pathological complete response and non- pathological complete response in different datasets.
Seventeen DEGs (GBP1, IGHM, IGKC, IGLC1, CXCL10, CXCL11, CXCL13, SP140, IGLJ3, IGK, UGT2B28, IGLL5, AC128677.4, IGKV1-17, IGKV1-37, IGKV1OR2-108, M24668) were identified, which showed lower expression in non-pCR patients (Table 2). These genes participate in the immune response, including B cell activation (IGKC, IGHM, IGLL5, IGLC1), cell chemotaxis (CXCL11, CXCL10, CXCL13), and phagocytosis (IGKV1-17, IGKC, IGLC1). These 17 genes might be classified as B-cell associated genes (IGHM, IGK, IGKC, IGKV1-17, IGKV1-37, IGKV1OR2-108, IGLC1, IGLJ3, IGLL5), T-cell associated signal transduction genes (GBP1, SP140), chemokines (CXCL10, CXCL11, CXCL13), and others (UGT2B28, AC128677.4, M24668). Four clusters could be developed based on these genes, and the higher total score was associated with higher pCR probability. The coefficient was calculated using lasso methods for each classification, and finally a formula was proposed:
y2 = (0.6×CXCL13 + 0.2×CXCL10 + 0.2×CXCL11) + (0.10×IGKV1OR2108 + 0.70×IGLJ3 + 0.20×IGKV117) + (0.5×GBP1 + 0.5×SP140) + (0.5×UGT2B28 + 0.5×AC128677.4).
This model showed a high predictive ability for pCR in the GSE37946 (AUC = 0.72, p = 0.008), GSE50948 (AUC = 0.71, p = 0.005), and GSE66305 (AUC = 0.90, p = 0.004) datasets. Using GSE130788 as a validation cohort, the AUC value was 0.72 (p = 0.04).
According to the ROC results, the best cutoff points were calculated. For those whose model score was less than 10, the pCR probability ranged from 0% to 28.6%, and for those whose model score was more than 20, the pCR probability ranged from 57.1% to 80%. After adjusting for ER and PR status, this model was still associated with pCR (Supplemental Table 16).
Each gene in genetic models was standardized from 0 to 10. There were four parts for genetic model, so the total score ranged from 0 to 40. There were two parts for clinical model (pathological and treatment factors), and the clinical models were standardized from 0 to 20. Multivariate logistic analyses were conducted to analyze the coefficient for clinical and genetic model (y1 and y2) in GSE22358, GSE37946, GSE50948 and GSE130788. The relative ratio (RR) between the coefficient for y2 and y1 ranged from 1 to 13. The best RR was 2, so a formula which combined clinical and genetic models was proposed:
y3=y1*20/3+y2*2/3.
The AUC values for the combination modes were 0.79 (p = 0.014) for GSE22358, 0.71 (p = 0.014) for GSE37946, 0.64 (p = 0.012) for GSE50948, 0.72 (p = 0.04) for GSE130788.
We further used all data in the GSE50948 and GSE130788 cohorts to analyze differences in the signatures and models between ER+PR+ and ER-PR- breast cancer patients (Supplemental Tables 17, 18). Based on these data, ER-PR- patients were associated with higher scores in B, T, NK cell, chemotaxis, immune set 2, and metabolic signatures. However, in the GSE37946 and GSE58984 datasets, no significant differences were observed between ER+PR+ and ER-PR- breast cancer patients (Supplemental Tables 19, 20), which suggested HR status and immune signatures were independent predictive factors for pCR. Analyzing the interaction across ER status, PR status, and the predictive model, no significant differences were found. Intratumoral lymphocytic infiltration and stromal lymphocytic infiltration was associated with increased scores in B, T, NK cell, chemotaxis, immune set 2, and metabolic signatures, as well as the model score in GSE58984 (Table 3).
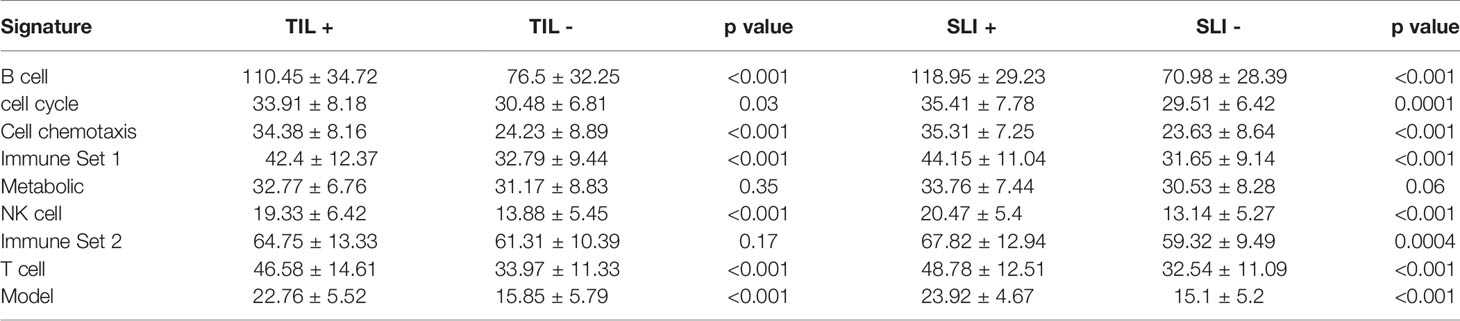
Table 3 The signatures in GSE58984 between different intratumoral lymphocytic infiltration (TIL) and stromal lymphocytic infiltration (SLI) statuses.
Our study developed a clinical model for pCR in HER2+ breast cancer patients receiving neoadjuvant chemotherapy and trastuzumab using real world data, which consisted of preoperative ER status, PR status, HER2+ expression, chemotherapy regimens, and completion of all predefined cycles of chemotherapy. Although this clinical model integrated more clinical variables and was validated by an independent cohort and other published cohorts, this model was limited in the ability for the prediction of pCR. We also explored the predictive values of genetic data on pCR, and developed genetic models for those who received trastuzumab. This genetic model has a wider applicability and allows greater generalization.
Years ago, women with HER2+ breast cancer were associated with worse prognosis (15). But trastuzumab drastically changed outcomes and substantial evidence has now shown that trastuzumab treatment could improve patient survival. A Cochrane review study based on 8 studies (11,991 patients) showed that trastuzumab-based regimens significantly improved OS (HR 0.66, 95% CI 0.57-0.77) and DFS (HR 0.60, 95% CI 0.50-0.71) (16). Tang et al. showed that 12 months of trastuzumab significantly reduced overall mortality (HR 0.71, 95% CI 0.62-0.81) as compared to those who received <12 months of therapy (17). All these data were about those who received adjuvant trastuzumab. Our study confirmed adequate (12 months) trastuzumab therapy could prolong the survival of HER2+ breast cancer patients in terms of DFS (HR 0.49, 95% CI 0.29-0.85) and OS (HR 0.30, 95% CI 0.15-0.60) in the neoadjuvant setting.
Substantial evidence from clinical trials confirmed that achieving pCR was correlated with long-term survival benefits in HER2+ breast cancer (18). Cortazar et al. showed that pCR was associated with long-term outcome (event-free survival (EFS): HR 0.39, 95% CI 0.31-0.50; OS: 0.34, 0.24-0.47) for HER2+ breast cancer patients (8). Our retrospective cohort of 600 HER2+ breast cancer cases in the neoadjuvant treatment setting confirmed pCR was associated with better survival irrespective of the patients’ clinical and pathological characteristics, and could be used to predict these patients’ prognosis.
For HER2+ breast cancer patients in neoadjuvant setting, the standard regimen to date was chemotherapy and trastuzumab (AC-TH, AC-PH, or TCbH), in which the pCR rates of 50% or more can be achieved (15, 19–21). TCbH has often been preferred by clinicians due to its lower cardiotoxicity profile, in which weekly regimens are much more effective than the three-week schedule (69% vs. 41%; p = 0.03) (15, 21). In our study, we separately analyzed the influence of neoadjuvant chemotherapy, trastuzumab use and completion of chemotherapy cycles on pCR, and found wPC, trastuzumab, and a sufficient number of cycles completed were associated with higher pCR rates. The weekly PC has higher pCR rate than the three-week schedule (46.70% vs. 35.53%, p=0.08). Interesting, the AC-T/P has a similar pCR rate as 3wPC (33.85% vs. 35.53%, p=0.84), which was lower than wPC (46.70% vs. 33.85%, p=0.06). The TAC regimen had the lowest pCR rate, which may have been due to the impossibility of trastuzumab use, which decreased the pCR rate. Our study showed that trastuzumab use could at least double the pCR rates (OR 3.47, 95% CI 1.93-6.22). Although the pCR rate achieved 39.8%, most patients did not achieve pCR. Thus, the identification of patients not sensitive to treatment is critical for improving their prognosis. Based on our study, ER and PR positivity, low Ki-67 and HER2 were associated with lower pCR. For those patients, how to improve the efficacy of NACT warrants further research.
Several nomograms have been developed to predict pCR for breast cancer patients, however, most considered HER2-negative patients alone (22). Using these to predict the pCR for HER2+ breast cancer patients might not be suitable. There were lots of studies that aimed to evaluate the predictive values of some clinical and pathological factors, such as hormone receptor (23, 24), trastuzumab use (24). The nomograms based on these factors were proposed (25, 26). The first nomogram to predict pCR for HER2+ breast cancer patients treatment with trastuzumab was developed by Jankowski et al. (25). This model was based on 101 patients and could be applied to those with trastuzumab without considering chemotherapy regimens and cycles. Thus, this model could not be generalized to conditions in which patients did not receive trastuzmab, had received different chemotherapy regimens, or to stage T4 patients. Fujii et al. developed a nomogram which consisted of ER, PR, HER2 FISH ratio, inflammatory breast cancer and neoadjuvant systemic therapy regimen based on 793 patients (26). The strength of this model is that it could be used to predict pCR for those who received pertuzumab, but this model did not consider different chemotherapy regimens. However, available models could not fully utilize available clinical and pathological information. Our model included ER, PR, and HER2 status, trastuzumab use, neoadjuvant chemotherapy, and chemotherapy cycles, which has a wider applicability and allows greater generalization. Our validation cohort and testing in the published cohort confirmed the stability of this model.
There were lots of studies that aimed to identify the biomarkers with the best association with pCR in response to trastuzumab (27–30), but there are currently no conclusive biomarkers for patient response to trastuzumab (31). Tanioka et al. developed a multi-dimensional genomic analysis to integrate DNA mutations, DNA copy number aberrations, and RNA transcriptional expression with clinical variables using prospectively collected frozen tissue samples from a Phase III trial to predict pCR (28). This model reported higher predictive values, but is difficult to apply into routine clinical practice. Fernandez-Martinez et al. (32) found a total of 215 genomic variables were significantly associated with pCR. Among these genetic models, cases that achieved a pCR had evidence of an activated immune response, including g T-Cell, B-Cell, and inflammatory signatures (33). However, these models were not easy to apply into routine clinical practice and need to be confirmed by other cohorts. Thus we analyzed the available RNA sequence databases in order to develop a genetic model which could be confirmed by other published cohorts. Seventy-four genes showed same trends in at least two databases, and 17 genes were higher in pCR patients in all three databases. These 17 genes could be classified as T, B, NK cell, cell proliferation, cell metabolism, and chemotaxis signatures. Across four different datasets, these immune signatures were higher in pCR patients than in non-pCR. The 17 DEGs identified were associated with the immune response in breast cancer patients, including B cell activation (IGKC, IGHM, IGLL5, IGLC1), cell chemotaxis (CXCL11, CXCL10, CXCL13), phagocytosis (IGKV1-17, IGKC, IGLC1). CXCL13, which might suppress regulatory T (Treg)-mediated immune response and activation of adaptive antitumor humoral responses (34) and thus, might be associated with higher pCR in neoadjuvant setting (35). The CXC subfamily chemokines (CXCL10, CXCL11, and CXCL13) might induce the migration mainly of T cells and B cells (36). IGHM and IGLJ3, B cell-specific immunoglobulin, have been reported as adaptive immunity effector genes (37). Studies have shown that higher CXCL13 and SP140 expression were associated with increased recurrence free survival (RFS) for those receiving adjuvant trastuzumab (38). However, the roles of these genes in trastuzumab resistance have not been studied. In our study, we developed a predictive model that also consisted of genes related to T, B cell and cell chemokines. Our model could predict the pCR and multivariate logistic analysis showed this model could predict pCR independently.
In our study, we identified HR status and immune cell infiltrations as two independent predictive factors for pCR in HER2+ breast cancer. Plenty of evidence has shown that ER is an independent predictive factor in breast cancer (28, 39–41) and inhibition of the ER might enhance responses to trastuzumab in HER2 positive breast cancer cells (40). However, clinical study did not confirm the additional benefits by concurrent targeting of ER and HER2. NSABP B-52 study randomly assigned 315 patients to receive neoadjuvant therapy consisting of docetaxel, carboplatin, trastuzumab, and pertuzumab with or without estrogen deprivation therapy. This study failed to show superior benefits in pCR (40.9% vs. 46.1%, p = 0.36) (42). Several immune-cell signatures have been reported, which were highly associated with pCR, but the models were not adequately cross-validated by independent cohorts or by other researchers (43, 44). Our model was validated in four datasets and showed stable predictive ability. Nonetheless, this model was not confirmed by real-time quantitative PCR validation. But we have an independent cohort which was used to test the model. Meanwhile, we found that the immune signatures and model scores were different for ER+PR+ and ER-PR- patients, which might suggest that HR status might be associated with immune cell infiltrations. We then analyzed GSE58984 and GSE37946 in this regard, this was not well confirmed. Likewise, no interaction between HR and immune signatures for pCR were found. This might be due to different lymphocytic infiltrations, whose levels were associated with a higher probability of pCR and immune signature scores. Continuous stromal-tumor-infiltrating lymphocytes (TIL)s (OR 1.03, 95% CI 1.02-1.05, p<0.001) and intratumoral-TILs (OR 1.09, 95% CI 1.04-1.15, p<0.001) were significantly associated with pCR (45). In our study, higher TILs were associated with higher immune signatures and model scores, which further validated the stability of our model. T and B cell infiltrations have often been reported in previous studies (43–45). In our study we found that NK cell activation might be associated with pCR. The NK cell signatures were higher in pCR patients across the three datasets tested. Recruitment of NK cells and subsequent induction of antibody-dependent cell-mediated cytotoxicity (ADCC) contributed to this beneficial effect (46).
For those who were unlikely to achieve pCR with standard treatment, how to improve the possibility of pCR was still required further research. NeoSphere study showed patients given pertuzumab and trastuzumab plus docetaxel had a highest pCR (49/107) compared with those given trastuzumab plus docetaxel (31/107, p=0.014) (30). PEONY study showed the pCR rates were 39.3% (86 of 219) in the pertuzumab and trastuzumab group and 21.8% (24 of 110) in the trastuzumab group (p = 0.001) (47). This study confirmed the additional benefits in pCR when adding pertuzumab for ER-PR- breast cancer patients, but not for ER+ and/or PR+ breast cancer patients. CALGB 40601 study (43) randomly assigned 305 patients to paclitaxel, trastuzumab, lapatinib (THL), paclitaxel plus trastuzumab (TH), or paclitaxel plus lapatinib (TL). The additional use of laptinib did not increase the pCR rate (56% vs. 46%, p = 0.13). Subgroup analysis confirmed similar results in HR+ breast cancer (41% vs. 41%), but THL was associated with higher pCR rates than TH (79% vs. 54%, p = 0.01). TEAL study (48) showed the proportion of patients with RCB 0 or I in the T-DM1, lapatinib, nab-paclitaxel group was higher than that in trastuzumab, pertuzumab and paclitaxel group (100% vs. 62.5%, p = 0.0035). ER- patients in both groups achieved RCB-0 or RCB-I, but all ER+ patients in the experimental arm achieved RCB 0-I versus 25% in the standard arm (p = 0.0035). In total, for ER- breast cancer patients, adding pertuzumab or lapatinib might improve the pCR rates, but for ER+ breast cancer, this might not work. For ER+ breast cancer patients, how to improve the pCR rates deserved further research. Although bispecific antibody (BsAb) simultaneously targeting both PD1 or PD-L1 and HER2 inhibited tumor growth (49–51), no clinical studies in neoadjuvant setting were conducted. Whether adding immunotherapy to enhance immune recognition of tumors with low immune markers deserved further research. Even for those who did not achieve pCR, adjuvant T-DM1 might reduce the risk of recurrence as compared to trastuzumab (52).
Our study was the largest retrospective cohort in Chinese HER2+ breast cancer patients in the neoadjuvant setting. We analyzed clinical factors that might affect the pCR and developed a model that could be used to predict pCR. Although some of the factors have already been used by other groups to build prediction models, our study was the first one that assessed and incorporated all relevant important clinical factors. All clinical variables were analyzed. In addition, we developed a genetic model to predict pCR, which showed stability across different databases. Nonetheless, our study also had its limitations. First, all data was collected retrospectively with the inherent biases deriving from data collection, although three co-authors verified the data. Our study was based on real-world data, and many unevaluated variables that might have affected the outcomes. For example, there was a high proportion (53%) of patients who did not complete all predefined NACT cycles and whose pCR rate was 33.96%. Second, although the genetic model was validated by other databases, its real value in clinical practice should be determined in future studies, alongside its further confirmation by RT-QPCR, in vitro or in vivo experiments. Third, this study developed clinical and genetic models for HER2+ breast cancer patients who received neoadjuvant chemotherapy and trastuzumab, so whether these models could predict pCR for those who received pertuzumab or tyrosine kinase inhibitors deserved further research. Meanwhile, the size of the training and validation cohort is small. The validation cohort is from Chinese population and the model needs to be tested with other ethnicity cohorts to have global use.
We determined that pCR was an independent prognostic factor for DFS and OS. Further, ER, PR, and HER2 status were associated with pCR, and use of wPC, and trastuzumab with a sufficient number of cycles completed could improve the possibility of achieving a pCR. The model we developed could be used for the prediction of pCR in HER2+ breast cancer patients. Genetic data including RNA expression evaluation might be helpful to guide clinical treatment and could be used for the prediction of clinical outcomes. However, both clinical and genetic models should be verified in other clinical contexts.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
The studies involving human participants were reviewed and approved by Ethics Committee in Fudan University Shanghai Cancer Center. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Study design: LL and JioW. Data collection: LL, MC, SZ, and HL. Data analysis: LL and JioW. Manuscript writing: LL, MC, SZ, WC, BX, QZ, JiaW, JH, HL, and JioW. All authors contributed to the article and approved the submitted version.
This study was funded by the Academic Leaders of Shanghai Science and Technology Commission (18XD1401300), the Open Fund of the Key Laboratory of Evidence Based Medicine and Knowledge Translation of Gansu Province (GSKL-EBM&KT-201902), and Youth Program of National Natural Science Foundation of China (82002797).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.592393/full#supplementary-material
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2018) 68(6):394–424. doi: 10.3322/caac.21492
2. Ahn S, Woo JW, Lee K, Park SY. HER2 Status in Breast Cancer: Changes in Guidelines and Complicating Factors for Interpretation. J Pathol Transl Med (2020) 54(1):34–44. doi: 10.4132/jptm.2019.11.03
3. Perez EA, Romond EH, Suman VJ, Jeong JH, Sledge G, Geyer CE Jr, et al. Trastuzumab Plus Adjuvant Chemotherapy for Human Epidermal Growth Factor Receptor 2-Positive Breast Cancer: Planned Joint Analysis of Overall Survival From NSABP B-31 and NCCTG N9831. J Clin Oncol (2014) 32(33):3744–52. doi: 10.1200/JCO.2014.55.5730
4. Spring LM, Fell G, Arfe A, Sharma C, Greenup RA, Reynolds KL, et al. Pathological Complete Response After Neoadjuvant Chemotherapy and Impact on Breast Cancer Recurrence and Survival: A Comprehensive Meta-Analysis. Clin Cancer Res (2020) 26(12):2838–48. doi: 10.1158/1078-0432.CCR-19-3492
5. Wuerstlein R, Harbeck N. Neoadjuvant Therapy for HER2-Positive Breast Cancer. Rev Recent Clin Trials (2017) 12(2):81–92. doi: 10.2174/1574887112666170202165049
6. LeVasseur N, Sun J, Gondara L, Diocee R, Speers C, Lohrisch C, et al. Impact of Pathologic Complete Response on Survival After Neoadjuvant Chemotherapy in Early-Stage Breast Cancer: A Population-Based Analysis. J Cancer Res Clin Oncol (2020) 146(2):529–36. doi: 10.1007/s00432-019-03083-y
7. Untch M, Fasching PA, Konecny GE, Hasmuller S, Lebeau A, Kreienberg R, et al. Pathologic Complete Response After Neoadjuvant Chemotherapy Plus Trastuzumab Predicts Favorable Survival in Human Epidermal Growth Factor Receptor 2-Overexpressing Breast Cancer: Results From the TECHNO Trial of the AGO and GBG Study Groups. J Clin Oncol (2011) 29(25):3351–7. doi: 10.1200/JCO.2010.31.4930
8. Cortazar P, Zhang L, Untch M, Mehta K, Costantino JP, Wolmark N, et al. Pathological Complete Response and Long-Term Clinical Benefit in Breast Cancer: The CTNeoBC Pooled Analysis. Lancet (9938) 2014). 384:164–72. doi: 10.1016/S0140-6736(13)62422-8
9. Plichta JK, Ren Y, Thomas SM, Greenup RA, Fayanju OM, Rosenberger LH, et al. Implications for Breast Cancer Restaging Based on the 8th Edition AJCC Staging Manua. Ann Surg (2020) 271(1):169–76. doi: 10.1097/SLA.0000000000003071
10. Liu JC, Voisin V, Bader GD, Deng T, Pusztai L, Symmans WF, et al. Seventeen-Gene Signature From Enriched Her2/Neu Mammary Tumor-Initiating Cells Predicts Clinical Outcome for Human HER2+:ERalpha- Breast Cancer. Proc Natl Acad Sci USA (2012) 109(15):5832–7. doi: 10.1073/pnas.1201105109
11. Prat A, Bianchini G, Thomas M, Belousov A, Cheang MC, Koehler A, et al. Research-Based PAM50 Subtype Predictor Identifies Higher Responses and Improved Survival Outcomes in HER2-Positive Breast Cancer in the NOAH Study. Clin Cancer Res (2014) 20(2):511–21. doi: 10.1158/1078-0432.CCR-13-0239
12. Guarneri V, Dieci MV, Frassoldati A, Maiorana A, Ficarra G, Bettelli S, et al. Prospective Biomarker Analysis of the Randomized CHER-LOB Study Evaluating the Dual Anti-HER2 Treatment With Trastuzumab and Lapatinib Plus Chemotherapy as Neoadjuvant Therapy for HER2-Positive Breast Cancer. Oncologist (2015) 20(9):1001–10. doi: 10.1634/theoncologist.2015-0138
13. Zoeller JJ, Press MF, Selfors LM, Dering J, Slamon DJ, Hurvitz SA, et al. Clinical Evaluation of BCL-2/XL Levels Pre- and Post- HER2-Targeted Therapy. PLoS One (2021) 16(5):e0251163. doi: 10.1371/journal.pone.0251163
14. Gluck S, Ross JS, Royce M, McKenna EF Jr, Perou CM, Avisar E, et al. TP53 Genomics Predict Higher Clinical and Pathologic Tumor Response in Operable Early-Stage Breast Cancer Treated With Docetaxel-Capecitabine +/- Trastuzumab. Breast Cancer Res Treat (2012) 132(3):781–91. doi: 10.1007/s10549-011-1412-7
15. van Ramshorst MS, van Werkhoven E, Mandjes IAM, Schot M, Wesseling J, Vrancken Peeters M, et al. Trastuzumab in Combination With Weekly Paclitaxel and Carboplatin as Neo-Adjuvant Treatment for HER2-Positive Breast Cancer: The TRAIN-Study. Eur J Cancer (2017) 74:47–54. doi: 10.1016/j.ejca.2016.12.023
16. Moja L, Tagliabue L, Balduzzi S, Parmelli E, Pistotti V, Guarneri V, et al. Trastuzumab Containing Regimens for Early Breast Cancer. Cochrane Database Syst Rev (2012) 4):CD006243. doi: 10.1002/14651858.CD006243.pub2
17. Tang M, Schaffer A, Kiely BE, Daniels B, Simes RJ, Lee CK, et al. Treatment Patterns and Survival in HER2-Positive Early Breast Cancer: A Whole-of-Population Australian Cohort Study (2007-2016). Br J Cancer (2019) 121(11):904–11. doi: 10.1038/s41416-019-0612-5
18. Colomer R, Saura C, Sanchez-Rovira P, Pascual T, Rubio IT, Burgues O, et al. Neoadjuvant Management of Early Breast Cancer: A Clinical and Investigational Position Statement. Oncologist (2019) 24(5):603–11. doi: 10.1634/theoncologist.2018-0228
19. Pomponio MK, Burkbauer L, Goldbach M, Nazarian SM, Xie F, Clark AS, et al. Refining the Indications for Neoadjuvant Chemotherapy for Patients With HER2+ Breast Cancer: A Single Institution Experience. J Surg Oncol (2020) 121(3):447–55. doi: 10.1002/jso.25814
20. Swain SM, Tang G, Lucas PC, Robidoux A, Goerlitz D, Harris BT, et al. Pathologic Complete Response and Outcomes by Intrinsic Subtypes in NSABP B-41, a Randomized Neoadjuvant Trial of Chemotherapy With Trastuzumab, Lapatinib, or the Combination. Breast Cancer Res Treat (2019) 178(2):389–99. doi: 10.1007/s10549-019-05398-3
21. Yu KD, Liu GY, Chen CM, Li JW, Wu J, Lu JS, et al. Weekly Paclitaxel/Carboplatin/Trastuzumab Therapy Improves Pathologic Complete Remission in Aggressive HER2-Positive Breast Cancers, Especially in Luminal-B Subtype, Compared With a Once-Every-3-Weeks Schedule. Oncologist (2013) 18(5):511–7. doi: 10.1634/theoncologist.2012-0057
22. Frati A, Chereau E, Coutant C, Bezu C, Antoine M, Chopier J, et al. Comparison of Two Nomograms to Predict Pathologic Complete Responses to Neoadjuvant Chemotherapy for Breast Cancer: Evidence That HER2-Positive Tumors Need Specific Predictors. Breast Cancer Res Treat (2012) 132(2):601–7. doi: 10.1007/s10549-011-1897-0
23. González-Santiago S, Saura C, Ciruelos E, Alonso JL, de la Morena P, Santisteban Eslava M, et al. Real-World Effectiveness of Dual HER2 Blockade With Pertuzumab and Trastuzumab for Neoadjuvant Treatment of HER2-Positive Early Breast Cancer (The NEOPETRA Study). Breast Cancer Res Treat (2020) 184(2):469–79. doi: 10.1007/s10549-020-05866-1
24. Tanioka M, Sasaki M, Shimomura A, Fujishima M, Doi M, Matsuura K, et al. Pathologic Complete Response After Neoadjuvant Chemotherapy in HER2-Overexpressing Breast Cancer According to Hormonal Receptor Status. Breast (Edinburgh Scotland) (2014) 23(4):466–72. doi: 10.1016/j.breast.2014.03.008
25. Jankowski C, Guiu S, Cortet M, Charon-Barra C, Desmoulins I, Lorgis V, et al. Predictive Factors of Pathologic Complete Response of HER2-Positive Breast Cancer After Preoperative Chemotherapy With Trastuzumab: Development of a Specific Predictor and Study of Its Utilities Using Decision Curve Analysis. Breast Cancer Res Treat (2017) 161(1):73–81. doi: 10.1007/s10549-016-4040-4
26. Fujii T, Kogawa T, Wu J, Sahin AA, Liu DD, Chavez-MacGregor M, et al. Nomogram to Predict Pathologic Complete Response in HER2-Positive Breast Cancer Treated With Neoadjuvant Systemic Therapy. Br J Cancer (2017) 116(4):509–14. doi: 10.1038/bjc.2016.444
27. Bianchini G, Kiermaier A, Bianchi GV, Im Y-H, Pienkowski T, Liu M-C, et al. Biomarker Analysis of the NeoSphere Study: Pertuzumab, Trastuzumab, and Docetaxel Versus Trastuzumab Plus Docetaxel, Pertuzumab Plus Trastuzumab, or Pertuzumab Plus Docetaxel for the Neoadjuvant Treatment of HER2-Positive Breast Cancer. Breast Cancer Res (2017) 19(1):16. doi: 10.1186/s13058-017-0806-9
28. Tanioka M, Fan C, Parker JS, Hoadley KA, Hu Z, Li Y, et al. Integrated Analysis of RNA and DNA From the Phase III Trial CALGB 40601 Identifies Predictors of Response to Trastuzumab-Based Neoadjuvant Chemotherapy in HER2-Positive Breast Cancer. Clin Cancer Res (2018) 24(21):5292–304. doi: 10.1158/1078-0432.CCR-17-3431
29. De Angelis C, Nagi C, Hoyt CC, Liu L, Roman K, Wang C, et al. Evaluation of the Predictive Role of Tumor Immune Infiltrate in Patients With HER2-Positive Breast Cancer Treated With Neoadjuvant Anti-HER2 Therapy Without Chemotherapy. Clin Cancer Res (2020) 26(3):738–45. doi: 10.1158/1078-0432.CCR-19-1402
30. Gianni L, Pienkowski T, Im YH, Roman L, Tseng LM, Liu MC, et al. Efficacy and Safety of Neoadjuvant Pertuzumab and Trastuzumab in Women With Locally Advanced, Inflammatory, or Early HER2-Positive Breast Cancer (NeoSphere): A Randomised Multicentre, Open-Label, Phase 2 Trial. Lancet Oncol (2012) 13(1):25–32. doi: 10.1016/S1470-2045(11)70336-9
31. Claret F, Vu T. Trastuzumab: Updated Mechanisms of Action and Resistance in Breast Cancer. Front Oncol (2012) 2(62):1–6. doi: 10.3389/fonc.2012.00062
32. Fernandez-Martinez A, Krop IE, Hillman DW, Polley MY, Parker JS, Huebner L, et al. Survival, Pathologic Response, and Genomics in CALGB 40601 (Alliance), a Neoadjuvant Phase III Trial of Paclitaxel-Trastuzumab With or Without Lapatinib in HER2-Positive Breast Cancer. J Clin Oncol (2020) 38(35):4184–93. doi: 10.1200/JCO.20.01276
33. Lesurf R, Griffith OL, Griffith M, Hundal J, Trani L, Watson MA, et al. Genomic Characterization of HER2-Positive Breast Cancer and Response to Neoadjuvant Trastuzumab and Chemotherapy-Results From the ACOSOG Z1041 (Alliance) Trial. Ann Oncol Off J Eur Soc Med Oncol (2017) 28(5):1070–7. doi: 10.1093/annonc/mdx048
34. Gu-Trantien C, Migliori E, Buisseret L, de Wind A, Brohee S, Garaud S, et al. CXCL13-Producing TFH Cells Link Immune Suppression and Adaptive Memory in Human Breast Cancer. JCI Insight (2017) 2(11):e91487. doi: 10.1172/jci.insight.91487
35. Denkert C, von Minckwitz G, Brase JC, Sinn BV, Gade S, Kronenwett R, et al. Tumor-Infiltrating Lymphocytes and Response to Neoadjuvant Chemotherapy With or Without Carboplatin in Human Epidermal Growth Factor Receptor 2-Positive and Triple-Negative Primary Breast Cancers. J Clin Oncol (2015) 33(9):983–91. doi: 10.1200/JCO.2014.58.1967
36. Triulzi T, Forte L, Regondi V, Di Modica M, Ghirelli C, Carcangiu ML, et al. HER2 Signaling Regulates the Tumor Immune Microenvironment and Trastuzumab Efficacy. Oncoimmunology (2019) 8(1):e1512942. doi: 10.1080/2162402X.2018.1512942
37. Vidova V, Stuchlikova E, Vrbova M, Almasi M, Klanova J, Thon V, et al. Multiplex Assay for Quantification of Acute Phase Proteins and Immunoglobulin A in Dried Blood Spot. J Proteome Res (2019) 18(1):380–91. doi: 10.1021/acs.jproteome.8b00657
38. Perez EA, Thompson EA, Ballman KV, Anderson SK, Asmann YW, Kalari KR, et al. Genomic Analysis Reveals That Immune Function Genes are Strongly Linked to Clinical Outcome in the North Central Cancer Treatment Group N9831 Adjuvant Trastuzumab Trial. J Clin Oncol (2015) 33(7):701–8. doi: 10.1200/JCO.2014.57.6298
39. McGuire A, Kalinina O, Holian E, Curran C, Malone CA, McLaughlin R, et al. Differential Impact of Hormone Receptor Status on Survival and Recurrence for HER2 Receptor-Positive Breast Cancers Treated With Trastuzumab. Breast Cancer Res Treat (2017) 164(1):221–9. doi: 10.1007/s10549-017-4225-5
40. McDermott MSJ, Canonici A, Ivers L, Browne BC, Madden SF, O’Brien NA, et al. Dual Inhibition of IGF1R and ER Enhances Response to Trastuzumab in HER2 Positive Breast Cancer Cells. Int J Oncol (2017) 50(6):2221–8. doi: 10.3892/ijo.2017.3976
41. Vignoli A, Muraro E, Miolo G, Tenori L, Turano P, Di Gregorio E, et al. Effect of Estrogen Receptor Status on Circulatory Immune and Metabolomics Profiles of HER2-Positive Breast Cancer Patients Enrolled for Neoadjuvant Targeted Chemotherapy. Cancers (2020) 12(2):314. doi: 10.3390/cancers12020314
42. Rimawi M, Cecchini R, Rastogi P, Geyer C, Fehrenbacher L, Stella P, et al. Abstract S3-06: A Phase III Trial Evaluating pCR in Patients With HR+, HER2-Positive Breast Cancer Treated With Neoadjuvant Docetaxel, Carboplatin, Trastuzumab, and Pertuzumab (TCHP)+/-Estrogen Deprivation: NRG Oncology/NSABP B-52. Cancer Res (2017) 77(4):S3–06 doi: 10.1158/1538-7445.SABCS16-S3-06
43. Carey LA, Berry DA, Cirrincione CT, Barry WT, Pitcher BN, Harris LN, et al. Molecular Heterogeneity and Response to Neoadjuvant Human Epidermal Growth Factor Receptor 2 Targeting in CALGB 40601, a Randomized Phase III Trial of Paclitaxel Plus Trastuzumab With or Without Lapatinib. J Clin Oncol (2016) 34(6):542–9. doi: 10.1200/JCO.2015.62.1268
44. Gavin PG, Song N, Kim SR, Pogue-Geile KL, Paik S. Immune Signature to Predict Trastuzumab Benefit: Potential and Pitfall. J Clin Oncol (2015) 33(31):3671–2. doi: 10.1200/JCO.2015.61.9650
45. Dieci MV, Prat A, Tagliafico E, Pare L, Ficarra G, Bisagni G, et al. Integrated Evaluation of PAM50 Subtypes and Immune Modulation of pCR in HER2-Positive Breast Cancer Patients Treated With Chemotherapy and HER2-Targeted Agents in the CherLOB Trial. Ann Oncol Off J Eur Soc Med Oncol (2016) 27(10):1867–73. doi: 10.1093/annonc/mdw262
46. Diessner J, Bruttel V, Becker K, Pawlik M, Stein R, Hausler S, et al. Targeting Breast Cancer Stem Cells With HER2-Specific Antibodies and Natural Killer Cells. Am J Cancer Res (2013) 3(2):211–20.
47. Shao Z, Pang D, Yang H, Li W, Wang S, Cui S, et al. Efficacy, Safety, and Tolerability of Pertuzumab, Trastuzumab, and Docetaxel for Patients With Early or Locally Advanced ERBB2-Positive Breast Cancer in Asia: The PEONY Phase 3 Randomized Clinical Trial. JAMA Oncol (2020) 6(3):e193692. doi: 10.1001/jamaoncol.2019.3692
48. Patel TA, Ensor JE, Creamer SL, Boone T, Rodriguez AA, Niravath PA, et al. A Randomized, Controlled Phase II Trial of Neoadjuvant Ado-Trastuzumab Emtansine, Lapatinib, and Nab-Paclitaxel Versus Trastuzumab, Pertuzumab, and Paclitaxel in HER2-Positive Breast Cancer (TEAL Study). Breast Cancer Res (2019) 21(1):100–. doi: 10.1186/s13058-019-1186-0
49. Gu CL, Zhu HX, Deng L, Meng XQ, Li K, Xu W, et al. Bispecific Antibody Simultaneously Targeting PD1 and HER2 Inhibits Tumor Growth via Direct Tumor Cell Killing in Combination With PD1/PDL1 Blockade and HER2 Inhibition. Acta Pharmacol Sin (2021) 1–9. doi: 10.1038/s41401-021-00683-8
50. Li H, Yuan W, Bin S, Wu G, Li P, Liu M, et al. Overcome Trastuzumab Resistance of Breast Cancer Using Anti-HER2 Chimeric Antigen Receptor T Cells and PD1 Blockade. Am J Cancer Res (2020) 10(2):688–703. doi: 10.3389/fonc.2020.01143
51. Mittal D, Vijayan D, Neijssen J, Kreijtz J, Habraken M, Van Eenennaam H, et al. Blockade of ErbB2 and PD-L1 Using a Bispecific Antibody to Improve Targeted Anti-ErbB2 Therapy. Oncoimmunology (2019) 8(11):e1648171. doi: 10.1080/2162402X.2019.1648171
Keywords: breast cancer, HER2, neoadjuvant chemotherapy, predictive model, immune signature, trastuzumab
Citation: Li L, Chen M, Zheng S, Li H, Chi W, Xiu B, Zhang Q, Hou J, Wang J and Wu J (2021) Clinical and Genetic Predictive Models for the Prediction of Pathological Complete Response to Optimize the Effectiveness for Trastuzumab Based Chemotherapy. Front. Oncol. 11:592393. doi: 10.3389/fonc.2021.592393
Received: 07 August 2020; Accepted: 24 June 2021;
Published: 15 July 2021.
Edited by:
Giuseppe Giaccone, Cornell University, United StatesReviewed by:
Sonia Jain, Mayo Clinic, United StatesCopyright © 2021 Li, Chen, Zheng, Li, Chi, Xiu, Zhang, Hou, Wang and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiong Wu, d3VqaW9uZzExMjJAdmlwLnNpbmEuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.