
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol. , 08 February 2021
Sec. Skin Cancer
Volume 10 - 2020 | https://doi.org/10.3389/fonc.2020.628367
This article is part of the Research Topic Mechanisms of Melanoma Tumor Progression View all 10 articles
Cancer initiation, progression, and metastasis leverage many regulatory agents, such as signaling molecules, transcription factors, and regulatory RNA molecules. Among these, regulatory non-coding RNAs have emerged as molecules that control multiple cancer types and their pathologic properties. The human microRNA-211 (MIR211) is one such molecule, which affects several cancer types, including melanoma, glioblastoma, lung adenocarcinomas, breast, ovarian, prostate, and colorectal carcinoma. Previous studies suggested that in certain tumors MIR211 acts as a tumor suppressor while in others it behaves as an oncogenic regulator. Here we summarize the known molecular genetic mechanisms that regulate MIR211 gene expression and molecular pathways that are in turn controlled by MIR211 itself. We discuss how cellular and epigenetic contexts modulate the biological effects of MIR211, which exhibit pleiotropic effects. For example, up-regulation of MIR211 expression down-regulates Warburg effect in melanoma tumor cells associated with an inhibition of the growth of human melanoma cells in vitro, and yet these conditions robustly increase tumor growth in xenografted mice. Signaling through the DUSP6-ERK5 pathway is modulated by MIR211 in BRAFV600E driven melanoma tumors, and this function is involved in the resistance of tumor cells to the BRAF inhibitor, Vemurafenib. We discuss several alternate but testable models, involving stochastic cell-to-cell expression heterogeneity due to multiple equilibria involving feedback circuits, intracellular communication, and genetic variation at miRNA target sties, to reconcile the paradoxical effects of MIR211 on tumorigenesis. Understanding the precise role of this miRNA is crucial to understanding the genetic basis of melanoma as well as the other cancer types where this regulatory molecule has important influences. We hope this review will inspire novel directions in this field.
MicroRNAs1 (miRNAs) are highly conserved small non-coding RNA molecules of approximately 22 nucleotides that control gene expression either by direct translational inhibition of protein synthesis or by affecting the degradation of the target mRNA (2). miRNA biogenesis occurs through biochemical pathways that are well conserved among metazoans (3), which is described briefly below [miRNA biogenesis has been reviewed recently (4)]. In animals, miRNA transcription begins by the binding of RNA polymerase II enzyme (Pol II) to transcriptional regulatory regions. The primary pol II transcript (Pri-miRNA) molecules, which are precursors of the mature miRNAs, often contain multiple and complex intramolecularly bases-paired looped structures and are subsequently processed by the nuclear enzyme Drosha to produce the intermediate precursor molecules termed Pre-miRNA. Pre-miRNA molecules generally possess a single hairpin loop. Pre-miRNAs are then exported to the cytoplasm by Exportin-5 (Ex-5) and are further processed to generate a mature miRNA duplex without a hairpin by the Dicer enzyme. Mature miRNA molecules, which can be either the 5′ (miRNA-5p) or the 3’ (miRNA-3p) component of the Pre-miRNA double-stranded stems of the hairpin, are loaded into the multi-protein RNA-induced silencing complex (RISC) to generate single-stranded guide miRNAs. Such guide miRNA molecules preloaded into the RISC complex interact with various target mRNAs by complementary base pairing over short regions of perfect or near-perfect sequence complementarity, most frequently at the 3’untranslated regions (3’UTR) of the target mRNAs (4). In animals, long 3’UTRs of target mRNAs are often recognized by multiple miRNAs (5, 6). Most such mRNAs with long 3’UTR are known to be involved in cellular fate-determination and developmental processes (4, 7), whereas target genes with relatively short 3’UTR regions generally encode proteins that participate in fundamental cellular biochemistry and stress response (7, 8). For example, genes encoding ribosomal RNAs and proteins have shorter 3’UTR sequences; therefore, these RNA molecules present less complex regulatory potential through the binding of a small number of different miRNAs than do genes encoding longer 3’UTRs, which are generally associated with mRNA of regulatory proteins. Less frequently, biologically important miRNA binding sites are also located in 5’UTR regions, coding regions and promoter regions of coding genes (9, 10). This dichotomy in 3’UTR or 5’UTR lengths have dynamical consequences: genes encoding proteins that participate in fundamental biochemistry/metabolism of the cells (“housekeeping” genes) are regulated quantitatively, as a rheostat does, through a graded response to regulation by miRNAs (11). By contrast, genes encoding regulatory proteins such as transcription factors, each exhibiting multiple miRNA targets, are regulated through complex analog-logical circuits that often result in a switch-like behavior in their expression levels. To understand the biological effects of a miRNA, therefore, it is important to understand how the expression of the miRNA is regulated as well as how that specific miRNA molecule regulates other genes. We illustrate some of these complexities with MIR211, which is one of the major regulators in human cancer, specifically in human melanomas (12–17). Specifically, this molecule can act both as a tumor suppressor and an oncogene based on the cellular context and thus demonstrates a paradoxical behavior in melanoma and other cancers.
MIR211 gene is located on chromosome 15q13.3, encoded within the sixth intron of the Transient Receptor Potential cation channel subfamily M member1 gene (TRPM1, Melastatin). The primary human melanocytes exhibit a high expression level of MIR211 (12). Normal human organs such as the eye (18) and the heart (19) also exhibit high expression levels of the primary TRPM1 transcript. As expected, the levels of TRPM1 and MIR211 expression are positively correlated (20).
MIR211 has now been recognized as an important player in the molecular pathogenesis of skin cancers. The Melanocyte Inducing Transcription Factor (MITF) regulates MIR211’s host gene TRPM1 by serving as its primary transcriptional regulator. MITF is one of the critical regulators of melanocyte differentiation and melanoma formation (12, 21, 22). A positive correlation of gene expression levels between MIR211 and other MITF target genes such as TYRP1, TYR, MLANA, CDK2, and SILV has also been reported, further supporting the role of MITF in regulating MIR211 expression (12, 23), and allowing the inclusion of MIR211 into the gene ontology (24) cluster of “melanosome pigment granule related genes” (FDR corrected P =4.36 × 10−8). An MITF-MIR211-BRN2 regulatory feedback loop has been demonstrated (Figure 1), and this regulatory mechanism may be important for cell state specification in both melanoblasts and melanoma cells (25–28). BRN2 (Brain-Specific Homeobox/POU Domain Protein class 3 transcription Factor 2 or POU3F2), a transcription factor, is associated with aggressive melanoma development, and MIR211 is a strong suppressor of BRN2 mediated invasiveness (25). In this feedback model BRN2 is a direct target of MIR211: predictably, BRN2 expression shows an inverse correlation with MITF expression. On the other hand, BRN2 has been shown to be a repressor of MITF transcription (29). This is predicted to induce bistable states in different subpopulations of cells (21): one in which MIR211 expression and in the other BRN2 expression would dominate, thus producing different cellular behaviors due to mutually exclusive expression of these two genes in the two subpopulations (see later for a significance of this feedback loop).
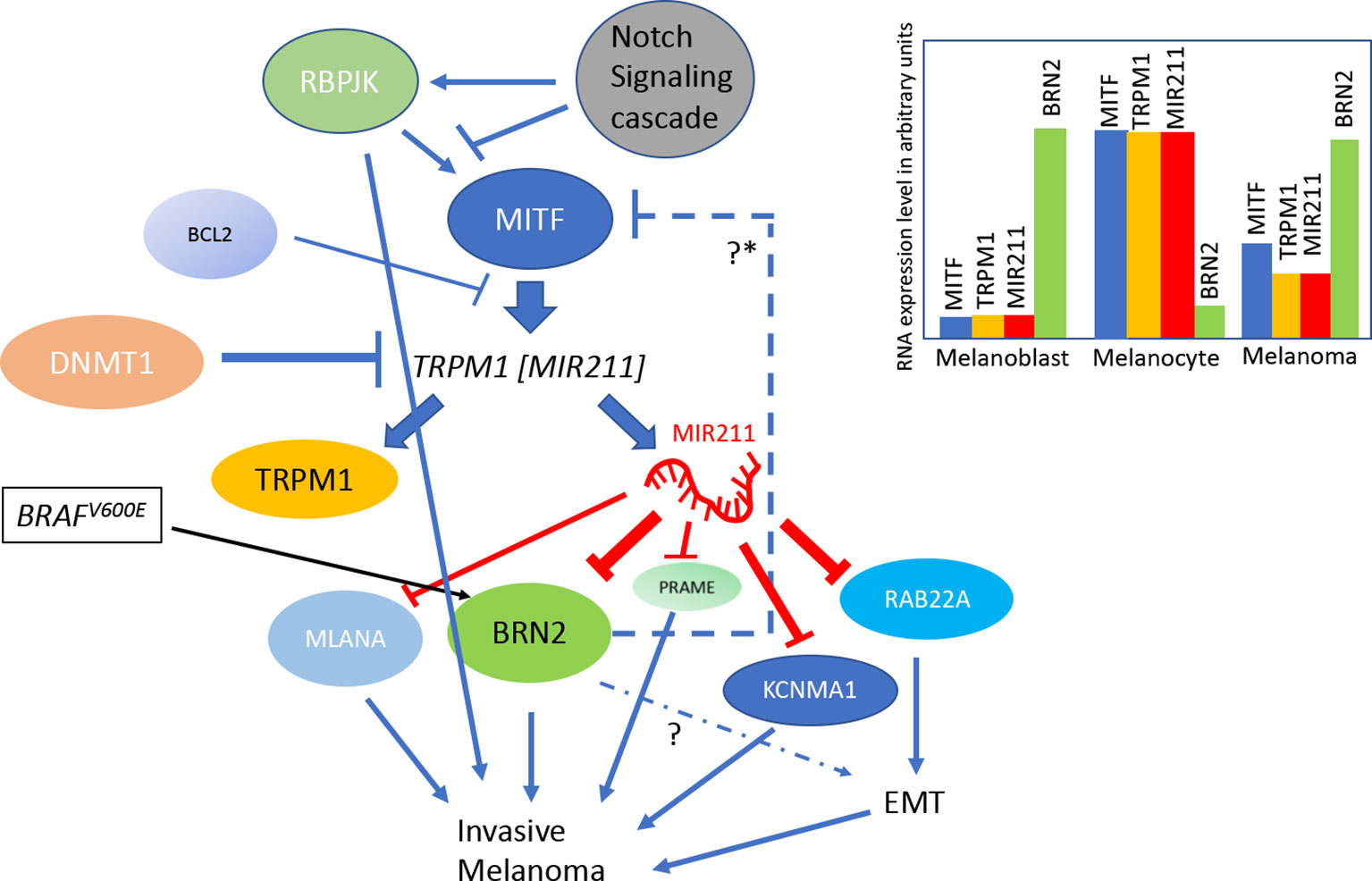
Figure 1 A model summarizing the regulation of MIR211 gene expression: MIR211 gene is nested within the TRMP1 transcript. TRPM1 transcription is promoted by the transcription factor MITF, and the latter’s transcription is in turn regulated by the Notch signaling pathway via the expression of RBPJK. The DNA methyl transferase DNMT1 regulates the epigenetic state of TRPM1 gene via promoter methylation. MIR211 negatively regulates various genes, including MLANA, PRAME, RABB22A, KCNMA1, and BRN2. Simultaneously BRN2 gene product induces MIR211expression through an unknown mechanism. Apart from that, MIR211 expression also negatively regulated by pro-apoptotic protein BCL2 through MITF inhibition. The BRAFV600E mutation, a frequent driver mutation for melanoma, activates BRN2, which is negatively regulated by MIR211. EMT is Epithelial Mesenchymal Transition. The inset is a color-coded summary of salient mRNA expression patterns in different cell types related to melanoma formation (blue, MITF; orange, TRPM1; red, MIR211; green, BRN2). The Y-axis is an arbitrary scale of gene expression levels, used as a schematic representation of the salient points on how MIR211 expression varies in various cell types. See text for evidence regarding these circuits and the gene expression levels from the following sources (12, 14, 25–33). *Proposed negative regulation.
The transcription regulatory gene Recombination signal Binding Protein for immunoglobulin kappa J region (RBPJ), one of the primary downstream molecular effectors of Notch signaling, shares a similar gene expression profile with that of MITF, suggesting a conserved biological function (30). Indeed, the expression levels of RBPJ and MITF exhibit a positive correlation (R=0.47, P<10−10) in the Genotype-Tissue Expression (GTEx) datasets for healthy skin (both sun-exposed and unexposed), but this correlation is absent in melanoma cells (34). Furthermore, RBPJ occupies and regulates the TRPM1 promoter in a MITF dependent manner and thereby increases the expression of MIR211 (30, 31). Bcl-2, a pro-apoptotic protein, regulates the expression of MIR211 by modulating MITF expression in melanoma cell lines, in addition to the reciprocal regulation of BCL2 by MITF (32). Figure 1 summarizes these pathways of regulation of MIR211 gene.
Transcriptional silencing associated with methylation of CpG islands is an important correlate that is thought to be responsible for the regulation of tumor suppressor genes (35–37). DNA methyltransferase 1 (DNMT1) mediated methylation of TRPM1/MIR211 promoter reduces MIR211 expression levels in melanoma cells (26). Decreased MIR211 levels were correlated with increased cell proliferation and Epithelial to Mesenchymal Transition (EMT, a morphogenetic state transition that is important for the development of numerous cancer types) of melanoma cells (26). The 3’UTR of the oncogenic Ras-related protein Rab-22A (RAB22A) gene’s mRNA was found to be a direct target MIR211 (33) suggesting that the suppressive effects of MIR211 on oncogenic properties might be at least partially through the inhibition of RAB22A expression. Similarly, epigenetic silencing of TRPM1/MIR211 gene by EZH2-induced (Enhancer Of Zeste Homolog 2 protein) histone H3K27 trimethylation and DNMT1 mediated promoter methylation were associated with cell proliferation, increased AKT/β-catenin signaling, and the induction of EMT, whereas restoration of MIR211 expression reduced cell proliferation, inhibited AKT/β-catenin signaling and reversed EMT in glioblastoma multiformi (38). Thus, the EZH2/DNMT1/MIR211/RAB22A axis might provide novel insights into the molecular pathogenesis of both melanoma and glioblastoma, particularly on the EMT processes in these two different cancers. In a separate study, Li N et al. reported that the silencing of MIR211 expression by methylation is associated with reduced cisplatin sensitivity in melanoma (39). This observation, however, may be interpreted as an indirect consequence of reduced EMT in MIR211 deficient cells, because increased EMT increases the resistance of cells to cisplatin (40).
As expected of a regulatory molecule with a potential tumor-suppressor activity, MIR211 was first identified as one of the most differentially under-expressed miRNA molecules in non-pigmented human melanoma cell lines and in a majority of clinical melanoma tissue samples relative to those expressed in normal melanocytes or in matched normal patient tissues, respectively (12, 14). Conversely, when MIR211 levels were increased artificially in melanoma cells, a significant inhibition of growth and in vitro invasive properties were observed, which could again be reversed by inhibiting MIR211 by an antagomir (12–14), fulfilling the criteria for a tumor-suppressor molecule. Consistently, several MIR211 targets genes, all of which were previously known to be oncogenic, were identified, including the calcium-activated potassium channel subunit a-1 (KCNMA1) (12), Insulin-like Growth Factor 2 Receptor (IGF2R) (14), TGF-beta Receptor 2 (TGFBR2) (41), Insulin Growth Factor Binding Protein 5 (IGFBP5) (42, 43), POU domain-containing transcription factor BRN2 (25), and Nuclear Factor of Activated T-cells 5 (NFAT5) (14). These evidences together are consistent with the proposal that MIR211 exerts its tumor suppressor properties by means of direct negative regulation of potentially oncogenic mRNAs (44). This proposal is also consistent with the tumor suppressor role of MITF as a directly activating transcription factor of MIR211 gene (12). Subsequently, a series of additional evidences solidified the proposal of miR-211 action as akin to a tumor suppressor for melanomas (13) as well as several other cancer types, including epithelial ovarian cancer (45), ovarian carcinoma (46), breast cancer (47), hepatocellular carcinoma (48), renal cancer (49), and thyroid tumors (50), among others (see Table 1 for a more complete list).
Paradoxically, when MIR211 expression was artificially induced in human melanoma cell lines, where its expression is generally reduced relative to those in melanocytes, which were xenografted into immunodeficient mice they resulted into aggressive tumor growth (68). Interestingly, this surprising observation is consistent with similar oncogenic effects associated with MIR211 expression in a number of other cancer types, including oral carcinoma (65, 69), head and neck carcinoma (67), Burkitt lymphoma (70), breast cancer cell lines (71), and non-small-cell lung cancer (66).
There is no a priori reason why a regulatory molecule cannot be a tumor suppressor in one cancer type but oncogenic in another because the identity of differentially regulated downstream target genes in different cancer types might be the obvious mechanism. Indeed, separate sets of target genes of MIR211 have been shown to be responsible for its tumor-enhancing role (66, 69, 71). However, such an explanation does not easily explain how a regulatory molecule can be both a tumor suppressor and an oncogene in the same cancer type, such as seen with MIR211 in melanoma. We propose below several mechanisms that should help resolve this important paradox.
We and others have reported that MIR211 expression is reduced in amelanotic melanoma cell lines (12) and in clinical melanoma samples (44, 72), driving efforts to use MIR211 as a clinical diagnostic test to discriminate melanomas from benign nevi (52). We also reported that the artificial over-expression of MIR211 in amelanotic melanoma cells (A375) increases mitochondrial respiration by inhibiting Pyruvate Dehydrogenase Kinase 4 (PDK4) mRNA levels, which was associated with reduced cell survival and lessened invasive properties under cell culture conditions (13). Consistently, miRNA profiling studies revealed that MIR211 expression is down-regulated not only in melanoma (12, 54), but also in other cancers such as glioma (73) and glioblastoma (38), prostate cancer (74), hepatocellular carcinoma (48), epithelial ovarian cancer (45), cervical cancer (16, 56), and breast cancer (75). These foregoing results together point towards a general tumor-suppressor role of MIR211. By contrast, MIR211 is also known exhibit high expression levels in certain melanoma subtypes and in other cancers: MIR211 expression is high in a majority (6/8) of melanoma lines in the NCI-60 cancer cell panel (76), and in 9/29 clinical melanoma samples (12), suggesting that either MIR211 level by itself is irrelevant to the tumor-like status or malignancy or that in certain cancer cells MIR211 levels might have tumor-promoting activity.
The dual role of MIRNA211 described above may hinge on the cellular context, similar to context-dependent regulation that was previously reported for other miRNA genes, in particular with MIR7 (77, 78), MIR125B (79) and MIR155 (80). This fascinating paradox—whether and how a particular miRNA could either inhibit or augment cancer growth and development depending on the particular cellular context—raises several questions concerning the importance of MIR211 in melanocyte dedifferentiation (43), melanoma genesis (25), aging and senescence (81), and cardiac development (19).
On the basis of the observations that MIR211 over-expression in the amelanotic melanoma cell line A375 decreased Warburg effect and simultaneously decreased cell proliferation and invasiveness (13), it was anticipated that these cells when transplanted as murine xenografts would also reduce tumor size relative to those by the control (normal MIR211 levels) A375 cells. Paradoxically, exactly the opposite results were observed (68), in which extraordinarily aggressive tumors were formed by the xenografted cells over-expressing MIR211. The PDK4 gene, the target gene of MIR211 which was previously thought to be an effector of the metabolic switch controlled by MIR211, was indeed downregulated in these xenografts, and therefore the response of this target gene was unable to explain the paradoxical behavior of MIR211 on tumorigenesis. Deletion of MIR211 gene in these xenografted cells produced much reduced tumors, supporting the conclusion that this MIR211 was directly responsible for the hypertrophic tumor phenotype. RNAseq analysis of mRNA molecules co-immunoprecipitated with the Argonaut-2 protein suggested that these mRNA molecules are associated with the RISC complex in cells with or without MIR211 (68). These studies further revealed several additional target mRNAs, including DUSP6 and BIRC2, which were then confirmed to be causally related to tumor hypertrophy via a modulation of the ERK5/MEK kinase signaling pathway (68) (Figure 2). These cells simultaneously became more resistant to the BRAF inhibitor vemurafenib and the MEK inhibitor cobimetinib, suggesting that the modulation of the DUSP6/ERK5 signaling pathway by MIR211 is related to the development of BRAF/MEK inhibitor resistant melanoma (68).
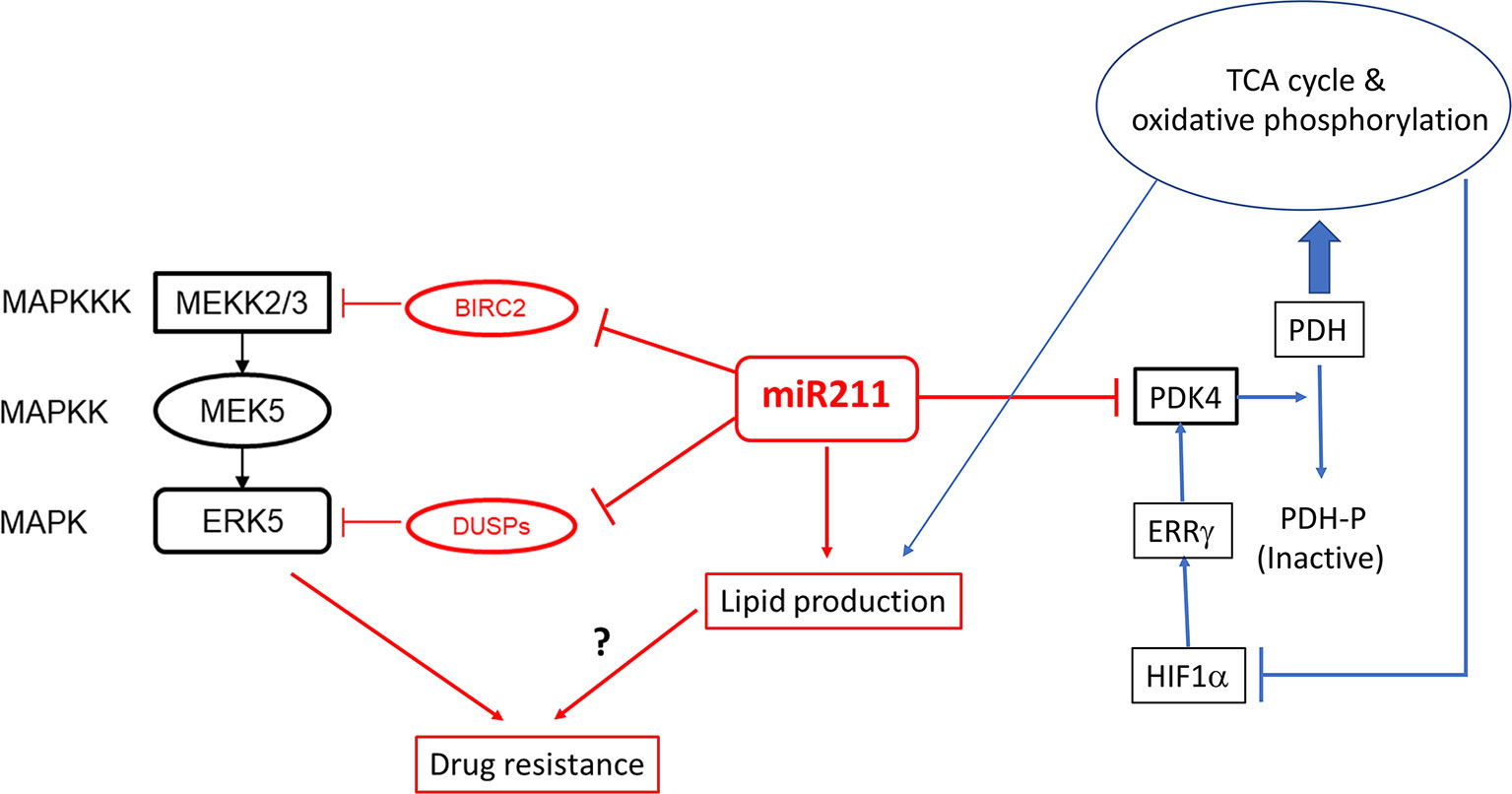
Figure 2 The regulatory cascade of MIR211 involving the BRAF/ERK5/DUSP and PDK4. MIR211 is a negative regulator of at least two negative regulators (BIRC2, DUSP6 and possibly other DUSP mRNAs) of the ERK5/MEK signaling pathway. The latter is involved in resistance of melanoma to vemurafenib and cobimetinib. PDK4 is a pyruvate dehydrogenase kinase, which inactivates pyruvate dehydrogenase (PDH) by phosphorylation, which therefore cannot catalyze the entry of pyruvate to tricarboxylic acid (TCA) cycle. Metabolites of the TCA cycle induces lipid biosynthesis and inhibits hypoxia inducing factor HIF1α, which is a transcriptional activator of ERRβ, which in turn is a transcriptional activator of PDK4 gene. Evidence for these pathways are obtained from Lee et al. (2016 and 2020) (68, 82).
As described above, MIR211 acts as a tumor suppressor in a variety of cancer types, it can also act as an oncogenic regulator in others. Therefore an understanding of the paradoxical behavior of MIR211 is of paramount importance for understanding cancer biology.
Though the functional role of MIR211 has not been studied extensively in primary human melanocytes, its tumor suppressor role in pigmented melanoma has reported widely and suggested to be due to a multitude of target genes [see citations above, and also (44, 83)]. Endoplasmic reticulum stress initiates the activation of three important signaling molecules like Ire-1α/β, ATF 6, and PERK, whose function is to enable adaptation to the ER stress. PERK (Eukaryotic translation initiation factor 2-alpha kinase 3, also known as protein kinase R (PKR)-like endoplasmic reticulum kinase), is an enzyme that in humans is encoded by the EIF2AK3 gene. PERK has been reported to induce the expression of MIR211, which in turn reduces the stress-dependent expression of GAD153/CHOP, a pro-apoptotic transcription factor at early stages of ER stress (84). If the ER stress is sustained, MIR211 expression is turned off and increases the expression of CHOP and thereby apoptosis (84). A reduction in the expression of MIR211 in multiple myeloma is associated with increased expression of pro-apoptotic factor CHOP which in turn triggers ER stress mediated apoptosis (55). In epithelial ovarian cancer (EOC) cells, MIR211 expression was down-regulated and increased the expression of cyclin D1 and CDK1 and thereby apoptosis (45). Levy et al. reported that MIR211 can directly inhibit the TGFBR2 and NFAT5 and can indirectly inhibit IGF2R, which leads to the reduced invasive potential of malignant melanomas (14).
EMT is a highly dynamic process that can convert nonmotile epithelial cells to motile mesenchymal cells (85), which involves molecular reprogramming and phenotypic changes that lead to tumor metastasis. EMT in cancer is associated with a decrease in the expression of epithelial cell markers such as γ-catenin and E cadherin, and with an increase in the expression of mesenchymal markers such as vimentin and N-cadherin (86). Mucin 4 (MUC4) is a glycoprotein that covers the epithelial cells and belongs to a large MUCIN family of glycoproteins. Mucin could be used as a lineage marker in benign cervical tissue, whose expression might be higher in cervical cancer (87). MIR211 could directly target MUC4 mRNA and thereby can inhibit the invasion and EMT in cervical cancer cells (57). In melanoma cells, MIR211 can modulate EMT by targeting RAB22A, a member of the RAB family of small GTPase, which regulates tumor invasion and metastasis in various cancer types (32).
Secreted protein acidic and rich in cysteine (SPARC) is a matricellular family of proteins, which modulates cell-matrix interactions and tissue remodeling and, thereby, EMT. In cervical cancer, MIR211 directly targets SPARC and thus acts as a tumor suppressor gene (16). MIR211 regulates Matrix metalloproteinase-9 (MMP-9) and thereby reduces EMT in gastric cancer (58). Sex-determining region Y-related high-mobility group box 4 (SOX4) is another crucial transcription factor that contributes to tumor cell survival, metastasis, and possibly to the maintenance of cancer stem cell properties. In gastric cancer cells, overexpression of MIR211 directly inhibits SOX4 expression and thereby down regulates tumor metastasis (88), suggesting the involvement of cancer stem cells. The Snail family transcriptional repressor 1 (SNAI1) has a significant role in regulating genes required for cell-cell interaction and EMT. By targeting SNAI1 mRNA, MIR211 directly regulates the metastatic behavior of tumor cells in renal cancer cells (49). PHF19 is a member of polycomb repressive complex 2 Complex, which mediates transcriptional repression of developmental regulatory genes and modulates embryonic stem cell differentiation. A recent report suggests that MIR211 can target PHF19 and thereby inhibit cell proliferation, migration and apoptosis in ovarian cancer (89). At the same time, MALAT1, a long non-coding RNA antagonizes the effect of MIR211 in ovarian cancer cells (46). The Secreted protein acidic and rich in cysteine (SPARC) is an oncogene, which is highly expressed in various tumors such as glioma, melanoma, prostate, and gastric carcinoma. SPARC belongs to the matricellular family and has a significant role in tissue repair and remodeling (90). MIR211 is a direct regulator of SPARC expression, and thereby down-regulates other metastasis-associated genes (62). ZEB1, a member of the zinc finger family protein, is reported to be upregulated in ovarian, breast, prostate, and endometrial cancer (91–94). MIR211 directly targets ZEB1 mRNA and reduces the proliferation and metastasis of cervical cancer cells (56).
Apart from regulating the apoptosis, Endoplasmic reticulum stress and EMT, MIR211 also regulates various other processes important for cancer, including angiogenesis, through influencing multiple signaling pathways. In hepatocellular carcinoma MIR211 down-regulates SATB2 (Human special AT-rich sequence-binding protein-2) and reduces its invasion and proliferation (48). Another important study demonstrated that in tongue squamous cell carcinoma the long non-coding RNA KCNQ1OT1 sponges MIR211 and thereby activates Ezrin/Fak/Src signaling pathways (95). Mazar et al. reported that MIR211 directly targets pyruvate dehydrogenase kinase 4 (PDK4) mRNA, and the ectopic expression of MIR211 reduced hypoxia-inducible factor 1α (HIF-1α) protein levels and decreased cell growth during hypoxia (13). Ribonucleotide reductase M2 (RRM2) is associated with tumor progression and metastasis; and in colorectal cancer k–ras mutation reduces the expression of MIR211 which thereby enhances the expression of RRM2 (96). In melanotic melanoma cells, MIR211 is induced by BRAFi/MEKi and favors their pro-pigmentation activity by targeting EDEM1, hence promoting TYR expression and melanin accumulation (53). In another interesting study with breast cancer cells it was suggested that MIR211 targeted CDC25B, CCNB1, and FOXM1 and thereby inhibited cell cycle and reduced genomic stability, proliferation, migration, and invasion in triple-negative breast cancer cells (63). It was recently shown that adipocytes secret IL6, which leads to downregulation of MIR211 in melanoma which further promotes melanoma invasion (51).
In contrast to the tumor suppressor activities of MIR211 mentioned above, increased expression of MIR211 is reportedly oncogenic [see above, and (67, 97)]. Some of its oncogenic properties can also be traced to its target genes. For example, MIR211 appears to up-regulate cell cycle progression by targeting the tumor suppressor gene CHD5 (64), thus modulating the p53 pathway, and via several protein kinases (98). MIR211 acts as an oncogene in Acute Myeloid Leukemia (97) by down-regulating the expression of BIN1 and by activating the EGFR/MAPK pathway. Increased expression of MIR211 in head and neck squamous cell carcinoma (HNSCC) inhibited TGFβRII and thus decreased the SMAD3 phosphorylation and increased c-myc expression (67). MIR211 contributes to melanoma progression not only by targeting genes of melanoma cells but also by modulating the tumor niche in melanoma microenvironment via communication through melanosomes (15, 51). Melanosomal-MIR211 released from melanoma cells are transferred into the surrounding primary skin fibroblasts and which then induce their reprogramming into cancer-associated fibroblasts (CAFs) by targeting the IGF2R mRNA and through regulating MAPK signaling (15). The target genes of MIR211 which are considered to be important in various cancer types are summarized in Table 1.
A second miRNA gene that belongs to the same family as MIR211 is MIR204, which is located within the TRPM3 gene at 9q21.12, and is believed to be a paralog of MIR211 (71). Both of these two mature MIRNAs share a similar seed sequence, but they differ in two nucleotides within their 3’ regions; these two MIRNAs share multiple common mRNA targets (99). Among these shared targets are the mRNAs of the following tumor-suppressing genes: XIAP Associated Factor 1 (XAF I) (71, 100, 101), whose loss of function mutation causes gastric carcinoma, HRAS-Like-Suppressor4 (or HRASLS4) (71, 101, 102), also associated with mutations in gastric cancer, Homeodomain Interacting Protein Kinase 2 (HIPK2) (71, 103), mutations in which gene are associated with keratocanthoma (a rare skin cancer), breast cancer, and Thioredoxin Interacting Protein (TXNIP) (71, 104), associated with exocervical carcinoma. In the opposite direction, TargetScan (www.targetscan.org) analysis of MIR211-5p and MIR204-5p also yields the following common oncogene or oncogene-like genes as high-confidence targets: Ras-Related Protein Rab-22A (RAB22A), Paired Like Homeobox 2B (PHOX2B) whose high expression is associated with neuroblastoma (105), and Adaptor Related Protein Complex 1 Subunit Sigma 2 (AP1S2) whose mRNA expression is enhanced in melanoma and is associated with significantly poorer survival of melanoma patients (although confers a better survival probability to ovarian and breast cancer patients). In summary, MIR211 and its paralog MIR204 together regulate a number of genes whose expression are related to both tumor suppressor and oncogenic activities in a number of different cancer types including melanoma. As a result, the complexity of phenotypes associated with MIR211 could also be due to the combined as well as single negative effects on a number of different oncogenic and tumor suppressor mRNA levels, which at least in theory may produce rather complex net dynamics depending on the expression signatures of a large number of genes and the relative levels of the two microRNAs.
The paradoxical behavior of MIR211 brings to mind the early controversy of p53, acting, as initially proposed, to promote tumor growth (106) but later understood to be a tumor suppressor (107). While as yet there is no direct relationship between p53 and MIR211, by reminding ourselves of the importance of paradoxes in science (108), especially concerning the p53 controversy, we suggest that MIR211 biology is important for understanding cancer biology.
There might be at least three different mechanisms that can explain the paradoxical behavior of a miRNA on a biological process: (a) differences in the allelic variants at miRNA targets; (b) differences in the cellular micro-environment, (c) stochastic fluctuations in the expression levels of the miRNA which set into motion different trajectories of regulatory influences in different cells. Figure 3 summarizes the three models. It is unclear at this time which of these models is true of MIR211. These models are offered as the rationale to guide future experiments. The models are general, in the sense that these could formally apply to all cases where the same miRNA has multiple, mutually opposite, biological effects depending on the cellular contexts.
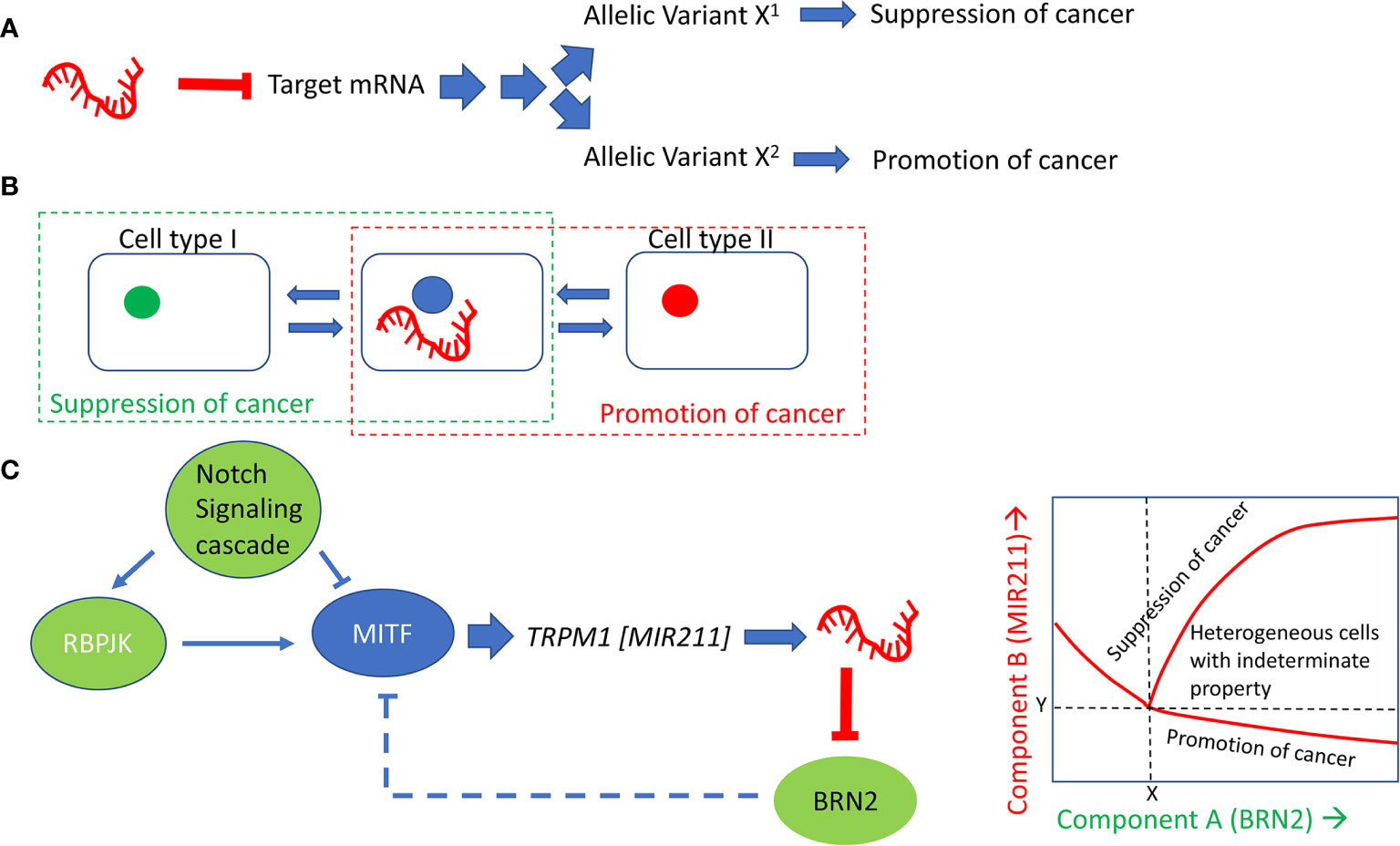
Figure 3 Models that explain the paradoxical effects of MIR211 on cancer development. (A) posits that allelic variants in MIR211 target genes explain the behavior of cells with respect to tumorigenesis. (B) assumes that the cellular type and the microenvironment, including cell-cell communication, determines the context for whether MIR211 will be tumorigenic or tumor suppressive. MIR211 secreted within exosomes, and/or signaling molecules or metabolites secreted by cells with high or low expression of MIR211 may influence adjacent tumor cells in concentration dependent manner, thus helping to establish distinct morphogenetic fields within the tumors and contribute to tumor heterogeneity. (C) explains the mutually opposite effects of MIR211 on the basis of cell to cell heterogeneity and dynamics of gene expression related to MIR211. In this last model, feed-forward and negative feedback with time delays set up opportunities for complex dynamical patterns of expression involving the two genes, MIR211 and BRN2. The inset shows a schematic of such a dynamical pattern, in which stochastic fluctuation in the initial levels of MIR211 or BRN2 could lead to the transition at the parameter values (X,Y) of a cell to a tumor progression (high BRN2/low MIR211) or to a tumor suppressive (high BRN2/high MIR211) state and thus lock in the characteristic state. Intermediate states (high BRN2/intermediate MIR211) could represent cells that could be driven to either direction through other (environmental) factors, which has been shown to occur with melanoma cells with respect to their vemurafenib resistance phenotype (109).
The allelic variant model (Figure 3A) proposes that nucleotide differences at the target site, especially within the seed sequences of the miRNA, are responsible for the cell-line to cell-line variability of its biological effects. Such variability might be inherited in the germline, or maybe due to somatic mutations. This model is consistent with several previous observations on miRNA target site sequence variations (110–112).
The micro-environment influence model (Figure 3B) posits that the expression of MIR211 triggers cell-nonautonomous events. Some cells in the tumor micro-environment sense the downstream effectors of MIR211, through either direct exosomal contribution of MIR211 to the neighboring cells or indirectly via cell-to-cell signaling and/or metabolites secreted by the primary cells. This proposal assumes the presence of morphogenetically heterogeneous cells within the tumor micro-environment, which takes into account the following previous observations: (i) Melanocytes are derived from non-pigmented melanocyte precursor stem cells, and non-pigmented melanomas are thought to be mainly derived from these stem cells through oncogenic transformation (113); (ii) pigmented melanoma cells are derived from dedifferentiation and transformation of melanocytes); (iii) MIR211 expression is low in undifferentiated precursor cells, is high in melanocytes, low in non-pigmented melanoma, but high in pigmented melanoma cells (12); (iv) MIR211 targets PDK4, which causes high NADP+/NADPH ratio (13); (v) High NADP+/NADPH ratio causes high ROS levels (114). Diffusion of these molecules is in principle sufficient to set up distinct morphogenetic fields within the tumors (115).
The stochastic single-cell expression model (Figure 3C) posits that MIR211 expression fluctuates stochastically from cell to cell (116), which leads to cell-to-cell fluctuations in the relative levels of MITF (component X) and BRN2 (component Y). A well-known consequence of such fluctuations of two regulatory components in a mutually negative feedback regulatory loop is the establishment of multiple distinct equilibrium regions of expression levels of the two components in which they are both low, both high or one high/one low. Tumor gene expression systems have been shown to operate under such multiple equilibria, producing heterogenous cell populations within the tumors, leading to vastly different biological consequences (117). Importantly, the three models proposed here are not mutually exclusive; elements of each can coexist with those of the other two in defining the full range of biological complexity associated with this regulatory molecule.
MIR211 shows both tumor suppressive and oncogenic activities in the same or different cancer types. Since MIR211 has also been shown to be associated with various human diseases apart from cancers, it is regarded as one of the most promising miRNA molecules for therapeutic applications. To our knowledge no clinical trial is currently on-going with this miRNA. Although numerous studies have illuminated the molecular mechanisms associated with the tumor-suppressive and oncogenic characteristics of MIR211, there are many important questions that need to be answered in the future, and we have provided three different frameworks for approaching these mechanisms. We suggest that further work on this interesting regulatory molecule should focus on (a) deciphering the source of mutually opposite behavior of MIR211 in cancer, (b) understanding the mechanisms of cell-to-cell expression variances of MIR211, and (c) how such variations could be utilized for diagnostic, prognostic or therapeutic applications.
AR and RP incepted the idea on the paradoxical behavior of the MIR-211 in melanoma and other cancers. All authors (AR, RP, and HK) contributed to the writing. All authors contributed to the article and approved the submitted version.
This study was supported by P30 CA006973 (JHU SKCCC) and Florida Department of Health, Bankhead-Coley Cancer Research Program (5BC08) to RP, and CDMRP/DTRA grant W81XWH-16-1-0170 to AR.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank Drs. Carmit Levy and Tamar Golan for their initial suggestion and comments on this manuscript.
1. Desvignes T, Batzel P, Berezikov E, Eilbeck K, Eppig JT, McAndrews MS, et al. microRNA nomenclature: A view incorporating genetic origins, biosynthetic pathways, and sequence variants. Trends Genet (2015) 31:613–26. doi: 10.1016/j.tig.2015.09.002
2. Dexheimer PJ, Cochella L. MicroRNAs: From Mechanism to Organism. Front Cell Dev Biol (2020) 8:1–18. doi: 10.3389/fcell.2020.00409
3. Meunier J, Lemoine F, Soumillon M, Liechti A, Weier M, Guschanski K, et al. Birth and expression evolution of mammalian microRNA genes. Genome Res (2013) 23:34–45. doi: 10.1101/gr.140269.112
5. Grimson A, Farh KK-H, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell (2007) 27:91–105. doi: 10.1016/j.molcel.2007.06.017
6. Sætrom P, Heale BSE, Snøve O, Aagaard L, Alluin J, Rossi JJ. Distance constraints between microRNA target sites dictate efficacy and cooperativity. Nucleic Acids Res (2007) 35:2333–42. doi: 10.1093/nar/gkm133
7. Zhang Y, Tang C, Yu T, Zhang R, Zheng H, Yan W. MicroRNAs control mRNA fate by compartmentalization based on 3′ UTR length in male germ cells. Genome Biology (2017) 18:105. doi: 10.1186/s13059-017-1243-x
8. Zheng D, Wang R, Ding Q, Wang T, Xie B, Wei L, et al. Cellular stress alters 3′UTR landscape through alternative polyadenylation and isoform-specific degradation. Nature Communication (2018) 9:2268. doi: 10.1038/s41467-018-04730-7
9. Gu W, Xu Y, Xie X, Wang T, Ko J-H, Zhou T. The role of RNA structure at 5′ untranslated region in microRNA-mediated gene regulation. RNA (2014) 20:1369–75. doi: 10.1261/rna.044792.114
10. Lee I, Ajay SS, Yook JI, Kim HS, Hong SH, Kim NH, et al. New class of microRNA targets containing simultaneous 5′-UTR and 3′-UTR interaction sites. Genome Res (2009) 19:1175–83. doi: 10.1101/gr.089367.108
11. Siciliano V, Garzilli I, Fracassi C, Criscuolo S, Ventre S, di Bernardo D. miRNAs confer phenotypic robustness to gene networks by suppressing biological noise. Nat Commun (2013) 4:2364. doi: 10.1038/ncomms3364
12. Mazar J, DeYoung K, Khaitan D, Meister E, Almodovar A, Goydos J, et al. The regulation of miRNA-211 expression and its role in melanoma cell invasiveness. PloS One (2010) 5:e13779. doi: 10.1371/journal.pone.0013779
13. Mazar J, Qi F, Lee B, Marchica J, Govindarajan S, Shelley J, et al. MicroRNA 211 Functions as a Metabolic Switch in Human Melanoma Cells. Mol Cell Biol (2016) 36:1090–108. doi: 10.1128/MCB.00762-15
14. Levy C, Khaled M, Iliopoulos D, Janas MM, Schubert S, Pinner S, et al. Intronic miR-211 assumes the tumor suppressive function of its host gene in melanoma. Mol Cell (2010) 40:841–9. doi: 10.1016/j.molcel.2010.11.020
15. Dror S, Sander L, Schwartz H, Sheinboim D, Barzilai A, Dishon Y, et al. Melanoma miRNA trafficking controls tumour primary niche formation. Nat Cell Biol (2016) 18:1006–17. doi: 10.1038/ncb3399
16. Qu X, Gao D, Ren Q, Jiang X, Bai J, Sheng L. miR-211 inhibits proliferation, invasion and migration of cervical cancer via targeting SPARC. Oncol Lett (2018) 16:853–60. doi: 10.3892/ol.2018.8735
17. Pei Y, Yao Q, Li Y, Zhang X, Xie B. microRNA-211 regulates cell proliferation, apoptosis and migration/invasion in human osteosarcoma via targeting EZRIN. Cellular & Molecular Biological Letter (2019) 24:48. doi: 10.1186/s11658-019-0173-x
18. Gagliano C, Russo A, Ragusa M, Caltabiano R, Puzzo L, Avitabile T, et al. MicroRNAs in vitreous humor from patients with ocular disease: preliminary results. Invest Ophthalmol Vis Sci (2013) 54:6365–5.
19. Thomas T, Galuppo P, Wolf C, Fiedler J, Kneitz S, van Laake LW, et al. MicroRNAs in the Human Heart. Circulation (2007) 116:258–67. doi: 10.1161/CIRCULATIONAHA.107.687947
20. Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, et al. The Genotype-Tissue Expression (GTEx) project. Nature Genetics (2013) 45:580–5. doi: 10.1038/ng.2653
21. Goding CR, Arnheiter H. MITF—the first 25 years. Genes Dev (2019) 33:1–25. doi: 10.1101/gad.324657.119
22. Levy C, Khaled M, Fisher DE. MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol Med (2006) 12:406–14. doi: 10.1016/j.molmed.2006.07.008
23. Wu G, Feng X, Stein L. A human functional protein interaction network and its application to cancer data analysis. Genome Biology (2010) 11:R53. doi: 10.1186/gb-2010-11-5-r53
24. The Gene Ontology Consortium. Gene Ontology Consortium: going forward. Nucleic Acids Res (2015) 43:D1049–56. doi: 10.1093/nar/gku1179
25. Boyle GM, Woods SL, Bonazzi VF, Stark MS, Hacker E, Aoude LG, et al. Melanoma cell invasiveness is regulated by miR-211 suppression of the BRN2 transcription factor. Pigment Cell Melanoma Research (2011) 24:525–37. doi: 10.1111/j.1755-148X.2011.00849.x
26. Yu H, Yang W. MiR-211 is epigenetically regulated by DNMT1 mediated methylation and inhibits EMT of melanoma cells by targeting RAB22A. Biochem Biophys Res Commun (2016) 476:400–5. doi: 10.1016/j.bbrc.2016.05.133
27. De Luca T, Pelosi A, Trisciuoglio D, D’Aguanno S, Desideri M, Farini V, et al. miR-211 and MITF modulation by Bcl-2 protein in melanoma cells. Mol Carcinog (2016) 55:2304–12. doi: 10.1002/mc.22437
28. Miller AJ, Du J, Rowan S, Hershey CL, Widlund HR, Fisher DE. Transcriptional Regulation of the Melanoma Prognostic Marker Melastatin (TRPM1) by MITF in Melanocytes and Melanoma. Cancer Res (2004) 64:509–16. doi: 10.1158/0008-5472.CAN-03-2440
29. Goodall J, Carreira S, Denat L, Kobi D, Davidson I, Nuciforo P, et al. Brn-2 represses microphthalmia-associated transcription factor expression and marks a distinct subpopulation of microphthalmia-associated transcription factor-negative melanoma cells. Cancer Res (2008) 68:7788–94. doi: 10.1158/0008-5472.CAN-08-1053
30. Tabach Y, Golan T, Hernández-Hernández A, Messer AR, Fukuda T, Kouznetsova A, et al. Human disease locus discovery and mapping to molecular pathways through phylogenetic profiling. Mol Syst Biol (2013) 9:692. doi: 10.1038/msb.2013.50
31. Golan T, Levy C. Negative Regulatory Loop between Microphthalmia-Associated Transcription Factor (MITF) and Notch Signaling. Int J Mol Sci (2019) 20. doi: 10.3390/ijms20030576
32. McGill GG, Horstmann M, Widlund HR, Du J, Motyckova G, Nishimura EK, et al. Bcl2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability. Cell (2002) 109:707–18. doi: 10.1016/s0092-8674<(>02<)>00762-6
33. Su F, Chen Y, Zhu S, Li F, Zhao S, Wu L, et al. RAB22A overexpression promotes the tumor growth of melanoma. Oncotarget (2016) 7:71744–53. doi: 10.18632/oncotarget.12329
34. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res (2017) 45:W98–W102. doi: 10.1093/nar/gkx247
35. Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet (1999) 21:163–7. doi: 10.1038/5947
36. Baylin SB, Esteller M, Rountree MR, Bachman KE, Schuebel K, Herman JG. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum Mol Genet (2001) 10:687–92. doi: 10.1093/hmg/10.7.687
37. Esteller M. CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene (2002) 21:5427–40. doi: 10.1038/sj.onc.1205600
38. Li W, Miao X, Liu L, Zhang Y, Jin X, Luo X, et al. Methylation-mediated silencing of microRNA-211 promotes cell growth and epithelial to mesenchymal transition through activation of the AKT/β-catenin pathway in GBM. Oncotarget (2017) 8:25167–76. doi: 10.18632/oncotarget.15531
39. Li N, Liu Y, Pang H, Lee D, Zhou Y, Xiao Z. Methylation-Mediated Silencing of MicroRNA-211 Decreases the Sensitivity of Melanoma Cells to Cisplatin. Med Sci Monit (2019) 25:1590–9. doi: 10.12659/MSM.911862
40. Ashrafizadeh M, Zarrabi A, Hushmandi K, Kalantari M, Mohammadinejad R, Javaheri T, et al. Association of the Epithelial-Mesenchymal Transition (EMT) with Cisplatin Resistance. Int J Mol Sci (2020) 21:4002–48. doi: 10.3390/ijms21114002
41. Dai X, Rao C, Li H, Chen Y, Fan L, Geng H, et al. Regulation of pigmentation by microRNAs: MITF-dependent microRNA-211 targets TGF-β receptor 2. Pigment Cell Melanoma Res (2015) 28:217–22. doi: 10.1111/pcmr.12334
42. Díaz-Martínez M, Benito-Jardón L, Alonso L, Koetz-Ploch L, Hernando E, Teixidó J. miR-204-5p and miR-211-5p Contribute to BRAF Inhibitor Resistance in Melanoma. Cancer Res (2018) 78:1017–30. doi: 10.1158/0008-5472.CAN-17-1318
43. Margue C, Philippidou D, Reinsbach SE, Schmitt M, Behrmann I, Kreis S. New target genes of MITF-induced microRNA-211 contribute to melanoma cell invasion. PloS One (2013) 8:e73473. doi: 10.1371/journal.pone.0073473
44. Xu Y, Brenn T, Brown ERS, Doherty V, Melton DW. Differential expression of microRNAs during melanoma progression: miR-200c, miR-205 and miR-211 are downregulated in melanoma and act as tumour suppressors. Br J Cancer (2012) 106:553–61. doi: 10.1038/bjc.2011.568
45. Xia B, Yang S, Liu T, Lou G. miR-211 suppresses epithelial ovarian cancer proliferation and cell-cycle progression by targeting Cyclin D1 and CDK6. Mol Cancer (2015) 14:57–70. doi: 10.1186/s12943-015-0322-4
46. Tao F, Tian X, Ruan S, Shen M, Zhang Z. miR-211 sponges lncRNA MALAT1 to suppress tumor growth and progression through inhibiting PHF19 in ovarian carcinoma. FASEB J (2018) 32:6330–43. doi: 10.1096/fj.201800495RR
47. Li X, Wang S, Li Z, Long X, Guo Z, Zhang G, et al. The lncRNA NEAT1 facilitates cell growth and invasion via the miR-211/HMGA2 axis in breast cancer. International Journal of Biological Macromolecules (2017) 105:346–53. doi: 10.1016/j.ijbiomac.2017.07.053
48. Jiang G, Cui Y, Yu X, Wu Z, Ding G, Cao L. miR-211 suppresses hepatocellular carcinoma by downregulating SATB2. Oncotarget (2015) 6:9457–66. doi: 10.18632/oncotarget.3265
49. Wang K, Jin W, Jin P, Fei X, Wang X, Chen X. miR-211-5p Suppresses Metastatic Behavior by Targeting SNAI1 in Renal Cancer. Mol Cancer Res (2017) 15:448–56. doi: 10.1158/1541-7786.MCR-16-0288
50. Wang L, Shen Y, Shi Z, Shang X, Jin D, Xi F. Overexpression miR-211-5p hinders the proliferation, migration, and invasion of thyroid tumor cells by downregulating SOX11. J Clin Lab Anal (2018) 32:e22293. doi: 10.1002/jcla.22293
51. Golan T, Parikh R, Jacob E, Vaknine H, Zemser-Werner V, Hershkovitz D, et al. Adipocytes sensitize melanoma cells to environmental TGF-β cues by repressing the expression of miR-211. Sci Signal (2019) 12:1–15. doi: 10.1126/scisignal.aav6847
52. Babapoor S, Horwich M, Wu R, Levinson S, Gandhi M, Makkar H, et al. microRNA in situ hybridization for miR-211 detection as an ancillary test in melanoma diagnosis. Modern Pathol (2016) 29:461–75. doi: 10.1038/modpathol.2016.44
53. Vitiello M, Tuccoli A, D’Aurizio R, Sarti S, Giannecchini L, Lubrano S, et al. Context-dependent miR-204 and miR-211 affect the biological properties of amelanotic and melanotic melanoma cells. Oncotarget (2017) 8:25395–417. doi: 10.18632/oncotarget.15915
54. Sakurai E, Maesawa C, Shibazaki M, Yasuhira S, Oikawa H, Sato M, et al. Downregulation of microRNA-211 is involved in expression of preferentially expressed antigen of melanoma in melanoma cells. Int J Oncol (2011) 39:665–72. doi: 10.3892/ijo.2011.1084
55. Cha JA, Song H-S, Kang B, Park MN, Park KS, Kim S-H, et al. miR-211 Plays a Critical Role in Cnidium officinale Makino Extract-Induced, ROS/ER Stress-Mediated Apoptosis in U937 and U266 Cells. Int J Mol Sci (2018) 19:865. doi: 10.3390/ijms19030865
56. Chen G, Huang P, Xie J, Li R. microRNA−211 suppresses the growth and metastasis of cervical cancer by directly targeting ZEB1. Mol Med Rep (2018) 17:1275–82. doi: 10.3892/mmr.2017.8006
57. Xu D, Liu S, Zhang L, Song L. MiR-211 inhibits invasion and epithelial-to-mesenchymal transition (EMT) of cervical cancer cells via targeting MUC4. Biochem Biophys Res Commun (2017) 485:556–62. doi: 10.1016/j.bbrc.2016.12.020
58. Wang X-D, Wen F-X, Liu B-C, Song Y. MiR-211 inhibits cell epithelial-mesenchymal transition by targeting MMP9 in gastric cancer. Int J Clin Exp Pathol (2017) 10:7551–8.
59. Geethadevi. An interplay between MicroRNA and SOX4 in the regulation of epithelial–mesenchymal transition and cancer progression. Available at: https://www.cancertm.com/article.asp?issn=2395-3977;year=2018;volume=4;issue=1;spage=17;epage=27;aulast=Geethadevi (Accessed December 15, 2020).
60. Liu D, Li Y, Luo G, Xiao X, Tao D, Wu X, et al. LncRNA SPRY4-IT1 sponges miR-101-3p to promote proliferation and metastasis of bladder cancer cells through up-regulating EZH2. Cancer Lett (2017) 388:281–91. doi: 10.1016/j.canlet.2016.12.005
61. Yang C, Zheng J, Xue Y, Yu H, Liu X, Ma J, et al. The Effect of MCM3AP-AS1/miR-211/KLF5/AGGF1 Axis Regulating Glioblastoma Angiogenesis. Front Mol Neurosci (2018) 10:1–14. doi: 10.3389/fnmol.2017.00437
62. Deng B, Qu L, Li J, Fang J, Yang S, Cao Z, et al. MiRNA-211 suppresses cell proliferation, migration and invasion by targeting SPARC in human hepatocellular carcinoma. Sci Rep (2016) 6:26679. doi: 10.1038/srep26679
63. Song G, Zhao Y. MicroRNA-211, a direct negative regulator of CDC25B expression, inhibits triple-negative breast cancer cells’ growth and migration. Tumour Biol (2015) 36:5001–9. doi: 10.1007/s13277-015-3151-6
64. Cai C, Ashktorab H, Pang X, Zhao Y, Sha W, Liu Y, et al. MicroRNA-211 Expression Promotes Colorectal Cancer Cell Growth In Vitro and In Vivo by Targeting Tumor Suppressor CHD5. PloS One (2012) 7:e29750. doi: 10.1371/journal.pone.0029750
65. Chang K-W, Liu C-J, Chu T-H, Cheng H-W, Hung P-S, Hu W-Y, et al. Association between High miR-211 microRNA Expression and the Poor Prognosis of Oral Carcinoma. J Dent Res (2008) 87:1063–8. doi: 10.1177/154405910808701116
66. Hong W, Yu S, Zhuang Y, Zhang Q, Wang J, Gao X. SRCIN1 Regulated by circCCDC66/miR-211 Is Upregulated and Promotes Cell Proliferation in Non-Small-Cell Lung Cancer. BioMed Res Int (2020) 2020:e5307641. doi: 10.1155/2020/5307641
67. Chu T-H, Yang C-C, Liu C-J, Lui M-T, Lin S-C, Chang K-W. miR-211 promotes the progression of head and neck carcinomas by targeting TGFβRII. Cancer Lett (2013) 337:115–24. doi: 10.1016/j.canlet.2013.05.032
68. Lee B, Sahoo A, Sawada J, Marchica J, Sahoo S, Layng FIAL, et al. MicroRNA-211 Modulates the DUSP6-ERK5 Signaling Axis to Promote BRAFV600E-Driven Melanoma Growth In Vivo and BRAF/MEK Inhibitor Resistance. J Invest Dermatol (2021) 141:385–94. doi: 10.1016/j.jid.2020.06.038
69. Chen Y-F, Yang C-C, Kao S-Y, Liu C-J, Lin S-C, Chang K-W. MicroRNA-211 Enhances the Oncogenicity of Carcinogen-Induced Oral Carcinoma by Repressing TCF12 and Increasing Antioxidant Activity. Cancer Res (2016) 76:4872–86. doi: 10.1158/0008-5472.CAN-15-1664
70. Bu Y, Yoshida A, Chitnis N, Altman BJ, Tameire F, Oran A, et al. A PERK–miR-211 axis suppresses circadian regulators and protein synthesis to promote cancer cell survival. Nat Cell Biol (2018) 20:104–15. doi: 10.1038/s41556-017-0006-y
71. Lee H, Lee S, Bae H, Kang H-S, Kim SJ. Genome-wide identification of target genes for miR-204 and miR-211 identifies their proliferation stimulatory role in breast cancer cells. Sci Rep (2016) 6:25287. doi: 10.1038/srep25287
72. Kozubek J, Ma Z, Fleming E, Duggan T, Wu R, Shin D-G, et al. In-depth characterization of microRNA transcriptome in melanoma. PloS One (2013) 8:e72699. doi: 10.1371/journal.pone.0072699
73. Zhang J, Lv J, Zhang F, Che H, Liao Q, Huang W, et al. MicroRNA-211 expression is down-regulated and associated with poor prognosis in human glioma. J Neurooncol (2017) 133:553–9. doi: 10.1007/s11060-017-2464-2
74. Hao P, Kang B, Yao G, Hao W, Ma F. MicroRNA-211 suppresses prostate cancer proliferation by targeting SPARC. Oncol Lett (2018) 15:4323–9. doi: 10.3892/ol.2018.7877
75. Guda MR, Asuthkar S, Labak CM, Tsung AJ, Alexandrov I, Mackenzie MJ, et al. Targeting PDK4 inhibits breast cancer metabolism. Am J Cancer Res (2018) 8:1725–38.
76. Gaur A, Jewell DA, Liang Y, Ridzon D, Moore JH, Chen C, et al. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res (2007) 67:2456–68. doi: 10.1158/0008-5472.CAN-06-2698
77. Chou Y-T, Lin H-H, Lien Y-C, Wang Y-H, Hong C-F, Kao Y-R, et al. EGFR promotes lung tumorigenesis by activating miR-7 through a Ras/ERK/Myc pathway that targets the Ets2 transcriptional repressor ERF. Cancer Res (2010) 70:8822–31. doi: 10.1158/0008-5472.CAN-10-0638
78. Kalinowski FC, Brown RAM, Ganda C, Giles KM, Epis MR, Horsham J, et al. microRNA-7: a tumor suppressor miRNA with therapeutic potential. Int J Biochem Cell Biol (2014) 54:312–7. doi: 10.1016/j.biocel.2014.05.040
79. Banzhaf-Strathmann J, Edbauer D. Good guy or bad guy: the opposing roles of microRNA 125b in cancer. Cell Commun Signal (2014) 12:30. doi: 10.1186/1478-811X-12-30
80. Svoronos AA, Engelman DM, Slack FJ. OncomiR or Tumor Suppressor? The Duplicity of MicroRNAs in Cancer. Cancer Res (2016) 76:3666–70. doi: 10.1158/0008-5472.CAN-16-0359
81. Bu H, Wedel S, Cavinato M, Jansen-Dürr P. MicroRNA Regulation of Oxidative Stress-Induced Cellular Senescence. Oxid Med Cell Longevity (2017) 2017:e2398696. doi: 10.1155/2017/2398696
82. Lee B, Sahoo A, Marchica J, Holzhauser E, Chen X, Li J-L, et al. The long noncoding RNA SPRIGHTLY acts as an intranuclear organizing hub for pre-mRNA molecules. Sci Adv (2017) 3:e1602505. doi: 10.1126/sciadv.1602505
83. Bell RE, Khaled M, Netanely D, Schubert S, Golan T, Buxbaum A, et al. Transcription factor/microRNA axis blocks melanoma invasion program by miR-211 targeting NUAK1. J Invest Dermatol (2014) 134:441–51. doi: 10.1038/jid.2013.340
84. Chitnis NS, Pytel D, Bobrovnikova-Marjon E, Pant D, Zheng H, Maas NL, et al. miR-211 is a prosurvival microRNA that regulates chop expression in a PERK-dependent manner. Mol Cell (2012) 48:353–64. doi: 10.1016/j.molcel.2012.08.025
85. Nisticò P, Bissell MJ, Radisky DC. Epithelial-Mesenchymal Transition: General Principles and Pathological Relevance with Special Emphasis on the Role of Matrix Metalloproteinases. Cold Spring Harb Perspect Biol (2012) 4(2):a011908. doi: 10.1101/cshperspect.a011908
86. Micalizzi DS, Farabaugh SM, Ford HL. Epithelial-mesenchymal transition in cancer: parallels between normal development and tumor progression. J Mammary Gland Biol Neoplasia (2010) 15:117–34. doi: 10.1007/s10911-010-9178-9
87. Munro EG, Jain M, Oliva E, Kamal N, Lele SM, Lynch MP, et al. Upregulation of MUC4 in cervical squamous cell carcinoma: pathologic significance. Int J Gynecol Pathol (2009) 28:127–33. doi: 10.1097/PGP.0b013e318184f3e0
88. Wang C-Y, Hua L, Sun J, Yao K-H, Chen J-T, Zhang J-J, et al. MiR-211 inhibits cell proliferation and invasion of gastric cancer by down-regulating SOX4. Int J Clin Exp Pathol (2015) 8:14013–20.
89. Ruan S, Zhang H, Tian X, Zhang Z, Huang H, Shi C, et al. PHD Finger Protein 19 Enhances the Resistance of Ovarian Cancer Cells to Compound Fuling Granule by Protecting Cell Growth, Invasion, Migration, and Stemness. Front Pharmacol (2020) 11:150. doi: 10.3389/fphar.2020.00150
90. Jayakumar AR, Apeksha A, Norenberg MD. Role of Matricellular Proteins in Disorders of the Central Nervous System. Neurochem Res (2017) 42:858–75. doi: 10.1007/s11064-016-2088-5
91. Hurt EM, Saykally JN, Anose BM, Kalli KR, Sanders MM. Expression of the ZEB1 (δEF1) transcription factor in human: additional insights. Mol Cell Biochem (2008) 318:89–99. doi: 10.1007/s11010-008-9860-z
92. Singh M, Spoelstra NS, Jean A, Howe E, Torkko KC, Clark HR, et al. ZEB1 expression in type I vs type II endometrial cancers: a marker of aggressive disease. Mod Pathol (2008) 21:912–23. doi: 10.1038/modpathol.2008.82
93. Graham TR, Zhau HE, Odero-Marah VA, Osunkoya AO, Kimbro KS, Tighiouart M, et al. Insulin-like growth factor-I-dependent up-regulation of ZEB1 drives epithelial-to-mesenchymal transition in human prostate cancer cells. Cancer Res (2008) 68:2479–88. doi: 10.1158/0008-5472.CAN-07-2559
94. Drake JM, Strohbehn G, Bair TB, Moreland JG, Henry MD. ZEB1 enhances transendothelial migration and represses the epithelial phenotype of prostate cancer cells. Mol Biol Cell (2009) 20:2207–17. doi: 10.1091/mbc.e08-10-1076
95. Zhang S, Ma H, Zhang D, Xie S, Wang W, Li Q, et al. LncRNA KCNQ1OT1 regulates proliferation and cisplatin resistance in tongue cancer via miR-211-5p mediated Ezrin/Fak/Src signaling. Cell Death Dis (2018) 9:1–16. doi: 10.1038/s41419-018-0793-5
96. Chang C-C, Lin C-C, Wang C-H, Huang C-C, Ke T-W, Wei P-L, et al. miR-211 regulates the expression of RRM2 in tumoral metastasis and recurrence in colorectal cancer patients with a k-ras gene mutation. Oncol Lett (2018) 15:8107–17. doi: 10.3892/ol.2018.8295
97. Narayan N, Morenos L, Yuen HLA, Chen M, Fornerod M, Zwaan CM, et al. Microrna-211 - a Novel Oncogene in Acute Myeloid Leukemia. Blood (2017) 130:2507–7. doi: 10.1182/blood.V130.Suppl_1.2507.2507
98. Zheng J, Wang J, Jia Y, Liu T, Duan Y, Liang X, et al. microRNA-211 promotes proliferation, migration, and invasion ability of oral squamous cell carcinoma cells via targeting the bridging integrator 1 protein. J Cell Biochem (2019) 120:4644–53. doi: 10.1002/jcb.27753
99. Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res (2004) 32:D109–11. doi: 10.1093/nar/gkh023
100. Liston P, Fong WG, Kelly NL, Toji S, Miyazaki T, Conte D, et al. Identification of XAF1 as an antagonist of XIAP anti-Caspase activity. Nat Cell Biol (2001) 3:128–33. doi: 10.1038/35055027
101. DiSepio D, Ghosn C, Eckert RL, Deucher A, Robinson N, Duvic M, et al. Identification and characterization of a retinoid-induced class II tumor suppressor/growth regulatory gene. Proc Natl Acad Sci USA (1998) 95:14811–5. doi: 10.1073/pnas.95.25.14811
102. Higuchi E, Chandraratna RAS, Hong WK, Lotan R. Induction of TIG3, a putative class II tumor suppressor gene, by retinoic acid in head and neck and lung carcinoma cells and its association with suppression of the transformed phenotype. Oncogene (2003) 22:4627–35. doi: 10.1038/sj.onc.1206235
103. Hofmann TG, Glas C, Bitomsky N. HIPK2: A tumour suppressor that controls DNA damage-induced cell fate and cytokinesis. Bioessays (2013) 35:55–64. doi: 10.1002/bies.201200060
104. Jiao D, Huan Y, Zheng J, Wei M, Zheng G, Han D, et al. UHRF1 promotes renal cell carcinoma progression through epigenetic regulation of TXNIP. Oncogene (2019) 38:5686–99. doi: 10.1038/s41388-019-0822-6
105. Thwin KKM, Ishida T, Uemura S, Yamamoto N, Lin KS, Tamura A, et al. Level of Seven Neuroblastoma-Associated mRNAs Detected by Droplet Digital PCR Is Associated with Tumor Relapse/Regrowth of High-Risk Neuroblastoma Patients. J Mol Diagn (2020) 22:236–46. doi: 10.1016/j.jmoldx.2019.10.012
106. Reich NC, Levine AJ. Growth regulation of a cellular tumour antigen, p53, in nontransformed cells. Nature (1984) 308:199–201. doi: 10.1038/308199a0
107. Finlay CA, Hinds PW, Levine AJ. The p53 proto-oncogene can act as a suppressor of transformation. Cell (1989) 57:1083–93. doi: 10.1016/0092-8674<(>89<)>90045-7
108. Popper K. The Logic of Scientific Discovery. London: Routledge (2005). doi: 10.4324/9780203994627
109. Shaffer SM, Dunagin MC, Torborg SR, Torre EA, Emert B, Krepler C, et al. Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature (2017) 546:431–5. doi: 10.1038/nature22794
110. Saunders MA, Liang H, Li W-H. Human polymorphism at microRNAs and microRNA target sites. Proc Natl Acad Sci USA (2007) 104:3300–5. doi: 10.1073/pnas.0611347104
111. Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer (2010) 10:389–402. doi: 10.1038/nrc2867
112. Piletic K, Kunej T. MicroRNA-Target Interactions Reloaded: Identification of Potentially Functional Sequence Variants Within Validated MicroRNA-Target Interactions. OMICS (2018) 22:700–8. doi: 10.1089/omi.2018.0159
113. Becker JC, zur Hausen A. Cells of Origin in Skin Cancer. J Invest Dermatol (2014) 134:2491–3. doi: 10.1038/jid.2014.233
114. Murphy MP. How mitochondria produce reactive oxygen species. Biochem J (2009) 417:1–13. doi: 10.1042/BJ20081386
115. Levin M. Morphogenetic fields in embryogenesis, regeneration, and cancer: non-local control of complex patterning. Biosystems (2012) 109:243–61. doi: 10.1016/j.biosystems.2012.04.005
116. Sherman MS, Lorenz K, Lanier MH, Cohen BA. Cell-to-cell variability in the propensity to transcribe explains correlated fluctuations in gene expression. Cell Syst (2015) 1:315–25. doi: 10.1016/j.cels.2015.10.011
Keywords: microRNA-211, melanoma, tumor-promoter, tumor suppresser, epigenetics, miRNA, bistability, MIR211
Citation: Ray A, Kunhiraman H and Perera RJ (2021) The Paradoxical Behavior of microRNA-211 in Melanomas and Other Human Cancers. Front. Oncol. 10:628367. doi: 10.3389/fonc.2020.628367
Received: 11 November 2020; Accepted: 21 December 2020;
Published: 08 February 2021.
Edited by:
Vladimir Spiegelman, Penn State Milton S. Hershey Medical Center, United StatesReviewed by:
Consuelo Amantini, University of Camerino, ItalyCopyright © 2021 Ray, Kunhiraman and Perera. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ranjan J. Perera, anBlcmVyYTJAamguZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.