
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Oncol. , 02 September 2020
Sec. Cancer Genetics
Volume 10 - 2020 | https://doi.org/10.3389/fonc.2020.01214
This article is part of the Research Topic Computational Methods in Inferring Cancer Tissue-of-Origin and Cancer Molecular Classification, Volume I View all 20 articles
 Wei-Han Zhang1†
Wei-Han Zhang1† Shou-Yue Zhang2†
Shou-Yue Zhang2† Qian-Qian Hou2†
Qian-Qian Hou2† Yun Qin3
Yun Qin3 Xin-Zu Chen1
Xin-Zu Chen1 Zong-Guang Zhou4
Zong-Guang Zhou4 Yang Shu2*
Yang Shu2* Heng Xu2*
Heng Xu2* Jian-Kun Hu1*‡
Jian-Kun Hu1*‡Objective: The objective of this study was to summarize the clinicopathological characteristics of the CLDN18-ARHGAP fusion gene in gastric cancer patients.
Background: The CLDN18-ARHGAP26 fusion gene is one of the most frequent somatic genomic rearrangements in gastric cancer, especially in the genomically stable (GS) subtype. However, the clinical and prognostic meaning of the CLDN18-ARHGAP fusion in gastric cancer patients is unclear.
Methods: Studies that investigated CLDN18-ARHGAP fusion gastric cancer patients were identified systematically from the PubMed, Cochrane, and Embase databases through the 28th of February 2020. A systematic review and meta-analysis were performed to estimate the clinical significance of CLDN18-ARHGAP fusion in patients.
Results: A total of five eligible studies covering 1908 patients were selected for inclusion in the meta-analysis based on specified inclusion and exclusion criteria. Several fusion patterns were observed linking CLDN18 and ARHGAP26 or ARHGAP6, with the most common type being CLDN18/exon5-ARHGAP26/exon12. The survival outcome meta-analysis of the CLDN18-ARHGAP fusion gene showed that it was associated with overall survival outcomes in gastric cancer (HR, 2.03, 95% CI 1.26–3.26, P < 0.01, random-effects). In addition, diffuse gastric cancer had a greater proportion of CLDN18-ARHGAP fusions than intestinal gastric cancer (13.3%, 151/1,138 vs. 1.8%, 8/442; p < 0.001). Moreover, gastric cancer patients with the CLDN18-ARHGAP fusion gene are more likely to be female or have a younger age, lymph node metastasis and advanced TNM stages.
Conclusion: The CLDN18-ARHGAP fusion is one of the molecular characteristics of diffuse gastric cancer and is also an independent prognostic risk factor for gastric cancer. In addition, it is also related to multiple clinical characteristics, including age, sex, lymph node metastasis and tumor stage. However, the mechanism of the CLDN18-ARHGAP fusion gene and potential targeted therapeutic strategies need further exploration.
Recurrent chromosomal translocations have been implicated in multiple tumor types. It has been demonstrated that frequent fusion genes are involved in oncogenesis and progression as driver events. The Philadelphia chromosome is well reported as the first cancer-associated chromosomal rearrangement, resulting in the BCR-ABL fusion, which was also identified as a diagnostic feature and therapeutic target of chronic myeloid leukemia patients. Thereafter, oncologic fusion genes were frequently identified in leukemia, lymphoma and sarcoma, but with a relatively low incidence rate in epithelial tumors (1). For epithelial tumors, the most well-known fusion gene is the EML4-ALK gene, which was detected in ~5–10% of non-small cell lung cancer patients (2, 3).
Regarding gastric cancer, The Cancer Genome Atlas (TCGA) project proposes the molecular classification of gastric cancers and divides them into four separate subtypes, and the CLDN18-ARHGAP26 fusion gene is highly enriched in the genomically stable (GS) subtype (4). Histologically, the majority of GS subtype cancers were the diffuse type according to the Lauren classification, with common somatic mutations located in the CDH1 and RHOA genes. It is notable that CLDN18-ARHGAP fusions were mutually exclusive with RHOA mutations in the classification of TCGA (4). A subsequent functional study indicated that the introduction of the CLDN18-ARHGAP26 fusion to tumor cells can direct the loss of the epithelial phenotype, epithelial-mesenchymal transition, and inhibition of the RHOA signaling pathway and contribute to tumor invasiveness in cancer cell lines (5). Our study group previously used a whole-genome sequencing approach to characterize the genomic features of signet ring cell gastric cancer and identified frequent CLDN18-ARHGAP fusions (6). More importantly, we linked multiple clinical characteristics with CLDN18-ARHGAP28/6 fusions, including the proportion of signet ring cell content, TNM stage, and poor prognosis with the current chemotherapy strategy (6). These findings were quickly validated by an independent Japanese study (7, 8). Meanwhile, a further Korean study found that the CLDN18-ARHGAP fusion gene can promote the invasion and migration capacity of gastric cancer cells (9). However, there is a lack of studies systematically evaluating the clinicopathological characteristics and prognostic meaning of CLDN18-ARHGAP fusions in gastric cancers.
Therefore, in this meta-analysis and systematic review, we will systematically summarize and assess the clinical significance and advances of the CLDN18-ARHGAP fusion gene in gastric cancer. The primary endpoint of the present study is the survival outcomes of patients with the CLDN18-ARHGAP fusion gene, and other endpoints are the relationship of the CLDN18-ARHGAP fusion gene with tumor-related clinicopathological characteristics, such as age, sex, tumor location and tumor stage.
We searched the Web of Knowledge, PubMed, Embase and Cochrane Collaborative Center Register of Controlled Trials databases on the 28th of February 2020 by using the terms “gastric cancer,” “gastric carcinoma,” “gastric neoplasm,” “stomach cancer,” “stomach carcinoma,” “stomach neoplasm,” “CLDN,” “claudin,” “ARHGAP,” “Rho GTPase-activating protein,” “oligophrenin-1-like” and “OPHN1L” and strictly restricted search results to titles, abstracts and keywords. We also searched previously published meta-analyses and systematic reviews. All of those articles were independently screened by two authors (WH Zhang and SYZ) based on the inclusion and exclusion criteria of the study. Because the studies included in this meta-analysis have been published, ethical approval was not needed from ethics committees. The results of this study were reported according to the PRISMA statement (10).
Those studies that reported the relationship between the CLDN18-ARHGAP fusion gene and the clinicopathological characteristics or survival outcomes of gastric cancer patients were included. The exclusion criteria included the following: (1) mixed benign disease of the stomach; (2) articles in languages other than English; and (3) incomplete data or duplicated data. For studies with more than one article and with duplicated data, only the article with the most complete data was included for analysis in this study.
Data from the included studies were independently extracted by two authors (WH Zhang and QQ Hou). For each study, we recorded the following information: name of the first author, year of publication, country of the study, study design, time period of the study and examined method for the CLDN18-ARHGAP fusion gene. Furthermore, the following clinicopathological characteristics were also extracted and included in the present study: fusion types of the CLDN18 and ARHGAP genes, age (years), sex (male or female), tumor location (upper third of stomach), tumor stage (T stage, N stage and TNM stage) and survival outcomes between CLDN18-ARHGAP fusion-positive and CLDN18-ARHGAP fusion-negative patients. The patients were divided into a CLDN18-ARHGAP fusion-positive group and a CLDN18-ARHGAP fusion-negative group according to the status of the expression of CLDN18-ARHGAP26/6 fusions.
The quality assessment of the included studies was evaluated by two authors (WH Zhang and QQ Hou) independently. Retrospective studies were assessed by the Newcastle–Ottawa Scale (NOS), which is a 9-point scale (11). Studies with NOS scores lower than 6 were deemed moderate or low-quality studies. Any disagreements regarding the quality assessment were resolved by discussion with supervisors (H Xu and JK Hu).
This study was performed according to the Cochrane guidelines (12). For studies that only reported the medians and ranges for continuous variables, the data were converted to means and standard deviations (SDs) with the method reported by Hozo et al. (13). Categorical variables are presented as ratios and were analyzed by the Mantel-Haenszel method, and continuous variables are presented as the mean ± SD and were analyzed by the inverse variance method. The odds ratio (OR) and mean difference (MD) were used to evaluate dichotomous and continuous data, respectively. The hazard ratio (HR) was used to evaluate survival outcomes. The OR, HR and MD were reported with 95% confidence intervals (CIs). Heterogeneity among studies was assessed by the I2 value. According to the I2 value, the studies were determined to have low (I2 < 30%), moderate (30–50%) or considerable (I2 ≥ 50%) heterogeneity. Begg's test was used to assess publication bias. For the survival analysis, we updated the survival information to Jan 2019 of our previous study (all 829 patients, Shu et al.) (6). In addition, individual survival information from the TCGA cohort was also updated according to a recent report from the TCGA research network (14). Survival information from other studies was extracted with the method reported by Tierney et al. by Engauge Digitizer software (Version 11.2) (15). A P-value < 0.05 was considered statistically significant for the present study. All of the statistical analyses were performed by R software (http://www.R-project.org/) with the “survival,” “survminer,” “ggplot2,” “meta,” and “metafor” packages.
We retrieved 395 records with 128 duplicates. After reading the titles and abstracts, 26 articles remained for reassessment according to their full texts. After reading the full texts of these articles, we included five studies that presented the relationship of clinicopathological characteristics and survival outcomes with CLDN18-ARHGAP fusion gene status (Figure 1, PRISMA Flow Diagram). We also evaluated the quality of all included studies with the Newcastle-Ottawa Scale, and the results showed that all studies had a score ≥ 6.
All 5 studies were from four countries (United States, Japan, Korea and China) and were published between 2014 and 2019. Finally, a total of 1,908 patients were included in the present study: 151 (7.9%) patients in the CLDN18-ARHGAP fusion-positive group and 1,757 (92.1%) patients in the CLDN18-ARHGAP fusion-negative group. General characteristics of those included five studies were summarized in Table 1. There were several fusion types in CLDN18-ARHGAP fusion gene gastric cancer patients. Also, the fusion types from the five reported studies between CLDN18 and ARHGAP genes are presented in Table 2.
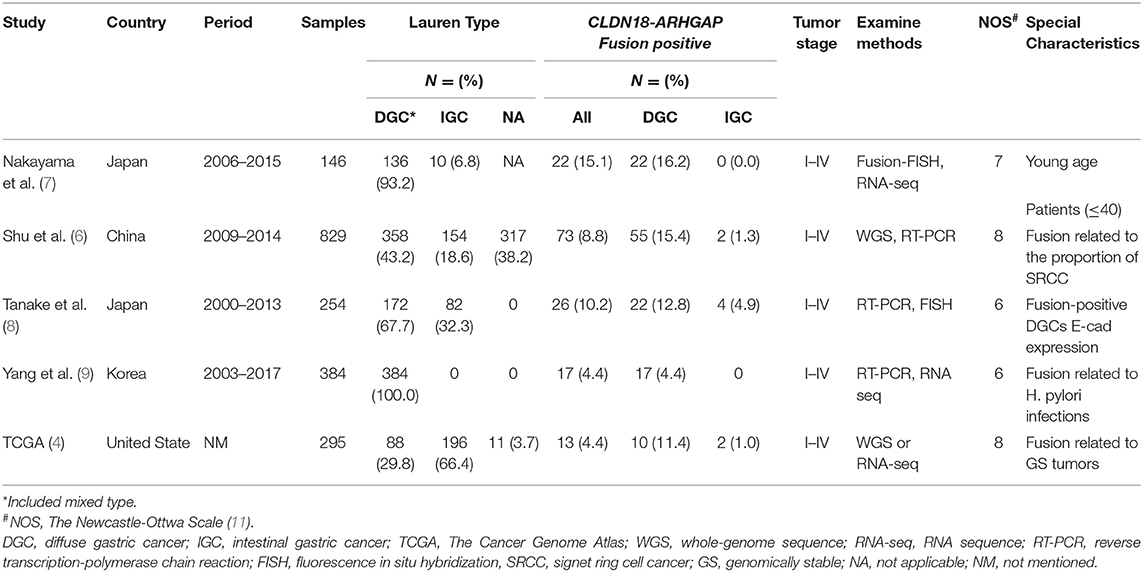
Table 1. Characteristics of studies reported clinicopathological characteristics between CLDN18-ARHGAP fusion and gastric cancers.
The CLDN18/exon5-ARHGAP26/exon12 fusion was the most common fusion pattern among present reports and is clearly understood. In addition, the mutation counts between the CLDN18-ARHGAP fusion-positive and CLDN18-ARHGAP fusion-negative patients in cohorts of Shu et al. and TCGA were analyzed (Supplementary Tables 1, 2). However, there was no overlap of the significant mutation counts gene between CLDN18-ARHGAP fusion-positive and -negative patients in the Shu and TCGA cohorts. Different pathological subtypes and the sample size difference of the two cohorts may be the reasons for the results. However, the study of Shu et al. was based on the whole genome sequencing, so we cannot analyze the RNA expression level between fusion positive and negative patients. Therefore, we analyzed copy number variation in the cohort of Shu et al. (Supplementary Table 3) and the RNA expression level (Supplementary Table 4) in the TCGA cohort between CLDN18-ARHGAP fusion-positive and CLDN18-ARHGAP fusion-negative gastric cancer patients.
In the meta-analysis of clinicopathological characteristics, the CLDN18-ARHGAP fusion gene-positive group had more patients with a younger age (MD: −5.85, 95% CI: −11.22 to −0.48, p = 0.03), a lower proportion of male patients (OR: 0.40, 95% CI 0.23–0.70, p = 0.001), patients with a more advanced N stage (OR: 3.41, 95% CI 2.00–5.82, p < 0.001) and patients with a more advanced TNM stage (OR: 3.07, 95% CI 1.56–6.05, p = 0.001) than the CLDN18-ARHGAP fusion gene-negative group (Table 3). However, tumor location (p = 0.43) and T stage (p = 0.07) were not significantly different between the two groups. Moreover, we found that diffuse gastric cancers had a greater proportion of CLDN18-ARHGAP fusion genes than intestinal gastric cancers (13.3%, 151/1,138 vs. 1.8%, 8/442; p < 0.001).
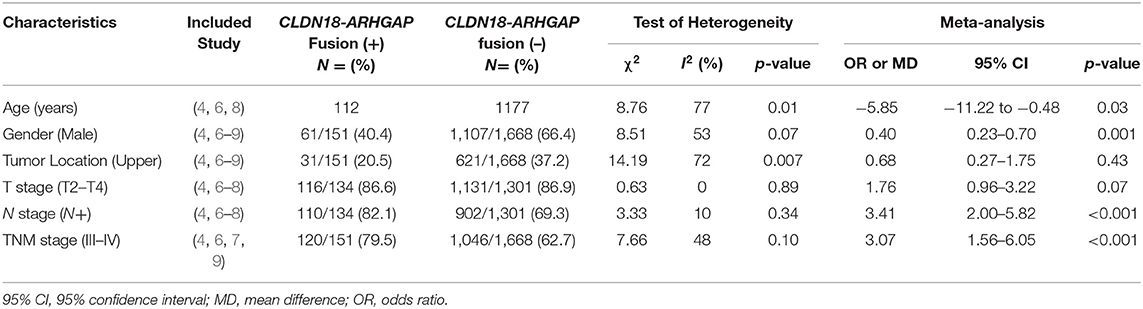
Table 3. The Meta-analysis of clinicopathological characteristics between patients with CLDN18-ARHGAP fusion positive and negative patients.
CLDN18-ARHGAP fusion-positive gastric cancer patients had significantly poorer overall survival outcomes than CLDN18-ARHGAP fusion-negative patients in the meta-analysis (HR: 2.03, 95% CI 1.26–3.26, p < 0.01, random effects) (Figure 2A), and the survival results in the meta-analysis were relatively stable in the sensitivity analysis (Figure 2B). Because there were only 4 studies included in the survival analysis, publication bias was only evaluated by Begg's test. The results demonstrated that there was no publication bias according to Begg's test with continuity correction (p = 0.555).
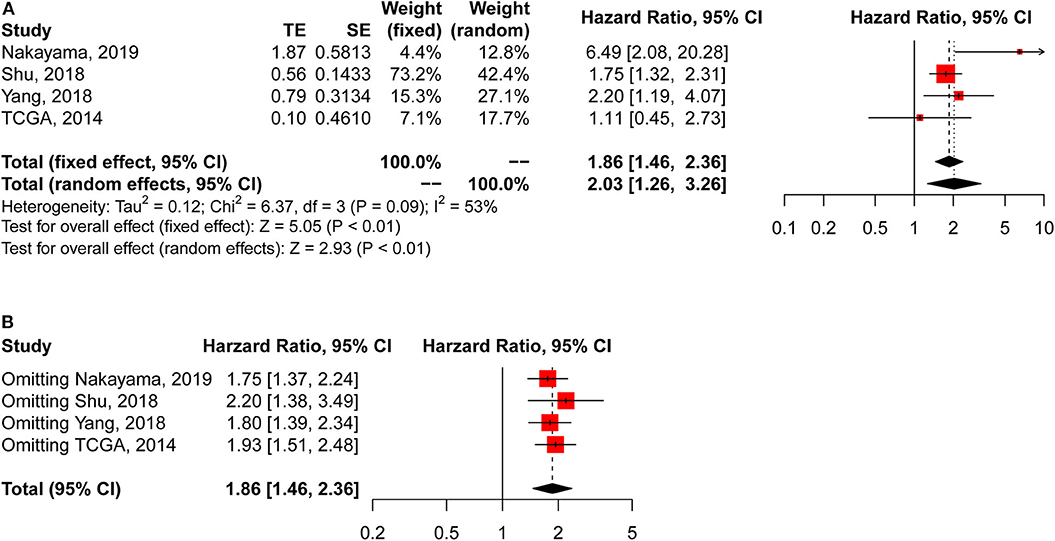
Figure 2. Meta-analysis of survival outcomes between CLDN18-ARHGAP fusion-negative and CLDN18-ARHGAP fusion-positive gastric cancer patients. (A) Survival outcomes of the CLDN18-ARHGAP26 fusion gene (positive vs. negative); (B) sensitivity analysis of the included studies.
In addition, we acquired updated individual survival information from the cohort of Shu et al. and the TCGA cohort. Therefore, the survival difference between CLDN18-ARHGAP fusion-positive and CLDN18-ARHGAP fusion-negative patients was evaluated in these two cohorts (Figures 3A,B). A significant survival difference was found between the CLDN-ARHGAP fusion gene-positive and CLDN-ARHGAP fusion gene-negative groups with the combination of the data from the two cohorts (p < 0.001) (Figure 3C). In addition, the Cox proportional hazards model was used to present the independent prognostic risk factors in the merged data of the Shu and TCGA cohorts (Table 4). In multivariate survival analysis, positive CLDN18-ARHGAP fusion (HR: 1.365, 95% CI 1.031–1.809, p = 0.030) and TNM stage (stage III vs. stage I, HR: 3.018, 95% CI 1.763–5.164, p < 0.001; stage IV vs. stage I, HR: 7.155, 95% CI 4.083–12.538, p < 0.001) were independent prognostic risk factors.
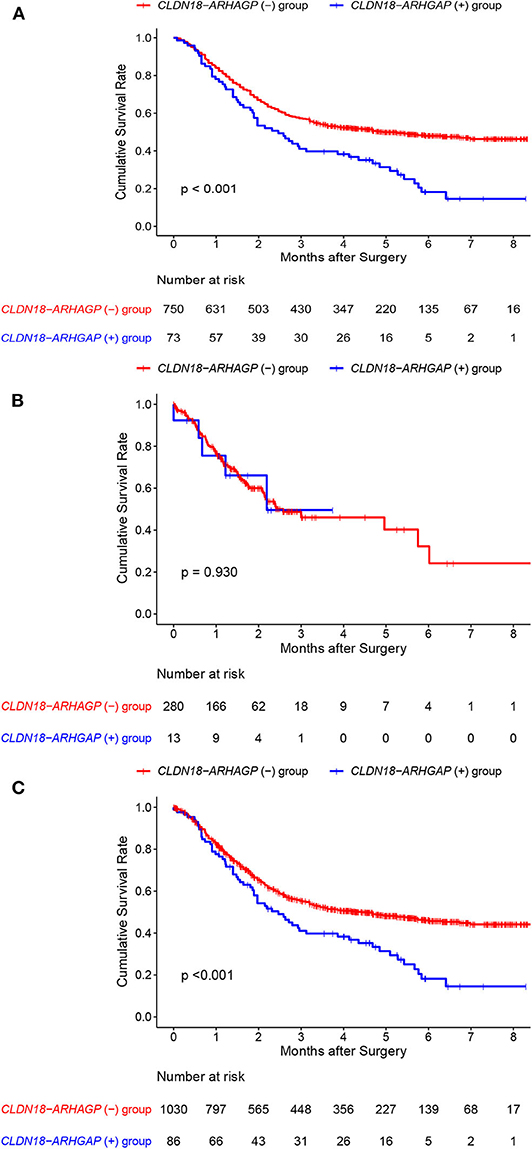
Figure 3. Individual overall survival outcomes of CLDN18-ARHGAP26/6 fusion gastric cancer patients in the Shu et al. and TCGA cohorts. (A) Overall survival outcomes in the Shu et al. cohort; (B) overall survival outcomes in the TCGA cohort; (C) overall survival outcomes in the two cohorts.
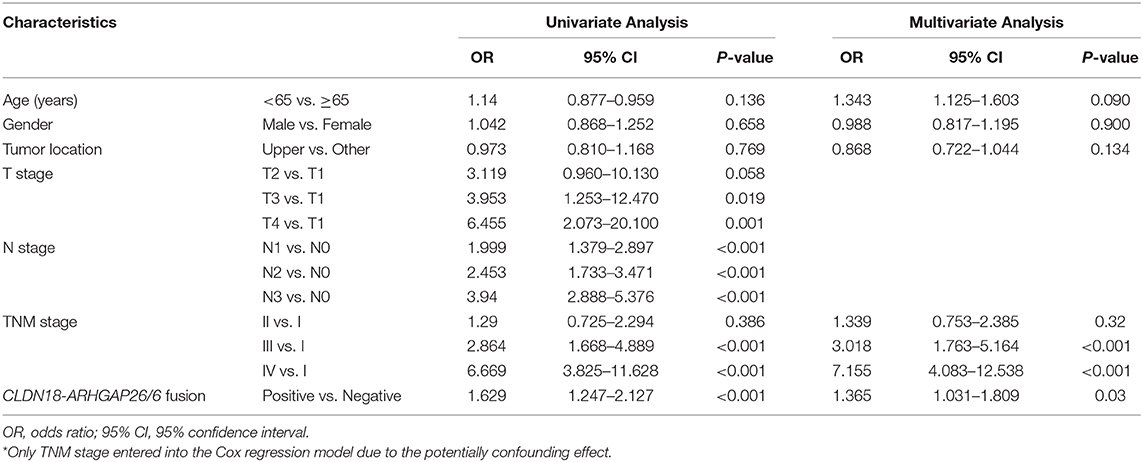
Table 4. Univariate and Multivariate survival analysis of CLDN18-ARHGAP26/6 fusion gene in TCGA and Shu cohort.
In the present study, a total of 5 cohort studies reported the presence of CLDN18-ARHGAP fusions and its relationship with clinicopathological characteristics and survival outcomes in gastric cancer patients. Multiple clinical characteristics were observed to be correlated with the frequency of CLDN18-ARHGAP fusions. Finally, significant enrichment of CLDN18-ARHGAP fusions was observed in female patients and patients with a younger age, diffuse gastric cancer by Lauren classification and more advanced tumor stages (N stage and TNM stage), but CLDN18-ARHGAP fusions were not related to the primary tumor location. Most importantly, the CLDN18-ARHGAP fusion was significantly related to poor survival outcomes in the meta-analysis (HR: 2.03, 95% CI 1.26–3.26, p < 0.01, random effects). Meanwhile, in the survival analysis with the combination of individual data from the Shu et al. and TCGA cohorts, the CLDN18-ARHGAP fusion was an independent prognostic risk factor for overall survival outcomes (HR: 1.365, 95% CI 1.031–1.809, p = 0.030).
The CLDN18-ARHGAP fusion gene is formed by chromosomal rearrangements of the CLDN18 and ARHGAP genes, mainly CLDN18-ARHGAP26 and CLDN18-ARHGAP6 fusions. CLDN18-ARHGAP26/6 contains a nearly full coding region of CLDN18 and the conserved domain of ARHGAP26/6. Functionally, the CLDN18 gene encodes the claudin-18 protein, which forms tight junctions in epithelial cells. The CLDN18-ARHGAP fusion protein may disrupt the structure of the wild-type CLDN18 protein, which may impact the cellular adhesion of cancer cells. The ARHGAP26 gene encodes the ARHGAP26 protein, a member of the Rho GTPase activating the protein, which is also known as the GTPase regulator associated with focal adhesion kinase (GRAF). GRAF not only regulates the activity of RHOGAP family proteins (16) but also coordinates membrane remodeling, which is necessary for the CLIC/GEEC endocytic pathway (17). Regev et al. suggested that the GRAF protein may play a role in the maintenance of the normal epithelial phenotype, the depletion of which can induce a neoplastic transformation-related epithelial-mesenchymal transition (EMT)-like process (18). For the fusion proteins of CLDN18-ARHGAP, the large segment of ARHGAP fuses to the carboxy terminus of CLDN18 and retains the carboxy-terminal GAP domain, which may affect ARHGAP's regulation of the RHOA pathway and/or the epithelial phenotype of gastric cancer cells. Several studies have indicated that the introduction of the CLDN18-ARHGAP26 fusion in cancer cells can increase their migration and invasion ability (5, 6), which can partially explain the advanced tumor stages in CLDN18-ARHGAP fusion-positive patients.
In the TCGA gastric cancer cohort, the CLDN18-ARHGAP26/6 fusion was enriched in patients with the genomically stable subtype, which has higher frequency of lower third tumors, patients with a younger age and more diffuse histological subtype tumors (4). In our previous study, we found that the CLDN18-ARHGAP26/6 fusion was significantly associated with the proportion of signet ring cancer cells and tumor stage (6). Tanaka et al. also observed a higher frequency of CLDN18-ARHGAP26/6 fusions in diffuse gastric cancers than in intestinal gastric cancers (22/172, 12.8% vs. 4/82, 4.8%) (8). In the present study, we found a significant difference in the frequency of the CLDN18-ARHGAP fusion gene between diffuse gastric cancer and intestinal gastric cancer (13.3%, 151/1,138 vs. 1.8%, 8/442; p < 0.001). Therefore, the CLDN18-ARHGAP fusion may be an important molecular characteristic of diffuse gastric cancer.
It is well reported that the diffuse subtype has a significantly poorer prognosis than the intestinal subtype of gastric cancer according to the Lauren classification, probably because of the more advanced tumor stages and potential resistance to traditional chemotherapy regimens of diffuse gastric cancers (19). We previously reported the prognostic value of the CLDN18-ARHGAP26/6 fusion, which is a risk factor for overall survival and confers postoperative chemotherapy resistance (6). The present study summarized the survival outcomes of previously reported studies focused on the CLDN18-ARHGAP fusion gene. Thereafter, the survival outcome meta-analysis showed that patients with CLDN18-ARHGAP fusion have a significantly poorer prognosis than patients without CLDN18-ARHGAP fusion. Due to the enrichment of the CLDN18-ARHGAP fusion in patients with more advanced stages, it is important to assess the factors independently associated with the CLDN18-ARHGAP fusion. Multivariate analyses of individual data from the Shu and TCGA cohorts presented a significant association of the CLDN18-ARHGAP fusion status with poor treatment outcomes after adjusting for tumor stage, indicating that the CLDN18-ARHGAP fusion is an independent prognostic factor for gastric cancers. In addition, it is necessary to mention that some of the included studies were not traditional clinical studies, and limited follow-up durations (such as that in the TCGA cohort) may increase the bias risk in the survival analysis above.
According to a previous study (6), gastric cancer patients with the CLDN18-ARHGAP26/6 fusion gene cannot obtain survival benefits from 5-FU/oxaliplatin-based chemotherapy, which may partially explain the poor prognosis of CLDN18-ARHGAP fusion patients. However, no other study analyzed the relationship between the CLDN18-ARHGAP26 fusion gene and chemotherapy drug therapeutic sensitivity. Mechanistically, resistance to these chemotherapy drugs was observed after the introduction of the CLDN18-ARHGAP26/6 fusion into cell lines (6). Because no reported gastric cancer cell lines carry CLDN18-ARHGAP26/6 fusions according to the Cancer Cell Line Encyclopedia database (20, 21), patient-derived xenograft (PDX) and organoid models may be the breakthrough point for future research to help validate drug resistance, screen fusion-targeted drugs, and guide personalized therapy (22–24). Yan et al. described a gastric cancer organoid model that can be used to assess the efficacy of chemotherapy (25). Unfortunately, no patient with CLDN18-ARHGAP26 fusion was captured in their gastric cancer organoid bank. Nakayama et al. established two CLDN18-ARHGAP26 fusion-positive cell lines from 125 gastric cancer PDXs (26). Collectively, these results suggest that the establishment of PDX and organoid models can help researchers conduct drug sensitivity screening and explore personalized medicine applications for therapy response testing in the future.
Specifically, the aberrant activation of claudin-18 splice variant 2 (claudin-18.2) was detected in multiple types of cancer compared with its limited expression in normal tissues. The rate of claudin-18.2-positive patients was more than 80% according to a Japanese study on gastric cancer, and more than 40% of patients had moderate-to-strong expression (≥ 2+ membrane staining intensity in ≥ 40% of tumor cells) in both the primary tumor and metastatic lymph nodes (27). Claudin-18.2 has been considered a novel druggable target for some epithelial tumors (28). Indeed, a chimeric monoclonal antibody drug has been recently developed (i.e., zolbetuximab, formerly known as IMAB362), which induces the immune-mediated lysis of CLDN18.2-positive cancer cells by activating immune effector mechanisms (29). Clinical trials evaluating the safety and efficacy of zolbetuximab for claudin-18.2-positive cancer patients are ongoing. The up-to-date evidence is promising but remains to be validated by high-quality clinical trials (30, 31). We also noticed that there is an ongoing clinical trial, which is focused on safety and efficacy of anti-claudin18.2 chimeric antigen receptor t-cell (CAR-T) immunotherapy in patients with advanced gastric cancer or pancreatic cancer (ClinicalTrials.gov Identifier: NCT03159819). The final results of the anti-claudin-18.2 CAR-T immunotherapy as a new anti-tumor targeted immunotherapy study are highly anticipated. Considering that a higher claudin-18.2 positive rate was observed in patients with the diffuse subtype of gastric cancer than in those with the intestinal type (57.5 vs. 39.0%) (27), an unsolved and important question is whether the CLDN18-ARHGAP fusion gene is correlated with claudin-18.2 protein expression and thus is suitable for zolbetuximab treatment, which requires solid clinical evidence.
According to previous studies, whole-genome sequencing, RNA sequencing, reverse transcription-polymerase chain reaction (RT-PCR) and fluorescence in situ hybridization (FISH) are all effective methods for the detection of CLDN18-ARHGAP fusions. Once the clinical significance of the CLDN18-ARHGAP fusion has been proven and potential target treatment regimens have been determined, establishing a stable and effective detection method for this fusion gene is particularly important. Considering the preservation of tumor tissue as well as the stability and economic cost of the examination, FISH may be the first choice for promotion in clinical practice. In addition, RNA in situ hybridization techniques may be useful in the detection of fusion genes. However, the sensitivity and specificity of the detection of fusion genes by such methods should be validated by clinical studies. In addition, oncogenic fusion circRNAs (f-circRNAs) derived from cancer-associated chromosomal translocations exhibit properties of tumor-promoting cellular transformation, cell viability and resistance to treatment (32, 33). Consistently, f-circRNAs derived from SLC32A2-ROS1 and EML4-ALK fusion genes, which have been determined as biomarkers for the use of targeted drugs in lung cancer, were also demonstrated to impact cell migration, invasion and cell proliferation in lung cancer cells (34, 35). More importantly, the f-circRNAs of EML4-ALK can be detected in the plasma of EML4-ALK-positive NSCLC patients (36). These results suggest the following: (1) f-circRNAs are involved in the mechanism of tumorigenesis, progression and therapy resistance; and (2) cell-free f-circRNAs could be a novel “liquid biopsy” biomarker to monitor the status of fusion genes in a noninvasive way. Therefore, we speculate and propose the following hypothesis: f-circRNAs of the CLDN18-ARHGAP fusion gene exist and can affect tumor function or act as a potential “liquid biopsy” biomarker for targeted drugs (e.g., zolbetuximab).
The present study also has some limitations. (1) This study only included five retrospective studies. Therefore, selection bias and quality deviation are likely among these studies, which may have an influence on the results of the meta-analysis. (2) The detection methods varied among the included studies. The potential false positive and false negative rates of CLDN18-ARHGAP fusion in the included studies may have influenced the results of the meta-analysis. (3) In addition, the limited follow-up duration of the included studies was another limitation of the presented studies. (4) Although previous studies successfully demonstrated that the CLDN18-ARHGAP26 fusion gene can induce EMT, the loss of the epithelial phenotype, and cell-cell and cell-extracellular matrix adhesion, as well as increase the invasion ability and resistance to chemotherapy drugs in cancer cell lines (5, 6), the specific molecular mechanisms by which CLDN18-ARHGAP26 regulates downstream molecules and pathways remain unclear. As different fusion modes generate various fusion proteins, it is difficult to design and develop antibodies to specifically target these fusion proteins, which may hinder mechanistic investigation of the CLDN18-ARHGAP fusion gene. Furthermore, large sample size multicenter studies are expected to validate the clinical significance and prognostic meaning of the CLDN18-ARHGAP26 fusion gene in gastric cancer patients.
The CLDN18-ARHGAP fusion gene is characterized as one of the features of diffuse gastric cancer. The CLDN18-ARHGAP fusion gene is correlated with advanced tumor stages in gastric cancer, as well as poor survival outcomes. Although CLDN18-ARHGAP fusion can increase the invasion and migration ability of gastric cancer cells in vitro, the molecular mechanism remains to be elucidated. Furthermore, the early detection of the CLDN18-ARHGAP fusion and targeted drugs for this fusion may potentially improve the survival outcomes of gastric cancer patients.
The raw data supporting the findings of this study are available from the corresponding author upon reasonable request.
W-HZ, HX, and J-KH designed the study. W-HZ, S-YZ, Q-QH, X-ZC, YQ, and YS collected information and analyzed and interpreted the data. Z-GZ, YS, HX, and J-KH supervised this study. All authors contributed to the article and approved the submitted version.
This study was funded by the National Natural Science Foundation of China (grant numbers: 81902437, 81673452, and 81973408); the 1.3.5 Project for Disciplines of Excellence, West China Hospital, Sichuan University (grant number: ZY2017304); the Post-Doctoral Research Project, West China Hospital, Sichuan University (grant numbers: 2018HXBH010 and 2018HXBH024); the China Postdoctoral Science Foundation (grant numbers: 2019M653418 and 2019M653428); and the Fostering Academic and Technical Leaders of Sichuan Province (No. [2016] 183-19).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.01214/full#supplementary-material
TCGA, The Cancer Genome Atlas; NOS, Newcastle–Ottawa Scale; SD, standard deviation; OR, odds ratio; MD, mean difference; HR, hazard ratio; CI, confidence intervals; GRAF, GTPase Regulator Associated with Focal Adhesion Kinase; EMT, Epithelial-Mesenchymal Transition; PDX, Patient-Derived Xenograft; RT-PCR, reverse transcription-polymerase chain reaction; FISH, Fluorescence in situ Hybridization.
1. Brien GL, Stegmaier K, Armstrong SA. Targeting chromatin complexes in fusion protein-driven malignancies. Nat Rev Cancer. (2019) 19:255–255 doi: 10.1038/s41568-019-0132-x
2. Koivunen JP, Mermel C, Zejnullahu K, Murphy C, Lifshits E, Holmes AJ, et al. EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin Cancer Res. (2008) 14:4275–427 doi: 10.1158/1078-0432.CCR-08-0168
3. Stransky N, Cerami E, Schalm S, Kim JL, Lengauer C. The landscape of kinase fusions in cancer. Nat Commun. (2014) 5:4846. doi: 10.1038/ncomms5846
4. Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. (2014) 513:202–20 doi: 10.1038/nature13480
5. Yao F, Kausalya JP, Sia YY, Teo AS, Lee WH, Ong AG, et al. Recurrent fusion genes in gastric cancer: CLDN18-ARHGAP26 induces loss of epithelial integrity. Cell Rep. (2015) 12:272–272 doi: 10.1016/j.celrep.2015.06.020
6. Shu Y, Zhang W, Hou Q, Zhao L, Zhang S, Zhou J, et al. Prognostic significance of frequent CLDN18-ARHGAP26/6 fusion in gastric signet-ring cell cancer. Nat Commun. (2018) 9:2447. doi: 10.1038/s41467-018-04907-0
7. Nakayama I, Shinozaki E, Sakata S, Yamamoto N, Fujisaki J, Muramatsu Y, et al. Enrichment of CLDN18-ARHGAP fusion gene in gastric cancers in young adults. Cancer Sci. (2019) 110:1352–135 doi: 10.1111/cas.13967
8. Tanaka A, Ishikawa S, Ushiku T, Yamazawa S, Katoh H, Hayashi A, et al. Frequent CLDN18-ARHGAP fusion in highly metastatic diffuse-type gastric cancer with relatively early onset. Oncotarget. (2018) 9:29336–293 doi: 10.18632/oncotarget.25464
9. Yang H, Hong D, Cho SY, Park YS, Ko WR, Kim JH, et al. RhoGAP domain-containing fusions and PPAPDC1A fusions are recurrent and prognostic in diffuse gastric cancer. Nat Commun. (2018) 9:4439. doi: 10.1038/s41467-018-06747-4
10. Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ. (2009) 339:b2535. doi: 10.1136/bmj.b2535
11. Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. (2010) 25:603–60 doi: 10.1007/s10654-010-9491-z
12. Higgins JPT, Thomas J, Chandler J, Cumpston M, Li T, Page MJ, et al. Cochrane Handbook for Systematic Reviews of Interventions version 6.0 (updated July 2019). Cochrane (2019). Available online at: https://training.cochrane.org/handbook
13. Hozo SP, Djulbegovic B, Hozo I. Estimating the mean and variance from the median, range, and the size of a sample. BMC Med Res Methodol. (2005) 5:13. doi: 10.1186/1471-2288-5-13
14. Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. (2018) 173:400–400tics doi: 10.1016/j.cell.2018.02.052
15. Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. (2007) 8:16. doi: 10.1186/1745-6215-8-16
16. Amin E, Jaiswal M, Derewenda U, Reis K, Nouri K, Koessmeier KT, et al. Deciphering the molecular and functional basis of RHOGAP family proteins: a systematic approach toward selective inactivation of RHO family proteins. J Biol Chem. (2016) 291:20353–203 doi: 10.1074/jbc.M116.736967
17. Lundmark R, Doherty GJ, Howes MT, Cortese K, Vallis Y, Parton RG, et al. The GTPase-activating protein GRAF1 regulates the CLIC/GEEC endocytic pathway. Curr Biol. (2008) 18:1802–18 doi: 10.1016/j.cub.2008.10.044
18. Regev M, Sabanay H, Kartvelishvily E, Kam Z, Bershadsky AD. Involvement of Rho GAP GRAF1 in maintenance of epithelial phenotype. Cell Adh Migr. (2017) 11:367–367 doi: 10.1080/19336918.2016.1227910
19. Cheng X, Yu S, Wang Y, Cui Y, Li W, Yu Y, et al. The role of oxaliplatin in the adjuvant setting of different Lauren's type of gastric adenocarcinoma after D2 gastrectomy: a real-world study. Gastric Cancer. (2019) 22:587–587 doi: 10.1007/s10120-018-0895-x
20. Ghandi M, Huang FW, Jane-Valbuena J, Kryukov GV, Lo CC, McDonald ER, et al. Next-generation characterization of the cancer cell line encyclopedia. Nature. (2019) 569:503–50 doi: 10.1038/s41586-019-1186-3
21. Li H, Ning S, Ghandi M, Kryukov GV, Gopal S, Deik A, et al. The landscape of cancer cell line metabolism. Nat Med. (2019) 25:850–850 doi: 10.1038/s41591-019-0404-8
22. Hidalgo M, Amant F, Biankin AV, Budinska E, Byrne AT, Caldas C, et al. Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer Discov. (2014) 4:998–998er doi: 10.1158/2159-8290.CD-14-0001
23. Tuveson D, Clevers H. Cancer modeling meets human organoid technology. Science. (2019) 364:952–95 doi: 10.1126/science.aaw6985
24. Kondo J, Inoue M. Application of cancer organoid model for drug screening and personalized therapy. Cells. (2019) 8:470. doi: 10.3390/cells8050470
25. Yan HHN, Siu HC, Law S, Ho SL, Yue SSK, Tsui WY, et al. A comprehensive human gastric cancer organoid biobank captures tumor subtype heterogeneity and enables therapeutic screening. Cell Stem Cell. (2018) 23:882–882 Ste doi: 10.1016/j.stem.2018.09.016
26. Nakayama I, Shinozaki E, Mashima T, Kawaguchi T, Sakata S, Yamamoto N, et al. Abstract 1954: functional analyses of CLDN18-ARHGAP26 fusion gene in gastric cancers. Cancer Res. (2018) 78(13 Suppl.):1954. doi: 10.1158/1538-7445.AM2018-1954
27. Rohde C, Yamaguchi R, Mukhina S, Sahin U, Itoh K, Tureci O. Comparison of Claudin 18.2 expression in primary tumors and lymph node metastases in Japanese patients with gastric adenocarcinoma. Jpn J Clin Oncol. (2019) 49:870–87 doi: 10.1093/jjco/hyz068
28. Sahin U, Koslowski M, Dhaene K, Usener D, Brandenburg G, Seitz G, et al. Claudin-18 splice variant 2 is a pan-cancer target suitable for therapeutic antibody development. Clin Cancer Res. (2008) 14:7624–762 doi: 10.1158/1078-0432.CCR-08-1547
29. Singh P, Toom S, Huang Y. Anti-claudin 18.2 antibody as new targeted therapy for advanced gastric cancer. J Hematol Oncol. (2017) 10:105. doi: 10.1186/s13045-017-0473-4
30. Tureci O, Sahin U, Schulze-Bergkamen H, Zvirbule Z, Lordick F, Koeberle D, et al. A multicentre, phase IIa study of zolbetuximab as a single agent in patients with recurrent or refractory advanced adenocarcinoma of the stomach or lower oesophagus: the MONO study. Ann Oncol. (2019) 30:1487–148 doi: 10.1093/annonc/mdz199
31. Sahin U, Schuler M, Richly H, Bauer S, Krilova A, Dechow T, et al. A phase I dose-escalation study of IMAB362 (Zolbetuximab) in patients with advanced gastric and gastro-oesophageal junction cancer. Eur J Cancer. (2018) 100:17–170 doi: 10.1016/j.ejca.2018.05.007
32. Guarnerio J, Bezzi M, Jeong JC, Paffenholz SV, Berry K, Naldini MM, et al. Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell. (2016) 165:289–289l doi: 10.1016/j.cell.2016.03.020
33. Oncogenic circular RNAs arise from chromosomal translocations. Cancer Discov. (2016) 6:OF20. doi: 10.1158/2159-8290.CD-RW2016-068
34. Tan S, Sun D, Pu W, Gou Q, Guo C, Gong Y, et al. Circular RNA F-circEA-2a derived from EML4-ALK fusion gene promotes cell migration and invasion in non-small cell lung cancer. Mol Cancer. (2018) 17:138. doi: 10.1186/s12943-018-0887-9
35. Wu K, Liao X, Gong Y, He J, Zhou JK, Tan S, et al. Circular RNA F-circSR derived from SLC34A2-ROS1 fusion gene promotes cell migration in non-small cell lung cancer. Mol Cancer. (2019) 18:98. doi: 10.1186/s12943-019-1028-9
Keywords: gastric cancer, fusion gene, CLDN18-ARHGAP, survival, therapy
Citation: Zhang W-H, Zhang S-Y, Hou Q-Q, Qin Y, Chen X-Z, Zhou Z-G, Shu Y, Xu H and Hu J-K (2020) The Significance of the CLDN18-ARHGAP Fusion Gene in Gastric Cancer: A Systematic Review and Meta-Analysis. Front. Oncol. 10:1214. doi: 10.3389/fonc.2020.01214
Received: 03 March 2020; Accepted: 15 June 2020;
Published: 02 September 2020.
Edited by:
Ling Kui, Harvard Medical School, United StatesReviewed by:
Hong Zheng, Stanford University, United StatesCopyright © 2020 Zhang, Zhang, Hou, Qin, Chen, Zhou, Shu, Xu and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yang Shu, c2h1eWFuZzE5ODZAZ21haWwuY29t; Heng Xu, eHVoZW5nODE5MTZAc2N1LmVkdS5jbg==; Jian-Kun Hu, aHVqa3djaEAxMjYuY29t
†These authors have contributed equally to this work
‡ORCID: Jian-Kun Hu orcid.org/0000-0002-3294-3471
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.