
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 20 May 2020
Sec. Cancer Genetics
Volume 10 - 2020 | https://doi.org/10.3389/fonc.2020.00785
 Qiang Cao1†
Qiang Cao1† Pengchao Li1†
Pengchao Li1† Pu Cao2†
Pu Cao2† Jian Qian1
Jian Qian1 Mulong Du3
Mulong Du3 Li Li4
Li Li4 Meilin Wang3
Meilin Wang3 Chao Qin1
Chao Qin1 Pengfei Shao1
Pengfei Shao1 Zhengdong Zhang3
Zhengdong Zhang3 Qiang Lu1*
Qiang Lu1* Zengjun Wang1*
Zengjun Wang1*The long non-coding RNA (lncRNA) H19 has been demonstrated to play a crucial role in carcinogenesis, including renal cell carcinoma (RCC). However, the impact of genetic variations in H19 gene on RCC has not been investigated before. In the present study, we sought to evaluate whether genetic polymorphisms in H19 are related to the susceptibility and mortality of RCC. We genotyped four widely studied polymorphisms in H19 and assessed their relationship with susceptibility and prognosis of RCC in a case-control study compromising 1,027 cases and 1,094 controls. The functionality of the important polymorphism was further investigated by real-time polymerase chain reaction and luciferase reporter assay. We found that H19 rs2839698 was significantly associated with risk and prognosis of RCC. Compared with the H19 rs2839698 CC genotype, the variant genotypes (CT/TT) were significantly associated with increased risk of RCC (P = 0.023, OR = 1.21; 95% CI = 1.03–1.45). Besides, patients with variant genotypes (CT/TT) were more likely to develop large tumor (P = 0.003, OR = 1.47; 95% CI = 1.16–1.85) and advanced disease (P = 0.010, OR = 1.59; 95% CI = 1.12–2.26); and had a significantly unfavorable overall survival than those with the rs2839698 CC genotype (CT/TT vs. CC: Log-rank P = 0.026, HR = 2.25, 95%CI = 1.07–4.75). Furthermore, the CT/TT genotypes were associated with significantly increased expression of H19 in renal tissue. The luciferase reporter assays revealed the potential effect of rs2839698 variant on the binding of microRNAs to H19. Our results suggest that the H19 rs2839698 variant may be a genetic predictor of susceptibility and mortality of RCC. The risk effects and the functional impact of the variant on H19 still need further validation.
Renal cell carcinoma (RCC) is the predominant type of kidney cancer, accounting for 5 and 3% of all malignancies in men and women, respectively (1, 2). It is estimated that up to 17% of patients show developed metastases at diagnosis and ~30% of the remaining patients will develop metastases even following surgical treatment (3, 4). Patients' prognosis is generally poor after detecting these metastases (3). Renal carcinogenesis is a complex process involving interactions between environmental and genetic factors (5). In recent years, genetic variations have been widely studied and demonstrated to influence the susceptibility, progression and prognosis of RCC (6–8).
Long non-coding RNAs (lncRNAs) are a class of endogenous cellular RNAs of more than 200 nucleotides in length that lack protein-coding capacity (9). Accumulating evidence suggests that lncRNAs play a crucial role in the occurrence and progression of various type of cancer (10, 11). LncRNA H19 is a paternally imprinted gene which is located in chromosome 11p15.5 in human (12), and participates in numerous important biological and pathological processes through regulating the function of miRNAs (13) and mediating the DNA methylation (14). H19 is closely linked to insulin-like growth factor-2 (IGF2) gene which is also located in the region subjected to imprinting by methylation and plays a crucial role in the normal growth and development of the fetus (15) and also important in cancer occurrence and progression (16). It has been suggested that aberrant H19 expression was involved in variety of malignancies including bladder cancer (17), breast cancer (18), esophageal cancer (19), and RCC (20). In RCC, Wang et al. demonstrated that lncRNA H19 was over-expressed in tumor tissues and correlated with tumor stage, lymph node metastasis, and distant metastasis. Besides, the expression of lncRNA H19 was proposed to be an independent predictor for the clinical outcome of RCC patients (20).
Emerging studies have consistently demonstrated that genetic polymorphisms in H19 gene are associated with the risk or prognosis of various cancers, including gastric cancer (21), colorectal cancer (22), bladder cancer (23, 24) and breast cancer (25). Most recently, a meta-analysis conducted by Hashemi et al. (26) on the association between H19 polymorphisms and cancer risk has demonstrated that H19 rs2839698 was associated with increased risk of gastrointestinal cancer. In light of the critical role of H19 in RCC, it is possible that genetic variants in H19 may have an effect on the risk and/or prognosis of RCC. However, to the best of our knowledge, no published study has yet investigated this issue. Therefore, in the present study, we selected four most widely studied polymorphisms in H19 (rs2839698, rs3024270, rs217727, and rs2735971), and evaluated their associations with RCC risk and prognosis in a two-stage case-control study comprising a total of 1,027 cases and 1,094 controls in Chinese population.
The present ongoing study was started in May 2004 and was approved by the Local Ethics Committees of the First Affiliated Hospital with Nanjing Medical University. The details of the inclusion criteria were described previously (7, 27). In brief, all subjects included in the study were ethnic Han Chinese individuals and genetically unrelated. Those patients who had received prior chemotherapy or radiotherapy, or had a different type of malignancy, were excluded from the present study. The controls were recruited from individuals who were seeking health examination at the outpatient department in the hospital and were frequency-matched to cases by sex and age (±5 years). Those subjects who were genetically related to the patients or had individual history of any cancer were excluded. The study was originally two-stage designed; the first set was recruited between May 2004 and October 2009 and a total of 355 RCC patients and 362 controls were included. The patients in this cohort were followed up prospectively every 6 months at the outpatient department or though patients' contacts. The endpoint was death or loss of follow-up which was considered censoring. Forty-four patients (12.4%) were excluded due to lack of adequate information for follow-up (41 cases) or low DNA quality (3 cases) for genotyping. The maximum follow-up time was 72 months and the median follow-up time was 25.1 months. The second set comprised of 672 RCC cases and 703 controls which was described previously (27). Before recruitment, a standard questionnaire was administered through face-to-face interviews by trained interviewers to obtain information on demographic data and related factors. The definition of smoker and drinker was described previously (28). At recruitment, information on demographic data and related factors of the participants were obtained by trained interviewers through face-to-face interviews. Each participant donated 5 ml venous blood collected in an EDTA tube after providing written informed consent at the interviews. In the present study, we genotyped and analyzed the four polymorphisms in the combination of both the set which comprised a total of 1,027 cases and 1,094 controls in the case-control analysis.
Genetic polymorphisms in both the H19 gene and its promoter located in human chromosome 11p15.5 (position 2016406-2021693) were identified using the UCSC browser (http://genome.ucsc.edu/) with the selection criterion of a minor allele frequency (MAF) >0.05 in the CHB and JPT population from the 1000 Genomes Project. The detail information on selecting of these four polymorphisms was reported previously (24). Four tagged polymorphisms including rs2839698, rs217727, rs3741216, and rs3741219 were finally selected and investigated in this study. Genomic DNA was prepared and stored as described previously (7, 27). The genotyping of these four polymorphisms was performed using TaqMan SNP Genotyping Assays (Applied Biosystems, Foster City, CA, USA), as described previously (24). The sequences of the primers and probes are listed in Table S1.
A total of 46 surgically removed renal cancer tissues with paired paratumor renal tissues (>2 cm away from tumor) were collected to test the expression level of lncRNA H19 in vivo. The tissues were immediately stored in liquid nitrogen after taken from the surgically removed samples. The RNA extraction and cDNA synthesis were described previously (29). The level of lncRNA H19 was measured by quantitative real-time reverse transcription (RT)-PCR on the ABI Prism 7900 sequence detection system (Applied Biosystems). β-actin was used as an internal reference gene. The primers used for lncRNA H19 were 5′-CCCACAACATGAAAGAAATGGTGC-3′(sense) and 5′-CACCTTCGAGAGCCGATTCC-3′(anti-sense), and the primers for β-actin were 5′-AGAAAATCTGGCACCACACC-3′(sense) and 5′-TAGCACAGCCTGGATAGCAA-3′(anti-sense). The reaction mixture contained 0.1M of each primer, 2 × SYBR Green PCR Master Mix (TaKaRa, Berkeley, CA, USA), and 1 mL of cDNA (1:10 dilution). The amplification was performed under the following conditions: 95°C for 30s, and 40 cycles of 95°C for 15s and 60°C for 30s. Each reaction was done in triplicate.
Reporter plasmids containing 160-bp lncRNA H19 exon region fragments flanking the rs2839698 polymorphism with either the C or T allele were synthesized based on the reporter vector psiCHECK-2 (Promega, Madison, WI, USA). The accuracy of the constructed plasmids was verified by DNA sequencing. The renal cell adenocarcinoma cell line (786-o) and Human Embryonic Kidney 293 cells (HEK-293) were used for cell transfection and luciferase assays, as reported previously (29). Cells were seeded (at the density of 1.5 × 104 cells/well) into 96-well plates containing culture medium, followed by 24-h incubation at the culture condition of 37°C, 100% humidity and 5% CO2. Using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA), reporter plasmids containing either rs2839698 C or T allele along with different miRNAs mimics or inhibitors were co-transfected into 786-o and HEK-293 cells. The non-miRNA-inserted plasmid was used as a negative control. Luciferase activity of the cells was measured with a Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) 24 h after the transfection. Experiments were performed independently in triplicate for each of the plasmid constructs.
Student's t-test or chi-square test was used to test the differences in the distribution of demographic factors, clinical parameters, and frequencies of genotypes between cases and controls depending on the type of the variables. A goodness-of-fit chi-square test was used to test against departure from Hardy-Weinberg equilibrium in the control group for all the polymorphisms. False discovery rate (FDR) based on the Benjamini-Hochberg method was used to adjust the P value for multiple comparisons of the four SNPs before further analysis. The associations were considered statistically significant when FDR-adjusted P values were <0.05. The relationships between H19 polymorphisms and RCC risk as well as tumor characteristics were assessed by computing odds ratios (ORs) and 95% confidence intervals (CIs) from unconditional logistic regression models adjusted for potential confounders. Survival time was calculated from the date of RCC diagnosis to the date of death or last follow-up. Specific survival curves according to clinical features and different H19 genotypes were evaluated by using the Kaplan-Meier method and compared by the log-rank test. Cox regression analysis was used to determine predictive factors of RCC prognosis by estimating the hazard ratios (HRs) and their 95% CIs with adjustment for potential confounders. Differences in luciferase reporter activity and H19 expression levels among different groups were evaluated using Student's t-test or ANOVA. All analyses were performed using SAS 9.1.3 (SAS Institute, Cary, NC, USA) with two-sided P values. P value of <0.05 was considered statistically significant.
Frequency distributions of selected characteristics among the cases and controls are presented in Table S2. This is the same population as we described previously (27). Briefly, no significant difference was noticed between cases and controls regarding to age, gender, and drinking status (all P > 0.05). There were more smokers, hypertensive patients and diabetics in case group compared to that of the control group (P = 0.017, < 0.001 and <0.001, respectively). Among the patients, 65.3% of the patients were in stage I, whereas 19.5, 7.1, and 8.1% were found to have stage II, III, and IV diseases, respectively. The percentage of nuclear grade from I to IV was 21.6, 51.1, 20.7, and 6.5, respectively.
The associations between H19 rs2839698, rs3024270, rs217727, and rs2735971 polymorphisms and risk of RCC are presented in Table 1. Genotype frequencies of the four polymorphisms in controls all conformed to HWE (all P > 0.05). As shown in Table 1, the genotypic distributions of rs2839698 in case and control groups was significantly different (P = 0.006), and remained significant after adjusting for multiple comparisons (FDR = 0.024). Compared with individuals carrying the rs2839698CC genotype, individuals harboring the rs2839698 variant TT genotype was associated with a significant increased RCC risk (P = 0.028, OR = 1.48, 95% CI = 1.16–2.42). Significant increased RCC risk was also noted when combining the CT and TT genotypes, and compared with rs2839698CC genotype (P = 0.023, OR = 1.21, 95%CI = 1.03–1.45). No significant evidence of association was found between any of the other polymorphisms and RCC risk. These results suggest that H19 rs2839698 may confer individuals' genetic susceptibility to RCC.
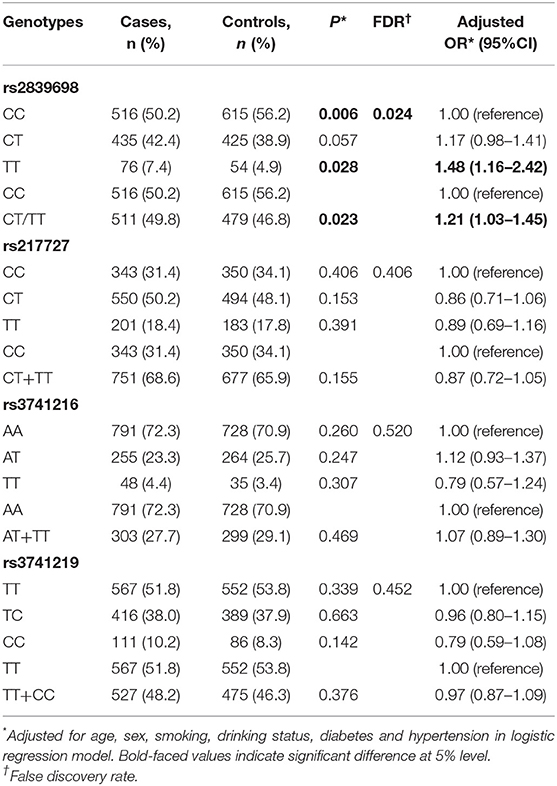
Table 1. Genotype frequencies of the H19 polymorphisms among the cases and controls and their associations with risk of RCC.
To test whether the H19 rs2839698 polymorphism could has effects on disease progression; we then analyze the association between rs2839698 genotypes and the tumor size, clinical stage and tumor grade of RCC in the patient group. As shown in Table 2, no significant association between rs2839698 genotypes and tumor grade was observed; however, the rs2839698 CT /TT genotypes were significantly more frequent in patients with tumor size more than 4 cm and those with advanced clinical stage (CT/TT vs. CC: P = 0.003, OR = 1.47; 95% CI = 1.16–1.85 and P = 0.010, OR = 1.59; 95% CI = 1.12–2.26, respectively). These results suggest that compare to patients with rs2839698 CC genotype, those harboring rs2839698 CT /TT genotypes were more prone to develop advanced disease. No significant association between any of the other polymorphisms and clinicopathological characteristics of RCC was found (data not show).
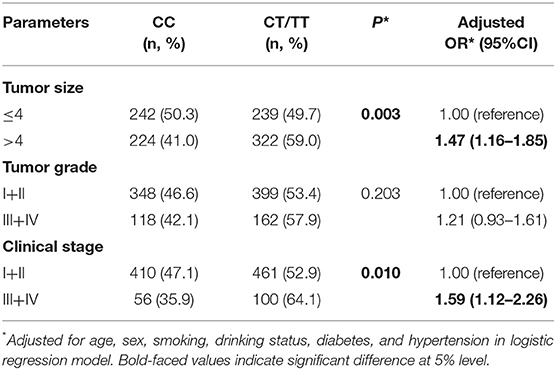
Table 2. Associations of lncRNA H19 rs2839698 polymorphism with clinicopathological characteristics of RCC.
As H19 rs2839698 was shown to contribute to RCC progression, we then sought to explore the association between rs2839698 polymorphism and survival of RCC patients using the Log-rank test and Kaplan-Meier analysis. As shown in Table 3 and Figures 1A,B, we found that the rs2839698 polymorphism was significantly associated with patients' survival (Log-Rank P = 0.006). The association remained significant after adjusting for multiple comparisons (FDR = 0.024). Compared with patients harboring the rs2839698CC genotype, those patients with rs2839698TT or CT/TT genotypes had an unfavorable RCC survival (HR = 4.29, 95%CI = 1.15–15.94 and HR=2.25, 95% CI=1.07–4.75, respectively). However, no significant evidence of association was found between the other polymorphisms and RCC survival as shown in Table 3. We then performed a univariate and multivariate Cox proportional hazard analysis for survival of RCC patients. As shown in Table 4 and Figures 1C,D, clinical stage, tumor grade, and H19 rs2839698 (CT/TT vs. CC) were associated with RCC survival in univariate analysis; in multivariate analysis, clinical stage was found to be the best prognostic factor for RCC survival, followed by tumor grade (P < 0.001, HR = 15.5; 95% CI = 5.94–40.34 and P = 0.004, HR = 3.98; 95% CI = 1.55–10.23, respectively). Interestingly, H19 rs2839698 (CT/TT vs. CC) was also an independent predictor of RCC survival (P = 0.33, HR = 2.25; 95% CI = 1.07–4.75) in the multivariate analysis.
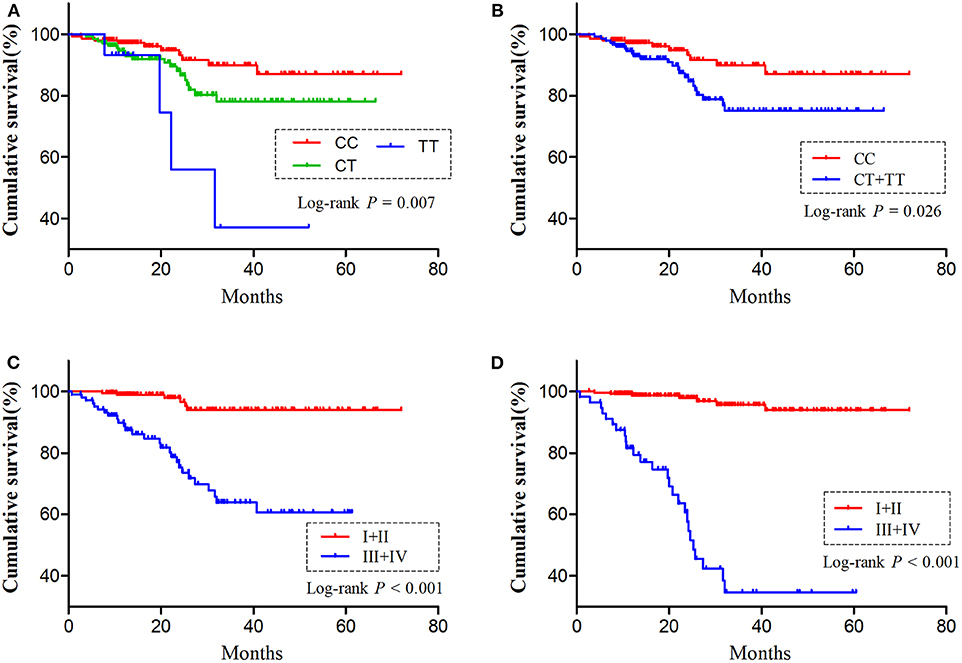
Figure 1. The influence of H19 rs2839698 polymorphism on RCC survival. Kaplan-Meier survival curves illustrate RCC overall survival according to (A,B) different H19 rs2839698 genotypes, (C) tumor grade (I + II vs. III + IV) and (D) clinical stage (I + II vs. III + IV).
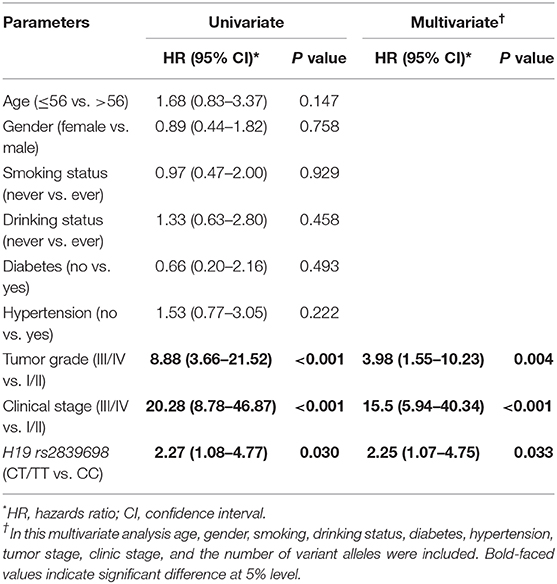
Table 4. Univariate and multivariate Cox proportional hazard analysis of death risk in patients with RCC.
To further explore the potential functionality of H19 rs2839698, we investigated the effect of this polymorphism on H19 expression in cancer tissues and paratumor normal renal tissues using real-time quantitative PCR. As shown in Figure 2A, the lncRNA H19 expression level in tumor tissues was significantly higher than that in the adjacent normal tissues (P < 0.01) which was consistent with previous reports (20, 30). Besides, in normal tissues, as shown in Figure 2B, compared with individuals carrying the rs2839698CC genotype, individuals carrying the variant T allele (CT/TT genotypes) had increased levels of H19 expression (P = 0.019, 0.004 and < 0.001 for CT vs. CC, TT vs. CC and CT/TT vs. CC respectively). These results suggested that overexpression of lncRNA H19 may contribute to renal carcinogenesis and that rs2839698 may be involved in the process by regulating the expression levels of lncRNA H19.
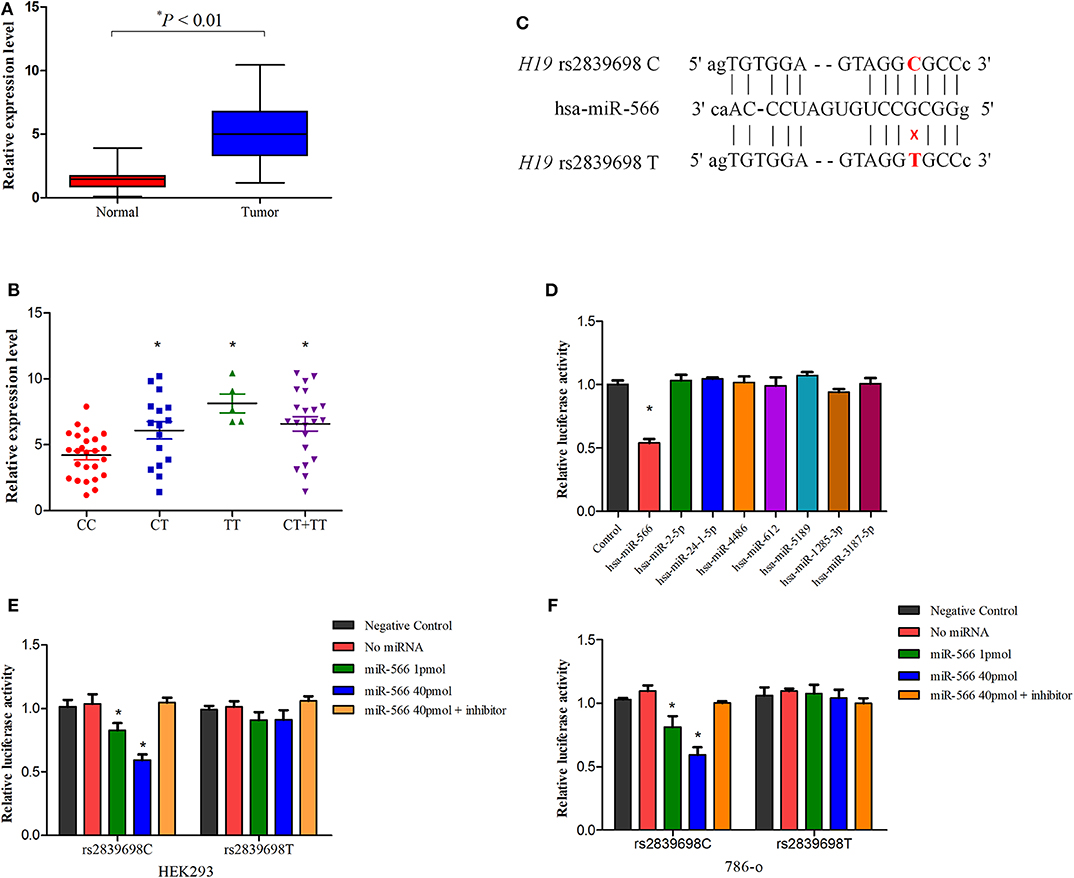
Figure 2. The influence of H19 rs2839698 variant on H19 expression. (A) Distribution and comparison of H19 expression in tumors and adjacent normal tissues (P < 0.01); (B) Association between the H19 expression in renal tissues and H19 rs2839698 genotypes (P = 0.019, 0.004, and 0.001 for CT vs. CC, TT vs. CC and CT/TT vs. CC, respectively); (C) Schematic image of binding interaction between miR-566 and H19 rs2839698 C and T alleles. (D) The psi-CHECK-2-H19-C-allele and miRNA mimics were co-transfected into HEK293 cell line. Relative luciferase activities in the cells were measured from three independently differentiated clones in each genotype. Only the luciferase activity in cells co-transfected with miR-566 was significantly decreased (*P < 0.05). The psi-CHECK-2-H19-C-allele or T allele construct as well as miR-566 mimic and inhibitor were co-transfected into (E) HEK293 and (F) 786-0 cell lines. *P < 0.05, compared with the psi-CHECK-2-rs2839698-C/T allele constructs co-transfected with miRNA control.
An in silico analyses was performed previously using RNAfold and SNPfold databases to explore whether biological functions of H19 would notably affected by the genetic variants (22). As reported previously, the secondary structure H19 was altered by rs2839698 variant (22). As rs2839698 is located in the 3′untranslated region of H19 gene, we speculate that the increased expression of H19 could be associated with alteration of miRNAs target. As predicted by bioinformatics mode (22), the change from a C allele to T allele at rs2839698 may disrupt the binding of hsa-miR-24-1-5p, hsa-miR-4486, hsa-miR-566, and hsa-miR-24-2-5p, and create binding site for hsa-miR-612, hsa-miR-5189, hsa-miR-1285-3p and hsa-miR-3187-5p. To test the prediction, we then introduced the luciferase reporter vectors (rs2839698 C or T allele) together with the mimics of the miRNAs into HEK-293 cell line. As shown in Figures 2C,D, in the presence of C allele, the luciferase activity of cells co-transfected with miR-566 was significantly decreased compared with the control in HEK-293 cells. No significant differences of the luciferase activity were observed for all the miRNAs in the presence of T allele in HEK-293 cells, as shown in Figure S1. To further confirm the prediction, the reporter vectors (psiCHECK-2-rs2839698C and psiCHECK-2- rs2839698T), miR-556 mimics and inhibitor were transiently co-transfected into 786-O and HEK-293 cell lines. In both cell lines, as shown in Figures 2E,F, luciferase activity was significant decreased in the presence of C allele when vectors co-transfected with miR-556 mimics; however, no significant alternations in luciferase activity were observed in the presence of the miR-556 inhibitor. No significant influence of miR-566 inhibitor alone on the luciferase activity of H19 rs2839698 variant, as shown in Figure S2.
In the preset study, we investigated the associations between polymorphisms in H19 and RCC susceptibility as well as prognosis in a Chinese population. Our results suggested that the H19 rs2839698 variant was associated with an increased risk of RCC. We also demonstrated that this variant was associated with larger tumor and advanced clinical stage of RCC, and was an independent prognostic predictor of patients' survival, along with clinical tumor stage and grade in multivariate analysis. The functional role of rs2839698 variant in elevating the H19 level was subsequently demonstrated in in vivo and in vitro assays. To the best of our knowledge, this is the first study to demonstrate a role of H19 rs2839698 variant in the etiology and prognosis of RCC.
Long non-coding RNA H19 is a paternally imprinted oncofetal gene located on chromosome 11p15.5 and acts as an oncogene involving in the process of occurrence and metastasis of malignancies (31, 32). Highly up-regulated of H19 was demonstrated in a variety of tumors including colorectal cancer (33), hepatocellular cancer (34), breast cancer (35), lung cancers (36), and RCC (20, 30). In RCC, Wang et al. demonstrated that relative level of H19 was significantly higher in renal tumors compared to the adjacent normal renal tissues. Besides, patients with higher H19 expression had more advanced clinical stage and poorer prognosis than those with lower expression (20). Consistent with this study, He et al. also showed that H19 was overexpressed in renal carcinoma and further demonstrated H19 was involved in the migration and invasion of RCC through miR-29a-3p/E2F1 pathway (30). In the present study, we also observed that the H19 level was significantly higher in RCC tissues than in normal renal tissues, which further confirmed a causative role of the H19 in RCC.
Accumulation of evidence has suggested that genetic variation in lncRNAs could play a crucial role in cancer susceptibility and prognosis. Given the important role of H19 in tumorigenesis, the influence of genetic polymorphisms in H19 on caner susceptibility and prognosis has been extensively investigated in various malignancies by previous studies (21–25). Consistent with our results, several other studies have also demonstrated H19 rs2839698 variant to be a risk factor for gastrointestinal cancer (26), colorectal cancer (22), and hepatocellular cancer (37). In hepatocellular cancer, Yang and colleagues found that H19 rs2839698 variant genotype not only confer an increased HCC risk, but also a potential genetic predictor for HCC prognosis in the subgroup of ever smokers (37). Similar to our findings, they found that patients with H19 rs2839698 variant genotype (CT) had a significant unfavorable prognosis (P = 0.035, HR = 5.19, 95% CI = 1.12–24.07).
Accumulating evidence has shown that genetic variations could exert function through altering both DNA and RNA secondary structures (38, 39), and then influencing the binding affinity of miRNAs to its' specific region (40, 41). As for lncRNAs, Zheng et al. demonstrated that a G>A change at rs11655237 in LINC00673 could create a target binding site for miR-1231, which diminished the effect of LINC00673 in an allele-specific manner and thus confers susceptibility to pancreatic cancer (42). Recently, in colorectal cancer, Wu et al. showed that the allele C>G change at rs664589 in MALAT1 altered the binding affinity of the miR-195-5p to the mutant region leading to increased MALAT1 expression and then promoting the colorectal cancer growth, and metastasis (43). However, to the best of our knowledge, no published studies have focused on the effect of H19 rs2839698 in regulating the H19 expression and function.
As predicted by in silico analyses using the SNPfold algorithm, the secondary structure of H19 was dramatically changed with rs2839698 C/T alleles, suggesting the rs2839698 variant might affect the susceptibility and prognosis of RCC by altering the secondary structure H19. It has been suggested that lncRNAs and miRNAs could negatively modulated each other by forming a reciprocal repression-regulatory loop (44, 45). For instance, Hirata et al. found that the interaction between MALAT1 and miRNA-205 had reciprocal effects in RCC (44). Using bioinformatics analysis and luciferase reporter analysis, we identified that hsa-miR-566 might lose the target in H19 in the presence of rs2839698 T allele. Interestingly, hsa-miR-566 was demonstrated to be an oncogene and a potential biomarker for prognosis in renal cell carcinoma (46). Therefore, we speculate that the rs2839698 T allele might block the binding of hsa-miR-566 to the 3'UTR of H19 and then disrupt the reciprocal repression-regulatory loop between hsa-miR-566 and H19, and subsequently, lead to up regulation of H19. This is also consistent with our findings that individuals with the rs2839698 TT genotype showed increased H19 level compared to individuals with the rs2839698 CC genotype. Given the oncogene role of H19, up-regulation of H19 due to the rs2839698 variant genotype may confer RCC susceptibility and prognosis, which may explain our findings in the case-control study. However, limitations should be concerned when interpreting our results. First, the present case-control study was hospital-based instead of population-based; therefore the possibility of the selection bias of individuals associated with a particular genotype could not be rule out. Besides, our functional results toward the H19 rs2839698 variant are only preliminary, the exact mechanism underlying the functional effects of this variant on H19 expression still requires further investigation.
In conclusion, this is the first study demonstrating a role of H19 rs2839698 polymorphism in RCC susceptibility and prognosis in a Chinese cohort. Our results, although preliminary, highlight a putatively functional effect of this variant on regulating the H19 expression, which might through disrupting the reciprocal repression-regulatory loop between hsa-miR-566 and H19. However, further validation in a larger population and functional studies are still warranted.
All datasets generated for this study are included in the article/Supplementary Material.
The studies involving human participants were reviewed and approved by the Local Ethics Committees of the First Affiliated Hospital with Nanjing Medical University. The patients/participants provided their written informed consent to participate in this study.
QC and PL proposed the hypothesis and designed the experiments. QC, JQ, MD, and CQ performed the experimental work and manuscript drafting. PS, MW, ZZ, and LL performed the analysis and manuscript drafting. PC, QC, and PL performed the experimental work. QL, ZW, and ZZ conceived the study, participated in its design and coordination, and helped edit the manuscript.
This work was supported by the National Natural Science Foundation of China (Grants No. 81772711, 81602235, and 81272832), Jiangsu Province's Key Provincial Talents Program (ZDRCA2016006), the 333 project of Jiangsu Province (LGY2016002), the Provincial Initiative Program for Excellency Disciplines of Jiangsu Province (Grant No. BE2016791).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.00785/full#supplementary-material
1. Capitanio U, Bensalah K, Bex A, Boorjian SA, Bray F, Coleman J, et al. Epidemiology of renal cell carcinoma. Eur Urol. (2019) 75:74–84. doi: 10.1016/j.eururo.2018.08.036
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. (2019) 69:7–34. doi: 10.3322/caac.21551
3. Cohen HT, McGovern FJ. Renal-cell carcinoma. N Engl J Med. (2005) 353:2477–90. doi: 10.1056/NEJMra043172
4. Capitanio U, Montorsi F. Renal cancer. Lancet. (2016) 387:894–906. doi: 10.1016/S0140-6736(15)00046-X
5. Semenza JC, Ziogas A, Largent J, Peel D, Anton-Culver H. Gene-environment interactions in renal cell carcinoma. Am J Epidemiol. (2001) 153:851–9. doi: 10.1093/aje/153.9.851
6. Purdue MP, Johansson M, Zelenika D, Toro JR, Scelo G, Moore LE, et al. Genome-wide association study of renal cell carcinoma identifies two susceptibility loci on 2p21 and 11q13.3. Nat Genet. (2011) 43:60–5. doi: 10.1038/ng.723
7. Qin C, Cao Q, Ju X, Wang M, Meng X, Zhu J, et al. The polymorphisms in the VHL and HIF1A genes are associated with the prognosis but not the development of renal cell carcinoma. Ann Oncol. (2012) 23:981–9. doi: 10.1093/annonc/mdr325
8. Wu X, Scelo G, Purdue MP, Rothman N, Johansson M, Ye Y, et al. A genome-wide association study identifies a novel susceptibility locus for renal cell carcinoma on 12p11.23. Hum Mol Genet. (2012) 21:456–62. doi: 10.1093/hmg/ddr479
9. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. (2009) 136:629–41. doi: 10.1016/j.cell.2009.02.006
10. Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. (2011) 10:38. doi: 10.1186/1476-4598-10-38
11. Sanchez Calle A, Kawamura Y, Yamamoto Y, Takeshita F, Ochiya T. Emerging roles of long non-coding RNA in cancer. Cancer Sci. (2018) 109:2093–100. doi: 10.1111/cas.13642
12. Gabory A, Jammes H, Dandolo L. The H19 locus: role of an imprinted non-coding RNA in growth and development. Bioessays. (2010) 32:473–80. doi: 10.1002/bies.200900170
13. Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. (2013) 52:101–12. doi: 10.1016/j.molcel.2013.08.027
14. Zhou J, Yang L, Zhong T, Mueller M, Men Y, Zhang N, et al. H19 lncRNA alters DNA methylation genome wide by regulating S-adenosylhomocysteine hydrolase. Nat Commun. (2015) 6:10221. doi: 10.1038/ncomms10221
15. Harris LK, Westwood M. Biology and significance of signalling pathways activated by IGF-II. Growth Factors. (2012) 30:1–12. doi: 10.3109/08977194.2011.640325
16. Lu L, Katsaros D, Wiley A, Rigault de la Longrais IA, Puopolo M, Schwartz P, et al. Promoter-specific transcription of insulin-like growth factor-II in epithelial ovarian cancer. Gynecol Oncol. (2006) 103:990–95. doi: 10.1016/j.ygyno.2006.06.006
17. Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. (2013) 333:213–21. doi: 10.1016/j.canlet.2013.01.033
18. Berteaux N, Lottin S, Monte D, Pinte S, Quatannens B, Coll J, et al. H19 mRNA-like noncoding RNA promotes breast cancer cell proliferation through positive control by E2F1. J Biol Chem. (2005) 280:29625–36. doi: 10.1074/jbc.M504033200
19. Hibi K, Nakamura H, Hirai A, Fujikake Y, Kasai Y, Akiyama S, et al. Loss of H19 imprinting in esophageal cancer. Cancer Res. (1996) 56:480–2.
20. Wang L, Cai Y, Zhao X, Jia X, Zhang J, Liu J, et al. Down-regulated long non-coding RNA H19 inhibits carcinogenesis of renal cell carcinoma. Neoplasma. (2015) 62:412–8. doi: 10.4149/neo_2015_049
21. Yang C, Tang R, Ma X, Wang Y, Luo D, Xu Z, et al. Tag SNPs in long non-coding RNA H19 contribute to susceptibility to gastric cancer in the Chinese Han population. Oncotarget. (2015) 6:15311–20. doi: 10.18632/oncotarget.3840
22. Li S, Hua Y, Jin J, Wang H, Du M, Zhu L, et al. Association of genetic variants in lncRNA H19 with risk of colorectal cancer in a Chinese population. Oncotarget. (2016) 7:25470–7. doi: 10.18632/oncotarget.8330
23. Verhaegh GW, Verkleij L, Vermeulen SH, den Heijer M, Witjes JA, Kiemeney LA. Polymorphisms in the H19 gene and the risk of bladder cancer. Eur Urol. (2008) 54:1118–26. doi: 10.1016/j.eururo.2008.01.060
24. Hua Q, Lv X, Gu X, Chen Y, Chu H, Du M, et al. Genetic variants in lncRNA H19 are associated with the risk of bladder cancer in a Chinese population. Mutagenesis. (2016) 31:531–8. doi: 10.1093/mutage/gew018
25. Sigurdson AJ, Bhatti P, Chang SC, Rajaraman P, Doody MM, Bowen L, et al. Polymorphisms in estrogen biosynthesis and metabolism-related genes, ionizing radiation exposure, and risk of breast cancer among US radiologic technologists. Breast Cancer Res Treat. (2009) 118:177–84. doi: 10.1007/s10549-009-0307-3
26. Hashemi M, Moazeni-Roodi A, Sarabandi S, Karami S, Ghavami S. Association between genetic polymorphisms of long noncoding RNA H19 and cancer risk: a meta-analysis. J Genet. (2019) 98:81. doi: 10.1007/s12041-019-1126-x
27. Cao Q, Liang C, Xue J, Li P, Li J, Wang M, et al. Genetic variation in IGF1 predicts renal cell carcinoma susceptibility and prognosis in Chinese population. Sci Rep. (2016) 6:39014. doi: 10.1038/srep39014
28. Cao Q, Qin C, Meng X, Ju X, Ding Q, Wang M, et al. Genetic polymorphisms in APE1 are associated with renal cell carcinoma risk in a Chinese population. Mol Carcinog. (2011) 50:863–70. doi: 10.1002/mc.20791
29. Cao Q, Ju X, Li P, Meng X, Shao P, Cai H, et al. A functional variant in the MTOR promoter modulates its expression and is associated with renal cell cancer risk. PLoS ONE. (2012) 7:e50302. doi: 10.1371/journal.pone.0050302
30. He H, Wang N, Yi X, Tang C, Wang D. Long non-coding RNA H19 regulates E2F1 expression by competitively sponging endogenous miR-29a-3p in clear cell renal cell carcinoma. Cell Biosci. (2017) 7:65. doi: 10.1186/s13578-017-0193-z
31. Raveh E, Matouk IJ, Gilon M, Hochberg A. The H19 Long non-coding RNA in cancer initiation, progression and metastasis - a proposed unifying theory. Mol Cancer. (2015) 14:184. doi: 10.1186/s12943-015-0458-2
32. Ye Y, Shen A, Liu A. Long non-coding RNA H19 and cancer: a competing endogenous RNA. Bull Cancer. (2019) 106:1152–9. doi: 10.1016/j.bulcan.2019.08.011
33. Tsang WP, Ng EK, Ng SS, Jin H, Yu J, Sung JJ, et al. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis. (2010) 31:350–8. doi: 10.1093/carcin/bgp181
34. Ye Y, Guo J, Xiao P, Ning J, Zhang R, Liu P, et al. Macrophages-induced long noncoding RNA H19 up-regulation triggers and activates the miR-193b/MAPK1 axis and promotes cell aggressiveness in hepatocellular carcinoma. Cancer Lett. (2000) 469:310–22. doi: 10.1016/j.canlet.2019.11.001
35. Zhang K, Luo Z, Zhang Y, Zhang L, Wu L, Liu L, et al. Circulating lncRNA H19 in plasma as a novel biomarker for breast cancer. Cancer Biomark. (2016) 17:187–94. doi: 10.3233/CBM-160630
36. Saito Y, Gopalan B, Mhashilkar AM, Roth JA, Chada S, Zumstein L, et al. Adenovirus-mediated PTEN treatment combined with caffeine produces a synergistic therapeutic effect in colorectal cancer cells. Cancer Gene Ther. (2003) 10:803–13. doi: 10.1038/sj.cgt.7700644
37. Yang ML, Huang Z, Wang Q, Chen HH, Ma SN, Wu R, et al. The association of polymorphisms in lncRNA-H19 with hepatocellular cancer risk and prognosis. Biosci Rep. (2018) 38:81. doi: 10.1042/BSR20171652
38. Lu L, Katsaros D, Mayne ST, Risch HA, Benedetto C, Canuto EM, et al. Functional study of risk loci of stem cell-associated gene lin-28B and associations with disease survival outcomes in epithelial ovarian cancer. Carcinogenesis. (2012) 33:2119–25. doi: 10.1093/carcin/bgs243
39. Lu L, Risch E, Deng Q, Biglia N, Picardo E, Katsaros D, et al. An insulin-like growth factor-II intronic variant affects local DNA conformation and ovarian cancer survival. Carcinogenesis. (2013) 34:2024–30. doi: 10.1093/carcin/bgt168
40. Novikova IV, Hennelly SP, Sanbonmatsu KY. Structural architecture of the human long non-coding RNA, steroid receptor RNA activator. Nucleic Acids Res. (2012) 40:5034–51. doi: 10.1093/nar/gks071
41. McGeary SE, Lin KS, Shi CY, Pham TM, Bisaria N, Kelley GM, et al. The biochemical basis of microRNA targeting efficacy. Science. (2019) 366:eaav1741. doi: 10.1126/science.aav1741
42. Zheng J, Huang X, Tan W, Yu D, Du Z, Chang J, et al. Pancreatic cancer risk variant in LINC00673 creates a miR-1231 binding site and interferes with PTPN11 degradation. Nat Genet. (2016) 48:747–57. doi: 10.1038/ng.3568
43. Wu S, Sun H, Wang Y, Yang X, Meng Q, Yang H, et al. MALAT1 rs664589 polymorphism inhibits binding to miR-194–5p, contributing to colorectal cancer risk, growth, and metastasis. Cancer Res. (2019) 79:5432–41. doi: 10.1158/0008-5472.CAN-19-0773
44. Hirata H, Hinoda Y, Shahryari V, Deng G, Nakajima K, Tabatabai ZL, et al. Long noncoding RNA MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and interacts with miR-205. Cancer Res. (2015) 75:1322–31. doi: 10.1158/0008-5472.CAN-14-2931
45. Li Q, Zhang C, Chen R, Xiong H, Qiu F, Liu S, et al. Disrupting MALAT1/miR-200c sponge decreases invasion and migration in endometrioid endometrial carcinoma. Cancer Lett. (2016) 383:28–40. doi: 10.1016/j.canlet.2016.09.019
Keywords: genetic variation, renal cell carcinoma, H19, susceptibility, mortality
Citation: Cao Q, Li P, Cao P, Qian J, Du M, Li L, Wang M, Qin C, Shao P, Zhang Z, Lu Q and Wang Z (2020) Genetic Variant in Long Non-Coding RNA H19 Modulates Its Expression and Predicts Renal Cell Carcinoma Susceptibility and Mortality. Front. Oncol. 10:785. doi: 10.3389/fonc.2020.00785
Received: 12 December 2019; Accepted: 22 April 2020;
Published: 20 May 2020.
Edited by:
Rengyun Liu, Johns Hopkins University, United StatesReviewed by:
Lingeng Lu, Yale University, United StatesCopyright © 2020 Cao, Li, Cao, Qian, Du, Li, Wang, Qin, Shao, Zhang, Lu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zengjun Wang, emVuZ2p1bndhbmcyMDAyQHNpbmEuY29t; Qiang Lu, ZG9jdG9ybHZxaWFuZ0BzaW5hLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.