- 1Office for Research Innovation and Commercialization, Lahore Garrison University, Lahore, Pakistan
- 2Department of Biotechnology, Faculty of Sciences, University of Sialkot, Sialkot, Pakistan
- 3School of Basic Medical Sciences, Lanzhou University, Lanzhou, China
- 4Department of Biotechnology, Balochistan University of Information Technology, Engineering and Management Sciences, Quetta, Pakistan
- 5Department of Life Sciences, University of Management and Technology, Lahore, Pakistan
- 6Chair and Department of Food and Nutrition, Medical University of Lublin, Lublin, Poland
- 7Department of Pharmacognosy, Medical University of Lublin, Lublin, Poland
- 8Department of Cosmetology, University of Information Technology and Management in Rzeszów, Rzeszów, Poland
- 9Department of Clinical Oncology, Queen Elizabeth Hospital, Kowloon, Hong Kong
- 10Phytochemistry Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 11Facultad de Medicina, Universidad del Azuay, Cuenca, Ecuador
Hedgehog (Hh) signaling aberrations trigger differentiation and proliferation in colorectal cancer (CRC). However, the current approaches which inhibit this vital cellular pathway provoke some side effects. Therefore, it is necessary to look for new therapeutic options. MicroRNAs are small molecules that modulate expression of the target genes and can be utilized as a potential therapeutic option for CRC. On the other hand, nanoformulations have been implemented in the treatment of plethora of diseases. Owing to their excessive bioavailability, limited cytotoxicity and high specificity, nanoparticles may be considered as an alternative drug delivery platform for the Hh signaling mediated CRC. This article reviews the Hh signaling and its involvement in CRC with focus on miRNAs, nanoformulations as potential diagnostic/prognostic and therapeutics for CRC.
Introduction
Colorectal cancer (CRC) is one of the leading causes of death globally with incidence rate around two million (1). A number of factors such as the dietary habits, family history, inflammatory bowel disease, elevated body mass index, socioeconomic status, environmental and genetic factors affect the likelihood of developing CRC (2). Despite advancements in the preclinical and clinical researches, devising a suitable cure for CRC still remains bleak. Thus, researchers keep pursuing on personalized and targeted therapeutic approaches, development of efficient diagnosis/prognosis biomarkers and clinical management which could inhibit CRC development. The molecular landscape of CRC is multifarious and governed by various signaling pathways such as the hedgehog (Hh) signaling and Wnt signaling pathways which orchestrate growth and development of tumor cells (3). Hh signaling play a crucial role in regeneration of adult tissues by regulating the stem cell behavior. It also interacts with other vital signaling molecular cascades to control cellular proliferation, polarity and differentiation (4). Aberrant expression of Hh signaling is reported to be the culprit of dysregulation in cellular behavior and contribute in the onset of many human malignancies (5). Hh signaling and aberrant expression of targeted molecules promote tumor microenvironment and induce stemness of cancer cells (6, 7). Aberrant expression of Hh signaling cascade has been reported to contribute in the cancer progression and metastasis including medulloblastoma, basal cell carcinoma, breast cancer, liver cancer, pediatric soft tissue cancer, prostate, stomach, pancreas, and colon cancer (8). Invertebrates and vertebrates share common signaling molecules and mechanism in general, involving Hh ligands, patched1/2 receptor, transcriptional factors GLI-1/2/3, smoothened (SMO) as a critical signal transducer and variety of regulatory molecules (8). In mammals three Hh genes have been identified namely, sonic hedgehog (SHH), Indian hedgehog (IHH), and Desert hedgehog (DHH) which play a vital role in the embryonic development and regeneration of different organs (9). Hh signaling pathway can modulate the self-renewal of cancer stem cells (CSCs) most commonly in hematological malignancies, breast cancer, and CRC (10). There has been significant progress regarding the development of small molecule inhibitors to block Hh signaling. Several of these molecules have been included in the clinical testing stage. Yet finding a sustainable small molecule inhibitor is still a challenge. On the other hand, microRNAs (miRNAs) are small molecules that effectively regulate and modulate the expression of target genes (11). Exploring miRNAs as diagnostic tool can aid in better clinical management of CRC. Nanoformulations have been investigated in many diseases for their efficient sustainability, limited cytotoxicity, increased bioavailability and few side effects. These features have urged scientists to explore these as a therapeutic option for different cancers. In this review, we delineate Hh signaling pathway as a vital therapeutic target for CRC and shed light on the role of miRNAs that may be used as potential diagnostic marker and therapeutic target for CRC. Furthermore, the role of nanoformulations as contenders for targeted delivery of Hh signaling inhibitors for the treatment of CRC is discussed.
Hedgehog Signaling in Cancer
Molecular link of Hh signaling with cancer was reported in basal cell carcinoma when mutation in human PTCH1 gene was observed (12, 13). It was confirmed that mutation in PTCH1 is responsible for SMO activation to trigger aberrant Hh cascade activation to induce carcinogenesis (Figure 1) (14, 15). Similarly, increased expression of Hh targeted gene was reported in different carcinomas including meningiomas, medulloblastoma (16), small cell lung carcinoma (SSLC), gastro-intestinal cancer (17), and colon cancer. Experimental work on genetically engineered mice models exhibits that in knock-out PTCH gene mice model organism, increased expression of SMO was observed with increased tumor size. The same experimental study designed to knock out SMO in mice models revealed reduction in tumor size and metastasis (6). However due to the complex behavior of cancer onset and variation in contributing factors, no significant molecular evidence was reported in KRAS associated onset of pancreas and prostate cancer (18). Hh signaling has been associated with cellular proliferation, tissue polarity, stem cell transformation and carcinogenesis. The first molecular association of Hh signaling with cancer was established in 1996 during experimental studies on Gorlin syndrome. Hh signaling was considered as a novel therapeutic target of cancer by clinical use of Hh inhibitors (erivedge/vismodegib) and was approved in 2012 by the FDA to treat basal cell carcinoma (BCC). In this article, Hh signal in carcinogenesis and recent molecular strategies to tackle cancer cell progression using Hh inhibitors (19) were discussed.
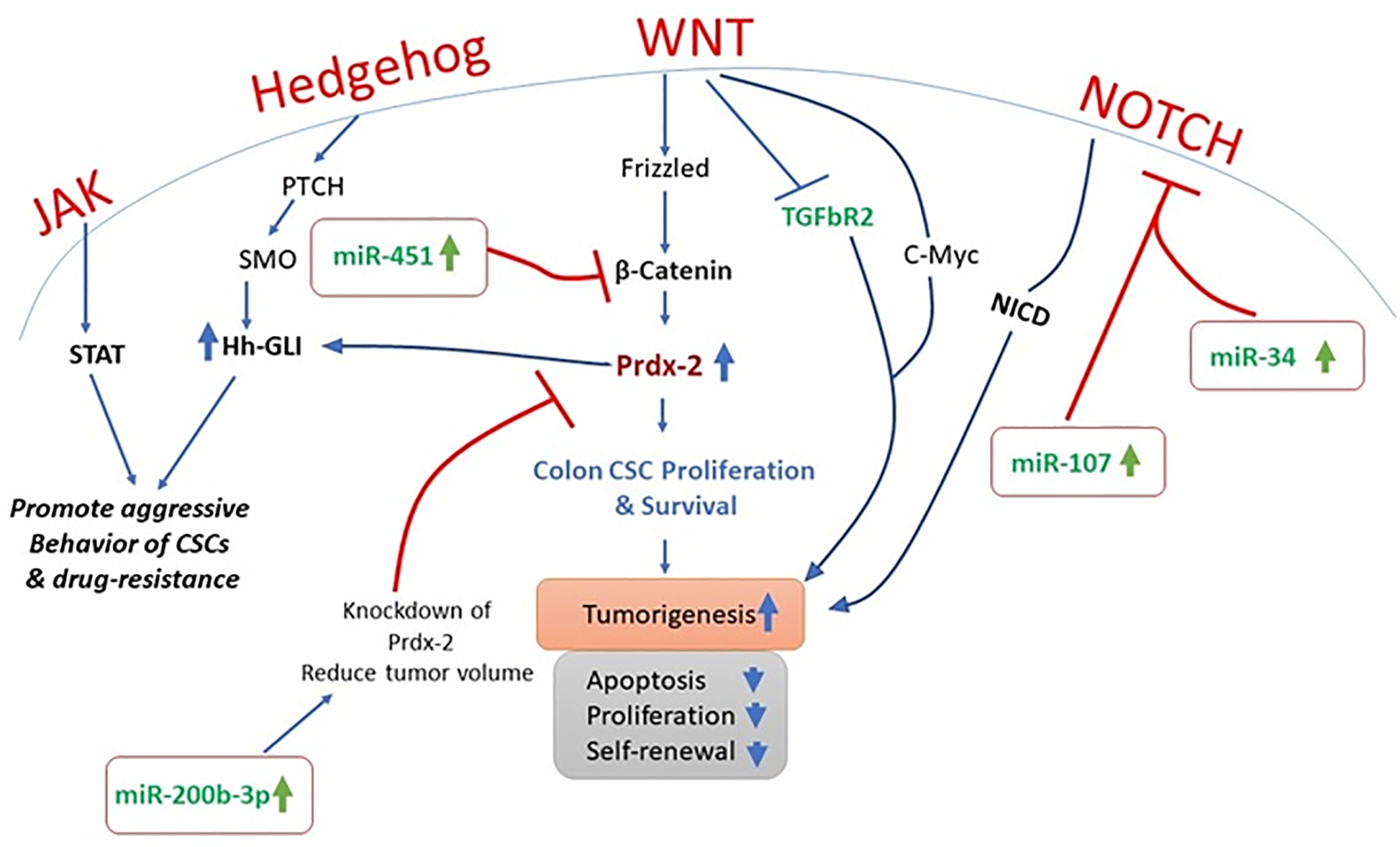
Cancer is a complex heterogenetic disorder that transform cellular microenvironment and involves multiple and complicated crosstalk of signaling pathways. In PTCH+/− mice models, inhibition of Hh signaling is an approach to limit cancer cell proliferation (20). It is also observed that STAT-3 knock-out significantly reduce the Hh-mediated delivery of BCC (21). It is widely accepted that Hh signaling has close association with the growth factor mediated pathways as Hh signaling is reported to regulate the platelet derived growth factor ᾳ (PDGFR-ᾳ). Furthermore, molecular crosstalk of Hh is reported to interplay with many other pathways including NOTCH, mTOR, Wnt, Muc5, EGF, IGF, TGF-β, RACK1, and PKC in different types of cancers (22–25). It has been observed that TGF-β regulates tumor microenvironment, while PDGFR-ᾳ and Notch play key role in triggering CSC (22, 25). Recent studies highlighted that inhibition of Hh signaling in cancer cells could be the iron gate for cancer therapy in many cancer types (21).
Aberrant Hh signaling is a distinguished feature of various human cancers (26). Gli1 and Gli2 are the two Hh pathway target genes that are overexpressed in the CRC (27). A gene expression microarray study conducted on 382 patients showed that Gli-1 was overexpressed in CRC patients. The expression of this target gene was responsible for the tumor recurrence and poor survival outcome in patients. In addition to this treatment of cell lines such as the HCT-116, SW480 and SW620 with SMO inhibitor GDC-0449 decreased the expression Gli target genes such as the PTCH1, HIP1, and MUC5AC. Furthermore, treatment of cell lines with GDC-0449 upregulated the expression of growth arrest gene p21 and downregulated the expression of cyclin D1 (28). The genetic silencing of SMO with 5E1, a specific antibody, prevented cell migration and invasion along with reduction in the expression of Hh target genes Gli-1 and Gli-2 (28). These findings suggest that Hh signaling affects cell plasticity, proliferation, invasion, and migration in CRC.
CSC functionality and polarity are dependent on the Hh signaling pathway (29). It has also been reported that Hh signaling induces chemotherapy and radiotherapy resistance in cancer cells (30, 31). However, Hh inhibitors are reported to promote delivery of chemo-therapeutic agent including IPI-926 (31).
Hh signaling pathway has its prime role in embryogenesis, i.e. cell differentiation and growth. It does not always active in all adults cells. However, in cells where stem cell development and growth is required, Hh signaling triggers on (32). Genes involved in the cellular differentiation i.e. proto-oncogenes and growth factors are targeted by Hh pathway, but if these pathways get activated by any mutation or if its regulation gets disturbed, then it may lead to tumor development (33). It has been observed that abnormal activation of the Hh pathway can lead to CRC. Molecular evidence realized that in CRC tissues, SHh ligand gets higher in number and increase the expression of all its downstream components, particularly SMO upregulates dramatically and difference in expression of GLI1 protein is observed. From different studies, it was revealed that SHh is a paracrine factor that works like aberrant p53 to inhibit anti-oncogenes (34). SMO activation in an abnormal way causes progression in colon cancer, and its expression was sharply upregulated in colon cancer tissues as compared to the non-cancerous colon tissues. It was observed that SMO expression is directly proportional to the stages of cancer so its level of expression can be used as an independent biomarker for liver postoperative metastasis to liver (35). Similarly, a different expression level of GLI1 was noted in normal tissue and cancerous tissue. Increased expressions of GLI1 cause activation of Hh signaling, which induce anti-apoptotic and anti-inflammatory effects on cancer cells. These alterations are potential driving forces for therapeutics to target GLI by molecular inhibitors to induce the cellular deaths of colon cancer cells. In one study it was reported that GLI1 regulation is exceptional in colon tissues and it is also related to lymph node metastasis (36). Recently, tumor suppressor gene RUNX3 is reported to play a decisive molecular role to limit endothelial proliferation in CRC. It has been observed that RUNX3 expression has inverse correlation with GLI-1 protein and it promotes GLI-1 ubiquitination in CRC. Molecular interplay of RUNX-3 gene to limit metastasis and stemness by targeting hedgehog signaling cascade could be a new contributing therapeutic agent to conquer the unbeatable fort of carcinogenesis (37). Inhibitors of hedgehog pathway are recognized in the scientific community as a therapeutic strategy for cancer treatment. Hh inhibitors hold promise for the development of a potential treatment option in CRC as its results have been proved to be very promising, suggesting that the targeting treatment of signaling pathway is a hopeful way for antitumor treatment. Therefore, the members of hedgehog signaling pathway are considered as significant therapeutic targets for the clinical treatment of colon cancer (38). The Hh signaling pathway has been seen to act as an antagonist to Wnt pathway, which is directly involved in the rapid increase of CRC cells. 90% of CRC have an active mutation in the Wnt pathway; particularly APC gene mutations are responsible agent, but Hh pathway mutations were not found as a molecular culprit in majority of CRC cells (39). There is mounting evidence that over-expression of SHh and SMO participates in the onset of multiple cancers, also recognized as SHh related carcinomas (40). Both these pathways have a significant relation between them in the occurrence and development of CRC and have numerous avenues for molecular crosstalk between the two pathways (41). The colon’s mucosa has a film of epithelial cells which gets replaced every week. It replaces large number of progenitor cells and generates plenty of new cells every day at the bottom of crypts i.e. small mucosal invaginations. Maintenance of the balance of cell is regulated by extrinsic signals. Morphogens, soluble proteins that make a long range of concentration gradients, produce cellular responses to target cells from a distance in a dose dependent way. It has been proved that morphogens are the main regulator in adult colon and support the notion that both Wnt and Hh pathways have significant roles in CRC (42). The metastatic transition of human colon carcinomas, which mainly occurs in the CD133+ epithelial tumor stem cell population, includes deregulation of the Wnt–TCF pathway and upregulation of the HH–GLI pathway (43). During this phase of metastatic transition all ligand-driven signaling pathways of Wnt are inhibited. In both CD133+ and CD133− cells of colon carcinoma signatures of expression of gene in various stages show that activity of Wnt–TCF i.e. non-metastatic stops at early stages in colon carcinomas and to become metastatic, the HH–GLI works actively in stem cells. The molecular linkage was established in the deregulation and upregulation of early adenoma-like Wnt–TCF and HH–GLI1 respectively. It was experimentally proved that upregulation of HH–GLI causes downregulation of TCF and thus results in low Wnt–TCF and high HH–GLI expression in metastatic colon carcinomas. It was also observed that silencing of TCF induces the HH–GLI signaling. The high regulation of Wnt–TCF causes transcription repressor GLI3 and high regulation of HH–GLI causes repression of Wnt–TCF and GLI3 (44).
Drug resistance, tumorigenesis, tumor progression, metastasis, and tumor recurrence are the key functions that are regulated by the CSC (45). These are subpopulations of cancer cells with the ability of self-renewal. The Hh signaling pathway has been reported to be involved in the activation of CSCs in various neoplastic tumors such as the glioblastoma, leukemia, and myeloma (46). The activated stem cells have been demonstrated to play a pivotal role in the progression, metastasis, and recurrence of tumors in colon, breast, liver, and pancreatic tissues (47). In addition to its involvement in regulating the CSCs, the Hh signaling along with the SMO and Gli signaling pathways promotes cell migration, growth, and self-sustenance of CSCs (48). The non-canonical Hh-signaling has been reported as a crucial mediator for the survival of CSCs (49). Both the canonical signaling and non-canonical signaling are pivotal in regulating the expression of key genes involved in growth and proliferation of cells (45). Accumulating lines of evidence have reported the fact the aberrant non-canonical hedgehog signaling can trigger uninterrupted cellular growth in CRC. Zhang et al. demonstrated that both SMO and Gli proteins were overexpressed in colon cancer cells and colonic adenoma tissues (38). The SMO expression has been related to prognosis and tumor status in CRC patients. The CSCs are pivotal in stemness and growth of CRC. New studies have begun to shed light on the fact that non-canonical Hh signaling and Wnt signaling are the two key molecular cascades that are disrupted in CRC stem cells. Both canonical and non-canonical Hh-signaling positively and negatively regulates the expression of Wnt in CRC stem cells. Regan et al. demonstrated that non-canonical Hh signaling had a positive role in maintaining growth and differentiation of CRC stem cells. Moreover, continuous overexpression of non-canonical Hh signaling promoted resistance in CRC stem cells and increased their survival in a PTCH1-dependant, Gli-independent manner. In addition to this, SMO dysregulation has been affiliated with CSC growth and differentiation targeting; the dysregulated SMO can be a potential target for the treatment of CRC (50). A specifically designed Hh signaling antagonist GDC-0449 (Vismodegib) has been reported to suppress growth and trigger apoptosis in colon cancer cells via downregulating the expression of Bcl-2 (51). Another study confirmed that GDC-0449 has the ability to initiate apoptosis, decrease cellular plasticity and invasiveness of CRC (28). Altogether these findings indicate that non-canonical Hh-signaling has a regulatory role in progression and spread of CRC via CSCs modulation. Cancer is a multifactorial disease. There are number of factors such as the age, genetic predisposition, alteration in the genetic framework, diet and habits that can trigger tumorigenesis (52). Studies over the past decades have evidenced the involvement of various mutations in the signaling machinery that contribute towards development of cancer (53). Development of CRC like several other tumors involves mutations in the signaling machinery. Mutations in KRAS, MYB, and BRAF are the most critical mutations that trigger tumorigenesis and can be targeted for therapeutic purposes (54–57). The role of Hh signaling in CRC is still questionable. The exact mechanism by which Hh signaling triggers growth and proliferation, invasiveness and metastasis in CRC still requires aggressive research. The scientific community seems divided on the role of Hh signaling in CRC. Accumulating lines of evidence have suggested that Hh signaling has the following implications in CRC: 1) Hh signaling is expressed variably in CRC, and CRCs as different components of the Hh signaling machinery are expressed differently. 2) Hh pathway can trigger mutations in CRC. 3) Hh signaling plays a role during the transformation of the cells from adenoma-to-adenocarcinoma. 4) The SMO has the most crucial role in the regulation of carcinogenesis of CRC (58, 59). Taken together, it can be evidenced here that the Hh role in CRC still requires plenty of research.
Role of miRNAs in Colorectal Cancers
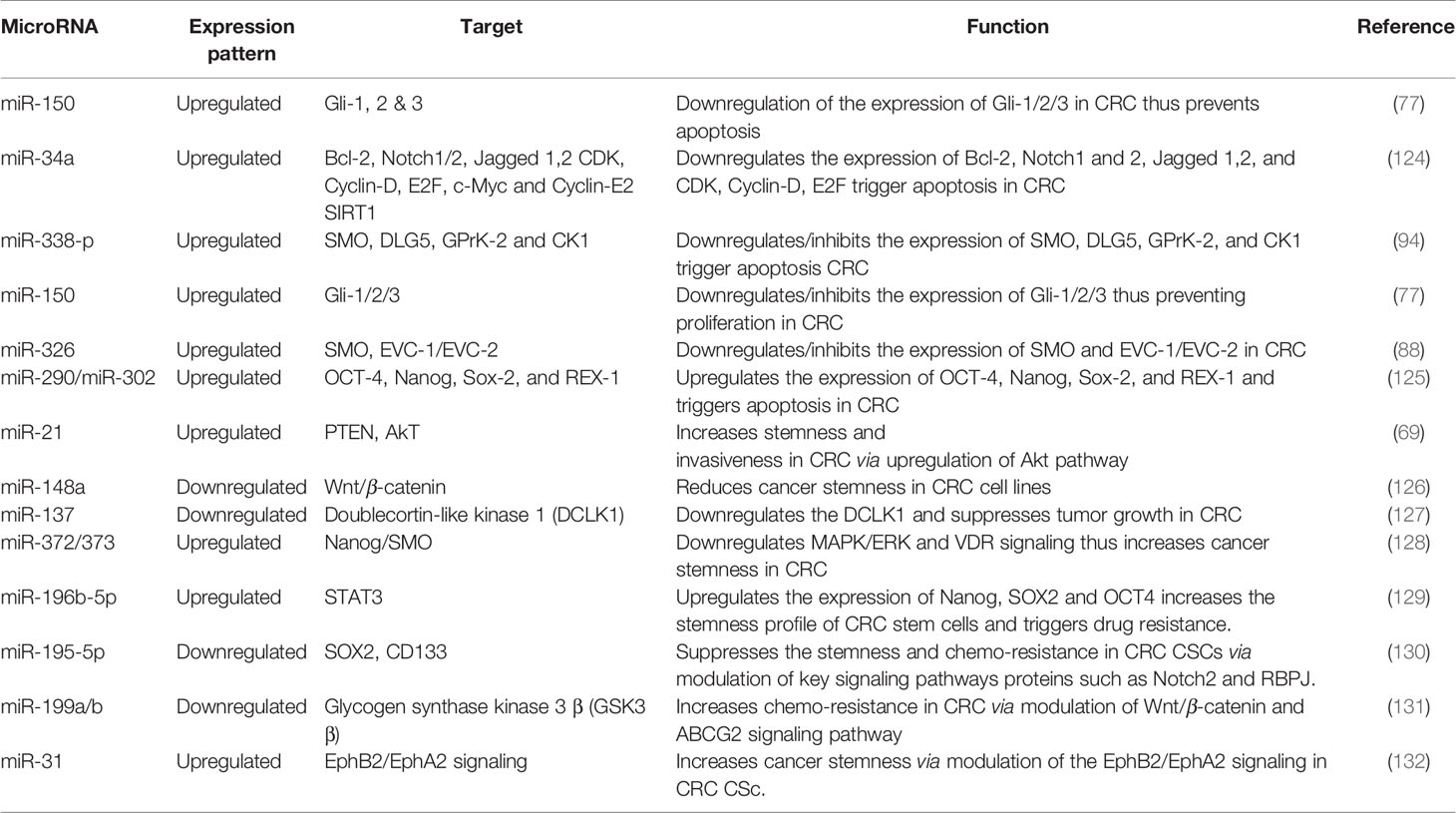
MiRNAs are short non-coding single-stranded nucleotide sequences (60), which affect almost all physiological processes in cells such as development (61, 62), proliferation (63), differentiation (64), apoptosis (65), signal transduction (66) and many more. The altered expression patterns of miRNAs are tightly linked with a wide range of anomalies including various cancers; thus miRNAs screening could serve a very good therapeutic and diagnostic tool in molecular biology (67). Till date, more than 25,000 miRNA sequences have been identified, and this number is growing fast amid current research interests in miRNAs all over the world. According to an estimation, 3–4% of human genome comprises of miRNAs (68). These miRNAs interfere with numerous key regulators of cellular processes by binding with post-transcriptional products. For this reason, miRNAs are considered as important biomarkers for many cancers including CRC (42). In this section, we shall focus on miRNAs which interact with Hh signaling and may affect CRC. There is a long list of miRNAs which affect CRC progression. More than 500 miRNAs have been found to be linked with CRC. Among these miRNAs few miRNAs such as miR-21 (69), miR-143, and miR-145 are reported most frequently and are summarized in (70). These miRNAs interact through various signaling pathways. For example miR-143 significantly inhibits KRAS which ultimately suppresses CRC (71). However, another study has shown the opposite phenomenon where reduced levels of miR-143 expression were detected in CRC tissues. Interestingly transfection of cells with transient miR-143 turns the cells to mimic SW480 cells, a CRC cell line, resulting in increased levels of cell proliferation and apoptosis (72). Thus, we may say that the role of particular miRNAs may also vary depending upon the cell type. On the other hand, the role of miR-145 remains much consistent as CRC suppressor in many studies. There has been a reverse interaction between erythroblast transformation-specific (ETS)-related gene (ERG) and miR-145 in CRC. Increased ERG results in decreased miR-145 levels and promotes CRC. The overexpression of miR-145 suppresses CRC by decreasing expression of ERG (73). A similar relation between P21-activated kinases 4 (PAK4), and miR-145 was also observed where miR-145 appeared to downregulate phosphorylation level of LIMK1 and cofilin in SW1116 cells through PAK4 (74). miR-224 activates the Wnt/β-catenin signaling by deregulation of GSK3β and SFRP2 to translocate β-catenin in CRC (75). Similarly, miR-361 is also downregulated in CSC (65). miR-150 is negatively correlated with circular RNA named Circ-ZNF609 and important transcription factor of hedgehog signaling i.e. Gli1 in HCT-116 cells (76). Another study stated that overexpression of miR-150 positively affect the EMT and subsequent downregulation of Gli1, further confirming the role of miR-150 in CRC through hedgehog signaling (77). Similarly, miR-142-3p appeared to promote cell invasion in CRC by upregulation of RAC1 (78). There are miRNAs also targeting other key regulators of hedgehog signaling. One such miRNA-378 inhibits SUFU and promotes cell survival and tumor growth (79). Another molecule, miR-146a, activates the Wnt pathway and stabilizes β-catenin, thereby promoting CRC by regulating the symmetrical cell division by a feedback loop of Snail-miRNA-146a-β-catenin (80). All variants of hedgehog pathways work upstream of epithelial-mesenchymal transition (EMT) (81). A number of miRNAs regulate EMT in CRC such as miR-29c which has been shown to be remarkably downregulated and also associated with metastasis and significantly shorter patient survival and this effect was reverted by transient expression of miR-29c (82).
MiRNAs are crucial molecular factors to regulate post-transcriptional processing, and more than 60% protein coding genes are expected to regulate miRNAs, and their dysregulation is often reported to trigger different human disorders including cancer. In recent years, many reports highlighted the significance of miR-34a as tumor suppressive molecular entity. It has been figured out that miR-34a has inverse relation with the cancer progression, and the expression of miR-34a declines with the increased progression of cancer and vice versa (83).
Scutellaria barbata (SB) is a natural compound and has been used for years as a potential compound among traditional Chinese medicines against multiple cancers. In vitro and in vivo clinical trials have proved that its ethanol extract of SB is an effective agent to induce apoptosis and limit cancer cell proliferation (84). Ethanol extract of SB has been found effective against human CRC HCT-8 cells and regulates miR-34a expression. Molecular assay confirmed that SB extract upregulates the miR-34a expression and negatively regulates the Bcl-2, Notch and Jagged-1 gene expression. miR-34a mediated down-stream targeted gene regulation plays a decisive role in apoptosis and limits cancer cell proliferation. In one of the studies, exogenous inactivation of miR-34a by using anti-miR-34a oligonucleotide triggers Bcl-2, Notch1/2, and Jagged-1 genes and promotes cancer growth (84, 85). Activation of miR-34a has been associated with regulation of various cellular processes including apoptosis, proliferation, and invasion (85). Molecular evidence also established a link of miR-34a with downregulation of Notch1/2 in colorectal CRCs (86).
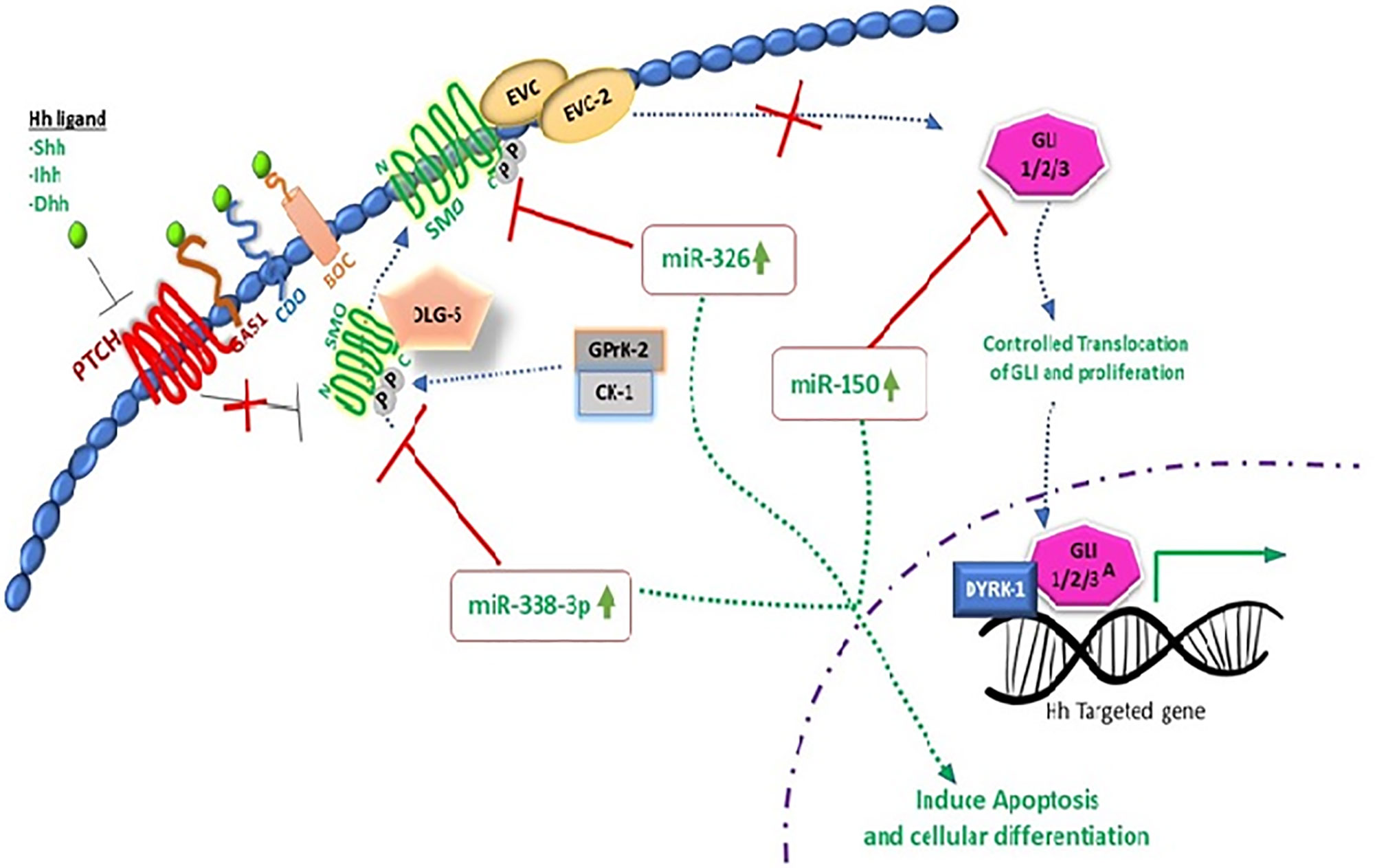
miR-449a has been documented as tumor suppressor gene and has been closely associated with SATB2 in different cancer types including CRC cells. SATB2 could be used as diagnostic marker for CRC and has comparative negative association with miR-449a. It has been noted that in CRC xenograft mouse models, increased expression of miR-449a promotes apoptosis by negatively regulating the expression of SATB2 (87). Similarly, molecular link has been established to understand the transcriptional deregulation of SMO by miR-326, and it was observed that upregulation of miR-326 negatively regulates SMO protein to induce apoptosis and limit cellular proliferation (88, 89).
Aberrant expression of GLI-1 (Glioma associated oncogene homolog 1) is a key culprit in the metastasis, invasion, and proliferation of various cancer cells. Ample lines of evidence have shown that expression of miR-150 declines with the pathogenesis of CRC. NCM-460 and SW-620 CRC cell lines were examined by using dual luciferase assay to decipher the molecular relation of miR-150 with GLI-1, and it has been noted that miR-150 inhibits the expression of GLI-1 protein in Hh signaling (77). Molecular evidence has proved that Hh is a cellular event responsible for structural development, cellular regeneration, and stemness. In multiple myeloma cancer (MMC), inverse relation of miR-324-5p and hedgehog signaling has been observed. Increased expression of miR-324-5p has significant inhibitory effect on SMO and GLi-1 and limits cancer stemness of cells (90). Pro-oncogenic effect of miR-212 is identified, and molecular relation was established that miR-212 induces pancreatic ductal adenocarcinoma (PDAC) by targeting PTCH-1 (91). miR-361-3P has been reported to have profound impact on different cancers including prostate cancer, breast cancer, lung cancer, and cervical cancer. To decipher the molecular interplay of miR-361-3p in retinoblastoma (RB) tissue and RB cell lines, Weri-Rb-1, and Y79, real-time PCR analysis was performed, and it is concluded that miR-361-3p expression is downregulated with cancer progression. Forced expression of miR-361-3p is reported to limit cancer cell proliferation by targeting GLI-1/3 and sonic hedgehog signaling (92). Multiple miRNA expressions have been associated with the onset of breast cancer including miR-454-3p, miR-130b-3p, miR-421, and miR-301a-3p. These miRNAs are noted to target PIEZO-2 gene. Downregulation of PIEZO-2 gene in breast cancer has been molecularly linked with estrogen and progesterone receptors which are responsible agents for Hh signaling cascade in breast cancer (93).
Extensive research work on miRNAs has been done in recent years to unfold the molecular complexity of carcinoma and to bridge the gap towards new and effective therapeutic approaches. miR-338-3p interaction with hedgehog pathway by using recombinant lentiviral vectors PLV-THM-miR-338-3p and PLV-THM-miR-338-3p inhibitor has been reported and successful transfection in SW-620 CRC cells was achieved. Increased expression of miR-338-3p was observed, it significantly suppresses SMO protein and inhibits proliferation ability. Molecular interplay of miR-338-3p is also confirmed by using PLV-THM-miR-338-3p inhibitor, and it was concluded that it upregulates the SMO protein expression to initiate hedgehog signaling pathway and induces CRC onset. miR-338-3p could be a therapeutic agent to suppress CRC growth by targeting SMO, (Figures 2 and 3) (94). Now, withstanding the fact that SMO is a possible target of miR-338-3p. 40 CRC tissue samples and 2 CRC cell lines, SW620 and SW480 were investigated to understand the corresponding protein expression of SMO and miR-338-3p by using semi-quantitative RT-PCR, western and northern blotting assays. It was established that miR-338-3p plays a significant role in metastasis and progression of CRC carcinoma (95).
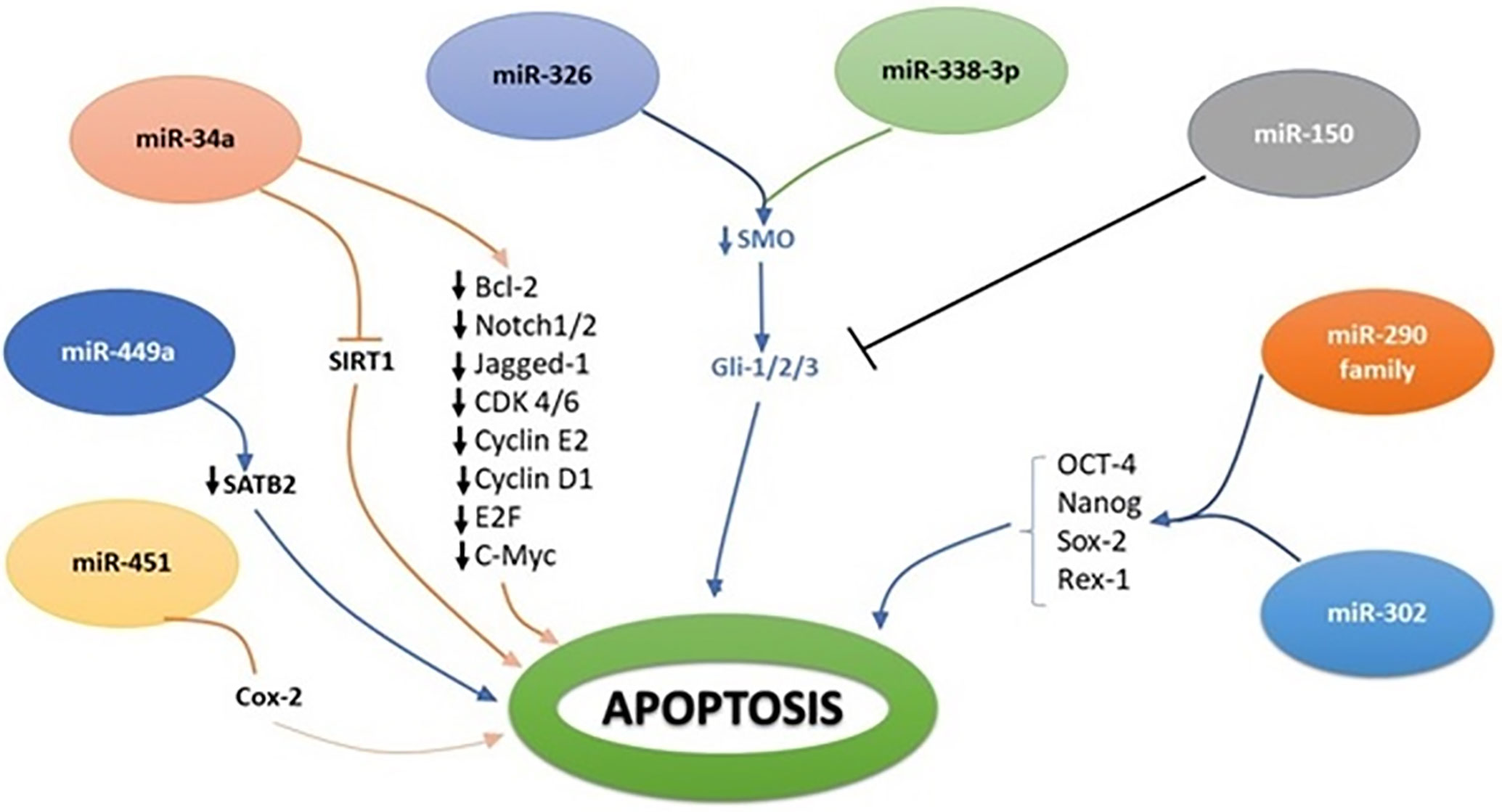
Figure 3 Molecular interplay between several miRNAs and their downstream target genes to induce apoptosis.
Accumulation of genetic and epigenetic errors can trigger the aberrant signaling cascades. miRNAs are the critical key players to fine-tune genetic expression upon exogenous factors including DNA hypermethylation, hypomethylation, histone modification, and deacetylation. Thus epigenetic-miRNA regulatory molecular cascades are the contributing agents for onset of different types cancers (96). Tumorigenic activation of SMO by over-expression of Shh ligand is reported as responsible agent in 40% cases of human hepatocarcinogenesis (97). Molecular balance between cellular proliferation, differentiation, and renewal is modulated by epigenetic regulatory network and miRNAs. miRNAs are the functional short RNAs that control stemness of cancer cells and promote stem cell self-renewal (97). CSCs are believed to be the critical source for tumor initiation. It has been reported that SHH signaling has reprograming potential for epigenetic memory power within CSCs to modulate cancer hallmarks. miR-302-367 clusters regulate cellular plasticity molecular cascade by engaging cyclin-D1, CDK-4, OCT-4 and SOX-2 genes (98).
MicroRNAs as Master Regulator of Stemness and Metastasis
Cancer stem cells are the cells with self-renewal potential within the tumor and are the key responsible agent for radio- and chemo- resistance behavior of cancer cells. Increasing evidence strongly suggests that CSCs are the responsible factor for the onset of carcinogenesis in many human cancer types as cancer cells have self-renewable stem cell like characteristics. Several microRNAs expression have been associated to regulate cancer stemness pathways and its downstream targeting genes (115). Tumor cells were believed to derived from normal stem cells or progenitor cells that undergo genetic or epigenetic modification and transform themselves into CSCs by attaining unlimited self-renewable and differentiation potential (116, 117). Recent findings provide striking evidence that dysregulation of miRNAs regulates CSC characteristics and induces tumorigenesis, and multi drug resistance behavior of cancer cells. The four basic stemness transcriptional factors, OCT4, Nanog, Sox2, and Rex1 are responsible entities for cellular pluripotency and differentiation (118, 119). Recent molecular evidence supports the notion that members of the miR-290 family provide protective defense again differentiation defects in ESCs and play a key role for OCT-4 stability (120). miR-302 family is reported to limit the self-renewal ability and cellular differentiation by regulating the expression of key genes in stem cells. miR-34 family members contribute effectively in P53 dependent reprograming of human ESCs, and it has been noted that loss of functional ability of miR-34 is associated with the upregulation of pluripotency genes including N-Myc, SOX2, and NANOG (115). miR-34a is also reported to modulate neural differentiation by targeting SIRT1. It has been widely accepted that miRNAs are the potential contributors to regulate stem cell properties and stemness of cancer cells (115).
To understand the molecular underpinning of cancer stemness, CSCs were isolated from SW-1116 colon cancer cell lines with both CD133+/CD44+ and CD133−/CD44− surface phenotype antigen for comparative analysis. Researcher found 62 differentially expressed miRNAs in cancerous and non-cancerous stem cells and noted 31 miRNAs overexpressed including miR-29a and miR-29b, as well as 31 miRNAs under-expressed including miR-449a, miR-4524, and miR-451 (117). Exogenous expression of miR-451 declines the self-renewable capacity of stem cells and decreases multi drug resistant potential of cancer cells. Induced expression of miR-451 negatively regulates COX-2 gene that plays a decisive role in Wnt cascade activation and is believed to act as complementary factor for CSC activation. Wnt pathway has a key association with intestinal stem cell regulation and reported to linked with colon cancer onset (117, 121). Inhibition of Wnt pathway leads to the degradation of β-catenin in the cytoplasm and is unable to initiate epithelial renewal. Increased expression of miR-21 is noted to induce stemness by regulating Wnt activity and thus initiates carcinogenesis by inhibiting the tumor suppressor gene, TGF-βR2, that is a key regulatory gene for cellular differentiation (122). One of the fundamentals signaling pathway to regulate colon stemness is Notch. Notch cascade activation is believed to induce cellular proliferation of progenitor cells. miR-34a is shown to downregulate Notch signaling activity and regulate cellular differentiation of targeted cells and colon stem cells (123) (Table 1).
Putative Markers for CRC
LGR5 is a member of G-protein coupled receptor that can interact and trigger activation of Wnt signaling via binding to furin-like repeat FU2 domain of R-spondin (133). LGR5 has been reported to be a putative marker for CRC stem cells. It has come to light less lately that LGR5 triggers the activation of both Wnt and TGF-β signaling in cancer stem cells. Overexpression of LGR5 increases drug resistance and cancer stemness in both brain tumors and CRC. LGR5 has been reported to have high expression in most of the CRC cell lines and adenomas but this overexpression has nothing to do with progression of tumor as presence of LGR5 increases cell-cell adhesion which in turn promotes stemness and hampers invasiveness and migration (134). Experimentation conducted on the triple positive cells having LGR5-positive subpopulations demonstrated peculiar characteristics of self-renewal, differentiation, colony formation, tumorigenicity, and stemness (135). These findings suggest that LGR5 is a putative marker of CRC stem cells. A transgenic mice experiment confirmed the status of LGR5 as CRC stem cell marker. Addition of suicide gene to a transgenic mice genome that was activated in the presence of overexpressed LGR5 and tamoxifen resulted in the death of LGR5 rich colorectal stem cells. The absence of tamoxifen resulted in differentiation of LGR5 CSCs (136). From these findings it can be concluded that LGR5 is a putative CSC marker that should be considered as potential target for advanced grade CRC and such CSCs must be eradicated before the start of any combinational therapies for CRC. CD44 is a surface protein responsible for interaction between cells and also plays vital role in the adhesion and migration of the cells (137). The CD44 has a specific binding site for hyaluronic acid which facilitates interaction with selectin, osteopontin, fibronectin, laminin, and collagen in the extracellular matrix (138). The binding of hyaluronic acid with CD44 results in the activation of epidermal growth factor receptor family kinases such as the MAPK and PI3/AkT that in turn promotes growth and proliferation in various cancers (138). Majority of CSCs population have CD44 surface markers along with other cell surface markers that increase invasiveness and stemness (139). Considering its abundance in CRC tumorous stem cells it has been reported as putative marker for the detection of invasiveness and metastasis. Cluster differentiation 24 is an emerging biomarker for CRC (140). Overexpression of CD24 is affiliated with tumor differentiation, invasion metastasis (141). In addition, CD24 overexpression also promotes poor survival rates in the patients with CRC. These findings suggest that CD24 involvement increases stemness in CRC stem cells and may be used as a prognostic marker for patients with CRC.
Nanotherapeutics as an Approach for the Treatment of CRC
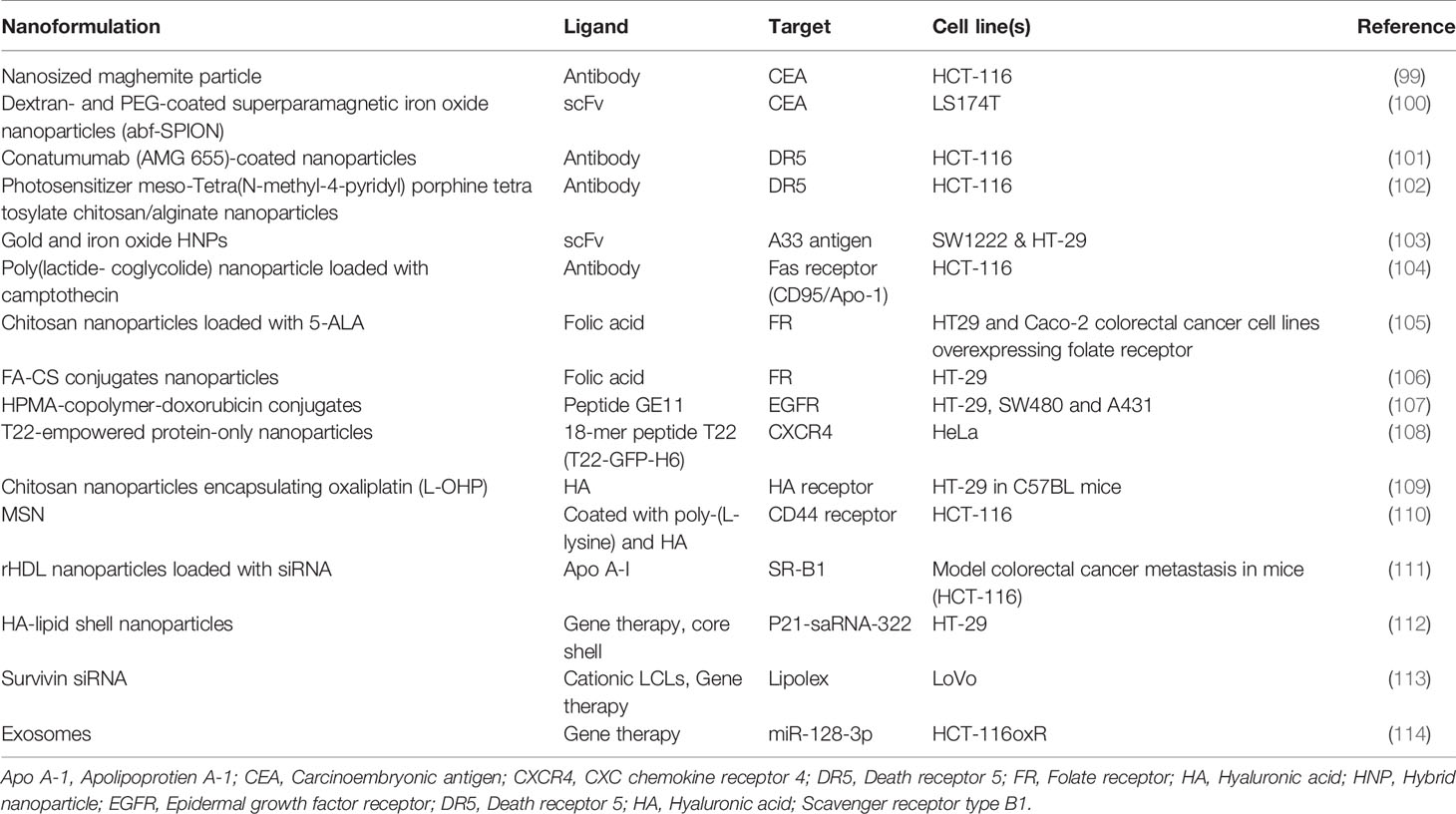
Hh pathway can be targeted by the specific inhibitors at various sites. Therefore, inhibitors of Hh pathway are useful anti-cancer agents. Till now, several small molecules inhibitors have been developed tested for their inhibitory effects on Hh signaling pathway. A natural alkaloid cyclopamine obtained from the corn lily V. californicum is the first Hh inhibitor reported. Cyclopamine impedes the functioning of smoothened via inhibiting its attachment to the heptahelical bundle (142). However, cyclopamine has several drawbacks such as the limited bioavailability, chemical instability, and shorter half-life. Therefore, it cannot be considered as potential therapeutic target. Several small molecule antagonists such as the SANT1, SANT2, SANT3, SANT4, CUR-61414, and GDC-0449 have been synthesized and evaluated in pre-clinical models for their anti-cancer activity in various solid tumors (143). The Hh small molecule inhibitors were first evaluated in basal cell carcinomas. Vismodegib a small molecule inhibitor of SMO is the first reported drug used for the treatment of basal cell carcinoma (144). In comparison to the cyclopamine, vismedegib was efficient to culminate cancer growth in both advanced and metastatic basal cell carcinomas. Over the years new therapeutic interventions in the development of SMO for Hh signaling antagonists such as LDE225 also known as the sonidegib has increased the drug efficacy via increasing tissue absorption and better penetration in the blood brain barrier for skin cancer and brain tumor respectively (144). There are several SMO antagonists designed to inhibit Hh pathway are in clinical trials that specifically target medulloblastoma, ovarian cancer, pancreatic cancer, and colon cancer (145). Yet the clinical success of these antagonists is still limited. In order to understand the effects of these SMOs on hedgehog dependent inhibition of CRC further investigation is required for finding suitable and effective drug. In recent years huge development in the field of nanotechnology has enabled us to devise efficient therapeutics for various diseases (146). In addition to this, nano-carriers have greater efficiency in delivering drug to target site with limited cytotoxicity. These observations have urged scientists to seek more efficient nano-drug delivery systems that can hamper cancer progression and increase apoptosis. There have been some serious drawbacks of utilizing SMO as inhibitors of Hh signaling (147). The SMO antagonists have poor bioavailability, drug resistance and non-specific activation of Gli (148). Nanoformulations can address these drawbacks by increasing bioavailability, reducing drug resistance and specific activation of Gli. Based on current data there are two types of nano-based Hh signaling inhibitors: Natural Inhibitors and synthetic inhibitors. Cyclopamine comes under the list of natural inhibitor that has faced severe criticism because of its limited bioavailability, poor solubility, and several side effects. However, nanoformulations of cyclopamine have reduced these obstacles. It has been reported that cycolopamine loaded lipid nanoparticles (NPs) efficiently reduced the growth of radiation therapy treated breast and pancreatic cells (149, 150). In another study, polymeric nanoparticles designed to carry both cyclopamine and doxorubicin reduced the growth in orthotropic breast cancer model (151). The polymeric nanoformulations of cyclopamine and paclitaxel successfully cured prostate cancer (152) and pancreatic cancer (153) in combination with chemotherapy in mice. A biomimetic nanoparticle delivery system having cyclopamine encapsulated in erythrocyte membrane camouflaged PLGA resulted in super enhanced bioavailability of cyclopamine. Moreover, a combination of biomimetic NPs with paclitaxel NPs increased the delivery of paclitaxel to the tumor tissue increased tumor profusion and inhibited tumor growth in vivo (154). Vismodegib an FDA approved natural inhibitor for Hh signaling pathway has limited solubility and bioavailability. The polymeric nanoformulations for vismodegib have resolved these issues. The encapsulation of vismodegib in SN38 pro-drug polymer which is an active metabolite of irinotecan to treat pancreatic ductal carcinoma resulted in decreased tumor growth and reduced fibrosis. In addition to this SN38 NPs facilitate the inhibition of Hh signaling which is crucial for the communication between tumor and stromal cells. SN38 NPs provided better diffusion for vismodegib thus prevented the drug resistance. SN38 NP encapsulation of vismodgib suppressed Gil1 expression in the tumor microenvironment of xenograft model suggesting the fact that SN38 NPs could aid in restoring normal drug resistance of the tumor cells (155). In another study, pH-responsive polymeric NPs containing vismodegib and gemcitabine inhibited growth of pancreatic cancer cells (156). From these findings, NP mediated drug delivery of Hh signaling inhibitors can be used a potential tactic to trigger chemotherapy. Apart from cyclopamine and vismodegib, several other natural Hh signaling inhibitors have been reported to be delivered by the nanofomulations. Anthothecol carrying PLGA nanoparticles have been reported to suppress proliferation and colony formation of pancreatic cancer stem cells through modulating the activity of the Gli-DNA binding (68). Another study confirmed that α-mangostin carrying PLGA nanoformulation disrupted the Gli-DNA binding activity in pancreatic cancer cells. This resulted in decreased growth, development, and metastasis of pancreatic cancer stem cells (157). Nanoformulation of glabrescione B has been reported to show tremendous anti-cancer activity in a Hh dependent manner (158). Nano-carriers have been employed in the delivery of the synthetic inhibitors of Hh signaling pathway. Quinacrine a synthetic inhibitor of Hh signaling when loaded into NP formulation triggered the recruitment of GSK-3β and PTEN which induced the apoptosis in cancer stem cells. In addition to this qunacrine loaded NPs also reduced the expression of Gli vital for the self-renewal of CSCs (159, 160). PLGA NPs encapsulating the HPI-1 a specific inhibitor of Gli1 prevented growth and metastasis of hepatocellular carcinoma mice model. Moreover, HPl-1 delivery reduced the expression of CD133+ cells a type of CSCs in hepatocellular carcinoma (161). A combination of the NPs and gemcitabine reduced cellular growth in xenograft model of pancreatic cancer in a ligand dependent paracrine activation of Hh signaling pathway (162). GANT61 a specific Gli1 inhibitor when encapsulated in PLGA NPs prevented the translocation of Gli-1 to the nucleus and reduced the growth of CSCs (163). Although there has been slight progress towards the utilization of nano-carriers as a module to treat Hh mediated CRC, these observations are in favor that nanoformulations could be used as small molecules Hh inhibitors to cancer. It has come to light less lately that nanoformulations can be used as a carrier for the targeted delivery of the miRNAs. There are different types of nanoparticle based formulations that are being used for this purpose. Lipid nanoparticles, extracellular vesicles, minicells (genetically developed from bacteria), dendrimers, polyamidoamine (PAMAM), and inorganic materials such as the silica, gold, and silver nanoparticles have been extensively synthesized for delivering specific miRNAs to targeted tumors. Oshima et al. have developed an in-vivo delivery system to target liver and CRC. They specifically designed nano-scale coordination polymers for the effective delivery of oligometastatic miR-655-3p. Their findings revealed that co-delivery of miR-655-3p along with oxaliplatin reduced tumor growth (164). Yang et al. successfully formulated polyethyleneimine/hyalouronic acid mesoporous silica nanoparticle loaded with oxaliplatin and miR-204-5p. This nanoformulation enhanced the apoptosis and therapeutic efficacy in HT-29 cell lines (165). Altogether these findings suggest that nanoformulations are a suitable platform for the delivery of miRNAs and can increase the therapeutic efficacy in CRC (Table 2).
Oral Nanotherapeutics
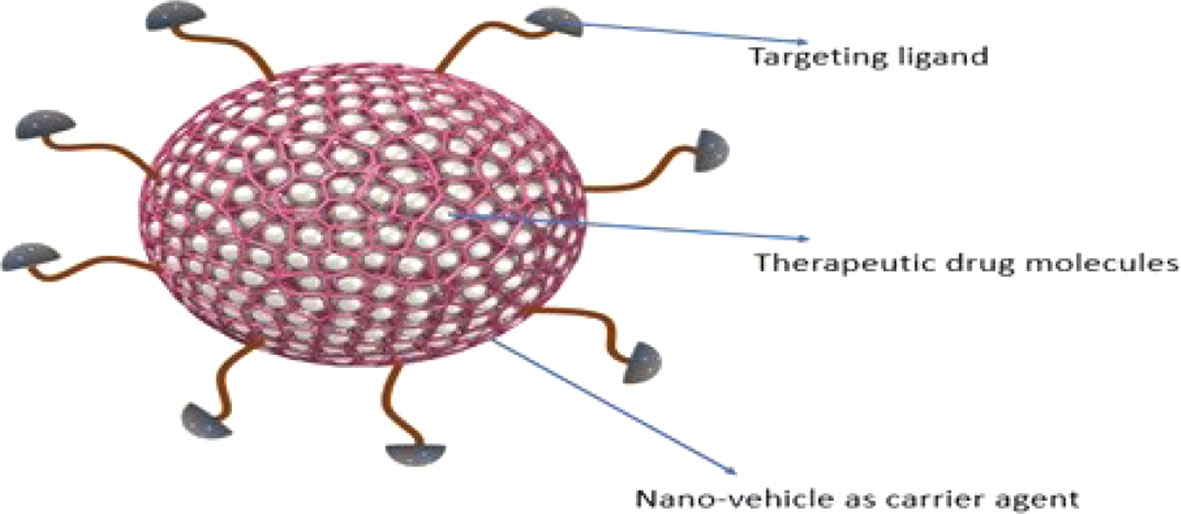
Nanotherapeutics is one of the promising strategies that offer dynamic surface functionalized modifications to improve targeted drug delivery and to limit the adverse effects. Nano-platform for drug designing offers potent routes to enhance drug profile. Nanotherapeutics are generally comprise of three core elements including nano-vehicle as carrier agent, target ligand and therapeutic drug molecule (Figures 3 and 4) (166). Different nanotherapeutics including lipid nanoparticles, organic and inorganic nanoparticles, polymeric nanoparticles, nanocrystals and plant derived nanomaterials have been used clinically including Taxel®, Lipo-Dox®, Abraxene®, Abraxene®, Marqib®, Onivyde® etc., and there is large number of nanotherapeutics in pre-clinical and clinical trials that can be fabricated to target Hh signaling cascade to reduce cancer cell proliferation (166, 167).
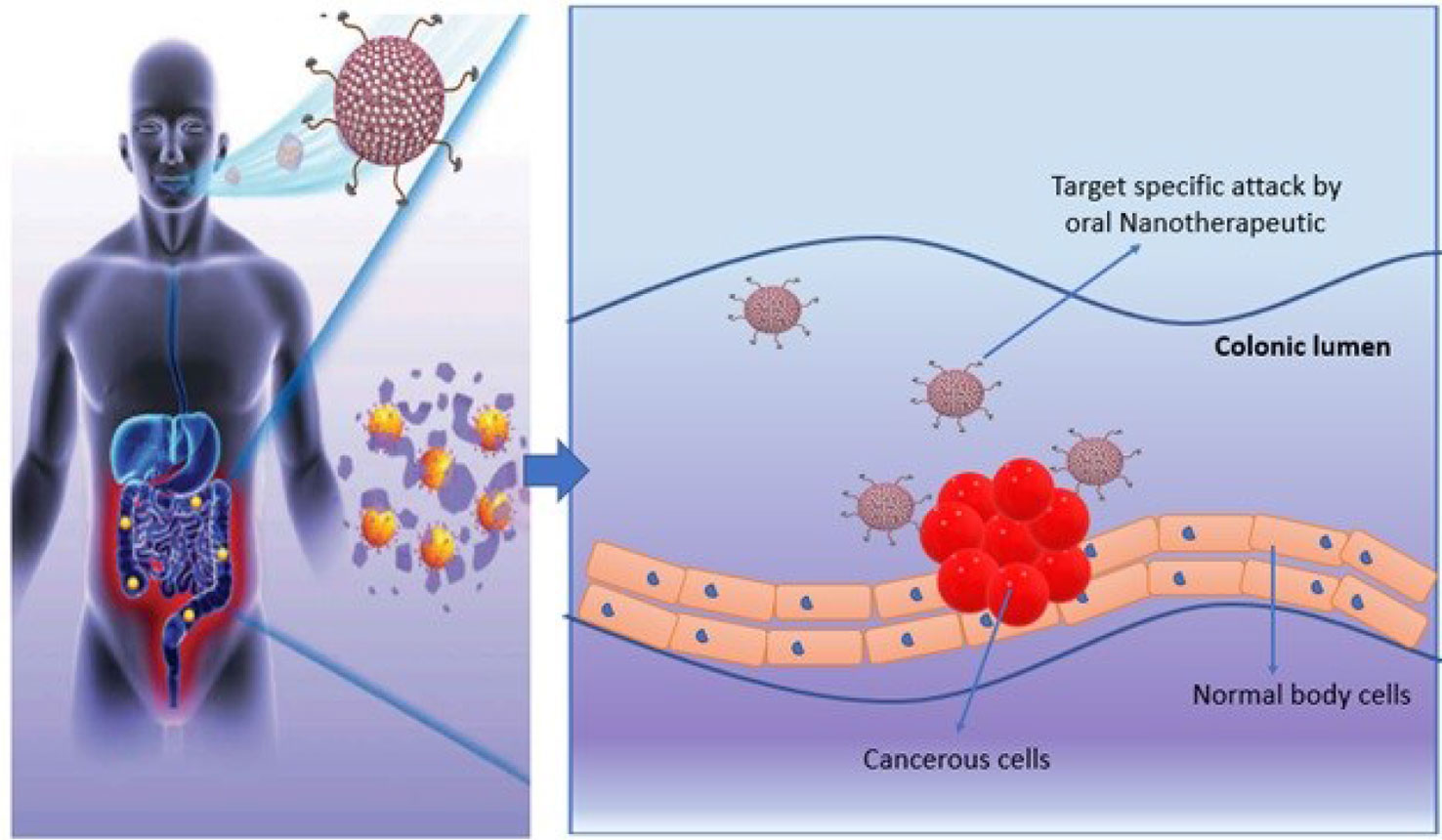
Oral chemotherapeutics are reported to have multiple limitations that demand novel alternative therapeutics for cancer treatment. Recently, the concept of oral nanotherapeutics paved the avenue in pharmaceutics towards more stable and high tumor targeted therapy with minimize adverse effects. A successful pre-clinical attempt has been performed by synthesizing redox nanoparticles (RNPs) for colon cancer treatment. This novel RNP contains nitroxide radicals for antioxidant activity and to scavenge ROS (reactive oxidative species) in cellular microenvironment. RNPs are specialized to accumulate in colonic mucosa and targeted cancer cells predominantly. These specialized oral-nanotherapeutics are also reported to limit cellular toxicity issues upon prolong exposure and is significant agent to inhibit tumor growth (Figure 5). It has been noticed that synergistic effect of RNP and conventional therapeutics can suppress adverse effects in gastro-intestinal tract and is an ideal candidate for future with significant potential in the existing pool for cancer treatment (168).
Various attempts have been made for successful oral delivery of targeted nanotherapeutics to treat cancer including bowel inflammatory cancer and colon cancers. Engineered chitosan based amphiphilic muco-adhesive drug-delivery strategies have been examined in in-vivo therapeutic studies. In one of the recent studies, SN38 (7-ethyl-10-hydroxycamptothecin) nanoparticles and water insoluble curcumin are proved to be a significant candidate to limit carcinogenesis and shrink tumor diameter >4 mm. Thus, bio-adhesive chitosan based stable colloidal nanotherapeutics is a novel and reliable approach to improve the outcome of colon cancer treatment (169).
There are a number of inhibitors related to Hh signaling namely sonidegib, saridegib, itraconazole, BMS-833923, LEQ-506, Taladegib, Glasdegib, TAK-441, Vismodegib, and several others (Table 3). But a very limited data is available regarding the use of these inhibitors for CRC. In addition to this majority of work done so far is on Wnt/β-catenin and mutations in this pathway. Therefore a lot of potential works need to be done against the implementation of such inhibitors in the clinical trials (Table 3).
Future Perspective
The Hh pathway is activated during regeneration and tissue repair in adults. Compelling pieces of evidence have indicated that inhibition of Hh pathway can prevent tumor progression and increased apoptosis. However, the clinical outcomes of such inhibition are unsatisfactory. The tumor heterogeneity and complex signaling crosstalk are the two major stumbling blocks that challenge the specificity of a specific Hh inhibitor. A tumor cell can trigger multiple signaling pathways simultaneously that can hamper the anti-proliferative abilities of a single inhibitor. Therefore, an outlook for new inhibitors of Hh signaling pathway that can hamper the activity of interconnected pathways is necessary. miRNAs have been reported to regulate the expression of vital genes involved in the proliferation and spread of CRC. They have been extensively examined for their putative role as diagnosis and prognosis markers for stratification of risk groups. Exploring the interplay between miRNAs and Hh signaling can aid in the development of therapeutics for Hh mediated CRCs. In addition to this miRNAs can regulate the cancer stem cells proliferation and metastasis (170). Therefore, they can be utilized as a probe to investigate cancer stemness and drug resistance in CRC stem cells. miRNAs modulating the expression of the proliferative genes is a hallmark in CRCs. A suitable drug delivery system can transfer miRNA modulating moieties to the target cell can impede the proliferative capabilities. Development of such delivery system will revolutionize therapeutics. Considering such scenario, nanoformulations can be a suitable platform for the treatment of various malignancies including cancer. Nanoformulations are advantageous because of their specificity, low toxicity, limited side effects, and enhanced bioavailability of the cargo such as various natural compounds i.e., berberine, paclitaxel curcumin, and SMOs. Hh signaling can be targeted with nanoparticles, but there are several drawbacks affiliated. Drug resistance is the major hurdle with the devising of NPs for Hh signaling. The complex interaction of Hh pathway makes it difficult to be targeted with nanoformulations of SMO. It has been reported that SMO nanoparticles were unable to hamper tumor growth when Gli was activated via non-canonical Hh signaling. In addition to this the interaction of Hh signaling with other molecular cascades such as the Wnt and Notch also affect the targeting capacity of small molecular inhibitors of Hh signaling. The nanoformulations have to overcome cellular resistance which is an effective barrier for suitable nano-drug delivery which can be overcome by the combination of nanoformulations such as pegylated liposomes and formulations that prevent quick release of cargo (Chitosan). Compelling research has dictated the fact that only 0.7% of the total nanoformulation reaches the solid tumor (171). This can lead to the development of side toxicity which is a major concern for most of the nanoformulations. However, recent advances in nano-drug delivery have culminated the side toxicity by implementing the use of biphosphonates. In addition, combining NP formulation can modulate the tumor microenvironment to enhance the drug delivery. A combination of cycolopamine and paclitaxel nanoformulation designed to impede the Hh signaling disrupted the extracellular matrix of the tumor cells and increased drug profusion. In addition choice of nanoformulation, size of the NPs and their diffusion in the cell, cost of production, clinical translation, and cancer cell resistance are the limiting barriers that need to be addressed for designing specific Hh inhibitors. Overcoming these challenges can improve treatment methods for cancer patients. Altogether miRNAs and natural compounds mediated regulation of Hh signaling might help us devising new diagnostic/prognosis and therapeutics for CRC.
Author Contributions
ZJ, MJ, and AR drafted the manuscript. HS, SR, AI, and WC revised the draft. HS, WK, WK-K, AG-L, JS-R, and WC conceptualized the study and reviewed it critically. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Javed Z, Khan K, Sadia H, Raza S, Salehi B, Sharifi-Rad J, et al. LncRNA & Wnt signaling in colorectal cancer. Cancer Cell Int (2020) 20(1):1–10. doi: 10.1186/s12935-020-01412-7
2. Johnson CM, Wei C, Ensor JE, Smolenski DJ, Amos CI, Levin B, et al. Meta-analyses of colorectal cancer risk factors. Cancer Causes Control (2013) 24(6):1207–22. doi: 10.1007/s10552-013-0201-5
3. Salaritabar A, Berindan-Neagoe I, Darvish B, Hadjiakhoondi F, Manayi A, Devi KP, et al. Targeting Hedgehog signaling pathway: paving the road for cancer therapy. Pharmacol Res (2019) 141:466–80. doi: 10.1016/j.phrs.2019.01.014
4. Tickle C, Towers M. Sonic hedgehog signaling in limb development. Front Cell Dev Biol (2017) 5:14. doi: 10.3389/fcell.2017.00014
5. Niewiadomski P, Niedziółka SM, Markiewicz Ł, Uśpieński T, Baran B, Chojnowska K. Gli proteins: Regulation in development and cancer. Cells (2019) 8(2):147. doi: 10.3390/cells8020147
6. Park K-S, Martelotto LG, Peifer M, Sos ML, Karnezis AN, Mahjoub MR, et al. A crucial requirement for Hedgehog signaling in small cell lung cancer. Nat Med (2011) 17(11):1504. doi: 10.1038/nm.2473
7. Bailey JM, Swanson BJ, Hamada T, Eggers JP, Singh PK, Caffery T, et al. Sonic hedgehog promotes desmoplasia in pancreatic cancer. Clin Cancer Res (2008) 14(19):5995–6004. doi: 10.1158/1078-0432.CCR-08-0291
8. Ingham PW, Placzek M. Orchestrating ontogenesis: variations on a theme by sonic hedgehog. Nat Rev Genet (2006) 7(11):841–50. doi: 10.1038/nrg1969
9. Skoda AM, Simovic D, Karin V, Kardum V, Vranic S, Serman L. The role of the Hedgehog signaling pathway in cancer: A comprehensive review. Bosnian J Basic Med Sci (2018) 18(1):8. doi: 10.17305/bjbms.2018.2756
10. Takebe N, Harris PJ, Warren RQ, Ivy SP. Targeting cancer stem cells by inhibiting Wnt, Notch, and Hedgehog pathways. Nat Rev Clin Oncol (2011) 8(2):97–106. doi: 10.1038/nrclinonc.2010.196
11. Wen D, Danquah M, Chaudhary AK, Mahato RI. Small molecules targeting microRNA for cancer therapy: Promises and obstacles. J Controlled Release (2015) 219:237–47. doi: 10.1016/j.jconrel.2015.08.011
12. Hahn H, Wicking C, Zaphiropoulos PG, Gailani MR, Shanley S, Chidambaram A, et al. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell (1996) 85(6):841–51. doi: 10.1016/S0092-8674(00)81268-4
13. Johnson RL, Rothman AL, Xie J, Goodrich LV, Bare JW, Bonifas JM, et al. Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science (1996) 272(5268):1668–71. doi: 10.1126/science.272.5268.1668
14. Couvé-Privat S, Bouadjar B, Avril MF, Sarasin A, Daya-Grosjean L. Significantly high levels of ultraviolet-specific mutations in the smoothened gene in basal cell carcinomas from DNA repair-deficient xeroderma pigmentosum patients. Cancer Res (2002) 62(24):7186–9.
15. Xie J, Murone M, Luoh S-M, Ryan A, Gu Q, Zhang C, et al. Activating Smoothened mutations in sporadic basal-cell carcinoma. Nature (1998) 391(6662):90–2. doi: 10.1038/34201
16. Clark VE, Erson-Omay EZ, Serin A, Yin J, Cotney J, Özduman K, et al. Genomic analysis of non-NF2 meningiomas reveals mutations in TRAF7, KLF4, AKT1, and SMO. Science (2013) 339(6123):1077–80. doi: 10.1126/science.1233009
17. Pelczar P, Zibat A, van Dop WA, Heijmans J, Bleckmann A, Gruber W, et al. Inactivation of Patched1 in mice leads to development of gastrointestinal stromal-like tumors that express Pdgfrα but not kit. Gastroenterology (2013) 144(1):134–44. e6. doi: 10.1053/j.gastro.2012.09.061
18. Mao J, Ligon KL, Rakhlin EY, Thayer SP, Bronson RT, Rowitch D, et al. A novel somatic mouse model to survey tumorigenic potential applied to the Hedgehog pathway. Cancer Res (2006) 66(20):10171–8. doi: 10.1158/0008-5472.CAN-06-0657
19. Xie J, Bartels CM, Barton SW, Gu D. Targeting hedgehog signaling in cancer: research and clinical developments. Onco Targ Ther (2013) 6:1425. doi: 10.2147/OTT.S34678
20. Kim J, Aftab BT, Tang JY, Kim D, Lee AH, Rezaee M, et al. Itraconazole and arsenic trioxide inhibit Hedgehog pathway activation and tumor growth associated with acquired resistance to smoothened antagonists. Cancer Cell (2013) 23(1):23–34. doi: 10.1016/j.ccr.2012.11.017
21. Gu D, Fan Q, Zhang X, Xie J. A role for transcription factor STAT3 signaling in oncogene smoothened-driven carcinogenesis. J Biol Chem (2012) 287(45):38356–66. doi: 10.1074/jbc.M112.377382
22. Xie J, Aszterbaum M, Zhang X, Bonifas JM, Zachary C, Epstein E, et al. A role of PDGFRα in basal cell carcinoma proliferation. Proc Natl Acad Sci (2001) 98(16):9255–9. doi: 10.1073/pnas.151173398
23. Wang Y, Ding Q, Yen C-J, Xia W, Izzo JG, Lang J-Y, et al. The crosstalk of mTOR/S6K1 and Hedgehog pathways. Cancer Cell (2012) 21(3):374–87. doi: 10.1016/j.ccr.2011.12.028
24. Johnson RW, Nguyen MP, Padalecki SS, Grubbs BG, Merkel AR, Oyajobi BO, et al. TGF-β promotion of Gli2-induced expression of parathyroid hormone-related protein, an important osteolytic factor in bone metastasis, is independent of canonical Hedgehog signaling. Cancer Res (2011) 71(3):822–31. doi: 10.1158/0008-5472.CAN-10-2993
25. Hsieh A, Ellsworth R, Hsieh D. Hedgehog/GLI1 regulates IGF dependent malignant behaviors in glioma stem cells. J Cell Physiol (2011) 226(4):1118–27. doi: 10.1002/jcp.22433
26. Niyaz M, Khan MS, Mudassar S. Hedgehog signaling: an Achilles’ heel in cancer. Trans Oncol (2019) 12(10):1334–44. doi: 10.1016/j.tranon.2019.07.004
27. Wu C, Zhu X, Liu W, Ruan T, Tao K. Hedgehog signaling pathway in colorectal cancer: function, mechanism, and therapy. OncoTarg Ther (2017) 10:3249. doi: 10.2147/OTT.S139639
28. Magistri P, Battistelli C, Strippoli R, Petrucciani N, Pellinen T, Rossi L, et al. SMO inhibition modulates cellular plasticity and invasiveness in colorectal cancer. Front Pharmacol (2018) 8:956. doi: 10.3389/fphar.2017.00956
29. Dierks C, Beigi R, Guo G-R, Zirlik K, Stegert MR, Manley P, et al. Expansion of Bcr-Abl-positive leukemic stem cells is dependent on Hedgehog pathway activation. Cancer Cell (2008) 14(3):238–49. doi: 10.1016/j.ccr.2008.08.003
30. Yoshikawa R, Nakano Y, Tao L, Koishi K, Matsumoto T, Sasako M, et al. Hedgehog signal activation in oesophageal cancer patients undergoing neoadjuvant chemoradiotherapy. Br J Cancer (2008) 98(10):1670–4. doi: 10.1038/sj.bjc.6604361
31. Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. nature (2001) 414(6859):105–11. doi: 10.1038/35102167
32. di Magliano MP, Hebrok M. Hedgehog signalling in cancer formation and maintenance. Nat Rev Cancer (2003) 3(12):903–11. doi: 10.1038/nrc1229
33. Scarpa M, Scarpa M. Hedgehog signaling in colorectal cancer: A spiny issue gets smoothened. Transl Cancer Res (2016) 5:1051S–S4. doi: 10.21037/tcr.2016.11.52
34. Curtin JC, Lorenzi MV. Drug discovery approaches to target Wnt signaling in cancer stem cells. Oncotarget (2010) 1(7):563. doi: 10.18632/oncotarget.191
35. Yoshimoto AN, Bernardazzi C, Carneiro AJV, Elia CC, Martinusso CA, Ventura GM, et al. Hedgehog pathway signaling regulates human colon carcinoma HT-29 epithelial cell line apoptosis and cytokine secretion. PloS One (2012) 7(9):e45332. doi: 10.1371/journal.pone.0045332
36. Ding Y-L, Zhou Y, Xiang L, Ji Z-P, Luo Z-H. Expression of glioma-associated oncogene homolog 1 is associated with invasion and postoperative liver metastasis in colon cancer. Int J Med Sci (2012) 9(5):334. doi: 10.7150/ijms.4553
37. Song I-S, Jeong YJ, Kim J, Seo K-H, Baek N-I, Kim Y, et al. Pharmacological inhibition of androgen receptor expression inducescell death in prostate cancer cells. Cell Mol Life Sci (2020) 77:4663–73. doi: 10.1007/s00018-019-03429-2
38. Zhang X, Zhang S-S, Wei G-J, Deng Z-M, Hu Y. Dysregulation of hedgehog signaling pathway related components in the evolution of colonic carcinogenesis. Int J Clin Exp Med (2015) 8(11):21379.
39. Miyaki M, Konishi M, Kikuchi-Yanoshita R, Enomoto M, Igari T, Tanaka K, et al. Characteristics of somatic mutation of the adenomatous polyposis coli gene in colorectal tumors. Cancer Res (1994) 54(11):3011–20.
40. Yoshikawa K, Shimada M, Miyamoto H, Higashijima J, Miyatani T, Nishioka M, et al. Sonic hedgehog relates to colorectal carcinogenesis. J Gastroenterol (2009) 44(11):1113. doi: 10.1007/s00535-009-0110-2
41. Song L, Li Z-Y, Liu W-P, Zhao M-R. Crosstalk between Wnt/β-catenin and Hedgehog/Gli signaling pathways in colon cancer and implications for therapy. Cancer Biol Ther (2015) 16(1):1–7. doi: 10.4161/15384047.2014.972215
42. To KK, Tong CW, Wu M, Cho WC. MicroRNAs in the prognosis and therapy of colorectal cancer: From bench to bedside. World J Gastroenterol (2018) 24(27):2949. doi: 10.3748/wjg.v24.i27.2949
43. Yochum GS, McWeeney S, Rajaraman V, Cleland R, Peters S, Goodman RH. Serial analysis of chromatin occupancy identifies β-catenin target genes in colorectal carcinoma cells. Proc Natl Acad Sci (2007) 104(9):3324–9. doi: 10.1073/pnas.0611576104
44. Varnat F, Siegl-Cachedenier I, Malerba M, Gervaz P, Ruiz i Altaba A. Loss of WNT-TCF addiction and enhancement of HH-GLI1 signalling define the metastatic transition of human colon carcinomas. EMBO Mol med (2010) 2(11):440–57. doi: 10.1002/emmm.201000098
45. Phi LTH, Sari IN, Yang Y-G, Lee S-H, Jun N, Kim KS, et al. Cancer stem cells (CSCs) in drug resistance and their therapeutic implications in cancer treatment. Stem Cells Int (2018) 2018. doi: 10.1155/2018/5416923
46. Sari IN, Phi LTH, Jun N, Wijaya YT, Lee S, Kwon HY. Hedgehog signaling in cancer: a prospective therapeutic target for eradicating cancer stem cells. Cells (2018) 7(11):208. doi: 10.3390/cells7110208
47. Yang L, Shi P, Zhao G, Xu J, Peng W, Zhang J, et al. Targeting cancer stem cell pathways for cancer therapy. Signal Transduct Targeted Ther (2020) 5(1):1–35. doi: 10.1038/s41392-020-0110-5
48. Jeng K-S, Sheen I, Leu C-M, Tseng P-H, Chang C-F. The Role of Smoothened in Cancer. Int J Mol Sci (2020) 21(18):6863. doi: 10.3390/ijms21186863
49. Pietrobono S, Gagliardi S, Stecca B. Non-canonical hedgehog signaling pathway in cancer: activation of GLI transcription factors beyond smoothened. Front Genet (2019) 10:556. doi: 10.3389/fgene.2019.00556
50. Regan JL, Schumacher D, Staudte S, Steffen A, Haybaeck J, Keilholz U, et al. Non-canonical hedgehog signaling is a positive regulator of the WNT pathway and is required for the survival of colon cancer stem cells. Cell Rep (2017) 21(10):2813–28. doi: 10.1016/j.celrep.2017.11.025
51. Wu C, Hu S, Cheng J, Wang G, Tao K. Smoothened antagonist GDC−0449 (Vismodegib) inhibits proliferation and triggers apoptosis in colon cancer cell lines. Exp Ther Med (2017) 13(5):2529–36. doi: 10.3892/etm.2017.4282
52. Rahman MS, Suresh S, Waly MI. Risk Factors for Cancer: Genetic and Environment. Bioactive Components Diet Med Treat Cancer Prevent: Springer (2018) 1–23. doi: 10.1007/978-3-319-75693-6_1
53. Williams JB, Li S, Higgs EF, Cabanov A, Wang X, Huang H, et al. Tumor heterogeneity and clonal cooperation influence the immune selection of IFN-γ-signaling mutant cancer cells. Nat Commun (2020) 11(1):1–14. doi: 10.1038/s41467-020-14290-4
54. Jeong WJ, Ro EJ, Choi KY. Interaction between Wnt/β-catenin and RAS-ERK pathways and an anti-cancer strategy via degradations of β-catenin and RAS by targeting the Wnt/β-catenin pathway. NPJ Precis Oncol (2018) 2(1):5. doi: 10.1038/s41698-018-0049-y
55. Wu WK, Wang XJ, Cheng AS, Luo MX, Ng SS, To KF, et al. Dysregulation and crosstalk of cellular signaling pathways in colon carcinogenesis. Crit Rev Oncol Hematol (2013) 86(3):251–77. doi: 10.1016/j.critrevonc.2012.11.009
56. Li D, Masiero M, Banham AH, Harris AL. The notch ligand JAGGED1 as a target for anti-tumor therapy. Front Oncol (2014) 4:254. doi: 10.3389/fonc.2014.00254
57. Liao W, Li G, You Y, Wan H, Wu Q, Wang C, et al. Antitumor activity of Notch−1 inhibition in human colorectal carcinoma cells. Oncol Rep (2018) 39(3):1063–71. doi: 10.3892/or.2017.6176
58. Katoh Y, Katoh M. WNT antagonist, SFRP1, is Hedgehog signaling target. Int J Mol Med (2006) 17(1):171–5. doi: 10.3892/ijmm.17.1.171
59. Wang H, Ke F, Zheng J. Hedgehog-glioma-associated oncogene homolog-1 signaling in colon cancer cells and its role in the celecoxib-mediated anti-cancer effect. Oncol Lett (2014) 8(5):2203–8. doi: 10.3892/ol.2014.2439
60. Finotti A, Fabbri E, Lampronti I, Gasparello J, Borgatti M, Gambari R. MicroRNAs and Long Non-coding RNAs in Genetic Diseases. Mol Diagn Ther (2019) 23(2):155–71. doi: 10.1007/s40291-018-0380-6
61. Cho KHT, Xu B, Blenkiron C, Fraser M. Emerging Roles of miRNAs in Brain Development and Perinatal BrainInjury. Front Physiol (2019) 10(227):200–27. doi: 10.3389/fphys.2019.00227
62. Wienholds E, Plasterk RHA. MicroRNA function in animal development. FEBS Lett (2005) 579(26):5911–22. doi: 10.1016/j.febslet.2005.07.070
63. Zhuang M, Qiu X, Cheng D, Zhu C, Chen L. MicroRNA-524 promotes cell proliferation by down-regulating PTEN expression in osteosarcoma. Cancer Cell Int (2018) 18(1):114. doi: 10.1186/s12935-018-0612-1
64. Li L, Miu K-K, Gu S, Cheung H-H, Chan W-Y. Comparison of multi-lineage differentiation of hiPSCs reveals novel miRNAs that regulate lineage specification. Sci Rep (2018) 8(1):9630. doi: 10.1038/s41598-018-27719-0
65. Slattery ML, Mullany LE, Sakoda LC, Wolff RK, Samowitz WS, Herrick JS. Dysregulated genes and miRNAs in the apoptosis pathway in colorectal cancer patients. Apoptosis (2018) 23(3-4):237–50. doi: 10.1007/s10495-018-1451-1
66. Barbu MG, Condrat CE, Thompson DC, Bugnar OL, Cretoiu D, Toader OD, et al. MicroRNA Involvement in Signaling Pathways During ViralInfection. Front Cell Dev Biol (2020) 8(143):1–22. doi: 10.3389/fcell.2020.00143
67. Dwivedi S, Purohit P, Sharma P. MicroRNAs and Diseases: Promising Biomarkers for Diagnosis and Therapeutics. Indian J Clin Biochem (2019) 34(3):243–5. doi: 10.1007/s12291-019-00844-x
68. Valinezhad Orang A, Safaralizadeh R, Kazemzadeh-Bavili M. Mechanisms of miRNA-Mediated Gene Regulation from Common Downregulation to mRNA-Specific Upregulation. Int J Genomics (2014) 2014:970607. doi: 10.1155/2014/970607
69. Zhao J, Zhang Y, Zhao G. Emerging role of microRNA-21 in colorectal cancer. Cancer Biomarkers (2015) 15(3):219–26. doi: 10.3233/CBM-150468
70. Cekaite L, Eide PW, Lind GE, Skotheim RI, Lothe RA. MicroRNAs as growth regulators, their function and biomarker status in colorectal cancer. Oncotarget (2016) 7(6):6476–505. doi: 10.18632/oncotarget.6390
71. Chen X, Guo X, Zhang H, Xiang Y, Chen J, Yin Y, et al. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene (2009) 28(10):1385–92. doi: 10.1038/onc.2008.474
72. Yang F, Xie Y-Q, Tang S-Q, Wu X-B, Zhu H-Y. miR-143 regulates proliferation and apoptosis of colorectal cancer cells and exhibits altered expression in colorectal cancer tissue. Int J Clin Exp Med (2015) 8(9):15308–12.
73. Li S, Wu X, Xu Y, Wu S, Li Z, Chen R, et al. miR-145 suppresses colorectal cancer cell migration and invasion bytargeting an ETS-related gene. Oncol Rep (2016) 36(4):1917–26. doi: 10.3892/or.2016.5042
74. Sheng N, Tan G, You W, Chen H, Gong J, Chen D, et al. MiR-145 inhibits human colorectal cancer cell migration and invasionvia PAK4-dependent pathway. Cancer Med (2017) 6(6):1331–40. doi: 10.1002/cam4.1029
75. Segditsas S, Tomlinson I. Colorectal cancer and genetic alterations in the Wntpathway. Oncogene (2006) 25(57):7531–7. doi: 10.1038/sj.onc.1210059
76. Wu L, Yang J, Shi Y, Xia H, Xiang X, Yu X, et al. Circ-ZNF609 promotes migration of colorectal cancer by inhibiting Gli1 expression via microRNA-150. J BUON (2018) 23(5):1343–9.
77. Fan H, Liu X, Zheng W, Zhuang Z, Wang C. MiR-150 alleviates EMT and cell invasion of colorectal cancer through targeting Gli1. Eur Rev Med Pharmacol Sci (2017) 21(21):4853–9.
78. Gao X, Xu W, Lu T, Zhou J, Ge X, Hua D. MicroRNA-142-3p Promotes Cellular Invasion of Colorectal CancerCells by Activation of RAC1. Technol Cancer Res Treat (2018) 17:1533033818790508. doi: 10.1177/1533033818790508
79. Lee DY, Deng C-H, Wang BB, Yang BB. MicroRNA-378 promotes cell survival, tumor growth, and angiogenesisby targeting SuFu and Fus-1 expression. Proc Natl Acad Sci (2007) 104(51):20350–5. doi: 10.1073/pnas.0706901104
80. Hwang W-L, Jiang J-K, Yang S-H, Huang T-S, Lan H-Y, Teng H-W, et al. MicroRNA-146a directs the symmetric division of Snail-dominant colorectal cancer stem cells. Nat Cell Biol (2014) 16(3):268–80. doi: 10.1038/ncb2910
81. Riaz SK, Ke Y, Wang F, Kayani MA, Malik MFA. Influence of SHH/GLI1 axis on EMT mediated migration and invasion of breast cancer cells. Sci Rep (2019) 9(1):6620. doi: 10.1038/s41598-019-43093-x
82. Zhang JX, Mai SJ, Huang XX, Wang FW, Liao YJ, Lin MC, et al. MiR-29c mediates epithelial-to-mesenchymal transition in humancolorectal carcinoma metastasis via PTP4A and GNA13 regulation of β-cateninsignaling. Ann Oncol (2014) 25(11):2196–204. doi: 10.1093/annonc/mdu439
83. Slabáková E, Culig Z, Remšík J, Souček K. Alternative mechanisms of miR-34a regulation in cancer. Cell Death Dis (2017) 8(10):e3100–e. doi: 10.1038/cddis.2017.495
84. Lin J, Chen Y, Cai Q, Wei L, Zhan Y, Shen A, et al. Scutellaria barbata D Don inhibits colorectal cancer growth via suppression of multiple signaling pathways. Integr Cancer Ther (2014) 13(3):240–8. doi: 10.1177/1534735413508811
85. Wei L-H, Lin J-M, Chu J-F, Chen H-W, Li Q-Y, Peng J. Scutellaria barbata D. Don inhibits colorectal cancer growth via suppression of Wnt/β-catenin signaling pathway. Chin J Integr med (2017) 23(11):858–63. doi: 10.1007/s11655-017-2775-3
86. Bu P, Chen K-Y, Chen JH, Wang L, Walters J, Shin YJ, et al. A microRNA miR-34a-regulated bimodal switch targets Notch in colon cancer stem cells. Cell Stem Cell (2013) 12(5):602–15. doi: 10.1016/j.stem.2013.03.002
87. Sun X, Liu S, Chen P, Fu D, Hou Y, Hu J, et al. miR-449a inhibits colorectal cancer progression by targeting SATB2. Oncotarget (2017) 8(60):100975. doi: 10.18632/oncotarget.10900
88. Babashah S, Sadeghizadeh M, Hajifathali A, Tavirani MR, Zomorod MS, Ghadiani M, et al. Targeting of the signal transducer Smo links microRNA-326 to the oncogenic Hedgehog pathway in CD34+ CML stem/progenitor cells. Int J Cancer (2013) 133(3):579–89. doi: 10.1002/ijc.28043
89. Sheybani Z, Rahgozar S, Ghodousi ES. The Hedgehog signal transducer Smoothened and microRNA-326: pathogenesis and regulation of drug resistance in pediatric B-cell acute lymphoblastic leukemia. Cancer Manage Res (2019) 11:7621. doi: 10.2147/CMAR.S214405
90. Tang B, Xu A, Xu J, Huang H, Chen L, Su Y, et al. MicroRNA-324-5p regulates stemness, pathogenesis and sensitivity to bortezomib in multiple myeloma cells by targeting hedgehog signaling. Int J Cancer (2018) 142(1):109–20. doi: 10.1002/ijc.31041
91. Ma C, Nong K, Wu B, Dong B, Bai Y, Zhu H, et al. miR-212 promotes pancreatic cancer cell growth and invasion by targeting the hedgehog signaling pathway receptor patched-1. J Exp Clin Cancer Res (2014) 33(1):54. doi: 10.1186/1756-9966-33-54
92. Zhao D, Cui Z. MicroRNA−361−3p regulates retinoblastoma cell proliferation and stemness by targeting hedgehog signaling. Exp Ther Med (2019) 17(2):1154–62. doi: 10.3892/etm.2018.7062
93. Lou W, Liu J, Ding B, Jin L, Xu L, Li X, et al. Five miRNAs-mediated PIEZO2 downregulation, accompanied with activation of Hedgehog signaling pathway, predicts poor prognosis of breast cancer. Aging (Albany NY) (2019) 11(9):2628. doi: 10.18632/aging.101934
94. Sun K, Deng H-J, Lei S-T, Dong J-Q, Li G-X. miRNA-338-3p suppresses cell growth of human colorectal carcinoma by targeting smoothened. World J Gastroenterol: WJG (2013) 19(14):2197. doi: 10.3748/wjg.v19.i14.2197
95. Sun K, Su G, Deng H, Dong J, Lei S, Li G. Relationship between miRNA-338-3p expression and progression andprognosis of human colorectal carcinoma. Chin Med J (2014) 127(10):1884–90. doi: 10a.3748/wjg.v19.i14.2197
96. Yu Z, Sze-Lok Cheng A. Epigenetic deregulation of microRNAs: new opportunities to target oncogenic signaling pathways in hepatocellular carcinoma. Curr Pharm Design (2013) 19(7):1192–200. doi: 10.2174/138161213804805757
97. Sicklick JK, Li Y-X, Jayaraman A, Kannangai R, Qi Y, Vivekanandan P, et al. Dysregulation of the Hedgehog pathway in human hepatocarcinogenesis. Carcinogenesis (2006) 27(4):748–57. doi: 10.1093/carcin/bgi292
98. Tandon I, Waghmode A, Sharma NK. Cancer stem cells equipped with powerful hedgehog signaling and better epigenetic memory: Avenues to look for cancer therapeutics. Curr Cancer Drug Targ (2019) 19(11):877–84. doi: 10.2174/1568009619666190808155432
99. da Paz MC, Santos Mde F, Santos CM, da Silva SW, de Souza LB, Lima EC, et al. Anti-CEA loaded maghemite nanoparticles as a theragnostic device for colorectal cancer. Int J Nanomed (2012) 7:5271–82. doi: 10.2147/IJN.S32139
100. Vigor KL, Kyrtatos PG, Minogue S, Al-Jamal KT, Kogelberg H, Tolner B, et al. Nanoparticles functionalized with recombinant single chain Fv antibody fragments (scFv) for the magnetic resonance imaging of cancer cells. Biomaterials (2010) 31(6):1307–15. doi: 10.1016/j.biomaterials.2009.10.036
101. Fay F, McLaughlin KM, Small DM, Fennell DA, Johnston PG, Longley DB, et al. Conatumumab (AMG 655) coated nanoparticles for targeted pro-apoptotic drug delivery. Biomaterials (2011) 32(33):8645–53. doi: 10.1016/j.biomaterials.2011.07.065
102. Abdelghany SM, Schmid D, Deacon J, Jaworski J, Fay F, McLaughlin KM, et al. Enhanced antitumor activity of the photosensitizer meso-Tetra(N-methyl-4-pyridyl) porphine tetra tosylate through encapsulation in antibody-targeted chitosan/alginate nanoparticles. Biomacromolecules (2013) 14(2):302–10. doi: 10.1021/bm301858a
103. Kirui DK, Rey DA, Batt CA. Gold hybrid nanoparticles for targeted phototherapy and cancer imaging. Nanotechnology (2010) 21(10):105105. doi: 10.1088/0957-4484/21/10/105105
104. McCarron PA, Marouf WM, Quinn DJ, Fay F, Burden RE, Olwill SA, et al. Antibody targeting of camptothecin-loaded PLGA nanoparticles to tumor cells. Bioconjugate Chem (2008) 19(8):1561–9. doi: 10.1021/bc800057g
105. Yang SJ, Lin FH, Tsai KC, Wei MF, Tsai HM, Wong JM, et al. Folic acid-conjugated chitosan nanoparticles enhanced protoporphyrin IX accumulation in colorectal cancer cells. Bioconjugate Chem (2010) 21(4):679–89. doi: 10.1021/bc9004798
106. Li P, Wang Y, Zeng F, Chen L, Peng Z, Kong LX. Synthesis and characterization of folate conjugated chitosan and cellular uptake of its nanoparticles in HT-29 cells. Carbohydr Res (2011) 346(6):801–6. doi: 10.1016/j.carres.2011.01.027
107. Kopansky E, Shamay Y, David A. Peptide-directed HPMA copolymer-doxorubicin conjugates as targeted therapeutics for colorectal cancer. J Drug Targeting (2011) 19(10):933–43. doi: 10.3109/1061186X.2011.632011
108. Unzueta U, Céspedes MV, Ferrer-Miralles N, Casanova I, Cedano J, Corchero JL, et al. Intracellular CXCR4+ cell targeting with T22-empowered protein-only nanoparticles. Int J Nanomed (2012) 7:4533. doi: 10.2147/IJN.S34450
109. Jain A, Jain SK, Ganesh N, Barve J, Beg AM. Design and development of ligand-appended polysaccharidic nanoparticles for the delivery of oxaliplatin in colorectal cancer. Nanomedicine (2010) 6(1):179–90. doi: 10.1016/j.nano.2009.03.002
110. Gary-Bobo M, Brevet D, Benkirane-Jessel N, Raehm L, Maillard P, Garcia M, et al. Hyaluronic acid-functionalized mesoporous silica nanoparticles for efficient photodynamic therapy of cancer cells. Photodiagn Photodynamic Ther (2012) 9(3):256–60. doi: 10.1016/j.pdpdt.2011.12.010
111. Shahzad MM, Mangala LS, Han HD, Lu C, Bottsford-Miller J, Nishimura M, et al. Targeted delivery of small interfering RNA using reconstituted high-density lipoprotein nanoparticles. Neoplasia (N Y NY) (2011) 13(4):309–19. doi: 10.1593/neo.101372
112. Xu J, Tam M, Samaei S, Lerouge S, Barralet J, Stevenson MM, et al. Mucoadhesive chitosan hydrogels as rectal drug delivery vessels to treat ulcerative colitis. Acta Biomater (2017) 48:247–57. doi: 10.1016/j.actbio.2016.10.026
113. Wang T, Liu Z, Zhang Z, Tang S, Yue M, Feng S, et al. Evaluation of antitumor activity of survivin short interfering RNA delivered by lipid nanoparticles in colon cancer in vitro and in vivo. Oncol Lett (2017) 14(2):2001–8. doi: 10.3892/ol.2017.6404
114. Liu T, Zhang X, Du L, Wang Y, Liu X, Tian H, et al. Exosome-transmitted miR-128-3p increase chemosensitivity of oxaliplatin-resistant colorectal cancer. Mol Cancer (2019) 18(1):43. doi: 10.1186/s12943-019-0981-7
115. Choi YJ, Lin C-P, Ho JJ, He X, Okada N, Bu P, et al. miR-34 miRNAs provide a barrier for somatic cell reprogramming. Nat Cell Biol (2011) 13(11):1353–60. doi: 10.1038/ncb2366
116. Sell S. Stem cell origin of cancer and differentiation therapy. Crit Rev Oncol (2004) 51(1):1–28. doi: 10.1016/j.critrevonc.2004.04.007
117. Li Q, Yao Y, Eades G, Liu Z, Zhang Y, Zhou Q. Downregulation of miR-140 promotes cancer stem cell formation in basal-like early stage breast cancer. Oncogene (2014) 33(20):2589–600. doi: 10.1038/onc.2013.226
118. Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell (2005) 122(6):947–56. doi: 10.1016/j.cell.2005.08.020
119. Loh Y-H, Wu Q, Chew J-L, Vega VB, Zhang W, Chen X, et al. The Oct4 and Nanog transcription network regulates pluripotency inmouse embryonic stem cells. Nat Genet (2006) 38(4):431–40. doi: 10.1038/ng1760
120. Sinkkonen L, Hugenschmidt T, Berninger P, Gaidatzis D, Mohn F, Artus-Revel CG, et al. MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat Struct Mol Biol (2008) 15(3):259. doi: 10.1038/nsmb.1391
121. Vermeulen L, Felipe De Sousa EM, Van Der Heijden M, Cameron K, De Jong JH, Borovski T, et al. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat Cell Biol (2010) 12(5):468–76. doi: 10.1038/ncb2048
122. Yu Y, Kanwar SS, Patel BB, Oh P-S, Nautiyal J, Sarkar FH, et al. MicroRNA-21 induces stemness by downregulating transforming growth factor beta receptor 2 (TGFβR2) in colon cancer cells. Carcinogenesis (2012) 33(1):68–76. doi: 10.1093/carcin/bgr246
123. Fre S, Huyghe M, Mourikis P, Robine S, Louvard D, Artavanis-Tsakonas S. Notch signals control the fate of immature progenitor cells in the intestine. Nature (2005) 435(7044):964–8. doi: 10.1038/nature03589
124. Zhang X, Ai F, Li X, Tian L, Wang X, Shen S, et al. MicroRNA-34a suppresses colorectal cancer metastasis by regulating Notch signaling. Oncol Lett (2017) 14(2):2325–33. doi: 10.3892/ol.2017.6444
125. Chakraborty S, Ghosh Z. MicroRNAs shaping cellular reprogramming. In: AGO-Driven Non-Coding RNAs. Elsevier (2019). p. 75–97. doi: 10.1016/B978-0-12-815669-8.00004-X
126. Shi L, Xi J, Xu X, Peng B, Zhang B. MiR-148a suppressed cell invasion and migration via targeting WNT10b and modulating β-catenin signaling in cisplatin-resistant colorectal cancer cells. Biomed Pharmacother (2019) 109:902–9. doi: 10.1016/j.biopha.2018.10.080
127. Razi S, Sadeghi A, Asadi-Lari Z, Tam KJ, Kalantari E, Madjd Z. DCLK1, a promising colorectal cancer stem cell marker, regulates tumor progression and invasion through miR-137 and miR-15a dependent manner. Clin Exp Med (2020) 1–9. doi: 10.1007/s10238-020-00665-w
128. Wang LQ, Yu P, Li B, Guo YH, Liang ZR, Zheng LL, et al. miR-372 and miR-373 enhance the stemness of colorectal cancer cellsby repressing differentiation signaling pathways. Mol Oncol (2018) 12(11):1–9. doi: 10.1002/1878-0261.12376
129. Ren D, Lin B, Zhang X, Peng Y, Ye Z, Ma Y, et al. Maintenance of cancer stemness by miR-196b-5p contributes to chemoresistance of colorectal cancer cells via activating STAT3 signaling pathway. Oncotarget (2017) 8(30):49807. doi: 10.18632/oncotarget.17971
130. Jin Y, Wang M, Hu H, Huang Q, Chen Y, Wang G. Overcoming stemness and chemoresistance in colorectal cancer through miR-195-5p-modulated inhibition of notch signaling. Int J Biol Macromol (2018) 117:445–53. doi: 10.1016/j.ijbiomac.2018.05.151
131. Zhu QD, Zhou QQ, Dong L, Huang Z, Wu F, Deng X. MiR-199a-5p Inhibits the Growth and Metastasis of Colorectal Cancer Cells by Targeting ROCK1. Technol Cancer Res Treat (2018) 17:1533034618775509–. doi: 10.1177/1533034618775509
132. De Robertis M, Mazza T, Fusilli C, Loiacono L, Poeta ML, Sanchez M, et al. EphB2 stem-related and EphA2 progression-related miRNA-based networks in progressive stages of CRC evolution: clinical significance and potential miRNA drivers. Mol Cancer (2018) 17(1):169. doi: 10.1186/s12943-018-0912-z
133. de Lau W, Peng WC, Gros P, Clevers H. The R-spondin/Lgr5/Rnf43 module: regulator of Wnt signal strength. Genes Dev (2014) 28(4):305–16. doi: 10.1101/gad.235473.113
134. Takahashi H, Ishii H, Nishida N, Takemasa I, Mizushima T, Ikeda M, et al. Significance of Lgr5+ ve cancer stem cells in the colon and rectum. Ann Surg Oncol (2011) 18(4):1166–74. doi: 10.1245/s10434-010-1373-9
135. Leng Z, Xia Q, Chen J, Li Y, Xu J, Zhao E, et al. Lgr5+ CD44+ EpCAM+ strictly defines cancer stem cells in human colorectal cancer. Cell Physiol Biochem (2018) 46(2):860–72. doi: 10.1159/000488743
136. Fumagalli A, Oost KC, Kester L, Morgner J, Bornes L, Bruens L, et al. Plasticity of Lgr5-negative cancer cells drives metastasis incolorectal cancer. Cell (2020) 26:569–78. doi: 10.1016/j.stem.2020.02.008
137. Chen L, Fu C, Zhang Q, He C, Zhang F, Wei Q. The role of CD44 in pathological angiogenesis. FASEB J (2020) 34(10):13125–39. doi: 10.1096/fj.202000380RR
138. Rios de la Rosa JM, Tirella A, Tirelli N. Receptor-Targeted Drug Delivery and the (Many) Problems We Know of: The Case of CD44 and Hyaluronic Acid. Advanced Biosyst (2018) 2(6):1800049. doi: 10.1002/adbi.201800049
139. Yan X, Han D, Chen Z, Han C, Dong W, Han L, et al. RUNX2 interacts with BRG1 to target CD44 for promoting invasion and migration of colorectal cancer cells. Cancer Cell Int (2020) 20(1):1–13. doi: 10.1186/s12935-020-01544-w
140. Chen Y-C, Lee T-H, Tzeng S-L. Reduced DAXX Expression Is Associated with Reduced CD24 Expression in Colorectal Cancer. Cells (2019) 8(10):1242. doi: 10.3390/cells8101242
141. Wang J-L, Guo C-R, Su W-Y, Chen Y-X, Xu J, Fang J-Y. CD24 Overexpression Related to Lymph Node Invasion and PoorPrognosis of Colorectal Cancer. Clinicallaboratory (2018) 64(4):497–505. doi: 10.7754/Clin.Lab.2017.171012
142. Ecke I, Rosenberger A, Obenauer S, Dullin C, Aberger F, Kimmina S, et al. Cyclopamine treatment of full-blown Hh/Ptch-associated RMS partially inhibits Hh/Ptch signaling, but not tumor growth. Mol Carcinogen: Published Cooperation Univ Texas MD Anderson Cancer Center (2008) 47(5):361–72. doi: 10.1002/mc.20394
143. Stanton BZ, Peng LF. Small-molecule modulators of the Sonic Hedgehog signaling pathway. Mol BioSyst (2010) 6(1):44–54. doi: 10.1039/B910196A
144. Jain S, Song R, Xie J. Sonidegib: mechanism of action, pharmacology, and clinical utility for advanced basal cell carcinomas. OncoTarg Ther (2017) 10:1645. doi: 10.2147/OTT.S130910
145. Rimkus TK, Carpenter RL, Qasem S, Chan M, Lo H-W. Targeting the sonic hedgehog signaling pathway: review of smoothened and GLI inhibitors. Cancers (2016) 8(2):22. doi: 10.3390/cancers8020022
146. Verma G, Shetake NG, Pandrekar S, Pandey B, Hassan P, Priyadarsini K. Development of surface functionalized hydroxyapatite nanoparticles for enhanced specificity towards tumor cells. Eur J Pharm Sci (2020) 144:105206. doi: 10.1016/j.ejps.2019.105206
147. Xin M, Ji X, De La Cruz LK, Thareja S, Wang B. Strategies to target the Hedgehog signaling pathway for cancer therapy. Med Res Rev (2018) 38(3):870–913. doi: 10.1002/med.21482
148. Hong I-S, Jang G-B, Lee H-Y, Nam J-S. Targeting cancer stem cells by using the nanoparticles. Int J Nanomed (2015) 10(Spec Iss):251. doi: 10.2147/IJN.S88310
149. Taipale J, Chen JK, Cooper MK, Wang B, Mann RK, Milenkovic L, et al. Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature (2000) 406(6799):1005–9. doi: 10.1038/35023008
150. You J, Zhao J, Wen X, Wu C, Huang Q, Guan F, et al. Chemoradiation therapy using cyclopamine-loaded liquid–lipid nanoparticles and lutetium-177-labeled core-crosslinked polymeric micelles. J Controlled Release (2015) 202:40–8. doi: 10.1016/j.jconrel.2015.01.031
151. Hu K, Zhou H, Liu Y, Liu Z, Liu J, Tang J, et al. Hyaluronic acid functional amphipathic and redox-responsive polymer particles for the co-delivery of doxorubicin and cyclopamine to eradicate breast cancer cells and cancer stem cells. Nanoscale (2015) 7(18):8607–18. doi: 10.1039/C5NR01084E
152. Yang R, Mondal G, Wen D, Mahato RI. Combination therapy of paclitaxel and cyclopamine polymer-drug conjugates to treat advanced prostate cancer. Nanomed: Nanotechnol Biol Med (2017) 13(2):391–401. doi: 10.1016/j.nano.2016.07.017
153. Zhao J, Wang H, Hsiao C-H, Chow DS-L, Koay EJ, Kang Y, et al. Simultaneous inhibition of hedgehog signaling and tumor proliferation remodels stroma and enhances pancreatic cancer therapy. Biomaterials (2018) 159:215–28. doi: 10.1016/j.biomaterials.2018.01.014
154. Jiang T, Zhang B, Zhang L, Wu X, Li H, Shen S, et al. Biomimetic nanoparticles delivered hedgehog pathway inhibitor to modify tumour microenvironment and improved chemotherapy for pancreatic carcinoma. Artif Cells Nanomed Biotechnol (2018) 46(sup1):1088–101. doi: 10.1080/21691401.2018.1445093
155. Wang L, Liu X, Zhou Q, Sui M, Lu Z, Zhou Z, et al. Terminating the criminal collaboration in pancreatic cancer: nanoparticle-based synergistic therapy for overcoming fibroblast-induced drug resistance. Biomaterials (2017) 144:105–18. doi: 10.1016/j.biomaterials.2017.08.002
156. Ray P, Confeld M, Borowicz P, Wang T, Mallik S, Quadir M. PEG-b-poly (carbonate)-derived nanocarrier platform with pH-responsive properties for pancreatic cancer combination therapy. Colloids Surfaces B: Biointerf (2019) 174:126–35. doi: 10.1016/j.colsurfb.2018.10.069
157. Verma RK, Yu W, Shrivastava A, Shankar S, Srivastava RK. α-Mangostin-encapsulated PLGA nanoparticles inhibit pancreatic carcinogenesis by targeting cancer stem cells in human, and transgenic (KrasG12D, and KrasG12D/tp53R270H) mice. Sci Rep (2016) 6(1):32743. doi: 10.1038/srep32743
158. Ingallina C, Costa PM, Ghirga F, Klippstein R, Wang JT, Berardozzi S, et al. Polymeric glabrescione B nanocapsules for passive targeting of Hedgehog-dependent tumor therapy in vitro. Nanomedicine (2017) 12(7):711–28. doi: 10.2217/nnm-2016-0388
159. Nayak A, Satapathy SR, Das D, Siddharth S, Tripathi N, Bharatam PV, et al. Nanoquinacrine induced apoptosis in cervical cancer stem cells through the inhibition of hedgehog-GLI1 cascade: Role of GLI-1. Sci Rep (2016) 6(1):20600. doi: 10.1038/srep20600
160. Nayak A, Siddharth S, Das S, Nayak D, Sethy C, Kundu CN. Nanoquinacrine caused apoptosis in oral cancer stem cells by disrupting the interaction between GLI1 and β catenin through activation of GSK3β. Toxicol Appl Pharmacol (2017) 330:53–64. doi: 10.1016/j.taap.2017.07.008
161. Xu Y, Chenna V, Hu C, Sun H-X, Khan M, Bai H, et al. Polymeric nanoparticle-encapsulated hedgehog pathway inhibitor HPI-1 (NanoHHI) inhibits systemic metastases in an orthotopic model of human hepatocellular carcinoma. Clin Cancer Res (2012) 18(5):1291–302. doi: 10.1158/1078-0432.CCR-11-0950
162. Chenna V, Hu C, Pramanik D, Aftab BT, Karikari C, Campbell NR, et al. A polymeric nanoparticle encapsulated small-molecule inhibitor of Hedgehog signaling (NanoHHI) bypasses secondary mutational resistance to Smoothened antagonists. Mol Cancer Ther (2012) 11(1):165–73. doi: 10.1158/1535-7163.MCT-11-0341
163. Nicolini C. Poly-lactic-co-glycolic acid nanoformulation of small moleculeantagonist GANT61 for cancer annihilation by modulating hedgehog pathway. NanoWorld J (2017) 1(2):1–10. doi: 10.17756/nwj.2017-038
164. Oshima G, Guo N, He C, Stack ME, Poon C, Uppal A, et al. In Vivo Delivery and Therapeutic Effects of a MicroRNA on Colorectal Liver Metastases. Mol Ther J Am Soc Gene Ther (2017) 25(7):1588–95. doi: 10.1016/j.ymthe.2017.04.005
165. Yang H, Liu Y, Qiu Y, Ding M, Zhang Y. MiRNA-204-5p and oxaliplatin-loaded silica nanoparticles for enhanced tumor suppression effect in CD44-overexpressed colon adenocarcinoma. Int J Pharmaceut (2019) 566:585–93. doi: 10.1016/j.ijpharm.2019.06.020
166. Zhang X, Song H, Canup BS, Xiao B. Orally delivered targeted nanotherapeutics for the treatment of colorectal cancer. Expert Opin Drug Deliv (2020) 1–10. doi: 10.1080/17425247.2020.1748005
167. Tyagi P, Subramony JA. Nanotherapeutics in oral and parenteral drug delivery: Key learningsand future outlooks as we think small. J Controlled Release (2018) 17(6):1–10. doi: 10.1016/j.jconrel.2018.01.009
168. Vong LB, Yoshitomi T, Matsui H, Nagasaki Y. Development of an oral nanotherapeutics using redox nanoparticles for treatment of colitis-associated colon cancer. Biomaterials (2015) 55:54–63. doi: 10.1016/j.biomaterials.2015.03.037
169. Han W, Xie B, Li Y, Shi L, Wan J, Chen X, et al. Orally deliverable nanotherapeutics for the synergistic treatment of colitis-associated colorectal cancer. Theranostics (2019) 9(24):7458. doi: 10.7150/thno.38081
170. Liu X, Fu Q, Du Y, Yang Y, C Cho W. MicroRNA as regulators of cancer stem cells and chemoresistance in colorectal cancer. Curr Cancer Drug Targets (2016) 16(9):738–54. doi: 10.2174/1568009616666151118114759
Keywords: Hedgehog signaling, miRNAs, nanoformulations, nanoparticle, therapeutics, colorectal cancer
Citation: Javed Z, Javed Iqbal M, Rasheed A, Sadia H, Raza S, Irshad A, Koch W, Kukula-Koch W, Głowniak-Lipa A, Cho WC and Sharifi-Rad J (2021) Regulation of Hedgehog Signaling by miRNAs and Nanoformulations: A Possible Therapeutic Solution for Colorectal Cancer. Front. Oncol. 10:607607. doi: 10.3389/fonc.2020.607607
Received: 17 September 2020; Accepted: 10 November 2020;
Published: 07 January 2021.
Edited by:
Marcelo Calderon, Polymat, SpainReviewed by:
Paul B. Fisher, Virginia Commonwealth University, United StatesMagnolia Laam Pak, University of Massachusetts Medical School, United States
Copyright © 2021 Javed, Javed Iqbal, Rasheed, Sadia, Raza, Irshad, Koch, Kukula-Koch, Głowniak-Lipa, Cho and Sharifi-Rad. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wojciech Koch, a29jaHdAaW50ZXJpYS5wbA==; William C. Cho, Y2hvY3NAaGEub3JnLmhr; Javad Sharifi-Rad, amF2YWQuc2hhcmlmaXJhZEBnbWFpbC5jb20=
 Zeeshan Javed
Zeeshan Javed Muhammad Javed Iqbal2
Muhammad Javed Iqbal2 Shahid Raza
Shahid Raza Wojciech Koch
Wojciech Koch Wirginia Kukula-Koch
Wirginia Kukula-Koch William C. Cho
William C. Cho Javad Sharifi-Rad
Javad Sharifi-Rad