- Department of Health Statistics, School of Public Health, China Medical University, Shenyang, China
Background: Hepatocellular carcinoma (HCC) is an intractable public health threat worldwide, representing the second leading cause of cancer-related mortality, with limited early detection and therapeutic options. Recent findings have revealed that the susceptibility of HCC is closely related to microRNA (miRNA). We performed this systematic review with a network meta-analysis to investigated four single nucleotide polymorphisms (SNPs) that most regularly reported in miRNAs, exploring their involvement in HCC susceptibility and interaction with hepatitis B virus (HBV).
Methods: Databases were reviewed for related studies published up to May 2019 to identify all studies that compared genotypes of miR-146a rs2910164, miR-149 rs2292832, miR-196a2 rs11614913, and miR-499 rs3746444 with no language and date restrictions. A pairwise meta-analysis was performed to estimate pooled odds ratios and 95% confidence intervals incorporating heterogeneity to assess the relationship between four miRNA polymorphisms and HCC. To further clarify the effect of polymorphisms on HCC, a Bayesian network meta-analysis was conducted to combine the effective sizes of direct and indirect comparisons. Calculations were performed by R version 3.6.1 and STATA 14.0. All steps were performed according to PRISMA guidelines.
Results: A total of 20 studies were enrolled in this network meta-analysis, providing 5,337 hepatocellular carcinoma cases and 6,585 controls. All included studies had an acceptable quality. Pairwise meta-analysis demonstrated that miR-196a2 rs11614913 was significantly associated with the susceptibility of HCC, while the other three SNPs were not found to have a significant association. In the analysis of HCC patients under different HBV infection status, only miR-196a2 revealed correlation of threefold risk. The network results showed no significant difference in the distribution of genotype frequencies except for miR-196a2, which appeared to have the highest superiority index when comparing and ranking four SNPs.
Conclusion: MiR-196a2 rs11614913 was significantly associated with the susceptibility of HCC, especially for HBV- related HCC, and that individuals with TC/CC were more susceptible. No significant association was found in the other three miRNA genes. MiR-196a2 could serve as the best predictor of susceptibility in HCC.
Introduction
Hepatocellular carcinoma (HCC), the most common form of primary liver cancer, has been an intractable public health threat worldwide (1). Approximately 700,000 new cases and 600,000 deaths are attributable to HCC annually (2), representing the sixth leading cause of cancer and the second leading cause of cancer-related mortality (3). Asia, Sub-Saharan Africa and the Middle East are high-risk regions with high incidence rates of HCC (4), and in some of these regions HCC ranks as the leading cause of death due to cancer (5). But, it is worth noting that the incidence and mortality have been increasing in North America and some areas of Europe (6, 7). The incidence in the United States has tripled over the past three decades (8). In the European Union, estimated by WHO, about 47,000 people die of liver cancer each year (9). In Canada, HCC has become the only cancer whose mortality rate is still on the rise. The incidence has been increasing rapidly and is projected to continue beyond 2020 (3). Multiple factors are responsible for its development including inborn diseases, chemicals, and viruses, of which Hepatitis B virus (HBV) is widely acknowledged. HCC has become a tremendous global burden (10), with the characteristics of high incidence, short duration, poor prognosis, high degree of malignancy, and five-year survival rate of 7% (11), yet remains one of the most ill-informed cancers and compounds by limited early detection and therapeutic options (12, 13). Therefore, exploring and clarifying the disease mechanism of HCC is conducive for effective prevention and treatment (14).
Genetic association studies are of great significance for epidemiological analyses, as they can identify candidate genome regions associated to specific diseases (15). Many findings have revealed that the presence of single nucleotide polymorphisms (SNPs) in some miRNA genes can alter the expression or maturation of miRNAs, making individuals more susceptible to certain types of cancer (16, 17). Several SNPs in miRNA genes can influence the development of HCC (18), providing a novel perspective of pathophysiological mechanism for the etiology of HCC (19).
MiR-146a (rs2910164), miR149 (rs2292832), miR-196a2 (rs11614913), and miR-499 (rs3746444) are well-established functional miRNAs (20–24). Researches have demonstrated that they can participate in essential regulatory processes related to cellular senescence, inflammation, immune response thus have potential value as biomarkers for many diseases (25, 26). The results on the association between genetic polymorphisms and HCC susceptibility remain inconsistent due to differences in race, disease stage, sample size, or other uncertainties. To further explore whether polymorphisms in these four SNPs might predispose to HCC, additional research and quantitative statistical studies are required to resolve discrepancies (27). Network meta-analyses (NMA) can be used to summarize and compare studies on multiple interventions (28), and combine direct and indirect evidence thus produce a result more precise (29). We conducted this systematic review with a network meta-analysis to provide more comprehensive information on the polymorphisms of four selected miRNAs and their involvement in HCC susceptibility and interaction with HBV.
Materials and Methods
Literature Search
In this systematic review and meta-analysis, we searched the database of PubMed, EMbase and the Cochrane Central Register to identify all eligible case-controlled trials that compared genotypes of the four selected miRNA genes in HCC patients with non-cancer control groups. All searches were performed in May 2019 and no language and date restrictions were set. The searching items were: “hepatocellular carcinoma”, “hepatoma”, “liver cancer”, “HCC”, and “microRNAs”, “miRNA”, and “polymorphism”, “allele”, “variation”, “SNP”.
Selection Criteria
Eligible studies met the following criteria: (1) Case-controlled trials of subjects with HCC and healthy participants without HCC; (2) Evaluate the relationship between four common SNPs of miRNA (miR-146a rs2910164, miR-149 rs2292832, miR-196a2 rs11614913, miR-499 rs3746444) and HCC risk; (3) Investigate at least two selected SNPs at the same time; (4) Either DNA sequencing or PCR is used as a genotypic method for detection.
Articles were excluded based on the criteria: (1) Duplicated articles or data; (2) Irrelevant cancers or SNPs; (3) Functional studies; (4) Lack of available genotype frequency.
Data Abstraction and Assessment of Bias
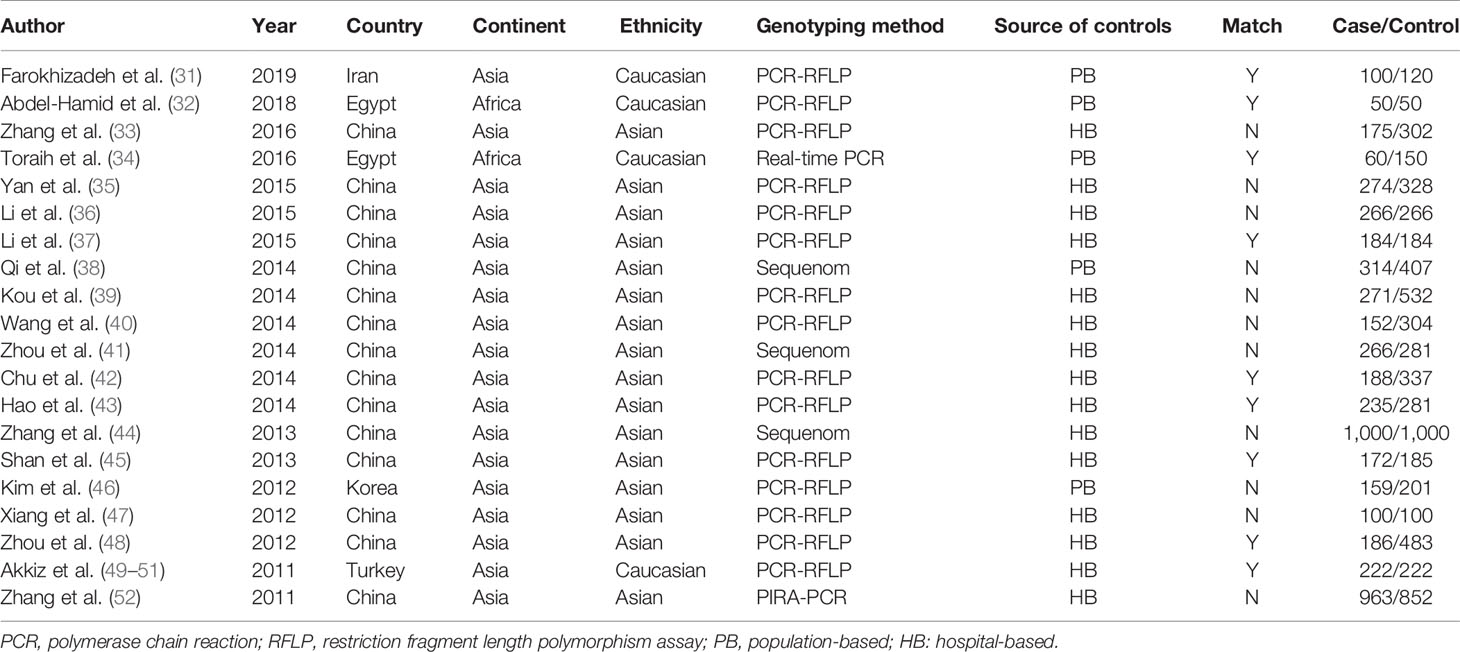
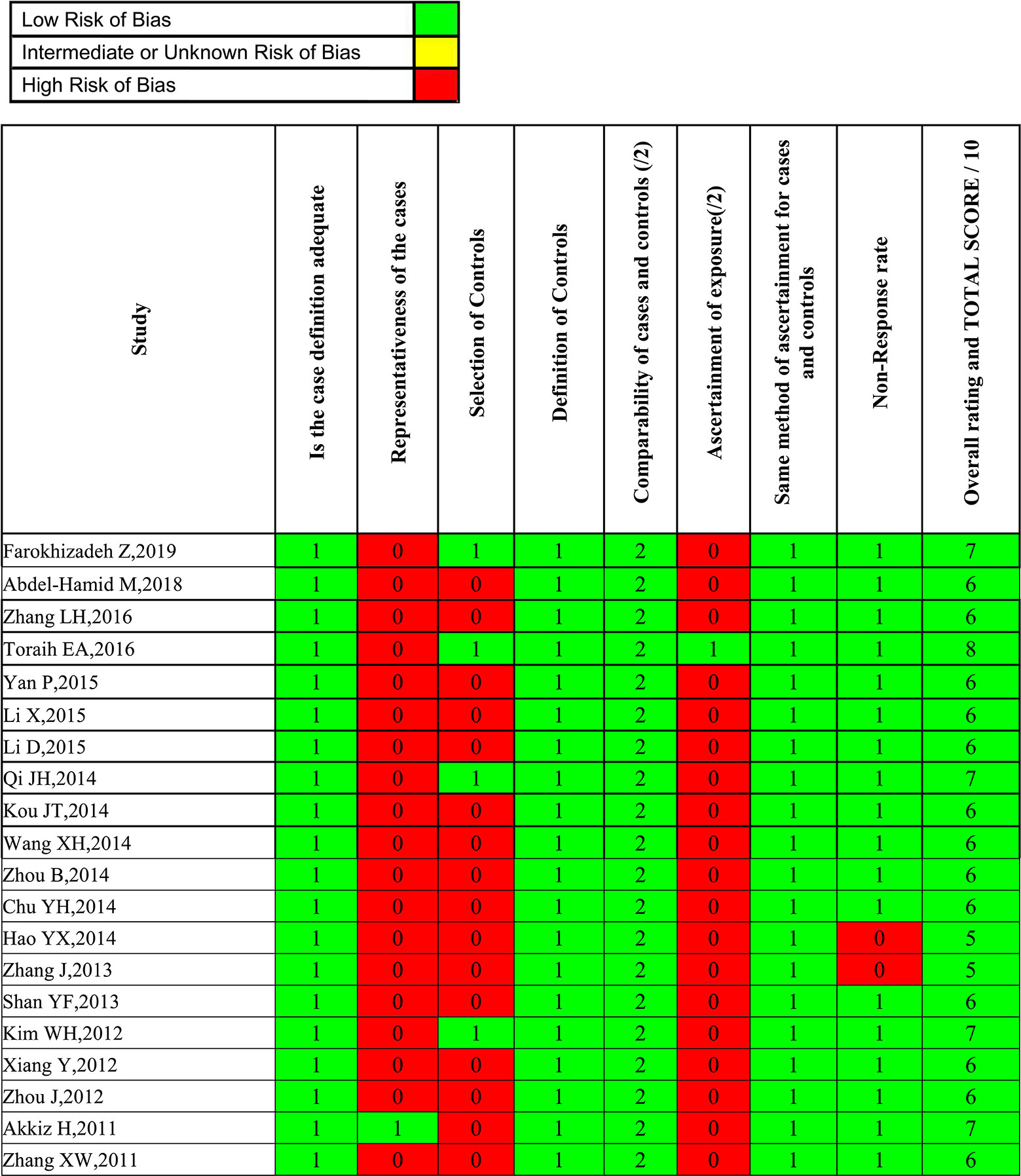
Two investigators independently abstracted the data on the studies. Discrepancies were resolved by consensus, referring back to the original study, or consulting a third reviewer. Besides genotype and frequency, the following data were also extracted from original studies: first author, year of publication, country, ethnicity, genotyping method, study design, case-control matching, sample size (cases/controls), and HBV infection status. To reduce the risk of bias due to individual studies, the Newcastle-Ottawa scale (NOS) score was applied to evaluate the methodological quality. The scale assesses three domains (selection bias, group comparability, and cohort exposure), based on “yes” or “no” answers to the following questions: (1) Is the case definition adequate; (2) Is there representativeness of the cases; (3) Is there selection of controls; (4) Is there a definition of controls; (5) Is there comparability of cases and controls; (6) Is there ascertainment of exposure; (7) Is the same method of ascertainment used for cases and controls; (8) Is there a non-response rate. The total score of NOS ranges from 0 to 9. A systematic analysis of the included studies was performed, and those with scores less than 5 were excluded. Two investigators independently performed the risk of bias assessments, with disagreement resolved by a third researcher when needed.
Statistical Analysis
A traditional pairwise meta-analysis was performed to estimate pooled odds ratios (ORs) and 95% confidence intervals (CIs) incorporating heterogeneity within and between studies. Statistical heterogeneity between each study was assessed with using the Chi-square tests and the inconsistency index I-square, with the values of 25%, 50%, and 75% denoting low, moderate and high heterogeneity, respectively. A random effects model was applied when I2 are over 50% (30). We went a step further and analyzed whether different genotypes might predispose to HCC under different HBV infection status. Meta-regression analysis was performed on the basis of the ethnicity, HWE, case-control match, and sample size to assess the heterogeneity that may have influences on the association between miRNA polymorphisms and HCC. The Begg’ s and Egger’ s tests were conducted to detect potential publication bias. Calculations and plotting were implemented by STATA 14.0 software.
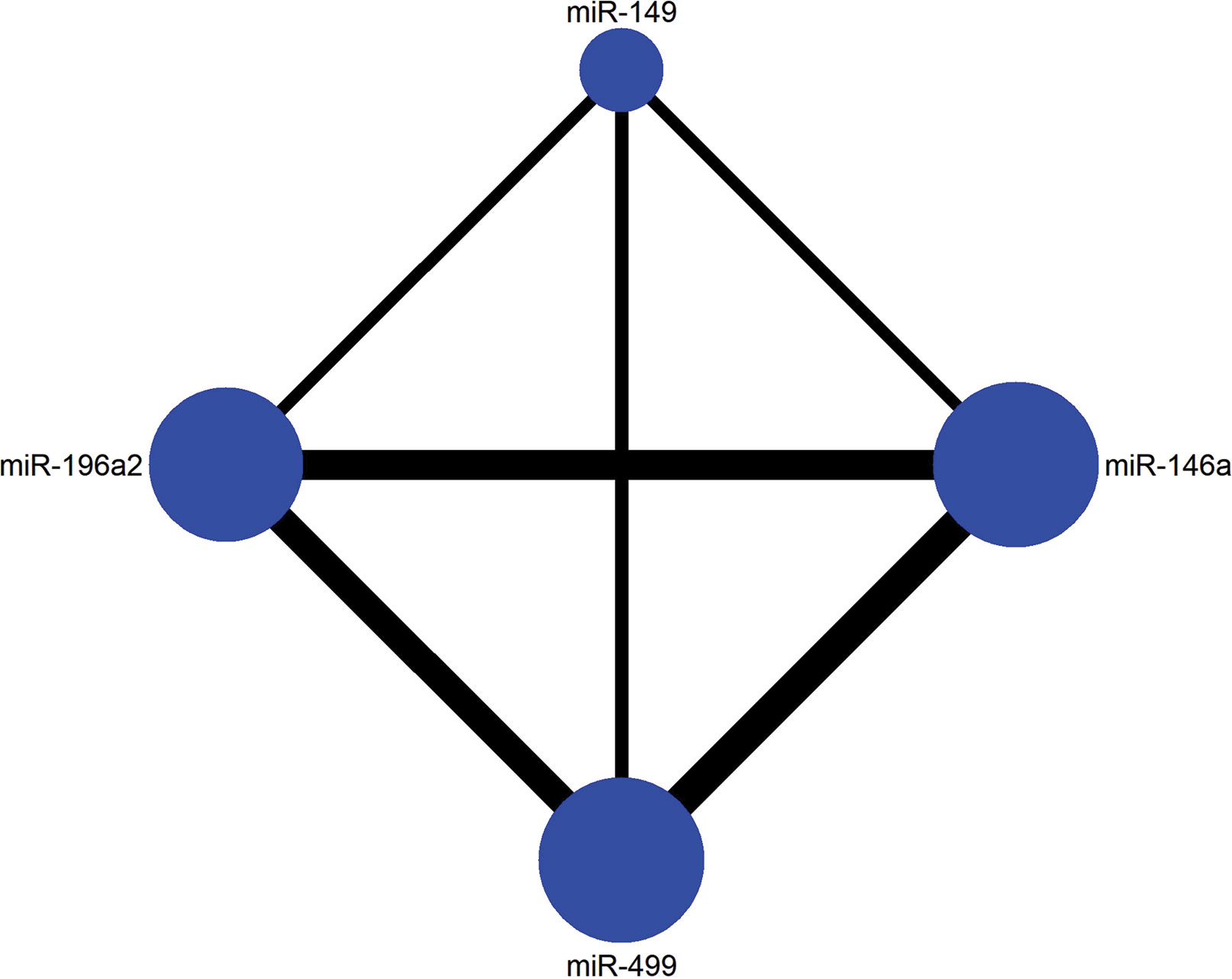
To further clarify the effect between four polymorphisms of miRNA on HCC, we conducted a Bayesian network meta-analysis. First a network plot depicting the connection within four SNPs was drawn. Every SNP was represented by a node, and the node size represented the number of studies of a corresponding SNP, the line thickness between two nodes represented the number of paired studies. Then, the analysis of variance (ANOVA) model was applied to combine the effective sizes of direct and indirect comparisons. The ability to rank interventions is an attractive feature of NMA compared to traditional analysis. The superiority index was calculated to rank competing polymorphisms. The superiority index ranges from 0 to ∞, which tends toward ∞ as the genetic model has a higher likelihood of predicting the risk of HCC and tends toward 1 indicating equal effect. Calculations were performed by R version 3.6.1 and STATA 14.0 was used to assist graphical functions.
Results
Characteristics and Bias of Enrolled Studies
Overall, 985 citations were identified using the search strategy. Among them, 269 citations were duplicates and 623 were excluded due to inappropriate tumor, functional studies, meta-analysis, reviews after assessing titles and abstracts. In the remaining 93 articles, there are two unhealthy controls, seven lack of sufficient data, 62 articles of irrelevant polymorphisms were removed. In Akkiz’s study, three articles investigating the same population and separately reporting three miRNA genes were considered as one. Therefore, 20 studies were enrolled providing a total of 5,337 HCC cases and 6,585 controls (Figure 1). The publication date of enrolled studies was from 2011 to 2019. The publications were mostly conducted in Asia, and two from Africa. In terms of ethnicity, 16 of the studies had Asian subjects and four studies had Caucasian subjects. Characteristics of included studies were presented in Table 1. The assessments of study quality were presented in Figure 2, and the NOS scale score result showed that all included studies had an acceptable quality.

Figure 1 PRISMA Flow diagram of the literature during the review process for the systematic review and meta-analysis.
Pairwise Meta-Analysis
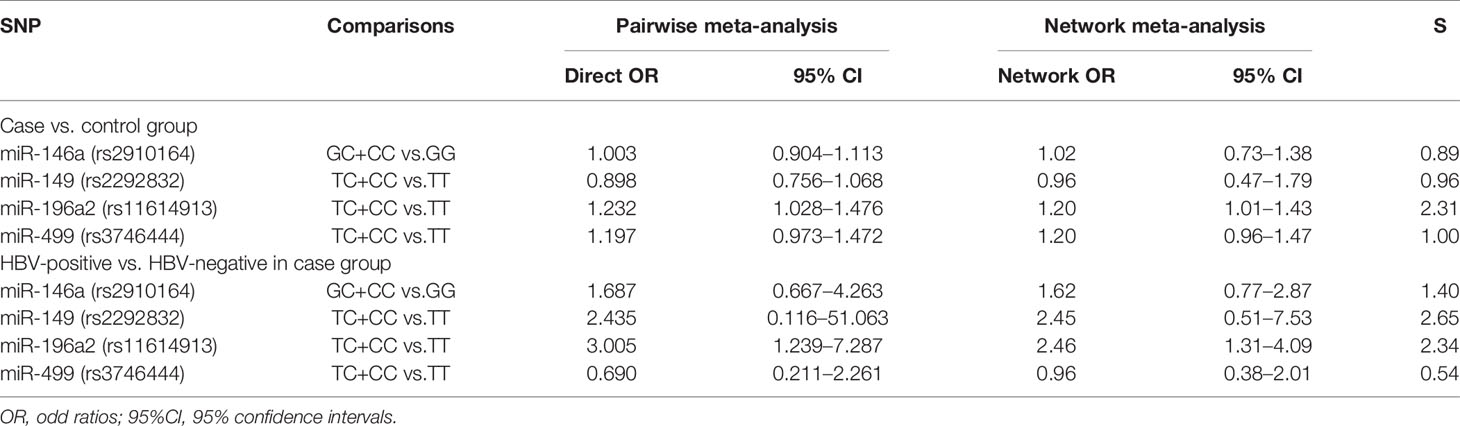
The forest plots of four miRNAs and their involvement in HCC susceptibility and relationship with HBV were explored and compared in Figures 3 and 4, respectively. The results indicated that miR-196a2 rs11614913 polymorphism was significantly associated with the susceptibility of HCC (miR-196a2 rs11614913: TC+CC vs. TT: OR=1.232, 95%CI=1.028–1.476), while the other three SNPs were not found to have a significant association (miR-146a rs2910164: GC+CC vs. GG: OR=1.003, 95%CI=0.904–1.113; miR-149 rs2292832: TC+CC vs. TT: OR=0.898, 95%CI=0.756–1.068; miR-499 rs3746444: TC+CC vs. TT: OR=1.197, 95%CI= 0.973–1.472, respectively). In the analysis of HCC patients under different HBV infection status, only miR-196a2 rs11614913 revealed significant correlation of threefold risk (miR-146a rs2910164: GC+CC vs. GG: OR=1.687, 95%CI=0.667–4.263; miR-149 rs2292832: TC+CC vs. TT: OR=2.435, 95%CI=0.116–51.063; miR-196a2 rs11614913: TC+CC vs. TT: OR=3.005, 95%CI=1.239–7.287; miR-499 rs3746444: TC+CC vs. TT: OR=0.690, 95%CI= 0.211–2.261, respectively). The results of meta-regression demonstrated that no overall significant heterogeneity was found in ethnicity, case-control match, and whether genotype distribution of controls was consistent with HWE or the sample size larger than 500 (Table 2). The results of Begg’s and Egger’ s tests were shown in Table 3, with the symmetrical distribution of effect sizes inside the Begg’s funnel plots (Figure 5), suggesting no significant publication bias among the included studies. Meta-regression and publication bias on miR-149 rs2292832 was not performed on account of insufficient studies.
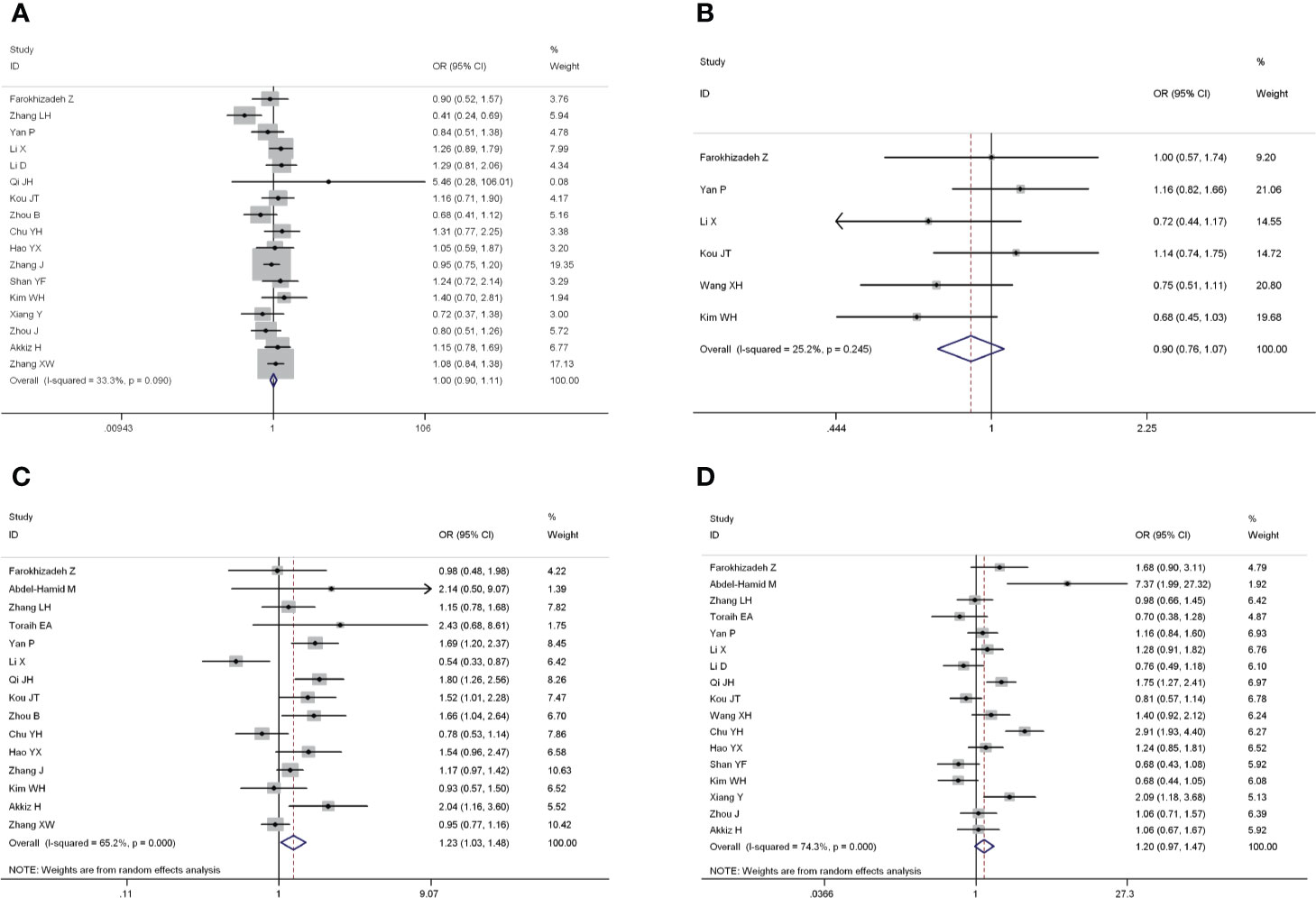
Figure 3 Forest plots of the association of four single nucleotide polymorphisms (SNPs) and hepatocellular carcinoma (HCC) risk. (A) miR-146a rs2910164; (B) miR-149 rs2292832; (C) miR-196a2 rs11614913; (D) miR-499 rs3746444.
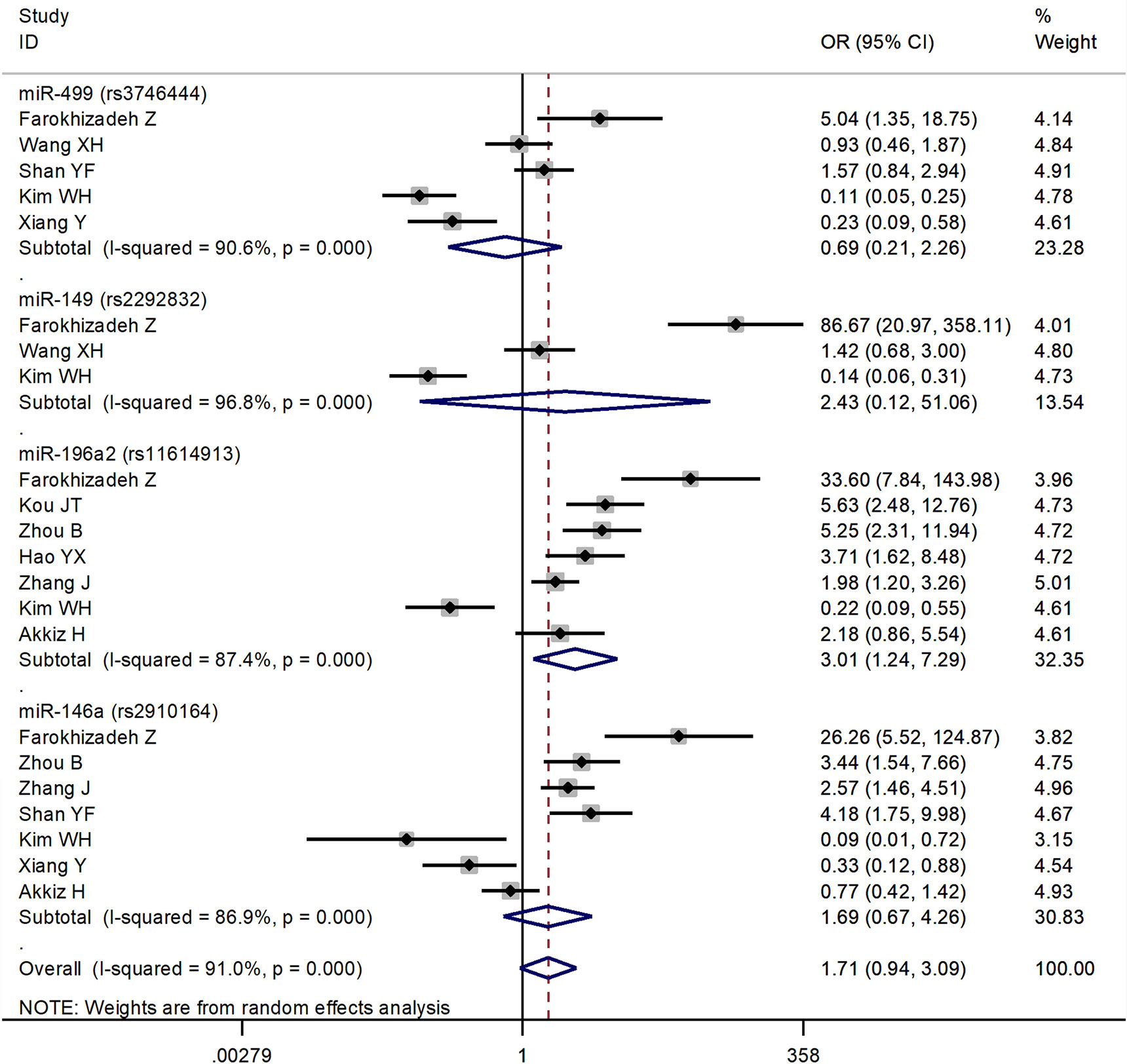
Figure 4 Forest plots of the association of four single nucleotide polymorphisms (SNPs) under different hepatitis B virus (HBV) infection status in hepatocellular carcinoma (HCC) patients.
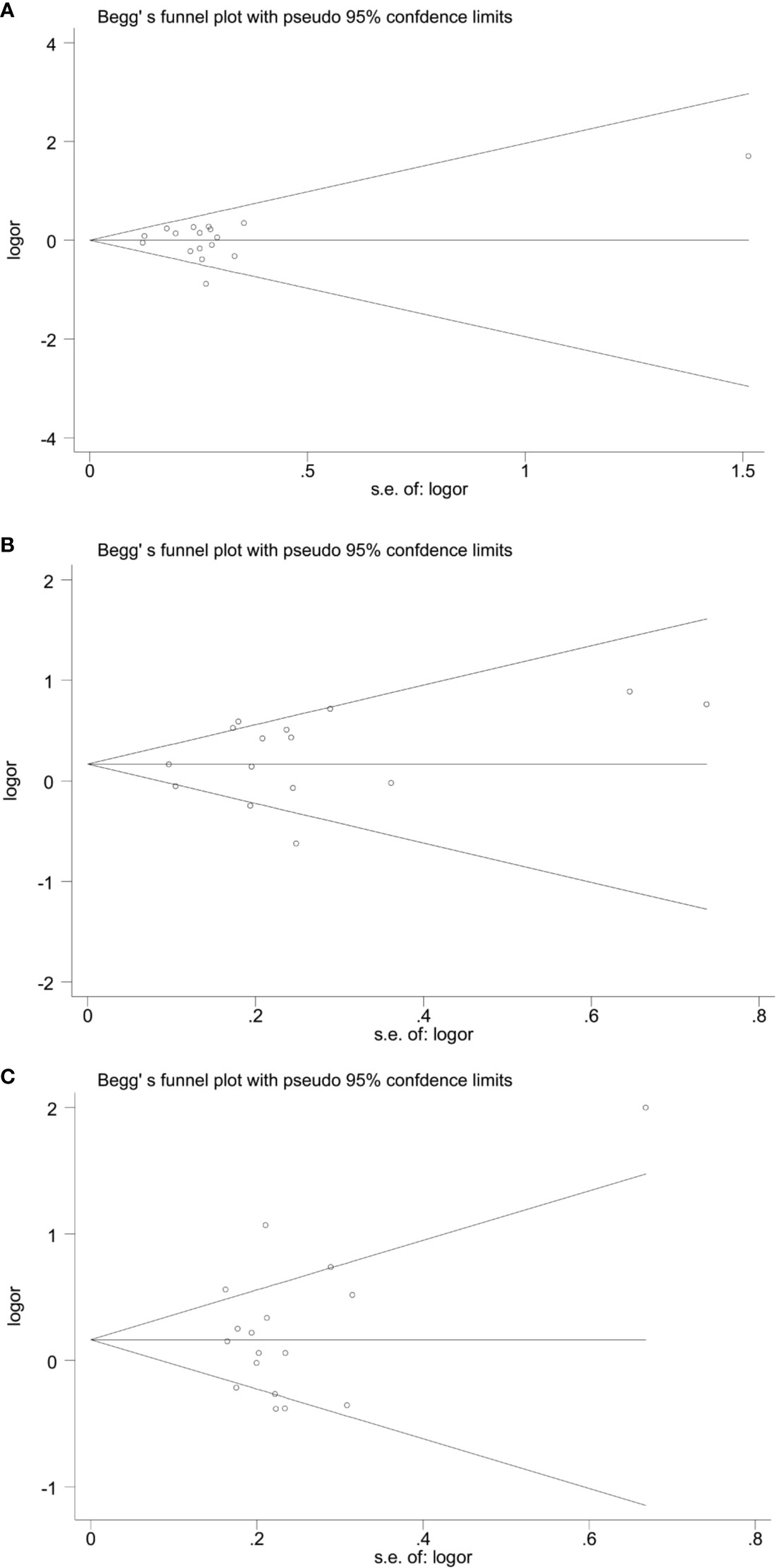
Figure 5 Begg’ funnel plots of publication bias. (A) miR-146a rs2910164; (B) miR-196a2 rs11614913; (C) miR-499 rs3746444.
Network Meta-Analysis
The current study contained four SNPs: miR-146a rs2910164, miR-149 rs2292832, miR-196a2 rs11614913, miR-499 rs3746444. It was observed from the network evidence that the number of direct comparisons of miR-146a vs. miR-499 was the largest, followed by miR-146a vs. miR-196a2, miR-196a2 vs. miR-499 (Figure 6). The predictive value of pairwise and network results of four miRNAs were explored and compared in Table 4. The NMA results showed no significant difference in the distribution of genotype frequencies except for miR-196a2 rs11614913, which appeared to have the highest superiority index when comparing and ranking four SNPs, further suggesting that it could be an effective indicator of the occurrence of HCC.
Discussion
MiRNAs play an important role in gene regulation of diseases (53), and have been proved to be tumor-suppressor genes as wells as oncogenes (54, 55). The dysregualtion of miRNA and its associated gene expression are involved in the occurrence and prognosis of HCC (56). The discovery of polymorphisms in miRNA genes has potential as new biomarkers for early diagnosis and prognosis in high-risk population, opening up new prospects for individualized treatment of HCC (57).
Of the 20 studies included in our research, the vast majority came from Asia, only two from Africa. Despite the rising incidence in North America and Europe, no enrolled studies came from either continent. The incidence of HCC varies widely within geographic locations. It is more common in low- and middle-income countries than in developed countries (58). It’s worth noting that, HCC incidence rates have been increasing in the United States, Europe and other developed areas (59). Obesity, smoking, diabetes, alcoholic cirrhosis and non-alcoholic steatosis are main causes of the increasing incidence of HCC (60–62). Currently, studies on the relationship between polymorphisms of miRNA and HCC are still lacking in relatively low-incidence areas. Our study illustrated the need for multi-ethnic, large-sample case-control studies that include data from a broad range of ethnic groups to obtain more stable and reliable results.
The pairwise results indicated that among the polymorphisms of miR-146a rs2910164, miR-149 rs2292832, miR-196a2 rs11614913, miR-499 rs3746444, only miR-196a2 was significantly associated with the susceptibility of HCC. When compared with TT genotype, CT or TT genotype in miR-196a2 carried a 1.232-fold increased risk of HCC. The network results were consistent with the direct results, with slight difference which was acceptable, indicating that our network evidence were robust. When comparing and ranking four SNPs, miR-196a2 rs11614913 appeared to have the highest superiority index. All of the above might come to the conclusion that miR-196a2 could serve as the best predictor of susceptibility in HCC.
HBV infection has been well established as one of the leading causes for the carcinogenesis of HCC (63). When comparing HBV-positive with HBV-negative HCC patients, a significant 3-fold increase in the frequencies of TC+CC versus TT was observed in miR-196a2 rs11614913. This indicated that the miR-196a2 rs11614913 polymorphism could be associated with the risk of HBV-related HCC. There have been studies that miRNAs could be involved in the development of HBV-related HCC. Previous reports have indicated that compared to normal liver, miRNA expression profiles were altered in chronic hepatitis B tissues (64). Wang et al. speculated that cellular miRNAs might function in HBV-related HCC, which affected HBV gene expression by binding to HBV transcripts or targeted cellular transcriptions factors that were necessary for HCC development (65). HBV infection could affect miRNA expression and contribute to enhanced viral replication and pathogenesis, and could ultimately lead to HCC (66).
MiR-196a2 rs11614913 is reported to be an important SNP associated with the etiology, progression and prognosis of several kinds of cancer. MiR-196a2 is located in the 3’passenger strand mature sequence of miR-196a2 (67), whose C to T mutation results in a G:C mutation to a G:U mismatch, leading to a decrease in the processing efficiency of the precursor of miRNAs to its mature form and ability to regulate target genes (68). The impacted expression level of the mature miR-196a2 can lead to genetic susceptibility and affect the survival of certain types of tumor. A number of studies has supported the proposition that the polymorphism of miR-196a2 rs11614913 may contribute to the susceptibility of several cancers (69–72). In the updated meta-analysis of Liu et al. (69), the link between miR-196a2 rs11614913 and a variety of cancers was explored and found that it was associated with HCC and lung cancer susceptibility. Hoffman’s research suggested that miR-196a2 had potential carcinogenic effects during the development of breast cancer (70). Hu et al. provided evidence that miR-196a2 variant homozygote was associated with a 1.76-fold-elevated HR, which was unfavorable to the overall survival of non-small cell lung cancer (71). In a case-control study conducted by Dikaiakos et al. (72), no significant association was found between miRNA-196a2 and colorectal cancer. Research has shown that the C allele of miRNA-196a2 increased the expression of mature miRNA-196a2 in HCC tissues (73). It is biologically plausible that miR-196a2 rs11614913 polymorphism may contribute to genetic susceptibility of HCC.
Although our results indicated that only miR-196a2 was associated with the susceptibility of HCC, and there was not enough evidence to support the association in miR-146, miR-149 or miR-499, the negative results still could not be ignored and should be interpreted cautiously. This is due to the occurrence of HCC is the result of multiple factors, in addition to complex genetic factors, there are hepatitis, aflatoxin exposure, Hepatitis C virus infection and other factors (74–76). Instead the incidence of HCC, gene variation may only cause increased susceptibility in a certain extent (77). Geography and ethnicity also need to be taken into account. Differences in populations are an important consideration in genetic association studies which may lead to inconsistent outcomes and difficulties in repetition (78, 79).
There are some limitations in our study. Only published studies were included, and those studies with negative results that could not be published were likely to be omitted, leading to incomplete studies. Secondly, selection bias could exist and impact on the results since the control group in most studies are hospital-based rather than population-based. Finally, although we found that miR-196a2 could be a potential indicator, how this might predispose to HCC are unclear and further functional studies are needed to clarify the mechanisms.
Conclusion
In conclusion, we found that the genetic polymorphism of miR-196a2 rs11614913 is significantly associated with the occurrence of HCC, especially for HBV- related HCC, and that individuals with TC/CC allele were more susceptible. No significant association was found in miR-146a rs2910164, miR-149 rs2292832, or miR-499 rs3746444. Our work could provide important information on the relationship between these four miRNAs and the susceptibility of HCC, suggesting potential novel diagnostic options. This would contribute to the reduction of mortality through early screening and diagnosis and improve the efficacy in HCC management.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary materials. Further inquiries can be directed to the corresponding author.
Author Contributions
QZ conducted the design of study, extracted the data, performed statistical analysis, and wrote the initial manuscript after consultation with the other authors. XX and HL improved the design, revised the manuscript, and approved the final version. TQ and SW checked the preliminary data. MW participated in the quality assessment adjudication. All authors contributed to the article and approved the submitted version.
Funding
This study was partly supported by the Social Sciences Foundation of Liaoning Province (No. L18ATJ001).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Rimassa L, Danesi R, Pressiani T, Merle P. Management of adverse events associated with tyrosine kinase inhibitors: Improving outcomes for patients with hepatocellular carcinoma. Cancer Treat Rev (2019) 77:20–8. doi: 10.1016/j.ctrv.2019.05.004
2. Saini A, Wallace A, Alzubaidi S, Knuttinen MG, Naidu S, Sheth R, et al. History and Evolution of Yttrium-90 Radioembolization for Hepatocellular Carcinoma. J Clin Med (2019) 8(1):55. doi: 10.3390/jcm8010055
3. Tang A, Hallouch O, Chernyak V, Kamaya A, Sirlin CB. Epidemiology of hepatocellular carcinoma: target population for surveillance and diagnosis. Abdominal Radiol (New York) (2018) 43:13–25. doi: 10.1007/s00261-017-1209-1
4. El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology (2012) 142:1264–73.e1261. doi: 10.1053/j.gastro.2011.12.061
5. Roayaie S, Llovet JM. Liver transplantation for hepatocellular carcinoma: is expansion of criteria justified? Clin Liver Dis (2005) 9:315–28. doi: 10.1016/j.cld.2004.12.007
6. Kulik L, El-Serag HB. Epidemiology and Management of Hepatocellular Carcinoma. Gastroenterology (2019) 156:477–91.e471. doi: 10.1053/j.gastro.2018.08.065
7. White DL, Thrift AP, Kanwal F, Davila J, El-Serag HB. Incidence of Hepatocellular Carcinoma in All 50 United States, From 2000 Through 2012. Gastroenterology (2017) 152:812–20.e815. doi: 10.1053/j.gastro.2016.11.020
8. Altekruse SF, McGlynn KA, Reichman ME. Hepatocellular carcinoma incidence, mortality, and survival trends in the United States from 1975 to 2005. J Clin Oncol Off J Am Soc Clin Oncol (2009) 27:1485–91. doi: 10.1200/jco.2008.20.7753
9. Blachier M, Leleu H, Peck-Radosavljevic M, Valla DC, Roudot-Thoraval F. The burden of liver disease in Europe: a review of available epidemiological data. J Hepatol (2013) 58:593–608. doi: 10.1016/j.jhep.2012.12.005
10. Yoshida H, Mamada Y, Taniai N, Uchida E. Spontaneous ruptured hepatocellular carcinoma. Hepatol Res Off J Japan Soc Hepatol (2016) 46:13–21. doi: 10.1111/hepr.12498
11. Raoul JL, Forner A, Bolondi L, Cheung TT, Kloeckner R, de Baere T. Updated use of TACE for hepatocellular carcinoma treatment: How and when to use it based on clinical evidence. Cancer Treat Rev (2019) 72:28–36. doi: 10.1016/j.ctrv.2018.11.002
12. Nakamoto Y. Promising new strategies for hepatocellular carcinoma. Hepatol Res Off J Japan Soc Hepatol (2017) 47:251–65. doi: 10.1111/hepr.12795
13. Sartorius K, Makarova J, Sartorius B, An P, Winkler C, Chuturgoon A, et al. The Regulatory Role of MicroRNA in Hepatitis-B Virus-Associated Hepatocellular Carcinoma (HBV-HCC) Pathogenesis. Cells (2019) 8:1504. doi: 10.3390/cells8121504
14. Dhanasekaran R, Nault JC, Roberts LR, Zucman-Rossi J. Genomic Medicine and Implications for Hepatocellular Carcinoma Prevention and Therapy. Gastroenterology (2019) 156:492–509. doi: 10.1053/j.gastro.2018.11.001
15. Cardon LR, Bell JI. Association study designs for complex diseases. Nat Rev Genet (2001) 2:91–9. doi: 10.1038/35052543
16. Dong LM, Potter JD, White E, Ulrich CM, Cardon LR, Peters U. Genetic susceptibility to cancer: the role of polymorphisms in candidate genes. Jama (2008) 299:2423–36. doi: 10.1001/jama.299.20.2423
17. Hirotsu Y, Zheng TH, Amemiya K, Mochizuki H, Guleng B, Omata M. Targeted and exome sequencing identified somatic mutations in hepatocellular carcinoma. Hepatol Res Off J Japan Soc Hepatol (2016) 46:1145–51. doi: 10.1111/hepr.12663
18. Xie KL, Zhang YG, Liu J, Zeng Y, Wu H. MicroRNAs associated with HBV infection and HBV-related HCC. Theranostics (2014) 4:1176–92. doi: 10.7150/thno.8715
19. Morishita A, Masaki T. miRNA in hepatocellular carcinoma. Hepatol Res Off J Japan Soc Hepatol (2015) 45:128–41. doi: 10.1111/hepr.12386
20. Zheng L, Zhuang C, Zhao J, Ming L. Functional miR-146a, miR-149, miR-196a2 and miR-499 polymorphisms and the susceptibility to hepatocellular carcinoma: An updated meta-analysis. Clin Res Hepatol Gastroenterol (2017) 41:664–76. doi: 10.1016/j.clinre.2017.03.005
21. Hashemi M, Moradi N, Ziaee SA, Narouie B, Soltani MH, Rezaei M, et al. Association between single nucleotide polymorphism in miR-499, miR-196a2, miR-146a and miR-149 and prostate cancer risk in a sample of Iranian population. J Adv Res (2016) 7:491–8. doi: 10.1016/j.jare.2016.03.008
22. Li CX, Weng H, Zheng J, Feng ZH, Ou JL, Liao WJ. Association Between MicroRNAs Polymorphisms and Risk of Ischemic Stroke: A Meta-Analysis in Chinese Individuals. Front Aging Neurosci (2018) 10:82. doi: 10.3389/fnagi.2018.00082
23. Guan X, Sturgis EM, Song X, Liu Z, El-Naggar AK, Wei Q, et al. Pre-microRNA variants predict HPV16-positive tumors and survival in patients with squamous cell carcinoma of the oropharynx. Cancer Lett (2013) 330:233–40. doi: 10.1016/j.canlet.2012.11.048
24. Du M, Lu D, Wang Q, Chu H, Tong N, Pan X, et al. Genetic variations in microRNAs and the risk and survival of renal cell cancer. Carcinogenesis (2014) 35:1629–35. doi: 10.1093/carcin/bgu082
25. Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer (2006) 6:259–69. doi: 10.1038/nrc1840
26. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer (2006) 6:857–66. doi: 10.1038/nrc1997
27. Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat Genet (2003) 33:177–82. doi: 10.1038/ng1071
28. Higgins JP, Welton NJ. Network meta-analysis: a norm for comparative effectiveness? Lancet (London England) (2015) 386:628–30. doi: 10.1016/s0140-6736(15)61478-7
29. Winter C, Kosch R, Ludlow M, Osterhaus A, Jung K. Network meta-analysis correlates with analysis of merged independent transcriptome expression data. BMC Bioinf (2019) 20:144. doi: 10.1186/s12859-019-2705-9
30. Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ (Clin Res ed) (2003) 327:557–60. doi: 10.1136/bmj.327.7414.557
31. Farokhizadeh Z, Dehbidi S, Geramizadeh B, Yaghobi R, Malekhosseini SA, Behmanesh M, et al. Association of MicroRNA Polymorphisms With Hepatocellular Carcinoma in an Iranian Population. Ann Lab Med (2019) 39:58–66. doi: 10.3343/alm.2019.39.1.58
32. Abdel-Hamid M, Elshaer S, Darwish A. Association of MicroRNA related single nucleotide polymorphisms 196A-2 and 499 with the risk of hepatocellular carcinoma in Egyptian patients. Meta Gene (2018) 16:139–42. doi: 10.1016/j.mgene.2018.02.007
33. Zhang LH, Hao BB, Zhang CY, Dai XZ, Zhang F. Contributions of polymorphisms in miR146a, miR196a, and miR499 to the development of hepatocellular carcinoma. Genet Mol Res GMR (2016) 15:399–407. doi: 10.4238/gmr.15038582
34. Toraih EA, Fawz MS, Elgazzaz MG, Hussein MH, Shehata RH, Daoud HG. Combined Genotype Analyses of Precursor miRNA196a2 and 499a Variants with Hepatic and Renal Cancer Susceptibility a Preliminary Study. Asian Pac J Cancer Prev (2016) 17:3369–75. doi: 10.14456/apjcp.2016.102
35. Yan P, Xia M, Gao F, Tang G, Zeng H, Yang S, et al. Predictive role of miR-146a rs2910164 (C>G), miR-149 rs2292832 (T>C), miR-196a2 rs11614913 (T>C) and miR-499 rs3746444 (T>C) in the development of hepatocellular carcinoma. Int J Clin Exp Pathol (2015) 8:15177–83.
36. Li X, Li K, Wu Z. Association of four common SNPs in microRNA polymorphisms with the risk of hepatocellular carcinoma. Int J Clin Exp Pathol (2015) 8:9560–6.
37. Li D, Peng JJ, Tan Y, Chen T, Wei D, Du M, et al. Genetic variations in microRNA genes and susceptibility to hepatocellular carcinoma. Genet Mol Res GMR (2015) 14:1926–31. doi: 10.4238/2015.March.20.2
38. Qi JH, Wang J, Chen J, Shen F, Huang JT, Sen S, et al. High-resolution melting analysis reveals genetic polymorphisms in microRNAs confer hepatocellular carcinoma risk in Chinese patients. BMC Cancer (2014) 14:643. doi: 10.1186/1471-2407-14-643
39. Kou JT, Fan H, Han D, Li L, Li P, Zhu J, et al. Association between four common microRNA polymorphisms and the risk of hepatocellular carcinoma and HBV infection. Oncol Lett (2014) 8:1255–60. doi: 10.3892/ol.2014.2257
40. Wang XH, Wang FR, Tang YF, Zou HZ, Zhao YQ. Association of miR-149C>T and miR-499A>G polymorphisms with the risk of hepatocellular carcinoma in the Chinese population. Genet Mol Res GMR (2014) 13:5048–54. doi: 10.4238/2014.July.4.20
41. Zhou B, Dong LP, Jing XY, Li JS, Yang SJ, Wang JP, et al. Association between miR-146aG>C and miR-196a2C>T polymorphisms and the risk of hepatocellular carcinoma in a Chinese population. J Korean Med Sci (2014) 35:7775–80. doi: 10.1007/s13277-014-2020-z
42. Chu YH, Hsieh MJ, Chiou HL, Liou YS, Yang CC, Yang SF, et al. MicroRNA gene polymorphisms and environmental factors increase patient susceptibility to hepatocellular carcinoma. PLoS One (2014) 9:e89930. doi: 10.1371/journal.pone.0089930
43. Hao YX, Wang JP, Zhao LF. Associations between three common MicroRNA polymorphisms and hepatocellular carcinoma risk in Chinese. Asian Pac J Cancer Prev (2014) 14:6601–4. doi: 10.7314/APJCP.2013.14.11.6601
44. Zhang J, Wang R, Ma YY, Chen LQ, Jin BH, Yu H, et al. Association between single nucleotide polymorphisms in miRNA196a-2 and miRNA146a and susceptibility to hepatocellular carcinoma in a Chinese population. Asian Pac J Cancer Prev (2013) 14:6427–31. doi: 10.7314/APJCP.2013.14.11.6427
45. Shan YF, Huang YH, Chen ZK, Huang KT, Zhou MT, Shi HQ, et al. miR-499A>G rs3746444 and miR-146aG>C expression and hepatocellular carcinoma risk in the Chinese population. Genet Mol Res GMR (2013) 12:5365–71. doi: 10.4238/2013.November.7.11
46. Kim WH, Min KT, Jeon YJ, Kwon CI, Ko KH, Park PW, et al. Association study of microRNA polymorphisms with hepatocellular carcinoma in Korean population. Gene (2012) 504:92–7. doi: 10.1016/j.gene.2012.05.014
47. Xiang Y, Fan S, Cao J, Huang S, Zhang LP. Association of the microRNA-499 variants with susceptibility to hepatocellular carcinoma in a Chinese population. Mol Biol Rep (2012) 39:7019–23. doi: 10.1007/s11033-012-1532-0
48. Zhou J, Lv R, Song X, Li D, Hu X, Ying B, et al. Association between two genetic variants in miRNA and primary liver cancer risk in the Chinese population. DNA Cell Biol (2012) 31:524–30. doi: 10.1089/dna.2011.1340
49. Akkiz H, Bayram S, Bekar A, Akgollu E, Uskudar O, Sandikci M. No association of pre-microRNA-146a rs2910164 polymorphism and risk of hepatocellular carcinoma development in Turkish population: a case-control study. Gene (2011) 486:104–9. doi: 10.1016/j.gene.2011.07.006
50. Akkiz H, Bayram S, Bekar A, Akgollu E, Uskudar O. Genetic variation in the microRNA-499 gene and hepatocellular carcinoma risk in a Turkish population: lack of any association in a case-control study. Asian Pac J Cancer Prev (2011) 12:3107–12.
51. Akkiz H, Bayram S, Bekar A, Akgollu E, Ulger Y. A functional polymorphism in pre-microRNA-196a-2 contributes to the susceptibility of hepatocellular carcinoma in a Turkish population: a case-control study. J Viral Hepat (2011) 18:e399–407. doi: 10.1111/j.1365-2893.2010.01414.x
52. Zhang XW, Pan SD, Feng YL, Liu JB, Dong J, Zhang YX, et al. [Relationship between genetic polymorphism in microRNAs precursor and genetic predisposition of hepatocellular carcinoma]. Zhonghua Yu Fang Yi Xue Za Zhi (2011) 45:239–43.
53. Morishita A, Masaki T. MicroRNAs as possible biomarkers for hepatocellular carcinoma. Hepatol Res Off J Japan Soc Hepatol (2018) 48:499–501. doi: 10.1111/hepr.13078
54. Volny O, Kasickova L, Coufalova D, Cimflova P, Novak J. microRNAs in Cerebrovascular Disease. Adv Exp Med Biol (2015) 888:155–95. doi: 10.1007/978-3-319-22671-2_9
55. Ma YY, Tao HQ. Microribonucleic acids and gastric cancer. Cancer Sci (2012) 103:620–5. doi: 10.1111/j.1349-7006.2011.02185.x
56. Yin W, Zhao Y, Ji YJ, Tong LP, Liu Y, He SX, et al. Serum/plasma microRNAs as biomarkers for HBV-related hepatocellular carcinoma in China. BioMed Res Int (2015) 2015:965185. doi: 10.1155/2015/965185
57. Wang Y, Tian Y. miRNA for diagnosis and clinical implications of human hepatocellular carcinoma. Hepatol Res Off J Japan Soc Hepatol (2016) 46:89–99. doi: 10.1111/hepr.12571
58. Baumeister SE, Schlesinger S, Aleksandrova K, Jochem C, Jenab M, Gunter MJ, et al. Association between physical activity and risk of hepatobiliary cancers: A multinational cohort study. J Hepatol (2019) 70:885–92. doi: 10.1016/j.jhep.2018.12.014
59. El-Serag HB, Davila JA, Petersen NJ, McGlynn KA. The continuing increase in the incidence of hepatocellular carcinoma in the United States: an update. Ann Internal Medicine (2003) 139:817–23. doi: 10.7326/0003-4819-139-10-200311180-00009
60. Akinyemiju T, Abera S, Ahmed M, Alam N, Alemayohu MA, Allen C, et al. The Burden of Primary Liver Cancer and Underlying Etiologies From 1990 to 2015 at the Global, Regional, and National Level: Results From the Global Burden of Disease Study 2015. JAMA Oncol (2017) 3:1683–91. doi: 10.1001/jamaoncol.2017.3055
61. Reeves HL, Zaki MY, Day CP. Hepatocellular Carcinoma in Obesity, Type 2 Diabetes, and NAFLD. Dig Dis Sci (2016) 61:1234–45. doi: 10.1007/s10620-016-4085-6
62. Saran U, Humar B, Kolly P, Dufour JF. Hepatocellular carcinoma and lifestyles. J Hepatol (2016) 64:203–14. doi: 10.1016/j.jhep.2015.08.028
63. Yao L, Zhou Y, Sui Z, Zhang Y, Liu Y, Xie H, et al. HBV-encoded miR-2 functions as an oncogene by downregulating TRIM35 but upregulating RAN in liver cancer cells. EBioMedicine (2019) 48:117–29. doi: 10.1016/j.ebiom.2019.09.012
64. Wakasugi H, Takahashi H, Niinuma T, Kitajima H, Oikawa R, Matsumoto N, et al. Dysregulation of miRNA in chronic hepatitis B is associated with hepatocellular carcinoma risk after nucleos(t)ide analogue treatment. Cancer Lett (2018) 434:91–100. doi: 10.1016/j.canlet.2018.07.019
65. Wang G, Dong F, Xu Z, Sharma S, Hu X, Chen D, et al. MicroRNA profile in HBV-induced infection and hepatocellular carcinoma. BMC Cancer (2017) 17:805–5. doi: 10.1186/s12885-017-3816-1
66. Naito Y, Hamada-Tsutsumi S, Yamamoto Y, Kogure A, Yoshioka Y, Watashi K, et al. Screening of microRNAs for a repressor of hepatitis B virus replication. Oncotarget (2018) 9:29857–68. doi: 10.18632/oncotarget.25557
67. Hussein MH, Toraih EA, Aly NM, Riad E, Fawzy MS. A passenger strand variant in miR-196a2 contributes to asthma severity in children and adolescents: A preliminary study. Biochem Cell Biol = Biochimie Biologie Cellulaire (2016) 94:347–57. doi: 10.1139/bcb-2016-0010
68. Chen C, Zhang Y, Zhang L, Weakley SM, Yao Q. MicroRNA-196: critical roles and clinical applications in development and cancer. J Cell Mol Med (2011) 15:14–23. doi: 10.1111/j.1582-4934.2010.01219.x
69. Liu Y, He A, Liu B, Zhong Y, Liao X, Yang J, et al. rs11614913 polymorphism in miRNA-196a2 and cancer risk: an updated meta-analysis. Onco Targets Ther (2018) 11:1121–39. doi: 10.2147/ott.s154211
70. Hoffman AE, Zheng T, Yi C, Leaderer D, Weidhaas J, Slack F, et al. microRNA miR-196a-2 and breast cancer: a genetic and epigenetic association study and functional analysis. Cancer Res (2009) 69:5970–7. doi: 10.1158/0008-5472.can-09-0236
71. Hu Z, Chen J, Tian T, Zhou X, Gu H, Xu L, et al. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J Clin Invest (2008) 118:2600–8. doi: 10.1172/jci34934
72. Dikaiakos P, Gazouli M, Rizos S, Zografos G, Theodoropoulos GE. Evaluation of genetic variants in miRNAs in patients with colorectal cancer. Cancer Biomarkers Section A Dis Markers (2015) 15:157–62. doi: 10.3233/cbm-140449
73. Yu JY, Hu F, Du W, Ma XL, Yuan K. Study of the association between five polymorphisms and risk of hepatocellular carcinoma: A meta-analysis. J Chin Med Assoc JCMA (2017) 80:191–203. doi: 10.1016/j.jcma.2016.09.009
74. Fatima N, Akhtar T, Sheikh N. Prebiotics: A Novel Approach to Treat Hepatocellular Carcinoma. Can J Gastroenterol Hepatol (2017) 2017:6238106. doi: 10.1155/2017/6238106
75. Marengo A, Rosso C, Bugianesi E. Liver Cancer: Connections with Obesity, Fatty Liver, and Cirrhosis. Annu Rev Medicine (2016) 67:103–17. doi: 10.1146/annurev-med-090514-013832
76. Chen Y, Wang X, Wang J, Yan Z, Luo J. Excess body weight and the risk of primary liver cancer: an updated meta-analysis of prospective studies. Eur J Cancer (2012) 48:2137–45. doi: 10.1016/j.ejca.2012.02.063
77. Perez-Losada J, Castellanos-Martin A, Mao JH. Cancer evolution and individual susceptibility. Integr Biol Quantitative Biosci nano to macro (2011) 3:316–28. doi: 10.1039/c0ib00094a
78. Colhoun HM, McKeigue PM, Davey Smith G. Problems of reporting genetic associations with complex outcomes. Lancet (London England) (2003) 361:865–72. doi: 10.1016/s0140-6736(03)12715-8
Keywords: hepatocellular carcinoma, microRNA, polymorphism, network meta-analysis, susceptibility, hepatitis B virus
Citation: Zhang Q, Xu X, Wu M, Qin T, Wu S and Liu H (2021) MiRNA Polymorphisms and Hepatocellular Carcinoma Susceptibility: A Systematic Review and Network Meta-Analysis. Front. Oncol. 10:562019. doi: 10.3389/fonc.2020.562019
Received: 14 May 2020; Accepted: 03 December 2020;
Published: 19 January 2021.
Edited by:
Rohini Mehta, BioReliance, United StatesReviewed by:
Amit Gupta, All India Institute of Medical Sciences, Rishikesh, IndiaAnjali Devi, JSS Academy of Higher Education and Research, India
Copyright © 2021 Zhang, Xu, Wu, Qin, Wu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongbo Liu, aGJsaXVAY211LmVkdS5jbg==
 Qimeng Zhang
Qimeng Zhang Xueying Xu
Xueying Xu Hongbo Liu
Hongbo Liu