- 1Department of Urology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
- 2Department of Thoracic and Cardiovascular Surgery, The First Affiliated Hospital of Guangxi Medical University, Nanning, China
- 3Department of Pharmacy, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
- 4Department of Medical Oncology, Fudan University Shanghai Cancer Center, Shanghai, China
PIK3CA is a key component of phosphatidylinositol 3-kinase (PI3K) pathway that its involvement in tumorigenesis has been revealed by previous research. However, its functions and potential mechanisms in bladder cancer are still largely undiscovered. Tissue microarray (TMA) with 66 bladder cancer patients was surveyed via immunohistochemistry to evaluate the level of PIK3CA and CUX1 and we found upregulation of PIK3CA in bladder cancer tissue and patients with higher level of PIK3CA presented with poorer prognosis. Overly expressed PIK3CA promoted growth, migration, invasion, and metastasis of bladder cancer cells and knockdown of PIK3CA had the opposite effect. Gain-of-function and loss-of-function studies showed that PIK3CA expression was facilitated by CUX1, leading to activation of epithelial-mesenchymal transition (EMT), accompanied by upregulated expression of Snail, β-catenin, Vimentin and downregulated expression of E-cadherin in the bladder cancer cell lines. Besides, over-expressed CUX1 could restore the expression of downregulated Snail, β-catenin, Vimentin and E-cadherin which was induced by PIK3CA knockdown. These results revealed that PIK3CA overexpression in bladder cancer was regulated by the transcription factor CUX1, and PIK3CA exerted its biological effects by activating EMT.
Introduction
Bladder cancer is one of the most frequent malignancies of the urogenital system, accounted for 199,922 deaths each year. Approximately 549,393 new cases reported every year ranks 12th among all types of cancer, according to 2018 Global Cancer Statistics report (1). The pathogenesis of bladder cancer is highly complex, and can be attributed to genetic factors and multiple environmental factors such as aromatic amines and tobacco (2, 3). Transitional cell carcinoma or urothelial carcinoma accounts for more than 90% of all pathological types of bladder cancer (4), there are only one-third belongs to muscle invasive bladder cancer, and the rest are characterized by a very high risk for distant metastasis (5, 6). Accumulated evidence has exhibited that genetic changes act as a vital role in the carcinogenesis and metastasis of bladder cancer, including changes in genes, such as PIK3CA, NPR3, and FGFR3, as well as non-coding RNAs, such as mir-93, mir-106b (7–12). Thus, exploring underlying mechanisms of bladder cancer progression in the molecular level is conducive to the investigation of corresponding targeted therapies.
PtdIns-3-kinase subunit P110-alpha (PIK3CA) is a critical regulator of carcinogenesis and metastasis (13). Various studies have ascertained that somatic mutations in PIK3CA can generally be found in cancers of the brain, breast, liver, and colon (14, 15). PIK3CA gene alteration and PIK3CA upregulation are frequent events for bladder cancer which promotes bladder cancer cell proliferation, invasion, and metastasis (9, 16). Previous researches have proved that the transcription factor FOXO3, can transactivate the catalytic subunit PIK3CA causing upregulated class PI3K-AKT1 activity (17). Other researches have exhibited that overly expressed miR-375 or miR-490-5p is able to decrease the expression of PIK3CA mRNA and protein, and consequently inhibit the PI3K/Akt signaling pathway (18, 19). It has been well established that PIK3CA can regulate tumor growth and progression through the PI3K pathway (13). However, more detailed analysis of the molecular mechanisms of PIK3CA-related functions in bladder cancer is still in urgent need.
In the present study, we identified Cut like homeobox1 (CUX1) as a crucial modulator of PIK3CA by mining public databases. Notably, recent researches have corroborated that the activation or overexpression of CUX1 in tumor progression is a crucial contributor to the growth, invasion, and metastasis of tumor cells (20–24). According to literature research, this is the first study demonstrating that PIK3CA expression is positively correlated with CUX1 in bladder cancer cells. In addition, epithelial-to-mesenchymal transition (EMT) is integral to cancer progression by reactivating wound healing, fibrosis and other cellular processes (25, 26). Previous studies implies that EMT may be an important factor in bladder cancer (27–29), that reduces expression of epithelial markers, such as E-cadherin, plakoglobin, and β-catenin, which have been correlated to both the grade and stage of bladder cancers (30, 31). In addition, abnormal expression of transcription factors such as Snail, Twist, Slug and ZEB1/ZEB2 has been demonstrated significant association with bladder cancer progression (32, 33).
Materials and Methods
Cell Lines and Culture
The human bladder cancer cell lines T24T and EJ were kindly provided by Professor Fuqing Zeng (Department of Urology, Union hospital, Tongji Medical College, Huazhong University of Science and Technology) in 2017, and subjected to DNA tests and authentication in previous studies (34, 35). Endothelial cell line HUVEC (CRL-1730) was purchased from American Type Culture Collection (ATCC, Manassas, VA, USA) in 2017. EJ and T24T cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS) (Gibco, Grand Island, NY, USA) and 1% penicillin–streptomycin (Wuhan Google Biotechnology Co., Ltd., Wuhan, China), while HUVEC cells were cultured in RPMI1640 medium. Cells were maintained at 37°C in a humidified atmosphere of 5% CO2.
Bladder Cancer Tissue Microarray and Immunohistochemistry
Tissue microarray (TMA) was consisted of 56 primary bladder cancer tissues and 10 paired non-tumor tissues obtained from Shanghai Outdo Biotech Co., Ltd. Although complete histologic data were available for tissues, but only limited and incomplete clinical information were available (Supplementary Table S1). The protein expression of PIK3CA was detected by immunohistochemistry (IHC) (PR8581C, 1:1000 dilution, Thermo Fisher Scientific, USA).
Chromatin Immunoprecipitation Assay
Chromatin immunoprecipitation (CHIP) assay was performed with EZ-CHIP kit (upstate Biotechnology) following instructions from the manufacturer with antibodies specific for CUX1. IgG as a negative control and histone H3 as a positive control. ABI Prism 7700 Sequence Detector was used for quantitative real-time PCR with SYBR Green PCR Master Mix (Applied Biosystems) using primer sets targeting gene promoters as shown in Supplementary Table S2. Chromatin immunoprecipitation assay was performed by using primary antibodies for histone H3 (ab1791, Abcam, 1:1000 dilution), IgG (ab150081, Abcam, 1:1000 dilution).
Dual-Luciferase Reporter Assay
Human PIK3CA luciferase reporter was established by annealing complementary oligonucleotides containing four canonical CUX1 binding sites (Supplementary Figures S3A, B) and inserting into pGL3-Basic (Promega). The PIKCA promoter region is −990 to +559, and the primer for luciferase is shown in Supplementary Table S2. Dual-luciferase reporter assay was carried out under manufacturer’s instructions (Promega, Madison, WI).
Western Blot
Proteins (40 μg) were collected and extracted from T24T and EJ cells, separated by gel electrophoresis, and then transferred to a PVDF membrane (Millipore, USA). Western blot analysis was performed by using primary antibodies for PIK3CA (PR8581C, 1:1000 dilution, Thermo Fisher Scientific, USA), CUX1(ab73885, 1:1000 dilution, Abcam, USA), E-cadherin (33–4000, 1:1000 dilution, Thermo Fisher Scientific, USA), Snail (PA1-86737, 1:1000 dilution, Thermo Fisher, USA), β-catenin (AB_2533982, 1:1000 dilution, Thermo Fisher, USA), Vimentin (AB_2544707, 1:1000 dilution, Thermo Fisher, USA), and β-actin (PA1-183-HRP, 1:2000 dilution, Thermo Fisher Scientific, USA); followed by goat anti-rabbit HRP-labeled secondary antibody (AB_2556786, 1:500 dilution, Thermo Fisher Scientific, USA).
Plasmids and shRNA
To construct PIK3CA and CUX1 overexpression plasmids, human PIK3CA and CUX1 cDNA were synthesized by TSINGKE (Wuhan, China), then inserted into pcDNA3.1 (Geenseed Biotech Co., Guangzhou, China). Short hairpin RNAs targeting human PIK3CA and CUX1 (Supplementary Table S3) were cloned into GV298 (Geenseed Biotech Co., Guangzhou, China). Lipofectamine 2000 (Life Technologies) was used for transfection according to the manufacturer’s instructions. The transfected cells were selected with G418 (Life Technologies) for 4 to 6 weeks to generate stable cell lines.
RT-PCR and Real-Time Quantitative RT-PCR
Total RNA was isolated using the RNeasy Mini Kit (Qiagen Inc., Valencia, CA). Reverse transcription reactions were conducted with Transcriptor First Strand cDNA Synthesis Kit (Roche, Indianapolis, IN). The PCR primers were designed using Premier Primer 5.0 software, and the primers for PIK3CA, CUX1, Snail, β-catenin, vimentin, E-cadherin, and β-actin are listed in Supplementary Table S2. Real-time PCR was performed with SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA). The fluorescent signals were collected during the extension phase, Ct values of the samples were calculated, and the transcript levels were analyzed using the 2−ΔΔCt method.
Cell Viability Assay
Cell Counting Kit-8 (CCK8) was purchased from Trans Gen Biotech, China. The treated cells were plated into 96-well plates at a density of 4 × 103 cells per well with 100 μl DMEM containing 10% fetal bovine serum and cultured at 37°C in a 5% CO2 humidified incubator. After 48 h, 10 μl of CCK-8 (Solarbo, China) was added to each well, and then the cells were incubated at 37°C for 4 h. The absorbance at 450 nm was measured using a micro-plate reader (Eon, BioTeck, USA). All experiments were done with 6–8 wells per experimental condition and repeated at least three times.
Wound Healing Assay
T24T and EJ cells were cultured in six-well plates and the cell layer was scratched with the fine end of 200 μl pipette tips (time, 0 h). Plates were washed twice with phosphate buffer saline to remove detached cells, and incubated with complete growth medium. Cell migration was photographed using10 high-power fields, at 0, 24, and 36 h after scratching. Remodeling was measured as reduction in the width of the scratch, normalized to the 0 h control, and expressed as relative migration (μm).
Cell Invasion Assay
Matrigel invasion assay was performed using membranes coated with Matrigel matrix (Invitrogen Incorporated, USA). Homogeneous single cell suspensions (1×105 cells/well) were added to the upper chambers and allowed to invade for 24 h at 37°C in a 5% CO2 incubator. The invasion rate was quantified by counting the invading cells in at least three random fields.
Colony Formation Assay
Treated T24T and EJ Bladder cancer cells (3 × 102) were seeded in six-well plates and allowed to grow at 37°C in a 5% CO2 incubator, and observed every 24 to 48 h. The medium was replenished regularly and cells were allowed to grow for 4 weeks. Cells were then fixed and stained with methylene blue or crystal violet, photomicrographs were captured, and colonies were counted.
Tube Formation Assay
Polymerization of 50 µl of growth factor-reduced Matrigel was polymerized in 96-well plates. HUVECs were starved for 24 h in incomplete RPMI1640 medium, suspended in tumor cell pretreated RPMI1640 medium, and then and 5 × 104 cells/well were added to Matrigel-coated wells, and incubated at 37°C for 18 h. Evaluation of angiogenic activity was calculated by measuring the length of the wall formed between discrete endothelial cells in each well of the control.
An Orthotopic Model of Murine Bladder Cancer
The 24G detaining needle, paraffin oil, and disinfectants required for the experiment were provided by the department of Urology, Wuhan Union Hospital. All animal experiments were approved by the Animal Care Committee of Tongji Medical College (approval number: 2016–0057). For the in vivo tumor growth studies, 4-week-old female BALB/c nude mice were randomly divided into two groups (n = 5 for each group). T24T cells stably transfected with sh-PIK3CA plasmids or control vector were prepared at a concentration of 1 × 106 cells/100 μl. Nude mice were anesthetized with 100 μl 10% chloral hydrate by intraperitoneal injection, and a transurethral indwelling catheter was placed using a 24G needle. The bladder was drained of urine, and then 100 μl 0.1 mmol/l HCL was injected into the bladder via the catheter, maintained for 15 s, and then followed by injection of 100 μl 0.1 mmol/l NAOH. Bladders were rinsed twice with sterile normal saline and 100 μl of T24T cells at a concentration of 1 × 106 cells/ml were then injected into the bladder via the catheter using a 1 ml syringe. The syringes were attached to the catheters to prevent efflux of the cell suspensions, and attachment was maintained for at least 5 min to allow the cells to attach to the burn sites. Nude mice were housed in a specific pathogen-free environment for 2 weeks and regularly monitored.
In Vivo Imaging of Small Animals
The in vivo Optical Imaging System (FX PRO, Alameda, CA) was used to acquire fluorescent images of orthotopic models of bladder cancer and lung metastases in nude mice. Before imaging, mice were anesthetized with 10% chloral hydrate by intraperitoneal injection. Red fluorescence was detected using 570 and 600 nm filter sets for excitation and emission, respectively. The Xenogen Living Image Software (Bruker MI SE. Alameda, CA) was used for collecting signals. Statistical analysis was performed using Origin8.1 (OriginLab, Northampton, MA) software. The bladder tissues and the lung samples were removed and prepared for histological examination (IHC or H&E staining).
Statistical Analysis
All analyses were completed using GraphPad Prism 7 or SPSS 20.0 software, with P < 0.05 considered statistically significant. Two-tailed unpaired t-test was used to evaluate statistical significance between the mean values of the two groups. Correlation analysis was assessed by Chi-square test and Pearson’s correlation test. Chi-square test was used to compare the difference in the proportion of high and low expression of PIK3CA in bladder cancer of different clinical stages.
Results
PIK3CA Was Expressed at High Levels in Bladder Cancer Tissues and Positively Associated With Poor Overall Survival Time of Bladder Cancer Cases
Mining the publicly available The Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov/) showed the top ten most commonly mutated genes (KMT2C, RYR2, PIK3CA, SYNE1, ARID1A, KDM6A, KMT2D, MUC16, TP53, TTN) ranked by the percent of cases affected (Figure 1A). Among the top ten genes, PIK3CA, RYR2, and KMT2C were highly expressed in the bladder cancer compared to normal bladder tissue (P < 0.001) (Figure 1B). The following seven genes showed no statistical significance: SYNE1, ARID1A, KDM6A, KMT2D, MUC16, TP53, TTN (Supplementary Figure S1). IHC assay indicated that PIK3CA was located in the cytoplasm of bladder cancer cells (Figure 1C), and displayed that PIK3CA protein was detected in 44/56 (78.6%) bladder cancer cases and with a higher positive rate than normal cases (P = 0.002). H&E staining of the TMA (Figure 1D) can also be exhibited (Figure 1G). PIK3CA immunoreactivity was notably higher in older patients with bladder cancer (P = 0.045), with more advanced clinical stage (P = 0.016), and higher rate positive lymph node metastasis (P = 0.031) (Figure 1E and Supplementary Table S1). Finally, Kaplan–Meier survival curves of 53 cases of bladder cancer patients derived from the TMA showed that patients with high expression of PIK3CA (n = 39) exhibited significantly worse survival as compared to those with low expression (n = 14) (P < 0.001) (Figure 1F). All the above findings indicate that PIK3CA is highly expressed in bladder cancer and positively related with poor clinical outcome. They have provided strong evidence that IPK3CA stands as a key component in the process of bladder cancer development.
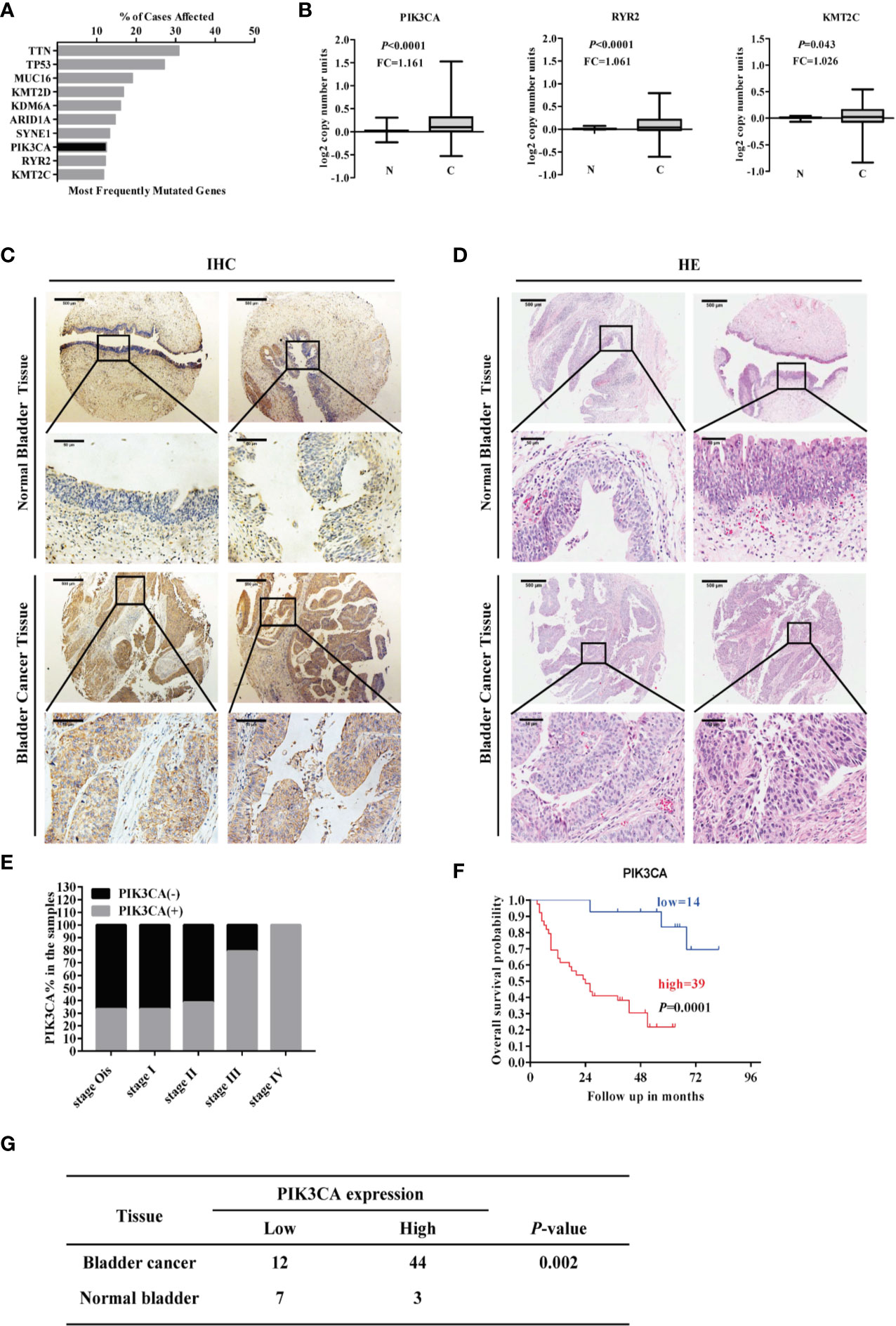
Figure 1 PIK3CA was highly expressed in bladder cancer tissues and positively correlated with poor overall survival time of patients with bladder cancer. (A) Top ten potential mutated genes in bladder cancer by mining TCGA database on December 01, 2018. (B) The RNA-sequence of the top ten mutated genes with statistical significance by mining TCGA database (PIK3CA, P<0.0001; RYR2, P<0.0001; KMT2C, P=0.043). (C, D) Representative immunohistochemical and H&E staining images of PIK3CA expression in bladder cancer tissues and adjacent normal bladder tissues on a TMA (magnification ×40 and × 400). Scale bars represent 500 μm (low magnification) and 50 μm (high magnification). (E) The expression of PIK3CA in different clinical stages of bladder cancer. (F) The survival curve of bladder cancer patients with low or high expression of PIK3CA (P=0.0001). (G) The amount of bladder cancer group and Normal control group with the low and high expression of PIK3CA respectively in TMA (P=0.002).
PIK3CA Promoted the Proliferation, Migration, Invasion, and Angiogenesis of Bladder Cancer In Vitro
To further probe the potential effects of PIK3CA in bladder cancer cells, we first transfected bladder cancer cell lines EJ and T24T with a PIK3CA-expressing vector and verified the expression of PIK3CA by qRT-PCR and Western blot (Supplementary Figures S2A, B). In CCK8 and colony formation assays, overexpression or knockdown of PIK3CA resulted in increased or decreased viability and growth of EJ and T24T cells, in comparison to control group (mock or sh-Scb) (Figure 2A and Supplementary Figures S3C, D). In a scratch assay, overexpression of PIK3CA promoted the migratory behaviors of EJ and T24T cells, meanwhile, knockdown of PIK3CA showed the opposite effect (Figure 2B). Transwell analysis demonstrated that PIK3CA up-regulation or knockdown made cells more invasive or less invasive, respectively (Figure 2C). Additionally, tube formation assay indicated similar effects on angiogenesis in EJ and T24T cells (Figure 2D).
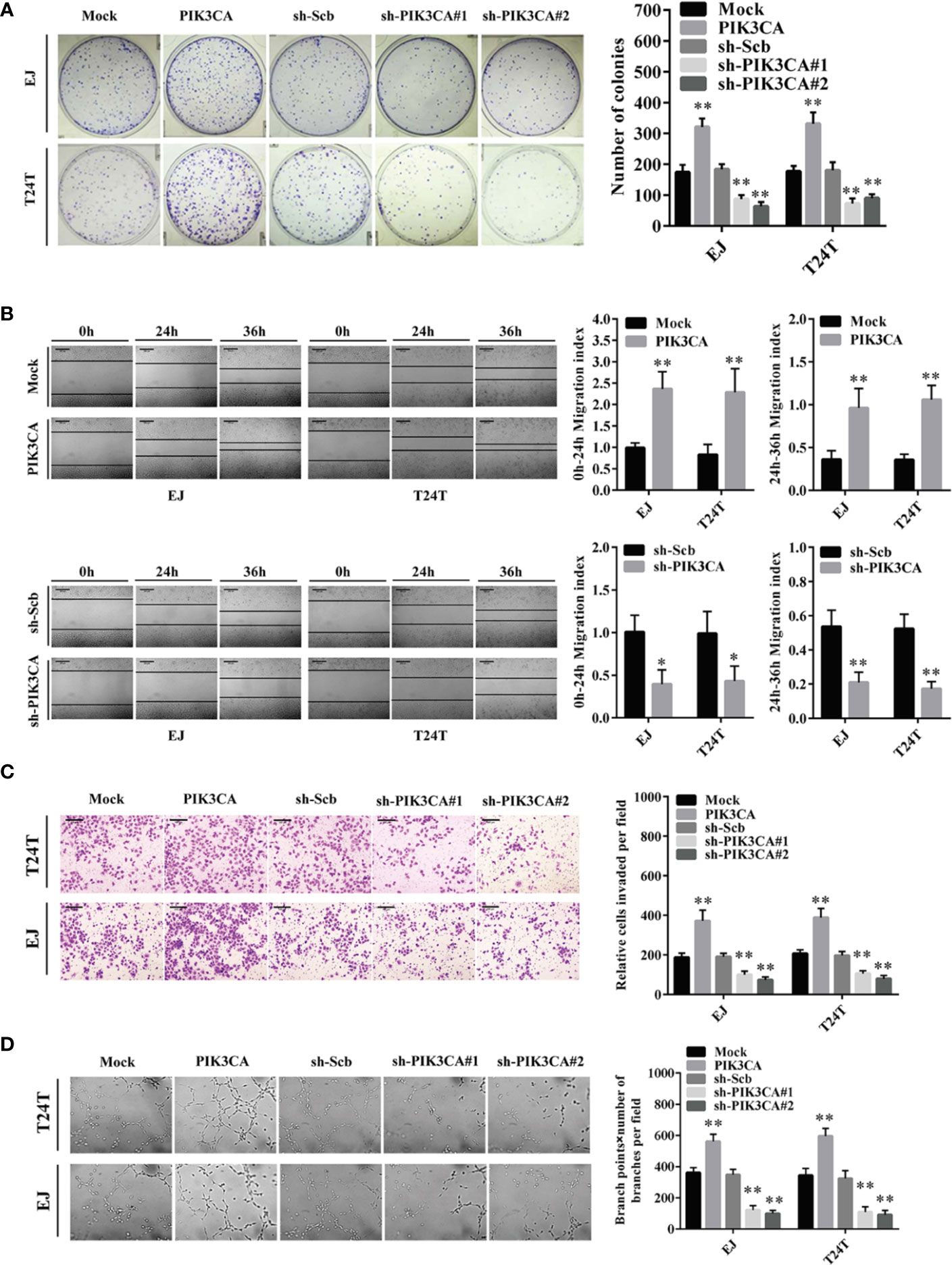
Figure 2 PIK3CA promoted the growth, migration, invasion and angiogenesis of bladder cancer cells in vitro. (A) Cell proliferation was detected by colony formation assay in EJ and T24T cells. (B) The effect of PIK3CA on cell migration was evaluated by wound healing assay in T24T and EJ cells. Scale bar indicates 200 μm. (C) Cell invasion of T24T and EJ cells was tested by transwell invasion assay. Scale bar indicates 100 μm. (D) Representation (left) and quantification (right) of tube formation assay showing the angiogenic capability of bladder cancer cells. Data are mean ± SEM, n = 3. *P < 0.05, **P < 0.01 (Student’s t-test).
In conclusion, these findings indicated that the tumor promoting roles of PIK3CA through regulatory effects on proliferation, migration, invasion, and angiogenesis of bladder cancer cells.
The Transcription Levels of PIK3CA Was Increased by CUX1 Regulation
We used the R2 database to analyze the correlation between the transcription factor collection and PIK3CA, and found that CUX1 and PIK3CA have a significant correlation, so CUX1 and PIK3CA were further explored. To verify the hypothesis that CUX1 may affect the expression of PIK3CA in bladder cancer, we performed computational assessment by transcription factor binding site analysis through the UCSC Database (https://genome.ucsc.edu/index.html) and Genomatix software (36). There are four potential binding sites of CUX1 located in the PIK3CA promoter (Supplementary Figures S3A, B). There was a positive correlation between PIK3CA and CUX1expression (P < 0.001) based on R2 Database (Tumor Bladder-Orntoft-60-MAS5.0-U133A, http://hgserver1.amc.nl/cgi-bin/r2/main.cgi) (Figure 3A). The results of 404 TCGA data also suggest that there may be a positive correlation between PIK3CA and CUX1 (P < 0.001) (Supplementary Figure S4A). Using TCGA bladder cancer data to perform survival analysis and disease free survival analysis on PIK3CA and CUX1, we found that the expression of PIK3CA is closely related to disease free survival, but the expression of CUX1 is not significantly related to patient survival and disease free survival. (Figure 3B, Supplementary Figure S4B). Trying to reveal the function of CUX1 in bladder cancer cells, we first transfected cell lines EJ and T24T with a CUX1-ovexpressing vector and verified the expression of CUX1 by qRT-PCR and Western blot (Supplementary Figures S4C, D). ChIP assay indicated that binding to the PIK3CA promoter could be increased or decreased with overexpression or knockdown of CUX1 in bladder cancer cells, respectively. Moreover, dual-luciferase reporter assay demonstrated that up-regulation of CUX1 facilitated the transcription activity of PIK3CA in bladder cancer cells (Figure 3C). To further identify the effect of CUX1 on the expression of PIK3CA in bladder cancer cell lines, we performed qRT-PCR and Western blot assay with overexpression or silencing of CUX1. We found that over-expression of CUX1 elevated mRNA and protein level of PIK3CA and CUX1 in EJ and T24T cells compared to control group (mock or sh-Scb). Additionally, knockdown of CUX1 induced decrease in mRNA and protein of PIK3CA and CUX1in EJ and T24T cells compared to control group (mock or sh-Scb) (Figures 3D–F). Together, the above results demonstrated that CUX1 stimulated transcription activity via direct interaction with the binding site of PIK3CA promoter.
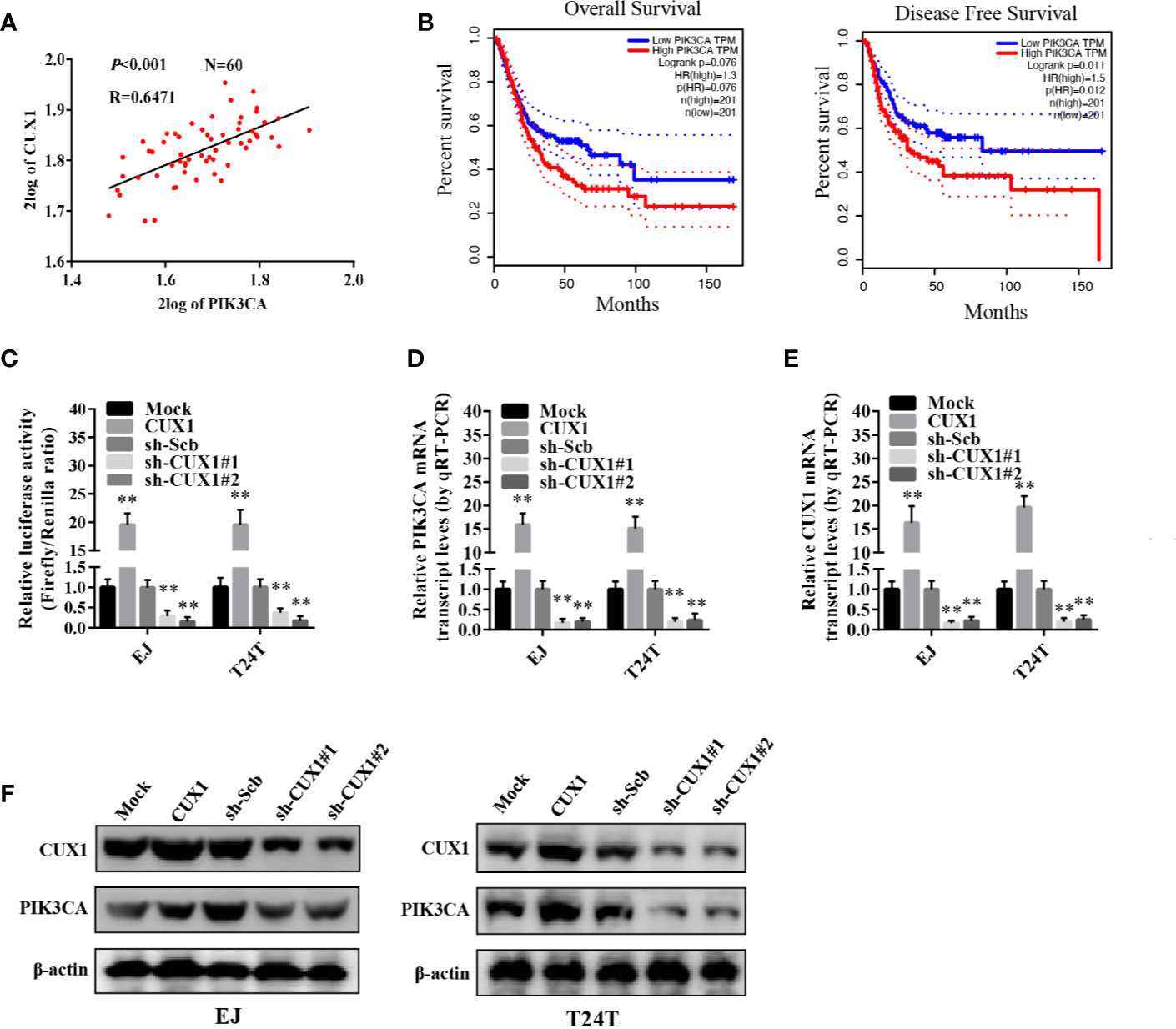
Figure 3 The transcription of PIK3CA was increased by CUX1 through direct binding to the PIK3CA promoter. (A) The relationship between PIK3CA and CUX1 was assessed by using the Genomics Analysis and Visualization Platform R2 Database (P<0.001). (B) The over survival and disease free survival curve of bladder cancer patients with low or high expression of PIK3CA (OS, P = 0.076, DFS, P = 0.012). (C) Dual-luciferase assay showing the activity of the PIK3CA promoter in bladder cancer cells stably transfected with empty vector (mock), CUX1, sh-Scb, and sh-CUX1. (D–F) The transcript and protein levels of PIK3CA were detected using Real-time PCR and Western blot, respectively in bladder cancer cells stably transfected with an empty vector, CUX1, sh-Scb, and sh-CUX1. **P < 0.01.
Overexpression of PIK3CA Restored the Proliferation, Migration, Invasion, and Angiogenesis of Bladder Cancer Cells Through Knockdown of CUX1
Given that PIK3CA has been reported participating in the proceeding of tumor proliferation, migration, and invasion (37, 38), and in combination with the evidence that the expression of PIK3CA can be directly regulated by CUX1, the effects of CUX1 knockdown and PIK3CA restoration on bladder cancer was further explored. Colony formation assay and CCK8 assay indicated suppressed anchorage-independent growth of EJ and T24T cells after CUX1 knock down in comparison to control group stably transfected with an empty vector (mock) (Figure 4A and Supplementary Figure S5F). Restoration of PIK3CA overexpression reversed the inhibitory effect on bladder cancer cell growth in vitro (Figure 4A and Supplementary Figure S5F). Transwell analysis showed that CUX1 knockdown attenuated invasion ability of EJ and T24T cells (Figure 4B). In a scratch assay, CUX1 knockdown attenuated migration ability of EJ and T24T cells (Figure 4C). Tube formation assay showed that cancer cells treated with medium preconditioned by CUX1-knockdown, significantly suppressed angiogenesis capabilities (Figure 4D). Additionally, under the condition of CUX knock down, restoration of PIK3CA overexpression showed revived migration, invasion and angiogenesis ability in EJ and T24T cells. (Figures 4B–D). It revealed same outcome like above that, PIK3CA knockdown weaken the proliferation, migration, invasion and angiogenesis capabilities of EJ and T24T cells, however restoration of CUX1 overexpression rescued bladder cancer cells from their defects resulted from knockdown of CUX1 (Supplementary Figures S5A–E). Our results confirmed a vital regulatory role of CUX1 in PIK3CA-induced aggressiveness and angiogenesis of bladder cancer cells
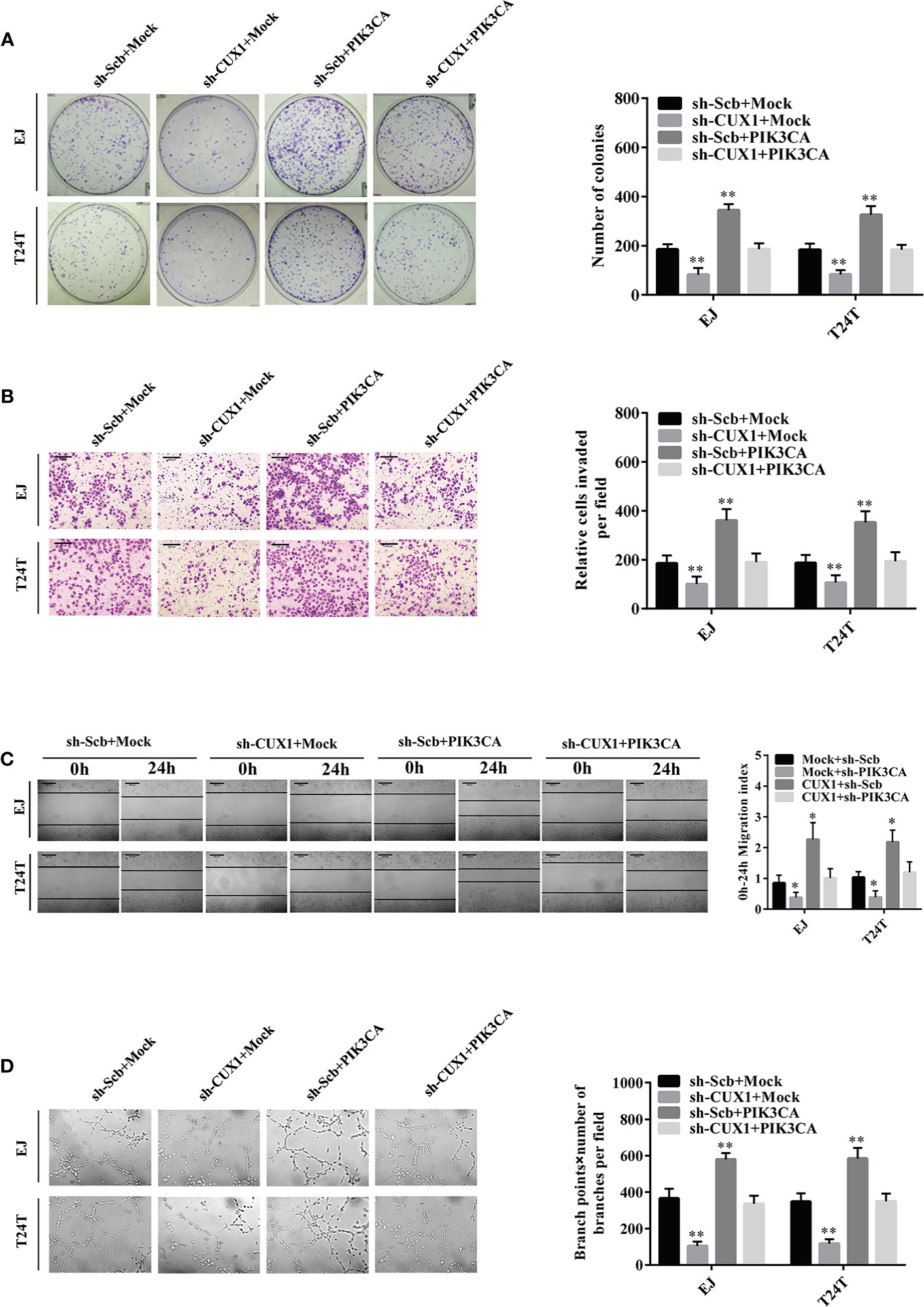
Figure 4 Overexpression of PIK3CA restored the growth, migration, invasion, and angiogenesis of bladder cancer cells through knockdown of CUX1. (A–D) Colony formation, transwell migration, Matrigel invasion, and tube formation assays were used to investigate the proliferation, migration, invasion, and angiogenic capacity, respectively, of bladder cancer cells stably transfected with mock, CUX1, sh-Scb, and sh-CUX1, and those co-transfected with PIK3CA and sh-PIK3CA. Data are mean ± SEM, n = 3. *P < 0.05, **P < 0.01 (Student’s t-test).
PIK3CA Promote Bladder Cancer Progression by Activating EMT Related Makers—Snail, E-cadherin, Vimentin, and β-Catenin
Although the functional role of PIK3CA in EMT has been investigated in several cancers (39), there are only a few reports in bladder cancer demonstrating involvement of PIK3CA in the process of EMT. Therefore, to investigate this potential functional relationship, we evaluated the expression of key EMT-related markers—Snail, β-catenin, Vimentin and E-cadherin using western blot and qRT-PCR. In EJ cells and T24T cells, over-expression of PIK3CA could upregulate the expression of Snail, β-catenin and Vimentin, while downregulate the expression of E-cadherin significantly (Figures 5A, B) Consistently, knock down of PIK3CA caused downregulation of Snail, β-catenin, and Vimentin, while E-cadherin was upregulated significantly (Figures 5A, B). In agreement with the results of a previous study (20), Western blot and qRT-PCR demonstrated that knocking down CUX1 also led to downregulation of Snail, β-catenin, and Vimentin, while upregulated the expression of E-cadherin (Figures 5C, D). In accordance with our expectation, over-expressed of PIK3CA could restore the expression level of Snail, β-catenin, Vimentin, and E-cadherin which decreased by CUX1 knockdown (Figures 5C, D). Meanwhile, we also found that knocking down PIK3CA resulted in down-regulation of Snail, β-catenin, and Vimentin, while up-regulation of E-cadherin (Supplementary Figures S6A, B). As expected, over-expressed of CUX1 could restore the expression level change of Snail, β-catenin, Vimentin, and E-cadherin which was induced by PIK3CA knockdown (Supplementary Figures S6A, B). These findings suggest that PIK3CA was targeted by CUX1 and the activation of CUX1/PIK3CA axis and consequently regulation of EMT pathway may contribute to promote bladder cancer cell progression.
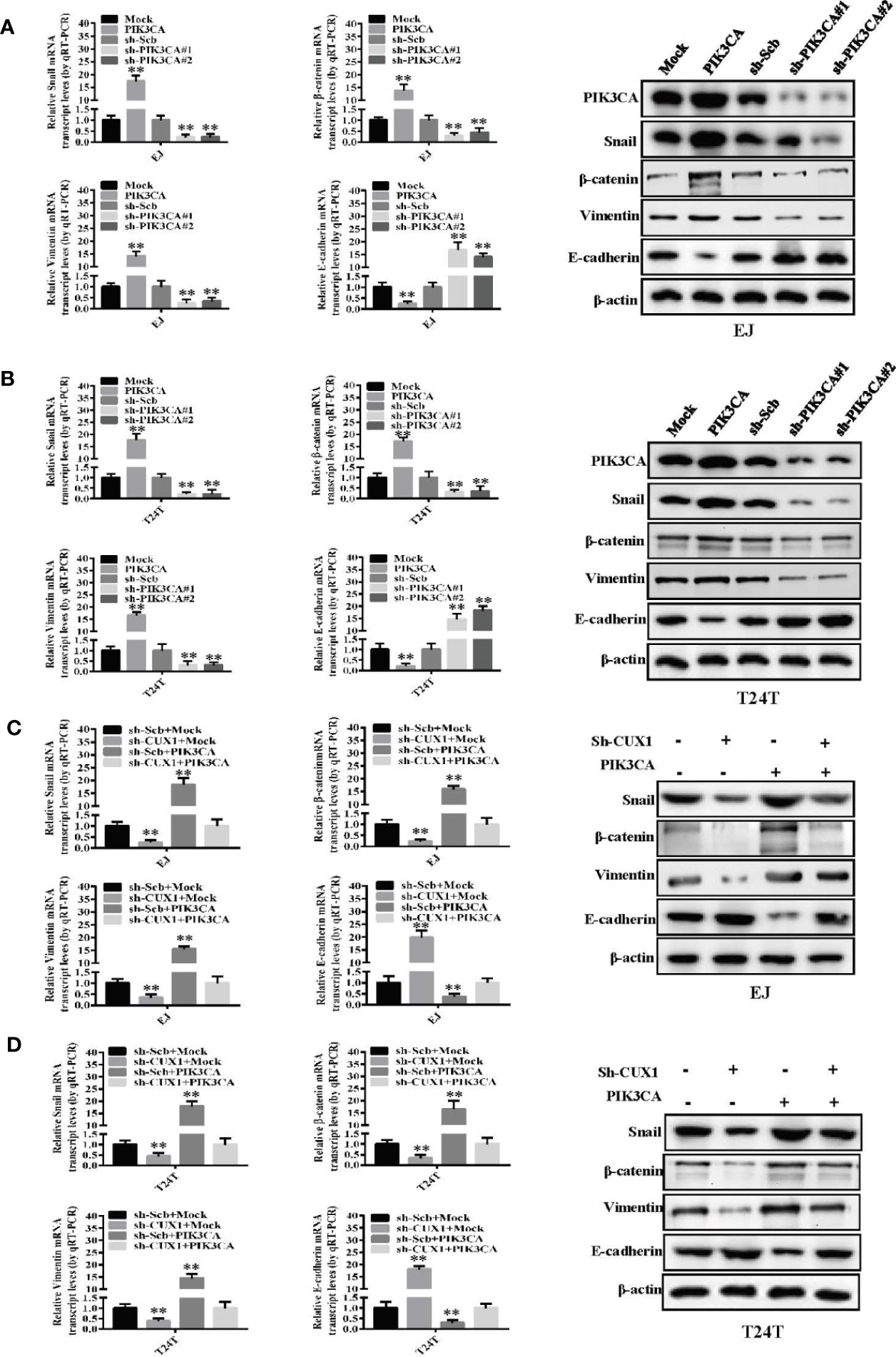
Figure 5 PIK3CA promoted bladder cancer progression by activating EMT related makers—Snail, β-catenin, vimentin and E-cadherin. (A, B) The mRNA expression and protein levels of Snail, β-catenin, vimentin and E-cadherin were determined using RT-PCR and Western blot, respectively, in EJ or T24T cells stably transfected with mock, PIK3CA, sh-Scb and sh-PIK3CA. (C, D) The transcript and protein levels of Snail, β-catenin, vimentin and E-cadherin in EJ or T24T cells stably transfected with mock, CUX1, sh-Scb, and sh-CUX1, and those co-transfected with PIK3CA and sh-PIK3CA, as detected by quantitative real-time PCR and Western blot. **P < 0.01.
Knockdown of PIK3CA Suppressed the Growth and Metastasis of Bladder Cancer Cells In Vivo
Next, we explored the effect of PIK3CA knockdown on tumor growth and metastasis in vivo. T24T cells treated with sh-PIK3CA were implanted into the bladders of BABL/C nude mice. Downregulated PIK3CA mice group (transfected with sh-PIK3CA) had a stronger fluorescence intensity detected in the bladder area, which showed that downregulated PIK3CA mice had a lower tumor cells than sh-Scb mice (transfected with an empty vector) (Figure 6A). Compared with sh-Scb mice tumor issue, the ratio of tumor cell with positive Ki67(proliferation related biomarker) was lower in sh-PIK3CA mice (Figures 6B, C). Consistent with the results of Ki67, sh-PIK3CA mice tumor issue had lower PIK3CA positive tumor cells than sh-Scb mice. The metastasis model was established by T24T cells stably transfected with sh-PIK3CA, showed that sh-PIK3CA group had fewer lung metastatic colonies than the sh-Scb group (Figures 6D, F). These findings proved that the bladder tumor growth, migration, invasion, and angiogenesis was substantially inhibited by knocking down PIK3CA in vitro.
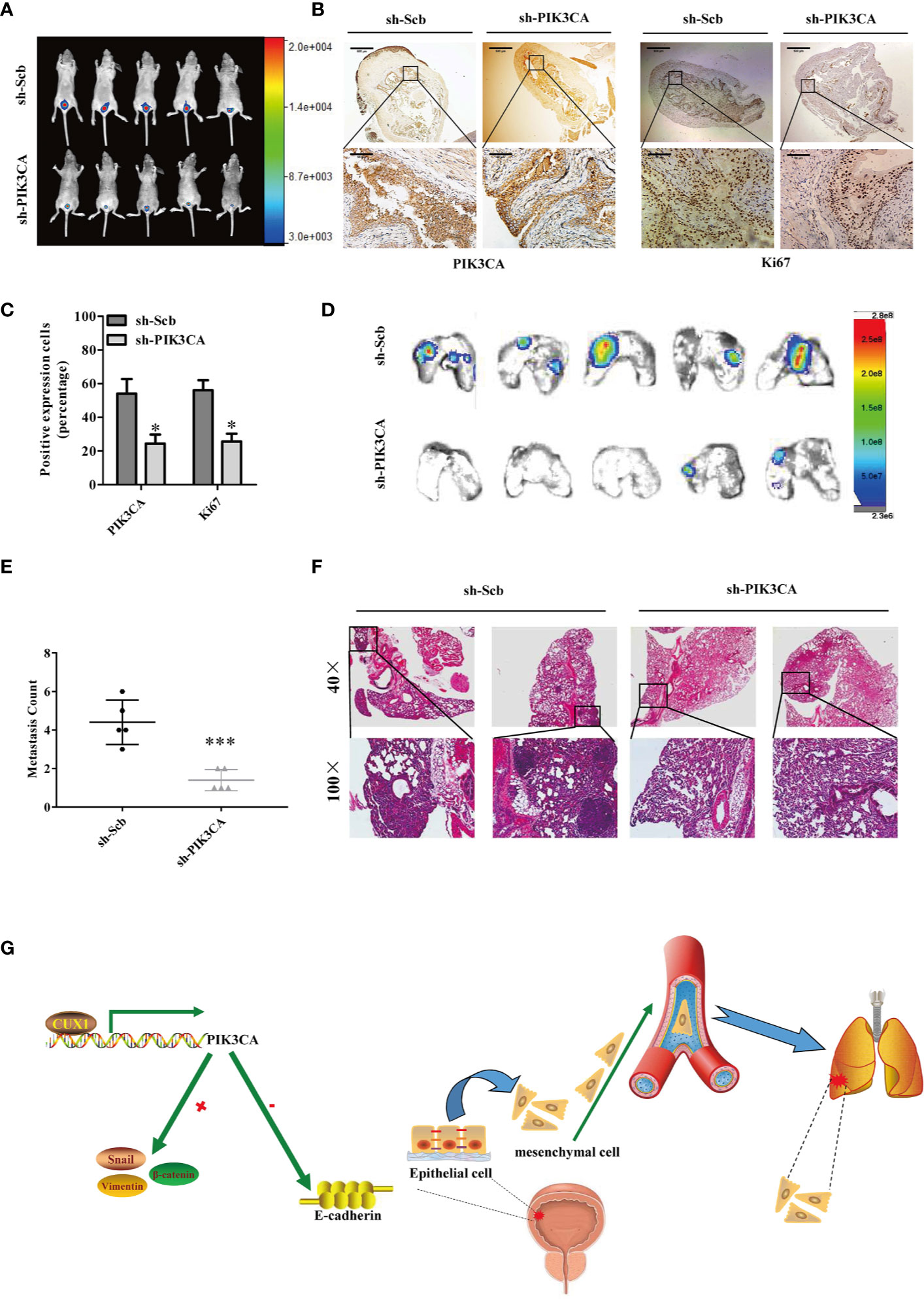
Figure 6 Knockdown of PIK3CA suppressed the growth and metastasis of bladder cancer cells in vivo. (A) Fluorescence intensity of sh-PIK3CA or negative control orthotopic bladder tumors in BALB/c nude mice. (B) Immunohistochemical staining of the expression of PIK3CA and Ki67 in bladder specimens. Original magnification, ×40 and ×400. Scale bars represent 500 μm (low magnification) and 50 μm (high magnification). (C) The positive expression of Ki67 and PIK3CA per microscopic field of the sh-Scb or sh-PIK3CA group. (D–F) Non-invasive bioluminescence imaging and H&E staining of the lung tissues were shown by in vivo tail vein metastatic assay. Original magnification, ×40 and ×400. Scale bars represent 500 μm (low magnification) and 50 μm (high magnification). (G) Schematic model of CUX1-PIK3CA-EMT oncoprotein axis is shown. CUX1 contributes to the upregulation of PIK3CA in bladder cancer by promoting transcriptional activity of PIK3CA. PIK3CA further activates EMT, thereby generating a CUX1-PIK3CA-EMT regulatory circuit to promote bladder cancer invasion and metastasis. Data are mean ± SEM, n = 3. *P < 0.05, ***P < 0.001 (Student’s t-test).
Discussion
PIK3CA, located at chromosome 3q26.3 encodes a 120-kDa protein, was first detected through in situ hybridization in 1994 by Volinia et al. (40–42). Former research has revealed that abnormally increased PIK3CA contributes to the growth and invasion of multiple cancer cell line, including breast, ovarian, colorectal, and gastric cancers (14, 15). In addition, the amplification of PIK3CA is a common event in many kinds of human tumors (43–45). Many studies have shown that the onset and progression of bladder cancer are related to the PIK3CA gene, and its mutations have been detected in in bladder cancer cases with different stages and grades (46, 47). In the current study, we revealed that PIK3CA is overexpressed in bladder cancer tissues, and PIK3CA participates in the proliferation, migration, and invasion of bladder cancer cells. It has been demonstrated that the primary molecular mechanisms regulating PIK3CA overexpression in cancer were mutation and amplification (43). Recently, studies have indicated that the PI3K/AKT/mTOR pathway may be commonly activated in multiple human cancers by molecular abnormalities such as PIK3CA mutations (48, 49). Furthermore, transcription factors such as FOXO3a can affect the expression of PIK3CA in CML (50).
Transcription factors (TF) play significant roles in carcinogenesis and cancer progression in diverse types of cancer, such as breast cancer (51), prostate cancer (52), pancreatic cancer (53), and other cancers (54). However, the detailed mechanism of TF function in regulating PIK3CA in bladder cancer is complex and not well understood. Using publicly available databases, we identified four potential CUX1 binding sites in the promoter region of PIK3CA. Interestingly, there are no literature reports of PIK3CA regulation by CUX1 in bladder cancer. CUX1 belongs to the homeodomain transcription factor family, And its involvement in the proliferation and progression of several cancers has been proved (55–57). Recently, studies have reported that the overexpression and activation of CUX1 may have a crucial impact on promoting cancer cell growth, invasion, and metastasis (20–22, 58, 59). CUX1 has been described as a transcriptional activator as well as a repressor, but its cancer-promoting effects have garnered increased research interest (57, 58). However, the potential mechanism of CUX1 regulation of PIK3CA in bladder cancer cells requires further investigation. In the current study, we revealed the positive correlation between CUX1 and PIK3CA expression in bladder cancer tumor samples and cell lines. Significantly, restoration of PIK3CA expression rescued bladder cancer cells from CUX1-knockdown inhibition of growth, aggressiveness, and angiogenesis. This suggests that CUX1 may exert its tumor promoting function through transcriptional regulation of PIK3CA in bladder cancer. The Ripka study has demonstrated that CUX1 expression was induced by activation of Akt/protein kinase B signaling, and decreased by PI3K inhibitors in pancreatic cancer (24). In contrast, our study claimed that the expression of PIK3CA is regulated by CUX1 in bladder cancer. Considering the inter-tumor heterogeneity between tumors, there are differences of genotypic and phenotypic in different types of tumors cells. Even in the same type of tumor, there is intra-tumor heterogeneity, that is, there are differences of genotypic and phenotypic in tumor cells. In addition, heterogeneity also exists between tumor microenvironment, between tumor cells and tumor circulating cells, and between different tumor cells in the same tumor tissue. Based on the above study, we think that in different tumor types, even the same signal pathway may be regulated by different genes or molecules. In the field of tumor research, the mutual regulation between the two molecules is common, so even our research is different from others (24), which precisely illustrates the complexity of the tumor itself and the tumor microenvironment. Of course, we will further explore the relationship between CUX and PIK3CA in bladder cancer. In summary, we determined that CUX1 is a vital TF of PIK3CA, regulating its expression and resulting changes in cellular functions in bladder cancer cells.
Epithelial-to-mesenchymal transition (EMT) is a biologic process that transforms epithelial cells into a mesenchymal cell phenotype (25). During EMT, cell junction proteins are downregulated, apical-basal polarity is lost and several biochemical changes will occur. All the processes stated above will allow the cells to migrate, invade, resist apoptosis, and remodel the extracellular matrix (ECM) (26, 60, 61). In recent years, mounting evidence has shown that cell migration and invasion are closely related to EMT, thereby promoting cancer cells to erode through surrounding tissues and translocate to distant tissues (62, 63). Previous studies have shown that malat1 promotes bladder cancer invasion and metastasis by activating TGF-β–induced EMT (64). In addition, DAB2IP can also be a down-stream target of miR-92b in bladder cancer cells to facilitate EMT and promote tumor cell migration or invasion (65). Additionally, activation of EMT is related to many factors, including an increase in the expression of Snail, and the decrease or loss of E-cadherin (66–68). In our study, we found activation of PIK3CA resulted in an increase in the expression of Snail and a decrease in the expression of E-cadherin at the mRNA and protein level, while inhibition of PIK3CA had the opposite effect. In our research, although we found that over-expression of PIK3CA could upregulate the expression of Snail, β-catenin, and Vimentin, while downregulate the expression of E-cadherin significantly (Figures 5A, B). However, the simple use of molecular markers to judge the occurrence of EMT in cells driven by PIK3CA requires further demonstration. With the continuous deepening of EMT research, EMT has been updated from the initial binary state to a process of continuous dynamic change (69). At the same time, epithelial–mesenchymal plasticity provides us with ideas for further understanding of tumor metastasis (70). Therefore, we need to pay attention to the specific EMT state when discussing EMT-related mechanisms. In addition, when exploring the influence mechanism of genes on EMT in cells, we need to explore the E/M phenotype of cells in depth.
Here, we propose a model of CUX1-PIK3CA-EMT oncoprotein axis, to illustrate how PIK3CA is activated and contributes to bladder cancer progression and metastasis (Figure 6G). It is entirely possible that PIK3CA may also be involved in the progression of bladder cancer through other regulatory pathways. Therefore, more studies are required to further investigate how PIK3CA act in bladder cancer development and progression. In addition, our study provides a possible new therapeutic target for the exploration of novel therapy of bladder cancer.
Our study showed that PIK3CA is up-regulation in bladder cancer and promoted the proliferation, migration, invasion and angiogenesis of bladder cancer in vitro. Considering that amplification and mutations of PIK3CA is a common event in human tumors, and PIK3CA mutations and/or PTEN loss cooperates with TP53 mutation to drive the development of bladder cancer (71). PTEN has a potential impact on response to mTOR inhibitors via the PI3K/Akt/mTOR pathway in the therapy of bladder cancer (72). Seront et al. document that PTEN loss was associated with resistance to the mTOR inhibitor in patients with advanced bladder cancer (73) Western blot and qRT-PCR.
The inhibition of PI3K/Akt can sensitize PTEN-deficient tumor cells to the effects of mTOR inhibition. In our study, whether the regulation of PIK3CA by CUX1 involves the loss of PTEN is worthy of further study. We will collect the tumor samples from patients with bladder cancer and the relationship between loss of PTEN and the mutation of PIK3CA can be observed by “Next-generation” sequencing technology (NGS) sequencing, laying a foundation for the treatment of bladder cancer.
In summary, our study demonstrates for the first time that PIK3CA is overexpressed in bladder cancer, and is regulated by the transcription factor CUX1. PIK3CA functions to promote proliferation and metastasis of bladder cancer by activating EMT. Furthermore, the expression of PIK3CA is an independent favorable prognostic factor for bladder cancer patients. Thus, as an oncogene, PIK3CA may serve as a potential target for the diagnosis and treatment of bladder cancer.
There are still many flaws in our research that need to be resolved. For example, the conclusion that CUX1 stimulates the expression of PIK3CA requires more experiments to further demonstrate. At the same time, our current findings suggest that PIK3CA may be related to EMT in regulating the occurrence and development of bladder cancer, but how PIK3CA regulates the dynamic process of EMT requires further research in the future.
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon reasonable request.
Ethics Statement
The animal study was reviewed and approved by the Animal Care Committee of Tongji Medical College (approval number: 2016-0057).
Author Contributions
ZW conceived of the study and performed the experiments. ZW, JS, and CZ wrote the manuscript. JS and ZW analyzed the data and mined the database. WJ and SL initiated, organized, and supervised the study. ZL revised the manuscript. HL and KH performed animal experiments. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by a grant from the National Natural Science Foundation of China (NSFC81660488). Bladder cancer cells were provided by Pro. Fuqing Zeng.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.536072/full#supplementary-material
Supplementary Figure 1 | RNA-sequence of genes in TCGA database. The RNA-sequence of the top ten mutated genes with no statistically significant by mining TCGA database (TTN, P=0.93; TP53, P=1.000; MUC16, P=0.985; KMT2D, P=0.070; KDM6A, ARID1A, P=0.804; SYNE1, P=1.000).
Supplementary Figure 2 | The exploration of the potential effects of PIK3CA in bladder cancer cells. (a) and (b) Screening the potential effects of PIK3CA in bladder cancer cells via RT-PCR and Western blot by transfection of bladder cancer cell lines with sh-Scb and sh-PIK3CA plasmids.
Supplementary Figure 3 | PIK3CA promoted the growth of bladder cancer cells in vitro. (a) and (b) Scheme of the potential binding site of CUX1 within the PIK3CA promoter. (c) and (d) CCK-8 assay showing the proliferation of EJ and T24T cells stably transfected with a PIK3CA overexpressing or silencing plasmid.
Supplementary Figure 4 | Screening the potential effects of CUX1 in bladder cancer cells. (a) Analysis of 404 bladder cancers in the TCGA database suggests that PIK3CA and CUX1 are significantly positively correlated. (b) In the TCGA 402 bladder cancer data, the expression of CUX1 has no obvious correlation with patient over survival and disease free survival. (c) and (d) Screening the potential effects of CUX1 in bladder cancer cells via RT-PCR and Western blot by transfection of bladder cancer cell lines with sh-Scb and sh-CUX1 plasmids.
Supplementary Figure 5 | Overexpression of CUX1 restored the growth, migration, invasion, and angiogenesis of bladder cancer cells through knockdown of PIK3CA. (a)–(d) Colony formation, transwell migration, Matrigel invasion, and tube formation assays were used to investigate the proliferation, migration, invasion, and angiogenic capacity, respectively, of bladder cancer cells stably transfected with mock, CUX1, sh-Scb, and sh-CUX1, and those co-transfected with PIK3CA and sh-PIK3CA.Data are mean ±SEM, n = 3. *P < 0.05, **P < 0.01, ***P < 0.001 (Student’s t-test). (e) The proliferation of EJ and T24T cells stably transfected with mock, PIK3CA, sh-Scb, and sh-PIK3CA, and those co-transfected with CUX1 and sh-CUX1 as detected by CCK-8 assay. (f) The proliferation of EJ and T24T cells stably transfected with mock, CUX1, sh-Scb, and sh-CUX1, and those co-transfected with PIK3CA and sh-PIK3CA as detected by CCK-8 assay.
Supplementary Figure 6 | Overexpression of CUX1 restored EMT-related makers. (a) and (b) The transcript and protein levels of Snail, and E-cadherin in EJ or T24T cells stably transfected with mock, PIK3CA, sh-Scb, and sh-PIK3CA, and those co-transfected with CUX1 and sh-CUX1 as detected by quantitative real-time PCR.
Abbreviations
IK3CA, PtdIns-3-kinase subunit P110-Alpha; CUX1, cut like homeobox 1; EMT, epithelial–mesenchymal transition; siRNA, small interfering; TMA, tissue microarray; ChIP, chromatin immunoprecipitation; RT-PCR, reverse transcription PCR; qRT-PCR, real-time polymerase chain reaction; IHC, immunohistochemistry; IF, immunofluorescence.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2018) 68(6):394–424. doi: 10.3322/caac.21492
2. Chen M, Cassidy A, Gu J, Delclos GL, Zhen F, Yang H, et al. Genetic variations in PI3K-AKT-mTOR pathway and bladder cancer risk. Carcinogenesis (2009) 30(12):2047–52. doi: 10.1093/carcin/bgp258
3. Ching CB, Hansel DE. Expanding therapeutic targets in bladder cancer: the PI3K/Akt/mTOR pathway. Lab Invest (2010) 90(10):1406–14. doi: 10.1038/labinvest.2010.133
4. Humphrey PA, Moch H, Cubilla AL, Ulbright TM, Reuter VE. The 2016 WHO Classification of Tumours of the Urinary System and Male Genital Organs-Part B: Prostate and Bladder Tumours. Eur Urol (2016) 70(1):106–19. doi: 10.1016/j.eururo.2016.02.028
5. Kaufman DS, Shipley WU, Feldman AS. Bladder cancer. Lancet (9685) 2009) 374:239–49. doi: 10.1016/s0140-6736(09)60491-8
6. Madka V, Mohammed A, Li Q, Zhang Y, Kumar G, Lightfoot S, et al. TP53 modulating agent, CP-31398 enhances antitumor effects of ODC inhibitor in mouse model of urinary bladder transitional cell carcinoma. Am J Cancer Res (2015) 5(10):3030–41.
7. Choi W, Ochoa A, McConkey DJ, Aine M, Hoglund M, Kim WY, et al. Genetic Alterations in the Molecular Subtypes of Bladder Cancer: Illustration in the Cancer Genome Atlas Dataset. Eur Urol (2017) 72(3):354–65. doi: 10.1016/j.eururo.2017.03.010
8. Juanpere N, Agell L, Lorenzo M, de Muga S, Lopez-Vilaro L, Murillo R, et al. Mutations in FGFR3 and PIK3CA, singly or combined with RAS and AKT1, are associated with AKT but not with MAPK pathway activation in urothelial bladder cancer. Hum Pathol (2012) 43(10):1573–82. doi: 10.1016/j.humpath.2011.10.026
9. Christensen E, Birkenkamp-Demtroder K, Nordentoft I, Hoyer S, van der Keur K, van Kessel K, et al. Liquid Biopsy Analysis of FGFR3 and PIK3CA Hotspot Mutations for Disease Surveillance in Bladder Cancer. Eur Urol (2017) 71(6):961–9. doi: 10.1016/j.eururo.2016.12.016
10. Li JK, Chen C, Liu JY, Shi JZ, Liu SP, Liu B, et al. Long noncoding RNA MRCCAT1 promotes metastasis of clear cell renal cell carcinoma via inhibiting NPR3 and activating p38-MAPK signaling. Mol Cancer (2017) 16(1):111. doi: 10.1186/s12943-017-0681-0
11. Li N, Miao Y, Shan Y, Liu B, Li Y, Zhao L, et al. MiR-106b and miR-93 regulate cell progression by suppression of PTEN via PI3K/Akt pathway in breast cancer. Cell Death Dis (2017) 8(5):e2796. doi: 10.1038/cddis.2017.119
12. Mao Y, Liu R, Zhou H, Yin S, Zhao Q, Ding X, et al. Transcriptome analysis of miRNA-lncRNA-mRNA interactions in the malignant transformation process of gastric cancer initiation. Cancer Gene Ther (2017) 24(6):267–75. doi: 10.1038/cgt.2017.14
13. Jaiswal BS, Janakiraman V, Kljavin NM, Chaudhuri S, Stern HM, Wang W, et al. Somatic mutations in p85alpha promote tumorigenesis through class IA PI3K activation. Cancer Cell (2009) 16(6):463–74. doi: 10.1016/j.ccr.2009.10.016
14. Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science (2004) 304(5670):554. doi: 10.1126/science.1096502
15. Bader AG, Kang S, Zhao L, Vogt PK. Oncogenic PI3K deregulates transcription and translation. Nat Rev Cancer (2005) 5(12):921–9. doi: 10.1038/nrc1753
16. Sathe A, Guerth F, Cronauer MV, Heck MM, Thalgott M, Gschwend JE, et al. Mutant PIK3CA controls DUSP1-dependent ERK 1/2 activity to confer response to AKT target therapy. Br J Cancer (2014) 111(11):2103–13. doi: 10.1038/bjc.2014.534
17. Zhou J, Liao W, Yang J, Ma K, Li X, Wang Y, et al. FOXO3 induces FOXO1-dependent autophagy by activating the AKT1 signaling pathway. Autophagy (2012) 8(12):1712–23. doi: 10.4161/auto.21830
18. Wang Y, Tang Q, Li M, Jiang S, Wang X. MicroRNA-375 inhibits colorectal cancer growth by targeting PIK3CA. Biochem Biophys Res Commun (2014) 444(2):199–204. doi: 10.1016/j.bbrc.2014.01.028
19. Chen K, Zeng J, Tang K, Xiao H, Hu J, Huang C, et al. miR-490-5p suppresses tumour growth in renal cell carcinoma through targeting PIK3CA. Biol Cell (2016) 108(2):41–50. doi: 10.1111/boc.201500033
20. Kedinger V, Sansregret L, Harada R, Vadnais C, Cadieux C, Fathers K, et al. p110 CUX1 homeodomain protein stimulates cell migration and invasion in part through a regulatory cascade culminating in the repression of E-cadherin and occludin. J Biol Chem (2009) 284(40):27701–11. doi: 10.1074/jbc.M109.031849
21. Liu KC, Lin BS, Zhao M, Wang KY, Lan XP. Cutl1: a potential target for cancer therapy. Cell Signal (2013) 25(1):349–54. doi: 10.1016/j.cellsig.2012.10.008
22. Krug S, Kuhnemuth B, Griesmann H, Neesse A, Muhlberg L, Boch M, et al. CUX1: a modulator of tumour aggressiveness in pancreatic neuroendocrine neoplasms. Endocr Relat Cancer (2014) 21(6):879–90. doi: 10.1530/ERC-14-0152
23. Ramdzan ZM, Nepveu A. CUX1, a haploinsufficient tumour suppressor gene overexpressed in advanced cancers. Nat Rev Cancer (2014) 14(10):673–82. doi: 10.1038/nrc3805
24. Ripka S, Neesse A, Riedel J, Bug E, Aigner A, Poulsom R, et al. CUX1: target of Akt signalling and mediator of resistance to apoptosis in pancreatic cancer. Gut (2010) 59(8):1101–10. doi: 10.1136/gut.2009.189720
25. Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol (2014) 15(3):178–96. doi: 10.1038/nrm3758
26. Davis FM, Azimi I, Faville RA, Peters AA, Jalink K, Putney JW Jr., et al. Induction of epithelial-mesenchymal transition (EMT) in breast cancer cells is calcium signal dependent. Oncogene (2014) 33(18):2307–16. doi: 10.1038/onc.2013.187
27. Wang X, Liang Z, Xu X, Li J, Zhu Y, Meng S, et al. miR-148a-3p represses proliferation and EMT by establishing regulatory circuits between ERBB3/AKT2/c-myc and DNMT1 in bladder cancer. Cell Death Dis (2016) 7(12):e2503. doi: 10.1038/cddis.2016.373
28. Chen Y, Peng Y, Xu Z, Ge B, Xiang X, Zhang T, et al. LncROR Promotes Bladder Cancer Cell Proliferation, Migration, and Epithelial-Mesenchymal Transition. Cell Physiol Biochem (2017) 41(6):2399–410. doi: 10.1159/000475910
29. Martinez VG, Rubio C, Martinez-Fernandez M, Segovia C, Lopez-Calderon F, Garin MI, et al. BMP4 Induces M2 Macrophage Polarization and Favors Tumor Progression in Bladder Cancer. Clin Cancer Res (2017) 23(23):7388–99. doi: 10.1158/1078-0432.CCR-17-1004
30. Baumgart E, Cohen MS, Silva Neto B, Jacobs MA, Wotkowicz C, Rieger-Christ KM, et al. Identification and prognostic significance of an epithelial-mesenchymal transition expression profile in human bladder tumors. Clin Cancer Res (2007) 13(6):1685–94. doi: 10.1158/1078-0432.CCR-06-2330
31. Jing Y, Cui D, Guo W, Jiang J, Jiang B, Lu Y, et al. Activated androgen receptor promotes bladder cancer metastasis via Slug mediated epithelial-mesenchymal transition. Cancer Lett (2014) 348(1-2):135–45. doi: 10.1016/j.canlet.2014.03.018
32. Fondrevelle ME, Kantelip B, Reiter RE, Chopin DK, Thiery JP, Monnien F, et al. The expression of Twist has an impact on survival in human bladder cancer and is influenced by the smoking status. Urol Oncol (2009) 27(3):268–76. doi: 10.1016/j.urolonc.2007.12.012
33. Schulte J, Weidig M, Balzer P, Richter P, Franz M, Junker K, et al. Expression of the E-cadherin repressors Snail, Slug and Zeb1 in urothelial carcinoma of the urinary bladder: relation to stromal fibroblast activation and invasive behaviour of carcinoma cells. Histochem Cell Biol (2012) 138(6):847–60. doi: 10.1007/s00418-012-0998-0
34. Wu X, Liu D, Tao D, Xiang W, Xiao X, Wang M, et al. BRD4 Regulates EZH2 Transcription through Upregulation of C-MYC and Represents a Novel Therapeutic Target in Bladder Cancer. Mol Cancer Ther (2016) 15(5):1029–42. doi: 10.1158/1535-7163.mct-15-0750
35. Li Y, Zheng F, Xiao X, Xie F, Tao D, Huang C, et al. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep (2017) 18(9):1646–59. doi: 10.15252/embr.201643581
36. Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, et al. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics (2005) 21(13):2933–42. doi: 10.1093/bioinformatics/bti473
37. Zheng S, Yang C, Lu M, Liu Q, Liu T, Dai F, et al. PIK3CA promotes proliferation and motility but is unassociated with lymph node metastasis or prognosis in esophageal squamous cell carcinoma. Hum Pathol (2016) 53:121–9. doi: 10.1016/j.humpath.2015.11.013
38. Zhao S, Cao Y, Liu SB, Wang XA, Bao RF, Shu YJ, et al. The E545K mutation of PIK3CA promotes gallbladder carcinoma progression through enhanced binding to EGFR. J Exp Clin Cancer Res (2016) 35(1):97. doi: 10.1186/s13046-016-0370-7
39. Salt MB, Bandyopadhyay S, McCormick F. Epithelial-to-mesenchymal transition rewires the molecular path to PI3K-dependent proliferation. Cancer Discovery (2014) 4(2):186–99. doi: 10.1158/2159-8290.CD-13-0520
40. Volinia S, Hiles I, Ormondroyd E, Nizetic D, Antonacci R, Rocchi M, et al. Molecular Cloning, cDNA Sequence, and Chromosomal Localization of the Human Phosphatidylinositol 3-Kinase p110α (PIK3CA) Gene. Genomics (1994) 24(3):472–7. doi: 10.1006/geno.1994.1655
41. Foukas LC, Claret M, Pearce W, Okkenhaug K, Meek S, Peskett E, et al. Critical role for the p110alpha phosphoinositide-3-OH kinase in growth and metabolic regulation. Nature (2006) 441(7091):366–70. doi: 10.1038/nature04694
42. Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer (2002) 2(7):489–501. doi: 10.1038/nrc839
43. Abe A, Minaguchi T, Ochi H, Onuki M, Okada S, Matsumoto K, et al. PIK3CA overexpression is a possible prognostic factor for favorable survival in ovarian clear cell carcinoma. Hum Pathol (2013) 44(2):199–207. doi: 10.1016/j.humpath.2012.05.005
44. Wang L, Shan L, Zhang S, Ying J, Xue L, Yuan Y, et al. PIK3CA gene mutations and overexpression: implications for prognostic biomarker and therapeutic target in Chinese esophageal squamous cell carcinoma. PLoS One (2014) 9(7):e103021. doi: 10.1371/journal.pone.0103021
45. Du L, Chen X, Cao Y, Lu L, Zhang F, Bornstein S, et al. Overexpression of PIK3CA in murine head and neck epithelium drives tumor invasion and metastasis through PDK1 and enhanced TGFbeta signaling. Oncogene (2016) 35(35):4641–52. doi: 10.1038/onc.2016.1
46. Lopez-Knowles E, Hernandez S, Malats N, Kogevinas M, Lloreta J, Carrato A, et al. PIK3CA mutations are an early genetic alteration associated with FGFR3 mutations in superficial papillary bladder tumors. Cancer Res (2006) 66(15):7401–4. doi: 10.1158/0008-5472.CAN-06-1182
47. Platt FM, Hurst CD, Taylor CF, Gregory WM, Harnden P, Knowles MA. Spectrum of phosphatidylinositol 3-kinase pathway gene alterations in bladder cancer. Clin Cancer Res (2009) 15(19):6008–17. doi: 10.1158/1078-0432.CCR-09-0898
48. Wang Z, Martin D, Molinolo AA, Patel V, Iglesias-Bartolome R, Degese MS, et al. mTOR co-targeting in cetuximab resistance in head and neck cancers harboring PIK3CA and RAS mutations. J Natl Cancer Inst (2014) 106(9):1–11. doi: 10.1093/jnci/dju215
49. Samuels Y, Diaz LA Jr., Schmidt-Kittler O, Cummins JM, Delong L, Cheong I, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell (2005) 7(6):561–73. doi: 10.1016/j.ccr.2005.05.014
50. Hui RC, Gomes AR, Constantinidou D, Costa JR, Karadedou CT, Fernandez de Mattos S, et al. The forkhead transcription factor FOXO3a increases phosphoinositide-3 kinase/Akt activity in drug-resistant leukemic cells through induction of PIK3CA expression. Mol Cell Biol (2008) 28(19):5886–98. doi: 10.1128/MCB.01265-07
51. Yoeli-Lerner M, Yiu GK, Rabinovitz I, Erhardt P, Jauliac S, Toker A. Akt blocks breast cancer cell motility and invasion through the transcription factor NFAT. Mol Cell (2005) 20(4):539–50. doi: 10.1016/j.molcel.2005.10.033
52. Fitzgerald KA, Evans JC, McCarthy J, Guo J, Prencipe M, Kearney M, et al. The role of transcription factors in prostate cancer and potential for future RNA interference therapy. Expert Opin Ther Targets (2014) 18(6):633–49. doi: 10.1517/14728222.2014.896904
53. Koenig A, Linhart T, Schlengemann K, Reutlinger K, Wegele J, Adler G, et al. NFAT-induced histone acetylation relay switch promotes c-Myc-dependent growth in pancreatic cancer cells. Gastroenterology (2010) 138(3):1189–99 e1-2. doi: 10.1053/j.gastro.2009.10.045
54. Muller MR, Rao A. NFAT. immunity and cancer: a transcription factor comes of age. Nat Rev Immunol (2010) 10(9):645–56. doi: 10.1038/nri2818
55. Neufeld EJ, Skalnik DG, Lievens PM, Orkin SH. Human CCAAT displacement protein is homologous to the Drosophila homeoprotein, cut. Nat Genet (1992) 1(1):50–5. doi: 10.1038/ng0492-50
56. Hulea L, Nepveu A. CUX1 transcription factors: from biochemical activities and cell-based assays to mouse models and human diseases. Gene (2012) 497(1):18–26. doi: 10.1016/j.gene.2012.01.039
57. Fan X, Wang H, Zhou J, Wang S, Zhang X, Li T, et al. The transcription factor CUTL1 is associated with proliferation and prognosis in malignant melanoma. Melanoma Res (2014) 24(3):198–206. doi: 10.1097/CMR.0000000000000064
58. Michl P, Ramjaun AR, Pardo OE, Warne PH, Wagner M, Poulsom R, et al. CUTL1 is a target of TGF(beta) signaling that enhances cancer cell motility and invasiveness. Cancer Cell (2005) 7(6):521–32. doi: 10.1016/j.ccr.2005.05.018
59. Truscott M, Harada R, Vadnais C, Robert F, Nepveu A. p110 CUX1 cooperates with E2F transcription factors in the transcriptional activation of cell cycle-regulated genes. Mol Cell Biol (2008) 28(10):3127–38. doi: 10.1128/MCB.02089-07
60. Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest (2009) 119(6):1420–8. doi: 10.1172/JCI39104
61. Jeanes A, Gottardi CJ, Yap AS. Cadherins and cancer: how does cadherin dysfunction promote tumor progression? Oncogene (2008) 27(55):6920–9. doi: 10.1038/onc.2008.343
62. Brabletz T, Jung A, Spaderna S, Hlubek F, Kirchner T. Migrating cancer stem cells - an integrated concept of malignant tumour progression. Nat Rev Cancer (2005) 5(9):744–9. doi: 10.1038/nrc1694
63. Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer (2002) 2(6):442–54. doi: 10.1038/nrc822
64. Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F, et al. TGF-beta-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clin Cancer Res (2014) 20(6):1531–41. doi: 10.1158/1078-0432.CCR-13-1455
65. Huang J, Wang B, Hui K, Zeng J, Fan J, Wang X, et al. miR-92b targets DAB2IP to promote EMT in bladder cancer migration and invasion. Oncol Rep (2016) 36(3):1693–701. doi: 10.3892/or.2016.4940
66. Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer (2009) 9(4):265–73. doi: 10.1038/nrc2620
67. Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, et al. The transcription factor Snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol (2000) 2(2):84–9. doi: 10.1038/35000034
68. Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, et al. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol (2000) 2(2):76–83. doi: 10.1038/35000025
69. Nieto MA, Huang RY, Jackson RA, Thiery JP. EMT: 2016. Cell (2016) 166(1):21–45. doi: 10.1016/j.cell.2016.06.028
70. Hari K, Sabuwala B, Subramani BV, La Porta CAM, Zapperi S, Font-Clos F, et al. Identifying inhibitors of epithelial-mesenchymal plasticity using a network topology-based approach. NPJ Syst Biol Appl (2020) 6(1):15. doi: 10.1038/s41540-020-0132-1
71. Puzio-Kuter AM, Castillo-Martin M, Kinkade CW, Wang X, Shen TH, Matos T, et al. Inactivation of p53 and Pten promotes invasive bladder cancer. Genes Dev (2009) 23(6):675–80. doi: 10.1101/gad.1772909
72. Cordes I, Kluth M, Zygis D, Rink M, Chun F, Eichelberg C, et al. PTEN deletions are related to disease progression and unfavourable prognosis in early bladder cancer. Histopathology (2013) 63(5):670–7. doi: 10.1111/his.12209
73. Seront E, Sautois B, Rottey S, D’Hondt LA, Canon J, Vandenbulcke J, et al. Phase II trial of everolimus monotherapy in the palliative treatment of patients with metastatic transitional cell carcinoma (TCC) after failure of platinum-based therapy: Activity and biomarkers. J Clin Oncol (2011) 29(15_suppl):4622–. doi: 10.1200/jco.2011.29.15_suppl.4622
Keywords: PIK3CA, CUX1, EMT, prognosis, bladder cancer
Citation: Wang Z, Shang J, Li Z, Li H, Zhang C, He K, Li S and Ju W (2020) PIK3CA Is Regulated by CUX1, Promotes Cell Growth and Metastasis in Bladder Cancer via Activating Epithelial-Mesenchymal Transition. Front. Oncol. 10:536072. doi: 10.3389/fonc.2020.536072
Received: 18 February 2020; Accepted: 29 October 2020;
Published: 03 December 2020.
Edited by:
Laura Pellegrini, Institute of Oncology Research (IOR), SwitzerlandReviewed by:
Matthias P. Wymann, University of Basel, SwitzerlandMohit Kumar Jolly, Indian Institute of Science (IISc), India
Copyright © 2020 Wang, Shang, Li, Li, Zhang, He, Li and Ju. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shikang Li, c2hpa2FuZ2xpQGhvdG1haWwuY29t; Wen Ju, anV3ZW51dUBzaW5hLmNvbQ==
†These authors have contributed equally to this work
 Zhongyu Wang1†
Zhongyu Wang1† Jun Shang
Jun Shang