- 1Department of Tumor Growth Biology, N.N. Petrov Institute of Oncology, St. Petersburg, Russia
- 2Department of Clinical Genetics, St.-Petersburg Pediatric Medical University, Saint Petersburg, Russia
- 3Institute of Oncology, Hadassah Moscow, Moscow, Russia
Comprehensive molecular testing plays a critical role in the choice of treatment for non-small lung cell cancer (NSCLC). The analysis of druggable alterations in EGFR, BRAF, MET, KRAS, ALK, ROS1, RET and NTRK1/2/3 genes is more or less standardized and can be achieved using a single diagnostic platform, e.g., next generation sequencing (NGS) or polymerase chain reaction (PCR). In contrast to above targets, PD-L1 testing requires the use of immunohistochemistry (IHC). There are multiple PD-L1 IHC assays, which utilize distinct antibodies and detection systems. These PD-L1 tests are tailored to distinct drugs, often rely on different thresholds and scoring guidelines, and are characterized by incomplete inter-laboratory and inter-observer reproducibility. Several studies evaluated the performance of PD-L1 RNA expression tests, as PCR-based RNA analysis is compatible with other NSCLC molecular testing platforms, can be performed in a semi-automated manner, and has a potential for proper standardization. These investigations revealed a correlation between PD-L1 protein and RNA expression; however, there were NSCLCs demonstrating decent amounts of PD-L1 transcript in the absence of PD-L1 IHC staining. Clinical studies are required to evaluate, which of the two PD-L1 testing approaches, i.e., RNA or protein expression measurement, has a better predictive value.
While only a decade ago the laboratory diagnosis of non-small cell lung cancer (NSCLC) required mainly conventional morphological analysis, the process of examination of NSCLC tissues is getting increasingly complex nowadays, thanks to the invention of new targeted drugs. EGFR tyrosine kinase inhibitors (TKIs) were the first to trigger the molecular profiling of lung cancer, as they demonstrated high response rates in tumors with EGFR exon 19 and 21 drug-sensitizing mutations. Subsequent advances were based on the discovery of ALK and ROS1 rearrangements, which also turned to be linked to the pronounced tumor sensitivity to corresponding TKIs. Interestingly, the development of gefitinib, erlotinib and crizotinib actually preceded the identification of their genuine molecular targets, so the incorporation of these drugs into the NSCLC management was somehow attributed to some chance discoveries. This is in stark contrast with the history of the invention of inhibitors of the mutated BRAF, which is clearly an output of a pre-planned research, starting from the systematic search for kinase activating mutations and eventually resulting in the intentional development of specific antagonists of the BRAF V600E protein. There is a multitude of new NSCLC drug targets, e.g., NTRK1-3 and RET gene fusions, MET exon 14 skipping mutations, KRAS G12C substitutions, etc. In addition, administration of several inhibitors of immune checkpoints involves testing for PD-L1 expression (1–3).
NSCLC diagnostic pipeline includes a spectrum of molecular assays which usually rely on distinct laboratory platforms. The analysis of EGFR, BRAF, and KRAS mutations usually requires allele-specific PCR and/or gene sequencing. The detection of ALK, ROS, RET, and NTRK1-3 rearrangements may be based on the immunohistochemistry (IHC) guided detection of the overexpression of the kinase portion of the corresponding protein or on the break-apart FISH assay (3). The methodology of MET testing remains to be standardized (4). PD-L1 expression analysis is apparently the most complicated assay for the time being. There are several approved antibodies for the PD-L1 status evaluation. These antibodies are tailored to particular diagnostic platforms, linked to the use of distinct therapeutic modulators of the PD-L1/PD1 pathway, have different scoring guidelines and utilize varying thresholds between “positive” and “negative” samples. The detailed listing of PD-L1 antibodies, detection systems, associated therapeutic compounds and staining patterns is provided in several reviews (5–7). Most importantly, while the majority of clinical trials involving PD-L1/PD1 pathway inhibitors generally demonstrate an association between PD-L1 expression and clinical benefit from the drug, there is a great variability across the NSCLC studies with regard to medical applicability of observed findings (Table 1).
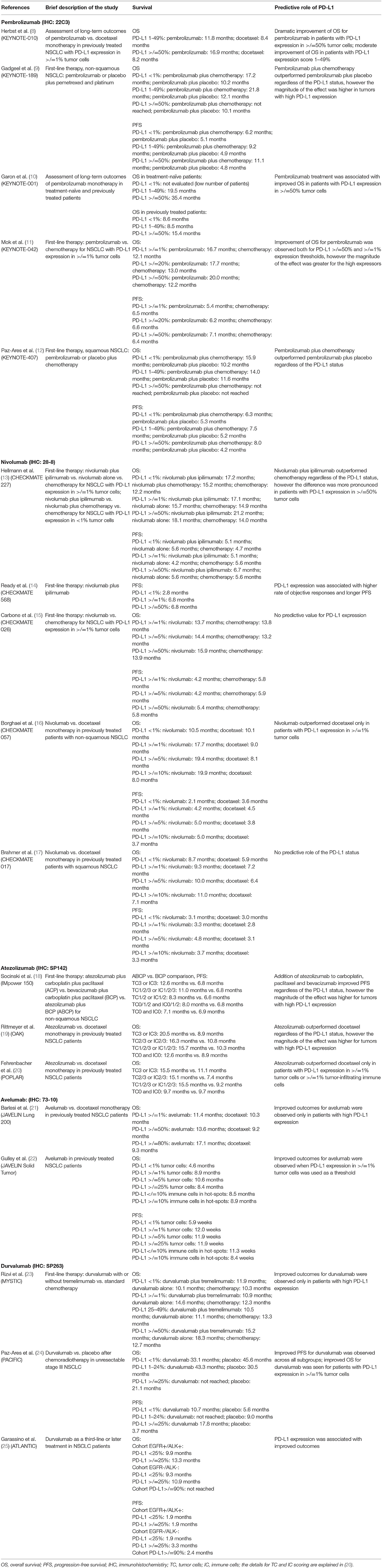
Table 1. Selected clinical studies on immune checkpoint inhibitors, which evaluated associations between clinical outcomes and the level of PD-L1 expression analysis.
Many NSCLCs are diagnosed as a metastatic disease, therefore tumor tissue material is represented by a single tiny biopsy sample. These samples must be divided for mutational analysis (PCR, sequencing) and visualization-based tests (IHC, FISH). There is a great need for a “one-for-all” approach, which would allow for a comprehensive NSCLC examination performed on a single platform. Next generation sequencing (NGS) provides a viable diagnostic opportunity, as it is capable of detecting all relevant genetic alterations within a single run. At the present time, NGS has significant limitations, such as relatively high cost, need for significant turn-around time, and requirement for sophisticated equipment (3, 26, 27). Furthermore, NGS is not yet fully compatible with a high-precision analysis of gene expression. There are ongoing efforts to utilize PCR for all types of NSCLC molecular analysis. These assays include simultaneous isolation of DNA and RNA in a single tube, synthesis of complementary DNA (cDNA) on the RNA template, conventional analysis of mutations and the test for 5′/3′-end unbalanced expression of rearranged kinases. The latter approach allows for identification of all druggable gene fusions irrespective of the translocation variant (28). PCR analysis is relatively non-expensive and is more flexible for the incorporation of new predictive tests, as exemplified by the development of the assay for detection of MET exon 14 skipping mutations (4).
While many NSCLC tests can be performed by a number of interchangeable approaches, PD-L1 analysis remains restricted to IHC technology. Use of IHC scoring is time-consuming and may be a subject of significant interobserver variability (5–7). Explicit analysis of the comparability of the existing IHC assays has been recently published by Koomen et al. (29). PD-L1 IHC comparative studies generally demonstrate acceptable results with regard to assays' interchangeability, inter-observer variability, and inter-laboratory agreement. However, it is necessary to keep in mind that the pathologists involved in research activities and scientific publishing are likely to have somewhat better standards of the laboratory practice, so the real-world inconsistencies in the IHC performance may be substantially higher when compared to pre-planned investigations. In addition, while the numerical comparisons of PD-L1 scores show good correlation, there is an alarming rate of discordance when clinically accepted thresholds are utilized (29). Consider a situation in which one pathologist determines the proportion of PD-L1 tumor cells slightly below 1%, while another pathologist determines this proportion to be slightly over 1%. When formal correlation coefficients for continuous numerical variables are calculated, these two results will be considered concordant; however, in clinical practice this difference may critically affect the access to immune therapy, as PD-L1 score of 1% is a commonly accepted threshold for consideration of immune therapies in several clinical scenarios.
Measurement of RNA expression of the gene of interest can offer advantages over IHC. In particular, PCR-based RNA expression analysis offers better reproducibility, as it evaluates not the quantity of the gene-specific transcript per se, but the ratio between the RNA messages of the gene-target and gene-referee. Furthermore, PCR tests are usually performed in a semi-automated manner, so they are less labor-consuming as compared to morphology-based analyses (30–32). However, RNA testing has several limitations. First, gene transcription is not always an equivalent of gene translation, as the production and decay of gene-specific RNAs and proteins involves different layers of regulation. Second, IHC analysis is capable to assess intracellular localization of the protein, while RNA assays evaluate only the bulk amount of gene product. Third, some analytical solutions, such as PD-L1 IHC assays, offer individual scoring for various cell types, for example, tumor cells and immune cells (6, 7). This advantage is not compatible with currently established PCR procedures. Several studies attempted to investigate in parallel the expression of PD-L1 on the level of RNA and protein in cell cultures and tumor tissues. These small-scale studies provided generally encouraging results indicating that the correlation between PD-L1 RNA and protein level does exist (30–36).
Recently published CLOVER study represents the first systematic attempt to evaluate the feasibility of PCR-based PD-L1 testing in comparison with IHC (37). The authors analyzed 437 NSCLC samples by three PD-L1 IHC assays (Ventana SP142, Ventana SP263, Dako 22C3) and by the laboratory-developed real-time PCR test for PD-L1 RNA expression. In agreement with other investigations, the CLOVER study showed significant concordance between the SP263 and the 22C3 IHC, while the SP142 produced lower rate of PD-L1 positive tumors. Indeed, the Blueprint Phase 1 study, which included 39 lung tumors stained with four different antibodies, showed that SP263 and 22C3 assays demonstrated similar IHC patterns in the majority of cases, while SP142 stained fewer number of tumor cells with generally lower intensity (38). The Blueprint Phase 2 study essentially replicated these results using a “real-world” series of 81 lung cancer specimens (39). The CLOVER study compared the performance of PCR-based PD-L1 expression measurement against conventional IHC assays. Strikingly, negative PCR tests appeared to be a reliable predictor for the lack of PD-L1 expression as determined by immunohistochemistry. This is an expected observation, given that the PCR is considered to be an ultrasensitive method for detection of biological molecules; so if the gene product cannot be detected by PCR it is unlikely to be seen by other methods. However, positive predictive value of PCR was low, as many PD-L1 RNA expressing tumors turned out to be PD-L1-negative by IHC analysis.
The results of the CLOVER study may potentially be relevant to already existing PCR diagnostic pipelines. It is relatively easy to add one more gene-specific assay to established PCR procedures, so if PCR is indeed helpful to identify PD-L1 non-expressors, its use may avoid unnecessary IHC tests. The reliability of this approach remains to be determined in subsequent studies. Overall, the CLOVER investigation calls for further efforts related to the harmonization of PD-L1 testing. Contrary to many studies, the CLOVER considered PD-L1 RNA measurement as a categorical variable by grouping tumors as “positive” and “negative” (37). It is essential to consider RNA expression as a continuous variable. Furthermore, the estimation of meaningful thresholds requires tedious consideration of various clinical and laboratory endpoints. The CLOVER study utilized a conditional threshold for the PCR test and did not adjust its value; so the additional efforts are needed to define the categories of PD-L1 expressors using biologically or clinically relevant criteria. Most importantly, the CLOVER investigation used IHC tests as a comparator for PCR assays. Ideally, studies of this type should consider treatment outcomes instead of surrogate markers; this is particularly true for PD-L1 testing, given that many PD-L1/PD1 targeted drugs show activity irrespective of the results of PD-L1 analysis (6, 7).
The instances of discordant results of PD-L1 expression measurement deserve a more systematic investigation on a case-by-case basis, given that the outcome of PD-L1 testing may dramatically influence clinical treatment decisions. There are examples of surprising discordance, when the same specimen is strongly positive by one antibody but clearly negative by another IHC assay (38, 39). Several factors may contribute to these discrepancies. Human error may be one of the factors when large series of tumors are analyzed. The Blueprint project revealed that the inter-observer variability may play a role in the interpretation of the results of PD-L1 staining (38). PD-L1 IHC assays calculate only the proportion of stained cells, while the intensity of the staining is not considered; therefore, the cut-off point between weak and absent staining may be defined differently. Intratumoral heterogeneity of PD-L1 expression may also contribute to these discrepancies, given that even serial sections may differ from each other with regard to the percentage of stained cells. The process of industrial development of distinct PD-L1 antibodies by definition involves distinct protein epitopes and distinct animals, so the individual antibody clones may differ in their ability to recognize various PD-L1 isoforms. The incorporation of the RNA testing adds complexity to this issue. It is not impossible that some tumor specimens lose their ability to interact with diagnostic antibodies during the archiving process; these samples may retain detectable PD-L1 RNA expression but show PD-L1 negativity by IHC.
There is a growing enthusiasm towards the use of PCR-based expression assays as a substitute or complement for IHC analysis. For example, Oncotype Dx breast cancer panel includes estrogen receptor, progesterone receptor and HER2 measurement to aid conventional IHC testing (40). Some studies demonstrate that Ki-67 RNA-based expression analysis has non-inferior or even better clinical performance as compared to conventional IHC tests (41, 42). There are several reported PCR-based biomarkers, which could assist the administration of immune checkpoint inhibitors (18, 43). It is highly likely that PD-L1 testing will undergo significant modification in a very near future.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This work has been supported by the Russian Science Foundation (Grant No. 17-75-30027).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Canon J, Rex K, Saiki AY, Mohr C, Cooke K, Bagal D, et al. The clinical KRAS (G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature. (2019) 575:217–23. doi: 10.1038/s41586-019-1694-1
2. Halliday PR, Blakely CM, Bivona TG. Emerging targeted therapies for the treatment of non-small cell lung cancer. Curr Oncol Rep. (2019) 21:21. doi: 10.1007/s11912-019-0770-x
3. Sokolenko AP, Imyanitov EN. Molecular diagnostics in clinical oncology. Front Mol Biosci. (2018) 5:76. doi: 10.3389/fmolb.2018.00076
4. Mitiushkina NV, Kholmatov MM, Tiurin VI, Romanko AA, Yatsuk OS, Sokolova TN, et al. Comparative analysis of expression of mutant and wild-type alleles is essential for reliable PCR-based detection of MET exon 14 skipping. Biochimie. (2019) 165:267–74. doi: 10.1016/j.biochi.2019.08.014
5. Cottrell TR, Taube JM. PD-L1 and emerging biomarkers in immune checkpoint blockade therapy. Cancer J. (2018) 24:41–6. doi: 10.1097/PPO.0000000000000301
6. Camidge DR, Doebele RC, Kerr KM. Comparing and contrasting predictive biomarkers for immunotherapy and targeted therapy of NSCLC. Nat Rev Clin Oncol. (2019) 16:341–55. doi: 10.1038/s41571-019-0173-9
7. Pirker R. Biomarkers for immune checkpoint inhibitors in advanced nonsmall cell lung cancer. Curr Opin Oncol. (2019) 31:24–8. doi: 10.1097/CCO.0000000000000496
8. Herbst RS, Garon EB, Kim DW, Cho BC, Perez-Gracia JL, Han JY, et al. Long-term outcomes and retreatment among patients with previously treated, programmed death-ligand 1–positive, advanced non–small-cell lung cancer in the KEYNOTE-010 study. J Clin Oncol. (2020) 38:1580–90. doi: 10.1200/JCO.19.02446
9. Gadgeel S, Rodríguez-Abreu D, Speranza G, Esteban E, Felip E, Dómine M, et al. Updated analysis from KEYNOTE-189: pembrolizumab or placebo plus pemetrexed and platinum for previously untreated metastatic nonsquamous non-small-cell lung cancer. J Clin Oncol. (2020) 38:1505–17. doi: 10.1200/JCO.19.03136
10. Garon EB, Hellmann MD, Rizvi NA, Carcereny E, Leighl NB, Ahn MJ, et al. Five-year overall survival for patients with advanced non–small-cell lung cancer treated with pembrolizumab: results from the phase I KEYNOTE-001 study. J Clin Oncol. (2019) 37:2518–27. doi: 10.1200/JCO.19.00934
11. Mok TSK, Wu YL, Kudaba I, Kowalski DM, Cho BC, Turna HZ, et al. Pembrolizumab versus chemotherapy for previously untreated, PD-L1-expressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): a randomised, open-label, controlled, phase 3 trial. Lancet. (2019) 393:1819–30. doi: 10.1016/S0140-6736(18)32409-7
12. Paz-Ares L, Luft A, Vicente D, Tafreshi A, Gümüş M, Mazières J, et al. Pembrolizumab plus chemotherapy for squamous non-small-cell lung cancer. N Engl J Med. (2018) 379:2040–51. doi: 10.1056/NEJMoa1810865
13. Hellmann MD, Paz-Ares L, Bernabe Caro R, Zurawski B, Kim SW, Carcereny Costa E, et al. Nivolumab plus ipilimumab in advanced non-small-cell lung cancer. N Engl J Med. (2019) 381:2020–31. doi: 10.1056/NEJMoa1910231
14. Ready N, Hellmann MD, Awad MM, Otterson GA, Gutierrez M, Gainor JF, et al. First-line nivolumab plus ipilimumab in advanced non-small-cell lung cancer (CheckMate 568): outcomes by programmed death ligand 1 and tumor mutational burden as biomarkers. J Clin Oncol. (2019) 37:992–1000. doi: 10.1200/JCO.18.01042
15. Carbone DP, Reck M, Paz-Ares L, Creelan B, Horn L, Steins M, et al. First-line nivolumab in stage IV or recurrent non-small-cell lung cancer. N Engl J Med. (2017) 376:2415–26. doi: 10.1056/NEJMoa1613493
16. Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, Ready NE, et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N Engl J Med. (2015) 373:1627–39. doi: 10.1056/NEJMoa1507643
17. Brahmer J, Reckamp KL, Baas P, Crinò L, Eberhardt WE, Poddubskaya E, et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med. (2015) 373:123–35. doi: 10.1056/NEJMoa1504627
18. Socinski MA, Jotte RM, Cappuzzo F, Orlandi F, Stroyakovskiy D, Nogami N, et al. Atezolizumab for first-line treatment of metastatic nonsquamous NSCLC. N Engl J Med. (2018) 378:2288–301. doi: 10.1056/NEJMoa1716948
19. Rittmeyer A, Barlesi F, Waterkamp D, Park K, Ciardiello F, von Pawel J, et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. Lancet. (2017) 389:255–65. doi: 10.1016/S0140-6736(16)32517-X
20. Fehrenbacher L, Spira A, Ballinger M, Kowanetz M, Vansteenkiste J, Mazieres J, et al. Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): a multicentre, open-label, phase 2 randomised controlled trial. Lancet. (2016) 387:1837–46. doi: 10.1016/S0140-6736(16)00587-0
21. Barlesi F, Vansteenkiste J, Spigel D, Ishii H, Garassino M, de Marinis F, et al. Avelumab versus docetaxel in patients with platinum-treated advanced non-small-cell lung cancer (JAVELIN lung 200): an open-label, randomised, phase 3 study. Lancet Oncol. (2018) 19:1468–79. doi: 10.1016/S1470-2045(18)30673-9
22. Gulley JL, Rajan A, Spigel DR, Iannotti N, Chandler J, Wong DJL, et al. Avelumab for patients with previously treated metastatic or recurrent non-small-cell lung cancer (JAVELIN solid tumor): dose-expansion cohort of a multicentre, open-label, phase 1b trial. Lancet Oncol. (2017) 18:599–610. doi: 10.1016/S1470-2045(17)30240-1
23. Rizvi NA, Cho BC, Reinmuth N, Lee KH, Luft A, Ahn MJ, et al. Durvalumab with or without tremelimumab vs standard chemotherapy in first-line treatment of metastatic non-small cell lung cancer: the MYSTIC phase 3 randomized clinical trial. JAMA Oncol. (2020) 6:661–74. doi: 10.1001/jamaoncol.2020.0237
24. Paz-Ares L, Spira A, Raben D, Planchard D, Cho BC, Özgüroglu M, et al. Outcomes with durvalumab by tumour PD-L1 expression in unresectable, stage III non-small-cell lung cancer in the PACIFIC trial. Ann Oncol. (2020) 31:798–806. doi: 10.1016/j.annonc.2020.03.287
25. Garassino MC, Cho BC, Kim JH, Mazières J, Vansteenkiste J, Lena H, et al. Durvalumab as third-line or later treatment for advanced non-small-cell lung cancer (ATLANTIC): an open-label, single-arm, phase 2 study. Lancet Oncol. (2018) 19:521–36. doi: 10.1016/S1470-2045(18)30144-X
26. Garinet S, Laurent-Puig P, Blons H, Oudart JB. Current and future molecular testing in NSCLC, what can we expect from new sequencing technologies? J ClinMed. (2018) 7:E144. doi: 10.3390/jcm7060144
27. Morganti S, Tarantino P, Ferraro E, D'Amico P, Duso BA, Curigliano G. Next Generation sequencing (NGS): a revolutionary technology in pharmacogenomics and personalized medicine in cancer. Adv Exp Med Biol. (2019) 1168:9–30. doi: 10.1007/978-3-030-24100-1_2
28. Iyevleva AG, Raskin GA, Tiurin VI, Sokolenko AP, Mitiushkina NV, Aleksakhina SN, et al. Novel ALK fusion partners in lung cancer. Cancer Lett. (2015) 362:116–21. doi: 10.1016/j.canlet.2015.03.028
29. Koomen BM, Badrising SK, van den Heuvel MM, Willems SM. Comparability of PD-L1 immunohistochemistry assays for non-small-cell lung cancer: a systematic review. Histopathology. (2020) 76:793–802. doi: 10.1111/his.14040
30. Vannitamby A, Hendry S, Makadia T, Danks J, Slavin J, Irving L, et al. A novel approach to detect programed death ligand 1 (PD-L1) status and multiple tumor mutations using a single non-small-cell lung cancer (NSCLC) bronchoscopy specimen. J Mol Diagn. (2019) 21:186–97. doi: 10.1016/j.jmoldx.2018.10.001
31. Vannitamby A, Hendry S, Irving L, Steinfort D, Bozinovski S. Novel multiplex droplet digital PCR assay for scoring PD-L1 in non-small cell lung cancer biopsy specimens. Lung Cancer. (2019) 134:233–7. doi: 10.1016/j.lungcan.2019.06.029
32. Brüggemann C, Kirchberger MC, Goldinger SM, Weide B, Konrad A, Erdmann M, et al. Predictive value of PD-L1 based on mRNA level in the treatment of stage IV melanoma with ipilimumab. J Cancer Res Clin Oncol. (2017) 143:1977–84. doi: 10.1007/s00432-017-2450-2
33. Shimoji M, Shimizu S, Sato K, Suda K, Kobayashi Y, Tomizawa K, et al. Clinical and pathologic features of lung cancer expressing programmed cell death ligand 1 (PD-L1). Lung Cancer. (2016) 98:69–75. doi: 10.1016/j.lungcan.2016.04.021
34. Le Goux C, Damotte D, Vacher S, Sibony M, Delongchamps NB, Schnitzler A, Terris B, et al. Correlation between messenger RNA expression and protein expression of immune checkpoint-associated molecules in bladder urothelial carcinoma: a retrospective study. Urol Oncol. (2017) 35:257–63. doi: 10.1016/j.urolonc.2017.01.014
35. Chen S, Crabill GA, Pritchard TS, McMiller TL, Wei P, Pardoll DM, et al. Mechanisms regulating PD-L1 expression on tumor and immune cells. J Immunother Cancer. (2019) 7:305. doi: 10.1186/s40425-019-0770-2
36. Gullo I, Oliveira P, Athelogou M, Gonçalves G, Pinto ML, Carvalho J, et al. New insights into the inflamed tumor immune microenvironment of gastric cancer with lymphoid stroma: from morphology and digital analysis to gene expression. Gastric Cancer. (2019) 22:77–90. doi: 10.1007/s10120-018-0836-8
37. Tsimafeyeu I, Imyanitov E, Zavalishina L, Raskin G, Povilaitite P, Savelov N, et al. Agreement between PDL1 immunohistochemistry assays and polymerase chain reaction in non-small cell lung cancer: CLOVER comparison study. Sci Rep. (2020) 10:3928. doi: 10.1038/s41598-020-60950-2
38. Hirsch FR, McElhinny A, Stanforth D, Ranger-Moore J, Jansson M, Kulangara K, et al. PD-L1 immunohistochemistry assays for lung cancer: results from phase 1 of the blueprint PD-L1 IHC assay comparison project. J Thorac Oncol. (2017) 12:208–22. doi: 10.1016/j.jtho.2016.11.2228
39. Tsao MS, Kerr KM, Kockx M, Beasley MB, Borczuk AC, Botling J, et al. PD-L1 Immunohistochemistry comparability study in real-life clinical samples: results of blueprint phase 2 project. J Thorac Oncol. (2018) 13:1302–11. doi: 10.1016/j.jtho.2018.05.013
40. McVeigh TP, Kerin MJ. Clinical use of the oncotype DX genomic test to guide treatment decisions for patients with invasive breast cancer. Breast Cancer. (2017) 9:393–400. doi: 10.2147/BCTT.S109847
41. Yamamoto S, Ibusuki M, Yamamoto Y, Fu P, Fujiwara S, Murakami K, et al. Clinical relevance of Ki67 gene expression analysis using formalin-fixed paraffin-embedded breast cancer specimens. Breast Cancer. (2013) 20:262–70. doi: 10.1007/s12282-012-0332-7
42. Gao W, Wu J, Chen X, Lin L, Fei X, Shen K, et al. Clinical validation of Ki-67 by quantitative reverse transcription-polymerase chain reaction (RT-PCR) in HR+/HER2- early breast cancer. J Cancer. (2019) 10:1110–6. doi: 10.7150/jca.29337
Keywords: non-small cell lung cancer, molecular testing, PD-L1, PCR, review
Citation: Imyanitov EN, Ivantsov AO and Tsimafeyeu IV (2020) Harmonization of Molecular Testing for Non-Small Cell Lung Cancer: Emphasis on PD-L1. Front. Oncol. 10:549198. doi: 10.3389/fonc.2020.549198
Received: 09 April 2020; Accepted: 14 August 2020;
Published: 30 September 2020.
Edited by:
Qing Zhou, Guangdong Provincial People's Hospital Lung Cancer Institute, ChinaReviewed by:
Haiyan Tu, Guangdong Provincial People's Hospital Lung Cancer Institute, ChinaXiaorong Dong, Huazhong University of Science and Technology, China
Copyright © 2020 Imyanitov, Ivantsov and Tsimafeyeu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ilya V. Tsimafeyeu, dHNpbWFmZXlldUBnbWFpbC5jb20=
 Evgeny N. Imyanitov
Evgeny N. Imyanitov Alexandr O. Ivantsov1,2
Alexandr O. Ivantsov1,2 Ilya V. Tsimafeyeu
Ilya V. Tsimafeyeu