
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 08 January 2021
Sec. Head and Neck Cancer
Volume 10 - 2020 | https://doi.org/10.3389/fonc.2020.565669
 Gang Chen1†
Gang Chen1† Zhexuan Li1†
Zhexuan Li1† Changhan Chen1
Changhan Chen1 Jiajia Liu2
Jiajia Liu2 Weiming Zhu3
Weiming Zhu3 Li She1
Li She1 Huimei Huang2
Huimei Huang2 Yuexiang Qin1
Yuexiang Qin1 Guancheng Liu1
Guancheng Liu1 Juncheng Wang1
Juncheng Wang1 Yong Liu1,4,5
Yong Liu1,4,5 Donghai Huang1,4,5
Donghai Huang1,4,5 Qinglai Tang2
Qinglai Tang2 Xin Zhang1,4,5
Xin Zhang1,4,5 Gangcai Zhu2*
Gangcai Zhu2*PRAS40 (Prolin-rich Akt substrate of 40 kDa) is a critical protein, which directly connects PI3K/Akt and mTORC1 pathway. It plays an indispensable role in the development of various diseases. However, the relationship between PRAS40 and head and neck squamous cell carcinoma (HNSCC) remains unclear. Here, our study indicated that high expression of PRAS40 mRNA is a favorable prognostic factor in HNSCC patients by analyzing 498 clinical and mRNA data. Moreover, we confirmed that CRISPR/Cas9 induced PRAS40-knockout would promote colony formation, cell migration, and invasion in several HNSCC cell lines. RNA-seq was employed to investigate the further possible mechanisms involving the above regulations by PRAS40 in HNSCC cells. The molecular landscape contributed by 253 differentially expressed mRNA after PRAS40-knockout was enriched in TGF-beta, PI3K-Akt, P53, mTOR, NF-κB signaling pathway. Partial molecular alternations within these pathways were validated by qPCR or Western blotting. Besides, we found that high expression of PRAS40 in HNSC patients would present more CD8+ T and T follicular helper cells, but less Th17 cells than the patients with low expression of PRAS40. The altered molecular pathways and tumor-infiltrating immune cells might associate with the mechanism of PRAS40 being a suppressor in HNSCC cells, which would provide a potential prognostic predictor and therapeutic target in HNSCC patients.
The head and neck squamous cell carcinoma (HNSCC) is the sixth most prevalent cancer worldwide (1), with an incidence of approximately 650,000 new patients annually (2). Although the diagnosis methods and therapeutic arsenal have been refined and improved in recent decades, the overall 5-year survival rate of HNSCC patients has been holding around 60% (3). Tumor metastasis and uncontrolled growth are the main reasons for the unsatisfied survival rate in HNSCC patients (4, 5). Tumor metastasis is a highly regulated and complicated process composed of local invasion, intravasation, extravasation, and metastatic colonization at distant sites (6). Abundant evidence has demonstrated that the Akt and mTOR pathway activations are critical to cancer growth, metastasis, and resistance to therapy (7, 8). Due to PIK3CA mutations or other growth stimuli, the constitutive activation of PI3K/AKT/mTOR signaling was observed in more than 90% of HNSCC patients (9). The drugs targeting AKT or mTOR pathway, such as MK2206 or Temsirolimus, presented a promising effect to improve the prognosis of HNSCC patients (10). However, the tumor would develop quick resistance to these targeted therapies in several ways. Identifying novel targets or a dual inhibitor of AKT and mTOR pathway may provide potential support for treating HNSCC patients.
PRAS40, a 40 kDa proline-rich Akt substrate protein, locates at 19q13.33 and functions as an intersection in PI3K/Akt and mTOR pathway. PRAS40 was initially discovered to be a phosphorylated Akt kinase substrate protein (11), which was also named AKT1S1. Subsequently, it was found as a constrained component of mTORC1 (12). Akt could phosphorylate the amino acid residue of PRAS40 on Thr246/198 or Ser183/221/88/92/116/202/203/211, mTORC1, insulin, and PDGF, which could relieve the restrained activity of mTORC1 (12–15). Besides, PRAS40 could interact with ribosomal protein L11 (RPL11) and modulate P53 expression in turn (16). Our previous study showed that PRAS40 could regulate NF-κB transcription activity through physical association with p65 (17). Given the vital effects of mTORC1, P53, and NF-κB on cells, PRAS40 would be expected to play a crucial role in tumor initiation and development.
However, the reports about PRAS40 in tumors seemed controversial. On the one hand, PRAS40 was considered as an oncogene in melanoma, prostate cancer, liver cancer, and Ewing sarcoma by PRAS40-knockdown investigations (18–20). On the other hand, PRAS40 could suppress tumor growth in gallbladder cancer, cervical squamous cell cancer, and colon cancer (21, 22). As far as we are aware, there is no study regarding the role of PRAS40 in head and neck cancer. Here, our study found that high expression of PRAS40 mRNA is a favorable prognosis factor in HNSCC patients. Moreover, we confirmed that CRISPR/Cas9 induced PRAS40-knockout would promote cell colony formation, migration, and invasion in several HNSCC cell lines. RNA-seq was employed to investigate the further possible mechanisms involving the above regulations by PRAS40 in HNSCC cells. The molecular landscape contributed by 253 differential expressed mRNAs after PRAS40-knockout was enriched in TGF-beta, PI3K-Akt, P53, mTOR, NF-κB signaling pathway. Partial molecular alternations within these pathways were validated by PCR or western blot. Besides, we found that high expression of PRAS40 in HNSC patients would present more CD8+ T and T follicular helper cells, but less Th17 cells than the patients with low expression of PRAS40. The altered molecular pathways and tumor-infiltrating immune cells might associate with the mechanism of PRAS40 being a suppressor in HNSCC cells, which would provide a potential prognosis predictor and therapeutic target in HNSCC patients.
All clinical follow-up and mRNA expression data (including 498 samples) were downloaded from the TCGA datahub (https://portal.gdc.cancer.gov/). The immune cell deconvolution applied to HNSC samples was performed by the CIBERSORT method (23).
Dr. Zhuo kindly provided Tu686 cell line, Georgia Chen (Winship Cancer Institute, Emory University School of Medicine, Atlanta, GA, USA) (24). The 6-10B cell line was purchased from the Cell Center of Central South University (Changsha, China). As we described previously (25), Tu686 cells were cultured in DMEM/F12 (1:1) (Cytiva, USA, Cat. No. SH30023.01); 6-10B cells were cultured in RPMI1640 medium (Cytiva, USA, Cat. No. SH30027.01). All media were supplemented with 10% fetal bovine serum (ThermoFisher, USA, Cat. No.16000044), 100 IU/ml penicillin, and 100 μg/ml streptomycin at 37°C with 5% CO2 in an incubator. Cells have been checked to ensure that all in free of contamination and exponential growth so that they were used for subsequent experiments.
Genomic sequences of human PRAS40 were retrieved from the NCBI Consensus Coding Sequence database (http://www.ncbi.nlm.nih.gov/CCDS/). For each consensus coding sequence (CCDS) entry, exons 2 were included as candidate exons for library design. Next, all possible Cas9 sgRNA sequences of the form (N)20NGG were listed as candidate targets for each candidate exon. Due to the limited laboratory conditions, the synthesis and screening of recombinant plasmids were completed by GeneCopoeia (Guangzhou, China, Cat No. HCP259164-CG04-3-10-a/b/c). The oligonucleotides used to respective sgRNAs were listed in Table S1. Enzyme digestion was performed to validate these plasmids.
The knockout of PRAS40 in Tu-686 and 6-10B cell lines was achieved by CRISPR/Case9 system. The sgRNA was pre-designed to the target site in the PRAS40. Briefly, cells were transfected at 50–60% confluency in a 6-well plate with 2 ug CRISPR/Cas9 expression vector and 6 ul Fugene transfection reagent (Promega, USA, Cat. No. E2311). After 48 h, the cells with green fluorescence were sorted by flow cytometry and significantly diluted into a 6-well plate. Seven lines of 6–10B (named as 6-10BPRAS40-KO-1 to 6-10BPRAS40-KO-7) and five lines of Tu686 single clone (labeled as Tu686PRAS40-KO-1 to Tu686PRAS40-KO-5) survived successfully. The PRAS40 protein expression was confirmed as being knockout by western blot (Supplementary Figure 1). Three PRAS40 knockout and corresponding wild type cell lines were the only sources used throughout the further functional and mechanistic investigations.
Regarding colony formation, a density of 300 cells per well was cultured in the 6-well plate. After 14 days, cells were fixed with 0.1% crystal violet and stained with 4% methanol. Colonies containing more than 50 cells were counted using the Image J software. The average number of colonies was determined from three duplicated wells.
For wound healing assay, cells were seeded in six-well plates and cultured to reach 90% confluency. Cells were disrupted with a disinfected micropipette tip for 48 h. A microscope captured the images of bruised areas in the wells. These experiments were performed in triplicates.
Transwell chambers were coated with 200 ug/ml of Matrigel (Corning, USA, Cat.No.356255). Cells were seeded on the top of Matrigel at a concentration of 2 × 104 cells/well. After 48 h of culture, the culture media and Matrigel were removed, while the cells that crossed the membrane of the insert were fixed with 0.1% crystal violet and stained with 4% methanol. The average number of cells within three random visions was counted under the light microscope. The experiment was repeated three times.
Thirty micrograms of total protein was separated by 10% SDS-PAGE for 1 h. These proteins were transferred to a polyvinylidene difluoride membrane for 90 min (Millipore, USA. Cat. No. IPFL85R). After the membrane was blocked, it was incubated with primary mouse antibodies against PRAS40 (1:1,000 dilution, Proteintech, USA, Cat. No. 21097-1-AP), mTOR (1:1,000 dilution, Abcam, USA, Cat. No. ab109268), p-mTOR (1:1,000 dilution, Abcam, USA, Cat. No. ab109268) or GAPDH (1:1,000 dilution, Proteintech, USA, Cat. No. 1E6D9) at 4°C overnight. After incubation, the membrane was washed with PBST (Phosphate Buffered Saline with Tween-20; Cytiva, USA, Cat. No. SH30256.01) and incubated with HRP-labeled goat anti-rabbit IgG or HRP-labeled goat anti-mouse IgG (1:3,000 dilution, TransGen Biotech, China, Cat.No. HS201-01) for 1 h at room temperature. Finally, the membrane was incubated with the HRP (ThermoFisher, USA, Cat.No.21130), and image the blot using X-ray film (BIO-RAD, USA). Each experiment was repeated in triplicate.
According to the manufacturer’s protocol, total RNA extracted from knockout and wild type cells were reverse transcripted to cDNA by All-in-One First-Strand cDNA Synthesis kit (GeneCopoeia, USA). The RNA sequencing service, such as sequencing, read alignment, and DEGseq analysis, was provided by a technical company (Reo Health Inc, China). After cDNA synthesis, quantitative PCR was carried out using All-in-One qPCR Mix (GeneCopoeia, MD, USA) on ABI 7500HT System (Applied Biosystems, Foster City, CA). The primers used in this study were summarized in Table S1.
The Gene Ontology (GO) enrichment analysis with the corrected p-value was applied for the differentially expressed genes indicated in the RNA sequencing results. KEGG (Kyoto Encyclopedia of Genes and Genomes) is a database resource for systematic analysis of gene function and genomic information. It helps researchers to study gene and expression information as a whole network (http://www.genome.jp/kegg/). We used an online tool (https://david.ncifcrf.gov/) to test the statistical enrichment of differential expression genes (DEGs) in KEGG pathways.
All data were evaluated using GraphPad Prism (version 6, San Diego, USA), or R (version 3.6.1). Chi-Squared test or one-way ANOVA (analysis of variance) was performed to assess the significance of differences between two groups of samples. A Dunnett’s test with ANOVA was applied to pairwise comparisons in more than two groups by GraphPad Prism. Categorical variables are expressed as absolute frequencies and percentages, and continuous data were analyzed with descriptive statistics (mean values). The Kaplan–Meier method and Log-rank test were applied for survival analysis, and identification of relevant prognostic factors was performed by univariate and multivariate Cox regression analysis. The best cut-off for PRAS40 high expression was determined by the “survival” package (https://github.com/therneau/survival). P < 0.05 was considered statistically significant. Two-tailed tests were performed. The codes used in this study are available for the asking.
A total of 498 cases of HNSCC patients were included in our analysis. The correlation between the main clinical parameters and PRAS40 expression in HNSCC patients are summarized in Table 1. Briefly, the expression of PRAS40 was noticed to have no association with age, gender, tumor stage, node status, and clinical stage in HNSCC patients. As shown in Figure 1A, Kaplan–Meier survival analysis implied that the five-year overall survival (OS) tended to favor patients with high PRAS40 expression (p = 0.036). Given the association of PRAS40 expression and survival in HNSCC patients was confirmed, we are led to clarify the independent factors affecting overall survival in these patients. As exhibited in Table 2, variables including age, gender, tumor size (T stage), neck nodal metastasis, clinical stage, and PRAS40 expression are significantly associated with survival hazard ratios of 5 years’ death in univariate cox analysis (all p < 0.05). According to the multivariate cox model analysis, PRAS40 expression was identified as independent factors predicting overall survival in HNSCC patients (Figure 1B). The hazard ratios of patients with high PRAS40 expression over PRAS40 low expression is 0.567 (0.322–0.997, p = 0.048) after adjusting age, tumor size, neck node metastasis.
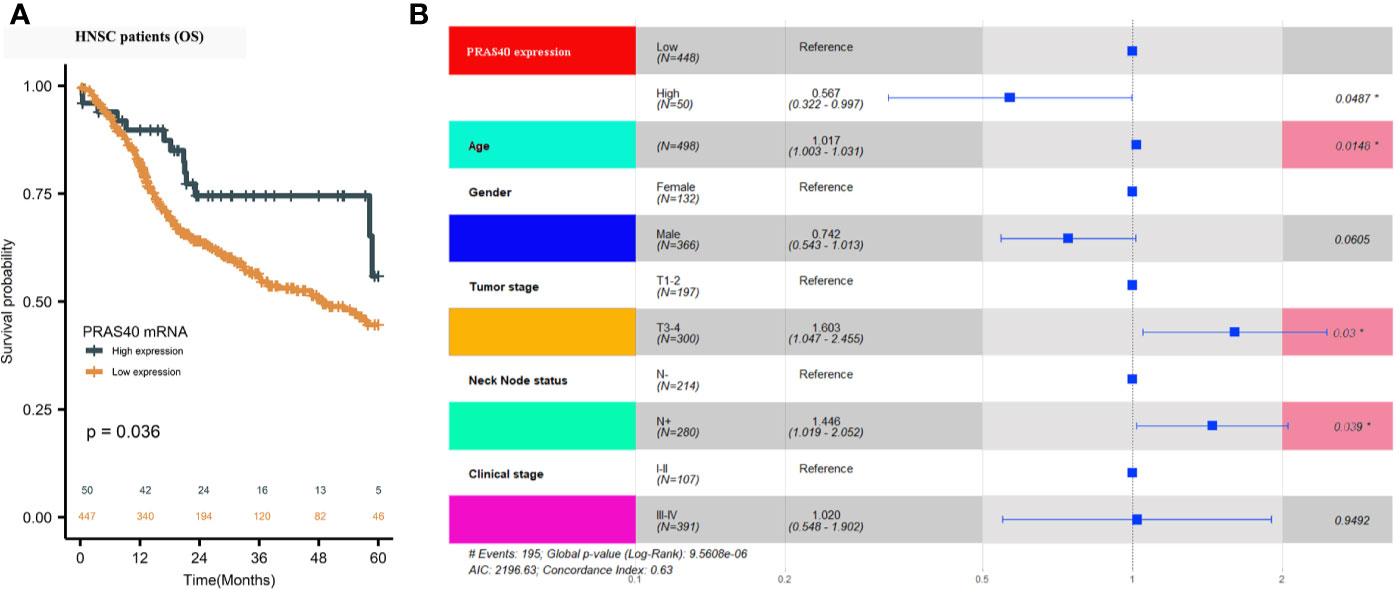
Figure 1 The clinical significance of PRAS40 expression in HNSCC. (A) Kaplan-Meier survival analysis implies that five-year overall survival tends to happen in favor of patients with high PRAS40 mRNA expression (p = 0.036). (B) Forest plot shows that the different PRAS40 mRNA levels could be an independent factor in HNSCC patients after age, gender, and clinical-stage adjustment (p = 0.0487).
To confirm the above finding that PRAS40 might confer a longer survival for HNSCC patients, we investigated the potential functions of PRAS40 in multiple HNSCC cell lines. Firstly, we established multiple HNSCC cell lines with PRAS40 knockout by CRISPR/Cas9. The PRAS40 protein expression was confirmed as no expression (Figure 2C) in all the knockout cell lines comparing to their parent cells. Compared to the wild type cell lines, the colony formation assays showed cells with PRAS40 knockout would significantly increase colony formation capability. As presented in Figure 2A, the mean ± SD of cell colonies raised from 75 ± 3 to 97 ± 3 and 108 ± 9 in 6–10B cell cohorts (p < 0.05). A similar tendency was observed in Tu686 related cell lines (123 ± 9 vs. 82 ± 8, p < 0.01, Figure 2B). These may indicate that PRAS40 could restrain the capability of HNSCC tumor formation.
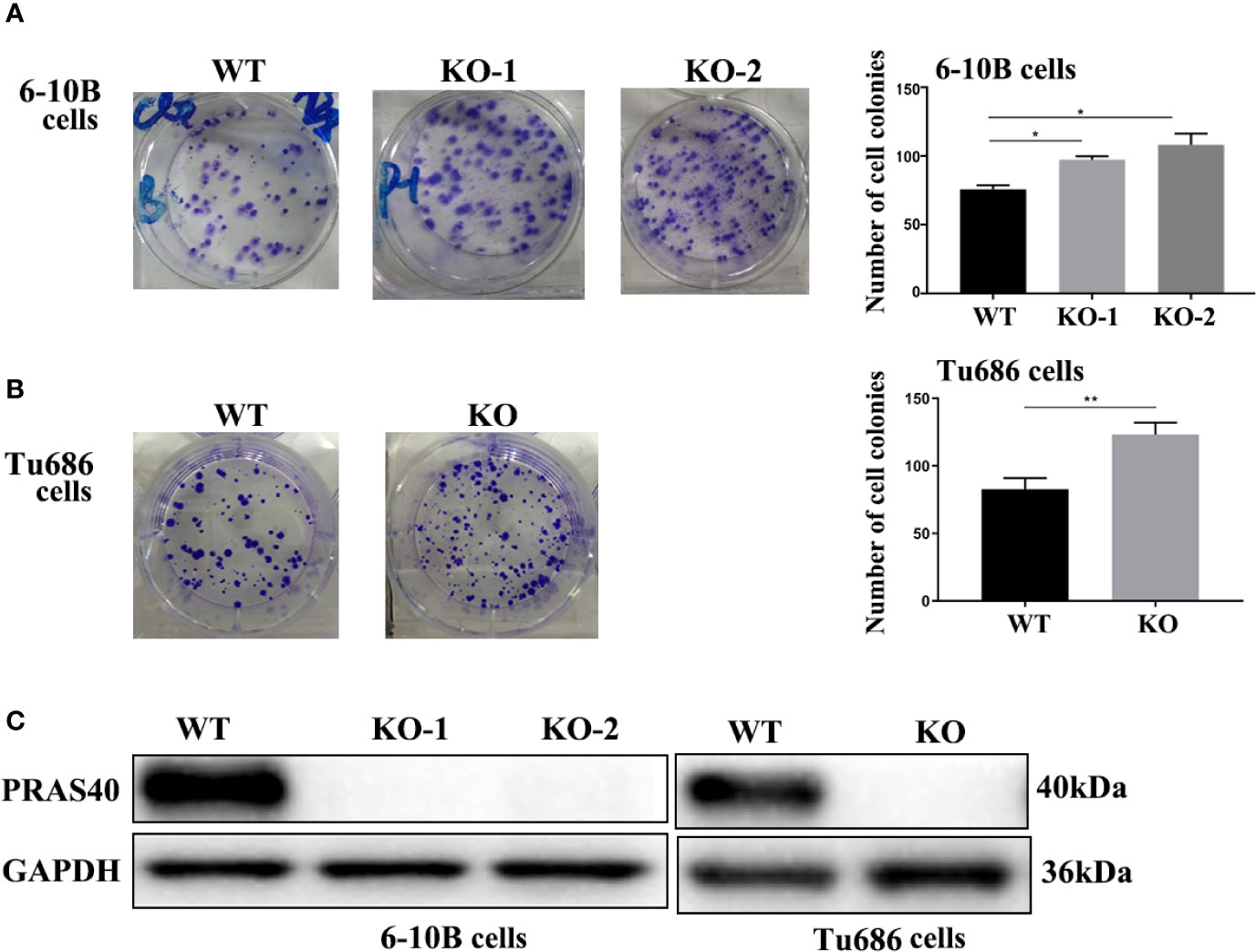
Figure 2 The knockout of PRAS40 promotes colony formation in HNSCC cell lines. (A, B) Colony formation assays show that knockout of PRAS40 increases the number of colony formation when comparing to their parent cells (6-10B and Tu686). Quantitative data for triplicated experiments are presented in the right panel. The mean ± SD of cell colonies raises 75 ± 3 to 97 ± 3 and 108 ± 9 in 6-10B cell cohorts (p<0.05). A similar tendency is observed in Tu686 related cell lines (123 ± 9 vs. 82 ± 8, p<0.01). (C) PRAS40 proteins are double-checked to confirm there is no PRAS40 expression in our experimental knockout 6-10B and Tu686 cell lines. WT means wildtype. KO means knockout. *p < 0.05 , **p < 0.01.
To further investigate the function of PRAS40 in the progression of HNSCC cells, wound healing and transwell assays were performed. Wound healing assay demonstrated that cells with PRAS40 knockout migrated more quickly than the wild type cells (Figure 3, 6–10B: 37.4 ± 7.1%, 6–10B-KO-1: 58.7 ± 12.7%, 6–10B-KO-2: 58.1 ± 6.6%, p < 0.05; Tu686: 35.3 ± 6.3%, Tu686-KO: 58.4 ± 6.1%, p < 0.05). Similar results were also observed in the number of cells crossing the transwell chamber (6–10B: 56 ± 6, 6–10B-KO-1: 111 ± 11, 6–10B-KO-2: 125 ± 7, p < 0.05; Tu686: 82 ± 8, Tu686-KO: 123 ± 9, p < 0.05), which implied that PRAS40 might inhibit HNSCC cell metastasis.
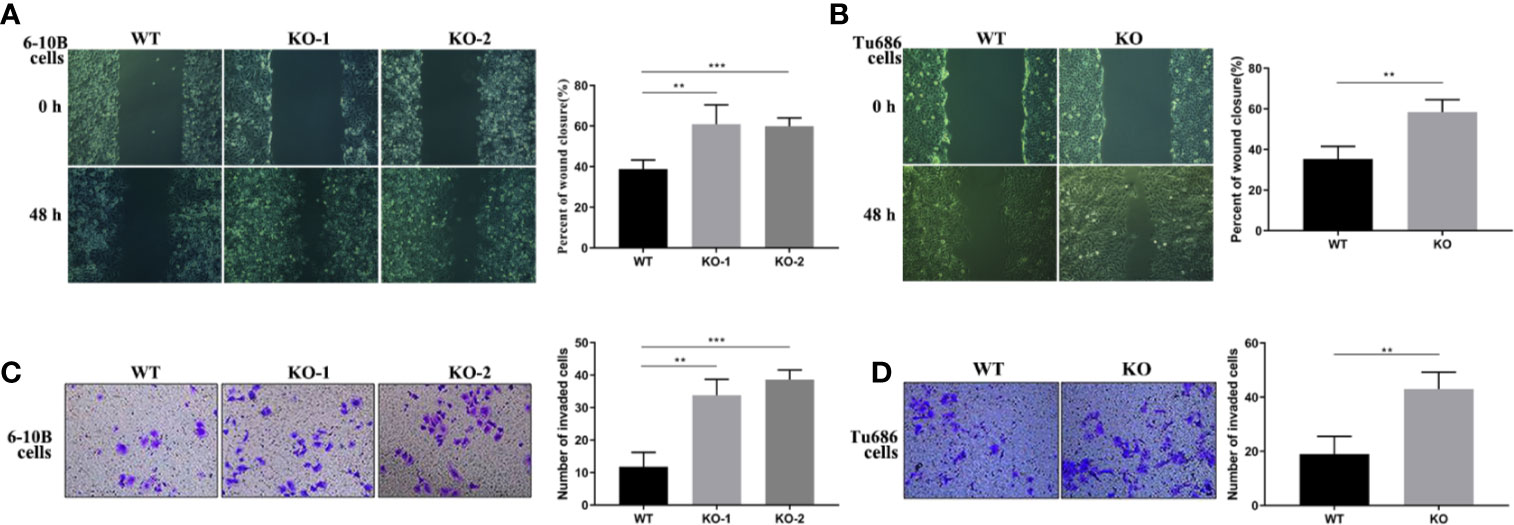
Figure 3 Knockout of PRAS40 increases the invasion and metastasis in HNSCC cell lines. Wounding-healing (A, B) and transwell chamber assays (C, D) were used to measure the changes following knockout of PRAS40 in 6-10B and Tu686 regarding migration and invasion. It indicates the ability of invasion and metastasis in the knockout of PRAS40 cells is higher than parent cells, respectively. Quantitative data for triplicated experiments are presented in the corresponding right XY plot (**P < 0.01, ***P < 0.005). WT means wildtype. KO means knockout.
To discover the potential molecular mechanism of the above PRAS40-regulating cell malignant behaviors, we employed RNA sequencing in two PRAS40 knockout cell lines as well as their parent cell line. The global mRNA expression profile was compared between wild type and PRAS40 KO cells and overlapped to establish the differential expressions. As shown in Figure 4A, a total of 253 genes were differentially expressed in the cells with PRAS40 knockout comparing to wild type, of which 97 mRNAs significantly declined, including IFI27, UGT1A7, SGK1, COL17A1, EMP1, and TXNIP. And there were 156 mRNAs NLRP1, TGFβI, MMP2, AR, GNAO1, HOXB9, and CA13 upregulated significantly. These 253 mRNAs constructed the expression of a heatmap showing the distinct molecular expression pattern between PRAS40 knockout cells and their parent cells (Figure 4B).
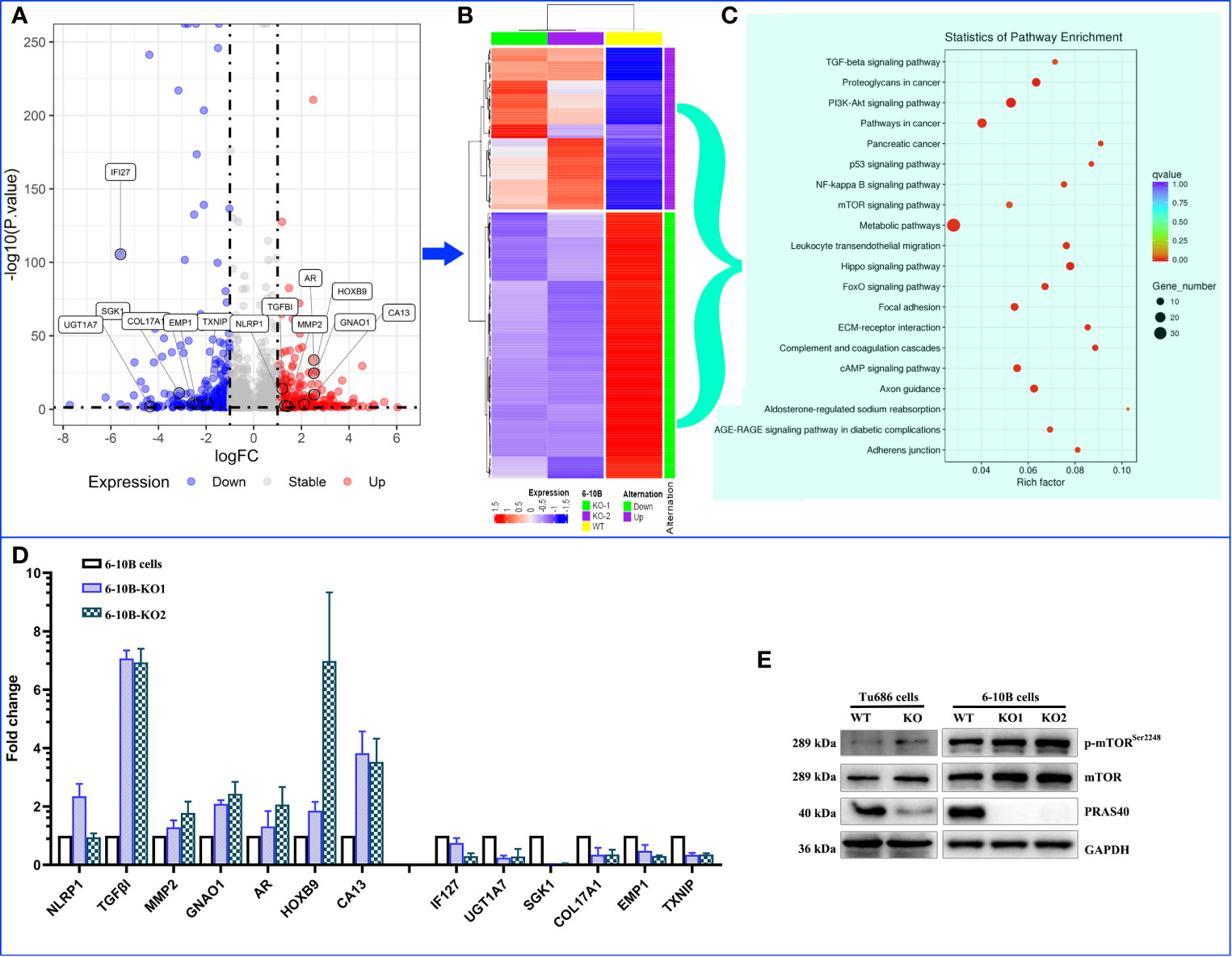
Figure 4 The alteration of molecular landscape induced by PRAS40 in HNSCC. (A) Volcano plot shows the logic fold change (logFC on the x-axis) and statistic reliability (p-value on the y-axis) for the global mRNA expression in PRAS40 knockout cells over wildtype cells. The red and blue dots represent up-regulated and down-regulated mRNAs in the PRAS40 knockout cells. Any dot beyond the horizontal dotted line means p <0.05. (B) The differential expressed mRNAs from (A) were entered to establish a heatmap showing the distinct molecular expression pattern between knockout of PRAS40 cells and their parent cells. (C) The top 20 signaling pathways for these differentially expressed genes by the KEGG signaling pathway enrichment analysis. (D) qPCR confirms the same changing directions for randomly selected mRNAs from the above RNA-sequencing results(A), all p <0.05. (E) Western blot shows the increased phosphorylation of mTOR in PRAS40 knockout cells as compared to their wildtype paraments cells. WT means wildtype. KO means knockout.
Inputting these 253 genes to KEGG pathway enrichment analysis, we found the top 20 pathways, such as TGF-β, PI3K-Akt, P53, mTOR, NF-kB signaling pathways, which may shoot the undermined molecular mechanism for PRAS40-regulating cell behaviors (Figure 4C). Moreover, 13 of 20 randomly selected mRNAs were validated by qPCR, consistently with our RNA sequencing results (Figure 4D). Additionally, the mTOR pathway was activated after the PRAS40 knockout in HNSCC cell lines (Figures 4E).
According to the above results, the downregulation of PRAS40 may involve the TGF-β signal pathway, a crucial pathway controlling immune cell fate and functions. Here, we deconvoluted 22 kinds of tumor-infiltrating immune cells based on the mRNA expression in TCGA HNSCC patients with high or low-expression of PRAS40. The comparison of the immune cell landscape was shown in Figure 5A, which implied that there were differential proportions of immune cells between patients with high and low expression of PRAS40. For instance, we found that high expression of PRAS40 in HNSCC patients would present more CD8+ T and T follicular helper cells, but less Th17 cells than the patients with low expression of PRAS40 (Figure 5B).
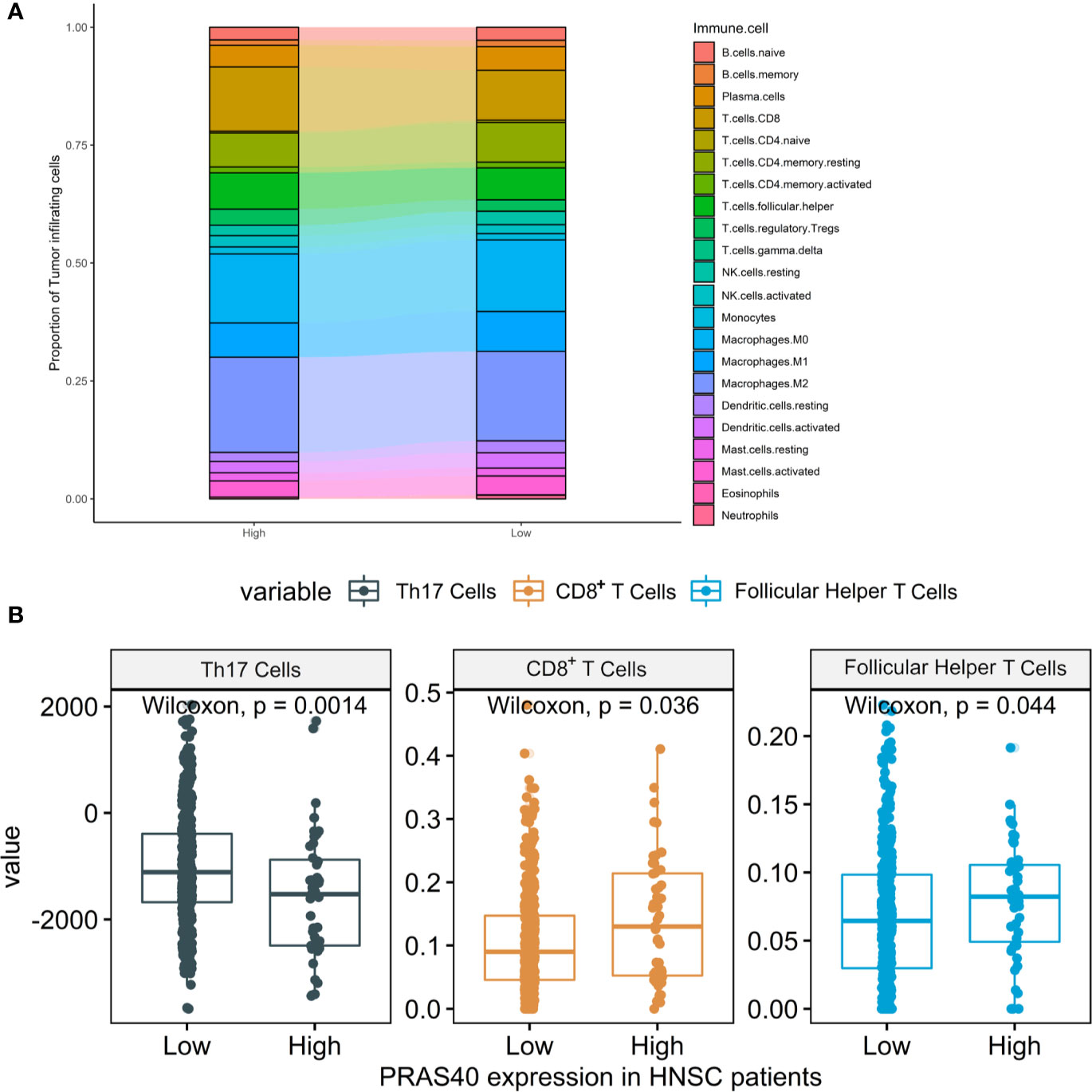
Figure 5 The immune cell signatures in HNSCC patients with different expressions of PRAS40. (A) The proportions of 22 tumor-infiltrating immune cells deconvoluted based on the mRNA expression show the landscape alterations of tumor-infiltrating immunity between the HNSC patients with high and low PRAS40 expression. Different color represents a unique type of immune cell, and y-axis value is the mean proportion of immune cell in the group of HNSC patients with different PRAS40 expression. (B) High expression of PRAS40 in HNSC patients would present more CD8+ T and T follicular helper cells, but less Th17 cells than the patients with low expression of PRAS40 (all p<0.05).
In the present study, we found that high expression of PRAS40 mRNA is an independent favorable factor for HNSCC patients. To our best knowledge, this is the first report showing the relationship between the PRAS40 expression and survival of HNSCC patients. Previous investigations indicated that PRAS40 could be either an oncogene or a suppressed gene in other types of cancer, such as melanoma, bladder cancer, and cervical cancer (17, 20). Here, our founding stands by PRAS40 might be a suppressor in HNSCC, which keeps pace with the concept that PRAS40 is a potent inhibitor to the mTORC1 activity (26). Decreased expression of PRAS40 would allow the downstream protein of mTORC1, including 4E-BP1 and P70S6K1, to be phosphorylated and activated, which could promote global mRNA transcription and protein synthesis (27). Moreover, our data showed that PRAS40 knockout in several HNSCC cell lines promoted cell colony formation, migration, and invasion. Our results implied that PRAS40 played as a restrainer on HNSCC formation and metastasis, consistent with our clinical data finding that PRAS40 may be a favorable factor for HNSCC prognosis. To ensure the efficiency and accuracy of CRISPR/Cas9 plasmid targeting PRAS40, we had validated the completely lost expression of PRAS40 by DNA sequences and western blot in all our experimental mono-clone cell lines. Therefore, it was highly confident to confirm that reduced PRAS40 expression could predict a worse outcome in HNSCC patients by analyzing both bench and bedside data in our study.
In order to investigate the possible mechanism underlying the regulations of the above biological behaviors by PRAS40 in HNSCC, RNA sequencing technology was applied in our study. We found 253 differentially expressed mRNA when comparing PRAS40-knockout and wild type of HNSCC cell lines. Most of the randomly selected mRNAs were validated by qPCR, making our RNA-seq results more confident to go further analysis. We found that TGF-β, PI3K-Akt, P53, mTOR, NF-kB signaling pathways are on the top enriched pathways as these 253 differential expressed genes input in KEGG analysis.
The misregulation of TGF-β signaling pathway is a vital player in cancer development (28). Our previous work showed that TGF-β could promote cell migration and invasion by induction of epithelial-mesenchymal transition in HNSCC cell line (14). Here, we confirmed that transforming growth factor beta-induced (TGFBI) mRNA was remarkably upregulated in HNSCC cells when PRAS40 was knockout. It was reported that TGFBI expressed higher in tumors than adjacent normal specimens within head and neck cancer patients (29). TGFBI may increase tumor metastasis by activating focal adhesion kinase signaling pathway through its binding to integrin αVβ5 (30), which may give some hints about the mechanism of PRAS40- regulating metastasis in HNSCC cell lines.
Numerous publications and our previous work showed the crucial role of the PI3K/Akt pathway in HNSCC initiation and development (31). Interestingly, our unpublished data indicated that PRAS40 could interact with FOXO1 directly, which may lead PRAS40 back to regulate the PI3K/Akt pathway. Additionally, the FOXO pathway was also enriched in our differential expressed mRNAs. However, whether PRAS40 and AKT would competitively bind to FOXO1 still under investigation. If the hypothesis is confirmed in the future, the biological behavior regulated by PRAS40 may be illustrated to some extent that PRAS40 knockout would allow more Akt binds and promotes FOXO1 transcription activity.
Both protein and mRNA expression of the mTORC1 pathway was upregulated after the PRAS40 knockout in our HNSCC cell lines. The first discovered function of PRAS40 was restraining mTORC1 activity (26). When PRAS40 expression was reduced, the over-activation of mTORC1 would not be surprisedly noticed, resulting in tumor cell colony formation and invasion in turn. This mechanism supported our finding that PRAS40 knockout could boost cell colony formation, migration, and invasion in HNSCC cells. Moreover, the NF-κB pathway also was included in the enrichments. That is includes evading growth suppressors, resisting cell death, and enabling replicative immortality. Also, inducing angiogenesis and activating invasion and metastasis acquired during the development of human tumors (32–35). In addition to cancer cell colony formation and survival, NF-κB related inflammation could regulate cancer patients’ local and systemic immunity (36, 37). It is unclear how much the NF-κB pathway plays in the PRAS40-regulating HNSCC cell growth and metastasis. However, our previous work indicated that PRAS40 could directly interact with P65 and promote NF-κB transcriptional activity (17), including various types of downstream targets such as pro-apoptotic genes (Fas, TP53, P21, and DR4/5) and anti-apoptotic genes (Bcl2, XIAP).
The investigation of the complexity and diversity of tumor immune microenvironment may help us understand the potential phenotypes. Our study showed that the HNSC patients with high PRAS40 expression would present more CD8+ T and T follicular helper (Tfh) cells but less Th17 cells in tumor samples than the patients with low PRAS40 expression. Tfh cell is a distinct subpopulation of CD4+ T cells that differentiates in secondary lymphoid organ germinal centers and plays critical roles in helping antigen-specific B cells generate antibodies (38). Tfh cells were also identified as residents in the tertiary lymphoid structure within tumor specimens (39). Th17 cells (characterized by high secretion of IL-17) differentiated from T helper lymphocytes participate in antimicrobial immunity at mucosal and epithelial barriers (40). Th17 cells’ prevalence was accumulated in peripheral blood, tumors, and drain lymph nodes of HNSCC patients that may promote tumor growth by inducing angiogenesis (via IL-17) and exerting themselves immunosuppressive functions (41–43). From an immune perspective, the high proportions of CD8+ T and Tfh cells and low proportion of Th17 cells may be the reasons for the favorable survival time in the group of HNSCC patients with high PRAS40 expression. Regarding the possible mechanism of different distributions of CD8+ T, Tfh, and Th17 cells in HNSCC patients with high or low expressed PRAS40, there are some clues from the molecular landscape alternations in PRAS40 knockout HNSCC cells: the overall effects of TGF-β, PI3K-Akt, NF-κB, and mTORC1 signaling.
All in all, this is the first report to show that the PRAS40 expression would be an independent prognosis factor in HNSCC patients. Furtherly, PRAS40 was confirmed as a suppressor of cell colony formation, migration, and invasion in HNSCC cell lines. Finally, we described a molecular landscape alternation in PRAS40 knockout HNSCC cell lines, which was validated in this study and partially our previous work. The differential distributions of CD8+ T cells, Tfh cells, and Th17 cells associating with the prognosis of HNSCC patients with high or low expression of PRAS40 solidified our conclusion that PRAS40 could be a tumor suppressor in HNSCC patients.
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE163613.
XZ, GZ, and YL conceived and designed the experiments. GC and ZL performed most of the cell experiments. CC did PCR and data analysis. WZ, YQ, JL, and GL did TCGA analysis. GZ and X Z wrote the paper. DH, QT contributed to the establishment of cell lines with PRAS40 knockout. HH, LS, and JW reviewed the draft. All authors contributed to the article and approved the submitted version.
This research was supported by the Project of Hunan Health Commission (B2019165), National Natural Science Foundation of China (Nos. 81874133, 81602389 and 81772903), Natural Science Foundation of Hunan Province (Nos. 2020JJ4827, 2018JJ2630, 2017JJ3456, and 2018JJ3317), the Huxiang Youth Talent Project (No. 2018RS3024), and National Key Research and Development Project (Nos. 2020YFC1316900 and 2020YFC1316901).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We appreciate Isaac A. Bernstein for helping with English language editing.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.565669/full#supplementary-material
Supplementary Figure 1 | The expression of PRAS40 in mono-colonies cells with PRAS40-knockout. The expression of PRAS40 was confirmed in eight cell lines of 6-10B and six cell lines of Tu686. It showed the silencing expression of PRAS40 in 6-10B and Tu686 KO cells as compared to the corresponding parent cells, respectively. Especially 6-10B KO-1, 6-10B KO-2, and Tu686 KO1 (named Tu686-KO in Figures 2–4) cell lines, which are selected for the future cell culture and experiments.
TCGA, The Cancer Genome Atlas; NCBI, National Center for Biotechnology Information; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; DEGs, differential expression genes; ANOVA, analysis of variance; PRAS40, Prolin-rich Akt substrate of 40 kDa; HNSCC, head and neck squamous cell carcinoma; CCDS, consensus coding sequence; TGFBI, growth factor beta-induced; Tfh, T follicular helper; OS, overall survival.
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin (2015) 65(2):87–108. doi: 10.3322/caac.21262
2. Westra WH. The changing face of head and neck cancer in the 21st century: the impact of HPV on the epidemiology and pathology of oral cancer. Head Neck Pathol (2009) 3(1):78–81. doi: 10.1007/s12105-009-0100-y
3. Kaboodkhani R, Karimi E, Khorsandi Ashtiani MT, Kowkabi S, Firouzifar MR, Yazdani F, et al. Evaluation of the Correlation between CD44, Tumor Prognosis and the 5-Year Survival Rate in Patients with Oral Tongue SCC. Iran J Otorhinolaryngol (2016) 28(89):407–11.
4. Bhave SL, Teknos TN, Pan Q. Molecular parameters of head and neck cancer metastasis. Crit Rev Eukaryot Gene Expr (2011) 21(2):143–53. doi: 10.1615/CritRevEukarGeneExpr.v21.i2.40
5. Qin Y, Wang J, Zhu G, Li G, Tan H, Chen C, et al. CCL18 promotes the metastasis of squamous cell carcinoma of the head and neck through MTDH-NF-κB signalling pathway. J Cell Mol Med (2019) 23(4):2689–701. doi: 10.1111/jcmm.14168
6. Jiang WG, Sanders AJ, Katoh M, Ungefroren H, Gieseler F, Prince M, et al. Tissue invasion and metastasis: Molecular, biological and clinical perspectives. Semin Cancer Biol (2015) 35 Suppl:S244–S75. doi: 10.1016/j.semcancer.2015.03.008
7. Song M, Bode AM, Dong Z, Lee M-H. AKT as a Therapeutic Target for Cancer. Cancer Res (2019) 79(6):1019–31. doi: 10.1158/0008-5472.CAN-18-2738
8. Wang W, Shen T, Dong B, Creighton CJ, Meng Y, Zhou W, et al. MAPK4 overexpression promotes tumor progression via noncanonical activation of AKT/mTOR signaling. J Clin Invest (2019) 129(3):1015–29. doi: 10.1172/JCI97712
9. Fruman DA, Chiu H, Hopkins BD, Bagrodia S, Cantley LC, Abraham RT. The PI3K Pathway in Human Disease. Cell (2017) 170(4):605–35. doi: 10.1016/j.cell.2017.07.029
10. Bose P, Brockton NT, Dort JC. Head and neck cancer: from anatomy to biology. Int J Cancer (2013) 133(9):2013–23. doi: 10.1002/ijc.28112
11. Kovacina KS, Park GY, Bae SS, Guzzetta AW, Schaefer E, Birnbaum MJ, et al. Identification of a proline-rich Akt substrate as a 14-3-3 binding partner. J Biol Chem (2003) 278(12):10189–94. doi: 10.1074/jbc.M210837200
12. Wiza C, Nascimento EBM, Ouwens DM. Role of PRAS40 in Akt and mTOR signaling in health and disease. Am J Physiol Endocrinol Metab (2012) 302(12):E1453–E60. doi: 10.1152/ajpendo.00660.2011
13. Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science (2011) 332(6035):1317–22. doi: 10.1126/science.1199498
14. Yu C, Liu Y, Huang D, Dai Y, Cai G, Sun J, et al. TGF-β1 mediates epithelial to mesenchymal transition via the TGF-β/Smad pathway in squamous cell carcinoma of the head and neck. Oncol Rep (2011) 25(6):1581–7. doi: 10.3892/or.2011.1251
15. Chong ZZ, Shang YC, Wang S, Maiese K. PRAS40 is an integral regulatory component of erythropoietin mTOR signaling and cytoprotection. PloS One (2012) 7(9):e45456. doi: 10.1371/journal.pone.0045456
16. Havel JJ, Li Z, Cheng D, Peng J, Fu H. Nuclear PRAS40 couples the Akt/mTORC1 signaling axis to the RPL11-HDM2-p53 nucleolar stress response pathway. Oncogene (2015) 34(12):1487–98. doi: 10.1038/onc.2014.91
17. Zhu G, Qi Q, Havel JJ, Li Z, Du Y, Zhang X, et al. PRAS40 promotes NF-κB transcriptional activity through association with p65. Oncogenesis (2017) 6(9):e381. doi: 10.1038/oncsis.2017.80
18. Hu F, Deng X, Yang X, Jin H, Gu D, Lv X, et al. Hypoxia upregulates Rab11-family interacting protein 4 through HIF-1α to promote the metastasis of hepatocellular carcinoma. Oncogene (2015) 34(49):6007–17. doi: 10.1038/onc.2015.49
19. Lv D, Liu J, Guo L, Wu D, Matsumoto K, Huang L. PRAS40 deregulates apoptosis in Ewing sarcoma family tumors by enhancing the insulin receptor/Akt and mTOR signaling pathways. Am J Cancer Res (2016) 6(2):486–97.
20. Madhunapantula SV, Sharma A, Robertson GP. PRAS40 deregulates apoptosis in malignant melanoma. Cancer Res (2007) 67(8):3626–36. doi: 10.1158/0008-5472.CAN-06-4234
21. Subbannayya T, Leal-Rojas P, Zhavoronkov A, Ozerov IV, Korzinkin M, Babu N, et al. PIM1 kinase promotes gallbladder cancer cell proliferation via inhibition of proline-rich Akt substrate of 40 kDa (PRAS40). J Cell Commun Signal (2019) 13(2):163–77. doi: 10.1007/s12079-018-00503-5
22. Yu X, Zheng B, Chai R. Lentivirus-mediated knockdown of eukaryotic translation initiation factor 3 subunit D inhibits proliferation of HCT116 colon cancer cells. Biosci Rep (2014) 34(6):e00161. doi: 10.1042/BSR20140078
23. Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods (2015) 12(5):453–7. doi: 10.1038/nmeth.3337
24. Yu C, Liu Y, Tan H, Li G, Su Z, Ren S, et al. Metadherin regulates metastasis of squamous cell carcinoma of the head and neck via AKT signalling pathway-mediated epithelial-mesenchymal transition. Cancer Lett (2014) 343(2):258–67. doi: 10.1016/j.canlet.2013.09.033
25. Zhu G, Cai G, Liu Y, Tan H, Yu C, Huang M, et al. Quantitative iTRAQ LC-MS/MS Proteomics Reveals Transcription Factor Crosstalk and Regulatory Networks in Hypopharyngeal Squamous Cell Carcinoma. J Cancer (2014) 5(7):525–36. doi: 10.7150/jca.9207
26. Völkers M, Toko H, Doroudgar S, Din S, Quijada P, Joyo AY, et al. Pathological hypertrophy amelioration by PRAS40-mediated inhibition of mTORC1. Proc Natl Acad Sci U.S.A. (2013) 110(31):12661–6. doi: 10.1073/pnas.1301455110
27. Mi W, Ye Q, Liu S, She Q-B. AKT inhibition overcomes rapamycin resistance by enhancing the repressive function of PRAS40 on mTORC1/4E-BP1 axis. Oncotarget (2015) 6(16):13962–77. doi: 10.18632/oncotarget.3920
28. Wu MY, Hill CS. Tgf-beta superfamily signaling in embryonic development and homeostasis. Dev Cell (2009) 16(3):329–43. doi: 10.1016/j.devcel.2009.02.012
29. Shen Y, Liu J, Zhang L, Dong S, Zhang J, Liu Y, et al. Identification of Potential Biomarkers and Survival Analysis for Head and Neck Squamous Cell Carcinoma Using Bioinformatics Strategy: A Study Based on TCGA and GEO Datasets. Biomed Res Int (2019) 2019:7376034. doi: 10.1155/2019/7376034
30. Costanza B, Rademaker G, Tiamiou A, De Tullio P, Leenders J, Blomme A, et al. Transforming growth factor beta-induced, an extracellular matrix interacting protein, enhances glycolysis and promotes pancreatic cancer cell migration. Int J Cancer (2019) 145(6):1570–84. doi: 10.1002/ijc.32247
31. Marquard FE, Jücker M. PI3K/AKT/mTOR signaling as a molecular target in head and neck cancer. Biochem Pharmacol (2020) 172:113729. doi: 10.1016/j.bcp.2019.113729
32. Fan Y, Mao R, Yang J. NF-κB and STAT3 signaling pathways collaboratively link inflammation to cancer. Protein Cell (2013) 4(3):176–85. doi: 10.1007/s13238-013-2084-3
33. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell (2011) 144(5):646–74. doi: 10.1016/j.cell.2011.02.013
34. House CD, Jordan E, Hernandez L, Ozaki M, James JM, Kim M, et al. NFκB Promotes Ovarian Tumorigenesis via Classical Pathways That Support Proliferative Cancer Cells and Alternative Pathways That Support ALDH Cancer Stem-like Cells. Cancer Res (2017) 77(24):6927–40. doi: 10.1158/0008-5472.CAN-17-0366
35. Johnson RF, Perkins ND. Nuclear factor-κB, p53, and mitochondria: regulation of cellular metabolism and the Warburg effect. Trends Biochem Sci (2012) 37(8):317–24. doi: 10.1016/j.tibs.2012.04.002
36. Pasparakis M. Regulation of tissue homeostasis by NF-kappaB signalling: implications for inflammatory diseases. Nat Rev Immunol (2009) 9(11):778–88. doi: 10.1038/nri2655
37. Taniguchi K, Karin M. NF-κB, inflammation, immunity and cancer: coming of age. Nat Rev Immunol (2018) 18(5):309–24. doi: 10.1038/nri.2017.142
38. Wing JB, Ise W, Kurosaki T, Sakaguchi S. Regulatory T cells control antigen-specific expansion of Tfh cell number and humoral immune responses via the coreceptor CTLA-4. Immunity (2014) 41(6):1013–25. doi: 10.1016/j.immuni.2014.12.006
39. Germain C, Gnjatic S, Dieu-Nosjean M-C. Tertiary Lymphoid Structure-Associated B Cells are Key Players in Anti-Tumor Immunity. Front Immunol (2015) 6:67:67. doi: 10.3389/fimmu.2015.00067
40. Ouyang W, Kolls JK, Zheng Y. The biological functions of T helper 17 cell effector cytokines in inflammation. Immunity (2008) 28(4):454–67. doi: 10.1016/j.immuni.2008.03.004
41. Kesselring R, Thiel A, Pries R, Trenkle T, Wollenberg B. Human Th17 cells can be induced through head and neck cancer and have a functional impact on HNSCC development. Br J Cancer (2010) 103(8):1245–54. doi: 10.1038/sj.bjc.6605891
42. Kesselring R, Thiel A, Pries R, Wollenberg B. The number of CD161 positive Th17 cells are decreased in head and neck cancer patients. Cell Immunol (2011) 269(2):74–7. doi: 10.1016/j.cellimm.2011.03.026
Keywords: head and neck squamous cell carcinoma, prolin-rich Akt substrate of 40 kDa, CRISPR/Cas9, metastasis, The Cancer Genome Atlas, immunomicro-environment
Citation: Chen G, Li Z, Chen C, Liu J, Zhu W, She L, Huang H, Qin Y, Liu G, Wang J, Liu Y, Huang D, Tang Q, Zhang X and Zhu G (2021) The Molecular Landscape and Biological Alterations Induced by PRAS40-Knockout in Head and Neck Squamous Cell Carcinoma. Front. Oncol. 10:565669. doi: 10.3389/fonc.2020.565669
Received: 25 May 2020; Accepted: 13 November 2020;
Published: 08 January 2021.
Edited by:
Jiajia Zhang, Johns Hopkins University, United StatesReviewed by:
SrinivasVinod Saladi, Massachusetts Eye & Ear Infirmary and Harvard Medical School, United StatesCopyright © 2021 Chen, Li, Chen, Liu, Zhu, She, Huang, Qin, Liu, Wang, Liu, Huang, Tang, Zhang and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gangcai Zhu, cWlhbmh1ZG9jdG9yQGNzdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.