- 1Department of Pathology, Xiangya Hospital, Central South University, Changsha, China
- 2Department of General Surgery, Xiangya Hospital, Central South University, Changsha, China
- 3Department of Anesthesiology, Third Xiangya Hospital of Central South University, Changsha, China
Hepatocellular carcinoma (HCC) is the most common primary liver malignancy with no curative treatments. Plexin D1 (PLXND1) is a cellular receptor whose functions have been explored in several human cancers; however, the critical roles of PLXND1 in HCC have rarely been probed. Therefore, the present study attempted to elucidate the expression pattern, prognostic significance, and potential roles of PLXND1 in HCC. We found that PLXND1 expression in HCC tissues was significantly higher compared with normal liver tissue from Gene Expression Profiling Interactive Analysis (GEPIA) and Integrative Molecular Database of Hepatocellular Carcinoma (HCCDB) databases. This result was further validated by immunohistochemistry staining (IHC) using tissue microarrays, which contained 216 HCC cases collected from our hospital. Additionally, PLXND1 expression showed a significant correlation with several clinical characteristics, including tumor grade and tumor hemorrhage (TH). Moreover, TISIDB and GEPIA databases were used to investigate the roles of PLXND1 in tumor-immune system interactions in HCC. As an immunoinhibitor, transforming growth factor-beta (TGF-β1) displayed the greatest correlations with PLXND1 in HCC. Finally, Kaplan-Meier curves and Cox analysis were conducted to further examine the potential clinical value of PLXND1 in HCC. We described a subclassification of HCC based on PLXND1 and TGF-β1 expression, which could be used to predict clinical outcomes and patient prognosis. Taken together, the results of this study indicate that PLXND1 might be a promising prognostic biomarker and potential therapeutic target in HCC.
Introduction
Hepatocellular carcinoma (HCC) is the most common primary liver malignancy and the fourth leading cause of cancer death worldwide (1–3). More than 70% of patients are diagnosed at an advanced stage with a dismal prognosis. Surgical resection and chemotherapy are the conventional treatment for HCC, but the mortality rate of this cancer remains high (4). In recent years, immunotherapy is increasingly considered as one of the most promising approaches for identifying drug targets for therapeutic interventions (5, 6). The precise estimation of patient prognosis plays an important and irreplaceable role on the treatment decision of HCC patients. Hence, the exploration and identification of novel therapeutic biomarkers and reliable prognostic biomarkers are urgently required.
Plexins are a family of transmembrane proteins that serve as cellular receptors for semaphorins and have a range of critical functions in vascular patterning, axonal guidance, cell proliferation, migration, and immune cell regulation (7–9). Plexin D1 (PLXND1), a newly identified member of the plexin family of molecules, has been reported to be dysregulated in several cancers (7, 10). Emerging data have shown that PLXND1 could function as a tumor suppressor or a classical oncogene. Serving as a tumor suppressor, PLXND1 was reported to promote apoptosis in the absence of semaphorin 3E in breast cancer (10). Conversely, as an oncogene, PLXND1 could promote tumor development and progression by aiding in tumor metastasis and epithelial mesenchymal transition (EMT) (7, 11, 12). In addition, PLXND1 was found to be highly correlated with tumor hemorrhage (TH) in this study and plays a critical role in the tumor immune microenvironment (13, 14). Mumblat Yelena et al. showed that PLXND1 was involved in inducing the collapse of the cytoskeleton of lymphatic endothelial cells (LEC), and impacted tumor lymphangiogenesis and metastasis (13): however, until recently, little was known about the abnormal expression and function of PLXND1 in HCC. Thus, the detailed function of PLXND1 in the tumorigenesis and immune infiltration of HCC warrants further investigation.
Due to high mortality rate, limited treatment effects and poor prognosis of HCC, a novel therapeutic strategy with different mechanisms from conventional treatments are urgently needed (15). Immunotherapy has recently emerged as one of the most promising frontiers of cancer treatment, especially the development of immune checkpoint inhibitors, and has yielded promising therapeutic outcomes in many types of solid tumors, including HCC (16, 17). In spite of these advances, fundamental differences in patient responsiveness to these treatments are inexplicable (18). Hence, comprehensive research on the immune microenvironment of HCC needs to be further investigated.
Prior research on PLXND1 in cancer is limited to cancer types apart from liver cancer. For liver cancer, neither abnormal PLXND1 expression nor its correlation with the clinicopathological parameters of HCC have been reported. In this study, we first reported a higher expression of PLXND1 in HCC and its correlation with clinicopathological features. Additionally, PLXND1 expression was related with tumor-infiltrating lymphocytes (TILs) and immunoinhibitors, and TGF-β1 displayed the greatest correlations with PLXND1 expression in HCC. Then, Kaplan-Meier curves and Cox analysis were conducted to further examine the potential clinical significance of PLXND1 in HCC. Taken together, these results highlight the critical roles of PLXND1 in tumorigenesis, the tumor immune microenvironment, and clinical opportunities for cancer research.
Materials and Methods
Patient Samples
Written informed consent was obtained from all recruited patients prior to the study. All procedures in this study were approved by the Ethics Review Committee of Xiangya Hospital of Central South University (N201303033). The 216 paraffin-embedded archived liver cancer tissues used in this study were histopathologically and clinically diagnosed at the Xiangya Hospital between 2011 and 2013. No patients in this study received chemotherapy before surgery.
Immunohistochemistry
IHC staining was performed on tissue microarrays as previously described (19). The following antibodies were used: anti–PLXND1 (ab28762; Abcam, Cambridge, MA) and anti–TGF-β1 (ab25121; Abcam, Cambridge, MA). Expression of PLXND1 were semiquantitatively classified according to the immunoreactive H-score (HS; range 0–300). Subsequently, stratification scoring was conducted according to H-score as follow: HS < 80, scored as 0; 80 < HS <120, scored as 1; 120 < HS < 200, scored as 2; HS > 200, scored as 3. If the score was equal to or greater than 2, the tumor was considered to have high PLXND1 expression; otherwise, low PLXND1 expression was classified. The expression levels of target proteins in tissue were examined by two independent pathologists blinded to the clinical characteristics of the patients.
Bioinformatics Analysis
Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn/) is an interactive web server for cancer and normal gene expression profiling and interactive analyses (20). In this database, we compared the PLXND1 mRNA expression between tumors and their paired normal tissues. Additionally, GEPIA enabled us to analyze the link between PLXND1 expression and gene markers of different types of tumor infiltrating immune cells.
Integrative Molecular Database of Hepatocellular Carcinoma (HCCDB, http://lifeome.net/database/hccdb) was developed at Tsinghua University and was used to verify the repeatability of identification of HCC gene expression patterns and the annotation of transcripts (21). We use this tool to analyze the expression of PLXND1.
TISIDB (http://cis.hku.hk/TISIDB) is an integrated repository portal which integrates multiple types of data resources in oncoimmunology (22). By using the multidimensional profiling data for 30 cancer types from TCGA, TISIDB pre-calculated the relation between the immune features (e.g., lymphocytes, immunomodulator) and expression, copy number, methylation, and mutation of any gene. Therefore, users can check “lymphocyte,” “immunomodulator,” or “chemokine” tab to evaluate whether the selected gene might regulate immune features across cancers. The heatmap graphic outputs which show Spearman correlations between expression of the selected genes and immune features across human cancers are implemented with the heatmap module of ECharts (https://echarts.apache.org/). Visualization scatter plot for a Spearman correlation between the selected genes and a specific member of immune features in a specific cancer type can be generated through geom_point function of R package ggplot (R script, https://www.r-project.org/). In this database, we analyzed the correlation between PLXND1 expression and TILs and immunoinhibitors in HCC.
Statistical Analysis
The statistical analyses were performed with SPSS 23.0 for Mac (SPSS Inc., Chicago, IL). Quantitative values are presented as the mean ± SEM. Paired t tests and Student’s tests were used for paired and unpaired continuous data, respectively. The χ2 test was used for categorical data. The Mann-Whitney test was applied to analyze correlations between PLXND1 expression and clinicopathologic characteristics in HCC. Survival analysis was performed by Kaplan–Meier analysis and log-rank test. A cox proportional hazards regression model was used for univariate and multivariate survival analyses. Significant changes were indicated with asterisks: *= (P < 0.05); **= (P < 0.01); *** = (P < 0.001).
Results
Elevated Expression of PLXND1 Messenger RNA in Hepatocellular Carcinoma Tissues
For liver cancer, neither abnormal PLXND1 expression nor its association with clinicopathological parameters of HCC has been reported. Here, we investigated the differential expression of PLXND1 using the GEPIA and HCCDB databases. In GEPIA, PLXND1 mRNA expression between tumors and their paired normal tissues across 32 TCGA cancer types were compared. Significantly differential expression was found in 12 cancer types (Figure 1A), and elevated expression of PLXND1 mRNA was found in HCC compared with normal liver tissues (Figures 1A, B). Next, in HCCDB, we validated PLXND1 mRNA expression in the large cohorts of HCC patients. Compared with the adjacent normal liver tissues, significantly upregulated PLXND1 expression in HCC was found in 10 out of 12 clinical cohorts, suggesting that the upregulation of PLXND1 is mainly attributed to the transformation of normal hepatocytes to cancer cells (Figure 1C, Figure S1). Moreover, PLXND1 was expressed in various tissues, but its expression was low in normal liver tissues (Figure S2).
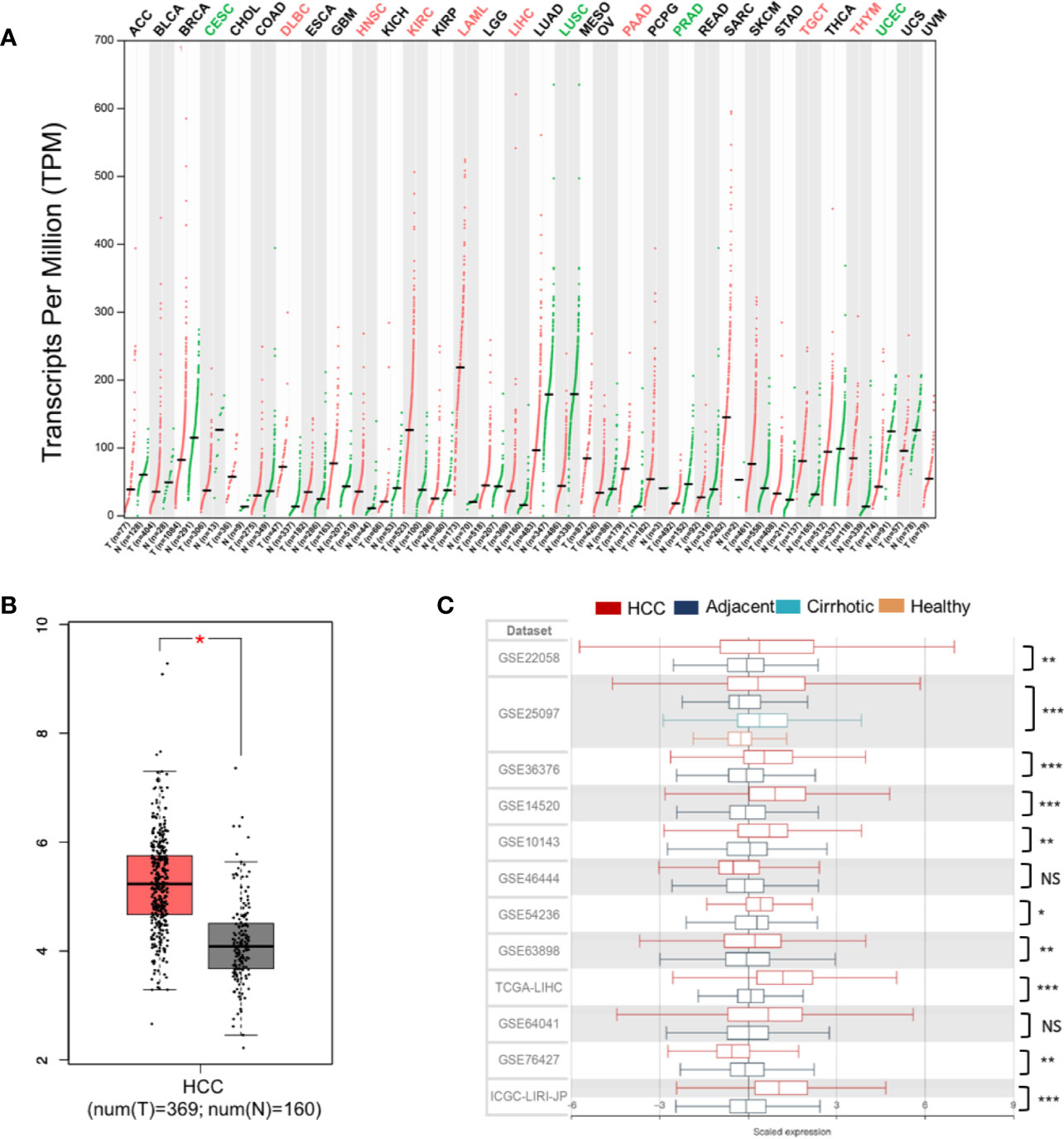
Figure 1 PLXND1 mRNA expression is elevated in hepatocellular carcinoma (HCC). (A) The expression of PLXND1 varied among different tissues and tumors across TCGA cancer types from Gene Expression Profiling Interactive Analysis (GEPIA), and the difference was significant in liver hepatocellular carcinoma (LIHC). (B) PLXND1 expression in HCC tissues was greater than normal tissues from GEPIA. (C) PLXND1 expression in 10 out of 12 clinical cohorts was significantly elevated in HCC tissues compared to the adjacent tissues. Twelve clinical cohorts contain both HCCs and the adjacent normal liver tissues. Only GSE25097 dataset contains tissues (tumor and adjacent non-tumor) from HCCs plus cirrhotic tissues and normal liver tissues. HCC, hepatocellular carcinoma; *P < 0.05, **P < 0.01 and ***P < 0.001; NS, not significant.
PLXND1 Protein Expression and Its Association With Clinicopathological Features
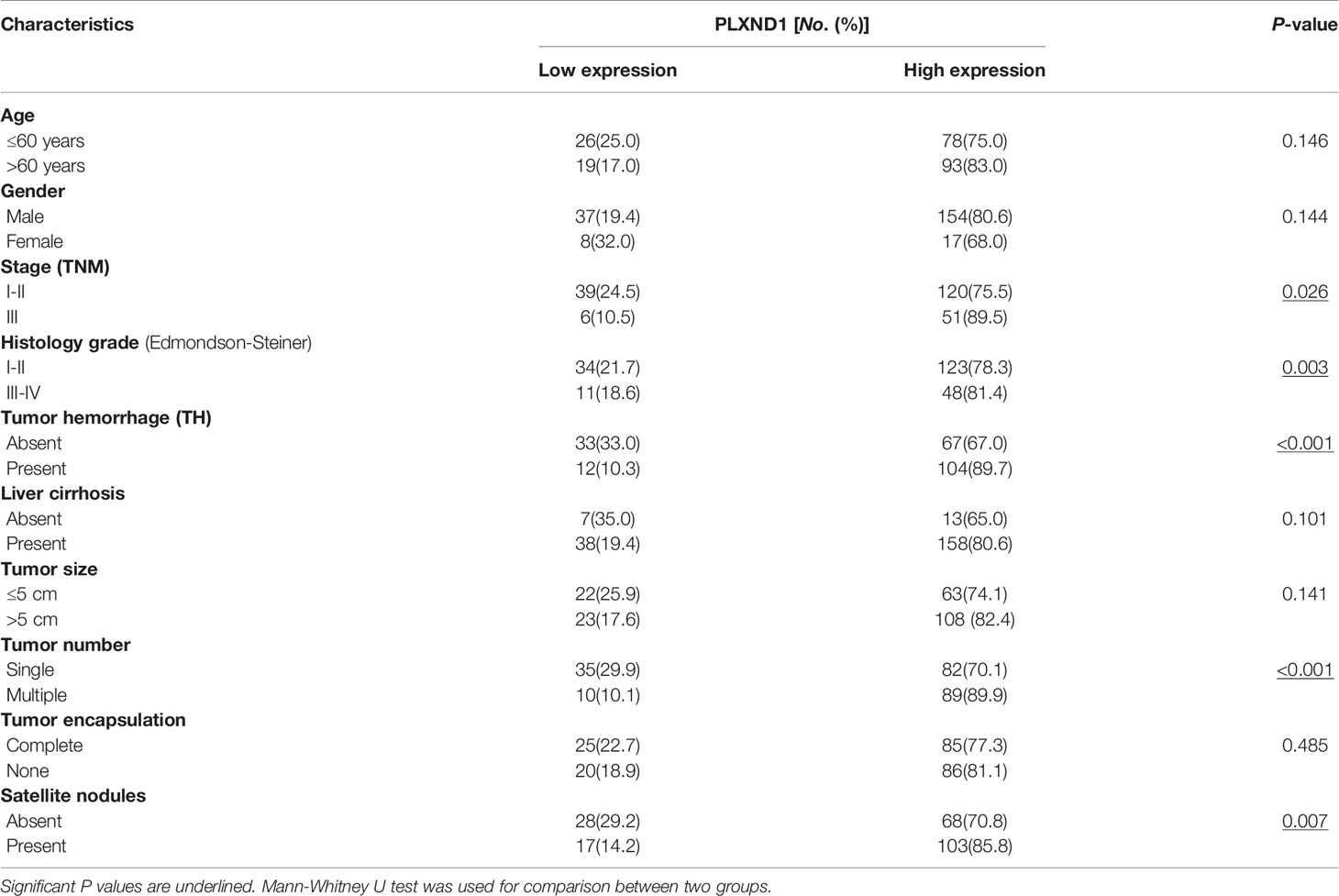
To verify the overexpression of PLXND1 protein in HCC, we explored the protein expression level of PLXND1 in HCC by immunohistochemistry staining (IHC) using tissue microarrays and analyzed its associations with clinicopathological variables. Representative images of PLXND1 IHC staining are shown in Figure 2A. In the clinical samples, significantly higher PLXND1 expression was found in higher-grade HCC samples (Figure 2), which was consistent with the HCC samples from the TCGA database (Figure S3). In addition, PLXND1 immunoreactivity was also found to be significantly associated with stage, histology grade, TH, tumor number, and satellite nodules, suggesting the potential oncogenic activity of PLXND1 in HCC (Table 1).
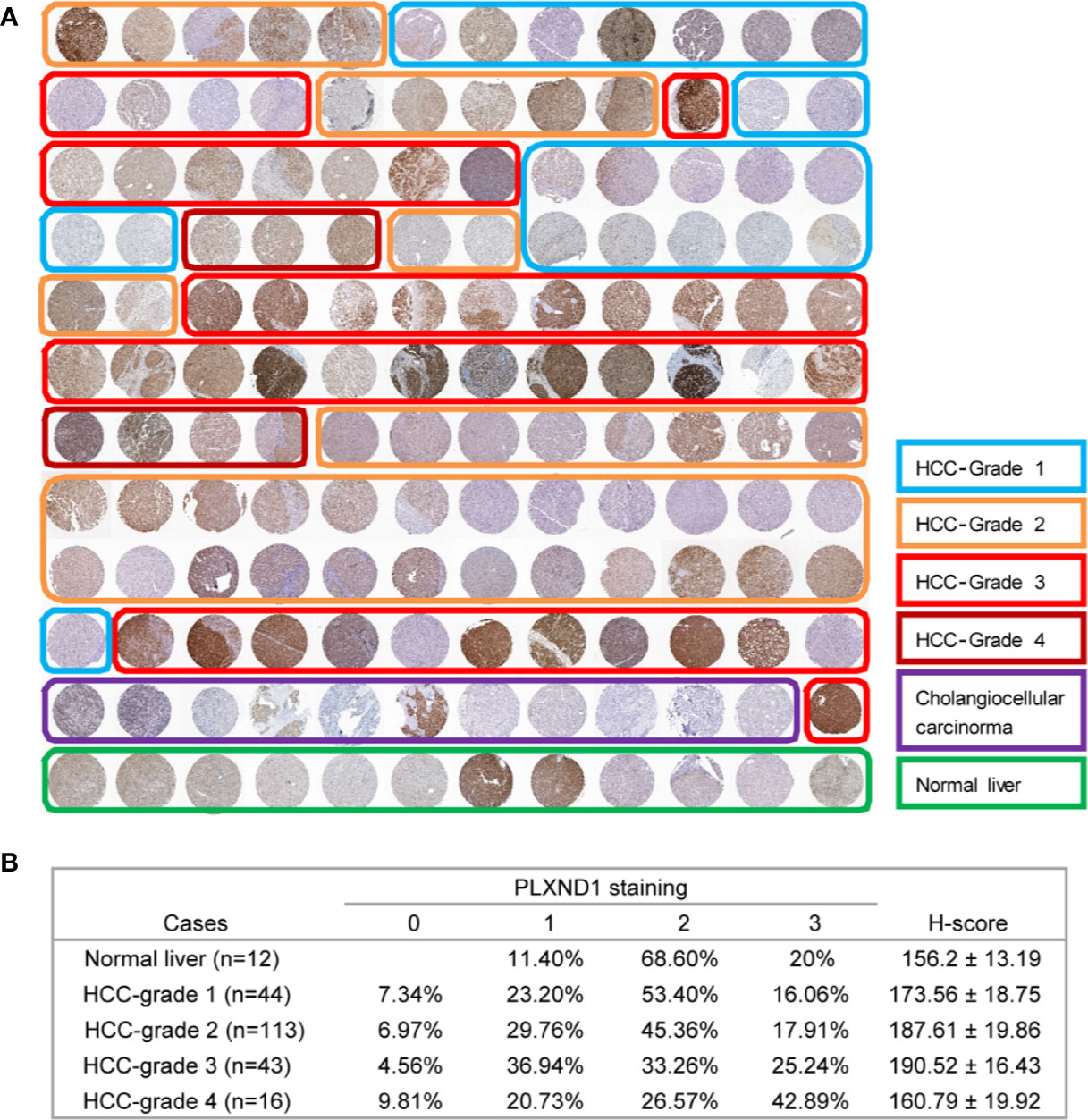
Figure 2 The expression of PLXND1 protein in hepatocellular carcinoma (HCC). (A) Tissue microarrays of HCC tumor tissues as well as normal liver tissues were stained for PLXND1 protein. (B) PLXND1 signal intensities in normal liver tissues versus different grades of HCC tissues were quantified by H-scores, H-score mean ± SD was showed. Blue square, well-differentiated HCC (grade 1); orange square, moderately differentiated HCC (grade 2); red square, poorly differentiated HCC (grade 3); purple square, undifferentiated HCC (grade 4); green square, normal liver. The normal liver tissues were the adjacent normal liver tissues from hepatic hemangioma and distributed in the bottom row of tissue microarray. The HCC tumor tissues were randomly distributed in the upper part of tissue microarray. All these tissues were embedded in the same paraffin block.
The Regulation of Immune Molecules by PLXND1
Based on the tumor hemorrhage (TH) status, we classified a cohort of patients with HCC into two subgroups, TH-negative and TH-positive. HCC tumors with TH showed even higher expression of PLXND1 as determined by the H-scores (Figure 3A). Our previous study reported that TH indicates a worse tumor immunological microenvironment, especially in HCC (23, 24). Thus, we further explored whether PLXND1 is a significant factor that relates to immune infiltration using the TISIDB and GEPIA databases. In TISIDB, we found that PLXND1 expression was correlated with several tumor-infiltrating lymphocytes (TILs) in HCC patients. Lymphocytes showing the greatest correlations included T follicular helper cells (Tfh; Spearman: r = 0.424, P < 2.2e−16), macrophages (Spearman: r = 0.415, P < 2.2e−16), central memory CD4 T cells (Tcm_CD4; Spearman: r = 0.411, P < 2.2e−16), and mast cells (Spearman: r = 0.4, P = 8.62e−17) (Figure 3B). We also investigated the correlation between PLXND1 expression and gene markers of different types of TILs, and the results showed that TGF-β1, as a marker of Tregs, was moderately associated with PLXND1 (Table S1). Moreover, as shown in Figure 3C, PLXND1 expression was associated with several immunoinhibitors in HCC. TGF-β1, a multifunctional cytokine involved in tumor invasion and progression, displayed the greatest correlations with PLXND1 expression in HCC (Spearman: r = 0.62, P < 2.2e−16). Therefore, these findings suggest that PLXND1 might exert a significant effect on immune fingerprinting in HCC.
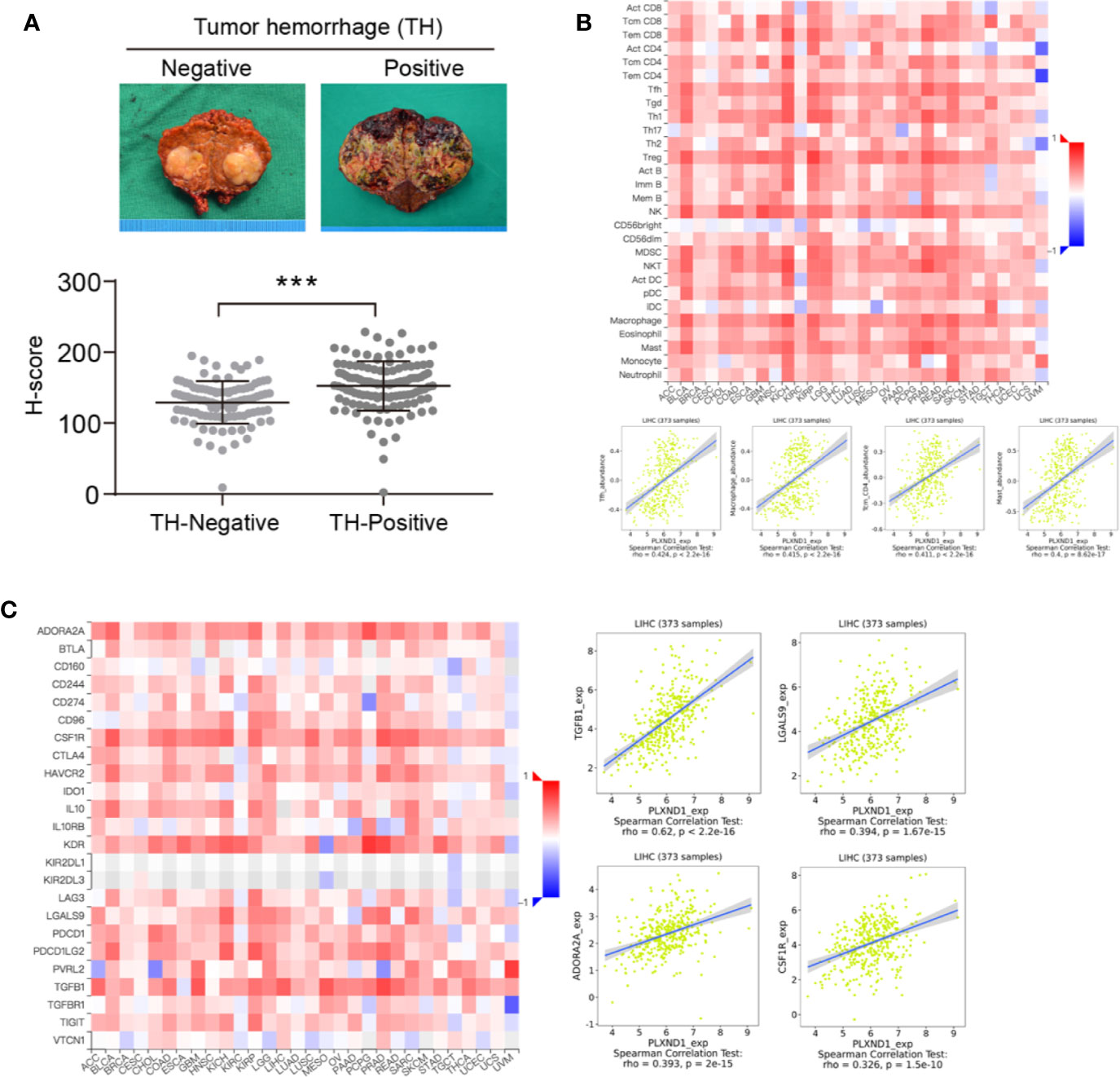
Figure 3 The correlation of PLXND1 expression with tumor hemorrhage (TH), lymphocytes, and immunoinhibitors in hepatocellular carcinoma (HCC). (A) Increased expression of PLXND1 protein was found in TH-positive HCC compared with to TH-negative HCC. Representative gross specimens of HCC with/without TH were also presented. (B) Upper panel: the correlation between PLXND1 expression and tumor-infiltrating lymphocytes (TILs) in pan-cancer; lower panel: in HCC, four TILs including Tfh cells, macrophages, Tcm_CD4 cells, and mast cells showed a kind of medium correlation with PLXND1 expression. (C) Left panel: the correlation between PLXND1 expression and immunoinhibitors in pan-cancer; right panel: in HCC, four immunoinhibitors was positively correlated with PLXND1 expression. TGF-β1 showed a relative strong correlation with PLXND1 expression. ***P < 0.001.
The Prognostic Value of PLXND1 and TGF-β1 in Patients With Hepatocellular Carcinoma
We have found that in HCC, PLXND1 expression was correlated with TGF-β1 (Figure 3C, Table S1), which is a well-known modulator involved in tumor formation, invasion, and metastasis. Here, we further explored whether the coexpression of PLXND1 and TGF-β1 was associated with patient prognosis. We quantified the expression of PLXND1 and TGF-β1 in HCC tissues by IHC, and a varied expression pattern and intensity among the HCC tissues was observed. Representative images of PLXND1 and TGF-β1 IHC staining are shown in Figure 4A. We then grouped patients into 3 categories: group I, double negative (PLXND1neg/TGF-β1neg); group II, PLXND1pos/TGF-β1neg and PLXND1neg/TGF-β1pos; and group III, double positive (PLXND1pos/TGF-β1pos). Survival analysis showed that the overall survival (OS) rates in group III were significantly lower than those in group II and group I (Figure 4B). A consistent result was obtained for the recurrence-free survival (RFS) analysis (Figure 4C). In addition, univariate analysis revealed the following features as prognostic factors related with both OS and RFS: the expression of PLXND1 and TGF-β1, stage, TH, liver cirrhosis, tumor size, tumor number, and satellite nodules (Figures 5A, B). Multivariate analysis further identified that only the expression of PLXND1 and TGF-β1, stage, TH, tumor number, and satellite nodules behaved as independent predictors for both OS and RFS (Figures 5C, D).
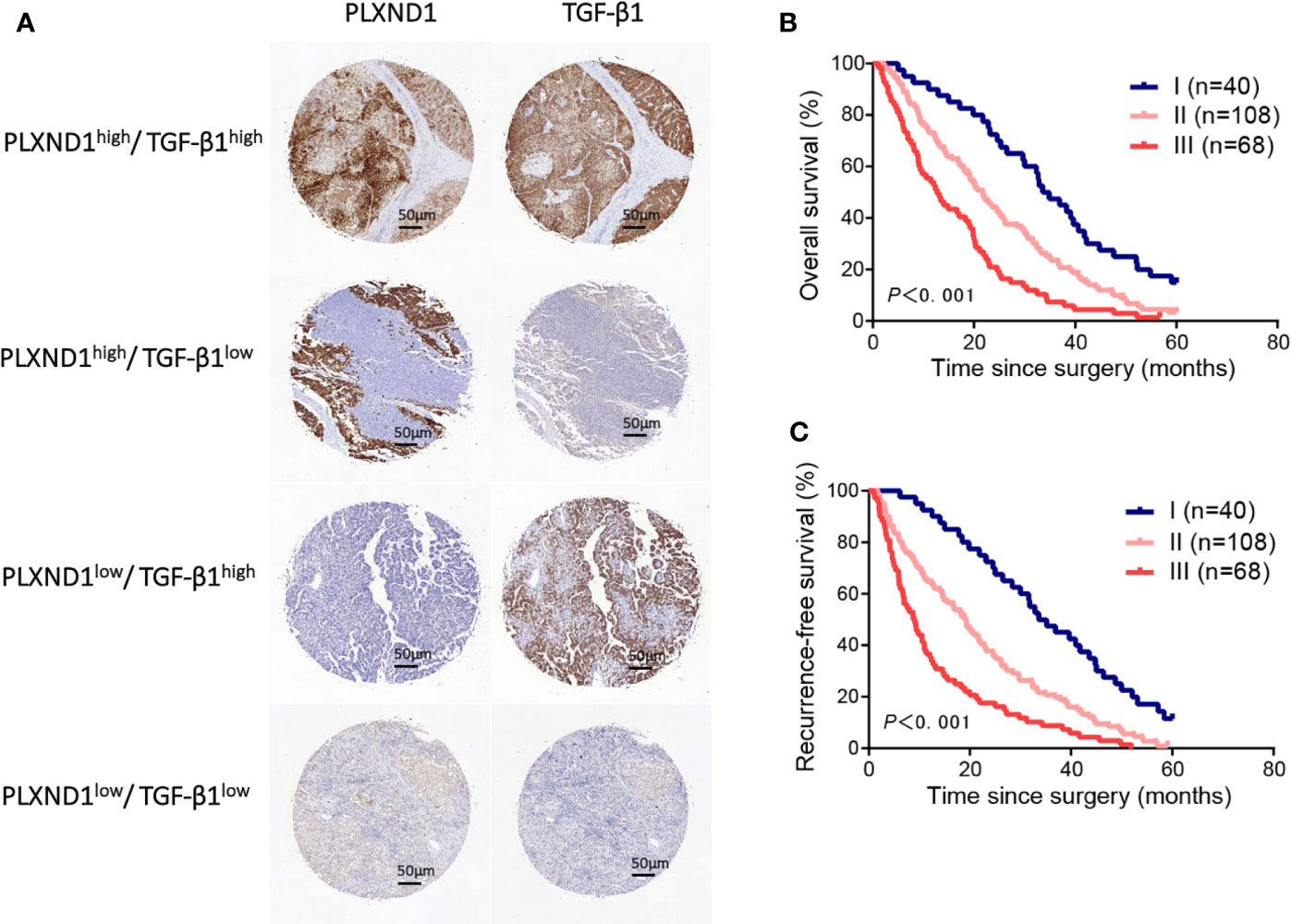
Figure 4 The prognostic value of PLXND1 and TGF-β1 in patients with hepatocellular carcinoma (HCC). Patient tumor tissues were immunostained for PLXND1 or TGF-β1 and classified into three groups: group I, double negative (PLXND1low/TGF-β1low); group II, PLXND1high/TGF-β1low and PLXND1low/TGF-β1high; and group III, double positive (PLXND1high/TGF-β1high). (A) Representative immunohistochemical staining characteristics of PLXND1 and TGF-β1 expression in each patient group. (B, C) Kaplan-Meier analysis of overall survival (OS) and recurrence-free survival (RFS) based on the expression of PLXND1 and TGF-β1 showed that the combination of PLXND1high and TGF-β1high indicated a worse clinical outcome for patients with HCC. High expression: the score was 2 or 3; low expression: the score was 1 or 0.
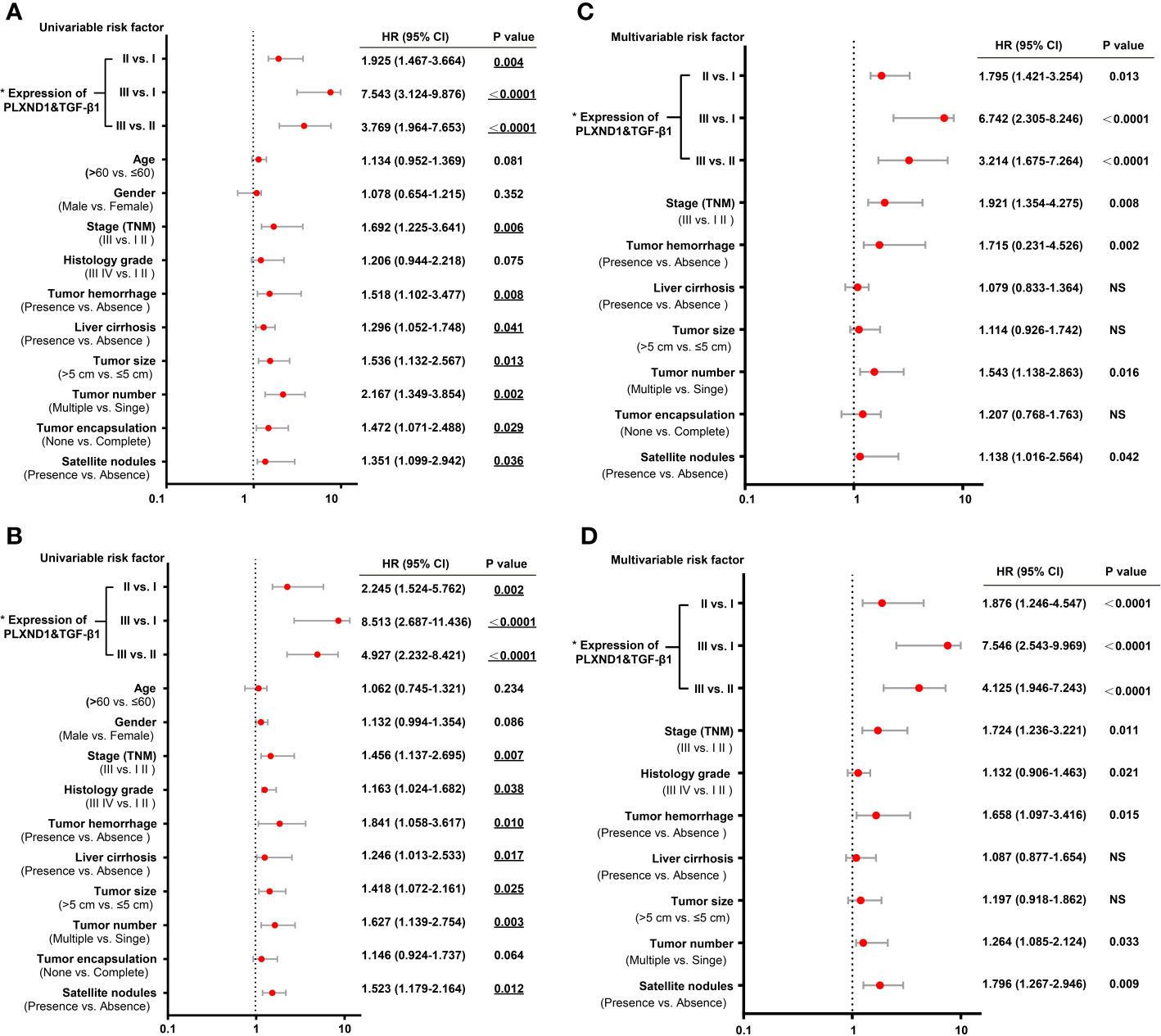
Figure 5 Univariate and multivariate analyses of prognostic factors with overall survival (OS) and recurrence-free survival (RFS) in hepatocellular carcinoma (HCC) patients. (A) The univariate analyses of prognostic factors with patient overall survival (OS) are presented in the forest plot. (B) The univariate analyses of prognostic factors with patient recurrence-free survival (RFS) are presented in the forest plot. (C) The multivariate analyses of prognostic factors with patient OS are presented in the forest plot. (D) The multivariate analyses of prognostic factors with patient RFS are presented in the forest plot. The expression of PLXND1 & TGF-β1 are indicated as follows: I, PLXND1low/TGF-β1low; II, PLXND1high/TGF-β1low and PLXND1low/TGF-β1high; and III, PLXND1high/TGF-β1high.
Discussion
In this study, we aimed to investigate critical and novel biomarkers involved in the development and progression of HCC. PLXND1 belongs to a family of single-pass transmembrane proteins called plexins and serves as a receptor for semaphorins (25–27). Semaphorin-plexin signaling is known to be essential for many cellular aspects of organogenesis, including cell proliferation, invasion, and survival (28, 29). Emerging data have shown that the expression of PLXND1 is often dysregulated in several types of cancer and acts either as a tumor suppressor or an oncogene. Some studies have shown that PLXND1 was overexpressed in tumor cells and promoted tumor development and progression in several cancers, including colon cancers (11), melanoma (30), pancreatic cancer (31), and ovarian cancer (12). Conversely, J. Luchino, M. et al. revealed that PLXND1 signaling could have antitumor activity as well and stimulated cell apoptosis in breast cancer (10): however, the detailed roles of PLXND1 in HCC have rarely been studied. In this study, we first revealed an abnormal expression level, prognostic value, and potential roles of PLXND1 in HCC biology. We found that PLXND1 expression in HCC tissues was significantly higher compared to normal liver tissues using data from the GEPIA and HCCDB databases. This result was further validated by IHC using tissue microarrays, which contained 216 HCC cases collected from our hospital. A high expression of PLXND1 was significantly correlated with several markers of HCC progression, including tumor grade and tumor hemorrhage (TH). These data suggest that PLXNDA may be a potential biomarker and therapeutic target in HCC.
Recently, increasing evidence has shown that the tumor microenvironment and immune infiltration plays a critical role in tumor development and progression (32–34). Our previous study revealed that tumor hemorrhage (TH) indicated a worse tumor immunological microenvironment, especially in HCC (23, 24). In this study, PLXND1 was found to be highly correlated with TH in HCC. These above findings inspired us to further investigate the correlation between PLXND1 and immunity in HCC development. We assessed the correlation between PLXND1 and the immune system using the TISIDB and GEPIA databases. The results showed that PLXND1 had the most significant correlation with tumor-infiltrating lymphocytes including Tfh cells, macrophages, Tcm_CD4 cells, and mast cells. Tfh cells are a subset of CD4+ helper T cells that play a crucial role in humoral immunity and protection against pathogens and in the generation of autoimmunity (32, 35). Macrophages, as the forefront of host defense, also play pivotal roles in tumor immunity (36, 37). Mast cells consistently infiltrate tumors and can be both a driving or an opposite force for tumor progression depending on the tumor microenvironment (38, 39). Additionally, TGF-β1 was revealed to be the most likely immune inhibitor associated with PLXND1 in HCC.
Transforming growth factor betas (TGF-βs) belong to a family of dimeric peptide growth factors and exists in three isoforms, TGF-β1, TGF-β2, and TGF-β3 (40–42). As a multifunctional cytokine, TGF-β1 was demonstrated to be both a tumor inhibitor, wherein it plays growth inhibitory and anti-inflammatory roles in the early stages of cancer; and a tumor promoter, wherein it promotes the development of aggressive growth characteristics and metastatic spread in late stages of cancer (43, 44). TGF-β1 is also involved in the cancer immune microenvironment and promotion of tumor progression (45). In cancers, especially HCC, immune responses and their modulation play critical roles on the outcome of liver malignancies (46–48). TGF-β1-induced epithelial-mesenchymal transition (EMT) was reported to maintain CD4+ CD25+ regulatory T cells (Tregs) and thus promote HCC invasion and metastasis (49). Interestingly, the present study found that TGF-β1 was the most likely immune inhibitor associated with PLXND1 in HCC. Thus, we further analyzed the patient samples and explored the value of the coexpression of PLXND1 and TGF-β1 as a marker for prognosis. We subclassified liver tumors based on PLXND1 and TGF-β1 expression, and the results showed that this classification could be used to predict clinical outcomes and patient prognosis. Survival analysis showed that the patients with double negative expression (PLXND1neg/TGF-β1neg) had the best outcome in HCC: H=however, patients with double positive expression (PLXND1pos/TGF-β1pos) had the worst survival prognosis in HCC. Both univariate and multivariate analyses revealed that the expression of PLXND1 and TGF-β1 could behave as prognostic predictors for both OS and RFS. These findings could be a step toward the development of new biomarkers and therapeutic approaches in HCC.
Nevertheless, several limitations in this study need to be noted. First, the PLXND1 mRNA expression data was extracted from the TCGA database, where ethnicities were mainly white and black, and thus more studies including other ethnic groups should be performed. Additionally, several attractive findings were revealed in this study, but more mechanistic experiments remain to be conducted to further validate the functional value of PLXND1 and TGF-β1 expression in HCC, these aspects are currently under investigation in our lab. Moreover, we mainly focused our analysis on the clinic value of PLXND1 and TGF-β1 for HCC. The analysis of PLXND1 and TGF-β1expression should be further investigated across multiple cancer types.
In conclusion, we first report that PLXND1 is overexpressed in HCC and shows a significant correlation with several clinicopathological features, including tumor stage and TH. Moreover, PLXND1 expression is related with tumor-infiltrating lymphocytes and immunoinhibitors, and TGF-β1 displays the greatest correlations with PLXND1 expression in HCC. Moreover, we further reveal that PLXND1 and TGF-β1 expression are associated with patient prognosis. Taken together, these findings provide a critical understanding of PLXND1 deregulation in HCC and reveal that PLXND1 likely displays an important role in the tumor immune microenvironment. Finally, the results show that PLXNDI is a promising prognostic biomarker in HCC.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Review Committee of Xiangya Hospital of Central South University (N201303033). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
Conception and design: YT and ZW. Writing, review, and/or revision of the manuscript: JL, KH, YT, and ZW. Administrative, technical, or material support: JL, LZ, and DH. All authors contributed to the article and approved the submitted version.
Funding
This study is supported by grants from the Natural Science Foundation of Hunan Province (2018JJ3820, 12JJ3118) and the National Natural Science Foundation of China (81372630).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.604131/full#supplementary-material
References
1. Caruso S, O’Brien DR, Cleary SP, Roberts LR, Zucman-Rossi J. Genetics of HCC: Novel approaches to explore molecular diversity. Hepatology (2020). doi: 10.1002/hep.31394
2. Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med (2018) 24(5):541–50. doi: 10.1038/s41591-018-0014-x
3. Fu Y, Liu S, Zeng S, Shen H. From bench to bed: the tumor immune microenvironment and current immunotherapeutic strategies for hepatocellular carcinoma. J Exp Clin Cancer Res (2019) 38(1):396. doi: 10.1186/s13046-019-1396-4
4. Hu B, Lin JZ, Yang XB, Sang XT. The roles of mutated SWI/SNF complexes in the initiation and development of hepatocellular carcinoma and its regulatory effect on the immune system: A review. Cell Prolif (2020) 53(4):e12791. doi: 10.1111/cpr.12791
5. Qiu MJ, He XX, Bi NR, Wang MM, Xiong ZF, Yang SL. Effects of liver-targeted drugs on expression of immune-related proteins in hepatocellular carcinoma cells. Clin Chim Acta (2018) 485:103–5. doi: 10.1016/j.cca.2018.06.032
6. Duffy MJ, Crown J. Biomarkers for Predicting Response to Immunotherapy with Immune Checkpoint Inhibitors in Cancer Patients. Clin Chem (2019) 65(10):1228–38. doi: 10.1373/clinchem.2019.303644
7. Vivekanadhan S, Mukhopadhyay D. Divergent roles of Plexin D1 in cancer. Biochim Biophys Acta Rev Cancer (2019) 1872(1):103–10. doi: 10.1016/j.bbcan.2019.05.004
8. Kong Y, Janssen BJ, Malinauskas T, Vangoor VR, Coles CH, Kaufmann R, et al. Structural Basis for Plexin Activation and Regulation. Neuron (2016) 91(3):548–60. doi: 10.1016/j.neuron.2016.06.018
9. Gay CM, Zygmunt T, Torres-Vazquez J. Diverse functions for the semaphorin receptor PlexinD1 in development and disease. Dev Biol (2011) 349(1):1–19. doi: 10.1016/j.ydbio.2010.09.008
10. Luchino J, Hocine M, Amoureux MC, Gibert B, Bernet A, Royet A, et al. Semaphorin 3E suppresses tumor cell death triggered by the plexin D1 dependence receptor in metastatic breast cancers. Cancer Cell (2013) 24(5):673–85. doi: 10.1016/j.ccr.2013.09.010
11. Casazza A, Finisguerra V, Capparuccia L, Camperi A, Swiercz JM, Rizzolio S, et al. Sema3E-Plexin D1 signaling drives human cancer cell invasiveness and metastatic spreading in mice. J Clin Invest (2010) 120(8):2684–98. doi: 10.1172/JCI42118
12. Tseng CH, Murray KD, Jou MF, Hsu SM, Cheng HJ, Huang PH. Sema3E/plexin-D1 mediated epithelial-to-mesenchymal transition in ovarian endometrioid cancer. PLoS One (2011) 6(4):e19396. doi: 10.1371/journal.pone.0019396
13. Mumblat Y, Kessler O, Ilan N, Neufeld G. Full-Length Semaphorin-3C Is an Inhibitor of Tumor Lymphangiogenesis and Metastasis. Cancer Res (2015) 75(11):2177–86. doi: 10.1158/0008-5472.CAN-14-2464
14. Holl EK, O’Connor BP, Holl TM, Roney KE, Zimmermann AG, Jha S, et al. Plexin-D1 is a novel regulator of germinal centers and humoral immune responses. J Immunol (2011) 186(10):5603–11. doi: 10.4049/jimmunol.1003464
15. Lu C, Rong D, Zhang B, Zheng W, Wang X, Chen Z, et al. Current perspectives on the immunosuppressive tumor microenvironment in hepatocellular carcinoma: challenges and opportunities. Mol Cancer (2019) 18(1):130. doi: 10.1186/s12943-019-1047-6
16. Greten TF, Wang XW, Korangy F. Current concepts of immune based treatments for patients with HCC: from basic science to novel treatment approaches. Gut (2015) 64(5):842–8. doi: 10.1136/gutjnl-2014-307990
17. Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med (2010) 363(8):711–23. doi: 10.1056/NEJMoa1003466
18. Lim CJ, Lee YH, Pan L, Lai L, Chua C, Wasser M, et al. Multidimensional analyses reveal distinct immune microenvironment in hepatitis B virus-related hepatocellular carcinoma. Gut (2019) 68(5):916–27. doi: 10.1136/gutjnl-2018-316510
19. Li J, Alvero AB, Nuti S, Tedja R, Roberts CM, Pitruzzello M, et al. CBX7 binds the E-box to inhibit TWIST-1 function and inhibit tumorigenicity and metastatic potential. Oncogene (2020) 39(20):3965–79. doi: 10.1038/s41388-020-1269-5
20. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res (2017) 45(W1):W98–102. doi: 10.1093/nar/gkx247
21. Lian Q, Wang S, Zhang G, Wang D, Luo G, Tang J, et al. HCCDB: A Database of Hepatocellular Carcinoma Expression Atlas. Genomics Proteomics Bioinf (2018) 16(4):269–75. doi: 10.1016/j.gpb.2018.07.003
22. Ru B, Wong CN, Tong Y, Zhong JY, Zhong SSW, Wu WC, et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics (2019) 35(20):4200–2. doi: 10.1093/bioinformatics/btz210
23. Wang D, Wang ZM, Zhang S, Wu HJ, Tao YM. Canopy Homolog 2 Expression Predicts Poor Prognosis in Hepatocellular Carcinoma with Tumor Hemorrhage. Cell Physiol Biochem (2018) 50(6):2017–28. doi: 10.1159/000495048
24. Hu K, Wang ZM, Li JN, Zhang S, Xiao ZF, Tao YM. CLEC1B Expression and PD-L1 Expression Predict Clinical Outcome in Hepatocellular Carcinoma with Tumor Hemorrhage. Transl Oncol (2018) 11(2):552–8. doi: 10.1016/j.tranon.2018.02.010
25. Oh WJ, Gu C. The role and mechanism-of-action of Sema3E and Plexin-D1 in vascular and neural development. Semin Cell Dev Biol (2013) 24(3):156–62. doi: 10.1016/j.semcdb.2012.12.001
26. Mehta V, Pang KL, Rozbesky D, Nather K, Keen A, Lachowski D, et al. The guidance receptor plexin D1 is a mechanosensor in endothelial cells. Nature (2020) 578(7794):290–5. doi: 10.1038/s41586-020-1979-4
27. Minchin JE, Dahlman I, Harvey CJ, Mejhert N, Singh MK, Epstein JA, et al. Plexin D1 determines body fat distribution by regulating the type V collagen microenvironment in visceral adipose tissue. Proc Natl Acad Sci USA (2015) 112(14):4363–8. doi: 10.1073/pnas.1416412112
28. Roodink I, Kats G, van Kempen L, Grunberg M, Maass C, Verrijp K, et al. Semaphorin 3E expression correlates inversely with Plexin D1 during tumor progression. Am J Pathol (2008) 173(6):1873–81. doi: 10.2353/ajpath.2008.080136
29. Roodink I, Raats J, van der Zwaag B, Verrijp K, Kusters B, van Bokhoven H, et al. Plexin D1 expression is induced on tumor vasculature and tumor cells: a novel target for diagnosis and therapy? Cancer Res (2005) 65(18):8317–23. doi: 10.1158/0008-5472.CAN-04-4366
30. Roodink I, Verrijp K, Raats J, Leenders WP. Plexin D1 is ubiquitously expressed on tumor vessels and tumor cells in solid malignancies. BMC Cancer (2009) 9:297. doi: 10.1186/1471-2407-9-297
31. Huang HY, Cheng YY, Liao WC, Tien YW, Yang CH, Hsu SM, et al. SOX4 transcriptionally regulates multiple SEMA3/plexin family members and promotes tumor growth in pancreatic cancer. PLoS One (2012) 7(12):e48637. doi: 10.1371/journal.pone.0048637
32. Yan Y, Liu W, Liu M, Gong Z, Xu Z. Immune Cell Infiltration Influences Long-Term Survivorship of Patients with SCLC. J Thorac Oncol (2019) 14(10):e241. doi: 10.1016/j.jtho.2019.06.017
33. Sun J, Zhang Z, Bao S, Yan C, Hou P, Wu N, et al. Identification of tumor immune infiltration-associated lncRNAs for improving prognosis and immunotherapy response of patients with non-small cell lung cancer. J Immunother Cancer (2020) 8(1):e000110. doi: 10.1136/jitc-2019-000110
34. Liu R, Hu R, Zeng Y, Zhang W, Zhou HH. Tumour immune cell infiltration and survival after platinum-based chemotherapy in high-grade serous ovarian cancer subtypes: A gene expression-based computational study. EBioMedicine (2020) 51:102602. doi: 10.1016/j.ebiom.2019.102602
35. Vinuesa CG, Linterman MA, Yu D, MacLennan IC. Follicular Helper T Cells. Annu Rev Immunol (2016) 34:335–68. doi: 10.1146/annurev-immunol-041015-055605
36. Kawakami T, Koike A, Amano F. Sodium bicarbonate regulates nitric oxide production in mouse macrophage cell lines stimulated with lipopolysaccharide and interferon gamma. Nitric Oxide (2018) 79:45–50. doi: 10.1016/j.niox.2018.07.008
37. Xiao J, Xie R, Li Q, Chen W, Zhang Y. Generation and characterization of bovine bone marrow-derived macrophage cell line. Cell Biol Int (2016) 40(5):603–8. doi: 10.1002/cbin.10599
38. Lv Y, Zhao Y, Wang X, Chen N, Mao F, Teng Y, et al. Increased intratumoral mast cells foster immune suppression and gastric cancer progression through TNF-alpha-PD-L1 pathway. J Immunother Cancer (2019) 7(1):54. doi: 10.1186/s40425-019-0530-3
39. Aponte-Lopez A, Fuentes-Panana EM, Cortes-Munoz D, Munoz-Cruz S. Mast Cell, the Neglected Member of the Tumor Microenvironment: Role in Breast Cancer. J Immunol Res (2018) 2018:2584243. doi: 10.1155/2018/2584243
40. Batlle E, Massague J. Transforming Growth Factor-beta Signaling in Immunity and Cancer. Immunity (2019) 50(4):924–40. doi: 10.1016/j.immuni.2019.03.024
41. Meng XM, Nikolic-Paterson DJ, Lan HY. TGF-beta: the master regulator of fibrosis. Nat Rev Nephrol (2016) 12(6):325–38. doi: 10.1038/nrneph.2016.48
42. Achyut BR, Yang L. Transforming growth factor-beta in the gastrointestinal and hepatic tumor microenvironment. Gastroenterology (2011) 141(4):1167–78. doi: 10.1053/j.gastro.2011.07.048
43. Zhu H, Luo H, Shen Z, Hu X, Sun L, Zhu X. Transforming growth factor-beta1 in carcinogenesis, progression, and therapy in cervical cancer. Tumour Biol (2016) 37(6):7075–83. doi: 10.1007/s13277-016-5028-8
44. Tang J, Gifford CC, Samarakoon R, Higgins PJ. Deregulation of Negative Controls on TGF-beta1 Signaling in Tumor Progression. Cancers (Basel) (2018) 10(6). doi: 10.3390/cancers10060159
45. Zarzynska JM. Two faces of TGF-beta1 in breast cancer. Mediators Inflamm (2014) 2014:141747. doi: 10.1155/2014/141747
46. Eggert T, Wolter K, Ji J, Ma C, Yevsa T, Klotz S, et al. Distinct Functions of Senescence-Associated Immune Responses in Liver Tumor Surveillance and Tumor Progression. Cancer Cell (2016) 30(4):533–47. doi: 10.1016/j.ccell.2016.09.003
47. Qasim W, Brunetto M, Gehring AJ, Xue SA, Schurich A, Khakpoor A, et al. Immunotherapy of HCC metastases with autologous T cell receptor redirected T cells, targeting HBsAg in a liver transplant patient. J Hepatol (2015) 62(2):486–91. doi: 10.1016/j.jhep.2014.10.001
48. Inada Y, Mizukoshi E, Seike T, Tamai T, Iida N, Kitahara M, et al. Characteristics of Immune Response to Tumor-Associated Antigens and Immune Cell Profile in Patients With Hepatocellular Carcinoma. Hepatology (2019) 69(2):653–65. doi: 10.1002/hep.30212
Keywords: plexin D1, hepatocellular carcinoma, tumor immune microenvironment, prognosis, transforming growth factor-beta
Citation: Li J, Hu K, He D, Zhou L, Wang Z and Tao Y (2021) Prognostic Value of PLXND1 and TGF-β1 Coexpression and Its Correlation With Immune Infiltrates in Hepatocellular Carcinoma. Front. Oncol. 10:604131. doi: 10.3389/fonc.2020.604131
Received: 08 September 2020; Accepted: 23 November 2020;
Published: 08 January 2021.
Edited by:
Divya P. Kumar, JSS Academy of Higher Education and Research, IndiaReviewed by:
Kelly Yi Ping Liu, University of British Columbia, CanadaPrasanna K. Santhekadur, JSS Academy of Higher Education and Research, India
Copyright © 2021 Li, Hu, He, Zhou, Wang and Tao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yiming Tao, eWltaW5ndGFvQGNzdS5lZHUuY24=
†These authors have contributed equally to this work
 Juanni Li
Juanni Li Kuan Hu2†
Kuan Hu2†