
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 23 October 2020
Sec. Hematologic Malignancies
Volume 10 - 2020 | https://doi.org/10.3389/fonc.2020.586567
This article is part of the Research Topic Insights Into Acute Leukemia Research: Genetic Predisposition and Mechanisms of Relapse View all 9 articles
 Satoshi Nishiwaki1*
Satoshi Nishiwaki1* Jeong Hui Kim2
Jeong Hui Kim2 Masafumi Ito3
Masafumi Ito3 Matsuyoshi Maeda4
Matsuyoshi Maeda4 Yusuke Okuno5
Yusuke Okuno5 Daisuke Koyama6
Daisuke Koyama6 Yukiyasu Ozawa7
Yukiyasu Ozawa7 Masaharu Gunji3
Masaharu Gunji3 Masahide Osaki7
Masahide Osaki7 Kunio Kitamura8
Kunio Kitamura8 Yoko Ushijima2
Yoko Ushijima2 Yuichi Ishikawa2
Yuichi Ishikawa2 Koichi Miyamura7
Koichi Miyamura7 Isamu Sugiura6
Isamu Sugiura6 Hitoshi Kiyoi2
Hitoshi Kiyoi2Background: Recently, various blood cell lineages expressing the BCR-ABL fusion gene in Philadelphia chromosome (Ph)-positive acute lymphoblastic leukemia (ALL) have been reported. However, the biological and clinical significance of these BCR-ABL lineages has not been established; therefore, we aimed to clarify the impacts of these different BCR-ABL-expressing lineages.
Patients: Multi-lineage BCR-ABL expression (multi-Ph) was defined as BCR-ABL expression outside of the B-lineage compartment, as determined by fluorescence in situ hybridization (FISH) in peripheral blood neutrophils and bone marrow clots, and flow cytometry-sorted polymerase chain reaction (PCR). We analyzed IKZF1 deletion patterns by PCR, examined gene expression profiles using RNA sequencing, and compared treatment outcomes across different BCR-ABL-expressing lineages.
Results: Among the 21 multi-Ph patients in our 59-patient cohort (36%), BCR-ABL expression was detected at the multipotential progenitor level. However, no IKZF1 deletion patterns or gene expression profiles were identified that were specific for multi-Ph. However, multi-Ph patients were found to have better survival rates than patients with uni-lineage BCR-ABL expression [event-free survival (EFS): 74 vs. 33%, P = 0.01; overall survival (OS): 79 vs. 44% at 4 years, P = 0.01]. In multivariate analyses, multi-Ph was identified as a good prognostic factor for both EFS and OS.
Conclusion: We confirmed that more than one-third of Ph+ALL patients could be classified as mutli-Ph. Although no specific molecular characteristics were identified for multi-Ph, this phenotype was associated with better treatment outcomes.
Cells that express BCR-ABL outside of the B-lineage compartment have been reported in a proportion of patients with Philadelphia chromosome-positive (Ph+) B-precursor acute lymphoblastic leukemia (ALL) (1–4). In most cases, the fusion gene BCR-ABL, which is the genetic abnormality associated with Ph+ALL, is expressed only in B-lymphocytes; however, in some cases, BCR-ABL was also expressed in the myeloid lineage, suggesting that these cells originated from a separate differentiation stage than B-lineage cells.
The expression of BCR-ABL by non-lymphocytic lineages is a relatively recent finding, and the biological and clinical significance of this finding has yet to be clarified. The differentiation stage during which the genetic abnormality occurs must be identified, and whether leukemic cells originating from various differentiation stages are molecularly heterogeneous must be determined. Furthermore, whether similar genetic abnormalities with different origins have varying influences on patient prognosis must also be determined. The occurrence of genetic abnormalities in cells other than leukemic cells may also affect the detection of minimal residual disease (MRD) (3, 4), which is currently crucial for determining an optimal treatment strategy for Ph+ALL (4–7). In this study, we confirmed two types of adult Ph+ALL that derive from different leukemic origins. In addition, we analyzed the genetic characteristics and therapeutic responses of Ph+ALL associated with genetic abnormalities in cells other than leukemic blast cells.
Fifty-nine adult Ph+ALL patients were deemed eligible for this retrospective study. Patients with de novo Ph+ALL were included if peripheral blood or bone marrow samples were available. No patients had any history of chronic myeloid leukemia (CML). Ph+ALL was confirmed through the detection of the Ph chromosome by fluorescence in situ hybridization (FISH) analysis and the positive detection of BCR-ABL fusion transcript expression by reverse-transcription-polymerase chain reaction (RT-PCR) analysis. The immunophenotyping of leukemic blast cells was performed using flow cytometry or immunohistochemistry, at individual hospitals (7). The copy numbers of BCR-ABL transcripts identified in each sample were normalized against the expression levels of GAPDH. The threshold for quantification was 50 copies/µg RNA, which corresponded to a minimal sensitivity of 10−5 (8). In the present study, the inability to detect BCR-ABL by RT-PCR was defined as complete molecular remission (CMR). All patients were treated with tyrosine kinase inhibitor (TKI) ± conventional chemotherapy. No criteria were established for TKI selection or the performance of allogeneic hematopoietic cell transplantation (allo-HCT). Treatment strategies depended on each physician’s judgments and the treatment policy of each hospital. Representative treatments were based on studies performed by the Japan Adult Leukemia Study Group for Ph+ALL [Ph+ALL202 (9), Ph+ALL208 (10), or Ph+ALL213 (11)], and written informed consent was obtained from each patient upon treatment and sample collection. Each hospital’s institutional review board approved the study.
To assess the expression of the BCR-ABL fusion gene outside of leukemic blast cells, we performed FISH analyses on two different specimens: peripheral blood (PB) neutrophils (PN-FISH) (12) and bone marrow clot sections (BM-FISH) (13). A Vysis LSI BCR/ABL ES Dual-Color Translocation Probe (Abbott Laboratories, Abbott Park, IL, USA) was hybridized with PB and BM samples (2, 14). The denaturation of the chromosome/probe, hybridization, and washing were performed according to the manufacturer’s instructions. Multi-Ph was defined as the detection of BCR-ABL fusion gene signals in segmented neutrophils, including of PB mononuclear cells and PB neutrophils. Similarly, the detection of BCR-ABL fusion gene signals in myeloperoxidase-positive cells and polynuclear megakaryocytes in 10% formalin-fixed paraffin-embedded bone marrow clot sections by BM-FISH was defined as multi-Ph.
The expression of BCR-ABL was examined by dividing bone marrow/peripheral blood cells into the following fractions, as defined in previous studies (1, 15): CD34+CD38–CD19–CD3– [hematopoietic stem cells (HSCs) and multipotent progenitor (MPP) cells], CD34+CD38+CD19–CD3– [myeloid and lymphoid progenitors (Pro)], CD34+CD19+CD20–CD3– [leukemia cells without CD20 coexpression; leukemia associated immunophenotype (LAIP) 20–], CD34+CD19+CD20+CD3– (leukemia cells with CD20 coexpression; LAIP 20+), CD34–CD19+CD20+ (mature B-cells), CD34–CD19–CD20–CD3+ (mature T-cells), CD34+CD19–CD13/33+CD10–CD16– (early myeloid compartment), CD34–CD19–CD13/33+CD10–CD16– (late myeloid compartment), and CD34–CD19–CD13/33+CD10–CD16+ (mature myeloid compartment). Fluorescence-activated cell sorting was performed using a FACS Aria II flow cytometer (BD Biosciences, San Jose, CA, USA), applying 4’,6-diamidino-2-phenylindole (DAPI) for live/dead cell discrimination. All antibodies were purchased from BD, Beckman Coulter (Indianapolis, IN, USA) or Biolegend (San Diego, CA, USA). Fluorochrome-conjugated antibodies included CD19 (J3-119), CD20 (2H7), CD34 (8G12), CD38 (HB7), CD10 (HI10a), CD3 (2H7), CD16 (3G8), CD13 (SJ1D1), and CD33 (P67-6) antibodies.
Total RNA was extracted from sorted cells, using an RNeasy MiniKit or a QIAamp RNA Blood Mini Kit (QIAGEN, Hilden, Germany), and reverse transcribed, using the SuperScript II Reverse Transcriptase Kit (Thermo Fisher Scientific, Waltham, MA, USA), according to the manufacturer’s instructions. BCR-ABL transcript expression levels were quantitated based on a real-time fluorescence detection method using an ABI Prism7300 Sequence Detection System and Taqman® Gene Expression Assay probe (Applied Biosystems, Foster City, CA, USA). GAPDH served as a control for cDNA quality. To detect IKZF1 isoforms, IKZF1 transcripts were amplified from cDNA, as previously described (16). BCR-ABL fusion transcripts that were detected outside of the lymphoid compartment, using a flow cytometry-sorted PCR method (FCM-PCR), were classified as multi-Ph.
All cryopreserved samples from which high-quality RNA could be extracted were submitted to RNA sequencing. We prepared non-directional sequencing libraries, using a NEBNext Ultra II RNA Prep Kit for Illumina and a NEBNext Poly(A) mRNA Magnetic Isolation Module (New England Biolabs, Ipswich, MA, USA), according to the manufacturer’s instructions. We ran the prepared libraries using a HiSeq 2500 next-generation sequencing platform (Illumina, San Diego, CA, USA) to obtain an average of 100 million reads per sample. The obtained reads were aligned against the hg19 reference genome, using TopHat2 (17), and fragments per kilobase of exon per million mapped reads (FPKM) values were calculated using Cufflinks (18). For clustering analysis, we used HTSeq (19) for raw read counting and DESeq2 (20) for variance stabilizing transformation. Cluster 3.0 (21) was used to filter genes with default parameters. Finally, we used pyclust (22) to perform average linkage hierarchical clustering using Pearson’s correlation coefficient. Bootstrapping was performed for 1,000 permutations to calculate the bootstrap probability and to obtain approximately unbiased values.
Primarily, we analyzed event-free survival (EFS), from the date of diagnosis to the date of any event, including death by any cause, failure to achieve remission after induction therapy, relapse in any site, and second malignancy (23–25); for patients without an event, the date of the last visit was used. EFS probabilities were estimated using the Kaplan–Meier method, and P-values were calculated using a log-rank test (26, 27). We also analyzed overall survival (OS), from the date of diagnosis to the date of death or the date of the last visit. Univariate and multivariate analyses were performed using a Cox proportional hazard regression model (28). A backward stepwise procedure was used to develop a final model, based on a P-value threshold of 0.05. The covariates considered were patient sex (male vs. female), patient age (<65 y vs. ≥65 y), white blood cell count at diagnosis (< 30,000/µl vs. ≥ 30,000/µl), BCR-ABL transcript [(e1a2 vs. b2a2 or b3a2 (b2a2/b3a2)], additional cytogenetic abnormality (no vs. yes), type of TKI (imatinib vs. dasatinib), allo-HCT (no vs. yes) and BCR-ABL lineage (uni-lineage vs. multi-lineage). A significance level of P <0.05 was used for all analyses.
In this study, patients were classified as multi-Ph through at least one of three approaches (PN-FISH, BM-FISH, and FCM-PCR). All other patients were defined as uni-lineage Ph leukemia (uni-Ph). PN-FISH was performed for 19 patients (Figure 1A), BM-FISH was performed for 49 patients (Figure 1B), and FCM-PCR was performed for six patients. A total of 21 patients (36%) were identified as multi-Ph; a discrepancy between detection methods was observed in only one patient (PN-FISH: uni-Ph, BM-FISH: multi-Ph). In two patients, BCR-ABL signals were also detected in megakaryocytes (Figure 1B-2). Four multi-Ph individuals expressed e1a2, whereas the remaining 17 individuals with multi-Ph expressed b2a2/b3a2. The frequency of multi-Ph was significantly higher among patients expressing b2a2/b3a2 than those expressing e1a2 (81 vs. 11%, P < 0.001, Table 1).
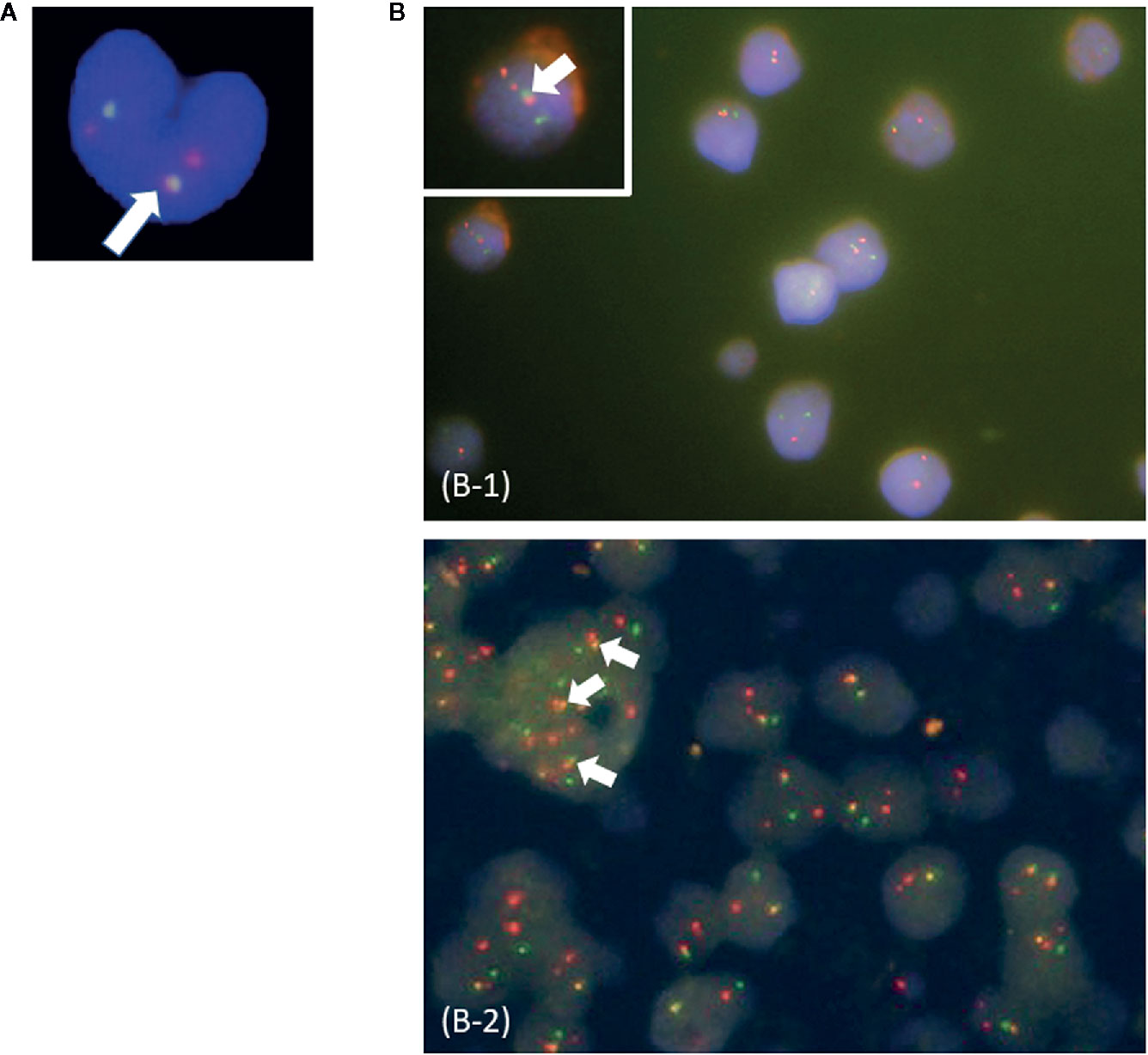
Figure 1 Fluorescence in situ hybridization (FISH) for the BCR-ABL fusion gene. (A) Peripheral blood neutrophil-FISH (PN-FISH); (B) FISH in bone marrow clot sections (BM-FISH). (B-1) shows MPO-positive myeloid cells. (B-2) shows a megakaryocyte. White arrows indicate BCR-ABL fusion signals. Red signals are from ABL probes, and green signals are from BCR probes.
FCM-PCR was evaluated in six patients (4 with e1a2 and 2 with b2a2/b3a2). BCR-ABL fusion transcripts were detected in the LAIP 20– and LAIP 20+ compartments, as expected, but the HSCs/MPP and Pro compartments were also BCR-ABL-positive in four patients (2 with e1a2 and 2 with b2a2/b3a2).
IKZF1 deletion was detected in five of six evaluated patients (4 with e1a2 and 1 with b2a2/b3a2). No deletion patterns were found associated with multi-Ph (Figure 2). In three patients, IKZF1 deletion was detected in the HSCs/MPP and Pro compartments, as well as in the LAIP 20– and LAIP 20+ compartments, which was consistent with the findings for BCR-ABL-positive compartments.
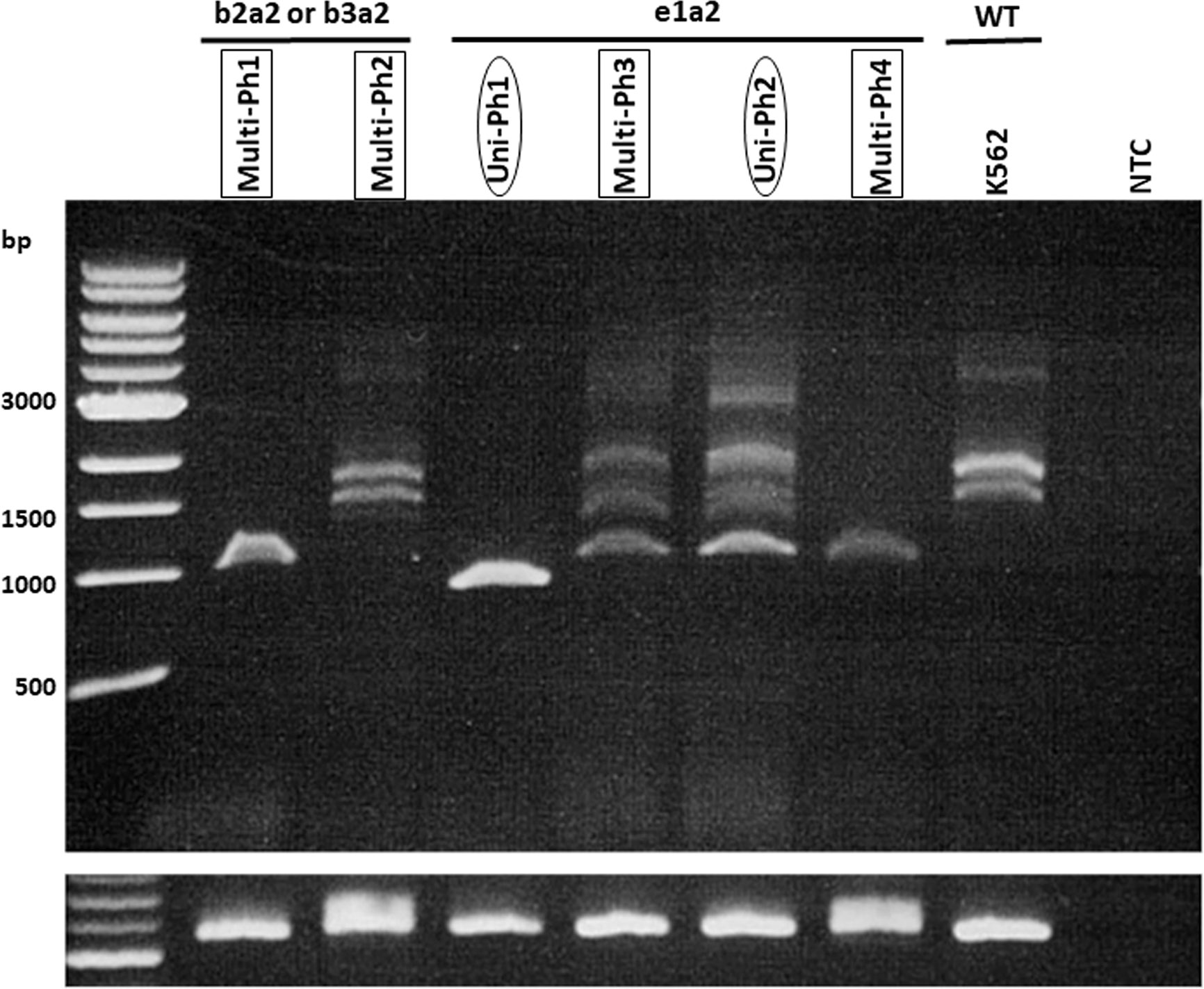
Figure 2 RT-PCR for IKZF1 transcripts. RT–PCR for IKZF1 transcripts (using exon 0- and 7-specific primers) in both uni-lineage Ph and multi-lineage Ph cases. K562 cell lines were used as the IKZF1 wild-type control.
The expression profiles of leukemic blast cells were assessed by performing a hierarchical clustering analysis, with bootstrapping, in ten patients (uni-Ph: 5, multi-Ph: 5). Blast cells derived from patients with multi-Ph were not grouped into a cluster that was distinguishable from those derived from patients with uni-Ph, suggesting that the global expression profiles of uni-Ph and multi-Ph blast cells were similar (Figure 3, Supplementary Table).
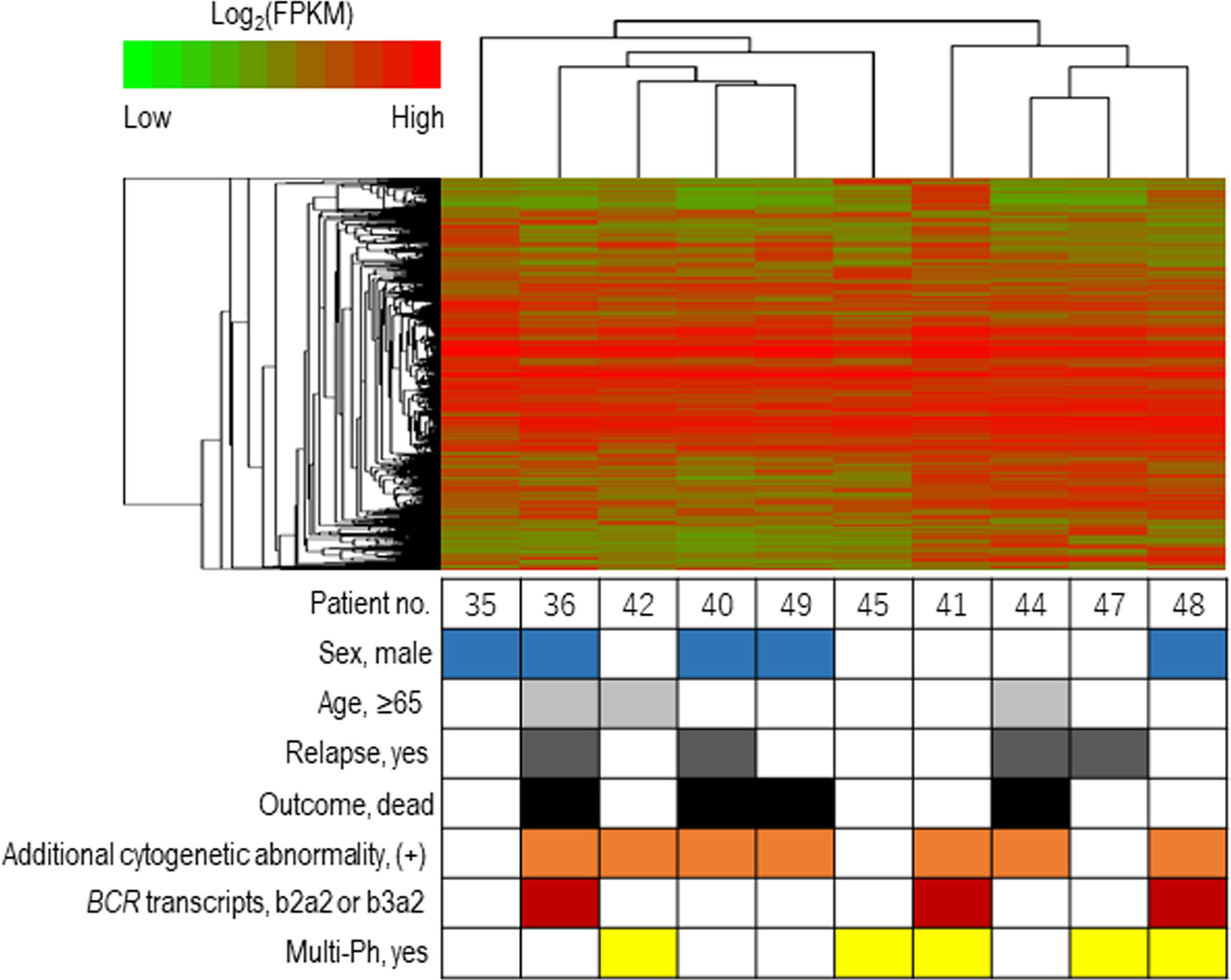
Figure 3 Expression profile-based clustering. Hierarchical clustering was performed based on the expression profiles obtained by RNA sequencing. The heat map shows the hierarchically clustered samples, with Pearson’s correlations, according to the significantly differentially expressed genes. Each column indicates an individual patient, and the lines at the top indicate the cluster dendrogram grouping of patients based on their similarities.
Both EFS and OS were significantly higher in patients with multi-Ph than in patients with uni-Ph (EFS: 74 vs. 33%, P = 0.01; OS: 79 vs. 44% at 4 years, P = 0.01, Figure 4). However, the CMR rate following the first induction therapy was significantly lower in patients with multi-Ph than in those with uni-Ph (11 vs. 56%, P = 0.003). In two patients with multi-Ph, BM-FISH at complete hematological remission showed positive BCR-ABL signals only outside of leukemic blast cells. In multivariate analyses, both BCR-ABL lineage and age were identified as significant prognostic factors for both EFS and OS (Table 2). Neither the type of TKI nor allo-HCT were found to be significant prognostic factors.
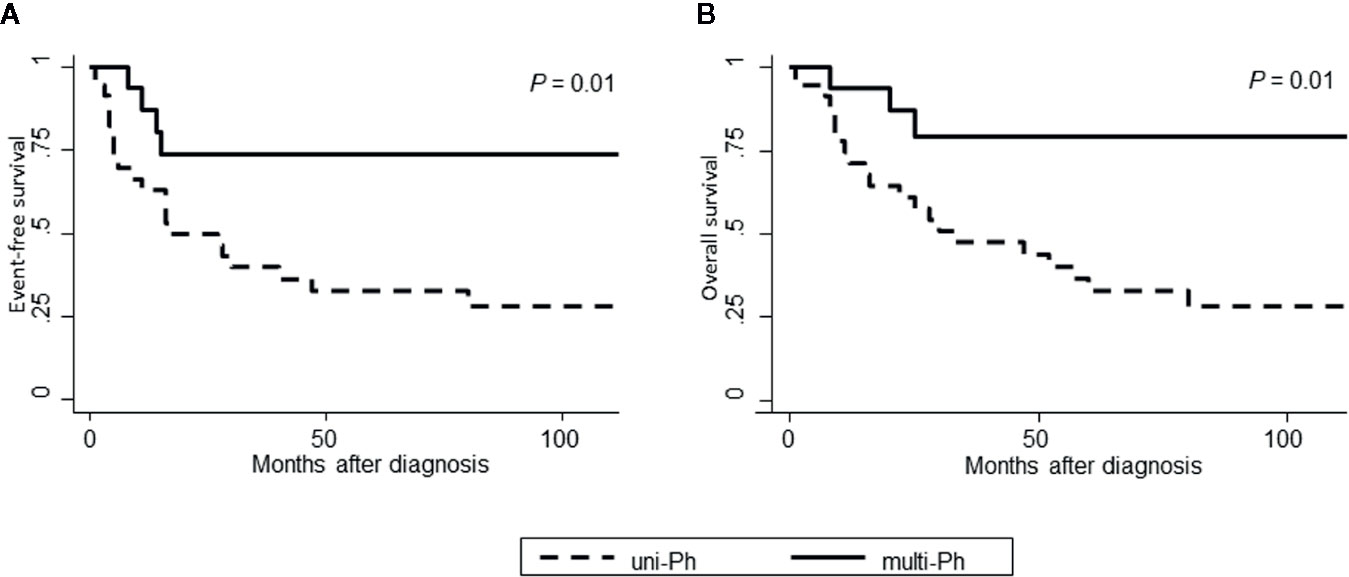
Figure 4 Patient outcomes, according to BCR-ABL-expressing lineage. (A) Event-free survival; (B) Overall survival.

Table 2 Univariate and multivariate analyses of factors influencing survival among patients with Ph+ALL.
In this study, we used multiple approaches to confirm the heterogeneity of adult Ph+ALL according to BCR-ABL expression lineages (1, 2). BCR-ABL signals were observed outside of leukemic blast cells in 36% of patients with Ph+ALL. We detected BCR-ABL signals not only in myeloid cells but also in megakaryocytes, which suggested the involvement of early differentiation stages in multi-Ph, including those that occur close to HSCs (29).
Three techniques were used to identify multi-Ph in this study: PN-FISH, BM-FISH, and FCM-PCR. Therefore, even in cases for which multi-Ph was difficult to determine using one method, the complementary use of multiple approaches allowed for the identification of multi-Ph. Most cells were classified as leukemic blast cells at diagnosis, and morphological judgments alone were not sufficient for the identification of fractions other than leukemic blast cells. Myeloperoxidase double staining was effective in such cases. Although FCM-PCR is a sensitive method, the number of cases we analyzed remained small because cryopreserved cells tended to feature too many dead cells to obtain an adequate signal.
To clarify the biological differences between uni-Ph and multi-Ph, we analyzed the deletion pattern of IKZF1, the most frequent genetic abnormality associated with Ph+ALL (16). However, no distinct IKZF1 deletion patterns were associated with multi-Ph. Gene expression analysis also failed to identify any clusters specific to multi-Ph. In addition to the limited number of samples included in this analysis, the lack of differences in gene expression may be associated with the use of stored samples, which were obtained at diagnosis and primarily contained leukemic blast cells. Because the only difference between uni-Ph and multi-Ph is the process through which the patient develops leukemia, any differences in the expression profile may disappear once a cell differentiates into a leukemia blast cell. Therefore, the analysis of other cell types or different differentiation stages between uni-Ph and multi-Ph may be necessary to observe genetic phenotypes.
Survival rates were significantly different between patients with uni-Ph and those with multi-Ph, consistent with a previous small-scale study (2). The better survival rate observed for patients with multi-Ph may be due to differences in the cell differentiation stage affected by TKI treatment between the two lineages, instead of specific genetic differences in leukemic cells. Leukemic blast cells are thought to derive from short-term HSC (ST-HSC) or the MPP level of development in multi-Ph, whereas they derive from common lymphoid progenitors (CLP)/lymphoid-primed multipotent progenitors (LMPP) in uni-Ph (30, 31). TKI treatment, alone, demonstrated a weak effect on the elimination of quiescent ST-HSCs/MPPs; however, when combined with chemotherapy, TKI may activate quiescent HSCs (32), resulting in the better eliminating of leukemic stem cells (33). In contrast, uni-Ph ALL, which is thought to originate from CLPs/LMPPs, includes ALL derived from aggressive B-1 progenitor cells, which is associated with poor TKI treatment outcomes (34, 35).
This study has several limitations. First, the number of samples that could be analyzed was limited. Because all available samples were used, the selection was not arbitrary; however, a limited selection of samples was available for data collection, such as few samples from remission cases. In addition, the MRD data were only based on PCR for BCR-ABL, and no MRD data associated with Immunoglobulin (Ig)/T-cell receptor (TCR) gene rearrangements were collected in this study. In Japan, Ig/TCR analysis is not commonly performed to detect MRD in adult Ph+ALL patients. Discrepancies in MRD data between BCR-ABL expression and Ig/TCR analyses can indicate the presence of BCR-ABL expression outside of lymphoblasts (3, 4). The histopathological distinction between blast cells and other blood cells using tissue FISH could complement the lack of Ig/TCR data by detecting the expression of BCR-ABL in cell types other than leukemic blast cells.
In conclusion, we confirmed BCR-ABL expression outside of leukemic blast cells in more than one-third of the Ph+ALL patients examined in this study. Although unique genetic patterns could not be identified, these patients revealed good prognoses following treatment, including TKI treatment.
All datasets generated for this study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
The studies involving human participants were reviewed and approved by Institutional review board of Nagoya University. The patients/participants provided their written informed consent to participate in this study.
SN, IS, and HK designed the research and interpreted the data. JHK, YU, and YI performed flow cytometric and PCR assays. MI, MM, and MG performed immunohistochemical assays. YOk performed RNA sequencing and bioinformatic analyses of the sequencing data. DK, YOz, MO, KK, and KM collected specimens and provide the data of patients. SN performed statistical analyses, and wrote the manuscript. All authors contributed to the article and approved the submitted version.
This study was supported in part by JSPS KAKENHI Grant Number JP17K16186 and JP20K08730.
HK received research funding from Chugai Pharmaceutical Co., Ltd., Kyowa Hakko Kirin Co., Ltd., Zenyaku Kogyo Co., Ltd., FUJIFILM Corporation, Daiichi Sankyo Co., Ltd., Astellas Pharma Inc., Otsuka Pharmaceutical Co., Ltd., Nippon Shinyaku Co., Ltd., Eisai Co., Ltd., Pfizer Japan Inc., Takeda Pharmaceutical Co., Ltd., Novartis Pharma K.K., Sumitomo Dainippon Pharma Co., Ltd., Sanofi K.K., and Celgene Corporation, consulting fees from Astellas Pharma Inc., Amgen Astellas Bio Pharma K.K., and Daiichi Sankyo Co., Ltd., and honoraria from Bristol-Myers Squibb, Astellas Pharma Inc., and Novartis Pharma K.K. These companies are not directly involved in any part of this study.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.586567/full#supplementary-material
1. Nagel I, Bartels M, Duell J, Oberg HH, Ussat S, Bruckmueller H, et al. Hematopoietic stem cell involvement in BCR-ABL1-positive ALL as a potential mechanism of resistance to blinatumomab therapy. Blood (2017) 130:2027–31. doi: 10.1182/blood-2017-05-782888
2. Kamoda Y, Izumi K, Iioka F, Akasaka T, Nakamura F, Kishimori C, et al. Philadelphia Chromosome-Positive Acute Lymphoblastic Leukemia Is Separated into Two Subgroups Associated with Survival by BCR-ABL Fluorescence in situ Hybridization of Segmented Cell Nuclei: Report from a Single Institution. Acta Haematol (2016) 136:157–66. doi: 10.1159/000445972
3. Zaliova M, Fronkova E, Krejcikova K, Muzikova K, Mejstrikova E, Stary J, et al. Quantification of fusion transcript reveals a subgroup with distinct biological properties and predicts relapse in BCR/ABL-positive ALL: implications for residual disease monitoring. Leukemia (2009) 23:944–51. doi: 10.1038/leu.2008.386
4. Hovorkova L, Zaliova M, Venn NC, Bleckmann K, Trkova M, Potuckova E, et al. Monitoring of childhood ALL using BCR-ABL1 genomic breakpoints identifies a subgroup with CML-like biology. Blood (2017) 129:2771–81. doi: 10.1182/blood-2016-11-749978
5. Lee S, Kim DW, Cho BS, Yoon JH, Shin SH, Yahng SA, et al. Impact of minimal residual disease kinetics during imatinib-based treatment on transplantation outcome in Philadelphia chromosome-positive acute lymphoblastic leukemia. Leukemia (2012) 26:2367–74. doi: 10.1038/leu.2012.164
6. Ravandi F, Jorgensen JL, Thomas DA, O’Brien S, Garris R, Faderl S, et al. Detection of MRD may predict the outcome of patients with Philadelphia chromosome-positive ALL treated with tyrosine kinase inhibitors plus chemotherapy. Blood (2013) 122:1214–21. doi: 10.1182/blood-2012-11-466482
7. Nishiwaki S, Imai K, Mizuta S, Kanamori H, Ohashi K, Fukuda T, et al. Impact of MRD and TKI on allogeneic hematopoietic cell transplantation for Ph+ALL: a study from the adult ALL WG of the JSHCT. Bone Marrow Transplant (2016) 51:43–50. doi: 10.1038/bmt.2015.217
8. Mizuta S, Matsuo K, Nishiwaki S, Imai K, Kanamori H, Ohashi K, et al. Pretransplant administration of imatinib for allo-HSCT in patients with BCR-ABL-positive acute lymphoblastic leukemia. Blood (2014) 123:2325–32. doi: 10.1182/blood-2013-11-538728
9. Yanada M, Takeuchi J, Sugiura I, Akiyama H, Usui N, Yagasaki F, et al. High complete remission rate and promising outcome by combination of imatinib and chemotherapy for newly diagnosed BCR-ABL-positive acute lymphoblastic leukemia: a phase II study by the Japan Adult Leukemia Study Group. J Clin Oncol (2006) 24:460–6. doi: 10.1200/JCO.2005.03.2177
10. Fujisawa S, Mizuta S, Akiyama H, Ueda Y, Aoyama Y, Hatta Y, et al. Phase II study of imatinib-based chemotherapy for newly diagnosed BCR-ABL-positive acute lymphoblastic leukemia. Am J Hematol (2017) 92:367–74. doi: 10.1002/ajh.24653
11. Sugiura I, Doki N, Hata T, Cho R, Ito T, Suehiro Y, et al. Dasatinib-Based Two-Step Induction Prior to Allogeneic Hematopoietic Cell Transplantation for Newly Diagnosed Philadelphia Chromosome-Positive Acute Lymphoblastic Leukemia: Results of the JALSG Ph+ALL213 Study. Blood (2019) 134:743–3. doi: 10.1182/blood-2019-123382
12. Takahashi N, Miura I, Kobayashi Y, Kume M, Yoshioka T, Otane W, et al. Fluorescence in situ hybridization monitoring of BCR-ABL-positive neutrophils in chronic-phase chronic myeloid leukemia patients during the primary stage of imatinib mesylate therapy. Int J Hematol (2005) 81:235–41. doi: 10.1532/ijh97.04095
13. Miranda RN, Mark HF, Medeiros LJ. Fluorescent in situ hybridization in routinely processed bone marrow aspirate clot and core biopsy sections. Am J Pathol (1994) 145:1309–14.
14. Primo D, Tabernero MD, Rasillo A, Sayagues JM, Espinosa AB, Chillon MC, et al. Patterns of BCR/ABL gene rearrangements by interphase fluorescence in situ hybridization (FISH) in BCR/ABL+ leukemias: incidence and underlying genetic abnormalities. Leukemia (2003) 17:1124–9. doi: 10.1038/sj.leu.2402963
15. Castor A, Nilsson L, Astrand-Grundstrom I, Buitenhuis M, Ramirez C, Anderson K, et al. Distinct patterns of hematopoietic stem cell involvement in acute lymphoblastic leukemia. Nat Med (2005) 11:630–7. doi: 10.1038/nm1253
16. Mullighan CG, Miller CB, Radtke I, Phillips LA, Dalton J, Ma J, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature (2008) 453:110–4. doi: 10.1038/nature06866
17. Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol (2013) 14:R36. doi: 10.1186/gb-2013-14-4-r36
18. Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc (2012) 7:562–78. doi: 10.1038/nprot.2012.016
19. Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics (2015) 31:166–9. doi: 10.1093/bioinformatics/btu638
20. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol (2014) 15:550. doi: 10.1186/s13059-014-0550-8
21. de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics (2004) 20:1453–4. doi: 10.1093/bioinformatics/bth078
22. Suzuki R, Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics (2006) 22:1540–2. doi: 10.1093/bioinformatics/btl117
23. Butturini AM, Dorey FJ, Lange BJ, Henry DW, Gaynon PS, Fu C, et al. Obesity and outcome in pediatric acute lymphoblastic leukemia. J Clin Oncol (2007) 25:2063–9. doi: 10.1200/jco.2006.07.7792
24. Nachman JB, La MK, Hunger SP, Heerema NA, Gaynon PS, Hastings C, et al. Young adults with acute lymphoblastic leukemia have an excellent outcome with chemotherapy alone and benefit from intensive postinduction treatment: a report from the children’s oncology group. J Clin Oncol (2009) 27:5189–94. doi: 10.1200/jco.2008.20.8959
25. Schultz KR, Carroll A, Heerema NA, Bowman WP, Aledo A, Slayton WB, et al. Long-term follow-up of imatinib in pediatric Philadelphia chromosome-positive acute lymphoblastic leukemia: Children’s Oncology Group study AALL0031. Leukemia (2014) 28:1467–71. doi: 10.1038/leu.2014.30
26. Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc (1958) 53:457–81. doi: 10.1080/01621459.1958.10501452
27. Peto R, Peto J. Asymptotically efficient rank invariant test procedures. J R Stat Soc A (1972) 135:185–207. doi: 10.2307/2344317
29. Woolthuis CM, Park CY. Hematopoietic stem/progenitor cell commitment to the megakaryocyte lineage. Blood (2016) 127:1242–8. doi: 10.1182/blood-2015-07-607945
30. Adolfsson J, Mansson R, Buza-Vidas N, Hultquist A, Liuba K, Jensen CT, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell (2005) 121:295–306. doi: 10.1016/j.cell.2005.02.013
31. Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature (2000) 404:193–7. doi: 10.1038/35004599
32. Duyvestyn JM, Taylor SJ, Dagger SA, Langdon WY. Dasatinib promotes the activation of quiescent hematopoietic stem cells in mice. Exp Hematol (2016) 44:410–421.e415. doi: 10.1016/j.exphem.2016.02.008
33. Dos Santos C, McDonald T, Ho YW, Liu H, Lin A, Forman SJ, et al. The Src and c-Kit kinase inhibitor dasatinib enhances p53-mediated targeting of human acute myeloid leukemia stem cells by chemotherapeutic agents. Blood (2013) 122:1900–13. doi: 10.1182/blood-2012-11-466425
34. Yoshimoto M. B-1 progenitor acute lymphoid leukemia. Blood (2019) 133:2557–8. doi: 10.1182/blood.2019001249
Keywords: BCR-ABL-expressing lineage, multi-lineage, multipotent progenitor, Philadelphia chromosome-positive acute lymphoblastic leukemia, uni-lineage
Citation: Nishiwaki S, Kim JH, Ito M, Maeda M, Okuno Y, Koyama D, Ozawa Y, Gunji M, Osaki M, Kitamura K, Ushijima Y, Ishikawa Y, Miyamura K, Sugiura I and Kiyoi H (2020) Multi-Lineage BCR-ABL Expression in Philadelphia Chromosome-Positive Acute Lymphoblastic Leukemia Is Associated With Improved Prognosis but No Specific Molecular Features. Front. Oncol. 10:586567. doi: 10.3389/fonc.2020.586567
Received: 23 July 2020; Accepted: 30 September 2020;
Published: 23 October 2020.
Edited by:
Ilaria Iacobucci, St. Jude Children’s Research Hospital, United StatesReviewed by:
Albrecht Reichle, University Medical Center Regensburg, GermanyCopyright © 2020 Nishiwaki, Kim, Ito, Maeda, Okuno, Koyama, Ozawa, Gunji, Osaki, Kitamura, Ushijima, Ishikawa, Miyamura, Sugiura and Kiyoi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Satoshi Nishiwaki, bi0zMTA0QHRmNy5zby1uZXQubmUuanA=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.