
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Oncol. , 27 January 2021
Sec. Gastrointestinal Cancers
Volume 10 - 2020 | https://doi.org/10.3389/fonc.2020.620352
Hepatocellular carcinoma (HCC) is a major cause of morbidity and mortality worldwide. Although therapeutic strategies have recently advanced, tumor metastasis and drug resistance continue to pose challenges in the treatment of HCC. Therefore, new molecular targets are needed to develop novel therapeutic strategies for this cancer. E-twenty-six (ETS) transcription family has been implicated in human malignancies pathogenesis and progression, including leukemia, Ewing sarcoma, gastrointestinal stromal tumors. Recently, increasing studies have expanded its great potential as functional players in other cancers, including HCC. This review focuses primarily on the key functions and molecular mechanisms of ETS factors in HCC. Elucidating these molecular details may provide novel potential therapeutic strategies for cancers.
Hepatocellular carcinoma (HCC) is a major cause of cancer-related death worldwide, placing a heavy burden on health systems. Although new advances in diagnostic methods and treatment over recent decades, the five-year survival rate of advanced HCC remains low mainly due to its characteristics of frequent metastasis and high drug resistance (1, 2). HCC has high molecular heterogeneity, and its development is a multi-stage biological process accompanying the accumulation of molecular alterations (3). Therefore, it is critically important for us to gain a more comprehensive and profound understanding of regulatory mechanisms in HCC.
The E-twenty-six (ETS) family of transcription factors, evolutionarily conserved and ubiquitously distributed throughout the metazoan lineage, were firstly discovered in the avian leukemia retrovirus E26 (4). Later, ETS1 and ETS2, the human homolog of virus-ETS, were identified on chromosome 11 and 21, respectively (5). To date, 12 subfamilies with 28 family members have been described in humans (6, 7). ETS transcription factors govern the diverse developmental process and vary from embryonic development to adult cells (8, 9). Further, many studies have focused on aberrant expression of the ETS family in diverse human malignancies. Mutations of ETV6 are frequently observed in hematopoietic diseases (10, 11). Besides, gene fusions or gene rearrangements of ETS factors are common driving events in Ewing’s sarcoma and prostate cancer (12, 13). The TMPRSS2/ERG rearrangement drives aberrant expression of ERG in 50% of prostate tumors (14). Moreover, research on targeting ETS fusions is in progress. Also, deregulation of ETS genes affected by inappropriate expression can occur in some other cancers, including breast, liver, colorectal, pancreatic cancer, and melanomas, which may involve different mechanisms and targeting strategies (6, 15–17). This review aims to conclude the roles of ETS family and discuss the regulatory mechanisms and possible therapeutic strategies involving HCC.
ETS family is one of the largest transcription families and is sub-classified by their sequence homology and different domains. All the members share a ~85-amino-acid conserved DNA-binding domain, known as the ETS domain, the family defining characteristic (7). It contains a stable winged helix-turn-helix structure (wHTH), recognizing a GGAA/T core motif. ETS factors bind to the genes regulatory regions to direct their expression when helix 3 inserts into the DNA backbone (18, 19). A subset of human ETS factors (11 of 28) is characterized by a pointed (PNT) domain comprising a 65~85-amino-acid region. It performs various functions, such as mediating protein-protein interaction and transcriptional regulation (20, 21). Through rigid body interaction, it plays a vital part in facilitating efficient phosphorylation of ETS1 by ERK2 (22). Moreover, the transactivation domain (TAD) is the key to activate the activity of transcription factors. ERK2 could phosphorylate TAD of ELK1 at multiple sites. The phosphorylation rates at 8 TAD sites are significantly different, which could either promote or inhibit transcriptional activation, thus ensuring a self-limiting response of ELK1 to ERK activation (23).
Different signal-transduction pathways and protein partners could modulate ETS factors. Stimulated by multiple growth factors, ETS factors are upregulated and activated by several signaling pathways, including the Ras/MAPK pathway and PI3K/AKT pathway. Recruiting CBP, the general coactivator of ETS1, the Ras/MAPK pathway could enhance the expression of ETS1 target genes (21, 24). Moreover, ETS factors could cooperate with AP-1 factors (JUN and FOS) whose motifs are close to the ETS binding sequence (13). Regulated by ETS factors, ETS/AP-1 enhancers could mediate promoters’ response to the Ras/ERK pathway, thus promoting ETS1’s own expression (25). And increasingly, a variety of other cooperative partners have been studied. For example, recent studies have found that ETS factors could cooperate with the FOX family as FOX-ETS pairs via DNA shape readout. R409 of ETS1 is vital for FOXO1-ETS1 interaction (26).
Although there are sequence similarity and functional redundancy among members, ETS proteins perform their distinct roles depending on the different contexts and regulatory mechanisms. ETS family has been related to various biological processes, such as cell migration, vasculogenesis, hematopoiesis, and homeostasis of epithelial cells. The function of ETS1 in regulating cell mobility is evolutionarily conserved. The mutation of ETS homolog in Drosophila reduces cell migratory potential; upregulation of ETS1 promotes cell migration of human vein endothelial cells (27, 28). ETV2, a master regulator of vasculogenesis, is dispensable for mesodermal lineage development (29). Transient re-expression of ETV2 could reset adult endothelial cells into an embryonic-like stage on which these cells are capable of vascularizing tissues for organ development (30). Several ETS factors, including ETS1, FLI1, PU.1, and SPIB, could control differentiation, localization, and homeostasis of B cells by regulating BCR signaling genes, BCL2 family, and positioning via a transcriptional network (31). Furthermore, modulating cell homeostasis is another important function of ETS proteins. In the liver, ERG promotes homeostasis of epithelial cells via controlling TGFβ-SMAD signaling. Ablation of ERG in mice leads to endothelial-to-mesenchymal transition and liver fibrosis (32). In addition, ETS1 regulates the antifibrotic properties of hepatic stellate cells (33). These studies suggest that some ETS factors play a protective role in the normal liver. However, when expressed in cancer, they display different abilities.
The aberrant expression of ETS factors is frequently observed in HCC, which is shown in Table 1. Of note, the expression of ETS1 in HCC tissues is higher than that in normal liver, whereas it is lower than that in noncancerous lesions (48). Besides, in terms of differently differentiated HCC tissues, the proportion of ETS1 expression is the highest in poorly differentiated HCC samples (49). These suggest a special character of ETS1 in tumorigenesis. Importantly, some ETS factors’ aberrant expression is associated with the clinicopathological characteristics and survival rate of HCC patients. Low PDEF expression was detected in 83.6% HCC patients and was related to larger tumor volume and more frequent microvascular invasion (39). The elevated expression of ELF3 has a positive correlation with larger tumor size and poor prognosis. Besides, it is an independent risk factor for survival of HCC patients (38). Additionally, the time to progression and overall survival is worse in HCC patients receiving sorafenib harboring high ETS1 level than those showing low ETS1 expression (50). These findings suggest that ETS factors might become potential prognostic markers for HCC patients, which would help stratify the patients and evaluate the therapeutic effect.
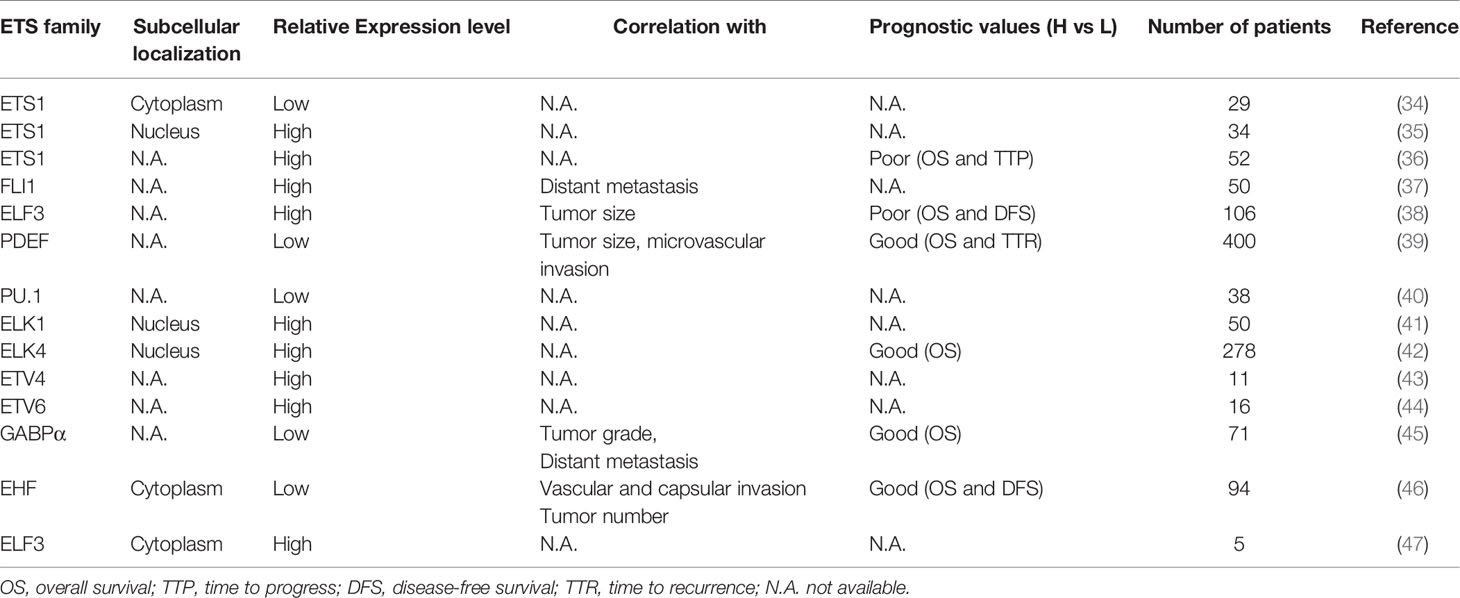
Table 1 The expression of the ETS transcription factors in HCC and their association with clinical-pathological features and patients’ survival.
Post-translational modifications are vital for these factors, such as phosphorylation. For instance, the Ras/Raf/MEK/ERK signaling cascade, one of the most important dysregulated pathways in HCC, plays a pivotal role in regulating the ETS superfamily. Activated ERK1/2 in the nucleus can nearly phosphorylate all the oncogenic ETS factors in vitro (13). ERK could phosphorylate ETS1 at T38 and S41 residues in various cell types, necessary for ETS1 to activate migration genes of epithelial cells modulated by Ras (51). Phosphorylation of ELK1 at Ser383 induced by sustained ERK activation promotes apoptotic cell death via upregulating death receptor 5 expression (52). What is more, ELK1 phosphorylation could be impeded by MDM2 binding protein via inhibiting nuclear translocation of phosphorylated ERK (53). It is worthy to note that ERK could indirectly promote ETS expression by regulating CIC, a tumor suppressor in various cancers, including HCC. CIC repression on PEA3 sub-group genes (ETV1, ETV4, and ETV5) is relieved following ERK phosphorylation (54).
Noteworthy, there are specific microRNAs (miRNAs) that participate in the regulation of ETS factors expression. MiR-324-5p negatively regulates ETS1 expression, thus inhibiting the modulation of MMP2 and MMP9 by ETS1 (55). Overexpression of miR-193b and miR-129-5p could, respectively, lead to decreased mRNA and protein levels of ETS1 via directly targeting its 3′-UTR (56, 57).
It has been recently shown that ETS1 could be modified by m6A in HCC, mediated by Wilms tumor 1-associated protein (WTAP) (34). WTAP, a vital component of the methyltransferase complex, increases m6A modification of ETS1 mRNA by interfering with the connection between HuR protein and ETS1 mRNA, and thus represses ETS1 expression.
Another interesting mechanism is that ETS factors can obtain transcriptional binding motif and actively bind to the region, provided by cis-acting mutations in the telomerase reverse transcriptase (TERT) promoter (6, 58). Somatic mutations of TERT promoter have been observed in some cancers, such as melanoma (71%) and HCC (60%) (59). It is one of the most common genetic alterations in HCC. Unfortunately, TERT is not clinically actionable in HCC at present (2, 60). Sequencing the promoter region of TERT in 305 HCC samples, Nault et al. found that mutations located at -124bp and -146bp from the ATG start site, consisting of G to A or G to T substitutions, generated typical ETS-binding motifs (61). GABAα could be selectively recruited to the mutant TERT core promoter (CCGGAA) in HepG2 cells harboring G228A mutation, activating TERT gene expression (58). In HCC, telomerase activation is critical in driving hepatocarcinogenesis. Recently, another novel cis-activating mechanism has been revealed: binding to the chimeric HBV EnhI at the TERT promoter, ELF4 could achieve TERT activation in TERT HBV-integrated HCC (62).
As cancer cells acquire a range of capabilities, such as sustained proliferation, resisting cell death, inducing angiogenesis, invasion, metastasis, and immune escape, they start to get out of control in human bodies (63). This process is brought by the changes in gene structure and function of cancer cells. Deregulation of ETS factors expression has been found to influence cellular proliferation, epithelial-mesenchymal transition (EMT), invasion, metastasis, and drug resistance of HCC cells (Figure 1A).
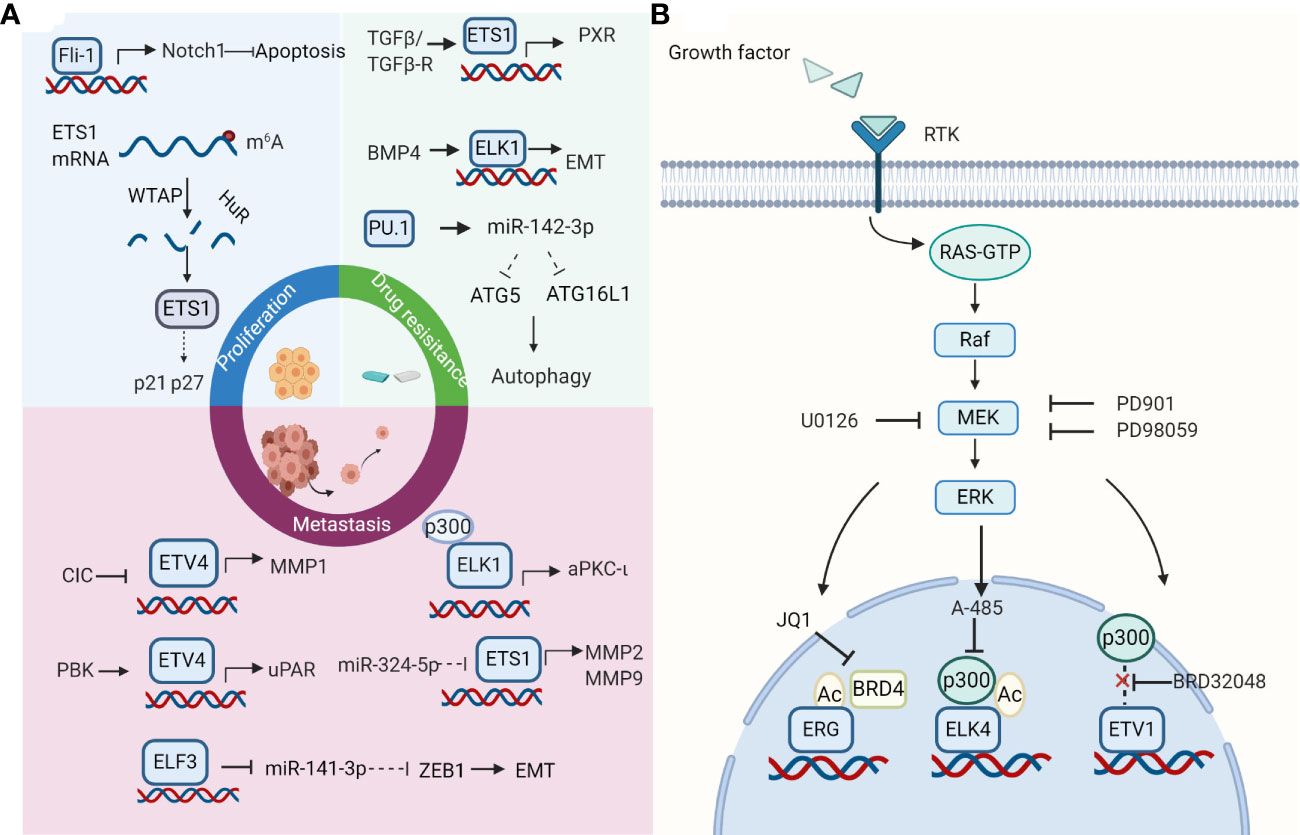
Figure 1 Roles and therapeutic strategies of the E-twenty-six (ETS) family. (A) Effects of ETS family on hepatocellular carcinoma (HCC) cells. ETS factors take part in cell proliferation, invasion, metastasis, and drug resistance in HCC. Selected examples of ETS factors and their upstream factors or genomic targets are shown. Diagram is adapted from reference (63). (B) ETS transcription factors as therapeutic targets. The strategies for targeting ETS factors include breaking its association with protein partners, inhibiting its regulatory pathways, using its small molecule inhibitors. The strategy of combination therapy to target the ETS family in cancers appears to be a promising way for future research. Created with BioRender.com.
Several studies have reported roles that ETS genes play in HCC proliferation. Chen et al. found that PDEF repressed cell proliferation by regulating their apoptosis and cell cycle in HCC (39). Moreover, Fli-1 inhibits HCC cell apoptosis and facilitates cell proliferation by regulating the levels of Bax and Bacl2 via the Notch1 pathway (37). EHF is mainly located in the cytoplasm and suppresses HepG2 cell growth both in vitro and in vivo (46). Interestingly, the different expression of ETS1 in the nucleus and cytoplasm may exhibit different roles in HCC cell proliferation. The elevated expression of ETS1 in the nucleus induces expression of HK1 and PFKFB3 and rescues the decrease of HCC cell proliferation induced by miR-139-5p in SK-Hep-1 and SMMC-7721 cells, indicating that ETS1 probably plays a carcinogenic role in HCC (35). On the contrary, decreased expression of ETS1 in cytoplasm mediated by WTAP leads to down-regulation of p21 and p27 and promotes HCC cell proliferation (34).
Studies have demonstrated a connection between high ETS expression and increased HCC cell invasion and metastasis. ETS1 could upregulate MMP2, MMP7, and MMP9 expression to enhance HCC cell invasive and metastatic capacity (55, 64). ETS2 promotes cell invasion by interacting with MLL to transactivate MMP1 and MMP3 (65). ELF3 represses E-cadherin and promotes EMT in HCC cells via suppressing miR-141-3p, thereby activating ZEB1 (38). ELK1 promotes invasion and EMT in HCC cells via upregulating aPKC-ι expression (66). Knocking down ETV4 completely blocks the facilitation of cell invasion and migration mediated by CIC deficiency (67). ETV4 can also modulate cell invasion and migration by directly binding to the core region of the uPAR promoter (68). Meanwhile, PDZ-binding kinase (PBK) could promote the binding of ETV4 to uPAR promoter. Combined, these studies indicate that targeting ETS factors could be a promising therapeutic strategy to treat HCC, especially in its metastatic forms.
Recently, some ETS factors have been recognized to be functional players in drug resistance. ETS1 could interact with the pregnane X receptor (PXR) to promote its accumulation in the nucleus, thus accelerating sorafenib clearance from hepatoma cells. In the meantime, ETS1 could improve the sorafenib-resistance of HCC in vivo tumor models (36). Another study further explored the upstream regulation mechanism of ETS1 in drug resistance (50). TGFβ1/ERK could promote drug resistance at the late stages of HCC. TGF-β pathway enhances the binding of ETS1 to the PXR promoter. Moreover, ERK inhibitor effectively blocks TGFβ - induced PXR expression. Furthermore, evidence suggests that acquired chemoresistance is closely related to EMT (69). ELK1 could mediate BMP4-induced oxaliplatin resistance in HCC, attributed to EMT initiation through E-cadherin (70). Blocking MEK/ERK/ELK1 could weaken EMT and improve the sensitivity of oxaliplatin (70). Finally, the cytoprotection of autophagy is another drug resistance mechanism, enabling cells to adapt to environmental stress and protect HCC cells treated with sorafenib from death. PU.1/miR-142-3p axis could target ATG5 and ATG16L1, two key autophagy-related proteins, to suppress protective autophagy and enhance apoptosis, thereby improving the sensitivity of HCC cells to sorafenib (40).
Transcription factors were long considered to be “undruggable” targets as their potential for off-target effects and insufficient delivery within the cell. Nevertheless, multiple attempts have been made to target ETS factors directly or indirectly, some of which appear promising. Moreover, interest in combination therapies is rapidly increasing as it may improve the outcomes in the front-line setting. Studies above suggest that ETS proteins may be a potential therapeutic target for HCC. Although the therapeutic strategy of targeting ETS factors in HCC has not been thoroughly studied, studies in other cancers may provide some ideas for the treatment of HCC (Figure 1B).
One way of targeting ETS factors is breaking interaction with their transcriptional co-factors. For example, CBP/p300 are major histone acetyltransferases (HATs) that could interact with and acetylate ETS transcription factors (21). A series of CBP/p300 inhibitors could inhibit the expression and activity of ETS members effectively. CBP30, a small‐molecule inhibitor of EP300, which is the vital constituent of the CBP/p300 complex, markedly inhibits the expression of ETV4 in HCC cells (71). A-485, an inhibitor of CBP/p300, could decrease the level of acetylated ELK4 and limit its transcriptional activity (72). Besides, acetylated ETS factors could be recognized by BET proteins. These BET proteins build transcriptional regulatory complexes by recognizing acetylated lysine and accumulating transcriptional activity elements (73). BET inhibitors, some of which have been in clinical trials, could suppress the transcriptional activity and expression of ETS factors (74, 75). For instance, transcriptional activation of ELK4 target genes could be inhibited by JQ1, a BET inhibitor, via disrupting the interaction between ELK4 and BRD2 in colorectal cancer (72). JQ1 could inhibit the function of acetylated ERG, which recruits BRD4 to promote transcriptional activation (76). Treatment with JQ1 or OTX015 represses the expression of ETV4 in HCC (71). However, the mechanism between BRD4 and ETV4 in HCC needs further research. Notably, a combination of A-485 and JQ1 has synergistic anti-tumor effects (77).
Alternatively, the pathway which regulates ETS factors could be targeted. For example, MEK/ERK inhibitor PD98059 could effectively decrease the expression of p-ETS1 in HCC cells treated with recombinant TGF-β (50). So far, numerous MEK inhibitors have been developed, but they have limited clinical benefits in solid tumors, including advanced HCC, as single-agent therapies. Researchers began to explore its efficacy as a combination drug. MEK and KIT inhibitors impede gastrointestinal stromal tumor (GIST) growth by interrupting the KIT–ETV1-positive feedback circuit in GIST (78). Moreover, MEK inhibitor U0126 exhibits a synergetic effect with BET inhibitor JQ1 via ELK4 activity inhibition in colorectal cancer (72). Besides, the combination of MEK inhibitor PD901 and JQ1 has a more significant inhibition on the expression of several ETS factors (ELK3, ETV1, ETV4, ETV5) than MEK inhibitor alone in lung cancer (79).
Encouragingly, plenty of small molecule inhibitors targeting ETS proteins such as VPC‐18005, YK‐4‐279 have been developed during the past few years and play a role in cell lines with chromosomal translocation (80). Notably, BRD32048, a tricyclic compound, can also be applied for ETV1-amplified cell lines. BRD32048 inhibits acetylation of ETV1 mediated by p300, thus facilitating its degradation. However, it is not clear whether BRD32048 blocks ETV1 residues K33 acetylation or affects some protein-protein interactions. Moreover, binding to ETV1 directly, BRD32048 suppresses the transcriptional activity of ETV1 on the MMP1 promoter to impede cancer cell invasion and proliferation (81). In addition, structure identification has determined protein 3D structures of many ETS factors, which helps develop structure‐based drugs (80). With the development of new technology and methods such as computer‐aided drug design, it will be possible to find more ETS inhibitors (13). Combinations of these ETS inhibitors and other types of inhibitors mentioned above could provide more strategies for HCC.
HCC is a highly invasive tumor, which is the major cause of the high recurrence rate after surgical resection. For patients with an unresectable tumor or at an advanced stage, drug resistance of tumor often leads to treatment failure. In this respect, identifying potential biomarkers and molecular mediators would help predict the prognosis of this malignancy.
The above studies have shown that the aberrant expression of some ETS factors are related to HCC, suggesting that they could be potential prognostic markers for HCC. ETS family regulates HCC cells, including but not limited to cell proliferation, invasion, metastasis, and drug resistance. Therefore, it is worthwhile to consider some ETS factors as potential targets of HCC. Furthermore, ETS factors expression and activity could be regulated at the transcriptional, post-transcriptional, and post-translational levels. The strategies for targeting ETS can be divided into (1) breaking its association with protein partners, (2) blocking its regulatory pathways, (3) directly inhibiting themselves. Epigenetic inhibitors such as HAT inhibitors and BET inhibitors repress the expression and activity of ETS factors and have enticing suppressive effects in cancers. However, the underlying mechanisms need to be further elucidated. Moreover, the Ras/Raf/MEK/ERK pathway is the most critical regulatory pathway for ETS factors, the inhibitors of which have limited efficacy as a single agent. Evidence suggests that the combination therapy of these inhibitors with additional agents will be of better effects. Also, small-molecule inhibitors targeting ETS proteins are in development, which is likely to serve as one component of combination therapy.
The studies about the ETS family in HCC are currently limited. Recently, emerging evidence has suggested that besides HCC, ETS factors function as important regulators in other gastrointestinal cancers. For instance, overexpression of ETV4, ELF3, and ETV5 facilitates tumor angiogenesis, growth, invasion and metastasis, and indicates poor prognosis (82–85). Besides, ETS1 could regulate cell viability via interfering glycolysis in pancreatic cancer (86); increased expression of ETS2 promotes drug resistance in colorectal cancer (87). Interestingly, unlike its tumor-suppressive role in colorectal cancer and HCC, PDEF is overexpressed in gastric cancer and promotes gastric cancer cell proliferation by regulating FoxM1 expression through a positive feedback loop (39, 88, 89). In conclusion, ETS family may offer vast potential for developing novel therapeutic strategies. With the deepness of research, it will be promising that targeting ETS factors is applied in HCC and other cancers.
TZ, DL, YW, and MS performed the literature search and manuscript drafting. LX supervised and revised the manuscript. All authors contributed to the article and approved the submitted version.
Research was supported by grants from the National Key Research and Development Program of China 2018YFC1312103 (L.X.), National Natural Science Foundation of China No. 81972237 (L.X.), and No. 81772623 (L.X.).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet (2018) 391:1301–14. doi: 10.1016/S0140-6736(18)30010-2
2. Llovet JM, Montal R, Sia D, Finn RS. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat Rev Clin Oncol (2018) 15:599–616. doi: 10.1038/s41571-018-0073-4
3. Craig AJ, von Felden J, Garcia-Lezana T, Sarcognato S, Villanueva A. Tumour evolution in hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol (2020) 17:139–52. doi: 10.1038/s41575-019-0229-4
4. Leprince D, Gegonne A, Coll J, de Taisne C, Schneeberger A, Lagrou C, et al. A putative second cell-derived oncogene of the avian leukaemia retrovirus E26. Nature (1983) 306:395–7. doi: 10.1038/306395a0
5. Watson DK, McWilliams-Smith MJ, Nunn MF, Duesberg PH, O’Brien SJ, Papas TS. The ets sequence from the transforming gene of avian erythroblastosis virus, E26, has unique domains on human chromosomes 11 and 21: both loci are transcriptionally active. Proc Natl Acad Sci U S A (1985) 82:7294–8. doi: 10.1073/pnas.82.21.7294
6. Sizemore GM, Pitarresi JR, Balakrishnan S, Ostrowski MC. The ETS family of oncogenic transcription factors in solid tumours. Nat Rev Cancer (2017) 17:337–51. doi: 10.1038/nrc.2017.20
7. Findlay VJ, LaRue AC, Turner DP, Watson PM, Watson DK. Understanding the role of ETS-mediated gene regulation in complex biological processes. Adv Cancer Res (2013) 119:1–61. doi: 10.1016/B978-0-12-407190-2.00001-0
8. Craig MP, Sumanas S. ETS transcription factors in embryonic vascular development. Angiogenesis (2016) 19:275–85. doi: 10.1007/s10456-016-9511-z
9. Pijuan-Sala B, Wilson NK, Xia J, Hou X, Hannah RL, Kinston S, et al. Single-cell chromatin accessibility maps reveal regulatory programs driving early mouse organogenesis. Nat Cell Biol (2020) 22:487–97. doi: 10.1038/s41556-020-0489-9
10. Gocho Y, Yang JJ. Genetic defects in hematopoietic transcription factors and predisposition to acute lymphoblastic leukemia. Blood (2019) 134:793–7. doi: 10.1182/blood.2018852400
11. Churpek JE, Bresnick EH. Transcription factor mutations as a cause of familial myeloid neoplasms. J Clin Invest (2019) 129:476–88. doi: 10.1172/JCI120854
12. Sbaraglia M, Righi A, Gambarotti M, Dei Tos AP. Ewing sarcoma and Ewing-like tumors. Virchows Arch (2020) 476:109–19. doi: 10.1007/s00428-019-02720-8
13. Nicholas TR, Strittmatter BG, Hollenhorst PC. Oncogenic ETS Factors in Prostate Cancer. Adv Exp Med Biol (2019) 1210:409–36. doi: 10.1007/978-3-030-32656-2_18
14. Blee AM, He Y, Yang Y, Ye Z, Yan Y, Pan Y, et al. Controls Luminal Epithelial Lineage and Antiandrogen Sensitivity in and -Mutated Prostate Cancer. Clin Cancer Res (2018) 24:4551–65. doi: 10.1158/1078-0432.CCR-18-0653
15. Heeg S, Das KK, Reichert M, Bakir B, Takano S, Caspers J, et al. ETS-Transcription Factor ETV1 Regulates Stromal Expansion and Metastasis in Pancreatic Cancer. Gastroenterology (2016) 151:540–53.e14. doi: 10.1053/j.gastro.2016.06.005
16. Buchwalter G, Hickey MM, Cromer A, Selfors LM, Gunawardane RN, Frishman J, et al. PDEF promotes luminal differentiation and acts as a survival factor for ER-positive breast cancer cells. Cancer Cell (2013) 23:753–67. doi: 10.1016/j.ccr.2013.04.026
17. Jané-Valbuena J, Widlund HR, Perner S, Johnson LA, Dibner AC, Lin WM, et al. An oncogenic role for ETV1 in melanoma. Cancer Res (2010) 70:2075–84. doi: 10.1158/0008-5472.CAN-09-3092
18. Sharrocks AD. The ETS-domain transcription factor family. Nat Rev Mol Cell Biol (2001) 2:827–37. doi: 10.1038/35099076
19. Vivekanand P. Lessons from Drosophila Pointed, an ETS family transcription factor and key nuclear effector of the RTK signaling pathway. Genesis (2018) 56:e23257. doi: 10.1002/dvg.23257
20. Luk IY, Reehorst CM, Mariadason JM. ELF3, ELF5, EHF and SPDEF Transcription Factors in Tissue Homeostasis and Cancer. Molecules (2018) 23:2191. doi: 10.3390/molecules23092191
21. Hollenhorst PC, McIntosh LP, Graves BJ. Genomic and biochemical insights into the specificity of ETS transcription factors. Annu Rev Biochem (2011) 80:437–71. doi: 10.1146/annurev.biochem.79.081507.103945
22. Piserchio A, Warthaka M, Kaoud TS, Callaway K, Dalby KN, Ghose R. Local destabilization, rigid body, and fuzzy docking facilitate the phosphorylation of the transcription factor Ets-1 by the mitogen-activated protein kinase ERK2. Proc Natl Acad Sci U S A (2017) 114:E6287–96. doi: 10.1073/pnas.1702973114
23. Mylona A, Theillet F-X, Foster C, Cheng TM, Miralles F, Bates PA, et al. Opposing effects of Elk-1 multisite phosphorylation shape its response to ERK activation. Science (2016) 354:233–7. doi: 10.1126/science.aad1872
24. Nelson ML, Kang HS, Lee GM, Blaszczak AG, Lau DK, McIntosh LP, et al. Ras signaling requires dynamic properties of Ets1 for phosphorylation-enhanced binding to coactivator CBP. Proc Natl Acad Sci U S A (2010) 107:10026–31. doi: 10.1073/pnas.0915137107
25. Dittmer J. The role of the transcription factor Ets1 in carcinoma. Semin Cancer Biol (2015) 35:20–38. doi: 10.1016/j.semcancer.2015.09.010
26. Ibarra IL, Hollmann NM, Klaus B, Augsten S, Velten B, Hennig J, et al. Mechanistic insights into transcription factor cooperativity and its impact on protein-phenotype interactions. Nat Commun (2020) 11:124. doi: 10.1038/s41467-019-13888-7
27. Lavenburg KR, Ivey J, Hsu T, Muise-Helmericks RC. Coordinated functions of Akt/PKB and ETS1 in tubule formation. FASEB J (2003) 17:2278–80. doi: 10.1096/fj.03-0040fje
28. Chen Z, Fisher RJ, Riggs CW, Rhim JS, Lautenberger JA. Inhibition of vascular endothelial growth factor-induced endothelial cell migration by ETS1 antisense oligonucleotides. Cancer Res (1997) 57:2013–9.
29. Koyano-Nakagawa N, Garry DJ. Etv2 as an essential regulator of mesodermal lineage development. Cardiovasc Res (2017) 113:1294–306. doi: 10.1093/cvr/cvx133
30. Palikuqi B, Nguyen DT, Li G, Schreiner R, Pellegata AF, Liu Y, et al. Adaptable haemodynamic endothelial cells for organogenesis and tumorigenesis. Nature (2020) 585:426–32. doi: 10.1038/s41586-020-2712-z
31. Wang H, Morse HC,3, Bolland S. Transcriptional Control of Mature B Cell Fates. Trends Immunol (2020) 41:601–13. doi: 10.1016/j.it.2020.04.011
32. Dufton NP, Peghaire CR, Osuna-Almagro L, Raimondi C, Kalna V, Chauhan A, et al. Dynamic regulation of canonical TGFbeta signalling by endothelial transcription factor ERG protects from liver fibrogenesis. Nat Commun (2017) 8:895. doi: 10.1038/s41467-017-01169-0
33. Liu X, Xu J, Rosenthal S, Zhang L-J, McCubbin R, Meshgin N, et al. Identification of Lineage-Specific Transcription Factors That Prevent Activation of Hepatic Stellate Cells and Promote Fibrosis Resolution. Gastroenterology (2020) 158:1728–44.e14. doi: 10.1053/j.gastro.2020.01.027
34. Chen Y, Peng C, Chen J, Chen D, Yang B, He B, et al. WTAP facilitates progression of hepatocellular carcinoma via m6A-HuR-dependent epigenetic silencing of ETS1. Mol Cancer (2019) 18:127. doi: 10.1186/s12943-019-1053-8
35. Hua S, Lei L, Deng L, Weng X, Liu C, Qi X, et al. miR-139-5p inhibits aerobic glycolysis, cell proliferation, migration, and invasion in hepatocellular carcinoma via a reciprocal regulatory interaction with ETS1. Oncogene (2018) 37:1624–36. doi: 10.1038/s41388-017-0057-3
36. Shao Z, Li Y, Dai W, Jia H, Zhang Y, Jiang Q, et al. ETS-1 induces Sorafenib-resistance in hepatocellular carcinoma cells via regulating transcription factor activity of PXR. Pharmacol Res (2018) 135:188–200. doi: 10.1016/j.phrs.2018.08.003
37. Wang H, Ou Y, Ou J, Jian Z. Fli−1 promotes metastasis by regulating MMP2 signaling in hepatocellular carcinoma. Mol Med Rep (2018) 17:1986–92. doi: 10.3892/mmr.2017.8047
38. Zheng L, Xu M, Xu J, Wu K, Fang Q, Liang Y, et al. ELF3 promotes epithelial-mesenchymal transition by protecting ZEB1 from miR-141-3p-mediated silencing in hepatocellular carcinoma. Cell Death Dis (2018) 9:387. doi: 10.1038/s41419-018-0399-y
39. Chen E-B, Zhou S-L, Pang X-G, Yin D, Miao P-Z, Yang Y, et al. Prostate-derived ETS factor improves prognosis and represses proliferation and invasion in hepatocellular carcinoma. Oncotarget (2017) 8:52488–500. doi: 10.18632/oncotarget.14924
40. Zhang K, Chen J, Zhou H, Chen Y, Zhi Y, Zhang B, et al. PU.1/microRNA-142-3p targets ATG5/ATG16L1 to inactivate autophagy and sensitize hepatocellular carcinoma cells to sorafenib. Cell Death Dis (2018) 9:312. doi: 10.1038/s41419-018-0344-0
41. Yan Q, Lou G, Qian Y, Qin B, Xu X, Wang Y, et al. SPAG9 is involved in hepatocarcinoma cell migration and invasion via modulation of ELK1 expression. OncoTargets Ther (2016) 9:1067–75. doi: 10.2147/OTT.S98727
42. Lee HS, Jung W, Lee E, Chang H, Choi JH, Kim HG, et al. SIRT7, H3K18ac, and ELK4 Immunohistochemical Expression in Hepatocellular Carcinoma. J Pathol Trans Medicine (2016) 50:337–44. doi: 10.4132/jptm.2016.05.20
43. Xiaohui C, Xin L, Dehua W. @ in vitro[E26 transformation-specific variant 4 promotes sorafenib and cisplatin resistance in hepatocellular carcinoma cells]. Nan fang yi ke da xue xue bao = J South Med University (2019) 39:875–82. doi: 10.12122/j.issn.1673-4254.2019.08.01
44. Guo C, Gao C, Zhao D, Li J, Wang J, Sun X, et al. A novel ETV6-miR-429-CRKL regulatory circuitry contributes to aggressiveness of hepatocellular carcinoma. J Exp Clin Cancer Res (2020) 39:70. doi: 10.1186/s13046-020-01559-1
45. Zhang S, Zhang K, Ji P, Zheng X, Jin J, Feng M, et al. GABPA predicts prognosis and inhibits metastasis of hepatocellular carcinoma. BMC Cancer (2017) 17:380. doi: 10.1186/s12885-017-3373-7
46. Lyu Z, Ma M, Xu Y, Wang X, Zhu Y, Ren W, et al. Expression and prognostic significance of epithelial tissue-specific transcription factor ESE3 in hepatocellular carcinoma. Int J Clin Oncol (2020) 25:1334–45. doi: 10.1007/s10147-020-01675-0
47. Wang F, Long Q, Gong Y, Hu L, Zhang H, Oettgen P, et al. Epithelium-Specific ETS (ESE)-1 upregulated GP73 expression in hepatocellular carcinoma cells. Cell Biosci (2014) 4:76. doi: 10.1186/2045-3701-4-76
48. Ito Y, Miyoshi E, Takeda T, Sakon M, Noda K, Tsujimoto M, et al. Expression and possible role of ets-1 in hepatocellular carcinoma. Am J Clin Pathol (2000) 114:719–25. doi: 10.1309/RAVV-8NM1-CJB7-GJFR
49. Kanda K, Nakayama T, Onizuka S, Tomioka T, Kanematsu T. Expression of the Ets-1 proto-oncogene is linked to cell differentiation of human hepatocellular carcinoma. Hepatogastroenterology (2002) 49:747–51.
50. Bhagyaraj E, Ahuja N, Kumar S, Tiwari D, Gupta S, Nanduri R, et al. TGF-β induced chemoresistance in liver cancer is modulated by xenobiotic nuclear receptor PXR. Cell Cycle (2019) 18:3589–602. doi: 10.1080/15384101.2019.1693120
51. Plotnik JP, Budka JA, Ferris MW, Hollenhorst PC. ETS1 is a genome-wide effector of RAS/ERK signaling in epithelial cells. Nucleic Acids Res (2014) 42:11928–40. doi: 10.1093/nar/gku929
52. Kuppusamy P, Nagalingam A, Muniraj N, Saxena NK, Sharma D. Concomitant activation of ETS-like transcription factor-1 and Death Receptor-5 via extracellular signal-regulated kinase in withaferin A-mediated inhibition of hepatocarcinogenesis in mice. Sci Rep (2017) 7:17943. doi: 10.1038/s41598-017-18190-4
53. Ranjan A, Iyer SV, Ward C, Link T, Diaz FJ, Dhar A, et al. MTBP inhibits the Erk1/2-Elk-1 signaling in hepatocellular carcinoma. Oncotarget (2018) 9:21429–43. doi: 10.18632/oncotarget.25117
54. Wong D, Yip S. Making heads or tails - the emergence of capicua (CIC) as an important multifunctional tumour suppressor. J Pathol (2020) 250:532–40. doi: 10.1002/path.5400
55. Cao L, Xie B, Yang X, Liang H, Jiang X, Zhang D, et al. MiR-324-5p Suppresses Hepatocellular Carcinoma Cell Invasion by Counteracting ECM Degradation through Post-Transcriptionally Downregulating ETS1 and SP1. PloS One (2015) 10:e0133074. doi: 10.1371/journal.pone.0133074
56. Xu C, Liu S, Fu H, Li S, Tie Y, Zhu J, et al. MicroRNA-193b regulates proliferation, migration and invasion in human hepatocellular carcinoma cells. Eur J Cancer (2010) 46:2828–36. doi: 10.1016/j.ejca.2010.06.127
57. Ma N, Chen F, Shen S-L, Chen W, Chen L-Z, Su Q, et al. MicroRNA-129-5p inhibits hepatocellular carcinoma cell metastasis and invasion via targeting ETS1. Biochem Biophys Res Commun (2015) 461:618–23. doi: 10.1016/j.bbrc.2015.04.075
58. Bell RJA, Rube HT, Kreig A, Mancini A, Fouse SD, Nagarajan RP, et al. The transcription factor GABP selectively binds and activates the mutant TERT promoter in cancer. Science (2015) 348:1036–9. doi: 10.1126/science.aab0015
59. Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA. Highly recurrent TERT promoter mutations in human melanoma. Science (2013) 339:957–9. doi: 10.1126/science.1229259
60. Schulze K, Imbeaud S, Letouzé E, Alexandrov LB, Calderaro J, Rebouissou S, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet (2015) 47:505–11. doi: 10.1038/ng.3252
61. Nault JC, Mallet M, Pilati C, Calderaro J, Bioulac-Sage P, Laurent C, et al. High frequency of telomerase reverse-transcriptase promoter somatic mutations in hepatocellular carcinoma and preneoplastic lesions. Nat Commun (2013) 4:2218. doi: 10.1038/ncomms3218
62. Sze KM-F, Ho DW-H, Chiu Y-T, Tsui Y-M, Chan L-K, Lee JM-F, et al. HBV-TERT Promoter Integration Harnesses Host ELF4 Resulting in TERT Gene Transcription in Hepatocellular Carcinoma. Hepatology (2020). doi: 10.1002/hep.31231
63. Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell (2016) 29:452–63. doi: 10.1016/j.ccell.2016.03.010
64. Ozaki I, Mizuta T, Zhao G, Yotsumoto H, Hara T, Kajihara S, et al. Involvement of the Ets-1 gene in overexpression of matrilysin in human hepatocellular carcinoma. Cancer Res (2000) 60:6519–25.
65. Takeda S, Liu H, Sasagawa S, Dong Y, Trainor PA, Cheng EH, et al. HGF-MET signals via the MLL-ETS2 complex in hepatocellular carcinoma. J Clin Invest (2013) 123:3154–65. doi: 10.1172/JCI65566
66. Ma C, Huang S, Xu L, Tian L, Yang Y, Wang J. Transcription coactivator P300 activates Elk1-aPKC-ι signaling mediated epithelial-to-mesenchymal transition and malignancy in hepatocellular carcinoma. Oncogenesis (2020) 9:32. doi: 10.1038/s41389-020-0212-5
67. Kim E, Kim D, Lee J-S, Yoe J, Park J, Kim C-J, et al. Capicua suppresses hepatocellular carcinoma progression by controlling the ETV4-MMP1 axis. Hepatology (2018) 67:2287–301. doi: 10.1002/hep.29738
68. Yang Q-X, Zhong S, He L, Jia X-J, Tang H, Cheng S-T, et al. PBK overexpression promotes metastasis of hepatocellular carcinoma via activating ETV4-uPAR signaling pathway. Cancer Lett (2019) 452:90–102. doi: 10.1016/j.canlet.2019.03.028
69. Song H, Liu D, Dong S, Zeng L, Wu Z, Zhao P, et al. Epitranscriptomics and epiproteomics in cancer drug resistance: therapeutic implications. Signal Transduct Target Ther (2020) 5:193. doi: 10.1038/s41392-020-00300-w
70. Ma J, Zeng S, Zhang Y, Deng G, Qu Y, Guo C, et al. BMP4 promotes oxaliplatin resistance by an induction of epithelial-mesenchymal transition via MEK1/ERK/ELK1 signaling in hepatocellular carcinoma. Cancer Lett (2017) 411:117–29. doi: 10.1016/j.canlet.2017.09.041
71. Tsang FH-C, Law C-T, Tang T-CC, Cheng CL-H, Chin DW-C, Tam W-SV, et al. Aberrant Super-Enhancer Landscape in Human Hepatocellular Carcinoma. Hepatology (2019) 69:2502–17. doi: 10.1002/hep.30544
72. Zhu Z, Song J, Guo Y, Huang Z, Chen X, Dang X, et al. LAMB3 promotes tumour progression through the AKT-FOXO3/4 axis and is transcriptionally regulated by the BRD2/acetylated ELK4 complex in colorectal cancer. Oncogene (2020) 39:4666–80. doi: 10.1038/s41388-020-1321-5
73. Zhang S, Chen Y, Tian C, He Y, Tian Z, Wan Y, et al. Dual-target inhibitors based on BRD4: Novel therapeutic approaches for cancer. Curr Med Chem (2020). doi: 10.2174/0929867327666200610174453
74. Cochran AG, Conery AR, Sims RJ,3. Bromodomains: a new target class for drug development. Nat Rev Drug Discovery (2019) 18:609–28. doi: 10.1038/s41573-019-0030-7
75. Gollavilli PN, Pawar A, Wilder-Romans K, Natesan R, Engelke CG, Dommeti VL, et al. EWS/ETS-Driven Ewing Sarcoma Requires BET Bromodomain Proteins. Cancer Res (2018) 78:4760–73. doi: 10.1158/0008-5472.CAN-18-0484
76. Roe J-S, Mercan F, Rivera K, Pappin DJ, Vakoc CR. BET Bromodomain Inhibition Suppresses the Function of Hematopoietic Transcription Factors in Acute Myeloid Leukemia. Mol Cell (2015) 58:1028–39. doi: 10.1016/j.molcel.2015.04.011
77. Zhang X, Zegar T, Lucas A, Morrison-Smith C, Knox T, French CA, et al. Therapeutic targeting of p300/CBP HAT domain for the treatment of NUT midline carcinoma. Oncogene (2020) 39:4770–9. doi: 10.1038/s41388-020-1301-9
78. Ran L, Sirota I, Cao Z, Murphy D, Chen Y, Shukla S, et al. Combined inhibition of MAP kinase and KIT signaling synergistically destabilizes ETV1 and suppresses GIST tumor growth. Cancer Discovery (2015) 5:304–15. doi: 10.1158/2159-8290.CD-14-0985
79. Guerra SL, Maertens O, Kuzmickas R, De Raedt T, Adeyemi RO, Guild CJ, et al. A Deregulated HOX Gene Axis Confers an Epigenetic Vulnerability in KRAS-Mutant Lung Cancers. Cancer Cell (2020) 37:705–19.e6. doi: 10.1016/j.ccell.2020.03.004
80. Hsing M, Wang Y, Rennie PS, Cox ME, Cherkasov A. ETS transcription factors as emerging drug targets in cancer. Med Res Rev (2020) 40:413–30. doi: 10.1002/med.21575
81. Pop MS, Stransky N, Garvie CW, Theurillat JP, Hartman EC, Lewis TA, et al. A small molecule that binds and inhibits the ETV1 transcription factor oncoprotein. Mol Cancer Ther (2014) 13:1492–502. doi: 10.1158/1535-7163.MCT-13-0689
82. Cai LS, Chen QX, Fang SY, Lian MQ, Lian MJ, Cai MZ. ETV4 promotes the progression of gastric cancer through regulating KDM5D. Eur Rev Med Pharmaco (2020) 24:2442–51. doi: 10.26355/eurrev_202003_20511
83. Mesci A, Taeb S, Huang XY, Jairath R, Sivaloganathan D, Liu SK. Pea3 expression promotes the invasive and metastatic potential of colorectal carcinoma. World J Gastroentero (2014) 20:17376–87. doi: 10.3748/wjg.v20.i46.17376
84. Wang JL, Chen ZF, Chen HM, Wang MY, Kong X, Wang YC, et al. Elf3 drives beta-catenin transactivation and associates with poor prognosis in colorectal cancer. Cell Death Dis (2014) 5:e1263. doi: 10.1038/cddis.2014.206
85. Cheng X, Jin Z, Ji X, Shen X, Feng H, Morgenlander W, et al. ETS variant 5 promotes colorectal cancer angiogenesis by targeting platelet-derived growth factor BB. Int J Cancer (2019) 145:179–91. doi: 10.1002/ijc.32071
86. Zhang X, Wu D, Aldarouish M, Yin X, Li C, Wang C. ETS-1: A potential target of glycolysis for metabolic therapy by regulating glucose metabolism in pancreatic cancer. Int J Oncol (2017) 50:232–40. doi: 10.3892/ijo.2016.3770
87. Wang H, Huang R, Guo W, Qin X, Yang Z, Yuan Z, et al. RNA-binding protein CELF1 enhances cell migration, invasion, and chemoresistance by targeting ETS2 in colorectal cancer. Clin Sci (2020) 134:1973–90. doi: 10.1042/cs20191174
88. Lo Y-H, Noah TK, Chen M-S, Zou W, Borras E, Vilar E, et al. SPDEF Induces Quiescence of Colorectal Cancer Cells by Changing the Transcriptional Targets of β-catenin. Gastroenterology (2017) 153:205–218.e208. doi: 10.1053/j.gastro.2017.03.048
Keywords: E-twenty-six transcription factor, hepatocellular carcinoma, metastasis, drug resistance, therapy
Citation: Zhang T, Liu D, Wang Y, Sun M and Xia L (2021) The E-Twenty-Six Family in Hepatocellular Carcinoma: Moving into the Spotlight. Front. Oncol. 10:620352. doi: 10.3389/fonc.2020.620352
Received: 22 October 2020; Accepted: 08 December 2020;
Published: 27 January 2021.
Edited by:
Prathibha Ranganathan, Centre for Human Genetics (CHG), IndiaReviewed by:
Manoj Kumar Kashyap, Amity University Gurgaon, IndiaCopyright © 2021 Zhang, Liu, Wang, Sun and Xia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Limin Xia, eGlhbGltaW5AdGpoLnRqbXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.