- 1Niguarda Cancer Center, Grande Ospedale Metropolitano Niguarda, Milan, Italy
- 2Department of Oncology and Hemato-Oncology, Università degli Studi di Milano, Milan, Italy
Background: Carcinomas of unknown primary (CUP) account for 3–5% of all malignancy and, despite a reduction in incidence, the overall survival has not improved over the last decade. Chemotherapy regimens have not provided encouraging results. New diagnostic technologies, such as next generation sequencing (NGS), could represent a chance to identify potentially targetable genomic alterations in order to personalize treatment of CUP and provide insights into tumor biology.
Methods: A systematic review of studies of patients with CUP, whose tumor specimen was evaluated through a NGS panel, has been performed on June 10th, 2019 according to PRISMA criteria from PubMed, ASCO meeting library and Clinicaltrial.gov. We have identified potentially targetable alterations for which approved/off-label/in clinical trials drugs are available. Moreover, we have included case reports about CUP patients treated with targeted therapies driven by NGS results in order to explore the clinical role of NGS in this setting.
Results: We have evaluated 15 publications of which eleven studies (9 full-text articles and 2 abstracts) have analyzed the genomic profiling of CUPs through NGS technology, with different platforms and with different patients cohorts, ranging from 16 to 1,806 patients. Among all these studies, 85% of patients demonstrated at least one molecular alteration, the most frequent involving TP53 (41.88%), KRAS (18.81%), CDKN2A (8.8%), and PIK3CA (9.3%). A mean of 47.3% of patients harbored a potentially targetable alteration for which approved/off-label/in clinical trials drugs were available. Furthermore, we have identified 4 case reports in order to evaluate the clinical relevance of a specific targeted therapy identified through NGS.
Conclusions: NGS may represent a tool to improve diagnosis and treatment of CUP by identifying therapeutically actionable alterations and providing insights into tumor biology.
Introduction
Carcinomas of unknown primary (CUPs) are a heterogeneous group of metastatic tumors for which a standardized diagnostic work-up fails to identify the site of origin at the time of diagnosis (1). CUPs account for 3–5% of all malignancies (2, 3) and disappointingly the overall survival in CUP population has not improved over the last decades, despite advancements in the knowledge of biology of solid tumors (4). This is partly due to a lack of therapeutic options, with chemotherapy regimens using either platinum or taxanes or both not having proved to prolong survival in patient with CUP (5). Based on the available categorization of CUPs into favorable and unfavorable groups according to histopathological and clinical patterns (1, 6), great efforts have been done to predict the organ tissue of origin of CUPs through the IHC, DNA sequencing and gene expression analyses with the aim to better customize therapy and possibly improve clinical outcome (7–9). Based on the assumption that a treatment directed to the molecularly predicted tissue of origin could improve clinical outcome (10), a recent randomized phase II trial comparing site-specific treatment based on gene expression profiling vs. carboplatin and paclitaxel for patients with CUP has been performed. This study however demonstrated that site-specific therapy does not result in a significant survival improvement compared with empirical chemotherapy (11), leaving a clear unmet need for this patient population. Clinical outcome of CUPs is unpredictable because, even if a primary tumor does exist, they behave and metastasize unpredictably from the known primary counterpart (6) and maybe this is their real secret: their unknown biology rather than their unknown primary. CUPs enigma is hidden in the molecular mechanism that causes a fast cellular dedifferentiation and spreading (12). The aim of this review is to describe genes and molecular pathways involved in CUP pathogenesis and focus on available data of targeted genotype-directed treatment in this setting.
Methods
Data Sources and Search Strategy
A systematic literature review was performed on June 10th, 2019 according to PRISMA Criteria of 2009 (13) (Figure 1). We reviewed PubMed, ASCO Meeting Library and ClinicalTrials.gov for ongoing trials. The search criteria were limited to human studies published in English language. The Medical Subject Headings terms used for the search in PubMed were [(CUP OR cancer of unknown primary) AND (NGS OR next generation sequencing OR genomic alterations OR targeted therapies)]. The Medical Subject Headings used for the search in ASCO Library were [(“CUP” OR “cancer of unknown primary”) AND (“NGS” OR “next generation sequencing” OR “genomic alterations” OR “targeted therapies”)]. The Medical Subject Headings terms used for the search in ClinicalTrials.gov were (“cancer of unknown primary site” as condition/disease).
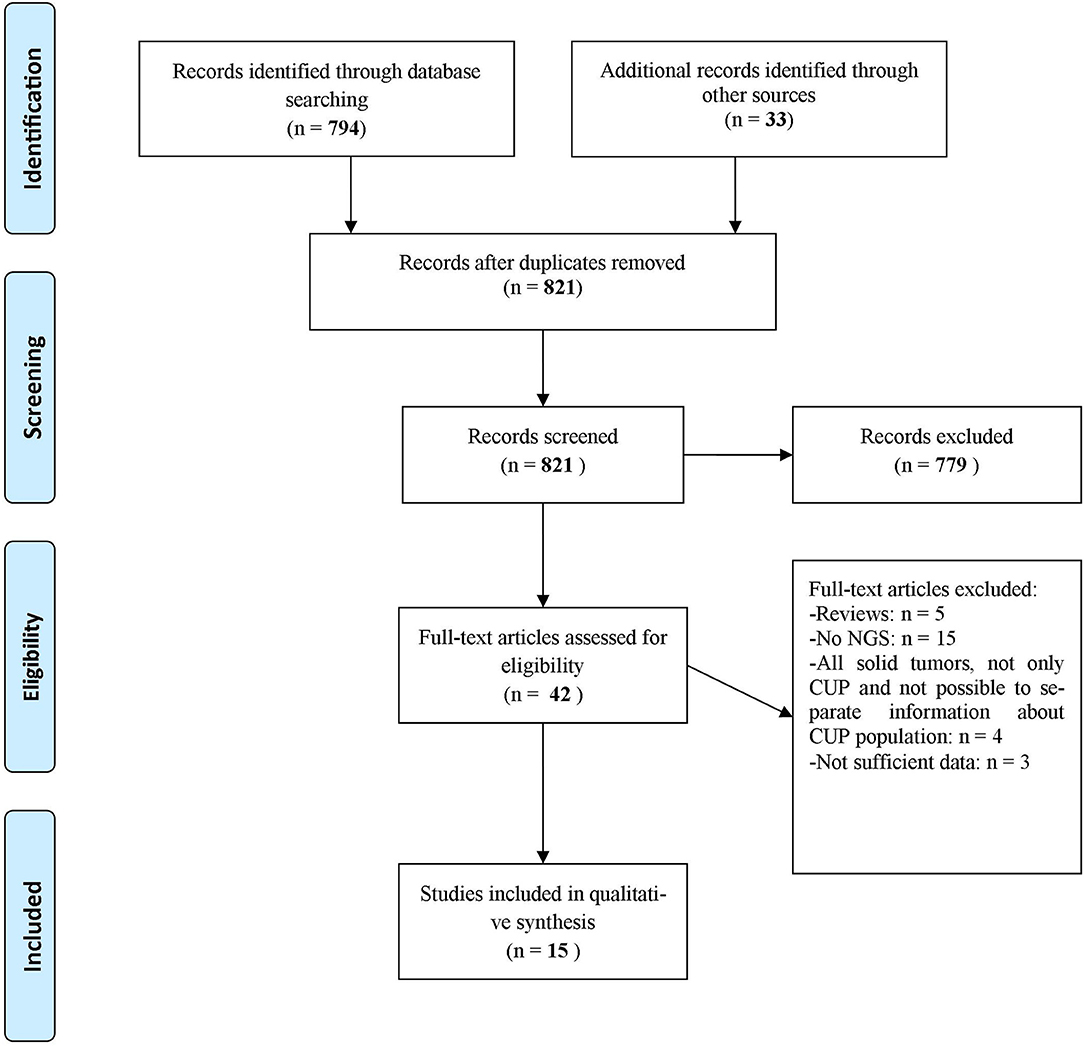
Figure 1. Flow diagram representing the systematic review process performed according to PRISMA Statement (13).
Selection Criteria
To be included in this review, a publication had to fulfill the following inclusion criteria: study performed in patients with cancer of unknown primary, whose tumor specimen could be evaluated through a NGS panel. We also included case reports and abstracts about CUP patients treated with targeted therapies based on the NGS results provided. The exclusion criteria were: publications written in language other than English and studies using a method of genomic analysis different from NGS.
Article Analysis
Among the articles included in the systematic review according to selection criteria, we evaluated all the studies performed in patients with cancer of unknown primary, comparing the results obtained through different NGS panels in each study, in order to understand how many and what are the genomic alterations that could be targeted with approved/off-label/in clinical trials drugs. We also included case reports about CUP patients treated with targeted therapies based on the NGS results provided, in order to understand if NGS could represent a valid therapeutic option in real life, improving the progression free survival and the response rate of the disease.
Results
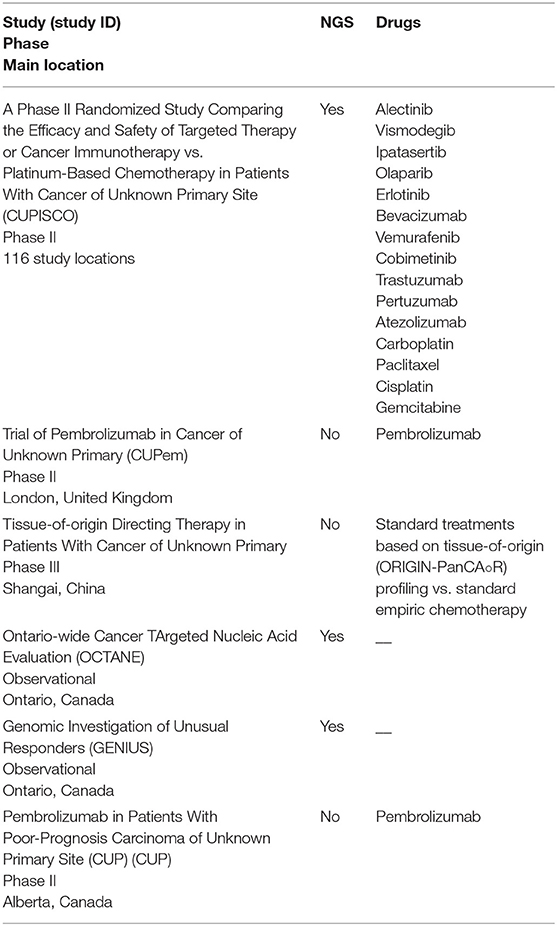
We identified 794 records through database searching and 33 additional records through other sources (i.e., online meeting library) (Figure 1). A total of 42 records were then screened to be included in the systematic review. Twenty-seven records were excluded for the following reasons: 5 were review articles (4, 9–12), 15 studies evaluated CUP molecular profile through techniques other than NGS (7, 14–27), 4 studies took into account all solid tumors and it was not possible to separate information about CUP population from other disease conditions (28–31), 1 study evaluated the validity of NGS in establishing a clonal relationship among metastasis and with an antecedent malignancy in a CUP population (32) and 2 abstracts contained few data to allow their inclusion in the review (33, 34). As a result, 15 publications were eligible and included in the systematic review of which 9 full-text articles studies about different CUP populations and their genomic alterations (35–43), 2 abstracts about CUPs and their genomic alterations (44, 45), 1 abstract about a case report in which NGS was performed with subsequent therapeutic decisions tailored on NGS results (46), 3 articles were case reports (47–49). Other 9 case reports were extrapolated from the abovementioned full-text articles. Table 1 displays studies included in the systematic review and Table 2 shows the case reports, each with its own genomic profile obtained through NGS and the subsequent response to targeted therapy.
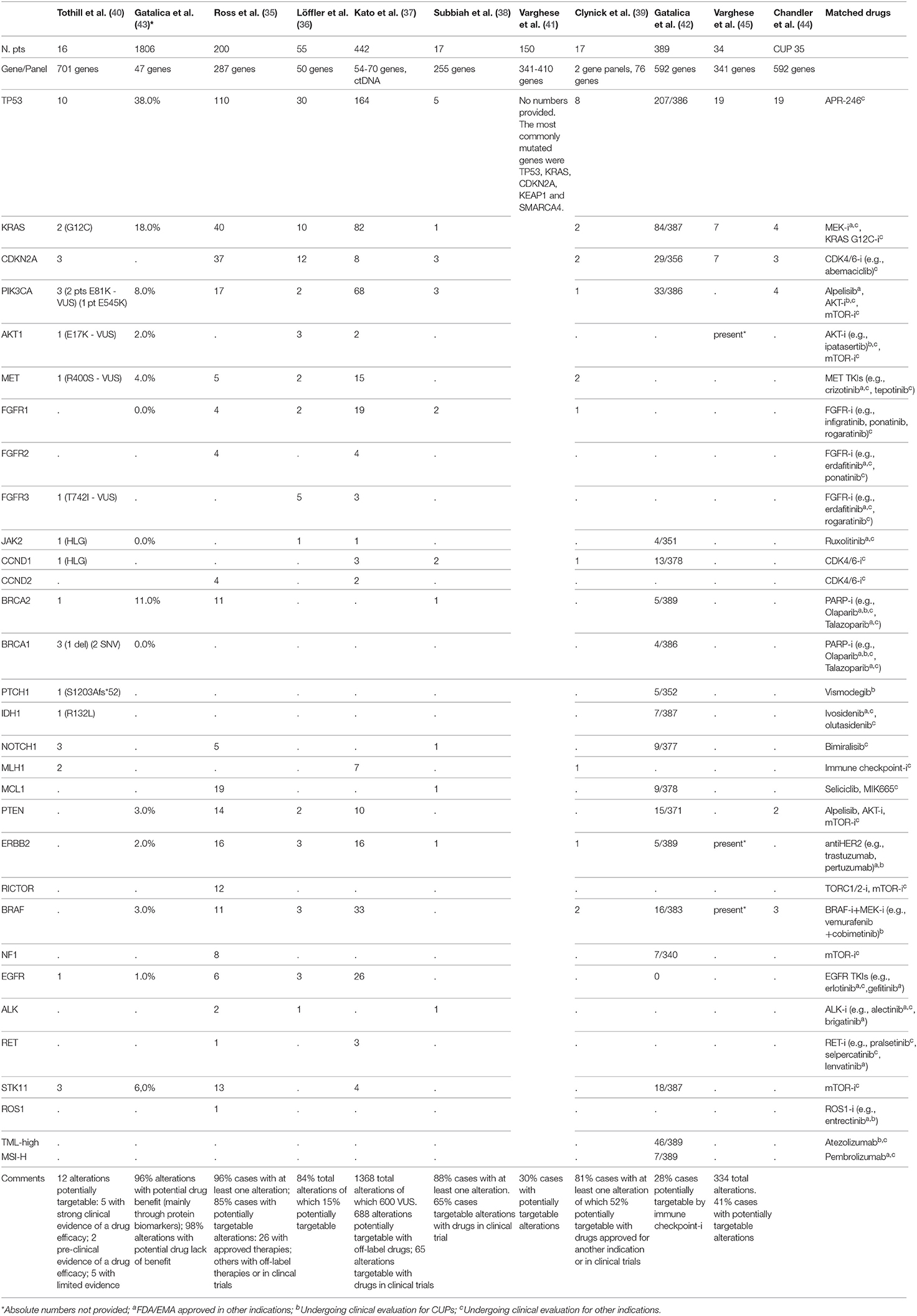
Table 1. Summary of full-text articles and abstracts of studies in CUP patients with reported genomic alterations as analyzed through different NGS panels and potentially matched drugs.
Eleven studies have analyzed the genomic profiling of CUPs through Next Generation Sequencing technology, with different platforms and with a different patients cohort, ranging from 16 to 1806 patients. Among all these studies (35–39), 85% of patients demonstrated at least one molecular alteration, with a mean of 47.3% of patients harboring a potentially targetable alteration for which approved/off-label/in clinical trials drugs were available. The most frequent alterations involved TP53 (41.88%), KRAS (18.81%), CDKN2A (8.8%), and PIK3CA (9.3%).
In the study by Ross et al. (35), one of the biggest in this setting including 200 patients, 96% of cases harbored at least one alteration and 85% of cases showed at least one genomic alteration that could be targeted. The most common clinically relevant alterations potentially targetable included KRAS (20%), CDKN2A(19%), MCL1 (10%), PTEN (7%), PIK3CA (9%), ERBB2 (8%). Twenty-six alterations were associated with targeted therapies approved in a known primary tumor type; in 14 cases there were alterations targetable with off-label drugs. Furthermore, this study identified 6 cases showing activating EGFR mutations.
In the study by Löffler et al. (36) the most frequently mutated genes in CUP population were TP53 (55%), KRAS (16%), CDKN2A (9%). In 15% of patients, they found alterations targetable by currently approved drugs. Collaterally, the investigators of this study observed that mutations of KRAS and CDKN2A were associated with poor PFS and females with wild type TP53 diseases had significantly better PFS and OS in comparison with male population.
In order to overcome both genomic heterogeneity between the primary tumor and all the metastatic lesions and temporal molecular changes occurring during sequential therapies, Kato et al. (37) analyzed the genomic profile of a CUP population of 442 patients using NGS applied on circulating tumor DNA (ctDNA). They found at least a genomic alteration in the 80% of cases, the most common of which interesting TP53 (37%), KRAS (18%) and PIK3CA (15%). Though, approximately 44% of the abovementioned alterations were variants of unknown significance (VUS). 50% of 1368 alterations were potentially targetable with off-label/in clinical trial drugs, whereas 63.8% of patients showed an alteration targetable with an FDA-approved agent. With this retrospective study Kato et al. demonstrated how the tumor molecular evolution during several lines of therapies could be pursued using NGS on ctDNA, with the possibility to customize the therapy time by time, also avoiding invasive biopsies.
In the study by Gatalica et al. (43) the most commonly mutated genes were TP53 and KRAS (38 and 18%, respectively), followed by BRCA2, PIK3CA, and STK11 (with a frequency ≥5%); the most commonly amplified genes were EGFR and HER2 (17 and 5%, respectively).
In other three studies by Tothill et al. (40), Subbiah et al. (38), and Clynick et al. (39), 75, 65, and 52%, potentially targetable alterations with approved/approved for another indication/in clinical trials drugs were found, respectively. The most common clinically relevant alterations detected in these studies included ERBB2, EGFR, KRAS, PIK3CA, and BRAF. Clynick et al. used two different gene panels, each one detecting a different number of alterations, 8 alterations were detected by both panels. Subbiah et al. also evaluated the clinical response in 7/17 patients who received a therapy matched to molecular aberrations (Table 2): the best tumor responses were stable disease lasting up to 6 months in 3 patients and 1 remaining on therapy for over 8 months.
In the study by Varghese et al. (41), including 150 patients, 54 potentially targetable alterations were identified in 45 patients. The most commonly mutated genes were TP53, KRAS, CDKN2A, KEAP1, and SMARCA4. Twenty-seven targetable alterations with FDA-approved drugs for another indication were found in 23 tumors (most common ERBB2 amplification and BRAF V600E mutation); 27 alterations targetable with drugs for which a clinical evidence exists but in another indication were found in 25 tumors (most common PIK3CA mutation); 32 alterations targetable with drugs for which preclinical evidence exists were found in 38 tumors (the most common KRAS mutation). Fifteen patients in the study received a targeted therapy shown to be active in patients with BRAF V600E mutations, ERBB2 amplification, KIF5B-ALK fusion and NCOA4-RET fusion. Among them, results in terms of time to treatment failure (TTF) were variable, ranging from <1 month to 14 months, and several patients remaining on therapy at the time of data cut-off.
Gatalica et al. (42) studied a CUP population of 389 patients in which tumor mutational load (TML) and microsatellite instability (MSI) were evaluated through NGS, while PD-L1 expression using immunohistochemistry (with 5% cut-off value). High TML was detected in approximately 12% of patients, MSI-high (MSI-H) in 2% of patients, expression of PD-L1 >5% in 22% of cases. Furthermore, predictive biomarkers of hyperprogression to immune checkpoint inhibitors, including MDM2 gene amplification and loss-of-function JAK2 gene mutations, were identified in 2 and 1% of cases, respectively.
Finally, in the study by Chandler et al. (44), in a population of different solid tumors of which 1,172 samples could be analyzed with NGS, CUPs represented the 3%, and among these, the most frequently alterations interested TP53 (54%), KRAS (11%), PIK3CA (11%), BRAF (9%). Also in the study by Varghese et al. (45), in a population of 34 patients, 334 alterations were identified, most commonly in TP53 (19/34), CDKN2A (7/34), KRAS (7/34); potentially targetable alterations were identified in 14/34 of patients and included BRAF V600E, ERBB2 S310F, and AKT1 E17K.
In the absence of prospective clinical trials, the clinical relevance of a specific targeted therapy identified through NGS, is coming from case reports. Table 2 displays case reports retrieved in this systematic review.
Discussion
Results of this systematic review from nine published studies and two abstracts show that 85% of patients with CUP harbor in their tumor at least one identified genomic alteration, including variants of uncertain significance (VUS), and that 47.3% of them present a potentially targetable alteration for which approved/off-label/in clinical trials drugs are available. The most frequent alterations were found in TP53, KRAS, CDKN2A, PIK3CA; interestingly none of the patients had two identical molecular profiles underlying the assumption that CUPs are an individually heterogeneous molecular and clinical entity.
Although there are no targeted therapies, TP53 is one of the most frequently altered gene in CUPs. This molecular alteration appears to be associated with high VEGF-A levels (50) and clinical data suggest that patients with TP53 mutations have better progression-free survival (51) and improved clinical outcome with anti-VEGF drugs (52) in comparison with patients with wild-type TP53. RAS-driven tumors are potentially targetable with MEK inhibitors (e.g., trametinib and cobimetinib) (53) and some ongoing trials are evaluating the activity of different drugs against KRAS G12C mutation (54, 55), while PIK3CA mutations have been shown to be associated with response in 45% in patients with advanced cancers when treated with a PI3K inhibitor or mTOR inhibitors (56); further, the specific inhibitor alpelisib has recently gained FDA-approval for breast cancer (57). Until now, measuring the clinical value of the genomic alterations identified in the CUP population has been elusive, because only case reports are available about targeted therapies customized according to individual genomic profiles (Table 2). The ongoing clinical trials will better elucidate the clinical validity of this approach for CUPs (Table 3).
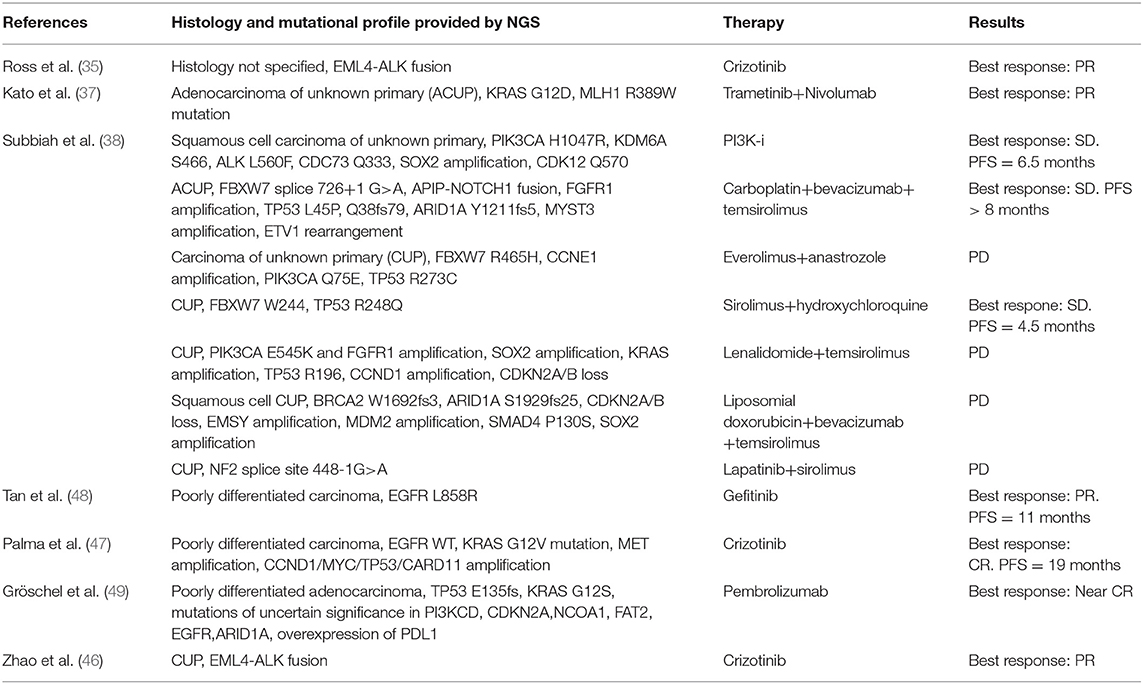
Table 2. Case reports of CUP patients with the genomic profile obtained through NGS panels and clinical outcome to matched targeted therapy.
Potential limitations of a tissue-agnostic therapeutic approach include that extrapolating therapeutic actionability from one cancer histology to another might provide uncertain results: for example, known differences exist in the clinical activity of BRAF inhibitors in melanoma and colorectal cancer (58, 59) and in the efficacy of HER2-directed treatments in breast as compared to gastric (60, 61) or colorectal cancers (62). Therefore, for CUP patients it would be still important to consider putative primary sites even when candidate actionable driver mutations are found. Finally, we should take into account that redundancy in activation of pathways of resistance does often take place as a mechanism of primary as well as secondary resistance (63, 64), for example through the co-activation of two or more antagonist pathways, affecting opportunities for a targeted pharmacological blockade (29). Liquid biopsies have a high sensitivity and specificity in reflecting the onset of resistance mutations as well as in detecting tumors alterations (65) and they could be used for these goals. Kato et al. (37) observed that NGS on blood-based biopsies can reveal clinically relevant genomic alterations in CUPs, occurring during different lines of therapy, enabling precision medicine, and liquid biopsy may also reflect the biological heterogeneity of the tumor in time and between the primary and the metastatic sites, providing more insights in CUPs biology and natural history.
Based on the assumption that, as far as they can evolve, metastatic lesions should retain the signature of the primary tumor (66), several trials have been performed using gene expression profiling techniques such as RT-PCR assay (10, 67) or using epigenetics by identifying the DNA methylation profile of the primary (15, 68), in order to identify the potential tissue of origin and drive therapy accordingly. However, based on the recent study by Hayashi et al. (11) it is still questionable whether a site-specific chemotherapy is beneficial and further studies are needed to define this point.
Multiple oncogenic pathways have been investigated in order to understand their contribution in pathogenesis of CUPs, their prognostic value and value as therapeutic targets. With this regard, we found that the most frequent alterations in CUPs involve TP53, KRAS, CDKN2A, PIK3CA. Pentheroudakis et al. (69) and Kampsorias et al. (70) did not find a prognostic value by analyzing the expression of TP53, RAS, C-Myc, and Bcl-2, as well as EGFR or HER2 overexpression; furthermore, they noticed that the incidence of tumor suppressor and DNA repair gene inactivation in CUPs is similar to that reported in other solid tumors with a known primary site. According to Löffler et al. (36), mutations of KRAS and CDKN2A are associated with poor PFS, while wild type TP53 in females has a positive prognostic value associated to a significantly better PFS and OS in comparison with male population. Karvasilis et al. (18) also studied the tissue expression of VEGF and TSP-1 in CUPs, the first with a role of activator of angiogenesis and the second as an endogenous inhibitor of the process and they found a negative association between TSP-1 expression and microvessel density, suggesting that TSP-1 can correlate with a favorable prognosis, furthermore, microvessel density was low in the group of favorable CUPs, but none of the abovementioned factors showed a prognostic value. Stella et al. (71) evaluated the role of MET in a heterogeneous population with a known or unknown primary. MET activation is a late event in tumorigenesis, after the onset of unfavorable microenvironmental conditions, and it was found to be mutated in all CUPs samples, associated with a negative prognosis; in our review we found that MET alterations are present in 2.7% of cases (35–37, 39, 40, 43) and, as demonstrated in a case report (35, 47), the targeted agent Crizotinib is associated with a better outcome in this setting. Gatalica et al. (43) noticed that the overexpression of two topoisomerases (Topo1 and Topo2alfa), identified through immunohistochemistry analysis, was associated with a potential benefit using cytotoxic therapies; on the contrary, the overexpression of multidrug resistance–associated protein 1 (MRP1) and the overexpression of breast cancer resistance protein (BCRP), a member of the ABC transporter proteins, were associated to a potential drug lack of benefit. Also the microenvironment could play a role in determining a dormancy of the primary tumor cells (72), as well as metalloproteinases allowing tumor cells migration (70) and Programmed Death-1 (PD-1) T cell co-receptor and its ligands, B7-H1/PD-L1 and B7-DC/PD-L2, maintaining an immunosuppressive tumor microenvironment (73). Gatalica et al. (42, 43) found that PD-L1 expression, detected through immunohistochemistry (with 5% cut-off value), was present in 22% of cases; while a high TML, detected through NGS, was expressed in 12% of patients and MSI-H status in 2% of patients. Ongoing clinical trials are evaluating the clinical impact of immune checkpoint inhibitors in CUP population (Table 3).
Altogether these data highlight the absence of an established distinguishing underlying molecular biology make-up of CUPs, confirming that their molecular pathogenesis is complex and heterogeneous. In order to overcome this lack of knowledge, NGS represents a chance, although not validated by clinical trials, to improve diagnosis and matched treatment of potential actionable molecular alterations.
Author Contributions
RL: writing of the article, analysis, and interpretation of data. AN, KB, VG, RR, FS, and FT: writing of the article and critical revision of the draft. SS: conception and design of the review, analysis and interpretation of data, critical revision of the draft, and final approval of the version to be submitted. AS-B: conception and design of the review, writing of the article, analysis and interpretation of data, critical revision of the draft, and final approval of the version to be submitted.
Funding
The authors are supported by grants from Associazione Italiana Ricerca Cancro grant AIRC 5 x mille [Project ID 51000] Special Program Molecular Clinical Oncology, AIRC Investigator Grant Project ID 20685 and 16788], and AIRC Special Program 5 per mille metastases [Project ID 21091]; IANG-CRC grant from Fondazione Regionale Ricerca Biomedica (FRRB) of Regione Lombardia; CORDIS Community Research and Development Information Service, Horizon 2020 [Project ID 635342] grant Molecularly Guided Trials with Specific Treatment Strategies in Patients with Advanced Newly Molecular Defined Subtypes of Colorectal Cancer (MoTriColor); Fondazione Oncologia Niguarda Onlus, grant Terapia Molecolare dei Tumori and grant Studies to Develop Therapies Against Colorectal Cancer in Young Adults.
Conflict of Interest
AS-B has acted as a consultant/advisory member for Amgen, Bayer, Lilly, and Merck- Serono. SS is advisory board member for Amgen, Bayer, BMS, Celgene, Incyte, Merck, Novartis, Roche, Seattle Genetics. RR is consultant/advisory member for Novartis, Pfizer, Janssen-Cilag, MSD, BMS, Sandoz, Sanofi Genzyme.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Fizazi K, Greco FA, Pavlidis N, Daugaard G, Oien K, Pentheroudakis G. Cancers of unknown primary site: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. (2015) 26:v133–8. doi: 10.1093/annonc/mdv305
2. Levi F, Te VC, Erler G, Randimbison L, La Vecchia C. Epidemiology of unknown primary tumours. Eur J Cancer. (2002) 38:1810–2. doi: 10.1016/S0959-8049(02)00135-1
3. Mnatsakanyan E, Tung W-C, Caine B, Smith-Gagen J. Cancer of unknown primary: time trends in incidence, United States. Cancer Causes Control. (2014) 25:747–57. doi: 10.1007/s10552-014-0378-2
4. Binder C, Matthes KL, Korol D, Rohrmann S, Moch H. Cancer of unknown primary-Epidemiological trends and relevance of comprehensive genomic profiling. Cancer Med. (2018) 7:4814–24. doi: 10.1002/cam4.1689
5. Golfinopoulos V, Pentheroudakis G, Salanti G, Nearchou AD, Ioannidis JPA, Pavlidis N. Comparative survival with diverse chemotherapy regimens for cancer of unknown primary site: multiple-treatments meta-analysis. Cancer Treat Rev. (2009) 35:570–3. doi: 10.1016/j.ctrv.2009.05.005
6. Pavlidis N, Khaled H, Gaafar R. A mini review on cancer of unknown primary site: a clinical puzzle for the oncologists. J Adv Res. (2015) 6:375–82. doi: 10.1016/j.jare.2014.11.007
7. Talantov D, Baden J, Jatkoe T, Hahn K, Yu J, Rajpurohit Y, et al. A quantitative reverse transcriptase-polymerase chain reaction assay to identify metastatic carcinoma tissue of origin. J Mol Diagn. (2006) 8:320–9. doi: 10.2353/jmoldx.2006.050136
8. Varadhachary GR, Talantov D, Raber MN, Meng C, Hess KR, Jatkoe T, et al. Molecular profiling of carcinoma of unknown primary and correlation with clinical evaluation. J Clin Oncol. (2008) 26:4442–8. doi: 10.1200/JCO.2007.14.4378
9. Rosenfeld N, Aharonov R, Meiri E, Rosenwald S, Spector Y, Zepeniuk M, et al. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol. (2008) 26:462–9. doi: 10.1038/nbt1392
10. Hainsworth JD, Rubin MS, Spigel DR, Boccia RV, Raby S, Quinn R, et al. Molecular gene expression profiling to predict the tissue of origin and direct site-specific therapy in patients with carcinoma of unknown primary site: a prospective trial of the Sarah cannon research institute. J Clin Oncol. (2013) 31:217–23. doi: 10.1200/JCO.2012.43.3755
11. Hayashi H, Kurata T, Takiguchi Y, Arai M, Takeda K, Akiyoshi K, et al. Randomized phase II trial comparing site-specific treatment based on gene expression profiling with carboplatin and paclitaxel for patients with cancer of unknown primary site. J Clin Oncol. (2019) 37:570–9. doi: 10.1200/JCO.18.00771
12. Stella GM, Senetta R, Cassenti A, Ronco M, Cassoni P. Cancers of unknown primary origin: current perspectives and future therapeutic strategies. J Transl Med. (2012) 10:12. doi: 10.1186/1479-5876-10-12
13. Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gøtzsche PC, Ioannidis JPA, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. PLOS Med. (2009) 6:e1000100. doi: 10.1371/journal.pmed.1000100
14. Soh KP, Szczurek E, Sakoparnig T, Beerenwinkel N. Predicting cancer type from tumour DNA signatures. Genome Med. (2017) 9:104. doi: 10.1186/s13073-017-0493-2
15. Moran S, Martinez-Cardús A, Boussios S, Esteller M. Precision medicine based on epigenomics: the paradigm of carcinoma of unknown primary. Nat Rev Clin Oncol. (2017) 14:682–94. doi: 10.1038/nrclinonc.2017.97
16. Selves J, Long-Mira E, Mathieu M-C, Rochaix P, Ilié M. Immunohistochemistry for diagnosis of metastatic carcinomas of unknown primary site. Cancers. (2018) 10:108. doi: 10.3390/cancers10040108
17. Speel E-JM, van de Wouw AJ, Claessen SMH, Haesevoets A, Hopman AHN, van der Wurff AAM, et al. Molecular evidence for a clonal relationship between multiple lesions in patients with unknown primary adenocarcinoma. Int J Cancer. (2008) 123:1292–300. doi: 10.1002/ijc.23616
18. Karavasilis V, Malamou-Mitsi V, Briasoulis E, Tsanou E, Kitsou E, Kalofonos H, et al. Angiogenesis in cancer of unknown primary: clinicopathological study of CD34, VEGF and TSP-1. BMC Cancer. (2005) 5:25. doi: 10.1186/1471-2407-5-25
19. Dova L, Pentheroudakis G, Golfinopoulos V, Malamou-Mitsi V, Georgiou I, Vartholomatos G, et al. Targeting c-KIT, PDGFR in cancer of unknown primary: a screening study for molecular markers of benefit. J Cancer Res Clin Oncol. (2008) 134:697–704. doi: 10.1007/s00432-007-0341-7
20. Lazaridis G, Pentheroudakis G, Fountzilas G, Pavlidis N. Liver metastases from cancer of unknown primary (CUPL): a retrospective analysis of presentation, management and prognosis in 49 patients and systematic review of the literature. Cancer Treat Rev. (2008) 34:693–700. doi: 10.1016/j.ctrv.2008.05.005
21. Honda A, Yoshimi A, Ushiku T, Shinoda Y, Kawano H, Toya T, et al. Successful control of carcinoma of unknown primary with axitinib, a novel molecular-targeted agent: a case report. Chemotherapy. (2014) 60:342–5. doi: 10.1159/000437135
22. Overby A, Duval L, Ladekarl M, Laursen BE, Donskov F. Carcinoma of unknown primary site (CUP) with Metastatic Renal-Cell Carcinoma (mRCC) histologic and immunohistochemical characteristics (CUP-mRCC): results from consecutive patients treated with targeted therapy and review of literature. Clin Genitourin Cancer. (2019) 17:e32–7. doi: 10.1016/j.clgc.2018.08.005
23. Kourie HR, Awada G, Awada AH. Unknown primary tumors: is there a future therapeutic role for immune checkpoint inhibitors? Future Oncol. (2016) 12:429–31. doi: 10.2217/fon.15.329
24. Greco FA, Lennington WJ, Spigel DR, Hainsworth JD. Molecular profiling diagnosis in unknown primary cancer: accuracy and ability to complement standard pathology. J Natl Cancer Inst. (2013) 105:782–90. doi: 10.1093/jnci/djt099
25. Kandalaft PL, Gown AM. Practical applications in immunohistochemistry: carcinomas of unknown primary site. Arch Pathol Lab Med. (2016) 140:508–23. doi: 10.5858/arpa.2015-0173-CP
26. Takei H, Monzon FA. Gene-expression assays and personalized cancer care: tissue-of-origin test for cancer of unknown primary origin. Pers Med. (2011) 8:429–36. doi: 10.2217/pme.11.37
27. Varadhachary G. New strategies for carcinoma of unknown primary: the role of tissue-of-origin molecular profiling. Clin Cancer Res. (2013) 19:4027–33. doi: 10.1158/1078-0432.CCR-12-3030
28. Kou T, Kanai M, Yamamoto Y, Kamada M, Nakatsui M, Sakuma T, et al. Clinical sequencing using a next-generation sequencing-based multiplex gene assay in patients with advanced solid tumors. Cancer Sci. (2017) 108:1440–6. doi: 10.1111/cas.13265
29. Millis SZ, Jardim DL, Albacker L, Ross JS, Miller VA, Ali SM, et al. Phosphatidylinositol 3-kinase pathway genomic alterations in 60,991 diverse solid tumors informs targeted therapy opportunities. Cancer. (2019) 125:1185–99. doi: 10.1002/cncr.31921
30. Alvarez RH, Thomas JW, Chalmers ZR, Schrock AB, Tapias C alvarez, Frampton GM, et al. Comparison of comprehensive genomic profiling (CGP) and hotspot next generation sequencing (NGS) assays in identifying treatment options for care of patients with metastatic cancer in in the community setting. J Clin Oncol. (2016) 34:e23120. doi: 10.1200/JCO.2016.34.15_suppl.e23120
31. Okuma HS, Kubo T, Ichikawa H, Kohno T, Mizuno T, Kojima Y, et al. Targeted-sequencing in rare cancers and the impact on patient treatment. J Clin Oncol. (2019) 37:e14755. doi: 10.1200/JCO.2019.37.15_suppl.e14755
32. Bochtler T, Endris V, Leichsenring J, Reiling A, Neumann O, Volckmar A-L, et al. Comparative genetic profiling aids diagnosis and clinical decision making in challenging cases of CUP syndrome. Int J Cancer. (2019) 145:2963–73. doi: 10.1002/ijc.32316
33. Krämer A, Losa F, Gay LM, Sokol E, Page DR, Frampton GM, et al. Genomic profiling of carcinomas of unknown primary (CUP) to support clinical decisions. J Clin Oncol. (2018) 36:e24162. doi: 10.1200/JCO.2018.36.15_suppl.e24162
34. Gay LM, Fabrizio D, Frampton GM, Connelly CF, Sun J, Daniel S, et al. Mutational burden of tumors with primary site unknown. J Clin Oncol. (2017) 35:3039. doi: 10.1200/JCO.2017.35.15_suppl.3039
35. Ross JS, Wang K, Gay L, Otto GA, White E, Iwanik K, et al. Comprehensive genomic profiling of carcinoma of unknown primary site: new routes to targeted therapies. JAMA Oncol. (2015) 1:40–9. doi: 10.1001/jamaoncol.2014.216
36. Löffler H, Pfarr N, Kriegsmann M, Endris V, Hielscher T, Lohneis P, et al. Molecular driver alterations and their clinical relevance in cancer of unknown primary site. Oncotarget. (2016) 7:44322–9. doi: 10.18632/oncotarget.10035
37. Kato S, Krishnamurthy N, Banks KC, De P, Williams K, Williams C, et al. Utility of genomic analysis in circulating tumor DNA from patients with carcinoma of unknown primary. Cancer Res. (2017) 77:4238–46. doi: 10.1158/0008-5472.CAN-17-0628
38. Subbiah IM, Tsimberidou A, Subbiah V, Janku F, Roy-Chowdhuri S, Hong DS. Next generation sequencing of carcinoma of unknown primary reveals novel combinatorial strategies in a heterogeneous mutational landscape. Oncoscience. (2017) 4:47–56. doi: 10.18632/oncoscience.352
39. Clynick B, Dessauvagie B, Sterrett G, Harvey NT, Allcock RJN, Saunders C, et al. Genetic characterisation of molecular targets in carcinoma of unknown primary. J Transl Med. (2018) 16:185. doi: 10.1186/s12967-018-1564-x
40. Tothill RW, Li J, Mileshkin L, Doig K, Siganakis T, Cowin P, et al. Massively-parallel sequencing assists the diagnosis and guided treatment of cancers of unknown primary. J Pathol. (2013) 231:413–23. doi: 10.1002/path.4251
41. Varghese AM, Arora A, Capanu M, Camacho N, Won HH, Zehir A, et al. Clinical and molecular characterization of patients with cancer of unknown primary in the modern era. Ann Oncol. (2017) 28:3015–21. doi: 10.1093/annonc/mdx545
42. Gatalica Z, Xiu J, Swensen J, Vranic S. Comprehensive analysis of cancers of unknown primary for the biomarkers of response to immune checkpoint blockade therapy. Eur J Cancer. (2018) 94:179–86. doi: 10.1016/j.ejca.2018.02.021
43. Gatalica Z, Millis SZ, Vranic S, Bender R, Basu GD, Voss A, et al. Comprehensive tumor profiling identifies numerous biomarkers of drug response in cancers of unknown primary site: analysis of 1806 cases. Oncotarget. (2014) 5:12440–7. doi: 10.18632/oncotarget.2574
44. Chandler JC, Vanderwalde AM, Somer BG, Vidal GA, Schwartzberg LS. Analysis of 1261 metastatic cancer patients evaluated by comprehensive molecular profiling (CMP) including next-gen sequencing (NGS) from a single institution. J Clin Oncol. (2016) 34:11570. doi: 10.1200/JCO.2016.34.15_suppl.11570
45. Varghese AM, Hyman DM, Vakiani E, Klimstra ES, Klimstra MF, et al. Prospective identification of potentially actionable molecular alterations in cancers of unknown primary. J Clin Oncol. (2015) 33:4110. doi: 10.1200/jco.2015.33.15
46. Zhao P, Peng L, Wu W, Zheng Y, Jiang W, Zhang H, et al. Carcinoma of unknown primary with EML4-ALK fusion response to ALK inhibitors. Oncologist. (2019) 24:449–54. doi: 10.1634/theoncologist.2018-0439
47. Palma NA, Ali SM, O'Connor J, Dutta D, Wang K, Soman S, et al. Durable response to crizotinib in a MET-amplified, KRAS-mutated carcinoma of unknown primary. Case Rep Oncol. (2014) 7:503–8. doi: 10.1159/000365326
48. Tan DS-W, Montoya J, Ng Q-S, Chan K-S, Lynette O, Sakktee Krisna S, et al. Molecular profiling for druggable genetic abnormalities in carcinoma of unknown primary. J Clin Oncol. (2013) 31:e237–9. doi: 10.1200/JCO.2012.44.3937
49. Gröschel S, Bommer M, Hutter B, Budczies J, Bonekamp D, Heining C, et al. Integration of genomics and histology revises diagnosis and enables effective therapy of refractory cancer of unknown primary with PDL1 amplification. Cold Spring Harb Mol Case Stud. (2016) 2:a001180. doi: 10.1101/mcs.a001180
50. Schwaederlé M, Lazar V, Validire P, Hansson J, Lacroix L, Soria J-C, et al. VEGF-A expression correlates with TP53 mutations in non-small cell lung cancer: implications for antiangiogenesis therapy. Cancer Res. (2015) 75:1187–90. doi: 10.1158/0008-5472.CAN-14-2305
51. Said R, Hong DS, Warneke CL, Lee JJ, Wheler JJ, Janku F, et al. P53 mutations in advanced cancers: clinical characteristics, outcomes, and correlation between progression-free survival and bevacizumab-containing therapy. Oncotarget. (2013) 4:705–14. doi: 10.18632/oncotarget.974
52. Wheler JJ, Janku F, Naing A, Li Y, Stephen B, Zinner R, et al. TP53 alterations correlate with response to VEGF/VEGFR inhibitors: implications for targeted therapeutics. Mol Cancer Ther. (2016) 15:2475–85. doi: 10.1158/1535-7163.MCT-16-0196
53. Falchook GS, Lewis KD, Infante JR, Gordon MS, Vogelzang NJ, DeMarini DJ, et al. Activity of the oral MEK inhibitor trametinib in patients with advanced melanoma: a phase 1 dose-escalation trial. Lancet Oncol. (2012) 13:782–9. doi: 10.1016/S1470-2045(12)70269-3
54. A Phase 1/2, Study Evaluating the Safety, Tolerability, PK, and Efficacy of AMG 510 in Subjects With Solid Tumors With a Specific KRAS Mutation. ClinicalTrials.gov. Available online at: https://clinicaltrials.gov/ct2/show/NCT03600883 (accessed July 13, 2019).
55. MRTX849 in Patients With Cancer Having a KRAS G12C Mutation. ClinicalTrials.gov. Available online at: https://clinicaltrials.gov/ct2/show/NCT03785249 (accessed July 13, 2019).
56. Janku F, Wheler JJ, Naing A, Falchook GS, Hong DS, Stepanek VM, et al. PIK3CA mutation H1047R is associated with response to PI3K/AKT/mTOR signaling pathway inhibitors in early-phase clinical trials. Cancer Res. (2013) 73:276–84. doi: 10.1158/0008-5472.CAN-12-1726
57. Research C for DE. FDA Approves Alpelisib for Metastatic Breast Cancer. FDA (2019) Available online at: http://www.fda.gov/drugs/resources-information-approved-drugs/fda-approves-alpelisib-metastatic-breast-cancer (accessed July 14, 2019).
58. Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. (2012) 483:100–3. doi: 10.1038/nature10868
59. Van Cutsem E, Cuyle P-J, Huijberts S, Yaeger R, Schellens JHM, Elez E, et al. BEACON CRC study safety lead-in (SLI) in patients with BRAFV600E metastatic colorectal cancer (mCRC): efficacy and tumor markers. J Clin Oncol. (2018) 36:627. doi: 10.1200/JCO.2018.36.4_suppl.627
60. Iqbal N, Iqbal N. Human epidermal growth factor receptor 2 (HER2) in cancers: overexpression and therapeutic implications. Mol Biol Int. (2014) 2014:852748. doi: 10.1155/2014/852748
61. Gomez-Martín C, Lopez-Rios F, Aparicio J, Barriuso J, García-Carbonero R, Pazo R, et al. A critical review of HER2-positive gastric cancer evaluation and treatment: from trastuzumab, and beyond. Cancer Lett. (2014) 351:30–40. doi: 10.1016/j.canlet.2014.05.019
62. Sartore-Bianchi A, Trusolino L, Martino C, Bencardino K, Lonardi S, Bergamo F, et al. Dual-targeted therapy with trastuzumab and lapatinib in treatment-refractory, KRAS codon 12/13 wild-type, HER2-positive metastatic colorectal cancer (HERACLES): a proof-of-concept, multicentre, open-label, phase 2 trial. Lancet Oncol. (2016) 17:738–46. doi: 10.1016/S1470-2045(16)00150-9
63. Sartore-Bianchi A, Marsoni S, Siena S. Human epidermal growth factor receptor 2 as a molecular biomarker for metastatic colorectal cancer. JAMA Oncol. (2018) 4:19–20. doi: 10.1001/jamaoncol.2017.3323
64. Siena S, Sartore-Bianchi A, Marsoni S, Hurwitz HI, McCall SJ, Penault-Llorca F, et al. Targeting the human epidermal growth factor receptor 2 (HER2) oncogene in colorectal cancer. Ann Oncol. (2018) 29:1108–19. doi: 10.1093/annonc/mdy100
65. Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. (2014) 6:224ra24. doi: 10.1126/scitranslmed.3007094
66. Pentheroudakis G, Golfinopoulos V, Pavlidis N. Switching benchmarks in cancer of unknown primary: from autopsy to microarray. Eur J Cancer. (2007) 43:2026–36. doi: 10.1016/j.ejca.2007.06.023
67. Yoon HH, Foster NR, Meyers JP, Steen PD, Visscher DW, Pillai R, et al. Gene expression profiling identifies responsive patients with cancer of unknown primary treated with carboplatin, paclitaxel, and everolimus: NCCTG N0871 (alliance). Ann Oncol. (2016) 27:339–44. doi: 10.1093/annonc/mdv543
68. Moran S, Martínez-Cardús A, Sayols S, Musulén E, Balañá C, Estival-Gonzalez A, et al. Epigenetic profiling to classify cancer of unknown primary: a multicentre, retrospective analysis. Lancet Oncol. (2016) 17:1386–95. doi: 10.1016/S1470-2045(16)30297-2
69. Pentheroudakis G, Pavlidis N. Perspectives for targeted therapies in cancer of unknown primary site. Cancer Treat Rev. (2006) 32:637–44. doi: 10.1016/j.ctrv.2006.08.004
70. Kamposioras K, Pentheroudakis G, Pavlidis N. Exploring the biology of cancer of unknown primary: breakthroughs and drawbacks. Eur J Clin Invest. (2013) 43:491–500. doi: 10.1111/eci.12062
71. Stella GM, Benvenuti S, Gramaglia D, Scarpa A, Tomezzoli A, Cassoni P, et al. MET mutations in cancers of unknown primary origin (CUPs). Hum Mutat. (2011) 32:44–50. doi: 10.1002/humu.21374
72. Bragado P, Sosa MS, Keely P, Condeelis J, Aguirre-Ghiso JA. Microenvironments dictating tumor cell dormancy. Recent Results Cancer Res. (2012) 195:25–39. doi: 10.1007/978-3-642-28160-0_3
Keywords: cancer of unknown primary (CUP), next generation sequencing (NGS), genomic alterations, comprehensive genomic profiling, targeted therapy
Citation: Lombardo R, Tosi F, Nocerino A, Bencardino K, Gambi V, Ricotta R, Spina F, Siena S and Sartore-Bianchi A (2020) The Quest for Improving Treatment of Cancer of Unknown Primary (CUP) Through Molecularly-Driven Treatments: A Systematic Review. Front. Oncol. 10:533. doi: 10.3389/fonc.2020.00533
Received: 02 October 2019; Accepted: 25 March 2020;
Published: 08 May 2020.
Edited by:
Giulia Maria Stella, San Matteo Hospital Foundation (IRCCS), ItalyReviewed by:
Nikolaus Christoph Netzer, University of Innsbruck, AustriaChiara Ambrogio, University of Turin, Italy
Copyright © 2020 Lombardo, Tosi, Nocerino, Bencardino, Gambi, Ricotta, Spina, Siena and Sartore-Bianchi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Salvatore Siena, c2FsdmF0b3JlLnNpZW5hQHVuaW1pLml0
 Roberta Lombardo
Roberta Lombardo Federica Tosi1,2
Federica Tosi1,2