
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Oncol. , 11 January 2021
Sec. Hematologic Malignancies
Volume 10 - 2020 | https://doi.org/10.3389/fonc.2020.611467
This article is part of the Research Topic Insights Into Acute Leukemia Research: Genetic Predisposition and Mechanisms of Relapse View all 9 articles
 Xin-Yue Zhang1,2†
Xin-Yue Zhang1,2† Hai-Ping Dai1,2†
Hai-Ping Dai1,2† Zheng Li1,2
Zheng Li1,2 Jia Yin1,2
Jia Yin1,2 Xing-Ping Lang3
Xing-Ping Lang3 Chun-Xiao Yang3
Chun-Xiao Yang3 Sheng Xiao4
Sheng Xiao4 Ming-Qing Zhu1,2
Ming-Qing Zhu1,2 Dan-Dan Liu1,2
Dan-Dan Liu1,2 Hong Liu1,2
Hong Liu1,2 Hong-Jie Shen1,2
Hong-Jie Shen1,2 De-Pei Wu1,2*
De-Pei Wu1,2* Xiao-Wen Tang1,2*
Xiao-Wen Tang1,2*Philadelphia chromosome-like B-lymphoblastic leukemia (Ph-like ALL) describes a group of genetically heterogeneous, Ph-negative entities with high relapse rates and poor prognoses. A Janus-kinase-2 (JAK2) rearrangement has been reported in approximately 7% of Ph-like ALL patients whose therapeutic responses to JAK inhibitors have been studied in clinical trials. Here, we report a novel STRBP-JAK2 fusion gene in a 21-year-old woman with Ph-like ALL. Although a normal karyotype was observed, a hitherto unreported JAK2 rearrangement was detected cytogenetically. STRBP-JAK2 fusion was identified by RNA sequencing and validated by Sanger sequencing. The Ph-like ALL proved refractory to traditional induction chemotherapy combined with ruxolitinib. The patient consented to infusion of autologous chimeric antigen receptor (CAR) T cells against both CD19 and CD22, which induced morphologic remission. Haplo-identical stem cell transplantation was then performed; however, she suffered relapse at just one month after transplantation. The patient subsequently received donor lymphocyte infusion after which she achieved and maintained a minimal residual disease negative remission. However, she succumbed to grade IV graft-versus-host disease 7 months post-transplant. In conclusion, this report describes a novel STRBP-JAK2 gene fusion in a Ph-like ALL patient with a very aggressive disease course, which proved resistant to chemotherapy combined with ruxolitinib but sensitive to immunotherapy. Our study suggests that CAR T-cell therapy may be a viable option for this type of leukemia.
Philadelphia chromosome-like B-lymphoblastic leukemia (Ph-like ALL) is characterized by a gene expression profile resembling Ph-positive ALL (Ph+ ALL) despite an absence of BCR-ABL1 rearrangement. Clinically, Ph-like ALL is deemed a poor-risk subtype of adult B-lymphoblastic leukemia due to the high possibility of induction failure, early relapse, and dismal outcome. Activation of the JAK-STAT pathway occurs in approximately 64% of Ph-like ALL patients, including 42% with CRLF2 rearrangements and 22% with non-CRLF2 rearrangements, both of which are therapeutically targeted by JAK inhibitors (1).
Chromosomal translocations, inversions, and deletions generating JAK2 fusion genes and JAK-STAT activation occur in 7% of Ph-like ALL patients (2). Hitherto, 23 different partner genes are reportedly rearranged with JAK2 in Ph-like ALL, including ATF71P, BCR, EBF1, ETV6, OFD1, PAX5, and ZBTB46 (3). Most reports of these fusion genes have only presented genetic profiles at the expense of clinical findings. Hence, there are only limited data detailing the efficacy of JAK inhibitors and immunotherapy in patients with different JAK2 rearrangements.
Here, we report a novel STRBP-JAK2 fusion gene in a patient with Ph-like ALL which proved resistant to standard induction chemotherapy and ruxolitinib, but achieved complete remission (CR) after chimeric antigen receptor (CAR) T-cell therapy, allogenic stem cell transplantation (allo-HSCT) and donor lymphocyte infusion (DLI).
FISH analyses were performed according to our institutional protocols (4). Accordingly, a commercial panel of FISH probes covering Ph-like ALL, including ABL1, ABL2, CRLF2, EPOR, and JAK2, was purchased (Vysis, Abbott/IL/USA). A positive rearrangement was reported when at least 3% of the nuclei showed break-apart split signals. Bone marrow (BM) samples at diagnosis and at 28 days after CAR T-cell therapy were analyzed by FISH (Olympus IX 71, Tokyo, Japan).
Total RNA was extracted from diagnostic BM samples using RNeasy Mini Kit (Qiagen, Hilden/Germany). RNA-sequencing libraries were prepared with 20–100 ng total RNA with the TruSight RNA Fusion Panel (Illumina, San Diego/CA/USA). All reads were independently aligned with STAR (version 2.3) against the human reference genome. For nested RT-PCR, sequences of the first pair of primers specific to STRBP were: (5′-AGAAAGTGGCAAAGGCGAGT-3′); and to JAK2 (5′-GCACATCTCCACACTCCCAA-3′). Primers for the second round of PCR specific to STRBP were: (5′-TTATCCCTCAGGCAAAGGGC-3′); and to JAK2: (5′-GGTCTTCAAAGGCACCAGAAAAC-3′).
CR was defined according to the consensus criteria (5). Minimal residual disease (MRD)-negative remission was defined as less than 0.01% of marrow blasts by multiparameter flow cytometry (FCM).
A 21-year-old woman presented with petechiae in the lower extremities in February 2018. Her complete blood count showed a white blood count of 55.4 × 109/L, hemoglobin 110 g/L, and platelets 18 × 109/L. BM morphology analysis detected 88% blasts (Figure 1A). FCM detected 61.45% blasts, which were positive for CD10, CD19, CD22, CD33, CD34, cCD79a, and HLA-DR (Figure 1B). Conventional karyotyping was normal. Results of multiplex PCR covering 41 fusion genes commonly detected in acute leukemia were negative. Targeted next-generation sequencing covering 172 genes reportedly mutated in hematological malignancies revealed NOTCH1 G881S and BCOR2 R1589H mutations: both germline.
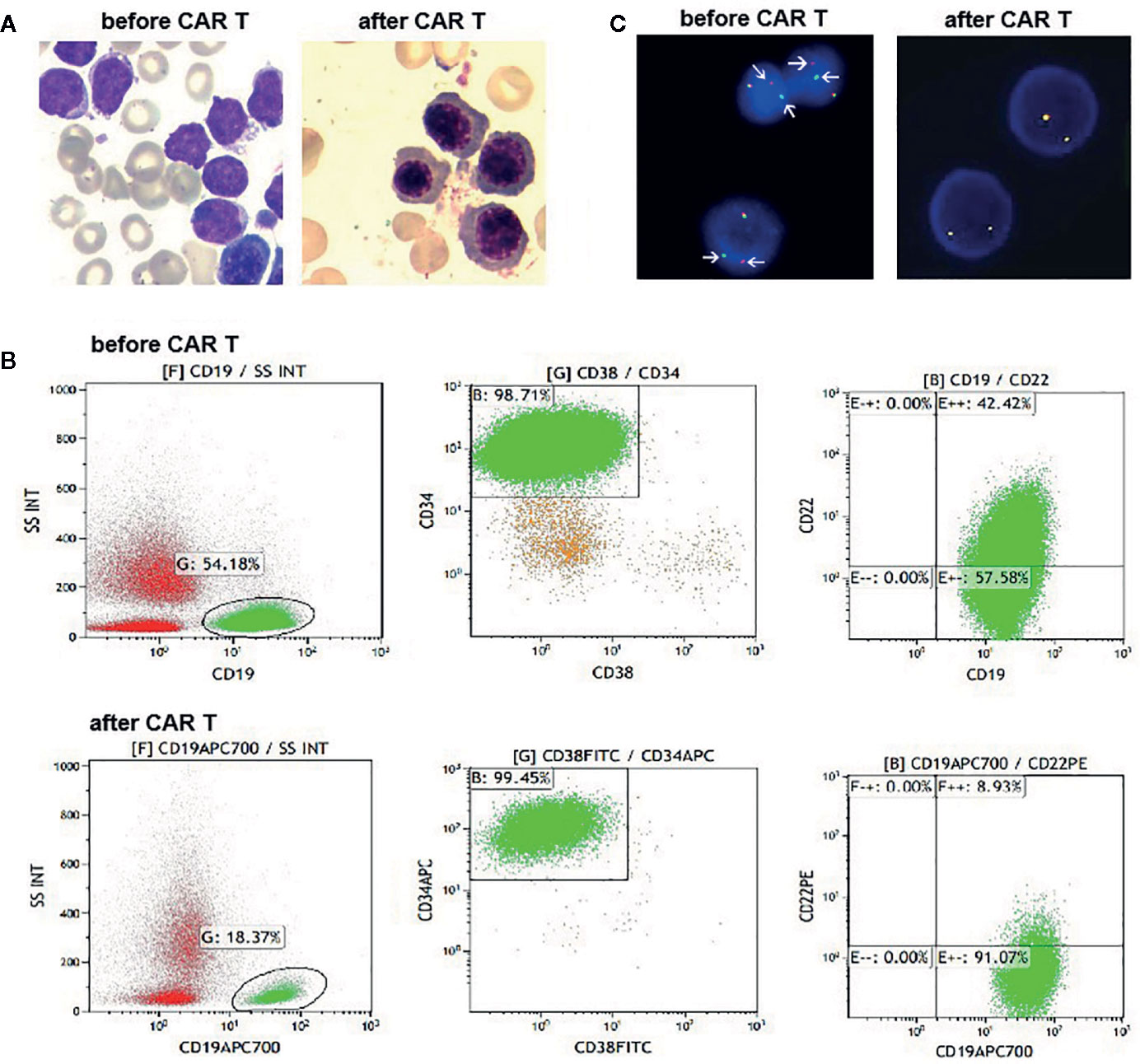
Figure 1 (A) Bone marrow (BM) aspirates before and after CAR T cells infusion. At day 28 post infusion, there was no evidence of blasts in the BM. (B) Flow cytometry (FCM) analysis of BM samples before and after CAR T cells infusion. (C) Fluorescence in situ hybridization results with a JAK2 break-apart probe, which detected split signals of JAK2 before infusion of CAR T cells (arrow), and normal JAK2 signals after infusion of CAR T cells.
The patient was treated with conventional induction chemotherapy, including idarubicin, vincristine, pegaspargase, and dexamethasone. Ruxolitinib (20 mg, twice daily) was administered after JAK2 rearrangement was detected by FISH. Unfortunately, BM analysis revealed 46.5% blasts at the end of the induction therapy. Accordingly, she was enrolled in the CAR T-cell clinical trial (NCT03614858). She received MVP (mitoxantrone 10 mg on day 1, vincristine 4 mg on day 1, and dexamethasone 14 mg on days 1–7) regimen to reduce her tumor burden, followed by lymphodepletion. Then, 1.5 × 107/kg tandem CD19/CD22 CAR T cells were infused via a dose-escalation schedule for five consecutive days (0.5 × 106/kg, 1.5 × 106/kg, 3 × 106/kg, 5 × 106/kg and 5 × 106/kg). She suffered grade 3 cytokine release syndrome, characterized by fever, hypotension and hypoxia. Symptoms of macrophage activated syndrome, including high fever, increase of ferritin, fibrinogen depletion and decreased NK cell activity, were also observed. All of the above complications were alleviated with dexamethasone (5 mg/d for 2 days), fluid boluses, vasopressors and supplemental oxygen. Typical symptoms of CAR T-cell related encephalopathy syndrome were not observed. Her changes of blood cell count and the persistence of the CAR T cells are shown in Supplementary Figures 1A, B. At day 28 after the CD19/CD22 CAR T-cell infusion, no blasts were detected on her BM smear (Figure 1A). However, 5.9% neoplastic cells were detected with FCM, which tested positive for CD10, CD19, CD33, CD34 but negative for CD22 (Figure 1B). She tested negative by FISH for JAK2 rearrangement (Figure 1C). Haplo-identical allo-HSCT was performed one month after the CAR T-cell therapy. Cyclosporin (3 mg/kg/d), mycophenolate mofetil (30 mg/kg/d) and low dose methotrexate (15mg/m2 at d1, 10 mg/m2 at d3, d6, and d10) were administered to prevent graft-versus-host disease. Unfortunately, early hematological relapse was detected at 30 days after allo-HSCT. Cyclosporin and mycophenolate mofetil were withdrawn at day 30 and day 28, respectively. Because of persisting blasts in the BM, donor lymphocytes were infused at day 60 after the allo-HSCT. MRD negative remission was achieved after the DLI; however, she died of grade IV graft-versus-host disease involving the intestine and liver at 7 months post-HSCT.
FISH analyses with split signal probes covering ABL1, ABL2, CRLF2, and EPOR all tested negative (data not shown). A split-signal of JAK2 was detected in 68% interphase nuclei from BM samples (Figure 1C). However, BM samples taken 28 days after CAR T-cell therapy showed normal intact signals for JAK2 (Figure 1C). RNA sequencing of diagnostic samples revealed in-frame fusion of exon 18 of STRBP with exon 19 of JAK2. The first round RT-PCR showed a fusion specific band, while the second round showed a sharp band, of a size (262bp) consistent with STRBP-JAK2 fusion, which was absent in placental control RNA template (Figure 2A). Sanger sequencing of the PCR band confirmed that the STRBP exon 18 was fused to JAK2 exon 19 (Figures 2B, C).
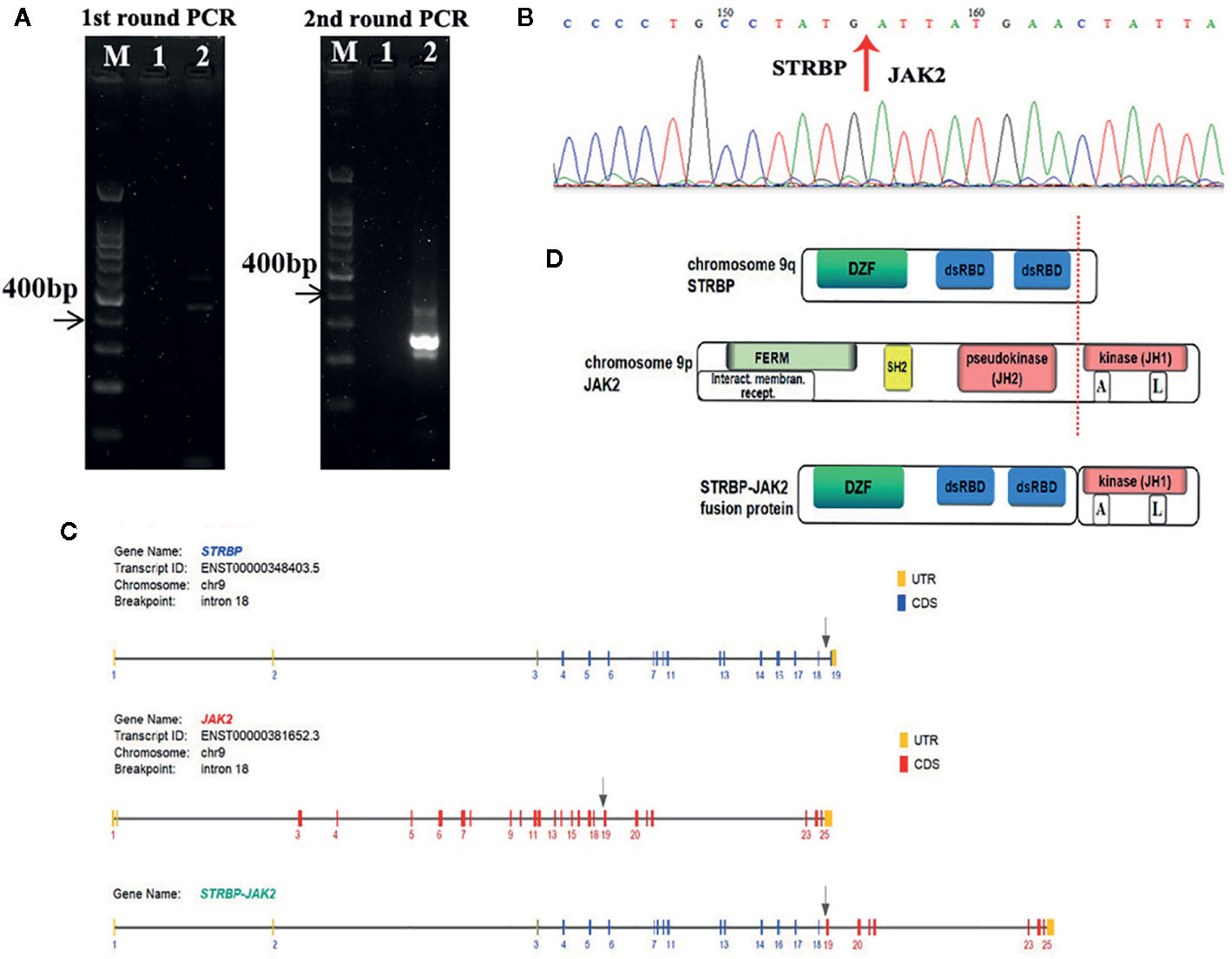
Figure 2 (A) Identification of the STRBP-JAK2 fusion transcripts by nested RT-PCR. Lane M, DNA ladder; Lane 1, PCR negative control; Lane 2, STRBP-JAK2. (B) Sanger sequencing result, which confirmed a fusion between exon 18 of STRBP and exon 19 of JAK2. (C) Schematic representation of the predicted STRBP-JAK2 fusion gene. (D) Schematic representation of the predicted STRBP-JAK2 fusion protein.
JAK2 is located on chromosome 9p24, and encodes a non-receptor tyrosine kinase that participates in the activation of the JAK-STAT pathway and plays a central role in regulating cell proliferation, differentiation, survival, and apoptosis during hematopoiesis (6). JAK2 contains an N-terminal FERM domain that is required for erythropoietin receptor association, a central Src homology 2 (SH2) domain that binds to STAT transcription factors, a pseudokinase domain (JH2) and a C-terminal tyrosine kinase domain (JH1) (Figure 2D). The JH2 domain can regulate activity of the JH1 domain. Deletion of the JH2 domain leads to enhance the tyrosine kinase activity of JAK2 and increase phosphorylation of intracellular signaling molecules, including STATs, PI3K/AKT/mTOR, and MAPKs (7). A high proportion of patients with myeloproliferative disorders carry dominant gain-of-function V617F mutations in the JH2 domain, which lead to constitutive tyrosine phosphorylation activation and excessive cell proliferation (8). JAK2 rearrangements such as PCM1-JAK2 are frequently detected in myeloproliferative diseases with eosinophilia (9). JAK2 rearrangements have also been observed in Ph-like ALL, chiefly chromosomal translocations resulting in fusion of JAK2 with EBF1, ETV6 and OFD1, or inversions resulting in fusion of JAK2 with PAX5 or RFX3 (10–14). Consequently, JAK2-related fusion genes served as driver mutations of Ph-like ALL, by causing hyperactivation of the JAK-STAT signaling pathway, deregulated cell proliferation and resistance to apoptosis (7).
STRBP, the spermatid perinuclear RNA-binding protein, is located on chromosome 9q33 and has widespread expression in lymph nodes, testes, and other tissues. Structurally, it contains a domain associated with zinc fingers (DZF domain) and two double-stranded RNA-binding domains (dsRBD) (Figure 2D). It is a double-stranded RNA binding protein which also associates with microtubules, and, accordingly, plays an important role in mammalian spermatogenesis (15). Reports of rearrangements involving STRBP have been hitherto limited in solid tumors, e.g. STRBP-ASTN2 observed in lung adenocarcinoma and STRBP-ATG14 observed in breast cancer (16). No functional analyses of these two fusion genes have been described so far. Consequently, this is the first report of STRBP involvement in a hematological malignancy. Indeed, this is the first case of STRBP-JAK2 fusion gene reported in Ph-like ALL. The STRBP-JAK2 fusion contained the 5′ STRBP, including DZF and dsRBD, and the 3′ JAK2 with an intact protein kinase domain (JH1) (Figure 2D). Because DZF and dsRBD domains of STRBP promote oligomerization in a wide variety of proteins, including STBRP itself (17), we propose an oncogenic model whereby STRBP-JAK2 homodimerization, driven by the DZF and dsRBD domains from STRBP, leads to auto- and trans-phosphorylation within the activation loop of the JAK2-JH1 domain and, thereby, constitutive activate JAK-STAT signaling.
JAK2 rearrangements universally result in an in-frame fusion of the carboxyl terminal portion of JAK2 with its intact tyrosine kinase domain, joining in-frame to the amino terminal portion of the partner gene. Structurally, STRBP-JAK2 fusion is congruent with previously reported fusion models (3). In this case, both JAK2 and STRBP are located on chromosome 9, implying that JAK2-STRBP fusion may result from inversion of chromosome 9. Both inv(9)(p24q33) and t(9;9)(p24;q33) are cryptic rearrangements, and due to the dearth of good metaphases in BM samples at diagnosis, we were unable to distinguish between these options in the affected patient.
In Ph-like ALL, JAK2 exhibits high levels of promiscuity, engaging with 23 different fusion partners reported in more than 50 patients hitherto. Most of these fusion genes were identified by RNA sequencing, but regrettably, little is known about their treatment responses and prognosis. Characteristics of 10 of these patients, where therapeutic details have been reported, are summarized in Supplementary Table 1 (including the current case). In addition to JAK2 fusion, five of the nine patients had concurrent deletions of IKZF1, which is consistent with the finding that an IKZF1 deletion is the most common secondary aberration in Ph-like ALL (2). Four of the 10 patients received combined treatments with ruxolitinib. The patient with GOLGA5-JAK2 achieved a CR with high dose ruxolitinib (40 mg/m2, bid) (18), while neither the patient with RNPC3-JAK2 (19) nor the current one with STRBP-JAK2 responded to ruxolitinib. In our patient, ruxolitinib was administered at the standard dosage used for treatment of myeloproliferative diseases (20 mg bid), and whether she would benefit from an increased dose remains unknown. Six patients were reported to have undergone allo-HSCT, without long-term follow-up data. Our patient achieved morphologic remission after CAR T-cell therapy and MRD negative remission after DLI in the setting of an early relapse after allo-HSCT, which implies that she benefited from immunotherapies.
In conclusion, we have characterized a novel STRBP-JAK2 fusion gene for the first time in a refractory Ph-like ALL patient who failed to respond to conventional chemotherapy plus ruxolitinib. Our study suggested that the CAR T-cell therapy might, however, prove a viable option in this entity.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
The studies involving human participants were reviewed and approved by the First Affiliated Hospital of Soochow University. The patients/participants provided their written informed consent to participate in this study.
H-PD, ZL, JY, D-PW and X-WT treated the patients. X-PL, C-XY, SX, HL, and H-JS designed the genetic analysis and interpreted the data. D-DL performed the morphology analysis. M-QZ performed flow cytometry analysis. X-YZ and H-PD wrote the manuscript. D-PW and X-WT critically read and revised the manuscript. All authors approved the final version of the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by research grants from the National Natural Science Foundation of China (81873443), National Science and Technology Major Project (2017ZX09304021), National Key R&D Program of China (2016YFC0902800), Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), Frontier Clinical Technical Project of the Science and Technology Department of Jiangsu Province (BE2017655), Jiangsu Provincial Medical Talent (ZDRCA2016045), Major Natural Science Research Projects in institutions of higher education of Jiangsu Province (19KJA210002), The Key Science Research Project of Jiangsu Commission of Health (K2019022), National Natural Science Foundation of China (81700139), and National Science Fund of Jiangsu Province (BK20170360).
X-PL and C-XY are employed by the company Sano Suzhou Precision Medicine Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.611467/full#supplementary-material
1. Roberts KG, Gu Z, Payne-Turner D, McCastlain K, Harvey RC, Chen IM, et al. High frequency and poor outcome of philadelphia chromosome-like acute lymphoblastic leukemia in adults. J Clin Oncol (2017) 35:394–401. doi: 10.1200/JCO.2016.69.0073
2. Roberts KG, Li Y, Payne-Turner D, Harvey RC, Yang YL, Pei D, et al. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N Engl J Med (2014) 371:1005–15. doi: 10.1056/NEJMoa1403088
3. Akkari YMN, Bruyere H, Hagelstrom RT, Kanagal-Shamanna R, Liu J, Luo M, et al. Evidence-based review of genomic aberrations in B-lymphoblastic leukemia/lymphoma: Report from the cancer genomics consortium working group for lymphoblastic leukemia. Cancer Genet (2020) 243:52–72. doi: 10.1016/j.cancergen.2020.03.001
4. Li TY, Xue YQ, Wu YF, Pan JL. FISH studies identify the i(20q-) anomaly as a der(20)del(20)(q11q13)idic(20)(p11). Genes Chromosomes Cancer (2006) 45:536–9. doi: 10.1002/gcc.20313
5. Lee DW, Kochenderfer JN, Stetler-Stevenson M, Cui YK, Delbrook C, Feldman SA, et al. T cells expressing CD19 chimeric antigen receptors for acute lymphoblastic leukaemia in children and young adults: a phase 1 dose-escalation trial. Lancet (2015) 385:517–28. doi: 10.1016/S0140-6736(14)61403-3
6. Leroy E, Constantinescu SN. Rethinking JAK2 inhibition: towards novel strategies of more specific and versatile Janus kinase inhibition. Leukemia (2017) 31:1023–38. doi: 10.1038/leu.2017.43
7. Santos FPS, Verstovsek S. JAK2 inhibitors: are they the solution? Clin Lymphoma Myeloma Leuk (2011) Suppl 1:S28–36. doi: 10.1016/j.clml.2011.02.007
8. Baxter EJ, Scott LM, Campbell PJ, East C, Fourouclas N, Swanton S, et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet (2005) 365:1054–61. doi: 10.1016/S0140-6736(05)71142-9
9. Gotlib J. World Health Organization-defined eosinophilic disorders: 2017 update on diagnosis, risk stratification, and management. Am J Hematol (2017) 92:1243–59. doi: 10.1002/ajh.24880
10. Roberts KG, Morin RD, Zhang J, Hirst M, Zhao Y, Su X, et al. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell (2012) 22:153–66. doi: 10.1016/j.ccr.2012.06.005
11. Zhou MH, Gao L, Jing Y, Xu YY, Ding Y, Wang N, et al. Detection of ETV6 gene rearrangements in adult acute lymphoblastic leukemia. Ann Hematol (2012) 91:1235–43. doi: 10.1007/s00277-012-1431-4
12. Yano M, Imamura T, Asai D, Kiyokawa N, Nakabayashi K, Matsumoto K, et al. Identification of novel kinase fusion transcripts in paediatric B cell precursor acute lymphoblastic leukaemia with IKZF1 deletion. Br J Haematol (2015) 171:813–7. doi: 10.1111/bjh.13757
13. Schinnerl D, Fortschegger K, Kauer M, Marchante JR, Kofler R, Den Boer ML, et al. The role of the Janus-faced transcription factor PAX5-JAK2 in acute lymphoblastic leukemia. Blood (2015) 125:1282–91. doi: 10.1182/blood-2014-04-570960
14. Harvey RC, Tasian SK. Clinical diagnostics and treatment strategies for Philadelphia chromosome-like acute lymphoblastic leukemia. Blood Adv (2020) 4:218–28. doi: 10.1182/bloodadvances.2019000163
15. Schumacher JM, Artzt K, Braun RE. Spermatid perinuclear ribonucleic acid-binding protein binds microtubules in vitro and associates with abnormal manchettes in vivo in mice. Biol Reprod (1998) 59:69–76. doi: 10.1095/biolreprod59.1.69
16. Yoshihara K, Wang Q, Torres-Garcia W, Zheng S, Vegesna R, Kim H, et al. The landscape and therapeutic relevance of cancer-associated transcript fusions. Oncogene (2015) 34:4845–54. doi: 10.1038/onc.2014.406
17. Coolidge CJ, Patton JG. A new double-stranded RNA-binding protein that interacts with PKR. Nucleic Acids Res (2000) 28:1407–17. doi: 10.1093/nar/28.6.1407
18. Ding YY, Stern JW, Jubelirer TF, Wertheim GB, Lin F, Chang F, et al. Clinical efficacy of ruxolitinib and chemotherapy in a child with Philadelphia chromosome-like acute lymphoblastic leukemia with GOLGA5-JAK2 fusion and induction failure. Haematologica (2018) 103:e427–e31. doi: 10.3324/haematol.2018.192088
Keywords: fusion gene, STRBP-JAK2, Ph-like, chimeric antigen receptor, B-lymphoblastic leukemia
Citation: Zhang X-Y, Dai H-P, Li Z, Yin J, Lang X-P, Yang C-X, Xiao S, Zhu M-Q, Liu D-D, Liu H, Shen H-J, Wu D-P and Tang X-W (2021) Identification of STRBP as a Novel JAK2 Fusion Partner Gene in a Young Adult With Philadelphia Chromosome-Like B-Lymphoblastic Leukemia. Front. Oncol. 10:611467. doi: 10.3389/fonc.2020.611467
Received: 29 September 2020; Accepted: 24 November 2020;
Published: 11 January 2021.
Edited by:
Annalisa Lonetti, University of Bologna, ItalyReviewed by:
Shimin Hu, University of Texas MD Anderson Cancer Center, United StatesCopyright © 2021 Zhang, Dai, Li, Yin, Lang, Yang, Xiao, Zhu, Liu, Liu, Shen, Wu and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: De-Pei Wu, ZHJ3dWRlcGVpQDE2My5jb20=; Xiao-Wen Tang, eHd0YW5nMTAyMEAxNjMuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.