- 1Department of Pathology, The Shengjing Hospital of China Medical University, Shenyang, China
- 2Department of Orthopedics, The Shengjing Hospital of China Medical University, Shenyang, China
Circular RNAs (circRNAs) are endogenous noncoding RNAs that are widely found in eukaryotic cells. They have been found to play a vital biological role in the development of human diseases. At present, circRNAs have been involved in the pathogenesis, diagnosis, and targeted treatment of multiple tumors. This article reviews the research progress of circRNAs in osteosarcoma (OSA) in recent years. The potential connection between circRNAs and OSA cell proliferation, apoptosis, metastasis, and chemotherapy sensitivity or resistance, as well as clinical values, is described in this review. Their categories and functions are generally summarized to facilitate a better understanding of OSA pathogenesis, and findings suggest novel circRNA-based methods may be used to investigate OSA and provide an outlook for viable biomarkers and therapeutic targets.
Introduction
Osteosarcoma (OSA) is the most common primary malignant bone tumor, accounting for 20% of all bone tumors and >5% of pediatric malignant tumors, with the highest incidence among children, adolescents, and the elderly (>50 years old) (1). The 5-year survival rate for nonmetastatic OSAs is 50–70%, but for metastatic OSAs (most commonly in the lungs), it is only 15–30% (2). So far, the pathogenesis and development of OSA remain unclear, and OSA treatment is still dominated by surgery and chemotherapy. For various reasons, outcomes for patients with OSA have not improved significantly in recent years mainly due to resistance of OSA cells to chemotherapy drugs (3). Therefore, further study into OSA pathogenesis is urgently needed, alongside the development of new and effective treatment regimens.
Less than 2% of the human genome's nucleic acid sequences encode proteins, and most genes are transcribed into noncoding RNA (ncRNA) (4). In recent years, circular RNA (circRNA) has become another research hit of ncRNA, following microRNA (miRNA) and long noncoding RNA (lncRNA). In 1976, Sanger et al. (5) and Kolakofsky (6) successfully discovered the existence of circRNA in plant viroids and sendai viruses. Electron microscopy was used in 1979 to clearly observe the circular structure of the circRNA in the cytoplasm of eukaryotic cells (7). However, at that time, circRNAs were regarded as abnormal RNA formed by the incorrect splicing of exon transcripts and, therefore, did not attract attention. In the 1990s, Nigro et al. (8) revealed that eukaryote protein-coding genes could form mature linear mRNA molecules. In addition, they discovered a special kind of reverse splicing reaction (back-splicing), which makes the exons sequence upstream and downstream (exon reverse cyclization), which eventually form a single closed loop structure with covalent bond connection (8). Rapid advances in RNA sequencing and bioinformatics have made large-scale analysis of transcriptome data a reality, and circRNAs have been found in a wide range of eukaryotes including humans, zebrafish, and fruit flies. Without A 3' end, 5' end, and poly A tail structure, circRNAs can escape the shear action of nucleic acid exonuclease such as RNase R, which is evolutionary conservative and stable than linear RNA, and more than 400 circRNAs can be detected even in human saliva (9–11). In addition, circRNAs also show cell specificity, tissue specificity, and sequence specificity (12). Studies on the biological functions of circRNAs are still at the exploratory stage. It has been found that circRNAs can act as competing endogenous RNAs, namely, miRNA sponges, to regulate the expression of target genes and can also act as transcription regulators or RNA-binding proteins to indirectly regulate genes at the posttranscriptional level (13, 14). Under certain conditions, circRNAs can even translate proteins directly (15). The main mechanisms of circRNA in tumor pathogenesis are illustrated in Figure 1.
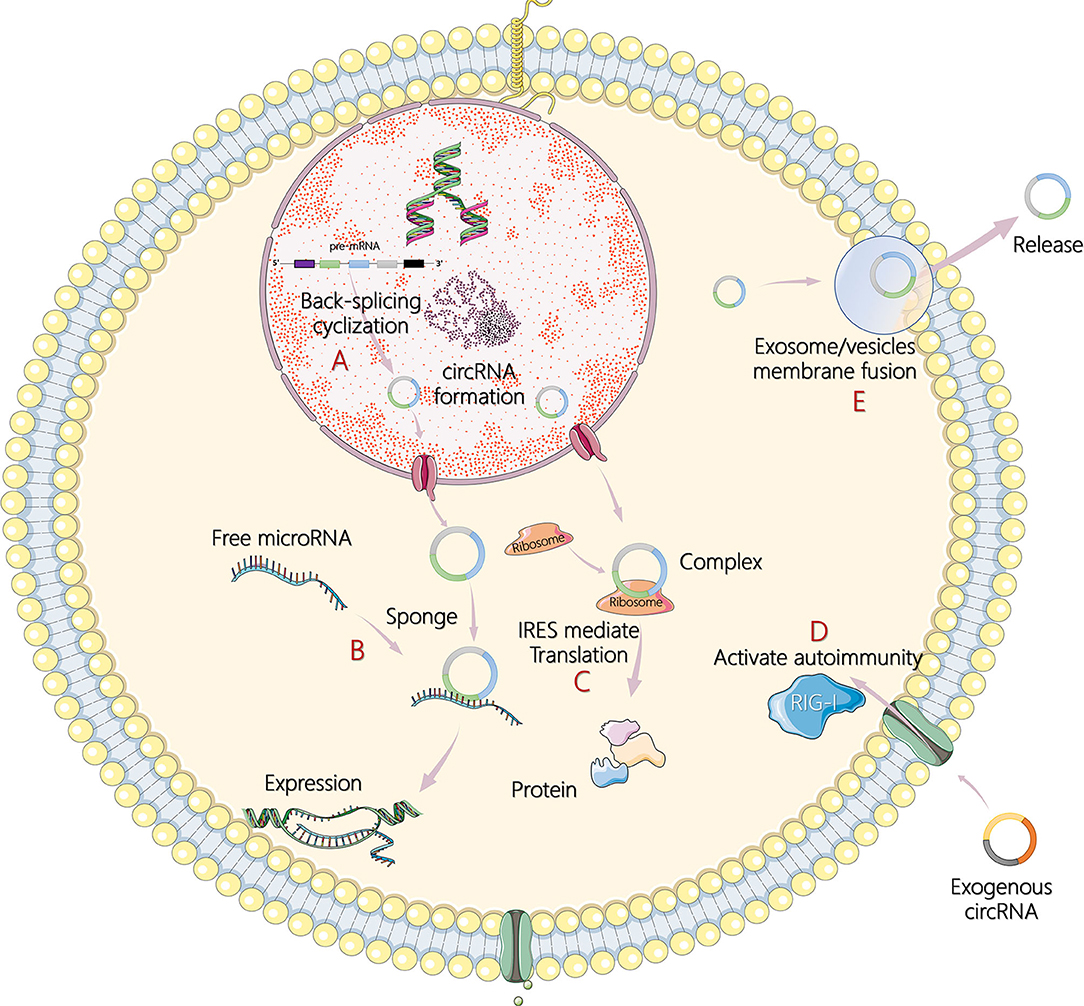
Figure 1. The main mechanisms of circular RNA (circRNA) in tumor pathogenesis. (A) Formation process of circRNA by back-splicing cyclization in cells. (B) CircRNAs act as sponges of microRNAs (miRNAs) and subsequently affect target gene expression. (C) CircRNAs encode itself by translation of proteins in an internal ribosome entry site (IRES)-mediated way. (D) Recognition of exogenous circRNAs activates retinoic acid inducible gene I (RIG-I)-mediated autoimmunity. (E) CircRNAs are transported outside cells to regulate responses through exosomes or extracellular vesicles by membrane fusion and release.
Recently, researchers have made many new advances in multidisciplinary fields by using second-generation sequencing technology and gene chip analysis to screen differential circRNAs and validate their biological functions. It is now recognized that modulation of circRNA levels can result in a variety of molecular and physiological phenotype changes in cells, including effects on miRNAs, innate immunity, and many disease-relevant pathways (16). However, their characteristics in OSA remain far less documented. In this review, we will discuss the cellular mechanisms of the circRNAs in OSA tumorigenesis and progression.
CircRNA and Cancer
CircRNAs have been studied in a variety of human tumor types, including those affecting the reproductive and urinary systems, digestive system, nervous system, respiratory system, etc. (17–21). Currently, the enormous potential for circRNA use in targeted therapy and as a noninvasive biomarker has garnered much attention from the broader scientific community (22). Numerous studies have shown that some circRNAs are involved in the pathogenesis of cancer and can be regarded as disease biomarkers or therapeutic targets (23). Noteworthy studies concerning the function of circRNAs in OSA pathogenesis and drug resistance have successively been reported, suggesting that they may provide valuable biomarkers for diagnosis or prognosis and show promise for the development of novel therapeutic approaches.
CircRNA and Osteosarcoma
To investigate the differentially expressed circRNAs in OSA, researchers usually conduct a circRNA microarray screening, an analysis based on OSA/paired adjacent normal tissue or OSA/normal osteoblast cell lines. These approaches allow the identification of novel circRNAs that may be involved in the biological process of OSA progression. CircRNA-targeted miRNAs and miRNA-targeted genes can also be screened by microarray or bioinformatics analysis performed by software such as circRNA Interactome, miRDB, and Targetscan, etc. (24–26). CircRNAs could act as miRNA sponges to compete with endogenous RNAs in regulating posttranscriptional levels of gene expression (27). Functional bioinformatics analysis was selected for further research followed by experimental validation (28).
In this review, we summarize the roles of circRNAs in the proliferation, apoptosis, metastasis, and chemotherapy sensitivity or resistance, as well as the prognosis of OSA. We also summarized the emerging OSA–circRNAs identified and collate circRNA symbols corresponding to circRNA ID in the circBase database. In addition, the possible mechanisms of action of circRNAs are characterized.
Regulation Mechanism of CircRNA on Osteosarcoma
Most aberrantly expressed circRNAs may serve as crucial regulators of OSA progression through the modulation of multifarious cancer hallmarks, functioning to deregulate cellular energetics, sustain proliferative signaling, promote invasion and metastasis, induce angiogenesis, and promote tumor immunology (29). Upregulated and downregulated circRNAs as well as their various mechanisms in OSA are represented in Tables 1, 2, respectively.
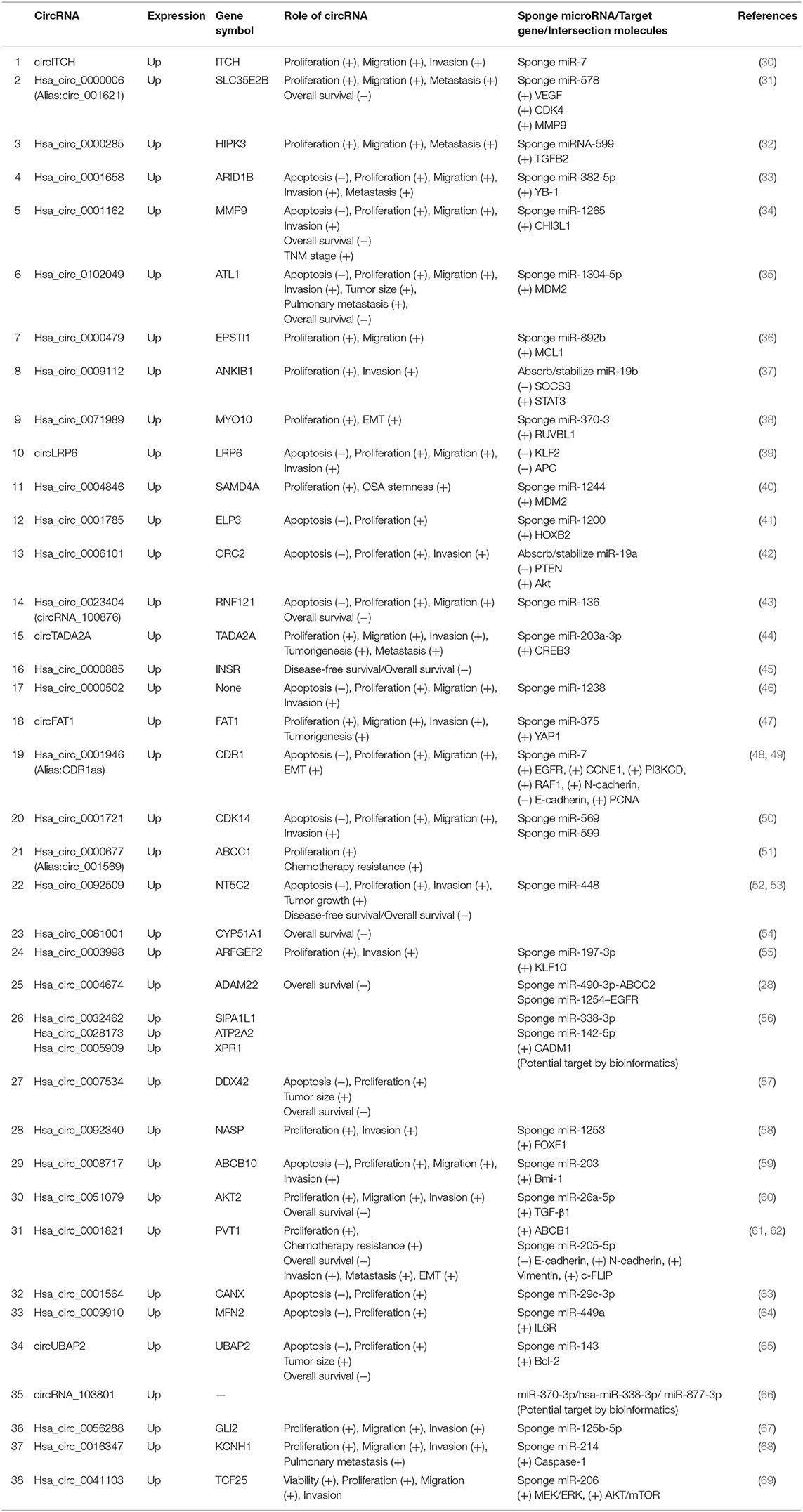
Table 1. Characterization of upregulated circular RNAs (circRNAs) as potential diagnostic biomarkers of osteosarcoma (OSA).
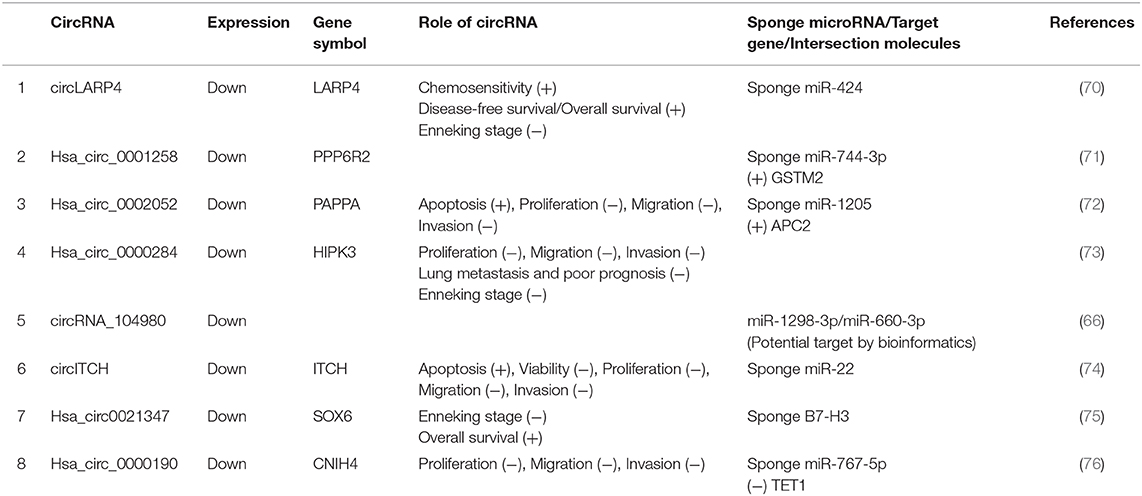
Table 2. Characterization of downregulated circular RNAs (circRNAs) as potential diagnostic biomarkers of osteosarcoma (OSA).
Onco-CircRNA (Upregulated CircRNA)
At present, most circRNA studies conducted are based on elevated circRNAs in OSA. Oncogenic circRNAs can participate in inducing the progression of OSA. Their oncogenic function involves promotion of cell proliferation, colony formation, migration, and invasion, as well as affecting the rate of apoptosis. Some circRNAs have also been identified as closely correlated with OSA prognosis. For example, compared with adjacent tissues, circ_0001658 displayed a significantly higher expression in OSA tissues. Hsa_circ_0001658 could impede apoptosis by sponging miR-382-5p and positively modulating Y-box binding protein 1 (YB-1) expression to facilitate the proliferation, migration, and invasion of OSA cells (33). Wu et al. (44) revealed that increased circTADA2A expression in OSA tissue and cells promotes the progression and metastasis of OSA by sponging miR-203a-3p and by targeting oncogene cyclic AMP-responsive element-binding protein 3 (CREB3), both functionally and mechanistically. CircRNAs shsa_circ_0032462, hsa_circ_0005909, and hsa_circ_0028173 were found to be overexpressed in human OSA and to promote cell adhesion molecule 1 (CADM1) expression by functioning as miRNA sponges (56). Similar regulatory mechanisms were found in other studies, such as for circCANX (63), hsa_circ_0009910 (64), and hsa_circ_0056288 (67).
Of note, a study conducted by Du et al. (37) found that circANKIB1 could play an absorbing role with miR-19b, and that both molecules were upregulated in OSA cells (77). This study further found that circANKIB1 promoted miR-19b expression through absorption, thereby inhibiting the expression of SOCS3, a downstream target gene, and activating the signal transducer and activator of transcription 3 (STAT3) pathway to promote OSA progression. Another similar study indicated that circORC2 could adsorb miR-19a to stabilize its inhibitory function on target gene phosphatase and tensin homolog (PTEN) expression and activate downstream Akt pathway (42). Currently, most studies have shown that circRNAs exert effects on target genes by competitive binding to miRNAs (78). However, circRNAs stabilize miRNA functions through adsorption and enhance regulation of target genes by reducing the degradation of miRNAs (79), which may represent a novel mechanism. These studies have provided a new research method based on circRNA–miRNA–target gene axis and demonstrate the potential of circRNAs as OSA-targeted therapies.
The Enneking surgical staging system has been used for classification of musculoskeletal tumors by surgeons around the world. It is characterized by reliability, reproducibility, and prognostic importance for musculoskeletal sarcomas, especially for those originating in the axial skeleton. Some circRNAs well reflected the stage of OSA, which indicated its clinicopathological features (80). Hsa_circ_0000885 expression was significantly increased in tissue and serum samples from patients with OSA compared with controls, and expression levels increased with Enneking stage IIB and III OSA compared with early-stage OSA. Receiver operating characteristic (ROC) curve analysis suggested that hsa_circ_0000885 may act as a good diagnostic biomarker for OSA (45).
Tumor-Suppressor CircRNA (Downregulated CircRNA)
CircRNAs can also act as tumor suppressants to inhibit tumor growth. The study of tumor-suppressor circRNA will provide a new direction for the diagnosis, treatment, and prognosis of OSA. In OSA, circRNA can act as a miRNA sponge to indirectly downregulate the expression of target genes, thus playing a role in cancer inhibition. For instance, underexpressed circRNA hsa_circ_0002052 was screened out and validated in OSA tissues and in OSA cells, which might be a potential therapeutic target for OSA intervention. Hsa_circ_0002052 suppressed Wnt/β-catenin activation by promoting APC2 expression through sponging miR-1205, which led to delayed OSA progression (72). McEachron et al. (81) and Wang et al. (82) concluded that hsa_circ0021347 was selected and validated to be significantly downregulated in OSA tissues, and cell lines showed a strong negative relationship with B7-H3, which served as a negative regulator of osteoimmunology, helping tumor cells escape immune surveillance. Furthermore, the hsa_circ0021347–miR-646-NOB1 axis was suggested to be involved in promoting tumor differentiation and invasion in OSA; however, this needs to be validated in future studies (75).
Research has found hsa_circ_0000190 to exhibit an obvious reduction in extracellular nanovesicles (EVs) and tissues of OSA patients. Most hsa_circ_0000190 was discovered to be encapsulated in EVs. EVs containing hsa_circ_0000190 in OSA cells transported from normal cells could block biological malignant behaviors by inhibiting the migration, proliferation, and invasion both in vitro and in vivo. In addition, EVs containing hsa_circ_0000190 might induce miR-767-5p to modulate TET1 and impede OSA progression. Li et al. (76) also offer a new concept for circRNA therapy based on cell–cell communication by packaging into circRNAs. Several other downregulated circRNAs, listed in Table 2, will be analyzed in details in the subsequent sections.
CircRNA and Osteosarcoma Metastasis
The metastasis of OSA depends on many pathological processes and regulation of cytokines. Epithelial-mesenchymal transition (EMT) refers to the transformation of epithelial cells into cells with mesenchymal phenotype. The main characteristic of EMT is that epithelial cells lose their original polarity but gain mesenchymal characteristics. The EMT biological process plays an important role in tumor metastasis, with tumor cells that lose the bond among cells being much more able to invade and metastasize (83). Chen et al. (38) showed that circMYO10 regulated EMT and activated Wnt/β-catenin signaling, thereby regulating the miR-370-3p/RUVBL1 axis to promote H4K16Ac at the promoter region of β-catenin/LEF1 target genes. It was also found that cerebellar degeneration-related protein 1 (CDR1) knockdown led to the inhibition of transforming growth factor-β (TGF-β)-induced EMT by upregulating the mesenchymal phenotype with increased N-cadherin and downregulated the epithelial phenotype with reduced E-cadherin (48, 49). EMT occurrence and development are always accompanied by multiple molecular interactions and signaling pathways, including TGF-β, E-cadherin, and Wnt/β-catenin signaling pathway, etc. (84). TGF-β is a crucial member of the TGF-β superfamily that is involved in EMT to regulate cell growth and differentiation (85). TGF-β1 was validated as a putative target of miR-26a-5p, which is bioinformatically analyzed to be sponged by circ_0051079 (60). The Wnt/β-catenin signaling pathway is one of the predisposing factors of EMT, which directs cancer cell migration, adhesion, invasion, and metastasis and is closely bound up with degradation of the extracellular matrix and tumor angiogenesis. This has been widely confirmed in many studies (38, 51, 72).
The ability of circRNA to affect tumor angiogenesis is usually directly related to tumor metastasis based on vascular endothelial growth factor (VEGF). Research suggests that circ_001621 augments the progression of OSA cells by abolishing the inhibition of VEGF by miR-578. The VEGF–CDK4–MMP9 axis was extended to be a novel VEGF-related pathway, which remained to be completely elucidated (31). Functional analysis by Liu et al. (66) found that circRNA_103801 as an miRNA sponge was involved in VEGF to promote tumor angiogenesis and tumor metastasis. Anoikis apoptosis is programmed cell death caused by the loss of contact between the extracellular matrix and other cells (86). Anoikis resistance was identified as a factor facilitating the progression of OSA, as metastatic OSA cells are able to colonize and survive in other sites (87). CircRNA has been found to play a role in anoikis resistance, involving Wnt pathway regulation (88). In addition, Bcl-2 and its related proteins as well as EMT processes are also involved in regulating anoikis (89). These findings are in line with studies on circUBAP2 and circ_0007534 (57, 65). Moreover, circ_0001785 showed a marked downregulation effect on antiapoptotic genes in Bcl-2 family (Bcl-W, Bcl-A1, and Bcl-2) and conversely upregulated the proapoptotic gene Bad (41).
CircRNA and Osteosarcoma Chemotherapy Resistance
Although existing targeted chemotherapies play a role in the treatment of OSA, the emergence of drug resistance causes OSA patients to fall into an impasse. Therefore, it is important to deeply understand the mechanism of drug resistance and open new therapeutic theories. It is evident that some circRNAs participate in regulating the mechanisms of drug resistance in OSA cells. In some studies, whole-transcriptome sequencing (RNA sequencing) and next-generation sequencing technologies were performed in paired multidrug chemoresistant and chemosensitive OSA samples. Circ_0001258 inhibited the doxorubicin (DXR) resistance of OSA cell lines through upregulation of glutathione S-transferase mu 2 (GSTM2) expression via sponging hsa-miR-744-3p (71). Overexpression of circ_0004674 has also been observed in chemoresistant OSA cell lines and OSA patients and is negatively correlated with prognosis (28). Furthermore, Hu et al. (70) suggested that circLARP4 might elevate chemosensitivity to cisplatin and DXR via sponging miR-424 in OSA, closely correlated with decreased Enneking stage, better histological response, and prolonged survival profiles. Kun-Peng et al. (61) reported that circPVT1 resensitizes OSA cells to the chemotherapy drugs DXR and cisplatin by reducing the expression of the classical multidrug resistance-related gene, ABCB1. CircPVT1 could also function as a sponge for miR-205-5p to promote c-FLIP expression, thereby enhancing EMT and inducing OSA invasion and metastasis (61, 62). Hsa_circ_001569 has also been shown to enhance cell resistance to cisplatin in OSA by activating Wnt/β-catenin signaling (51). Researchers found that hsa_circ_0081001 varied in paired chemosensitive and chemoresistant OSA cell lines. It was identified as significantly upregulated in OSA cell lines, tissues, and serums and was related to poor prognosis of OSA patients. The ROC curve analysis showed that it could be used as a promising biomarker and may be a better prognostic indicator than alkaline phosphatase (ALP) and lactate dehydrogenase (LDH) (54).
CircRNA and Osteosarcoma Prognosis
To determine the relationship between circRNA expression and prognosis in OSA, correlation analysis has shown that the expression of circRNA in OSA was related to certain prognostic factors, such as Enneking stage, tumor size, and the occurrence of distant metastasis. Meanwhile, the relationship between the expression of circRNAs and survival rates, including overall survival (OS) time and disease-free survival (DFS) time, was detected by Kaplan–Meier (KM) analysis.
Involvement of circRNAs in cancer progression has influenced the prognosis of OSA in many studies. Clinical pathologic characteristics of OSA patients and related literature were analyzed retrospectively. Xiao-Long et al. (75) found that circHIPK3 was consistently downregulated in OSA cell lines, tissues, and plasmas compared with control. Lung metastasis and advanced cancer or poor prognosis were negatively associated with lower expression levels of circHIPK3. CircHIPK3 may be used as a novel indicator for OSA with high degrees of sensitivity, specificity, and accuracy based on ROC curve analyses (73). Hsa_circ0021347 also showed the same prognostic trend (75). In contrast, research performed by Pan et al. (34) suggested that overexpression of circMMP9 was correlated with advanced tumor stage and predicted a low survival rate by KM analysis. Circ_0102049 was remarkably correlated with patients' poor OS analyzed by KM curves in a study of 76 OSA patients (35). Two studies jointly reported the role of circ-NT5C2 in OSA from the aspect of clinical and molecular biological mechanisms. Patients with a high expression of circ-NT5C2 had a shorter OS (p = 0.006) and DFS (p = 0.001) than those with a low expression of circ-NT5C2. High circ-NT5C2 expression was thought to be an independent prognostic parameter to predict poor prognosis by sponging miR-448 (52, 53). Additional upregulated circRNAs included circUBAP2 (65), hsa_circ_0000885 (45), hsa_circ_0081001 (54), hsa_circ_0004674 (28), hsa_circ_0000006 (31), hsa_circ_0051079 (60), hsa_circ_0007534 (57), and PVT1 (61).
Osteosarcoma-Related CircRNA in Other Cancers
With advancement of circRNA research, expression of different circRNAs was detected in normal and cancerous tissue. It is worth noting that some circRNAs show similarities and participate simultaneously in different tumor types, which is similar to the basic idea of pan-cancer (90). These circRNAs are of guiding significance on the progression of multiple cancer types.
Li et al. (30) showed that the expression of circ-ITCH in OSA cancer cell lines was significantly upregulated compared with hFOB1.19. Further mechanistic studies revealed that circ-ITCH could promote the growth, migration, and invasion of OSA cells and even enhance epidermal growth factor receptor expression by reducing levels of miR-7 (30). However, another study carried out by Ren et al. (74) stated the opposite result—that circ-ITCH had lower expression in OSA cells and was identified in clinical human OSA and para-tumor tissues. Their data showed that overexpression of circ-ITCH led to reduced SP-1 expression via PTEN/phosphoinositide 3-kinase (PI3K)/AKT pathways, which in turn suppressed proliferation, migration, and invasion by downregulating miR-22 (74). This is in concert with the conclusion that circ-ITCH might serve as an anti-oncogene via sponging multiple oncogenic miRNAs in multiple tumors, including ovarian cancer, prostate cancer, melanoma, gastric cancer, glioma, breast cancer, bladder cancer, papillary thyroid cancer, lung cancer, hepatocellular carcinoma, esophageal squamous cell carcinoma, and colorectal cancer (91, 92).
Hsa_circ-0000285 has also been acknowledged to be abundantly expressed in human cells and relevant to human multicancer progression (93, 94). Hsa_circ-0000285 was believed to regulate OSA by affecting the miRNA-599/TGFB2-axis (32). It was established that hsa_circ_0000285 might act as an oncogene in laryngocarcinoma. CircPVT1 seemed to be a potential candidate of oncogenic interest. Per available literature, so far, circPVT1 has been studied to induce malignancy of different tumors including non-small-cell lung carcinoma, gastric cancer, and acute lymphoblastic leukemia (95). Additionally, many other circRNAs have been shown to participate in a variety of tumors, as follows: hsa_circ_0000190, hsa_circ_0000285, circMMP9, circEPSTI1, circ-LARP4, hsa_circ_0001785, hsa_circ_100876, circTADA2A, hsa_circ_0000502, circFAT1, CDR1as, hsa_circ_001569, hsa_circ_0003998, hsa_circ_0007534, hsa_circ_0009910, and circUBAP2. These OSA-associated circRNAs exhibit tumor-regulatory characteristics and are expected to be biomarkers in the future.
Summary and Future Gazing
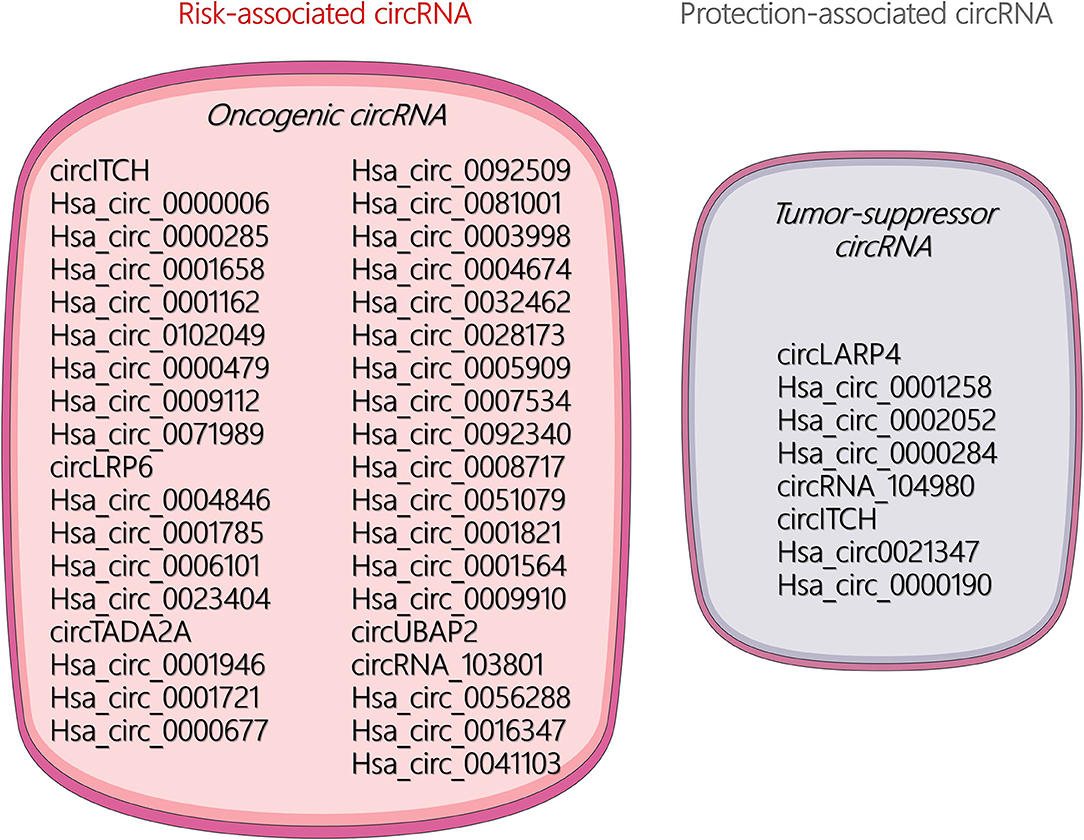
The unique circular structure of circRNA offers distinct functions and better stability than miRNA and lncRNA. Increasing research has found that circRNAs play a significant role in the occurrence, malignant progression, and metastasis in many tumors. Current research on circRNA in OSA has mainly focused on its role as an endogenous competitive RNA that acts as a molecular sponge to absorb miRNAs and thus affects the transcription of target genes. Available literature demonstrated that certain circRNAs can be regarded as risk- and protection-associated circRNAs for diagnosis of OSA characteristics, which are illustrated in Figure 2. In addition, some other biological mechanisms of circRNAs remain under investigation. For example, circRNAs may play multiple roles in the tumor microenvironment or act as noninvasive biomarkers for the early detection of cancers.
It is difficult to distinguish the source of the translation product of circRNAs because of protein-encoded exons overlapping. High-throughput analytical and detection methods such as ribosome profiling have technical challenges (96, 97). Researchers are also confronted with numerous challenges, such as difficulty in obtaining substantial tumor specimens because of the complex genetic background, extensive heterogeneity between or in tumor tissues, as well as the low morbidity. More efforts should be invested in the comprehensive evaluation of the mechanisms of circRNAs in OSA pathogenesis and chemoresistance in order to make them available for specific diagnosis and targeted gene therapy. With the research going on, we are convinced that circulating circRNAs might be used as canonical biomarkers for cancers in the future.
Author Contributions
JL, SL, and QF reviewed relevant literature and drafted the manuscript. JL, LY, and SL conducted all data collection. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Outstanding Scientific Fund of Shengjing Hospital (MD31) and the National Natural Science Foundation of China (no. 81370981), as well as the 345 Talent Project of Shengjing Hospital.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors want to show gratitude to the professors in the department who provided ideas for improvement in this project.
References
1. Coindre JM. [New WHO classification of tumours of soft tissue and bone]. Ann Pathol. (2012) 32(5 Suppl):S115–6. doi: 10.1002/cncr.26079
2. Ottaviani G, Jaffe N. The etiology of osteosarcoma. Cancer Treat Res. (2009) 152:15–32. doi: 10.1007/978-1-4419-0284-9_2
3. Ma W, Yang L, Liu H, Chen P, Ren H, Ren P. PAXX is a novel target to overcome resistance to doxorubicin and cisplatin in osteosarcoma. Biochem Biophys Res Commun. (2020) 521:204–11. doi: 10.1016/j.bbrc.2019.10.108
4. Consortium EP, Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. (2007) 447:799–816. doi: 10.1038/nature05874
5. Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA. (1976) 73:3852–6. doi: 10.1073/pnas.73.11.3852
6. Kolakofsky D. Isolation and characterization of Sendai virus DI-RNAs. Cell. (1976) 8:547–55. doi: 10.1016/0092-8674(76)90223-3
7. Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. (1979) 280:339–40. doi: 10.1038/280339a0
8. Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, et al. Scrambled exons. Cell. (1991) 64:607–13. doi: 10.1016/0092-8674(91)90244-S
9. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. (2013) 495:333–8. doi: 10.1038/nature11928
10. Bahn JH, Zhang Q, Li F, Chan TM, Lin X, Kim Y, et al. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem. (2015) 61:221–30. doi: 10.1373/clinchem.2014.230433
11. Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. (2015) 12:381–8. doi: 10.1080/15476286.2015.1020271
12. Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. (2013) 9:e1003777. doi: 10.1371/journal.pgen.1003777
13. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. (2013) 19:141–57. doi: 10.1261/rna.035667.112
14. Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. (2016) 44:2846–58. doi: 10.1093/nar/gkw027
15. Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, et al. Translation of CircRNAs. Mol Cell. (2017) 66:9–21 e27. doi: 10.1016/j.molcel.2017.02.021
16. Xiao MS, Ai Y, Wilusz JE. Biogenesis and functions of circular RNAs come into focus. Trends Cell Biol. (2020) 30:226–40. doi: 10.1016/j.tcb.2019.12.004
17. Su Y, Feng W, Shi J, Chen L, Huang J, Lin T. circRIP2 accelerates bladder cancer progression via miR-1305/Tgf-beta2/smad3 pathway. Mol Cancer. (2020) 19:23. doi: 10.1186/s12943-019-1129-5
18. Qian W, Huang T, Feng W. Circular RNA HIPK3 promotes emt of cervical cancer through sponging miR-338-3p to Up-Regulate HIF-1alpha. Cancer Manag Res. (2020) 12:177–87. doi: 10.2147/CMAR.S232235
19. Long X, Shi Y, Ye P, Guo J, Zhou Q, Tang Y. MicroRNA-99a Suppresses Breast Cancer Progression by Targeting FGFR3. Front Oncol. (2019) 9:1473. doi: 10.3389/fonc.2019.01473
20. Yao J, Xu G, Zhu L, Zheng H. circGFRA1 enhances NSCLC progression by sponging miR-188-3p. Onco Targets Ther. (2020) 13:549–58. doi: 10.2147/OTT.S230795
21. Wang N, Lu K, Qu H, Wang H, Chen Y, Shan T, et al. CircRBM33 regulates IL-6 to promote gastric cancer progression through targeting miR-149. Biomed Pharmacother. (2020) 125:109876. doi: 10.1016/j.biopha.2020.109876
22. Zhang Q, Wang W, Zhou Q, Chen C, Yuan W, Liu J, et al. Roles of circRNAs in the tumour microenvironment. Mol Cancer. (2020) 19:14. doi: 10.1186/s12943-019-1125-9
23. Liu J, Li D, Luo H, Zhu X. Circular RNAs: The star molecules in cancer. Mol Aspects Med. (2019) 70:141–52. doi: 10.1016/j.mam.2019.10.006
24. Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. (2016) 13:34–42. doi: 10.1080/15476286.2015.1128065
25. Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. (2015) 4:5. doi: 10.7554/eLife.05005
26. Chen Y, Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. (2020) 48:D127–D31. doi: 10.1093/nar/gkz757
27. Cui X, Wang J, Guo Z, Li M, Li M, Liu S, et al. Emerging function and potential diagnostic value of circular RNAs in cancer. Mol Cancer. (2018) 17:123. doi: 10.1186/s12943-018-0877-y
28. Kun-Peng Z, Xiao-Long M, Lei Z, Chun-Lin Z, Jian-Ping H, Tai-Cheng Z. Screening circular RNA related to chemotherapeutic resistance in osteosarcoma by RNA sequencing. Epigenomics. (2018) 10:1327–46. doi: 10.2217/epi-2018-0023
29. Bach DH, Lee SK, Sood AK. Circular RNAs in Cancer. Mol Ther Nucleic Acids. (2019) 16:118–29. doi: 10.1016/j.omtn.2019.02.005
30. Li H, Lan M, Liao X, Tang Z, Yang C. Circular RNA cir-ITCH promotes osteosarcoma migration and invasion through cir-ITCH/miR-7/EGFR pathway. Technol Cancer Res Treat. (2020) 19:1533033819898728. doi: 10.1177/1533033819898728
31. Ji X, Shan L, Shen P, He M. Circular RNA circ_001621 promotes osteosarcoma cells proliferation and migration by sponging miR-578 and regulating VEGF expression. Cell Death Dis. (2020) 11:18. doi: 10.1038/s41419-019-2204-y
32. Zhang Z, Pu F, Wang B, Wu Q, Liu J, Shao Z. Hsa_circ_0000285 functions as a competitive endogenous RNA to promote osteosarcoma progression by sponging hsa-miRNA-599. Gene Ther. (2019) doi: 10.1038/s41434-019-0112-5
33. Wang L, Wang P, Su X, Zhao B. Circ_0001658 promotes the proliferation and metastasis of osteosarcoma cells via regulating miR-382-5p/YB-1 axis. Cell Biochem Funct. (2020) 38:77–86. doi: 10.1002/cbf.3452
34. Pan G, Hu T, Chen X, Zhang C. Upregulation Of circMMP9 promotes osteosarcoma progression via targeting miR-1265/CHI3L1 axis. Cancer Manag Res. (2019) 11:9225–31. doi: 10.2147/CMAR.S226264
35. Jin Y, Li L, Zhu T, Liu G. Circular RNA circ_0102049 promotes cell progression as ceRNA to target MDM2 via sponging miR-1304-5p in osteosarcoma. Pathol Res Pract. (2019) 215:152688. doi: 10.1016/j.prp.2019.152688
36. Tan X, Tan D, Li H, Lin Y, Wen Z, Zeng C. circEPSTI1 acts as a ceRNA to regulate the progression of osteosarcoma. Curr Cancer Drug Targets. (2020) 20:288–94. doi: 10.2174/1568009619666191107140948
37. Du YX, Guo LX, Pan HS, Liang YM, Li X. Circ_ANKIB1 stabilizes the regulation of miR-19b on SOCS3/STAT3 pathway to promote osteosarcoma cell growth and invasion. Hum Cell. (2020) 33:252–260. doi: 10.1007/s13577-019-00298-6
38. Chen J, Liu G, Wu Y, Ma J, Wu H, Xie Z, et al. CircMYO10 promotes osteosarcoma progression by regulating miR-370-3p/RUVBL1 axis to enhance the transcriptional activity of beta-catenin/LEF1 complex via effects on chromatin remodeling. Mol Cancer. (2019) 18:150. doi: 10.1186/s12943-019-1076-1
39. Zheng S, Qian Z, Jiang F, Ge D, Tang J, Chen H, et al. CircRNA LRP6 promotes the development of osteosarcoma via negatively regulating KLF2 and APC levels. Am J Transl Res. (2019) 11:4126–38.
40. Yanbin Z, Jing Z. CircSAMD4A accelerates cell proliferation of osteosarcoma by sponging miR-1244 and regulating MDM2 mRNA expression. Biochem Biophys Res Commun. (2019) 516:102–11. doi: 10.1016/j.bbrc.2019.05.182
41. Li S, Pei Y, Wang W, Liu F, Zheng K, Zhang X. Circular RNA 0001785 regulates the pathogenesis of osteosarcoma as a ceRNA by sponging miR-1200 to upregulate HOXB2. Cell Cycle. (2019) 18:1281–91. doi: 10.1080/15384101.2019.1618127
42. Li X, Sun XH, Xu HY, Pan HS, Liu Y, He L. Circ_ORC2 enhances the regulatory effect of miR-19a on its target gene PTEN to affect osteosarcoma cell growth. Biochem Biophys Res Commun. (2019) 514:1172–8. doi: 10.1016/j.bbrc.2019.04.188
43. Jin J, Chen A, Qiu W, Chen Y, Li Q, Zhou X, et al. Dysregulated circRNA_100876 suppresses proliferation of osteosarcoma cancer cells by targeting microRNA-136. J Cell Biochem. (2019) 120:15678–87. doi: 10.1002/jcb.28837
44. Wu Y, Xie Z, Chen J, Chen J, Ni W, Ma Y, et al. Circular RNA circTADA2A promotes osteosarcoma progression and metastasis by sponging miR-203a-3p and regulating CREB3 expression. Mol Cancer. (2019) 18:73. doi: 10.1186/s12943-019-1007-1
45. Zhu K, Niu L, Wang J, Wang Y, Zhou J, Wang F, et al. Circular RNA hsa_circ_0000885 levels are increased in tissue and serum samples from patients with osteosarcoma. Med Sci Monit. (2019) 25:1499–505. doi: 10.12659/MSM.914899
46. Qi H, Sun Y, Jiang Y, Li X. Upregulation of circular RNA circ_0000502 predicts unfavorable prognosis in osteosarcoma and facilitates cell progression via sponging miR-1238. J Cell Biochem. (2018) doi: 10.1002/jcb.28134. [Epub ahead of print].
47. Liu G, Huang K, Jie Z, Wu Y, Chen J, Chen Z, et al. CircFAT1 sponges miR-375 to promote the expression of Yes-associated protein 1 in osteosarcoma cells. Mol Cancer. (2018) 17:170. doi: 10.1186/s12943-018-0917-7
48. Xu B, Yang T, Wang Z, Zhang Y, Liu S, Shen M. CircRNA CDR1as/miR-7 signals promote tumor growth of osteosarcoma with a potential therapeutic and diagnostic value. Cancer Manag Res. (2018) 10:4871–80. doi: 10.2147/CMAR.S178213
49. Erratum: CircRNA CDR1as/miR-7 signals promote tumor growth of osteosarcoma with a potential therapeutic and diagnostic value [Corrigendum]. Cancer Manag Res. (2018) 10:5867. doi: 10.2147/CMAR.S193081
50. Li L, Guo L, Yin G, Yu G, Zhao Y, Pan Y. Upregulation of circular RNA circ_0001721 predicts unfavorable prognosis in osteosarcoma and facilitates cell progression via sponging miR-569 and miR-599. Biomed Pharmacother. (2019) 109:226–32. doi: 10.1016/j.biopha.2018.10.072
51. Zhang H, Yan J, Lang X, Zhuang Y. Expression of circ_001569 is upregulated in osteosarcoma and promotes cell proliferation and cisplatin resistance by activating the Wnt/beta-catenin signaling pathway. Oncol Lett. (2018) 16:5856–62. doi: 10.3892/ol.2018.9410
52. Nie WB, Zhao LM, Guo R, Wang MX, Ye FG. Circular RNA circ-NT5C2 acts as a potential novel biomarker for prognosis of osteosarcoma. Eur Rev Med Pharmacol Sci. (2018) 22:6239–44. doi: 10.26355/eurrev_201810_16030
53. Liu X, Zhong Y, Li J, Shan A. Circular RNA circ-NT5C2 acts as an oncogene in osteosarcoma proliferation and metastasis through targeting miR-448. Oncotarget. (2017) 8:114829–38. doi: 10.18632/oncotarget.22162
54. Kun-Peng Z, Chun-Lin Z, Jian-Ping H, Lei Z. A novel circulating hsa_circ_0081001 act as a potential biomarker for diagnosis and prognosis of osteosarcoma. Int J Biol Sci. (2018) 14:1513–20. doi: 10.7150/ijbs.27523
55. Wang L, Du ZG, Huang H, Li FS, Li GS, Xu SN. Circ-0003998 promotes cell proliferative ability and invasiveness by binding to miR-197-3p in osteosarcoma. Eur Rev Med Pharmacol Sci. (2019) 23:10638–46. doi: 10.26355/eurrev_201912_19761
56. Chen G, Wang Q, Yang Q, Li Z, Du Z, Ren M, et al. Circular RNAs hsa_circ_0032462, hsa_circ_0028173, hsa_circ_0005909 are predicted to promote CADM1 expression by functioning as miRNAs sponge in human osteosarcoma. PLoS ONE. (2018) 13:e0202896. doi: 10.1371/journal.pone.0202896
57. Li B, Li X. Overexpression of hsa_circ_0007534 predicts unfavorable prognosis for osteosarcoma and regulates cell growth and apoptosis by affecting AKT/GSK-3beta signaling pathway. Biomed Pharmacother. (2018) 107:860–6. doi: 10.1016/j.biopha.2018.08.086
58. Jin P, Huang Y, Zhu P, Zou Y, Shao T, Wang O. CircRNA circHIPK3 serves as a prognostic marker to promote glioma progression by regulating miR-654/IGF2BP3 signaling. Biochem Biophys Res Commun. (2018) 503:1570–4. doi: 10.1016/j.bbrc.2018.07.081
59. Zhou X, Natino D, Qin Z, Wang D, Tian Z, Cai X, et al. Identification and functional characterization of circRNA-0008717 as an oncogene in osteosarcoma through sponging miR-203. Oncotarget. (2018) 9:22288–300. doi: 10.18632/oncotarget.23466
60. Zhang Z, Zhao M, Wang G. Hsa_circ_0051079 functions as an oncogene by regulating miR-26a-5p/TGF-beta1 in osteosarcoma. Cell Biosci. (2019) 9:94. doi: 10.1186/s13578-019-0355-2
61. Kun-Peng Z, Xiao-Long M, Chun-Lin Z. Overexpressed circPVT1, a potential new circular RNA biomarker, contributes to doxorubicin and cisplatin resistance of osteosarcoma cells by regulating ABCB1. Int J Biol Sci. (2018) 14:321–30. doi: 10.7150/ijbs.24360
62. Liu YP, Wan J, Long F, Tian J, Zhang C. circPVT1 Facilitates Invasion and Metastasis by Regulating miR-205-5p/c-FLIP Axis in Osteosarcoma. Cancer Manag Res. (2020) 12:1229–40. doi: 10.2147/CMAR.S231872
63. Song YZ, Li JF. Circular RNA hsa_circ_0001564 regulates osteosarcoma proliferation and apoptosis by acting miRNA sponge. Biochem Biophys Res Commun. (2018) 495:2369–75. doi: 10.1016/j.bbrc.2017.12.050
64. Deng N, Li L, Gao J, Zhou J, Wang Y, Wang C, et al. Hsa_circ_0009910 promotes carcinogenesis by promoting the expression of miR-449a target IL6R in osteosarcoma. Biochem Biophys Res Commun. (2018) 495:189–96. doi: 10.1016/j.bbrc.2017.11.028
65. Zhang H, Wang G, Ding C, Liu P, Wang R, Ding W, et al. Increased circular RNA UBAP2 acts as a sponge of miR-143 to promote osteosarcoma progression. Oncotarget. (2017) 8:61687–97. doi: 10.18632/oncotarget.18671
66. Liu W, Zhang J, Zou C, Xie X, Wang Y, Wang B, et al. Microarray expression profile and functional analysis of circular RNAs in osteosarcoma. Cell Physiol Biochem. (2017) 43:969–85. doi: 10.1159/000481650
67. Li JF, Song YZ. Circular RNA GLI2 promotes osteosarcoma cell proliferation, migration, and invasion by targeting miR-125b-5p. Tumour Biol. (2017) 39:1010428317709991. doi: 10.1177/1010428317709991
68. Jin H, Jin X, Zhang H, Wang W. Circular RNA hsa-circ-0016347 promotes proliferation, invasion and metastasis of osteosarcoma cells. Oncotarget. (2017) 8:25571–81. doi: 10.18632/oncotarget.16104
69. Wang Y, Shi S, Zhang Q, Dong H, Zhang J. MicroRNA-206 upregulation relieves circTCF25-induced osteosarcoma cell proliferation and migration. J Cell Physiol. (2020) doi: 10.1002/jcp.29570. [Epub ahead of print].
70. Hu Y, Gu J, Shen H, Shao T, Li S, Wang W, et al. Circular RNA LARP4 correlates with decreased Enneking stage, better histological response, and prolonged survival profiles, and it elevates chemosensitivity to cisplatin and doxorubicin via sponging microRNA-424 in osteosarcoma. J Clin Lab Anal. (2020) 34:e23045. doi: 10.1002/jcla.23045
71. Zhu KP, Zhang CL, Ma XL, Hu JP, Cai T, Zhang L. Analyzing the interactions of mRNAs and ncRNAs to predict competing endogenous RNA networks in osteosarcoma chemo-resistance. Mol Ther. (2019) 27:518–30. doi: 10.1016/j.ymthe.2019.01.001
72. Wu Z, Shi W, Jiang C. Overexpressing circular RNA hsa_circ_0002052 impairs osteosarcoma progression via inhibiting Wnt/beta-catenin pathway by regulating miR-1205/APC2 axis. Biochem Biophys Res Commun. (2018) 502:465–71. doi: 10.1016/j.bbrc.2018.05.184
73. Xiao-Long M, Kun-Peng Z, Chun-Lin Z. Circular RNA circ_HIPK3 is down-regulated and suppresses cell proliferation, migration and invasion in osteosarcoma. J Cancer. (2018) 9:1856–62. doi: 10.7150/jca.24619
74. Ren C, Liu J, Zheng B, Yan P, Sun Y, Yue B. The circular RNA circ-ITCH acts as a tumour suppressor in osteosarcoma via regulating miR-22. Artif Cells Nanomed Biotechnol. (2019) 47:3359–67. doi: 10.1080/21691401.2019.1649273
75. Wang L, Zhang GC, Kang FB, Zhang L, Zhang YZ. hsa_circ0021347 as a Potential Target Regulated by B7-H3 in Modulating the Malignant Characteristics of Osteosarcoma. Biomed Res Int. (2019) 2019:9301989. doi: 10.1155/2019/9301989
76. Li S, Pei Y, Wang W, Liu F, Zheng K, Zhang X. Extracellular nanovesicles-transmitted circular RNA has_circ_0000190 suppresses osteosarcoma progression. J Cell Mol Med. (2020) 24:2202–2214. doi: 10.1111/jcmm.14877
77. He C, Gao H, Fan X, Wang M, Liu W, Huang W, et al. Identification of a novel miRNA-target gene regulatory network in osteosarcoma by integrating transcriptome analysis. Int J Clin Exp Pathol. (2015) 8:8348–57.
78. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. (2013) 495:384–8. doi: 10.1038/nature11993
79. Piwecka M, Glazar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A, et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. (2017) 357:8526. doi: 10.1126/science.aam8526
80. Scott SM, Reiman HM, Pritchard DJ, Ilstrup DM. Soft tissue fibrosarcoma. A clinicopathologic study of 132 cases. Cancer. (1989) 64:925–31. doi: 10.1002/1097-0142(19890815)64:4<925::AID-CNCR2820640427>3.0.CO;2-T
81. McEachron TA, Triche TJ, Sorenson L, Parham DM, Carpten JD. Profiling targetable immune checkpoints in osteosarcoma. Oncoimmunology. (2018) 7:e1475873. doi: 10.1080/2162402X.2018.1475873
82. Wang L, Zhang Q, Chen W, Shan B, Ding Y, Zhang G, et al. B7-H3 is overexpressed in patients suffering osteosarcoma and associated with tumor aggressiveness and metastasis. PLoS ONE. (2013) 8:e70689. doi: 10.1371/journal.pone.0070689
83. Chakraborty S, Mir KB, Seligson ND, Nayak D, Kumar R, Goswami A. Integration of EMT and cellular survival instincts in reprogramming of programmed cell death to anastasis. Cancer Metastasis Rev. (2020) 39:553–66. doi: 10.1007/s10555-020-09866-x
84. Nieto MA, Huang RY, Jackson RA, Thiery JP. Emt: 2016. Cell. (2016) 166:21–45. doi: 10.1016/j.cell.2016.06.028
85. Batista KP, De Pina KAR, Ramos AA, Vega IF, Saiz A, Alvarez Vega MA. The role of contextual signal TGF-beta1 inducer of epithelial mesenchymal transition in metastatic lung adenocarcinoma patients with brain metastases: an update on its pathological significance and therapeutic potential. Contemp Oncol (Pozn). (2019) 23:187–94. doi: 10.5114/wo.2019.91543
86. Coates JM, Galante JM, Bold RJ. Cancer therapy beyond apoptosis: autophagy and anoikis as mechanisms of cell death. J Surg Res. (2010) 164:301–8. doi: 10.1016/j.jss.2009.07.011
87. Sun T, Zhong X, Song H, Liu J, Li J, Leung F, et al. Anoikis resistant mediated by FASN promoted growth and metastasis of osteosarcoma. Cell Death Dis. (2019) 10:298. doi: 10.1038/s41419-019-1532-2
88. Taddei ML, Giannoni E, Fiaschi T, Chiarugi P. Anoikis: an emerging hallmark in health and diseases. J Pathol. (2012) 226:380–93. doi: 10.1002/path.3000
89. Cao Z, Livas T, Kyprianou N. Anoikis and EMT: Lethal “Liaisons” during cancer progression. Crit Rev Oncog. (2016) 21:155–68. doi: 10.1615/CritRevOncog.2016016955
90. Xue JM, Liu Y, Wan LH, Zhu YX. Comprehensive analysis of differential gene expression to identify common gene signatures in multiple cancers. Med Sci Monit. (2020) 26:e919953. doi: 10.12659/MSM.919953
91. Li Y, Ge YZ, Xu L, Jia R. Circular RNA ITCH: a novel tumor suppressor in multiple cancers. Life Sci. (2019) 2019:117176. doi: 10.1016/j.lfs.2019.117176
92. Wang ST, Liu LB, Li XM, Wang YF, Xie PJ, Li Q, et al. Circ-ITCH regulates triple-negative breast cancer progression through the Wnt/beta-catenin pathway. Neoplasma. (2019) 66:232–9. doi: 10.4149/neo_2018_180710N460
93. Xie Y, Yuan X, Zhou W, Kosiba AA, Shi H, Gu J, et al. The circular RNA HIPK3 (circHIPK3) and its regulation in cancer progression: review. Life Sci. (2020) 2020:117252. doi: 10.1016/j.lfs.2019.117252
94. Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. (2016) 7:11215. doi: 10.1038/ncomms11215
95. Adhikary J, Chakraborty S, Dalal S, Basu S, Dey A, Ghosh A. Circular PVT1: an oncogenic non-coding RNA with emerging clinical importance. J Clin Pathol. (2019) 72:513–9. doi: 10.1136/jclinpath-2019-205891
96. King HA, Gerber AP. Translatome profiling: methods for genome-scale analysis of mRNA translation. Brief Funct Genomics. (2016) 15:22–31. doi: 10.1093/bfgp/elu045
Keywords: circular RNAs, osteosarcoma, prognosis, metastasis, epithelial mesenchymal transition
Citation: Liu J, Yang L, Fu Q and Liu S (2020) Emerging Roles and Potential Biological Value of CircRNA in Osteosarcoma. Front. Oncol. 10:552236. doi: 10.3389/fonc.2020.552236
Received: 21 April 2020; Accepted: 21 September 2020;
Published: 28 October 2020.
Edited by:
Matiullah Khan, AIMST University, MalaysiaReviewed by:
Pierpaolo Correale, Azienda Ospedaliera ‘Bianchi-Melacrino-Morelli’, ItalyYannan Geng, Nankai University, China
Copyright © 2020 Liu, Yang, Fu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shengye Liu, Y211bHN5MDQwNkAxNjMuY29t
 Jiamei Liu1
Jiamei Liu1 Qin Fu
Qin Fu Shengye Liu
Shengye Liu