
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol. , 07 April 2020
Sec. Molecular and Cellular Oncology
Volume 10 - 2020 | https://doi.org/10.3389/fonc.2020.00472
This article is part of the Research Topic Impact of Cancer Plasticity on Drug Resistance and Treatment in Solid Tumors. View all 33 articles
 Qiao-ru Guo1,2†
Qiao-ru Guo1,2† Hui Wang3†
Hui Wang3† Ying-da Yan1†
Ying-da Yan1† Yun Liu1
Yun Liu1 Chao-yue Su1
Chao-yue Su1 Hu-biao Chen4
Hu-biao Chen4 Yan-yan Yan5
Yan-yan Yan5 Rameshwar Adhikari6
Rameshwar Adhikari6 Qiang Wu2,7*
Qiang Wu2,7* Jian-ye Zhang1,2*
Jian-ye Zhang1,2*Exosomes affect the initiation and progression of cancers. In the tumor microenvironment, not only cancer cells, but also fibroblasts and immunocytes secrete exosomes. Exosomes act as a communicator between cells by transferring different cargos and microRNAs (miRNAs). Drug resistance is one of the critical factors affecting therapeutic effect in the course of cancer treatment. The currently known mechanisms of drug resistance include drug efflux, alterations in drug metabolism, DNA damage repair, alterations of energy programming, cancer stem cells and epigenetic changes. Many studies have shown that miRNA carried by exosomes is closely associated with the development of drug resistance mediated by the above-mentioned mechanisms. This review article will discuss how exosomal miRNAs regulate the drug resistance.
Chemotherapy, radiotherapy, surgery, and targeted therapy are important modalities of cancer treatment. However, the emergence of drug resistance leads to dismal prognosis in cancer patients. As an emerging therapeutic target and diagnostic biomarker, exosomal miRNAs play vital roles in tumor invasion, metastasis and progression. Studies have found that the occurrence and development of drug resistance is closely related to miRNA carried by exosomes (1–5). In this review article we discuss several classic mechanisms that exosomal miRNA involves in drug resistance. Also, we summarize the role of exosomal miRNA mediated drug resistance in different types of cancers.
Exosomes are extracellular vesicles (EV) with the size ranges between 30 and 100 nm. Exosomes are released to the extracellular environment after the fusion of the multivesicular body (MVB) or late endosomes with the plasma membrane (6). It was first described in 1983 as “seems to be akin to reverse endocytosis” (7), and has gradually recognized an important factor in oncology research (8). Many studies have shown that exosome can promote the intercellular communication (9) by transferring varieties of cargos, such as nucleic acid, proteins and metabolites (10–16).
microRNA (miRNA) is an important cargo that delivered by exosomes (17). miRNAs usually consist of 19–25 nucleotides. It can regulate post-transcriptional silence of target genes. Following the transcription of miRNA gene, a small hairpin-shaped RNA called pre-miRNA is generated. The pre-miRNA is exported into cytoplasm and processed by Dicer, a kind of RNase III-type endonuclease, and subsequently releases a small RNA duplex (18, 19). The RNA duplex will unwind after loading onto an Argonaute (AGO) protein and forming RNA-induced silencing complex (RISC) (20). Once binding to a RISC, the miRNA is complementary pairing with the mRNA. Depending on whether miRNA and mRNA are fully bind, two different mechanisms occur: (1) mRNA specifies cleavage if miRNA is sufficiently complementary to mRNA; (2) the productive translation is inhibited when miRNA is insufficiently complementary to mRNA (21, 22). Therefore, miRNA can regulate various physiological and pathological activities (Figure 1).
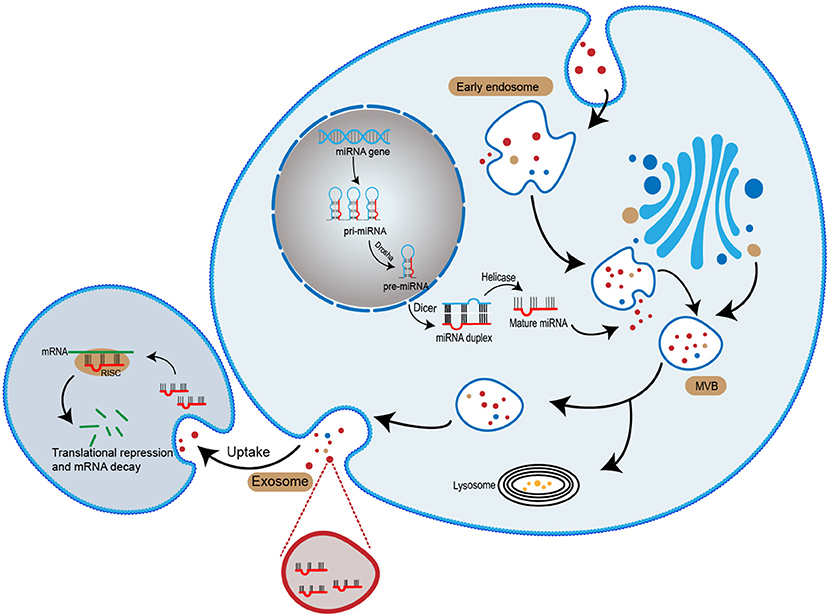
Figure 1. The biogenesis of exosomal miRNA. miRNA genes are transcribed into primary miRNA (pri-miRNA). The pri-miRNA is cleaved by Dorsha complex to produce precursor miRNA (pre-miRNA) and exported into the cytoplasma. Subsequently, pre-miRNA is processed by Dicer complex to become mature miRNA. Mature miRNA is sorted into exosomes and released to the extracellular space. With the recipient cell uptake exosomes, the miRNA entered the cell and mediates gene suppression by targeted translational repression and mRNA degradation.
Cancer drug resistance can be divided into intrinsic and acquired resistance. Intrinsic resistance occurs before receiving therapy, which limits the use of anticancer drugs. Acquired resistance may develop during treatment even if some drugs have anticancer effects at the early stage (23). Drug resistance seriously impacts the effectiveness of chemotherapy and molecular targeted therapies, ultimately leading dismal prognosis and tumor relapse.
Because of genomic instability, tumors may include a diverse collection of cells that possess different sensitivity to treatment (24). The positive selection of drug-resistant tumor subpopulation causes drug resistance. Therefore, accurate assessment of tumor heterogeneity is important to address drug resistance (25). The application of high-throughput screening technology facilitates the identification of genotype and helps predict drug response, providing convenience to individual therapy. In the past few years, microfluidic chips show tremendous promise in the study of tumor heterogeneity and the establishment of preclinical models (26). Grosselin et al. (27) set up a high-throughput droplet microfluidics platform. On this platform, the single cell chromatin landscapes of thousand cells can be profiled. They used the patient-derived xenograft models of acquired resistance to chemotherapy and target therapies in breast cancer and found a common chromatin signature between drug-sensitive and resistant cells (27). This technique paves the way to study the role of chromatin heterogeneity.
The limited cancer models hinder the clinical prediction of drug efficacy. Therefore, it is urgent to establish more reasonable, advanced and high-throughput cancer models to deal with drug resistance and explore the underlying factor of heterogeneous patient responses. Gao et al. (28) established about 1,000 patient-derived tumor xenograft models (PDXs) with a diverse set of driver mutation using high-throughput screening technology. It has been demonstrated that these PDXs have the potential to predict patient response to targeted therapies and perform in vivo compound screens (28). Furthermore, a 3D model of tumor tissue made up of numerous different cell types can better mimic tumor microenvironment and provide the similar information about clinical response. Kather et al. developed a 3D model of tumor tissue which reproduced key features of colorectal cancer (CRC) and based on the individual patient data, yielding in silico tumor explant (29).
Combinations of drugs are also the effective way to overcome or bypass drug resistance (30). Epidermal growth factor receptor tyrosine kinase inhibitor (EGFR TKI) is beneficial for the treatment of non-small cell lung cancer with EGFR mutation (31). However, after treatment with EGFR TKI for 10–14 months, the efficacy declines (32), the primary and acquired drug resistance limits their clinical benefit (33). To combat resistance, in addition to developing new drugs, drugs combinations through a so-called bypass signaling mechanism, is an excellent choice (34). In addition, nanomedicine approach can be used to encapsulate and co-delivery drugs in specific materials to improve their bioavailability and thus overcome drug resistance (35, 36). The application of high-throughput drug screening can identify the effective drug combination regimens. Using high-throughput screening technology, researchers identified that potassium antimony tartrate in combination with topotecan can significantly enhance the sensitivity of non-small cell lung cancer and colorectal cancer to cis-diamminedichloroplatinum/cisplatin (CDDP). It was found that topotecan impairs the ability to repair CDDP-induced DNA damage (37). DNA damage repair is a classic mechanism by which cells develop drug resistance, as detailed later in this article.
Cancer biomarkers are present in tumor tissue or serum that help to detect cancers in their early stage, simplified the prognosis of cancer development (38). Cancer biomarkers help stratify patients to receive specific therapeutics. Biomarker could be DNA, mRNA, protein and various cellular metabolites (39). Over the past years, many advances have been made in the detection and evaluation of cancer biomarkers (40–44). In the next section, we will detail the development of exosomal miRNA as cancer biomarkers.
Exosomal miRNA is involved in the proliferation, invasion, migration and drug resistance of various cancers. Therefore, exosomal miRNA has the potential to be the biomarkers for cancer diagnosis and treatment. In Table 1, we summarize some recent researches on various exosomal miRNA as regulators and biomarkers in various cancers.
Breast cancer (BC) is a highly prevalent cancer and the second leading cause of cancer-related death among women (95, 96). MiR-9 is a classic miRNA in cancer development. Baroni et al. indicated that exosomal miR-9 has the ability to induce human breast fibroblasts to have cancer associated fibroblasts (CAFs)-like properties (45). Exosome transferring miR-222 can promote BC cells migration and invasion by activating Nuclear factor-κB (NF-κB) (46). Exosomal miRNA also regulate tumor growth by influencing the metabolic reprogramming of BC cells. Yan et al. suggested that BC cells secrete exosomal miR-105 to promote tumor growth through the regulation of metabolic reprogramming in stromal cells (47).
Exosomal miRNA regulates tumor growth in other cancers as well. In colorectal cancer (CRC), transforming growth factor-beta (TGF-β) significantly contributes to the upregulation of exosome-meditated miR-200b, which promotes colorectal cancer cell proliferation by suppressing the expression of p27 in target cells. (51). Li et al. demonstrated that the absence of exosomal miR-148b derived from CAFs is the cause of invasion and metastasis in endometrial cancer (97). In lung cancer, Wu et al. indicated that exosomal miR-96 is associated with proliferation, migration and drug resistance by directly binding to wild-type LMO7 gene (60).
The expression of some exosomal miRNA in cancers is specific. The specificity enables exosomal miRNA to become cancer biomarkers. Sohn et al. proposed that serum exosomal miRNAs have the potential to become novel biomarkers for hepatocellular carcinoma (66). Moreover, Huang et al. extracted serum-derived exosome from patients with gastric non-cardia adenocarcinoma and detected the expression of miRNA. They identified the expression of miR-195-5p, miR-20a-3p, and miR-196-5p in exosomes and found that these miRNAs significantly increased. This finding provided a reference for clinical application and diagnosis using exosomal miRNAs (56).
The mechanism of drug resistance is complex, the currently known mechanisms of drug resistance include drug efflux (98), mutation of drug target (99, 100), alterations in drug metabolism (101), DNA damage repair (102, 103), alterations of energy programming, cancer stem cells and epigenetic changes (104, 105). Most of these processes were regulated by exosomal miRNA (Figure 2). Different treatments have been developed to circumvent these resistance mechanisms (106–112). In this section we associate several mechanisms of drug resistance with miRNAs.
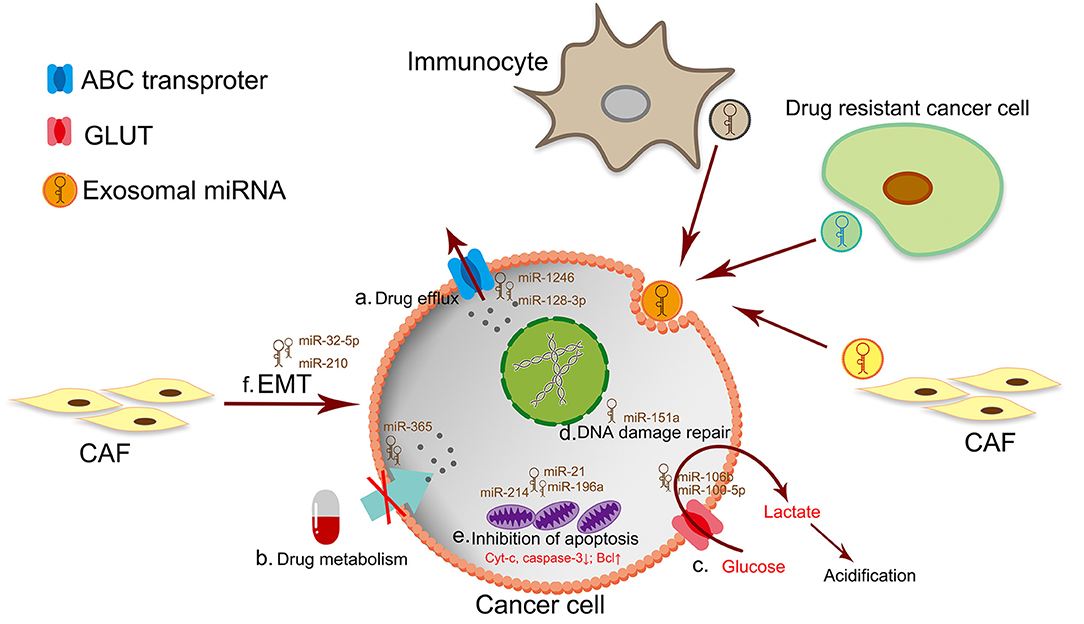
Figure 2. The mechanism of drug resistance in cancer cells. a. Drug efflux by the ATP-binding cassette (ABC) transporter superfamily. b. The dysfunction of drug metabolism by drug inactivation or lack of drug activation. c. The excessive production of lactate leads to TME acidosis. The acidic TME makes cancer cells a strong survival advantage and drug resistance. d. DNA damage repair is a classic mechanism for drug resistant. e. Drug resistant-cancer cells are often accompanied by downregulation of intracellular apoptotic proteins or up-regulation of anti-apoptotic proteins. f. Targeting therapy of CSCs by inhibiting EMT has become an effective way to anticancer and prevent drug resistance.
Drug resistance is always accompanied by the dysfunction of pharmacokinetic factors, that are absorption, distribution, metabolism and elimination (ADME) of drugs. Exosomal miRNA participate drug resistance by interfering drug efflux and metabolism as well.
The excessive drug efflux is a classic mechanism of drug resistance. The human ATP-binding cassette (ABC) transporter superfamily is closely associated with the excessive efflux of drug. In the ABC transporter superfamily, several ATP-driven efflux transporters are the classic regulator of drug efflux: ABCB1 (P-gp/MDR1), ABCC1 (MRP1), ABCG2 (BCRP), ABCC2, MDR4 and MDR5 (113–118). The efficacy of drugs is closely related to the concentration of the drug inside the cells. In drug resistant-tumor cells, overexpression of these ABC transporters pumps anticancer drug out of cells, decreasing the concentration of drugs.
Tumor-derived exosomal miRNA cargo regulates the expression of ABC transporters and facilitates drug resistance in tumor cells. ABCB1 is one of the ABC transporters, some researchers reported that ABCB1 enriched in microvesicles and exosomes shed by drug-resistant cells (119). These EVs transfer ABCB1 to drug-sensitive cells, making the recipient cells express functional ABCB1 and acquiring drug resistance. However, the half-life of ABCB1 is shorter than 24 h and the transfer of ABCB1 is unstable (120). The resistant mechanism of drug-sensitive cells cannot be merely explained by the transfer of ABCB1. Sousa conjectured in his review that ABCB1 may co-transport with miRNA so that ABCB1 can be expressed stably for a long time (121). After that, exosomal miRNA modulates transcripts in recipient cells to acquire resistance phenotype (122). For example, exosomal miR-1246 secreted by ovarian cancer (OC) cells inhibits the expression of Cav1 and upregulates ABCB1 expression to induce tumor-promoting phenotype and drug resistance. Based on the preclinical experiments in vivo, miR-1246 inhibitor treatment in combination with chemotherapy shows great potential in the treatment of OC (12). Exosomal miRNA is a double-edged sword in the occurrence and development of drug resistance. Some miRNAs have positive effects in drug resistance, while some can enhance the chemosensitivity of cancer cells. Liu et al. found that exosome-transmitted miR-128-3p down-regulates the expression of MDR5, decreasing oxaliplatin efflux and improving chemosensitivity of oxaliplatin-resist cells in colorectal cancer (123).
The activation of drug is related to the corresponding enzymes in body. The prodrug is activated by enzyme action, or the drug is metabolized into an inactive form due to some enzymes in vivo. For example, gemcitabine is metabolized by deoxycytidine kinase (dCK) and incorporate with nucleosides in DNA and RNA, preventing DNA from replicating properly. Cytidine deaminase (CDA) is an enzyme that metabolizes gemcitabine to become an inactive form. When the tumor emerges drug resistance, it is often accompanied by the inactivation of dCK or activation of CDA, leading to gemcitabine degradation or inactivation and eventually causing drug resistance (78, 124, 125). Exosomal miRNA is involved in drug metabolism. In pancreatic adenocarcinoma, tumor associated macrophages secrete exosomes that transferring miR-365 to induce gemcitabine-resistance (79). The specific mechanism is that macrophage-derived exosomes (MDE) transfer miR-365 into pancreatic ductal adenocarcinoma (PDAC) cells. Once miR-365 entered PDAC cells, the concentration of triphosphate-nucleotide (NTP) is increasing. NTP can compete with phosphorylated gemcitabine for DNA incorporation, so that it prevents activation of gemcitabine. Moreover, exosomal miR-365 upregulates the expression of CDA and promotes the inactivation of gemcitabine leading to gemcitabine resistance (79).
Energy reprogramming has been accepted as a hallmark of cancer (126). In order to maintain survival, proliferation and dissemination, cancer cells need to reprogram their metabolism to ensure the increasing energy demand (127–129). Mitochondrial oxidative phosphorylation (OXPHOS) and glycolysis are two major metabolic pathways to generate adenosine triphosphate (ATP) to support physiological activities in our daily life. A common characteristic in primary and metastatic cancer is the upregulation of glycolysis (130). Glycolysis usually occurs in an anoxic condition. However, even in aerobic conditions, cancer cells undergo aerobic glycolysis by reprogramming the glucose metabolism and glycolysis is still widespread in TME. This phenomenon is called Warburg Effect. An important reason of this effect is that during glycolysis, glucose is metabolized into pyruvate and lactate. In cancer cells, excessive production of lactate leads to TME acidosis (131). The acidic TME largely contributes to the immunologic escape, because the decrease in extracellular pH leads to the reduction of cytotoxic T- cell function, thus the cancer cells can acquire a strong survival advantage which promotes cancer metastasis, invasion and drug resistance (132–134).
Regulation of glycolysis is one of the ways to inhibit cancer drug resistance (134). The GLUT family is closely related to glucose transport into cells. GLUT1, one of the family members in GLUT, is upregulated in many malignant tumors (135). The upregulation of GLUT1 is associated with mammalian target of rapamycin (mTOR) and the activation of mTOR increases glycolysis and promotes drug resistance (132). The decreased expression of miR-100 is involved in drug resistance in several cancer. mTOR is a target gene of miR-100-5p which binds to the 3′UTR directly and decreases the expression of mTOR and enhances chemo-sensitive of cancer cells. Qin et al. (136) indicated in their study that the expressing of miR-100-5p is not only related to the cell itself, but also to the extracellular microenvironment. Exosome as a messenger for intracellular communication, the concentration of miR-100-5p in exosomes is reflected the content in surrounding microenvironment. The downregulation of miR-100-5p in microenvironment leads to cisplatin resistance in lung cancer cells (136). In addition, TP53INP1 is also a stress protein, which has been indicated to play a tumor suppressive role by regulating metabolic homeostasis (137). Fang et al. showed that CAF derives exosomal miR-106b, which promotes gemcitabine resistance by directly targeting TP53INP1 (138).
As a target of anticancer drugs, DNA damage induces cancer cell death. Genotoxic agents are designed for damaging DNA or preventing the synthesis of new DNA to inhibit cell proliferation. Genotoxic agents are classified as direct damage, such as cisplatin; and indirect damage, such as topoisomerase inhibitors (24). However, in addition to cell death, DNA damage response (DDR) includes the DNA damage repair (139).
DNA damage repair is originally a way to maintain genomic stability in cells. However, DNA damage repair has also been found to be a resistance mechanism because of the widespread use of genotoxic agents (140). DNA repair mechanisms can be briefly divided into the following four categories: (a) Nucleotide excision repair (NER): NER works in a way that is suitable for repairing bulky DNA lesions by using DNA ligase to attach repair patch to the damage DNA regions, which is associated with platinum agent resistance. (b) Base excision repair (BER): BER works through repairing a small number of bases and performing some modification, such as alkylation and oxidative lesions. This repair mechanism is related to the resistance of genotoxic agents nitrosoureas. (c) Mismatch repair (MMR): MMR participates in the modification of oxidation and methylation by bypassing the lesions to replicate. (d) DNA double-strand break repair: Double-strand break (DSB): DSB is the most toxic form of DNA damage. Two main repair pathways of DSB are non-homologous end joining (NHEJ) and homologous recombination (HR) (139, 141). Briefly, these two repair pathways have their own characteristics. NHEJ is more rapid, while HR is more complex and accurate. This mechanism is applicable to the damage induced by topoisomerase inhibitors, temozolomide (TMZ) and some alkylating agents (142).
Exosomal miRNA is a regulator to inhibit DNA damage repair. XRCC4 is a major participator of NHEJ, which forms a heterodimer with DNA ligase IV and covalently joins the broken DNA (143). There have been reports of XRCC4 linked to TMZ resistance in earlier years. XRCC4 is a direct target of miR-151a, the low expression of which leads to the upregulation of XRCC4 and triggers the DNA repair that makes cell resistant to TMZ. To investigate the effects of exosomal miR-151a on cancer cells, researchers incubated glioblastoma multiforme (GBM) receptor cells with exosomes secreted by TMZ-resistant cells and TMZ-sensitive cells. The result shows that GBM receptor cells co-cultured with TMZ-resistant exosomes have stronger resistance to TMZ. However, when researchers restore miR-151a in TMZ-resistant exosomes, the TMZ resistance of GBM recipient cells is significantly decreases (144). This study shows that exosomes have the ability to transfer chemoresistance to sensitive cancer cells and exosomal miR-151a has the potential to become a prognostic factor in GBM treatment.
Resisting cell death is a characteristic of cancers, which leads the unlimited proliferation of cancer cells and the development of drug resistance (145, 146). Drug resistant-cancer cells are often accompanied by downregulation of intracellular apoptotic proteins or up-regulation of anti-apoptotic proteins.
Exosome secreted by drug-resistant cells can transmit the resistance to neighboring cells. Zhang et al. (147) indicated that exosomal miR-214 mediates gefitinib resistance in non-small cell lung cancer (NSCLC). Compared with sensitive cancer cells, the miR-214 in exosomes secreted by gefitinib resistant-cells is significantly increased. Gefitinib resistant-cells secreted exosomal miR-214 could confer gefitinib resistance in NSCLC by suppressing cell apoptosis (147). In addition to cancer cells, exosomes secreted by stroma cells also act on resistant targets by transferring miRNA, making cancer cells to acquire drug resistance. Paclitaxel is a common agent for the treatment of ovarian cancer. However, the efficacy of paclitaxel treatment is greatly reduced if the ovarian cancer cells develop resistance to paclitaxel. By using sequencing technology, Au Yeung et al. (148) identified that miR-21 isomiRNAs have higher expression level in the exosomes of cancer-associated adipocytes (CAAs) and CAFs than in those from ovarian cancer cells. After exosomal miR-21 transship to ovarian cancer cells, miR-21 binds to apoptotic protease activating factor 1 (APAF1) and the expression of APAF1 is downregulated (148). APAF1 combined with cytochrome c (Cyt-c) and dATP to form apoptosomes, increasing caspase-9 and caspase-3, leading to massive mitochondrial damage and finally inducing cell apoptosis (149). Therefore, the decrease of APAF1 has the ability to suppress apoptosis and eventually cause drug resistance in cancer cells. This result showed that in omental tumor microenvironment, cancer cells have a negative effect on neighboring stromal-derived exosomal miR-21 and acquire malignant phenotype, including drug resistance (148). Moreover, exosomal miR-196a derived from CAFs confers cisplatin resistance in head and neck cancer (HNC). In order to explore the mechanism of exosomal miR-196a in HNC cells, Qin et al. (150) used miRecords algorithm and finally found the target of exosomal miR-196a: CDKN1B and ING5. CDKN1B and ING5 exhibit different functions in miR-196a-mediated cisplatin resistance. ING5 gene is a major gene to regulate apoptosis. Therefore, they proposed that exosomal miR-196a promote cisplatin-resistance in HNC cells by suppressing apoptosis of cancer cells (150).
In the process of cancer growth, genetic and non-genetic factors induce biological heterogeneity, resulting in phenotypic difference of tumors. The phenotypic diversity of malignant cancers is considered as a significant driver that induces drug resistance.
The cancer stem cells (CSCs) concept provides a good explanation for the association between heterogeneity and the resistance of cancer cells. Because of the renewal properties and genomic instability, CSCs are closely related to the proliferation, metastasis, and recurrence of cancer (151). Epigenetic regulation has a great contribution to the behaviors of cancer cells. Epigenetic differences between CSCs and non-CSCs have a great possibility that caused by epithelial-to-mesenchymal transition (EMT) (152). When epithelial cells transform into mesenchymal cells, cancer cells acquire the properties of migration and invasion and even drug resistance (153–155). What's interesting lies on the study that shows that EMT only occurs in tumors with CSCs (156–158). In tumor microenvironment, CSCs comprise a small proportion of total cells in tumor, most of the cancer cells are non-CSCs (157). However, the traditional cancer treatment merely kills most of the non-CSCs and the CSCs are retained. These residual CSCs eventually induce tumor recurrence and drug resistance through differentiation (159–162).
In recent years, targeting therapy of CSCs by inhibiting EMT has become an effective way to treat cancers and prevent drug resistance (163–167). EMT is an effective target to affect drug resistance. Exosomal miR-32-5p is proved to induce multidrug resistance in hepatocellular carcinoma via the PI3K/AKT pathway to promote EMT and angiogenesis (168). CSCs themselves also secret exosomes to induce drug resistance. MiR-155 is a classic and multifunctional modulating miRNA which is overexpressed in multiple malignant cancers (169). Santos et al. (170) carried out a study which supports a putative mechanism of exosomal miRNA transmission between cancer cells: miR-155 is enriched in exosomes secreted by CSCs and drug resistant cells. In addition, they observed the downregulation of E-Cadherin (E-Cad) and upregulation of mesenchymal biomarkers, which demonstrated that CSCs and drug resistant cells have the ability to trigger the EMT process in recipient cells by transferring exosomal miR-155 and eventually lead to the recipient cells possess resistance (170). In pancreatic cancer cells, the gemcitabine-resistant CSCs can secret miR-210 enriched exosomes. Gemcitabine-resistant CSCs enhance drug resistant by transferring exosomal miR-210 to gemcitabine-sensitive cells (171).
During these years, more and more studies have revealed that different types of cells secrete exosomal miRNA in tumor microenvironment and participate in the process of drug resistance. The drug resistant mechanism of exosomal miRNA on several common anticancer chemotherapeutic agents and molecular targeted agents are summarized in Table 2.
Drug resistance is an eternal topic in cancer treatment. In this article, we discussed the role of exosomal miRNA in different mechanisms of drug resistance. Some of them act as “communicators” and some of them “biomarkers” that facilitate communication between cancer cells with other cancer cells or cancer cells with tumor microenvironment, enriching the knowledge background about the diagnosis of cancer. However, drug resistance in cancer is not caused by only one or several mechanisms, it is the combined action of the intrinsic (such as mutation) and extrinsic (such as drug inactivation) factors. Although progress has been made in suppressing the emergence of drug resistance, there is still a long way to go to eradicate the problem of drug resistance. Nevertheless, the knowledge of exosomal miRNA will provide some clues to help exploring the secret of cancer drug resistance.
QG, QW, and JZ conceived the review. QG, YL, and CS searched the literature and drafted the manuscript. YaY, RA, and HC critically appraised the literature. YiY, HW, and CS edited the manuscript. All authors approved the final version of the manuscript.
This work was supported by National Natural Science Foundation of China (81773888, U1903126 and 81902152), Guangdong Basic and Applied Basic Research Foundation (2020A1515010005, 2020A1515010605), Fund from Guangzhou Institute of Pediatrics/Guangzhou Women and Children's Medical Center (No: IP-2018-012).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Gao D, Jiang L. Exosomes in cancer therapy: a novel experimental strategy. Am J Cancer Res. (2018) 8:2165−75.
2. Ha D, Yang N, Nadithe V. Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: current perspectives and future challenges. Acta Pharm Sin B. (2016) 6:287–96. doi: 10.1016/j.apsb.2016.02.001
3. Huang W, Yan Y, Liu Y, Lin M, Ma J, Zhang W, et al. Exosomes with low miR-34c-3p expression promote invasion and migration of non-small cell lung cancer by upregulating integrin α2β1. Signal Transduct Target Ther. (2020).
4. Liu F, Bu Z, Zhao F, Xiao D. Increased T-helper 17 cell differentiation mediated by exosome-mediated microRNA-451 redistribution in gastric cancer infiltrated T cells. Cancer Sci. (2018) 109:65–73. doi: 10.1111/cas.13429
5. Yousafzai NA, Wang H, Wang Z, Zhu Y, Zhu L, Jin H, et al. Exosome mediated multidrug resistance in cancer. Am J Cancer Res. (2018) 8:2210–26.
6. Hessvik NP Llorente A. Current knowledge on exosome biogenesis and release. Cell Mol Life Sci. (2018) 75:193–208. doi: 10.1007/s00018-017-2595-9
7. Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J Biol Chem. (1987) 262:9412–20.
8. Harding C, Heuser J, Stahl P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J Cell Biol. (1983) 97:329–39. doi: 10.1083/jcb.97.2.329
9. Wan Z, Gao X, Dong Y, Zhao Y, Chen X, Yang G, et al. Exosome-mediated cell-cell communication in tumor progression. Am J Cancer Res. (2018) 8:1661−73.
10. Bayraktar R, Van Roosbroeck K, Calin GA. Cell-to-cell communication: microRNAs as hormones. Mol Oncol. (2017) 11:1673–86. doi: 10.1002/1878-0261.12144
11. Devhare PB, Sasaki R, Shrivastava S, Di Bisceglie AM, Ray R, Ray RB. Exosome-mediated intercellular communication between hepatitis c virus-infected hepatocytes and hepatic stellate cells. J Virol. (2017) 91:e02225–16. doi: 10.1128/JVI.00349-17
12. Kanlikilicer P, Bayraktar R, Denizli M, Rashed MH, Ivan C, Aslan B, et al. Exosomal miRNA confers chemo resistance via targeting Cav1/p-gp/M2-type macrophage axis in ovarian cancer. EBioMed. (2018) 38:100–12. doi: 10.1016/j.ebiom.2018.11.004
13. Qin Y, Peng Y, Zhao W, Pan J, Ksiezak-Reding H, Cardozo C, et al. Myostatin inhibits osteoblastic differentiation by suppressing osteocyte-derived exosomal microRNA-218: a novel mechanism in muscle-bone communication. J Biol Chem. (2017) 292:11021–33. doi: 10.1074/jbc.M116.770941
14. Teng Y, Ren Y, Hu X, Mu J, Samykutty A, Zhuang X, et al. MVP-mediated exosomal sorting of miR-193a promotes colon cancer progression. Nat Commun. (2017) 8:14448. doi: 10.1038/ncomms14448
15. Yang F, Ning Z, Ma L, Liu W, Shao C, Shu Y, et al. Exosomal miRNAs and miRNA dysregulation in cancer-associated fibroblasts. Mol Cancer. (2017) 16:148. doi: 10.1186/s12943-017-0718-4
16. Zheng P, Chen L, Yuan X, Luo Q, Liu Y, Xie G, et al. Exosomal transfer of tumor-associated macrophage-derived miR-21 confers cisplatin resistance in gastric cancer cells. J Exp Clin Cancer Res. (2017) 36:53. doi: 10.1186/s13046-017-0528-y
17. Li Y, Yin Z, Fan J, Zhang S, Yang W. The roles of exosomal miRNAs and lncRNAs in lung diseases. Signal Transduct Target Ther. (2019) 4:47. doi: 10.1038/s41392-019-0080-7
18. Achkar NP, Cambiagno DA, Manavella PA. miRNA biogenesis: a dynamic pathway. Trends Plant Sci. (2016) 21:1034–44. doi: 10.1016/j.tplants.2016.09.003
19. Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. (2003) 425:415–9. doi: 10.1038/nature01957
20. Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. (2014) 15:509–24. doi: 10.1038/nrm3838
21. Bartel DP. microRNAs: genomics, biogenesis, mechanism, and function. Cell. (2004) 116:281–97. doi: 10.1016/S0092-8674(04)00045-5
22. Zeng Y, Cullen BR. Sequence requirements for micro RNA processing and function in human cells. RNA. (2003) 9:112–23. doi: 10.1261/rna.2780503
23. Zahreddine H, Borden KLB. Mechanisms and insights into drug resistance in cancer. Front Pharmacol. (2013) 4:28. doi: 10.3389/fphar.2013.00028
24. Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer. (2013) 13:714–26. doi: 10.1038/nrc3599
25. Dagogo-Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol. (2018) 15:81–94. doi: 10.1038/nrclinonc.2017.166
26. Dhiman N, Kingshott P, Sumer H, Sharma CS, Rath SN. On-chip anticancer drug screening - recent progress in microfluidic platforms to address challenges in chemotherapy. Biosens Bioelectron. (2019) 137:236–54. doi: 10.1016/j.bios.2019.02.070
27. Grosselin K, Durand A, Marsolier J, Poitou A, Marangoni E, Nemati F, et al. High-throughput single-cell ChIP-seq identifies heterogeneity of chromatin states in breast cancer. Nat Genet. (2019) 51:1060–6. doi: 10.1038/s41588-019-0424-9
28. Gao H, Korn JM, Ferretti S, Monahan JE, Wang Y, Singh M, et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat Med. (2015) 21:1318–25. doi: 10.1038/nm.3954
29. Kather JN, Charoentong P, Suarez-Carmona M, Herpel E, Klupp F, Ulrich A, et al. High-throughput screening of combinatorial immunotherapies with patient-specific in silico models of metastatic colorectal cancer. Cancer Res Cell Death Dis. (2018) 78:5155–63. doi: 10.1158/0008-5472.CAN-18-1126
30. Yuan M, Huang L-L, Chen J-H, Wu J, Xu Q. The emerging treatment landscape of targeted therapy in non-small-cell lung cancer. Signal Transduct Target Ther. (2019) 4:61. doi: 10.1038/s41392-019-0099-9
31. Vyse S, Huang PH. Targeting EGFR exon 20 insertion mutations in non-small cell lung cancer. Signal Transduct Target Ther. (2019) 4:5. doi: 10.1038/s41392-019-0038-9
32. Wu S, Shih J. Management of acquired resistance to EGFR TKI-targeted therapy in advanced non-small cell lung cancer. Mol Cancer. (2018) 17:38. doi: 10.1186/s12943-018-0777-1
33. Yang L, Ying S, Hu S, Zhao X, Li M, Chen M, et al. EGFR TKIs impair lysosome-dependent degradation of SQSTM1 to compromise the effectiveness in lung cancer. Signal Transduct Target Ther. (2019) 4:25. doi: 10.1038/s41392-019-0059-4
34. Tong CWS, Wu WKK, Loong HHF, Cho WCS, To KKW. Drug combination approach to overcome resistance to EGFR tyrosine kinase inhibitors in lung cancer. Cancer Lett Cancer Res. (2017) 405:100–10. doi: 10.1016/j.canlet.2017.07.023
35. Han W, Shi L, Ren L, Zhou L, Li T, Qiao Y, et al. A nanomedicine approach enables co-delivery of cyclosporin A and gefitinib to potentiate the therapeutic efficacy in drug-resistant lung cancer. Signal Transduct Target Ther. (2018) 3:16. doi: 10.1038/s41392-018-0019-4
36. Senapati S, Mahanta AK, Kumar S, Maiti P. Controlled drug delivery vehicles for cancer treatment and their performance. Signal Transduct Target Ther. (2018) 3:7. doi: 10.1038/s41392-017-0004-3
37. Sun Y, Jiang W, Lu W, Song M, Liu K, Chen P, et al. Identification of cisplatin sensitizers through high-throughput combinatorial screening. Int J Oncol. (2018) 53:1237–46. doi: 10.3892/ijo.2018.4447
38. Sharma AK. Emerging trends in biomarker discovery: ease of prognosis and prediction in cancer. Semin Cancer Biol. (2018) 52:iii–iv. doi: 10.1016/j.semcancer.2018.05.008
39. Wu L, Qu X. Cancer biomarker detection: recent achievements and challenges. Chem Soc Rev. (2015) 44:2963–97. doi: 10.1039/C4CS00370E
40. Bhoola NH, Mbita Z, Hull R, Dlamini Z. Translocator protein (TSPO) as a potential biomarker in human cancers. Int J Mol Sci. (2018) 19:2176. doi: 10.3390/ijms19082176
41. Folprecht G. Tumor mutational burden as a new biomarker for PD-1 antibody treatment in gastric cancer. Cancer Commun. (2019) 39:74–74. doi: 10.1186/s40880-019-0417-1
42. Han X, Wang J, Sun Y. Circulating tumor DNA as biomarkers for cancer detection. Genomics Proteomics Bioinformatics. (2017) 15:59–72. doi: 10.1016/j.gpb.2016.12.004
43. Patel SP, Kurzrock R. PD-L1 expression as a predictive biomarker in cancer immunotherapy. Mol Cancer Ther. (2015) 14:847–56. doi: 10.1158/1535-7163.MCT-14-0983
44. Wang H, Peng R, Wang J, Qin Z, Xue L. Circulating microRNAs as potential cancer biomarkers: the advantage and disadvantage. Clin Epigenet. (2018) 10:59. doi: 10.1186/s13148-018-0492-1
45. Baroni S, Romero-Cordoba S, Plantamura I, Dugo M, D'Ippolito E, Cataldo A, et al. Exosome-mediated delivery of miR-9 induces cancer-associated fibroblast-like properties in human breast fibroblasts. Cell Death Dis. (2016) 7:e2312. doi: 10.1038/cddis.2016.224
46. Ding J, Xu Z, Zhang Y, Tan C, Hu W, Wang M, et al. Exosome-mediated miR-222 transferring: An insight into NF-κB-mediated breast cancer metastasis. Exp Cell Res. (2018) 369:129–38. doi: 10.1016/j.yexcr.2018.05.014
47. Yan W, Wu X, Zhou W, Fong MY, Cao M, Liu J, et al. Cancer-cell-secreted exosomal miR-105 promotes tumour growth through the MYC-dependent metabolic reprogramming of stromal cells. Nat Cell Biol. (2018) 20:597–609. doi: 10.1038/s41556-018-0083-6
48. Singh R, Pochampally R, Watabe K, Lu Z, Mo Y-Y. Exosome-mediated transfer of miR-10b promotes cell invasion in breast cancer. Mol Cancer. (2014) 13:256. doi: 10.1186/1476-4598-13-256
49. Fong MY, Zhou W, Liu L, Alontaga AY, Chandra M, Ashby J, et al. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat Cell Biol. (2015) 17:183–94. doi: 10.1038/ncb3094
50. Zhai L, Li M, Pan W, Chen Y, Li M, Pang J, et al. In situ detection of plasma exosomal microRNA-1246 for breast cancer diagnostics by a Au nanoflare probe. ACS Appl Mater Interfaces. (2018) 10:39478–86. doi: 10.1021/acsami.8b12725
51. Zhang Z, Xing T, Chen Y, Xiao J. Exosome-mediated miR-200b promotes colorectal cancer proliferation upon TGF-β1 exposure. Biomed Pharmacother. (2018) 106:1135–43. doi: 10.1016/j.biopha.2018.07.042
52. Cheng W, Liao T, Lin C, Yuan LE, Lan H, Lin H, et al. RAB27B-activated secretion of stem-like tumor exosomes delivers the biomarker microRNA-146a-5p, which promotes tumorigenesis and associates with an immunosuppressive tumor microenvironment in colorectal cancer. Int J Cancer. (2019) 145:2209–24. doi: 10.1002/ijc.32338
53. Wang J, Yan F, Zhao Q, Zhan F, Wang R, Wang L, et al. Circulating exosomal miR-125a-3p as a novel biomarker for early-stage colon cancer. Sci Rep. (2017) 7:4150. doi: 10.1038/s41598-017-04386-1
54. Zeng Z, Li Y, Pan Y, Lan X, Song F, Sun J, et al. Cancer-derived exosomal miR-25-3p promotes pre-metastatic niche formation by inducing vascular permeability and angiogenesis. Nat Commun. (2018) 9:5395. doi: 10.1038/s41467-018-07810-w
55. Liu X, Pan B, Sun L, Chen X, Zeng K, Hu X, et al. Circulating exosomal miR-27a and miR-130a act as novel diagnostic and prognostic biomarkers of colorectal cancer. Cancer Epidemiol Biomark Prev. (2018) 27:746–54. doi: 10.1158/1055-9965.EPI-18-0067
56. Huang Z, Zhu D, Wu L, He M, Zhou X, Zhang L, et al. Six serum-based miRNAs as potential diagnostic biomarkers for gastric cancer. Cancer Epidemiol Biomark Prev. (2017) 26:188–96. doi: 10.1158/1055-9965.EPI-16-0607
57. Xu G, Zhang B, Ye J, Cao S, Shi J, Zhao Y, et al. Exosomal miRNA-139 in cancer-associated fibroblasts inhibits gastric cancer progression by repressing MMP11 expression. Int J Biol Sci. (2019) 15:2320–9. doi: 10.7150/ijbs.33750
58. Yang H, Zhang H, Ge S, Ning T, Bai M, Li J, et al. Exosome-Derived miR-130a activates angiogenesis in gastric cancer by targeting C-MYB in vascular endothelial cells. Mol Ther. (2018) 26:2466–75. doi: 10.1016/j.ymthe.2018.07.023
59. Yang H, Fu H, Wang B, Zhang X, Mao J, Li X, et al. Exosomal miR-423-5p targets SUFU to promote cancer growth and metastasis and serves as a novel marker for gastric cancer. Mol Carcinogenesis. (2018) 57:1223–36. doi: 10.1002/mc.22838
60. Wu H, Zhou J, Mei S, Wu D, Mu Z, Chen B, et al. Circulating exosomal microRNA-96 promotes cell proliferation, migration and drug resistance by targeting LMO7. J Cell Mol Med. (2017) 21:1228–36. doi: 10.1111/jcmm.13056
61. Hsu YL, Hung JY, Chang WA, Lin YS, Pan YC, Tsai PH, et al. Hypoxic lung cancer-secreted exosomal miR-23a increased angiogenesis and vascular permeability by targeting prolyl hydroxylase and tight junction protein ZO-1. Oncogene. (2017) 36:4929–42. doi: 10.1038/onc.2017.105
62. Grimolizzi F, Monaco F, Leoni F, Bracci M, Staffolani S, Bersaglieri C, et al. Exosomal miR-126 as a circulating biomarker in non-small-cell lung cancer regulating cancer progression. Sci Rep. (2017) 7:15277. doi: 10.1038/s41598-017-15475-6
63. Wei F, Ma C, Zhou T, Dong X, Luo Q, Geng L, et al. Exosomes derived from gemcitabine-resistant cells transfer malignant phenotypic traits via delivery of miRNA-222-3p. Mol Cancer. (2017) 16:132. doi: 10.1186/s12943-017-0694-8
64. Zhang L, Hao C, Zhai R, Wang D, Zhang J, Bao L, et al. Downregulation of exosomal let-7a-5p in dust exposed- workers contributes to lung cancer development. Respir Res. (2018) 19:235. doi: 10.1186/s12931-018-0949-y
65. Yang Y, Liu Q, Lu J, Adah D, Yu S, Zhao S, et al. Exosomes from Plasmodium-infected hosts inhibit tumor angiogenesis in a murine Lewis lung cancer model. Oncogenesis. (2017) 6:e351. doi: 10.1038/oncsis.2017.52
66. Sohn W, Kim J, Kang SH, Yang SR, Cho J-Y, Cho HC, et al. Serum exosomal microRNAs as novel biomarkers for hepatocellular carcinoma. Exp Mol Med. (2015) 47:e184. doi: 10.1038/emm.2015.68
67. Fang T, Lv H, Lv G, Li T, Wang C, Han Q, et al. Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat Commun. (2018) 9:191. doi: 10.1038/s41467-017-02583-0
68. Wang F, Li L, Piontek K, Sakaguchi M, Selaru FM. Exosome miR-335 as a novel therapeutic strategy in hepatocellular carcinoma. Hepatology. (2018) 67:940–54. doi: 10.1002/hep.29586
69. Zhang Z, Li X, Sun W, Yue S, Yang J, Li J, et al. Loss of exosomal miR-320a from cancer-associated fibroblasts contributes to HCC proliferation and metastasis. Cancer Lett. (2017) 397:33–42. doi: 10.1016/j.canlet.2017.03.004
70. Fang J, Zhang Z, Shang L, Luo Y, Lin Y, Yuan Y, et al. Hepatoma cell-secreted exosomal microRNA-103 increases vascular permeability and promotes metastasis by targeting junction proteins. Hepatology. (2018) 68:1459–75. doi: 10.1002/hep.29920
71. Kobayashi M, Salomon C, Tapia J, Illanes SE, Mitchell MD, Rice GE. Ovarian cancer cell invasiveness is associated with discordant exosomal sequestration of Let-7 miRNA and miR-200. J Transl Med. (2014) 12:4. doi: 10.1186/1479-5876-12-4
72. Zhou J, Li X, Wu X, Zhang T, Zhu Q, Wang X, et al. Exosomes released from tumor-associated macrophages transfer miRNAs that induce a Treg/Th17 cell imbalance in epithelial ovarian cancer. Cancer Immunol Res. (2018) 6:1578–92. doi: 10.1158/2326-6066.CIR-17-0479
73. He L, Zhu W, Chen Q, Yuan Y, Wang Y, Wang J, et al. Ovarian cancer cell-secreted exosomal miR-205 promotes metastasis by inducing angiogenesis. Theranostics. (2019) 9:8206–20. doi: 10.7150/thno.37455
74. Kim S, Choi MC, Jeong J-Y, Hwang S, Jung SG, Joo WD, et al. Serum exosomal miRNA-145 and miRNA-200c as promising biomarkers for preoperative diagnosis of ovarian carcinomas. J Cancer. (2019) 10:1958–67. doi: 10.7150/jca.30231
75. Chen X, Ying X, Wang X, Wu X, Zhu Q, Wang X. Exosomes derived from hypoxic epithelial ovarian cancer deliver microRNA-940 to induce macrophage M2 polarization. Oncol Rep. (2017) 38:522–8. doi: 10.3892/or.2017.5697
76. Kanlikilicer P, Rashed MH, Bayraktar R, Mitra R, Ivan C, Aslan B, et al. Ubiquitous release of exosomal tumor suppressor miR-6126 from ovarian cancer cells. Cancer Res. (2016) 76:7194–207. doi: 10.1158/0008-5472.CAN-16-0714
77. Pan C, Stevic I, Müller V, Ni Q, Oliveira-Ferrer L, Pantel K, et al. Exosomal microRNAs as tumor markers in epithelial ovarian cancer. Mol Oncol. (2018) 12:1935–48. doi: 10.1002/1878-0261.12371
78. Patel GK, Khan MA, Bhardwaj A, Srivastava SK, Zubair H, Patton MC, et al. Exosomes confer chemoresistance to pancreatic cancer cells by promoting ROS detoxification and miR-155-mediated suppression of key gemcitabine-metabolising enzyme, DCK. Br J Cancer. (2017) 116:609–19. doi: 10.1038/bjc.2017.18
79. Binenbaum Y, Fridman E, Yaari Z, Milman N, Schroeder A, Ben David G, et al. Transfer of miRNA in macrophage-derived exosomes induces drug resistance in pancreatic adenocarcinoma. Cancer Res. (2018) 78:5287–99. doi: 10.1158/0008-5472.CAN-18-0124
80. Nakamura S, Sadakari Y, Ohtsuka T, Okayama T, Nakashima Y, Gotoh Y, et al. Pancreatic juice exosomal microRNAs as Biomarkers for detection of pancreatic ductal adenocarcinoma. Ann Surg Oncol. (2019) 26:2104–11. doi: 10.1245/s10434-019-07269-z
81. Shang S, Wang J, Chen S, Tian R, Zeng H, Wang L, et al. Exosomal miRNA-1231 derived from bone marrow mesenchymal stem cells inhibits the activity of pancreatic cancer. Cancer Med. (2019) 8:7728–40. doi: 10.1002/cam4.2633
82. Madhavan B, Yue S, Galli U, Rana S, Gross W, Müller M, et al. Combined evaluation of a panel of protein and miRNA serum-exosome biomarkers for pancreatic cancer diagnosis increases sensitivity and specificity. Int J Cancer. (2015) 136:2616–27. doi: 10.1002/ijc.29324
83. Wang X, Luo G, Zhang K, Cao J, Huang C, Jiang T, et al. Hypoxic tumor-derived exosomal miR-301a mediates M2 macrophage polarization via PTEN/PI3Kγ to promote pancreatic cancer metastasis. Cancer Res. (2018) 78:4586–98. doi: 10.1158/0008-5472.CAN-17-3841
84. Huang X, Yuan T, Liang M, Du M, Xia S, Dittmar R, et al. Exosomal miR-1290 and miR-375 as prognostic markers in castration-resistant prostate cancer. Eur Urol. (2015) 67:33–41. doi: 10.1016/j.eururo.2014.07.035
85. Malla B, Aebersold DM, Dal Pra A. Protocol for serum exosomal miRNAs analysis in prostate cancer patients treated with radiotherapy. J Transl Med. (2018) 16:223. doi: 10.1186/s12967-018-1592-6
86. Rodríguez M, Bajo-Santos C, Hessvik NP, Lorenz S, Fromm B, Berge V, et al. Identification of non-invasive miRNAs biomarkers for prostate cancer by deep sequencing analysis of urinary exosomes. Mol Cancer. (2017) 16:156. doi: 10.1186/s12943-017-0726-4
87. Bhagirath D, Yang TL, Bucay N, Sekhon K, Majid S, Shahryari V, et al. microRNA-1246 is an exosomal biomarker for aggressive prostate cancer. Cancer Res. (2018) 78:1833–44. doi: 10.1158/0008-5472.CAN-17-2069
88. Sun L, Xu K, Cui J, Yuan D, Zou B, Li J, et al. Cancer-associated fibroblast-derived exosomal miR-382-5p promotes the migration and invasion of oral squamous cell carcinoma. Oncol Rep. (2019) 42:1319–28. doi: 10.3892/or.2019.7255
89. Sakha S, Muramatsu T, Ueda K, Inazawa J. Exosomal microRNA miR-1246 induces cell motility and invasion through the regulation of DENND2D in oral squamous cell carcinoma. Sci Rep. (2016) 6:38750. doi: 10.1038/srep38750
90. Li L, Li C, Wang S, Wang Z, Jiang J, Wang W, et al. Exosomes derived from hypoxic oral squamous cell carcinoma cells deliver miR-21 to normoxic cells to elicit a prometastatic phenotype. Cancer Res. (2016) 76:1770–80. doi: 10.1158/0008-5472.CAN-15-1625
91. Li Y, Tao Y, Gao S, Li P, Zheng J, Zhang S, et al. Cancer-associated fibroblasts contribute to oral cancer cells proliferation and metastasis via exosome-mediated paracrine miR-34a-5p. EBioMedicine. (2018) 36:209–20. doi: 10.1016/j.ebiom.2018.09.006
92. Bao L, You B, Shi S, Shan Y, Zhang Q, Yue H, et al. Metastasis-associated miR-23a from nasopharyngeal carcinoma-derived exosomes mediates angiogenesis by repressing a novel target gene TSGA10. Oncogene. (2018) 37:2873–89. doi: 10.1038/s41388-018-0183-6
93. Ye S, Zhang H, Cai T, Liu Y, Ni J, He J, et al. Exosomal miR-24-3p impedes T-cell function by targeting FGF11 and serves as a potential prognostic biomarker for nasopharyngeal carcinoma. J Pathol. (2016) 240:329–40. doi: 10.1002/path.4781
94. Lu J, Liu Q, Wang F, Tan J, Deng Y, Peng X, et al. Exosomal miR-9 inhibits angiogenesis by targeting MDK and regulating PDK/AKT pathway in nasopharyngeal carcinoma. J Exp Clin Cancer Res. (2018) 37:147. doi: 10.1186/s13046-018-0814-3
95. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. (2017) 67:7–30. doi: 10.3322/caac.21387
96. Tong CWS, Wu M, Cho WCS, To KKW. Recent advances in the treatment of breast cancer. Front Oncol. (2018) 8:227. doi: 10.3389/fonc.2018.00227
97. Li B, Lu W, Qu J, Ye L, Du G, Wan X. Loss of exosomal miR-148b from cancer-associated fibroblasts promotes endometrial cancer cell invasion and cancer metastasis. J Cell Physiol. (2019) 234:2943–53. doi: 10.1002/jcp.27111
98. Fletcher JI, Williams RT, Henderson MJ, Norris MD, Haber M. ABC transporters as mediators of drug resistance and contributors to cancer cell biology. Drug Resist Updat. (2016) 26:1–9. doi: 10.1016/j.drup.2016.03.001
99. Alsop K, Fereday S, Meldrum C, deFazio A, Emmanuel C, George J, et al. BRCA mutation frequency and patterns of treatment response in BRCA mutation-positive women with ovarian cancer: a report from the Australian Ovarian Cancer Study Group. J Clin Oncol. (2012) 30:2654–63. doi: 10.1200/JCO.2011.39.8545
100. Cardona AF, Arrieta O, Zapata MI, Rojas L, Wills B, Reguart N, et al. Acquired resistance to Erlotinib in EGFR mutation-positive lung adenocarcinoma among hispanics (CLICaP). Target Oncol. (2017) 12:513–23. doi: 10.1007/s11523-017-0497-2
101. Mazerska Z, Mróz A, Pawłowska M, Augustin E. The role of glucuronidation in drug resistance. Pharmacol Ther. (2016) 159:35–55. doi: 10.1016/j.pharmthera.2016.01.009
102. Nientiedt C, Heller M, Endris V, Volckmar A-L, Zschäbitz S, Tapia-Laliena MA, et al. Mutations in BRCA2 and taxane resistance in prostate cancer. Sci Rep. (2017) 7:4574. doi: 10.1038/s41598-017-04897-x
103. Sakai W, Swisher EM, Karlan BY, Agarwal MK, Higgins J, Friedman C, et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature. (2008) 451:1116–20. doi: 10.1038/nature06633
104. Leon G, MacDonagh L, Finn SP, Cuffe S, Barr MP. Cancer stem cells in drug resistant lung cancer: targeting cell surface markers and signaling pathways. Pharmacol Ther. (2016) 158:71–90. doi: 10.1016/j.pharmthera.2015.12.001
105. Lytle NK, Barber AG, Reya T. Stem cell fate in cancer growth, progression and therapy resistance. Nat Rev Cancer. (2018) 18:669–80. doi: 10.1038/s41568-018-0056-x
106. Fumarola C, Bonelli MA, Petronini PG, Alfieri RR. Targeting PI3K/AKT/mTOR pathway in non small cell lung cancer. Biochem Pharmacol. (2014) 90:197–207. doi: 10.1016/j.bcp.2014.05.011
107. Li L, Han R, Xiao H, Lin C, Wang Y, Liu H, et al. Metformin sensitizes EGFR-TKI-resistant human lung cancer cells in vitro and in vivo through inhibition of IL-6 signaling and EMT reversal. Clin Cancer Res. (2014) 20:2714–26. doi: 10.1158/1078-0432.CCR-13-2613
108. Mitchell R, Hopcroft LEM, Baquero P, Allan EK, Hewit K, James D, et al. Targeting BCR-ABL-independent TKI resistance in chronic myeloid leukemia by mTOR and autophagy inhibition. J Natl Cancer Inst. (2018) 110:467–78. doi: 10.1093/jnci/djx236
109. Rodrik-Outmezguine VS, Okaniwa M, Yao Z, Novotny CJ, McWhirter C, Banaji A, et al. Overcoming mTOR resistance mutations with a new-generation mTOR inhibitor. Nature. (2016) 534:272–6. doi: 10.1038/nature17963
110. Wang L, Dong X, Ren Y, Luo J, Liu P, Su D, et al. Targeting EHMT2 reverses EGFR-TKI resistance in NSCLC by epigenetically regulating the PTEN/AKT signaling pathway. Cell Death Dis. (2018) 9:129. doi: 10.1038/s41419-017-0120-6
111. Xiao B, Zhang M, Viennois E, Zhang Y, Wei N, Baker MT, et al. Inhibition of MDR1 gene expression and enhancing cellular uptake for effective colon cancer treatment using dual-surface-functionalized nanoparticles. Biomaterials. (2015) 48:147–60. doi: 10.1016/j.biomaterials.2015.01.014
112. Zhang S, Anjum R, Squillace R, Nadworny S, Zhou T, Keats J, et al. The Potent ALK inhibitor Brigatinib (AP26113) overcomes mechanisms of resistance to first- and second-generation ALK inhibitors in preclinical models. Clin Cancer Res. (2016) 22:5527–38. doi: 10.1158/1078-0432.CCR-16-0569
113. Chen L, Manautou JE, Rasmussen TP, Zhong X-B. Development of precision medicine approaches based on inter-individual variability of BCRP/ABCG2. Acta Pharm Sin B. (2019) 9:659–74. doi: 10.1016/j.apsb.2019.01.007
114. Chen Z, Shi T, Zhang L, Zhu P, Deng M, Huang C, et al. Mammalian drug efflux transporters of the ATP binding cassette (ABC) family in multidrug resistance: A review of the past decade. Cancer Lett. (2016) 370:153–64. doi: 10.1016/j.canlet.2015.10.010
115. Valinezhad Sani F, Palizban A, Mosaffa F, Jamialahmadi K. Glucosamine reverses drug resistance in MRP2 overexpressing ovarian cancer cells. Eur J Pharmacol. (2020) 868:172883. doi: 10.1016/j.ejphar.2019.172883
116. Toyoda Y, Takada T, Suzuki H. Inhibitors of human ABCG2: from technical background to recent updates with clinical implications. Front Pharmacol. (2019) 10:208. doi: 10.3389/fphar.2019.00208
117. Reid G, Wielinga P, Zelcer N, De Haas M, Van Deemter L, Wijnholds J, et al. Characterization of the transport of nucleoside analog drugs by the human multidrug resistance proteins MRP4 and MRP5. Mol Pharmacol. (2003) 63:1094–103. doi: 10.1124/mol.63.5.1094
118. Němcová-Fürstová V, Kopperová D, Balušíková K, Ehrlichová M, Brynychová V, Václavíková R, et al. Characterization of acquired paclitaxel resistance of breast cancer cells and involvement of ABC transporters. Toxicol Appl Pharmacol. (2016) 310:215–28. doi: 10.1016/j.taap.2016.09.020
119. Lopes-Rodrigues V, Seca H, Sousa D, Sousa E, Lima RT, Vasconcelos MH. The network of P-glycoprotein and microRNAs interactions. Int J Cancer. (2014) 135:253–63. doi: 10.1002/ijc.28500
120. Levchenko A, Mehta BM, Niu X, Kang G, Villafania L, Way D, et al. Intercellular transfer of P-glycoprotein mediates acquired multidrug resistance in tumor cells. Proc Natl Acad Sci USA. (2005) 102:1933–8. doi: 10.1073/pnas.0401851102
121. Sousa D, Lima RT, Vasconcelos MH. Intercellular transfer of cancer drug resistance traits by extracellular vesicles. Trends Mol Med. (2015) 21:595–608. doi: 10.1016/j.molmed.2015.08.002
122. Bebawy M, Combes V, Lee E, Jaiswal R, Gong J, Bonhoure A, et al. Membrane microparticles mediate transfer of P-glycoprotein to drug sensitive cancer cells. Leukemia. (2009) 23:1643–9. doi: 10.1038/leu.2009.76
123. Liu T, Zhang X, Du L, Wang Y, Liu X, Tian H, et al. Exosome-transmitted miR-128-3p increase chemosensitivity of oxaliplatin-resistant colorectal cancer. Mol Cancer. (2019) 18:43. doi: 10.1186/s12943-019-0981-7
124. Gebregiworgis T, Bhinderwala F, Purohit V, Chaika NV, Singh PK, Powers R. Insights into gemcitabine resistance and the potential for therapeutic monitoring. Metabolomics. (2018) 14:156. doi: 10.1007/s11306-018-1452-7
125. Nakano Y, Tanno S, Koizumi K, Nishikawa T, Nakamura K, Minoguchi M, et al. Gemcitabine chemoresistance and molecular markers associated with gemcitabine transport and metabolism in human pancreatic cancer cells. Br J Cancer. (2007) 96:457–63. doi: 10.1038/sj.bjc.6603559
126. Hanahan D Weinberg RA. Hallmarks of cancer: the next generation. Cell. (2011) 144:646–74. doi: 10.1016/j.cell.2011.02.013
127. Sancho P, Barneda D, Heeschen C. Hallmarks of cancer stem cell metabolism. Br J Cancer. (2016) 114:1305–12. doi: 10.1038/bjc.2016.152
128. Yang Z, Cappello T, Wang L. Emerging role of microRNAs in lipid metabolism. Acta Pharm Sin B. (2015) 5:145–50. doi: 10.1016/j.apsb.2015.01.002
129. Yoshida GJ. Metabolic reprogramming: the emerging concept and associated therapeutic strategies. J Exp Clin Cancer Res. (2015) 34:111. doi: 10.1186/s13046-015-0221-y
130. Gatenby RA Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. (2004) 4:891–9. doi: 10.1038/nrc1478
131. Hirschhaeuser F, Sattler UGA, Mueller-Klieser W. Lactate: a metabolic key player in cancer. Cancer Res. (2011) 71:6921–5. doi: 10.1158/0008-5472.CAN-11-1457
132. Bhattacharya B, Low SHH, Soh C, Kamal Mustapa N, Beloueche-Babari M, Koh KX, et al. Increased drug resistance is associated with reduced glucose levels and an enhanced glycolysis phenotype. Br J Pharmacol. (2014) 171:3255–67. doi: 10.1111/bph.12668
133. Fischer K, Hoffmann P, Voelkl S, Meidenbauer N, Ammer J, Edinger M, et al. Inhibitory effect of tumor cell-derived lactic acid on human T cells. Blood. (2007) 109:3812–9. doi: 10.1182/blood-2006-07-035972
134. Morandi A Indraccolo S. Linking metabolic reprogramming to therapy resistance in cancer. Biochim Biophys Acta Rev Cancer. (2017) 1868:1–6. doi: 10.1016/j.bbcan.2016.12.004
135. Guo Q, Li C, Zhou W, Chen X, Zhang Y, Lu Y, et al. GLUT1-mediated effective anti-miRNA21 pompon for cancer therapy. Acta Pharm Sin B. (2019) 9:832–42. doi: 10.1016/j.apsb.2019.01.012
136. Qin X, Yu S, Zhou L, Shi M, Hu Y, Xu X, et al. Cisplatin-resistant lung cancer cell-derived exosomes increase cisplatin resistance of recipient cells in exosomal miR-100-5p-dependent manner. Int J Nanomed. (2017) 12:3721–33. doi: 10.2147/IJN.S131516
137. Saadi H, Seillier M, Carrier A. The stress protein TP53INP1 plays a tumor suppressive role by regulating metabolic homeostasis. Biochimie. (2015) 118:44–50. doi: 10.1016/j.biochi.2015.07.024
138. Fang Y, Zhou W, Rong Y, Kuang T, Xu X, Wu W, et al. Exosomal miRNA-106b from cancer-associated fibroblast promotes gemcitabine resistance in pancreatic cancer. Exp Cell Res. (2019) 383:111543. doi: 10.1016/j.yexcr.2019.111543
139. Goldstein M Kastan MB. The DNA damage response: implications for tumor responses to radiation and chemotherapy. Ann Rev Med. (2015) 66:129–43. doi: 10.1146/annurev-med-081313-121208
140. Jackson SP Bartek J. The DNA-damage response in human biology and disease. Nature. (2009) 461:1071–8. doi: 10.1038/nature08467
141. Hnízda A Blundell TL. Multicomponent assemblies in DNA-double-strand break repair by NHEJ. Curr Opin Struct Biol. (2019) 55:154–60. doi: 10.1016/j.sbi.2019.03.026
142. Salehan MR Morse HR. DNA damage repair and tolerance: a role in chemotherapeutic drug resistance. Br J Biomed Sci. (2013) 70:31–40. doi: 10.1080/09674845.2013.11669927
143. Kang Y Yan CT. Regulation of DNA repair in the absence of classical non-homologous end joining. DNA Repair. (2018) 68:34–40. doi: 10.1016/j.dnarep.2018.06.001
144. Zeng A, Wei Z, Yan W, Yin J, Huang X, Zhou X, et al. Exosomal transfer of miR-151a enhances chemosensitivity to temozolomide in drug-resistant glioblastoma. Cancer Lett. (2018) 436:10–21. doi: 10.1016/j.canlet.2018.08.004
145. Lin M, Tang S, Zhang C, Chen H, Huang W, Liu Y, et al. Euphorbia factor L2 induces apoptosis in A549 cells through the mitochondrial pathway. Acta Pharm Sin B. (2017) 7:59–64. doi: 10.1016/j.apsb.2016.06.008
146. Pistritto G, Trisciuoglio D, Ceci C, Garufi A, D'Orazi G. Apoptosis as anticancer mechanism: function and dysfunction of its modulators and targeted therapeutic strategies. Aging. (2016) 8:603–19. doi: 10.18632/aging.100934
147. Zhang Y, Li M, Hu C. Exosomal transfer of miR-214 mediates gefitinib resistance in non-small cell lung cancer. Biochem Biophys Res Commun. (2018) 507:457–64. doi: 10.1016/j.bbrc.2018.11.061
148. Au Yeung CL, Co N-N, Tsuruga T, Yeung T-L, Kwan S-Y, Leung CS, et al. Exosomal transfer of stroma-derived miR21 confers paclitaxel resistance in ovarian cancer cells through targeting APAF1. Nat Commun. (2016) 7:11150. doi: 10.1038/ncomms11150
149. Gortat A, Sancho M, Mondragón L, Messeguer À, Pérez-Payá E, Orzáez M. Apaf1 inhibition promotes cell recovery from apoptosis. Protein Cell. (2015) 6:833–43. doi: 10.1007/s13238-015-0200-2
150. Qin X, Guo H, Wang X, Zhu X, Yan M, Wang X, et al. Exosomal miR-196a derived from cancer-associated fibroblasts confers cisplatin resistance in head and neck cancer through targeting CDKN1B and ING5. Genome Biol. (2019) 20:12. doi: 10.1186/s13059-018-1604-0
151. Dawson MA Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. (2012) 150:12–27. doi: 10.1016/j.cell.2012.06.013
152. Polyak K Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. (2009) 9:265–73. doi: 10.1038/nrc2620
153. Aiello NM, Maddipati R, Norgard RJ, Balli D, Li J, Yuan S, et al. EMT subtype influences epithelial plasticity and mode of cell migration. Dev Cell. (2018) 45:681–95.e4. doi: 10.1016/j.devcel.2018.05.027
154. Pearson G W. Control of invasion by epithelial-to-mesenchymal transition programs during metastasis. J Clin Med. (2019) 8:646. doi: 10.3390/jcm8050646
155. Wang J, Zhang Z, Li R, Mao F, Sun W, Chen J, et al. ADAM12 induces EMT and promotes cell migration, invasion and proliferation in pituitary adenomas via EGFR/ERK signaling pathway. Biomed Pharmacother. (2018) 97:1066–77. doi: 10.1016/j.biopha.2017.11.034
156. Chen L, Li Y, Wu L, Yu G, Zhang W, Huang C, et al. TRAF6 regulates tumour metastasis through EMT and CSC phenotypes in head and neck squamous cell carcinoma. J Cell Mol Med. (2018) 22:1337–49. doi: 10.1111/jcmm.13439
157. Shibue T Weinberg RA. EMT, CSCs, drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol. (2017) 14:611–29. doi: 10.1038/nrclinonc.2017.44
158. Xu M, Wang S, Wang Y, Wu H, Frank JA, Zhang Z, et al. Role of p38γ MAPK in regulation of EMT and cancer stem cells. Biochim Biophys Acta Mol Basis Dis. (2018) 1864:3605–17. doi: 10.1016/j.bbadis.2018.08.024
159. Eyler CE Rich JN. Survival of the fittest: cancer stem cells in therapeutic resistance and angiogenesis. J Clin Oncol. (2008) 26:2839–45. doi: 10.1200/JCO.2007.15.1829
160. Heng WS, Gosens R, Kruyt FAE. Lung cancer stem cells: origin, features, maintenance mechanisms and therapeutic targeting. Biochem Pharmacol. (2019) 160:121–33. doi: 10.1016/j.bcp.2018.12.010
161. Steinbichler TB, Dudás J, Skvortsov S, Ganswindt U, Riechelmann H, Skvortsova I-I. Therapy resistance mediated by cancer stem cells. Semin Cancer Biol. (2018) 53:156–67. doi: 10.1016/j.semcancer.2018.11.006
162. Vander Linden C, Corbet C. Therapeutic targeting of cancer stem cells: integrating and exploiting the acidic niche. Front Oncol. (2019) 9:159. doi: 10.3389/fonc.2019.00159
163. Bekaii-Saab T El-Rayes B. Identifying and targeting cancer stem cells in the treatment of gastric cancer. Cancer. (2017) 123:1303–12. doi: 10.1002/cncr.30538
164. Ghasemi F, Sarabi PZ, Athari SS, Esmaeilzadeh A. Therapeutics strategies against cancer stem cell in breast cancer. Int J Biochem Cell Biol. (2019) 109:76–81. doi: 10.1016/j.biocel.2019.01.015
165. Rosa R, D'Amato V, De Placido S, Bianco R. Approaches for targeting cancer stem cells drug resistance. Exp Opin Drug Discov. (2016) 11:1201–12. doi: 10.1080/17460441.2016.1243525
166. Talukdar S, Emdad L, Das SK, Sarkar D, Fisher PB. Evolving strategies for therapeutically targeting cancer stem cells. Adv Cancer Res. (2016) 131:159–91. doi: 10.1016/bs.acr.2016.04.003
167. Teeuwssen M, Fodde R. Wnt signaling in ovarian cancer stemness, EMT, and therapy resistance. J Clin Med. (2019) 8:1658. doi: 10.3390/jcm8101658
168. Fu X, Liu M, Qu S, Ma J, Zhang Y, Shi T, et al. Exosomal microRNA-32-5p induces multidrug resistance in hepatocellular carcinoma via the PI3K/Akt pathway. J Exp Clin Cancer Res. (2018) 37:52–52. doi: 10.1186/s13046-018-0677-7
169. Bayraktar R, Van Roosbroeck K. miR-155 in cancer drug resistance and as target for miRNA-based therapeutics. Cancer Metastasis Rev. (2018) 37:33–44. doi: 10.1007/s10555-017-9724-7
170. Santos JC, Lima N d S, Sarian LO, Matheu A, Ribeiro ML, Derchain SFM. Exosome-mediated breast cancer chemoresistance via miR-155 transfer. Sci Rep. (2018) 8:829–9. doi: 10.1038/s41598-018-19339-5
171. Yang Z, Zhao N, Cui J, Wu H, Xiong J, Peng T. Exosomes derived from cancer stem cells of gemcitabine-resistant pancreatic cancer cells enhance drug resistance by delivering miR-210. Cell Oncol. (2019) 43:123–36. doi: 10.1007/s13402-019-00476-6
172. Ma Y, Yuwen D, Chen J, Zheng B, Gao J, Fan M, et al. Exosomal transfer of cisplatin-induced miR-425-3p confers cisplatin resistance in NSCLC through activating autophagy. Int J Nanomed. (2019) 14:8121–32. doi: 10.2147/IJN.S221383
173. Bliss SA, Sinha G, Sandiford OA, Williams LM, Engelberth DJ, Guiro K, et al. Mesenchymal stem cell-derived exosomes stimulate cycling quiescence and early breast cancer dormancy in bone marrow. Cancer Res. (2016) 76:5832–44. doi: 10.1158/0008-5472.CAN-16-1092
174. Xu Y, Zhu M. Novel exosomal miR-46146 transfer oxaliplatin chemoresistance in colorectal cancer. Clin Transl Oncol. (2019). doi: 10.1007/s12094-019-02237-1. [Epub ahead of print].
175. Liu X, Lu Y, Xu Y, Hou S, Huang J, Wang B, et al. Exosomal transfer of miR-501 confers doxorubicin resistance and tumorigenesis via targeting of BLID in gastric cancer. Cancer Lett. (2019) 459:122–34. doi: 10.1016/j.canlet.2019.05.035
176. Wang M, Qiu R, Yu S, Xu X, Li G, Gu R, et al. Paclitaxel-resistant gastric cancer MGC-803 cells promote epithelial-to-mesenchymal transition and chemoresistance in paclitaxel-sensitive cells via exosomal delivery of miR-155-5p. Int J Oncol. (2019) 54:326–38. doi: 10.3892/ijo.2018.4601
177. Min Q, Wang X, Zhang J, Chen Q, Li S, Liu X, et al. Exosomes derived from imatinib-resistant chronic myeloid leukemia cells mediate a horizontal transfer of drug-resistant trait by delivering miR-365. Exp Cell Res. (2018) 362:386–93. doi: 10.1016/j.yexcr.2017.12.001
Keywords: exosome, miRNA, cancer, drug resistance, mechanism
Citation: Guo Q-r, Wang H, Yan Y-d, Liu Y, Su C-y, Chen H-b, Yan Y-y, Adhikari R, Wu Q and Zhang J-y (2020) The Role of Exosomal microRNA in Cancer Drug Resistance. Front. Oncol. 10:472. doi: 10.3389/fonc.2020.00472
Received: 14 January 2020; Accepted: 16 March 2020;
Published: 07 April 2020.
Edited by:
Dong-Hua Yang, St. John's University, United StatesReviewed by:
Xiumei Huang, Indiana University Bloomington, United StatesCopyright © 2020 Guo, Wang, Yan, Liu, Su, Chen, Yan, Adhikari, Wu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiang Wu, d3VxaWFuZzAwMTAwMUBhbGl5dW4uY29t; Jian-ye Zhang, amlhbnllekAxNjMuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.