
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CASE REPORT article
Front. Oncol. , 08 May 2020
Sec. Hematologic Malignancies
Volume 10 - 2020 | https://doi.org/10.3389/fonc.2020.00730
This article is part of the Research Topic The Molecular Biology of T Cell Lymphomas/Leukemia View all 5 articles
Background: Anaplastic lymphoma kinase (ALK)-positive anaplastic large cell lymphoma (ALCL) is a rare T-cell neoplasm, accounting for approximately 3% of adult non-Hodgkin lymphomas. Although NPM1 is the most common fusion partner with ALK, many others have been described, necessitating break-apart FISH studies for confirmation of the diagnosis. TNF receptor-associated factor 1 (TRAF1) is a rare ALK partner that is thought to confer a worse prognosis in patients. We describe the utility of next-generation sequencing (NGS) RNA analysis in detection of this uncommon ALK partner.
Case Description: A 42-year-old male with cervical lymphadenopathy presented for excisional biopsy. Following a tissue diagnosis of ALCL, ALK+, RNA from the biopsy was extracted from Formalin-fixed paraffin-embedded (FFPE) tissue and prepared for Anchored Multiplex PCR using the Archer® FusionPlex® v2 assay, which employs unidirectional gene-specific primers using NGS to detect novel or unknown gene partners.
Results: Histologic evaluation of the excised lymph node showed atypical cells, including “horseshoe/kidney”-shaped nuclei. Neoplastic cells were immunoreactive against CD30, ALK (diffuse, cytoplasmic), CD2, CD4, granzyme B, and TIA-1. A diagnosis of ALCL, ALK+ was made. The pattern of ALK immunostaining suggested a non-NPM1-associated ALK translocation pattern, prompting further investigation. NGS fusion analysis showed a translocation involving exon 7 of TRAF1 and exon 20 of ALK.
Conclusion: ALK positivity suggests an overall favorable prognosis of ALCL as compared to ALK-negative cases. However, in the rare published cases of TRAF1-ALK, an aggressive clinical course has been observed, which may reflect the aggressive propensity of this particular fusion, as these cases appear to be refractory to standard chemotherapy and also to the first generation ALK inhibitors. This study highlights the advantage of using NGS in RNA-based fusion assays to detect rare translocations, which can be of some clinical importance in detecting rare but aggressive fusion partners of ALK. As these technologies become more available, there is potential to identify such changes and effectively stratify the prognosis of ALCL patients.
In 1995, Stein and colleagues described a set of lymphomas characterized by bizarre morphologic features with large cells and positivity for the Ki-1 (CD30) antigen (1). It was later discovered that a subset of cases harbored the t(2,5)(p23;q35) translocation, juxtaposing the NPM1 gene to the ALK gene (2). An antibody against this translocation was subsequently developed and found to be immunoreactive against the variant translocations involving ALK (3). The antibody is also immunoreactive against the chimeric ALK protein harboring other partners (4).
Although the majority of ALK+ ALCL exhibit the NPM1-ALK reciprocal translocation, many variant translocations have been identified. Morphologically and immunohistochemically, there is no significant difference between the NPM1-associated ALK+ ALCL and those with the variant translocations (5). With respect to clinical outcome, most agree that there is no significant difference in survival between the different translocations (6). However, recent reports have characterized an ALK+ ALCL with the tumor necrosis factor-1 associated factor (TRAF1) gene translocation. Although only a few cases have been reported, some have exhibited more aggressive clinical features. We highlight in this report another TRAF1-ALK-associated ALK+ ALCL and describe the clinical course of this case. This case also highlights the utility of RNA-based next-generation sequencing (NGS) analysis to detect novel and rare translocations involving genes such as ALK.
A 42-year-old Caucasian male presented history of progressive fatigue, chills, night sweats, and unwanted weight loss. He was found to have left cervical lymphadenopathy. Magnetic resonance imaging (MRI) revealed widespread adenopathy, innumerable small sclerotic lesions throughout the bones and one small lucent lesion in the right side of the pelvis. He underwent excisional biopsy of the cervical lymph node.
Histologic evaluation of the excised lymph node showed effaced architecture with many atypical cells, including those with “horseshoe/kidney”-shaped nuclei. The neoplastic cells were immuno-reactive against CD30, ALK (diffuse, cytoplasmic), CD2, CD4, granzyme B, and TIA-1 (Figure 1). Many background CD68+ histiocytes were noted. A diagnosis of ALCL, ALK+ was made. The pattern of ALK immunostaining suggested a non-NPM1-associated ALK translocation pattern, prompting further investigation. The increased histiocytes were suggestive of a histiocyte-rich variant.
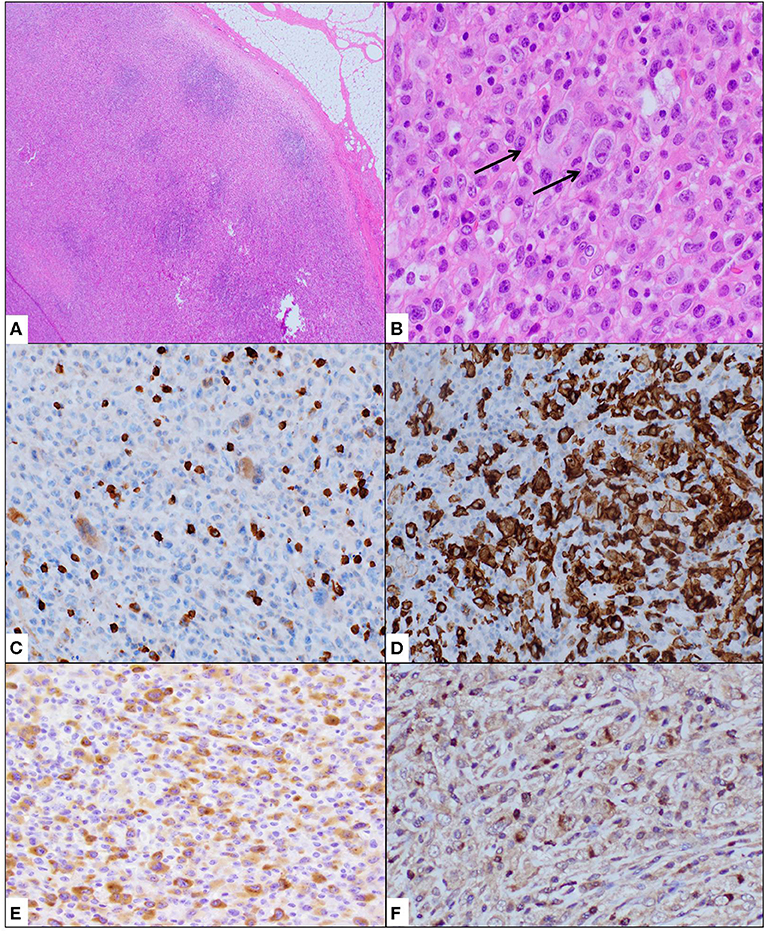
Figure 1. Cervical lymph node biopsy showed effacement of architecture (A) with atypical cells, often containing “hallmark” cells with kidney-shaped nuclei (B). Immunohistochemical stains showed these neoplastic cells to be weak or negative for CD3 (C) and strongly positive for CD30 (D). ALK1 immunostain showed granular cytoplasmic positivity (E) while TIA1 was variably positive (F). Staining for histiocyte CD68 showed increased histiocytes.
Next-generation sequencing (NGS) fusion analysis utilizing RNA from the sample was performed as described previously (7). Briefly, the tissue was prepared from 10-μM sections of formalin-fixed, paraffin-embedded tissue placed on charged slides. Following deparaffinization, PinPoint RNA Isolation System II (Zymo Research Corp.) was applied to a 1 mm2 area of the slide. After drying, this area was removed from the slide and transferred to a sterile tube. The product was treated with proteinase K, ethanol, and RNA extraction buffer. The RNA concentration of the final product was determined by the Qubit fluorometric quantification system (Life Technologies). The target-enriched library was prepared with the Archer FusionPlex Lung/Thyroid Panel (ArcherDX, Boulder, CO) as per manufacturer's instructions (8). This kit contains targets for the ALK gene. Using the anchored multiplex PCR (AMP) design, gene rearrangements, including novel variants, can be detected (9). Sequencing was performed using the IonTorrent S5 system (ThermoFisher, Waltham, MA).
Following confirmation of the diagnosis with NGS translocation analysis (Figure 2), a bone marrow biopsy was performed and showed extensive disease (Figure 3). The patient received 6 cycles of CHOEP chemotherapy, consisting of the following chemotherapy drug(s): Cyclophosphamide, Doxorubicin, Vincristine and Etoposide. This was followed by a repeat bone marrow biopsy at 4 months, which showed a cellular bone marrow with trilineage hematopoiesis without morphologic evidence of residual disease.
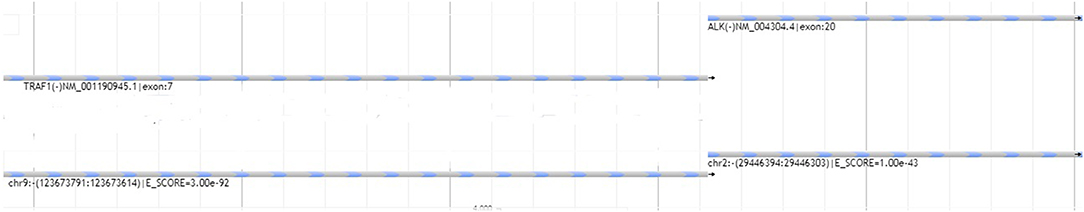
Figure 2. Graphical representation of next-generation sequencing translocation analysis. Reads aligned with exon 20 of ALK1 were found to be adjacent with reads aligned with exon 7 of the TRAF1 gene. E-score represents a confidence score whereby lower values reflect higher confidence of sequence alignment in the reads.
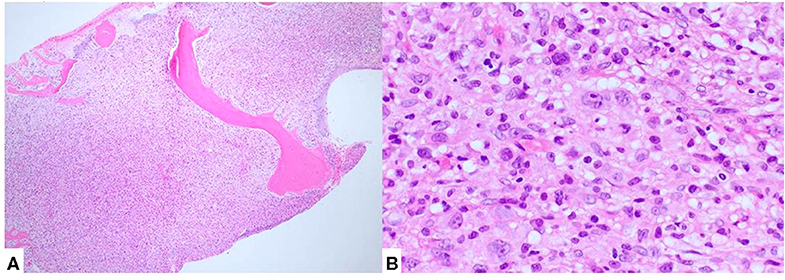
Figure 3. Bone marrow biopsy showed diffuse involvement by lymphoma (A). Atypical cells, morphologically similar to those seen in the lymph node biopsy, were also present (B).
To detect molecular residual disease, TRG gene rearrangement studies were performed on the bone marrow from the aspirate sample. DNA extraction was performed using Qiagen PureGene kits as described previously (10) (Qiagen, Germantown, MD). TRG gene rearrangement analysis was performed by next-generation sequencing using the Lymphotrack NGS assay (Invivoscribe, San Diego, CA) as previously described (11) following the manufacturer's protocol. Briefly, PCR amplification was performed using master mixes with primers derived from barcoded sequence adaptors. The library sequencing was performed using the IonTorrent platform. Results were analyzed using Lymphotrack S5-PGM Software version 2.4.5 (Invivoscribe, Inc.). Determination of clonality in these cases was based on current expert opinion outlined previously (12). Briefly, the total number of sequence reads should exceed 100,000, the dominant sequences should be >2.5% of all reads, and >10-times the polyclonal background.
The initial biopsy showed two dominant clones at 22.5% and 8.6% in an otherwise polyclonal background. The subsequent biopsy showed a mostly polyclonal pattern of rearrangements. Further analysis identified the identical sequences of the dominant clones of the original biopsies present in the re-biopsy at 0.011 and 0.003%, respectively.
The patient was later aggressively treated, starting on high dose BEAM chemotherapy which involved carmustine, etoposide, cytarabine, and melphalan with autologous stem cell reinfusion. The patient is overall doing well 9 months post-transplant.
In 1994, Bullrich and colleagues found that the NPM1 gene was recurrently translocated with the ALK gene in Ki-1 (CD30) positive lymphomas (13). The rearrangement results in an NPM1-ALK fusion protein that leads to constitutive activation of the ALK kinase. The consequence of this is constitutive activation of the ALK kinase and deregulation of multiple signaling pathways downstream of ALK, culminating in increased cellular proliferation and survival (14).
The translocation in question, t(2,5)(p23;q35), juxtaposes the chromosomal region 5q35, which includes the NPM1 gene, with 2p23 which includes the gene encoding for ALK, a receptor tyrosine kinase. This leads to increased expression of ALK. The translocation results in a chimeric gene that encodes an 80-kD fusion protein. This protein combines the N-terminal portion of NPM1 to the entire cytoplasmic domain of the receptor tyrosine kinase ALK (10). Antibodies against ALK used as immunohistochemical stains are reactive against the NPM1-ALK fusion protein as well as the full-length ALK protein, and have been used to detect lymphomas with ALK-associated translocations (15).
In ALK-positive lymphomas with NPM1-ALK translocation, strong immunohistochemical staining for ALK protein is not only seen in the cytoplasm of the lymphoma cells, but also within their nuclei and nucleoli. This is likely due to the association with NPM1, which functions to shuttle proteins to the nucleolus. Specifically, wild-type NPM provides nuclear localization signals, whereby the NPM-ALK protein can enter the nucleus (2, 16). In the t(2,5)(p23;q35) translocation, the particular cytoplasmic, nuclear, and nucleolar staining can be explained by the formation of dimers between wild-type NPM and the fusion NPM-ALK protein. When the NPM1-ALK fusion protein forms homodimers, the dimerization process mimics ligand binding. This leads to autophosphorylation of the tyrosine kinase domain of ALK and constitutive activation, resulting in the oncogenic properties seen in anaplastic lymphomas (10, 17). After its dimerization, the NPM1-ALK fusion protein activates various signaling pathways, including phosphatidylinositol-3 kinase(PI3K)/AKT/mTOR, phospholipase C (PLCγ), JAK/signal transducer and activator of transcription 3 (STAT3), and RAS/ERK (18–20). The activation of these multiple pathways by the NPM1-ALK fusion protein leads to cell-cycle progression, proliferation, cell survival and anti-apoptotic functions (21).
However, not all ALK-positive lymphomas of carry the NPM1-ALK fusion gene and other fusions have since been described. In these variant translocations, often only cytoplasmic ALK staining is observed and nuclear staining is negative. Besides the t(2,5) translocation, at least 11 variant translocations involving the ALK gene at p23 have been recognized. In all these translocations, the ALK gene is placed under the control of the promoter of genes that are constitutively expressed in lymphoid cells leading to elevated ALK gene expression. The most common variant translocation is t(1,2)(q25;p23), in which the TPM3 gene, located on chromosome 1, is fused to the ALK catalytic domain (4). However, in cases associated with the TPM3-ALK translocation, which express the TPM3-ALK fusion protein, measuring 104kDa, ALK staining is often restricted to the cytoplasm of the lymphoma cells. Additionally, most cases show stronger immunostaining of the cell membrane. The t(1,2) translocation is found in about 15–20% of ALK+ anaplastic lymphoma cases.
A less common translocation, the t(2,3)(p23;q11), involves the TFG gene. It produces two different fusion transcripts, of 85 and 97 kDa. These are referred to as TFG-ALKshort and TFG-ALKlong (22). The inv (2)(p23q35) translocation involves the ATIC gene (formerly known as pur-H), which codes for a protein involved in purine biosynthesis (26). In both TFG-ALK–positive and ATIC-ALK–positive ALCL cases, ALK staining is limited to the cytoplasm in a diffuse pattern, without nuclear staining. In rare cases of ALCL showing a unique granular ALK cytoplasmic staining pattern, the fusion gene partner is often implicated to be the CLTC gene the codes for clathrin heavy chain 1, a protein involved in intracellular trafficking of receptors (27).
Falini et al. described the immunohistologic and clinicopathologic features of 15 ALK-positive lymphoma cases that express ALK fusion protein(s) other than NPM1-ALK. They found that although there are some morphologic differences between non-NPM1-ALK fusion cases from the classical ALK+ ALCL with NPM1-ALK1 fusion, the clinical features are largely equivalent. These cases mostly occurred in children or young adults who presented with advanced disease associated with extranodal involvement and B symptoms (especially fever). Additionally, the overall prognosis of the variant ALK-positive lymphomas was similar to the classical NPM1-ALK cases with regard to relatively superior survival rates. The variant fusion cases were also significantly different in terms of prognosis from ALK-negative ALCLs (17). Other studies have also concluded that no significant differences have been found between NPM1-ALK-positive lymphomas and those showing variant fusions involving ALK and partners other than NPM1 (6) (Table 1).
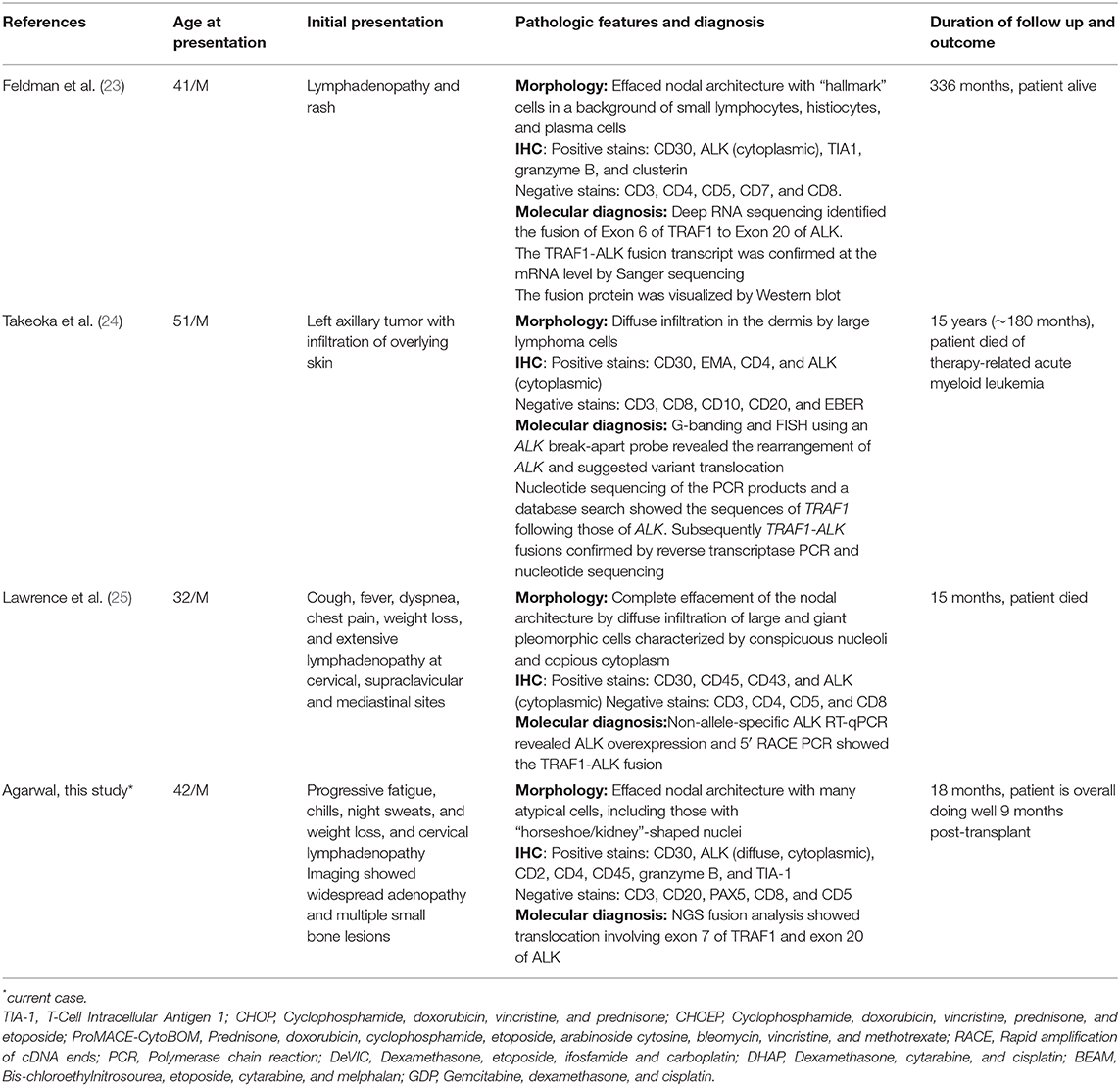
Table 1. Clinical summary and pathology of previously published and current case with ALK-TRAF fusion.
Another non–NPM fusion partner which has been described recently is the tumor necrosis factor receptor (TNFR)-1 associated factor (TRAF1) gene. Feldman et al. described a case of a 41-year-old male who suffered from an almost 30-year history of relapses of his lymphoma. He achieved remission only after high-dose chemotherapy and autologous stem cell transplantation (23). Another case was described by Lawrence et al. of a 32-year-old Caucasian male ALCL patient whose lymphoma was refractory to standard chemotherapy and autologous stem cell transplantation. This patient also showed poor response to ALK inhibitor targeted therapy (25). A third case described by Takeoka et al. was of a 51-year-old man with multiple skin tumors who also had a poor response and eventually died (24).
We describe a fourth case of TRAF1-ALK translocation. In our case, the patient underwent intensive chemotherapy and early bone marrow transplantation due to aggressive features of his disease. This highlights the importance of elucidating all ALK partner, as some of these cases can have worse prognosis. Despite their potential clinical and prognostic value, there is a paucity of information in the literature about TRAF1-ALK.
The TRAF family of proteins functions as adaptor molecules that interact with tumor necrosis factor (TNF) receptors. Once they link with TNFRs, they lead to activation of downstream kinase pathways. Binding of TNF to its receptor leads to recruitment of various TRAF proteins to result in activation of transcription factors such as nuclear factor κB (NF-κB) and activator protein-1 (AP-1) (28–30). To date, there are six members of the TRAF family, TRAF1-6. Yet another interaction is with CD30; the cytoplasmic tail of this protein interacts with TRAFs 1-3 and TRAF5 to result in activation of NF-κB (31, 32). The downstream result of this pathway activation is transcription of anti-apoptotic genes, leading to increased cell survival, protection from apoptosis, and proliferation (31).
In this regard, Abate et al. have studied the properties and functions of the TRAF1-ALK fusion protein. The N-terminal of the TRAF1 region and its interaction with ALK leads to alterations in NF-kB function, contributing to tumorigenesis and maintenance of the neoplastic phenotype (33). The chimeric protein can also associate with TRAF2 in order to alter the activation of NF-kB through the CD30 protein. This study also presented a gene data set that was able to stratify anaplastic large cell lymphoma based on NF-kB, regardless of ALK status. This intriguing model suggests that certain subgroups of ALCL may have different outcomes clinically. Data from such studies may be utilized in the future to target poor responders in ALK+ ALCL. One such option is the use of NF-kB inhibitors to target lymphoma cells or the host environment. The latter target is also significant as some studies suggest the NF-kB pathway may be relevant in host cells as well. Thus, targeting the tumor microenvironment is also a potential strategy in treating ALCL patients (34).
The popular notion that all ALK+ ALCLs behave in a similar fashion clinically has precluded the practical need for identification of the ALK partner gene. In this regard, most diagnoses of ALK+ ALCL can be confirmed by break-apart FISH analysis, such as in the more common t(2,5) translocation involving NPM1-ALK. One methodology that has been developed in cases when the identification of the partner gene is desired is rapid amplification of cDNA ends (RACE) (23). This technology utilizes a 50-gene panel to identify most ALK partner genes and has successfully identified some partners that are often cytogenetically cryptic, as in the case of the ATIC-ALK translocation (26).
The advent of next-generation sequencing and other deep sequencing platforms have allowed the rapid discovery of unique and novel translocations in cancer. However, this technology has not been utilized widely in the detection of translocations in ALK+ ALCLs. Here, we employed next-generation sequencing in an RNA-based fusion assay to detect this rare translocation partner. The advantage of the Archer assay is the ability to detect novel translocation partners for ALK in the case of ALCLs and lung carcinomas as well as other translocations in many other tumors. As NGS is employed increasingly in clinical and research settings, one may expect further studies investigating the biologic and clinical significance of these genetic lesions. Based on a cost prospective, it may be prohibitive to employ such testing on all ALK+ cases. It may also be possible to design a dual-fusion FISH probe to detect the specific TRAF1-ALK fusion. However, the utility of NGS is the discovery of novel fusions which may, in the future, be associated with adverse prognosis.
ALK positivity suggests an overall favorable prognosis of ALCL as compared to ALK-negative cases. However, in the rare published cases of TRAF1-ALK, an aggressive clinical course has been observed, which may reflect the aggressive propensity of this particular fusion, as these cases appear to be refractory to standard chemotherapy and also to first generation ALK inhibitors. In our case, aggressive chemotherapy and bone marrow transplantation likely aided in remission of disease. The current modalities to detect ALK translocation are IHC and FISH, but neither would identify these rare ALK partners. This study highlights the utility of using NGS in RNA-based fusion assays to detect rare translocations, which can be of clinical importance in detecting rare but aggressive fusion partners of ALK, e.g., TRAF. As these technologies become more available, there is potential to identify such changes and effectively stratify the prognosis of ALCL patients.
The original contributions presented in the study are publicly available. This data can be found here: the NCBI GeneBank (www.ncbi.nlm.nih.gov/nuccore/MT163661).
MA and IA contributed to the design of the study, review of relevant clinical and pathologic data, and development of the manuscript. LS provided technical review of the data and editing of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Stein H, Mason DY, Gerdes J, O'Connor N, Wainscoat J, Pallesen G, et al. The expression of the Hodgkin's disease associated antigen Ki-1 in reactive and neoplastic lymphoid tissue: evidence that Reed-Sternberg cells and histiocytic malignancies are derived from activated lymphoid cells. Blood. (1985) 66:848–58. doi: 10.1182/blood.V66.4.848.848
2. Mason DY, Bastard C, Rimokh R, Dastugue N, Huret JL, Kristoffersson U, et al. CD30-positive large cell lymphomas ('Ki-1 lymphoma') are associated with a chromosomal translocation involving 5q35. Br J Haematol. (1990) 74:161–8. doi: 10.1111/j.1365-2141.1990.tb02560.x
3. Shiota M, Nakamura S, Ichinohasama R, Abe M, Akagi T, Takeshita M, et al. Anaplastic large cell lymphomas expressing the novel chimeric protein p80NPM/ALK: a distinct clinicopathologic entity. Blood. (1995) 86:1954–60. doi: 10.1182/blood.V86.5.1954.bloodjournal8651954
4. Lamant L, Dastugue N, Pulford K, Delsol G, Mariamé B. A new fusion gene TPM3-ALK in anaplastic large cell lymphoma created by a (1;2)(q25;p23) translocation. Blood. (1999) 93:3088–95. doi: 10.1182/blood.V93.9.3088.409k30_3088_3095
5. Savage KJ, Harris NL, Vose JM, Ullrich F, Jaffe ES, Connors JM, et al. ALK- anaplastic large-cell lymphoma is clinically and immunophenotypically different from both ALK+ ALCL and peripheral T-cell lymphoma, not otherwise specified: report from the International peripheral T-Cell lymphoma project. Blood. (2008) 111:5496–504. doi: 10.1182/blood-2008-01-134270
6. Lamant-Rochaix L, Feldman AL, Delsol G, Brousset P. Anaplastic large cell lymphoma, ALK positive and ALK negative. In: Jaffe E, Arber DA, Campo E, Harris NL, Quintanilla-Martinez L, editors. Hematopathology. Philadelphia, PA: Elsevier (2016). p. 673–91.
7. Alberghini M, Benini S, Gamberi G, Cocchi S, Zanella L. RNA extraction from decalcified and non-decalcified formalin-fixed paraffin-embedded tissues. In: Stanta G, editor. Guidelines for Molecular Analysis in Archive issues. Berlin: Springer (2011). p. 63-6.
8. Zheng Z, Liebers M, Zhelyazkova B, Cao Y, Panditi D, Lynch KD, et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med. (2014) 20:1479–84. doi: 10.1038/nm.3729
9. Vendrell JA, Taviaux S, Béganton B, Godreuil S, Audran P, Grand D, et al. Detection of known and novel ALK fusion transcripts in lung cancer patients using next-generation sequencing approaches. Sci Rep. (2017) 7:12510. doi: 10.1038/s41598-017-12679-8
10. Aplenc R, Orudjev E, Swoyer J, Manke B, Rebbeck T. Differential bone marrow aspirate DNA yields from commercial extraction kits. Leukemia. (2002) 16:1865–6. doi: 10.1038/sj.leu.2402681
11. Kansal R, Grody WW, Zhou J, Dong L, Li X. The value of T-Cell receptor γ (TRG) clonality evaluation by next-generation sequencing in clinical hematolymphoid tissues. Am J Clin Pathol. (2018) 150:193–223. doi: 10.1093/ajcp/aqy046
12. Arcila ME, Yu W, Syed M, Kim H, Maciag L, Yao J, et al. Establishment of Immunoglobulin Heavy (IGH) chain clonality testing by next-generation sequencing for routine characterization of b-cell and plasma cell neoplasms. J Mol Diagn. (2019) 21:330–42. doi: 10.1016/j.jmoldx.2018.10.008
13. Morris SW, Kirstein MN, Valentine MB, Dittmer KG, Shapiro DN, Saltman DL, et al. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin's lymphoma. Science. (1994) 263:1281–4. doi: 10.1126/science.8122112
14. Pearson JD, Lee JKH, Bacani JTC, Lai R, Ingham RJ. NPM-ALK: the prototypic member of a family of oncogenic fusion tyrosine kinases. J Signal Transduct. 2012:123253. doi: 10.1155/2012/123253
15. Pulford K, Lamant L, Morris SW, Butler LH, Wood KM, Stroud D, et al. Detection of anaplastic lymphoma kinase (ALK) and nucleolar protein nucleophosmin (NPM)-ALK proteins in normal and neoplastic cells with the monoclonal antibody ALK1. Blood. (1997) 89:1394–404. doi: 10.1182/blood.V89.4.1394
16. Bischof D, Pulford K, Mason DY, Morris SW. Role of the nucleophosmin (NPM) portion of the non-Hodgkin's lymphoma-associated NPM-anaplastic lymphoma kinase fusion protein in oncogenesis. Mol Cell Biol. (1997) 17:2312–25. doi: 10.1128/MCB.17.4.2312
17. Falini B, Bigerna B, Fizzotti M, Pulford K, Pileri SA, Delsol G, et al. ALK expression defines a distinct group of T/null lymphomas (“ALK lymphomas”) with a wide morphological spectrum. Am J Pathol. (1998) 153:875–86. doi: 10.1016/S0002-9440(10)65629-5
18. Zamo A, Chiarle R, Piva R, Howes J, Fan Y, Chilosi M, et al. Anaplastic lymphoma kinase (ALK) activates Stat3 and protects hematopoietic cells from cell death. Oncogene. (2002) 21:1038–47. doi: 10.1038/sj.onc.1205152
19. Slupianek A, Nieborowska-Skorska M, Hoser G, Morrione A, Majewski M, Xue L, et al. Role of phosphatidylinositol 3-kinase-Akt pathway in nucleophosmin/anaplastic lymphoma kinase-mediated lymphomagenesis. Cancer Res. (2001) 61:2194–9.
20. Bai RY, Dieter P, Peschel C, Morris SW, Duyster J. Nucleophosmin-anaplastic lymphoma kinase of large-cell anaplastic lymphoma is a constitutively active tyrosine kinase that utilizes phospholipase C-gamma to mediate its mitogenicity. Mol Cell Biol. (1998) 18:6951–61. doi: 10.1128/MCB.18.12.6951
21. Chiarle R, Voena C, Ambrogio C, Piva R, Inghirami G. The anaplastic lymphoma kinase in the pathogenesis of cancer. Nat Rev Cancer. (2008) 8:11–23. doi: 10.1038/nrc2291
22. Hernández L, Pinyol M, Hernández S, Beà S, Pulford K, Rosenwald A, et al. TRK-fused gene (TFG) is a new partner of ALK in anaplastic large cell lymphoma producing two structurally different TFG-ALK translocations. Blood. (1999) 94:3265–8. doi: 10.1182/blood.V94.9.3265
23. Feldman AL, Vasmatzis G, Asmann YW, Davila J, Middha S, Eckloff BW, et al. Novel TRAF1-ALK fusion identified by deep RNA sequencing of anaplastic large cell lymphoma. Genes Chromosomes Cancer. 52:1097–102. doi: 10.1002/gcc.22104
24. Takeoka K, Okumura A, Honjo G, Ohno H. Variant translocation partners of the Anaplastic Lymphoma Kinase (ALK) gene in two cases of anaplastic large cell lymphoma, identified by inverse cdna polymerase chain reaction. J Clin Exp Hematop. (2014) 54:225–35. doi: 10.3960/jslrt.54.225
25. Lawrence K, Berry B, Handshoe J, Hout D, Mazzola R, Morris SW, et al. Detection of a TRAF1-ALK fusion in an anaplastic large cell lymphoma patient with chemotherapy and ALK inhibitor-resistant disease. BMC Res Notes. (2015) 8:308. doi: 10.1186/s13104-015-1277-7
26. Trinei M, Lanfrancone L, Campo E, Pulford K, Mason DY, Pelicci PG, et al. A new variant anaplastic lymphoma kinase (ALK)-fusion protein (ATIC-ALK) in a case of ALK-positive anaplastic large cell lymphoma. Cancer Res. (2000) 60:793–8.
27. Touriol C, Greenland C, Lamant L, Pulford K, Bernard F, Rousset T, et al. Further demonstration of the diversity of chromosomal changes involving 2p23 in ALK-positive lymphoma: 2 cases expressing ALK kinase fused to CLTCL (clathrin chain polypeptide-like). Blood. (2000) 95:3204–7. doi: 10.1182/blood.V95.10.3204.010k04_3204_3207
28. Baud V, Liu ZG, Bennett B, Suzuki N, Xia Y, Karin M. Signaling by proinflammatory cytokines: oligomerization of TRAF2 and TRAF6 is sufficient for JNK and IKK activation and target gene induction via an amino-terminal effector domain. Genes Dev. (1999) 13:1297–308. doi: 10.1101/gad.13.10.1297
29. Malinin NL, Boldin MP, Kovalenko AV, Wallach D. MAP3K-related kinase involved in NF-kappaB induction by TNF, CD95 and IL-1. Nature. (1997) 385:540–4. doi: 10.1038/385540a0
30. Nishitoh H, Saitoh M, Mochida Y, Takeda K, Nakano H, Rothe M, et al. ASK1 is essential for JNK/SAPK activation by TRAF2. Mol Cell. (1998) 2:389–95. doi: 10.1016/S1097-2765(00)80283-X
31. Horie R, Watanabe T. CD30: expression and function in health and disease. Semin Immunol. (1998) 10:457–70. doi: 10.1006/smim.1998.0156
32. Schneider C, Hübinger G. Pleiotropic signal transduction mediated by human CD30: a member of the tumor necrosis factor receptor (TNFR) family. Leuk Lymphoma. (2002) 43:1355–66. doi: 10.1080/10428190290033288
33. Abate F, Todaro M, van der Krogt J-A, Boi M, Landra I, Machiorlatti R, et al. A novel patient-derived tumorgraft model with TRAF1-ALK anaplastic large-cell lymphoma translocation. Leukemia. (2015) 29:1390–401. doi: 10.1038/leu.2014.347
Keywords: anaplastic T-cell lymphoma, ALK (anaplastic lymphoma kinase), next- generation sequencing, RNA based sequencing, lymphoma - diagnosis
Citation: Agarwal I, Sabatini L and Alikhan MB (2020) Diagnostic Capability of Next-Generation Sequencing Fusion Analysis in Identifying a Rare CASE of TRAF1-ALK-Associated Anaplastic Large Cell Lymphoma. Front. Oncol. 10:730. doi: 10.3389/fonc.2020.00730
Received: 30 October 2019; Accepted: 16 April 2020;
Published: 08 May 2020.
Edited by:
Laurence de Leval, CHU de Lausanne (CHUV), SwitzerlandReviewed by:
Roberto Chiarle, University of Turin, ItalyCopyright © 2020 Agarwal, Sabatini and Alikhan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mir B. Alikhan, bWFsaWtoYW4yQG5vcnRoc2hvcmUub3Jn
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.