
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 19 November 2019
Sec. Cancer Molecular Targets and Therapeutics
Volume 9 - 2019 | https://doi.org/10.3389/fonc.2019.01251
 Jun Ye1†
Jun Ye1† Jiacai Lei2†
Jiacai Lei2† Qingqing Fang3
Qingqing Fang3 Yimin Shen4
Yimin Shen4 Wenjie Xia5
Wenjie Xia5 Xiaoge Hu3
Xiaoge Hu3 Qiuran Xu3
Qiuran Xu3 Hongjun Yuan5
Hongjun Yuan5 Jian Huang1,6*
Jian Huang1,6* Chao Ni1,3,6*†
Chao Ni1,3,6*†Quiescent caner stem cells are identified as a subpopulation of colon cancer cells in dormant state and possess strong stem-cell like characteristics. Previously, we have identified this subpopulation in colorectal cancer (quiescent colon cancer stem cells, QCCSCs), and find QCCSCs are sensitive to the apoptotic effect of IFN-γ, which is attributed to their high IFN-γR expression levels. Microarray and bioinformatic analysis indicate miR-4666-3p is low expressed in QCCSCs and target IFN-γR1/2, which is proved by luciferase assay and western-blot. Furthermore, we find miR-4666-3p could also target TGF-βR1 to block the activation of TGF-β1/Smad pathway, therefore function as a tumor suppressor gene to inhibit the stemness of colon cancer cells. Besides, compared with QCCSCs, we find the TGF-β1 expression also decreased with the weakening of stemness properties. In terms of mechanism, our result reveal TGF-β1 is the target gene of miR-329, which is also high expressed in non-QCCSCs. Thereafter, we perform gain- and loss- function experiments to confirm the synergistic effect between miR-4666-3p and miR-329 in blocking the activation of TGF-β/Smad pathway. Finally, we evaluate the expression of both miR-4666-3p and miR-329 in 73 tumor specimens and paired normal tissue, and find both two miRNAs are related to unfavorable prognosis and advanced tumor stage in colorectal cancer. Our study revealed a novel epigenetic regulation mechanism in colon cancer stem cells, which could be exploited as a novel therapeutic strategy for cancer treatment.
The existence of quiescent cancer stem cells (QCSCs) has been reported in various solid tumors and QCSCs are defined as a distinct subset of cells in quiescent status or undergoing very slow proliferation that possess a strong capacity for tumorigenesis and a resistance to cytotoxic drugs (1). Previously, we identified a population of quiescent colon cancer cells with strong stemness by PKH staining and a serum-free medium cultivation assay (quiescent colon cancer stem cells, QCCSCs) and reported that QCCSCs are resistant to chemotherapy but sensitive to the apoptotic effect of IFN-γ, which is caused by their high expression of IFN-γ receptors (IFNGRs) (2). Herein, we want to illuminate the mechanisms underlying the different expression patterns of QCCSCs and non-QCCSCs.
Cancer is composed of many genetically distinct subclones that are produced by both intrinsic and therapeutic effects. However, several studies have found that differences in biological behaviors among cancer cells cannot be ascribed to genetic mutations alone. A more in-depth understanding of CSC (cancer stem cell) dynamics has revealed that cells in many tumors exhibit phenotypic plasticity, suggesting a transition between the CSC and non-CSC states. Iliopoulos et al. reported a mutual transition between the CD44high CSC phenotype and the CD44low non-CSC phenotype (3). Roesch et al. described the presence of a population of slow-cycling stem-like melanoma cells, which were characterized by high expression of JARID1B (Jumonji/AT-rich interactive domain-containing protein 1B), but the non-stem-like cells with low expression level of JARID1B could turn on high levels of the protein through epigenetic modulation (4). Based on the observation that some isolated populations could generate other populations to restore equilibrium, the transition from CSCs to non-CSCs must involve epigenetic changes.
Increasing evidence has demonstrated that microRNAs play important roles in maintaining the characteristics of CSCs, such as self-renewal, invasiveness and chemoresistance, and miRNAs could also be used as therapeutic targets or prognostic markers. A previous review indicated that approximately 164 miRNAs were altered in colon cancer. Some play pro-tumorigenic roles, such as miRNA-17, which induces epithelial-mesenchymal transition and promotes the formation of a stem cell-like population in colon cancer cells (5), and some act as tumor suppressive genes, such as miRNA-93, which suppresses the proliferation and tumorigenesis of human colon cancer stem cells (6). Besides, these microRNAs are revealed as important regulators of some key signaling pathway in maintaining stemness of CSCs. Among them, TGF-β/Smad pathway is acknowledged holding a key position in cancer stem cells, and reported there is a dual directional regulation mechanism with microRNAs: TGF-β could induce epithelial-to-mesenchymal transition (EMT) and stemness in pancreatic ductal adenocarcinoma via induces miR-100 and miR-125b but blocks let-7a (7); meanwhile, TGF-β pathway could also be regulated by microRNAs: TrkC-miR2 and miR-140 inhibit TGF-β pathway via direct targeting smad3 (8, 9).
Here, we report that miR-4666-3p and miR-329 are both expressed at low levels in QCCSCs, and both of their expression levels increased with cell division. Furthermore, we found that these two miRNAs work in concert to suppress tumorigenesis and stemness via the TGF-β/Smad pathway, which has been shown to play vital roles in stemness maintenance in colon CSCs (CCSCs). Understanding the mechanisms of how microRNAs regulate the stemness of colon cancer stem cells establishes a strong rationale for their application as prognostic markers and therapeutic markers.
P1 cancer cells were established from liver metastasis lesions of colon cancer (10) and were cultured in RPMI 1640 (Invitrogen, Grand Island, NY) medium containing 10% FBS. The colon cancer cell line SW480 was maintained in Leibovitz L-15 (Invitrogen, Grand Island, NY) medium with 10% FBS; 293T cells were maintained in DMEM (Invitrogen, Grand Island, NY), supplement with NaHCO3 1.5 g/L, medium with 10% FBS. The SW480 and 293T cell line was obtained from the Cell Bank of Chinese Academy of Science, Shanghai. Tumor samples were collected from patients undergoing surgery at the Department of Zhejiang Provincial People's Hospital.
This study was approved by the ethics committees of the Zhejiang Provincial People's Hospital (KY2014-AF-09). Informed consent was obtained from all of the enrolled subjects, and the study was performed in full compliance with all principles of the Helsinki Declaration.
For isolating PKHhi cells, colon cancer cells were suspended in Diluent C (~1 × 107 cells/ml) and then labeled with PKH26/67 (2 × 10−6 M, 5 min, Sigma-Aldrich, USA). The staining efficiency was confirmed with FCM (flow cytometry). Thereafter, 1,000 cells/well were cultured in serum-free medium with F-12, basic fibroblast growth factor, N2 and heparin, and the cells were cultured for 2 weeks in ultralow attachment six-well plates. PKHhi subpopulation cells were defined as previously reported and isolated by FACS (Fluorescence Activated Cell Sorting) (2).
In the series sphere formation assay, single cells were obtained from spheroids digested with Accutase (Invitrogen, Grand Island, NY) (37°C, 5 min) and filtered through a 40 μm strainer and then seeded into ultralow attachment plates. The ratio of spheroid formation was calculated according to the primary seed cell numbers. The cells were seeded into ultralow attachment plates at a density of 1,000 cells per well.
Antibodies against IFN-γR1 (BB1E2, 1:1000), IFN-γR2 (GTX81601, 1:1000), TGF-βR1 (E161, 1:1000), TGF-β1 (7F6, 1:1000), Smad2/3 (GTX111123, 1:1000), p-smad2 (GTX133614, 1:1000), and p-smad3 (GT1017, 1:1000) were obtained from GeneTex (Irvine, USA).
Flow cytometry was performed according to the manufacturer's protocols. Briefly, to detect the PKH26 fluorescence intensity and TGF-β1 expression (7F6, 1:400, GeneTex, Irvine, USA) in PKHhi/low/neg cells, spheroids were isolated into single cell suspension with Accumax (Invitrogen, San Diego, USA), then permeabilize cells with Intracellular Staining Permeabilization Wash Buffer (Biolegend, USA) and stained with antibody.
Western blot analysis was performed according to the previously reported (11). Briefly, cells were lysed in a RIPA buffer (Cell Signaling Technology, Danvers, MA, USA) and centrifuged. Protein products were separated by 10% SDS-PAGE electrophoresis and transferred to nitrocellulose membranes (Millipore, Billerica, MA, USA). Membranes were blocked with 1.5% BSA in Tris-buffered saline buffer with 0.1% Tween-20 (TBST) for 2 h and incubated with primary antibodies overnight at 4°C. Then membranes were incubated with the corresponding horseradish peroxidase (HRP)-conjugated secondary antibodies for 2 h at room temperature. Detection was performed using the electrochemiluminescence (ECL) kit (Pierce, Rockford, IL, USA).
Total RNA from colon rectal cancer (CRC) cells was obtained with Trizol reagent (Invitrogen), and total RNA from paraffin-embedded tissues was isolated by the RecoverAll™ Total Nucleic Acid Isolation Kit (Applied Biosystems). Taqman assays (KLF4, OCT4, NANOG, SOX2, miR-4666-3p, and miR-329) were purchased from Applied Biosystems (Life Technologies Co.), performed with the TaqMan Universal PCR Master Mix and analyzed with an ABI Step One Plus System (Applied Biosystems). Because of the extremely low amount of total RNA extracted, especially from the PKHhi cells, we used the Taqman MicroRNA Cells to CT™ Kit (Life Technologies Co.). All samples were run in triplicate, and microRNA expression levels in each sample were normalized to U6 levels.
Total RNA was isolated from QCCSCs (PKHhi cells) and non-QCCSCs (PKHlow and PKHneg cells) with Trizol reagent. The quantity of RNA was verified with a Nanodrop spectrophotometer (Thermo Fisher Scientific, Worcester, MA, USA). The miRNA expression profile of each sample was assessed with a miRNA microarray produced by JOIN GENOME (Hangzhou, China).
The pmirGLO Dual-Luciferase miRNA Target Expression vectors (Promega) contained the TGFBR1 mRNA 3′UTR with the miR-4666-3p target site or a mutated miR-4666-3p target site and the TGFβ1 mRNA 3′UTR with the miR-329 target site or a mutated miR-329 target site. The pRL-TK plasmids (Promega) were cotransfected into 293T cells with the negative control mimic (5′-UUCUCCGAACGUGUCACGUTT-3′), miR-4666-3p mimic (5′- CAUACAAUCUGACAUGUAUUU-3′), anti-miR-4666-3p mimic (5′- AAAUACAUGUCAGAUUGUAUG-3′), miR-329 mimic (5′- AACACACCUGGUUAACCUCUUU-3′), or anti-miR-329 mimic (5′-AAAGAGGUUAACCAGGUGUGUGUU-3′) (GenePharma Tech, Shanghai, China) using Lipofectamine 2000 (Invitrogen). The ratio of firefly to Renilla luciferase activity was determined with a luminometer using the Dual Luciferase Reporter Assay System (Promega) 48 h after transfection.
Cells were fixed with 4% paraformaldehyde for 20 min, blocked in 5% normal goat serum and incubated with a primary antibody against TGFBR1 (1:100, E161, Genetex, Irvine, USA), Smad2 (1:500, ab40855, Abcam, USA), or Smad3 (1:500, ab40854, Abcam, USA) (4°C, overnight), followed by Dylight488-conjugated goat anti-rabbit antibody (1:100, ab96983, Abcam, USA) incubation (37°C, 1 h). After immunolabeling, the cells were washed with PBS, stained with DAPI (Sigma, St. Louis, MO, USA) and then viewed under a laser confocal microscope (LCM, Leica SP-5, Germany).
Six-week-old nude mice were purchased from Shanghai Institutes for Biological Sciences and bred in specific pathogen-free conditions at the Laboratory Animal Research Center of Zhejiang Medical Academy with the permission of the local animal care and ethics committee.
The cells were suspended in 100 μl medium composed of DMEM F12 and Matrigel (BD Pharmingen, San Diego, CA, USA) and then injected subcutaneously into the dorsal side of nude mice to assess their ability to generate tumor xenografts. The volume of the tumors was measured weekly and calculated as length × width × height/2.
Other related methods, including IC50 determination, ELISA (enzyme-linked immunosorbent assays), plasmid construction, and the establishment of miR-4666-3p and miR-329 stable knockdown and overexpression cell lines are described in Supplementary Material.
All data are shown as one typical result from 3 independent experiments in similar conditions or as the mean ± SD of 3 independent experiments. Prism 7 software was used to generate graphs and to perform statistical analysis. The RT-PCR results from clinical samples (Figures 6A,B) were analyzed with a two-tailed paired Student's t-test, and the other results were analyzed with a two-tailed unpaired Student's t-test. P < 0.05 indicated statistical significance.
In our previous work, we found that a small subset of slow-cycling cells (PKHhi cells, Figure 1A, Figure S1) exhibited strong cancer stem cell-like features, including high stemness gene expression, great ability of spheroid and xenografts formation and resistance to chemotherapy, therefore, we considered PKHhi cells to be colon cancer stem cells (CCSCs). Meanwhile, we found PKHhi cells were sensitive to IFN-γ-induced apoptosis, which was attributed to their higher IFN-γR1/2 expression level (2). To explore whether microRNAs regulate the IFN-γ receptor levels in PKHhi cells, we performed a microarray analysis to compare the expression patterns of IFNGRs between PKHhi and the remaining cells. The results revealed four upregulated and eight downregulated microRNAs in the PKHhi population of P1 cells and four upregulated and fourteen downregulated microRNAs in the PKHhi population of SW480 cells (Fold change ≥ 2, Figure 1B). Through the comparison, we found that miR-4666-3p was downregulated in the PKHhi population of both cell lines.
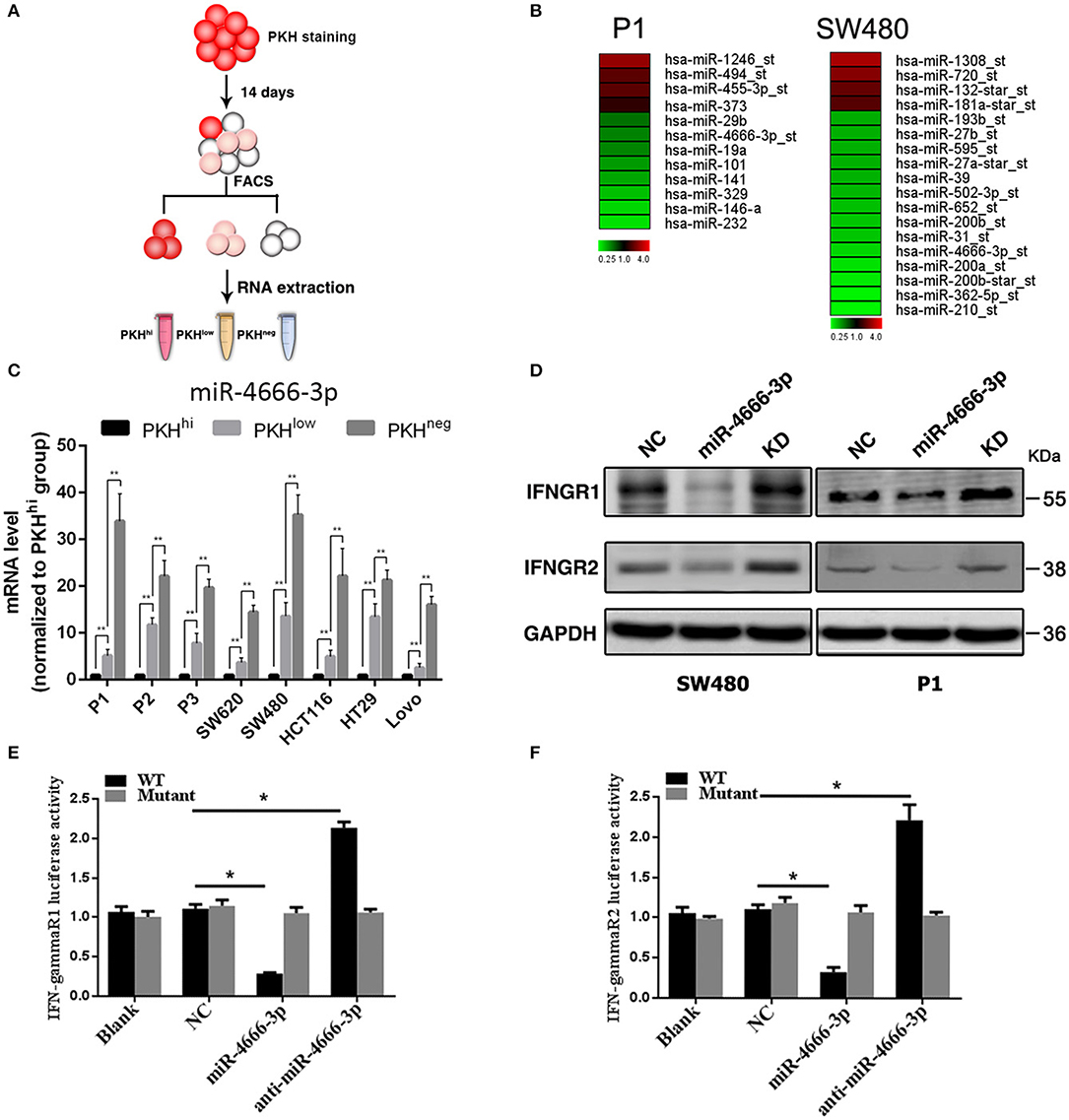
Figure 1. Low expression of miR-4666-3p in the PKHhi subpopulation and miR-4666-3p's effect on its target IFN-γR1/2. (A) Schematic model of sorting PKHhi/low/neg subpopulations; (B) Differentially expressed miRNA in PKHhi and the rest of the cell population. Red denotes high and green denotes low levels of expression; (C) The expression level of miR-4666-3p in different subpopulations of different colon cancer cell lines; (D) CRC cells were transfected with the NC, miR-4666-3p or anti-miR-4666-3p mimics, and the expression of IFN-γR1 and IFN-γR2 was detected by Western blotting, KD, knockdown; NC, negative control; (E,F) 293T cells were co-transfected with empty pmirGLO Dual-Luciferase reporter plasmids or IFN-γR1 and IFN-γR2 3'UTR firefly luciferase reporter plasmids, pRL-TK-luciferase plasmids, and the miR-4666-3p or anti-miR-4666-3p mimics. After 48 h, firefly luciferase activity was measured and normalized to that of Renilla luciferase. The data are shown as one typical result from three independent experiments with similar results or as the mean ± SD of three independent experiments. p < 0.05; p < 0.01. WT, wild type. *P < 0.05.
The target gene prediction by the Targetscan database (www.targetscan.org) showed that the IFN-γ receptor may be regulated by miR-4666-3p. RT-PCR confirmed that miR-4666-3p was expressed at low levels in PKHhi cells from various colon cancer cell lines, and it is interesting that the expression level gradually increased as the fluorescence intensity decreased (Figure 1C). We further identified both IFN-γR1 and IFN-γR2 as functional downstream targets of miR-4666-3p in SW480 and P1 cells. The protein levels of the IFNGRs were also influenced by transfection of a miR-4666-3p mimic or an anti-miR-4666-3p mimic (Figure 1D). In a dual luciferase reporter assay, we found that the activity of luciferase reporters containing the IFN-γR1 and IFN-γR2 3'UTRs decreased by 70 and 60%, respectively, upon co-transfected with the miR-4666-3p mimic, but both almost doubled upon co-transfected with the anti-miR-4666-3p mimic. Moreover, no change was identified upon co-transfected of the mutant reporter plasmid with either the miR-4666-3p or anti-miR-4666-3p mimic (Figures 1E,F). Finally, we assessed whether miR-4666-3p could regulate the IC50 of IFN-γ and oxaliplatin in colon cancer cell lines. The MTS assay showed that the miR-4666-3p mimic strongly increased the IC50 of IFN-γ in P1 and SW480 cells, while the anti-miR-4666-3p mimic slightly decreased the cell lines' sensitivities to IFN-γ (Figure S2A). In addition, we found that the IC50 of oxaliplatin was significantly increased by transfection of the anti-miR-4666-3p mimic and significantly decreased by transfection of the miR-4666-3p mimic (Figure S2B), which indicated that miR-4666-3p may function as a tumor suppressive gene in colon cancer cells.
Because the above data demonstrated that miR-4666-3p was expressed at low levels in PKHhi cells and acted as a tumor suppressor gene in CRC, we established both miR-4666-3p- and anti-miR-4666-3p-expressing stable cell lines to study miR-4666-3p's biological function by determining sphere formation in vitro and tumorigenesis in vivo. We first performed a series sphere formation assay in serum-free medium, and the results revealed that spheroid formation was significantly and inversely related to miR-4666-3p expression in both SW480 and P1 cells (Figures 2A,B). Thereafter, we determined if the expression of four stemness-related genes, OCT4, Nanog, BMI1, and SOX2, were affected by miR-4666-3p levels. Although not all the genes had their expression substantially changed by the miR-4666-3p level, the results showed that the expression of almost all of these genes was dramatically decreased in miR-4666-3p-overexpressing cells and increased in the anti-miR-4666-3p stable cell lines (Figure 2C). Thereafter, we evaluated if miR-4666-3p exert stemness suppressive function via targeting IFN-γ receptors. However, neither change the expression level of IFN-γR1 or IFN-γR2 could affect the spheroid formation ability or stemness gene expression in vitro (Figures S3A, B).
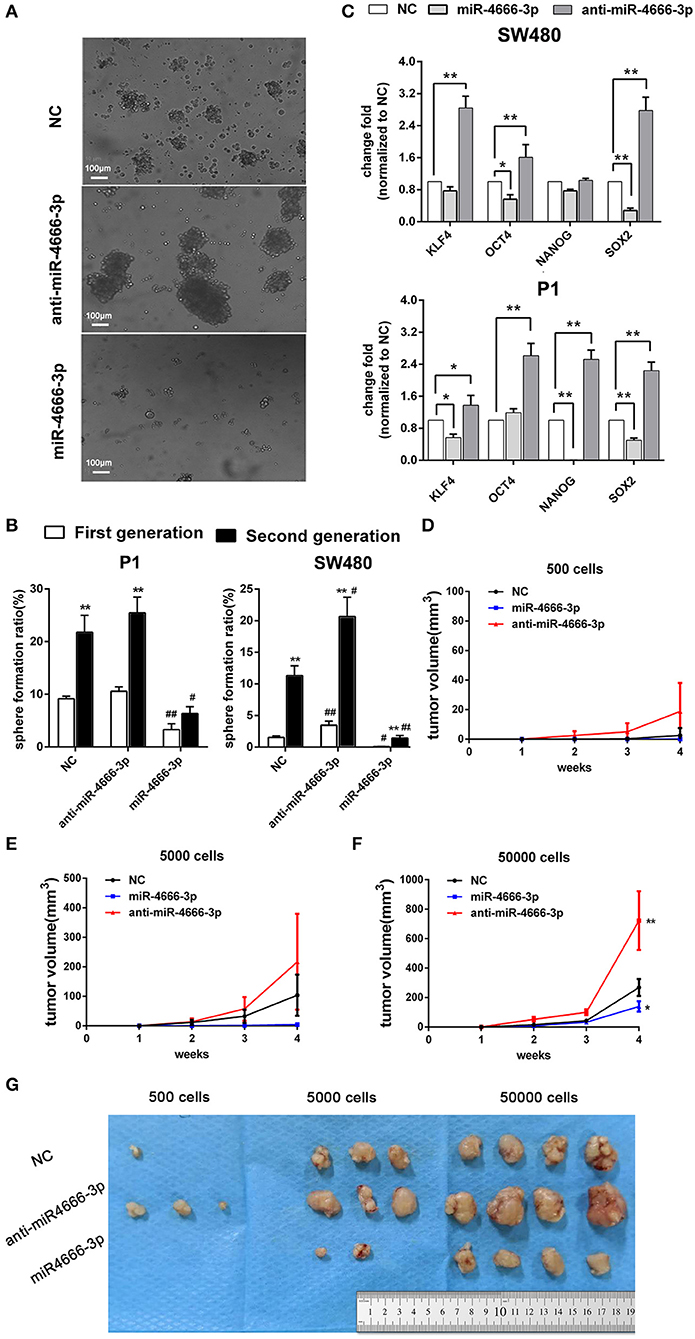
Figure 2. High expression of miR-4666-3p suppresses the stemness of CRC cells. (A) Representative images of sphere formation with control, miR-4666-3p mimic and anti-miR-4666-3p mimic transfected P1 cells; (B) Serial sphere formation with control, miR-4666-3p mimic and anti-miR-4666-3p mimic transfected CRC cells (* vs. first generation; # vs. NC group, scale bar = 100 μm); (C) The expression of stemness genes was assessed by qPCR in control, miR-4666-3p mimic and anti-miR-4666-3p mimic transfected CRC cells; (D–F) Comparison of in vivo tumorigenesis with the indicated numbers of cells (P1 cells); (G) Results from a tumorigenesis assay showing representative images of xenograft tumors in nude mice injected subcutaneously with the indicated numbers of cells (P1 cells). The data are shown as one typical result from three independent experiments with similar results or as the mean ± SD of 3 independent experiments. *,#P < 0.05, **,##P < 0.01.
In the tumorigenicity assay, the rate of tumor growth was also markedly reduced by miR-4666-3p overexpression in the 500, 5,000, or 50,000 cell groups (Figures 2D–F); besides, P1 cells with low miR-4666-3p expression revealed a much stronger tumor-initiating capacity than P1 cells overexpressing miR-4666-3p; only 500 low-expression P1 cells were needed to form xenografts in nude mice (3/4), while the threshold for xenograft formation in the control (3/4) and miR-4666-3p overexpression groups was ~5,000 cells (2/4) (Figure 2G). These data strongly suggested that miR-4666-3p suppresses tumor-initiating capacity and stemness in colon cancer cells.
Because miRNAs usually act as mediators that silence genes, we performed a bioinformatic analysis of the target genes of miR-4666-3p and attempted to elucidate the underlying mechanism of how miR-4666-3p suppresses tumor growth and stemness. First, we analyzed target genes of miR-4666-3p via TargetScan (www.targetscan.org), picTar (https://pictar.mdc-berlin.de/) and miRDB (www.mirdb.org), and 204 genes were found to be predicted by all three databases. Next, we functionally annotated these genes with GeneCodis and then subjected them to further enrichment analysis of cell signaling pathways using the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database (www.genome.jp/kegg/). Using this approach (Figure 3A), miR-4666-3p was predicted to target various cancer-related signaling pathways, including the TGF-β, mitogen-activated protein kinase (MAPK) and insulin signaling pathways, which comprised 33 target genes. The KEGG result and network diagram between these genes and miR-4666-3p are presented in Figure S4. Finally, we focused on TGF-βR1 because of its crucial function in the TGF-β pathway, which is one of the most common canonical signaling pathways to promote stemness and EMT in CSCs (12–14).
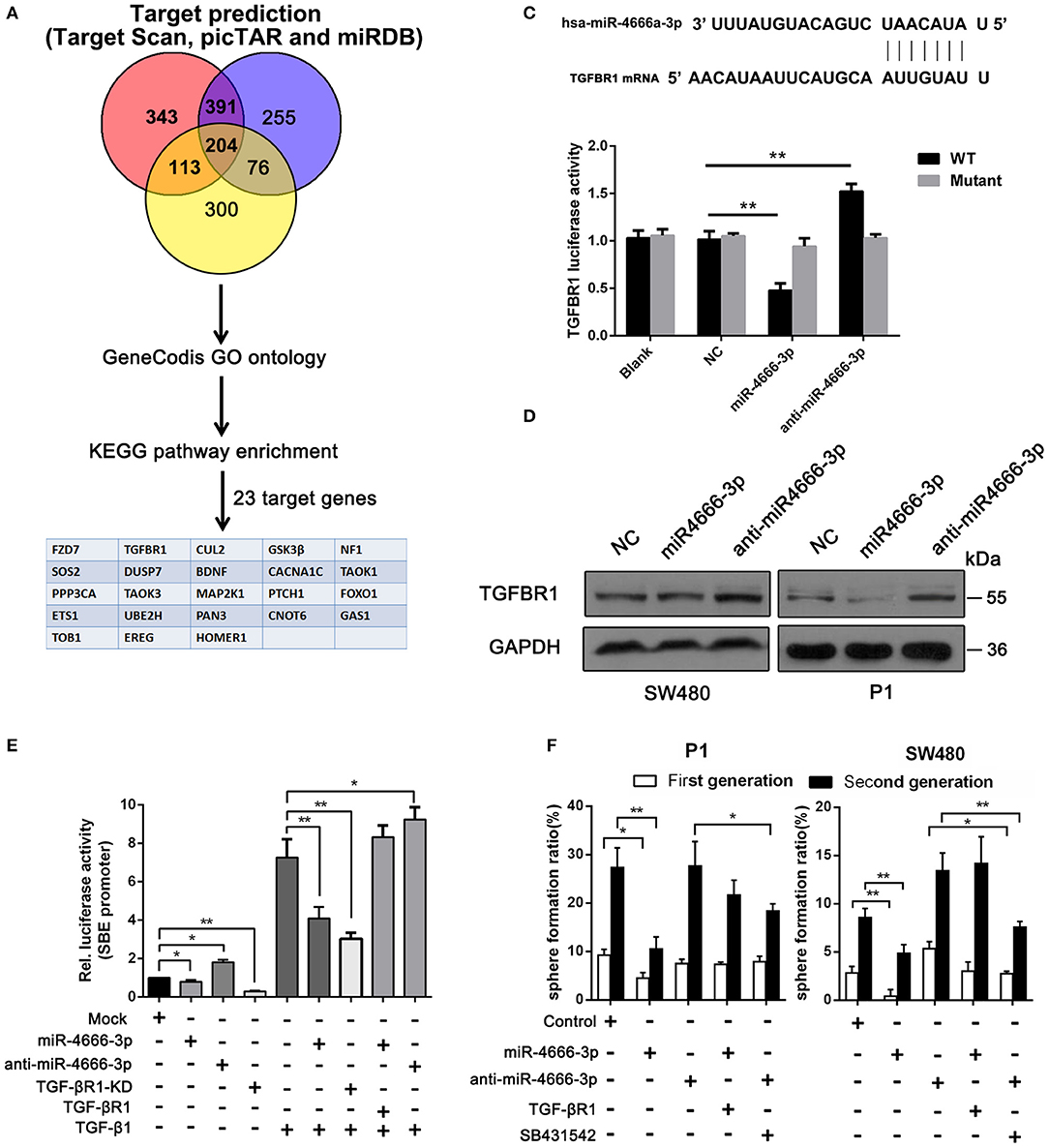
Figure 3. miR-4666-3p targets TGFBR1 in CRC cells. (A) Bioinformatics analysis of potential targets of miR-4666-3p; (B) TGFBR1 is predicted to be a novel target of miR-4666-3p; (C) 293T cells were cotransfected with empty pmirGLO Dual-Luciferase reporter plasmids or TGFBR1 3′UTR firefly luciferase reporter plasmids and pRL-TK-luciferase plasmids, together with the miR-329 or anti-miR-4666-3p mimic. After 48 h, firefly luciferase activity was measured and normalized to that of Renilla luciferase; (D) CRC cells were transfected with the NC, miR-4666-3p or anti-miR-4666-3p mimics, and expression of TGFBR1 was detected by Western blotting; (E) The activity of the TGF-β/Smad pathway was measured by the SBE luciferase reporter system in colon cancer cells (presented as P1 cells) with distinct treatment combination; (F) Serial sphere formation with different cells (control, miR-4666-3p, anti-miR-4666-3p stable expression and both miR-4666-3p and TGF-βR1 stable expression cells). The data are shown as one typical result from three independent experiments with similar results or as the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01.
Next, we identified TGF-βR1 as a functional downstream target of miR-4666-3p. The 3‘UTR of TGF-βR1 contained highly conserved miR-4666-3p binding sites (Figure 3B), which was proven in a luciferase reporter assay. Our results showed that the activity of a luciferase reporter containing the TGF-βR1 3'UTR decreased by 60% upon co-transfected with the miR-4666-3p mimic but increased by 50% upon co-transfected with the anti-miR-4666-3p mimic, and a negative result was found when the mutant reporter plasmid was co-transfected with either the miR-4666-3p mimic or anti-miR-4666-3p mimic (Figure 3C). Western blotting also confirmed that TGF-βR1 protein expression changed with miR-4666-3p overexpression or downregulation (Figure 3D).
Afterwards, we evaluated whether TGF-βR1 affected the stemness of CCSCs. Using a series spheroid formation assay, we found that the knockdown of TGF-βR1 suppressed sphere formation and stem gene expression, while overexpression of TGF-βR1 potentiated sphere formation and stemness gene expression (Figures S5A, B). TGF-βR1 is a key upstream part of activating TGF-β pathway, herein we assessed whether miR-4666-3p could suppress canonical TGF-β/Smad pathway with luciferase reporter gene assays using pGL3-SBE4-Luc plasmids, which report binding to SBE. The result revealed inhibition of TGF-β pathway with TGF-βR1 knockdown or selective TGF-βRIinhibitor (SB-431542, 10 μM) markedly decrease the luciferase activity and sphere formation ability (Figures 3E,F), meanwhile, overexpression of miR-4666-3p significantly inhibit the activity of TGF-β/Smad pathway induced by supplement of TGF-β1 (Figure 3E). Then we constructed TGF-βR1 overexpressing plasmid (without its 3'UTR) and performed rescue experiment. The cells were co-transfected with miR-4666-3p and TGF-βR1 plasmid, in parallel with control group. The restore of TGF-βR1 expression was confirmed with qRT-PCR (Figure S5C) and western-blot (Figure S5D). The re-expression of TGF-βR1 markedly restore the sphere formation ability (Figure 3F) and activity of TGF-β/Smad pathway induced by TGF-β1 (Figure 3E).
The above result indicate TGF-β/Smads pathway was activated with miR-4666-3p downregulation in colon cancer cells, and it has been reported that TGF-β1 is highly expressed in and secreted by cancer stem cells (15, 16), so it is rational to explore if there is a positive loop regulation mechanism exist here; herein, we evaluated TGF-β1 expression in PKHhi, PKHlow, and PKHneg cells by flow cytometry. The results showed that the TGF-β1 expression was highest in PKHhi cells (22.1 ± 4.8%) and decreased as PKH label intensity declined (PKHlow 3.3 ± 1.9%; PKHneg 0.8 ± 0.5%, Figure 4A). However, although TGF-β1 protein was upregulated in PKHhi cells, its mRNA levels did not differ significantly among three subsets (Figure S6D). This disparity between protein and mRNA in TGF-β expression suggests that a post-transcriptional mechanism is involved in TGF-β1 regulation.
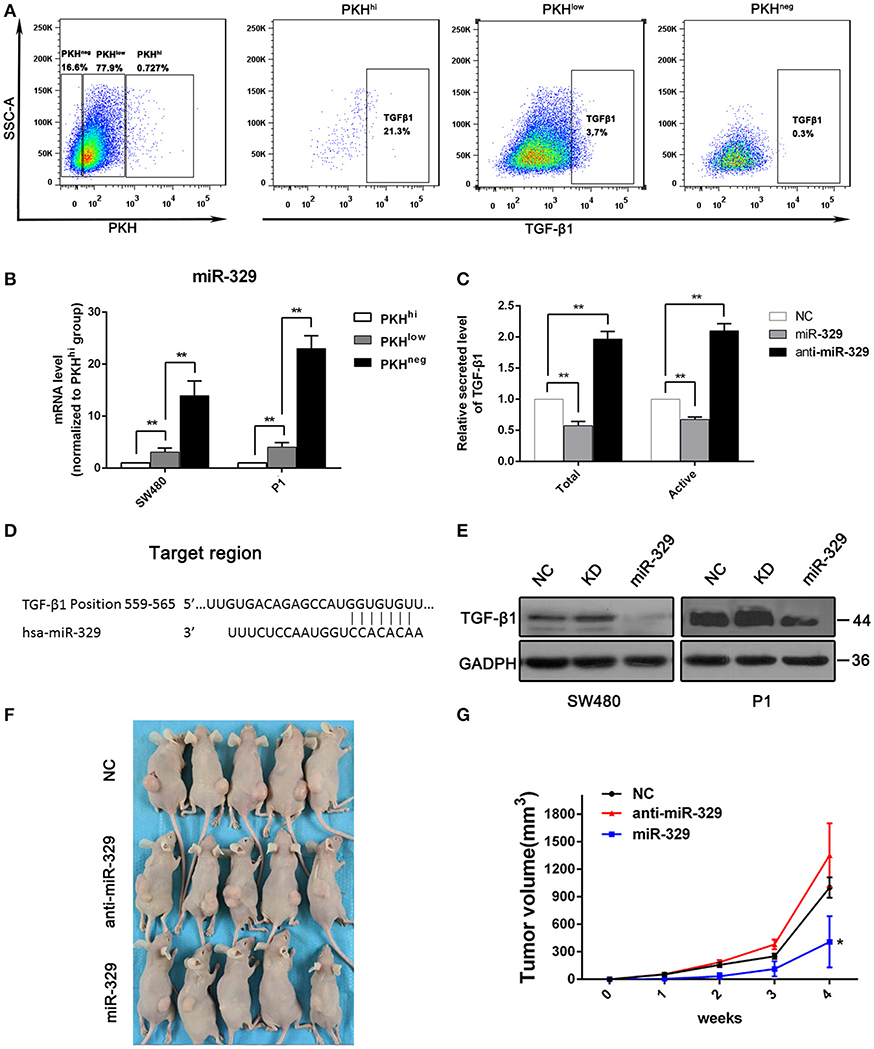
Figure 4. miR-329 is low expressed in PKHhi cells and targets TGFβ1. (A) Flow cytometry assessed the expression of TGF-β1 in PKHhi, PKHlow, and PKHneg subpopulations; (B) miR-329 expression in PKHhi, PKHlow, and PKHneg subpopulations assessed by qPCR; (C) colon cancer cells were transfected with the NC, miR-329 or anti-miR-329 mimic, and total and active TGF-β1 levels in culture medium were detected by ELISA, data was presented with P1 cells; (D) TGFβ1 is predicted to be a novel target of miR-329; (E) CRC cells were transfected with the NC, miR-329 or anti-miR-329 mimic and expression of TGFβ1 was detected by Western blotting; (F) Results from a tumorigenesis assay show representative images of xenograft tumors in nude mice injected subcutaneously with 1 × 104 P1 cells/mice; (G) Comparison of tumor growth in nude mice injected with 1 × 104 P1 cells/mice. The data are shown as one typical result from three independent experiments with similar results or as the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01.
miRNA-329 was reported to be a tumor suppressor gene that inhibits TGF-β1 (17), and it was interesting to find a relatively low expression level of miR-329 in the PKHhi population (Figure 1B). We then performed RT-PCR to verify the expression of miR-329 in different subsets of cells, and the results confirmed significant, high miR-329 levels in the PKHlow and PKHneg populations (Figure 4B), which indicated that the miR-329 level was negatively correlated with the stemness of CCSCs. To confirm that miR-329 regulated the production of TGF-β1, we performed ELISA with specific antibodies. The results showed that cells transfected with the miR-329 mimic increased the levels of both the total and active forms of TGF-β1 (Figure 4C). Afterwards, we determined that the 3‘UTR of TGF-β1 contained highly conserved miR-329 binding sites (Figure 4D), and the luciferase reporter assay found that the activity of a luciferase reporter containing the TGF-β1 3'UTR decreased by 60% upon co-transfection with the miR-329 mimic but increased by 100% upon co-transfection with the anti-miR-329 mimic (Figure S6A). Western blotting also confirmed that the expression of the TGF-β1 could be inhibited by miR-329 mimics (Figure 4E).
We also assessed the impact of miR-329 on the tumor initiation ability and stemness of colon cancer cells. We found that stable expression of the anti-miR-329 mimic potentiated spheroid formation, while overexpression of the miR-329 mimic inhibited sphere formation in both P1 and SW480 cells, which could be rescued by supplement with TGF-β1 (Figure S6B). In addition, in a xenograft tumor model, 1 × 104 cells from each group were injected subcutaneously, and the results revealed that the expression of miR-329 was negatively associated with tumor formation capacity (Figures 4F,G). All these data demonstrated miR-329 played as a tumor suppressive gene and negatively regulated TGF-β pathway in colon cancer stem cells.
Because the above evidence demonstrated that miR-4666-3p and miR-329 both acted as tumor suppressor genes, were expressed at low levels in PKHhi cells and were related to the TGF-β pathway, we tried to determine whether these two microRNAs acted synergistically to affect the stemness and TGF-β/Smad pathway. Since the above data already proved miR-4666-3p and miR-329 can individually suppress the sphere formation and the activity of TGF-β/smad pathway, the simultaneously introduction of both miR-4666-3p and miR-329 showed a cooperative repression function, while addition of 10 ng/ml TGF-β1 or transfected with TGF-βR1 overexpressing plasmid (without its 3′UTR) revealed a rescue effect (Figures 5A,B).
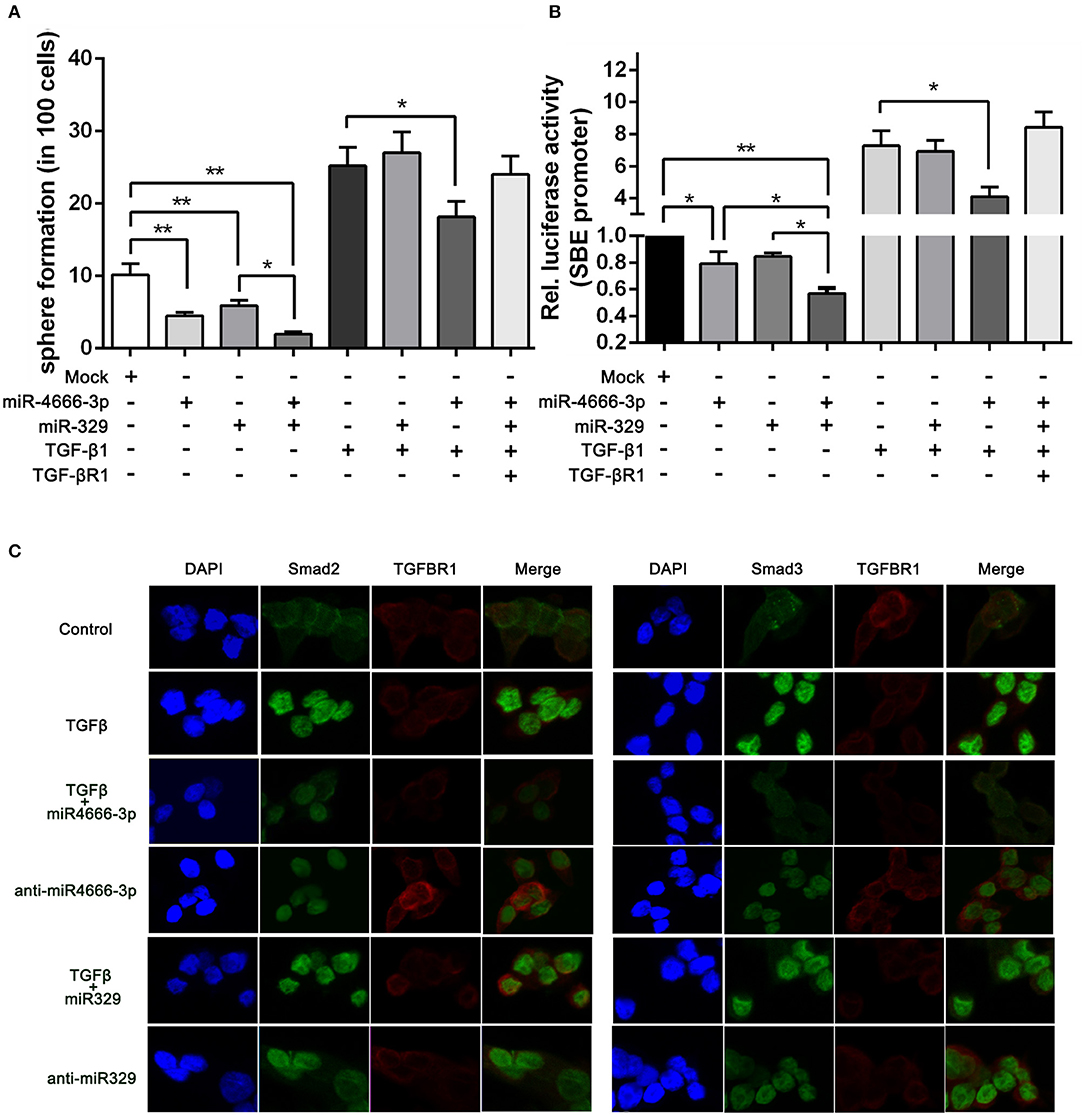
Figure 5. miR-4666-3p and miR-329 regulate the stemness of CCSCs via TGF-β/Smad pathway. (A) Sphere formation assay with distinct treatment combinations; (B) The activity of the TGF-β/Smad pathway was measured by the SBE luciferase reporter system (presented as P1 cells); (C) Immunofluorescence assay to assess the expression of TGFBR1 and nuclear expression of Smad2/3 in treated P1 cells. The data are shown as one typical result from three independent experiments with similar results or as the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01.
To further identify whether miR-329 and miR-4666-3p impeded activation of TGF-β/Smad pathway, we performed an immunofluorescence assay. The results revealed that TGF-β1 greatly potentiated the translocation of Smad2 and Smad3. This effect could be partially blocked by overexpression of miR-4666-3p, while the translocation of Smad2/3 was increased in the anti-miR-329 stable cell lines (Figure 5C). Besides, western-blot assay revealed anti-miR-4666-3p and anti-miR-329 increased Smad2/3 phosphorylation, and overexpression of miR-4666-3p could partially block the stimulation effect of TGF-β1 (10 ng/ml) on Smad2/3 phosphorylation (Figure S6C). The above results indicated that miR-329 and miR-4666-3p cooperated to regulate the stemness of CCSCs by inhibiting canonical TGF-β/Smad signaling.
To investigate the impact of miR-4666-3p and miR-329 expression on the clinical features of human CRC, we analyzed our data from 73 CRC patients (Table S1) and found that the expression level of both miR-4666-3p and miR-329 were much higher in normal tissue than in paired tumor specimens (p < 0.01, Figures 6A,B). Furthermore, miR-4666-3p and miR-329 expression was negatively correlated with the TNM (tumor, node, metastasis) stage (Figures 6C,D) and histological stage in CRC patients (Figures 6E,F). These findings suggested that miR-4666-3p and miR-329 was negatively associated with tumor invasiveness and progression, and that its expression level might be a prognostic factor in human CRC.
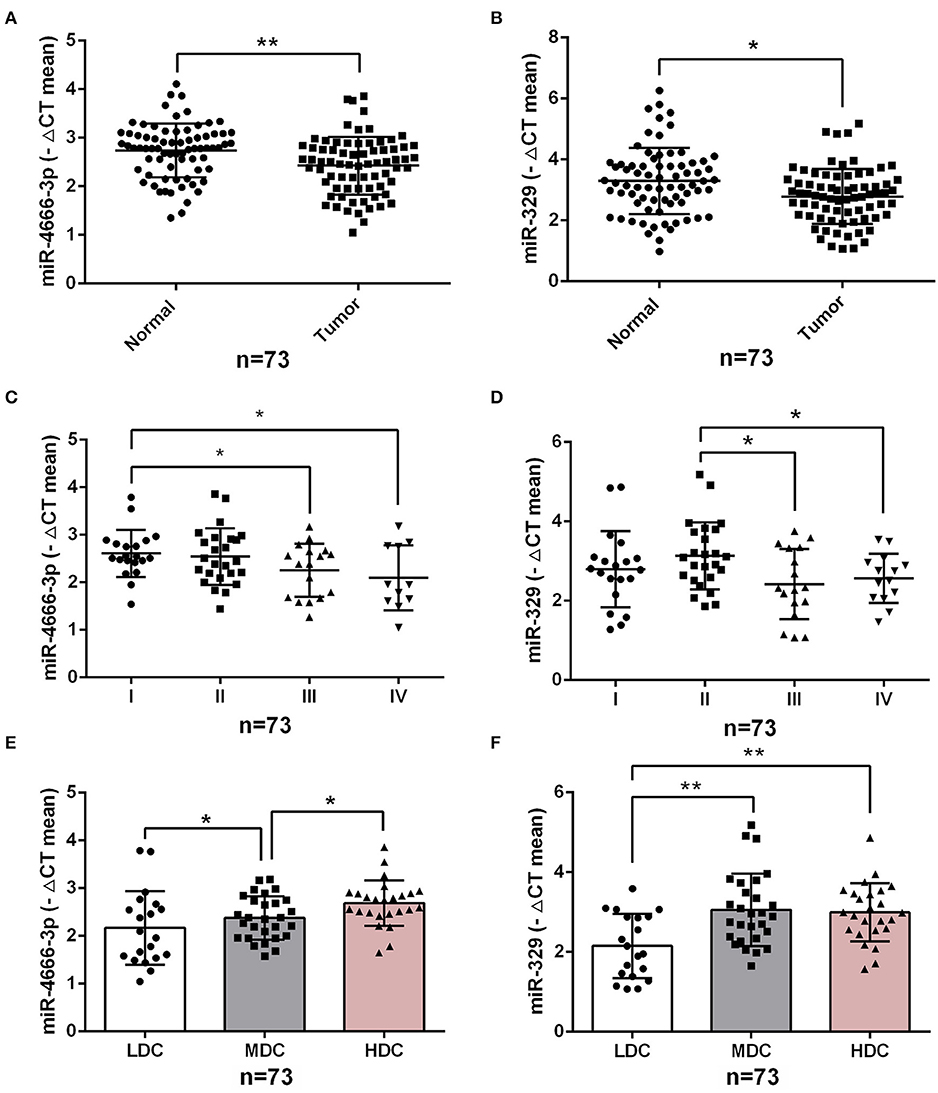
Figure 6. Low expression of miR-4666-3p and miR-329 are related to advanced clinicopathological features. (A,B) miR-4666-3p and miR-329 expression assessed by qRT-PCR in 73 pairs of paraffin-embedded CRC and adjacent normal tissue; (C,D) miR-4666-3p and miR-329 expression level stratified by different tumor stages; (E,F) miR-4666-3p and miR-329 expression level stratified by different tumor grades. LDC, low differentiated cancer; MDC, middle differentiated cancer; HDC, high differentiated cancer. All miR-4666-3p or miR-329 expression levels were presented as ΔCt value = Ct(U6)-Ct(miR-4666-3p or miR-329). Data are represented as the mean ± SD; *P < 0.05, **P < 0.01.
Here, we demonstrate that miR-4666-3p and miR-329 are expressed at low levels in colon cancer stem cells but are upregulated by cell division in non-CCSCs. Furthermore, we find that these two microRNAs work synergistically to inhibit the stemness of CCSCs by targeting the TGF-β/Smad pathway. It has been proven that microRNAs play a pivotal role in maintaining the stemness of CSCs, and some of them could be applied as therapeutic agents in cancer. Previously, we found that cholesterol-conjugated miR-27 mimics could target VEGF-C (Vascular endothelial growth factor C) in vivo to inhibit tumor progression and angiogenesis in colorectal cancer (18), and it has also been reported that miR-27 could target the Wnt pathway (19), which is important for stemness maintenance in CCSCs (20). In addition, several tumor suppressor miRNA mimics, such as the let-7 family, miR-34a, miR-143, miR-145, and miR-200 family, are being or have already been developed as therapeutics for clinical trials. It is worth noting that almost all of these miRNAs are also well-known for regulating cancer stem cells (21–23).
Cancer stem cells are considered the root of tumor initiation and metastasis. Abundant evidence suggests that CSCs comprise quiescent CSCs (QCSCs) and rapid cycling, or invasive CSCs, and rapid cycling CSCs can differentiate from QCSCs (2, 24). Here, we isolated quiescent cancer cells with a label staining assay (PKH dye) in serum-free culture medium, as we have previously demonstrated that PKHhi cells strongly exhibit stem cell-like properties (2). We analyzed the microRNA expression profile difference between PKHhi cells and the rest of the cancer cells. To the best of our knowledge, this is the first report screening the differences in expression of microRNAs between QCSCs and non-QCSCs, and by assessing the CRC cell line SW480 and primary CRC cells, we identified 29 differentially regulated miRNAs. We found that miR-4666-3p is the only miRNA downregulated in the PKHhi subpopulation of both SW480 and P1 cells. Here, we prove that miR-4666-3p could target IFN-γR1/2, which explains the higher sensitivity of PKHhi cells to the apoptotic effect of IFN-γ compared to the rest of the cancer cell population (2). Furthermore, we found that upregulation of miR-4666-3p could also suppress stemness gene expression, sphere formation and xenograft generation, but these effects could not be explained as alterations in the expression of IFN-γR (Figures S4A, B). Therefore, we performed bioinformatic analysis to investigate other potential targets that affect the stemness of CCSCs.
It has been reported that TGF-β signaling holds a key position in cancer stem cells. The activation of the TGF-β pathway is involved in regulating EMT (25), promoting invasiveness and metastasis of CCSCs (26), and inducing the secretion of angiogenesis-related factors, such as fibroblast growth factor 2 and vascular endothelial growth factor, by CCSCs (25, 27). TGF-β signaling is transduced by TGF-β receptors, which include TGF-βR1, 2, and 3. Studies have revealed that TGFBRs' expression is closely associated with cancer stem cells. Yusra et al. (28) found that the expression of TGFBR1 and TGFBR3 was higher in CD133-positive colon cancer stem cells, suggesting that CD133+ CCSCs have an increased sensitivity to TGF-β. The gene expression profile of mammary cancer stem-like cells revealed high expression of TGFBR2 (29). TGFBR1 was also reported as a therapeutic target to suppress stem-like features of these cells (12, 30). As we proved here by several methods, TGFBR1 is a functional downstream target of miR-4666-3p, and alterations in miR-4666-3p and TGFBR1 expression could affect the stemness of colon cancer cells and activation of TGF-β/Smad pathway.
Besides, it is noted that autocrine of TGF-β1 by cancer cells play important part in regulation of TGF-β/Smad pathway. Abundant evidence has highlighted the importance of the TGF-β autocrine mechanism in CSCs. Liu et al. (31) reported that TGF-β autocrine signaling greatly enhanced the ability of cells to grow anchorage independently in serum-free medium and potentiated the stemness and invasiveness of CSCs. Yeh et al. (32) found that elevated expression of PSPC1 (paraspeckle component 1) potentiated the TGF-β1 autocrine pathway and activated the TGF-β/Smad pathway, which promoted stemness, EMT and invasiveness in various cancer cells. Here, we found that PKHhi cells produce higher levels of TGF-β1 than PKHlow and PKHneg cells, which could be attributed to the low miR-329 expression level in PKHhi cells. miR-329 has been reported as a tumor suppressor gene and is related to favorable clinicopathological features in various cancers (17, 33, 34). TGF-β1 has been revealed to be a target of miR-329 (17), and here we also identified it as a functional target of miR-329. In addition, transfection of miR-329 mimics greatly inhibited the spheroid and xenograft formation abilities of colorectal cancer cells. Furthermore, our luciferase and immunofluorescence assays showed that miR-329 and miR-4666-3p mimics synergistically inhibit the spheroid formation and activation of TGF-β/Smad signaling. Here, although miR-4666-3p and miR-329 target on different gene, here we demonstrated both expressed at low levels in CCSCs and cooperated on the same signaling pathway (Figure 7).
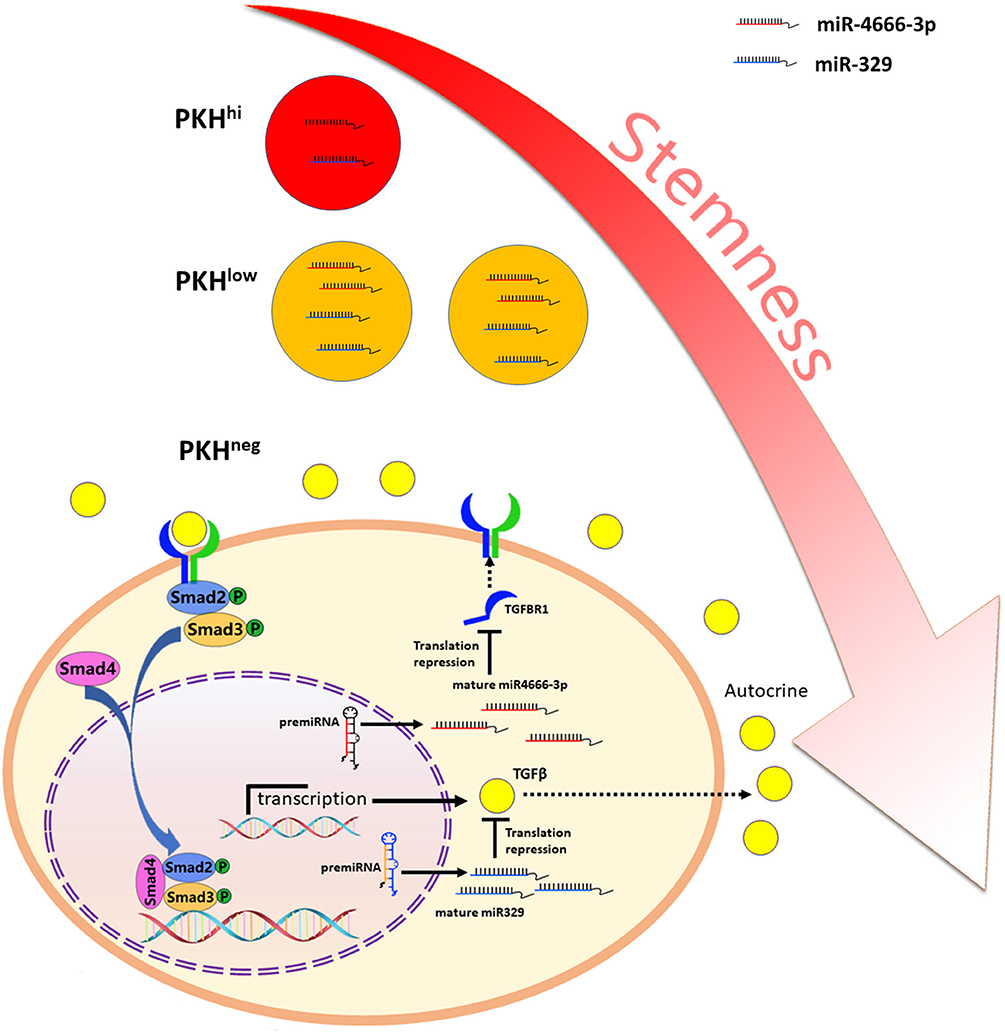
Figure 7. Schematic illustration of miR-4666-3p and miR-329 are both low expressed in non-QCCSCs and synergistically regulate TGF-β/Smad pathway.
However, the dysregulated expression of miRNAs in cancer cells is still not fully understood, and several studies indicate that underlying causes may contribute to the aberrant regulation of specific pathways. For example, activation of the MAPK and NF-κB (nuclear factor kappa-B) pathways could upregulate the transcriptional factor C/EBPβ, thereby increasing the expression of let-7f and miR-146 (35, 36). Here, we acknowledge that the mechanism behind the increase in the expression levels of miR-4666-3p and miR-329 that occurs during stem cell division is still undetermined, and future studies are needed to elucidate the detailed mechanism.
To our knowledge, this is the first study to report this specific and novel mechanism by which miRNAs cooperate to suppress the TGF-β/Smad pathway in colorectal cancer. Our results suggest that targeting multiple miRNAs in a single therapy could be more comprehensive and effective for cancer treatment.
All datasets generated for this study are included in the article/Supplementary Material.
This study was approved by the Ethical Committee of the Zhejiang Provincial People's hospital, Hangzhou Medical College. The patients were interviewed prior to inclusion in the study and were provided with written and verbal information regarding the study. All patients provided written informed consent. All methods were performed in accordance with the relevant guidelines and regulations. This study was approved by the ethics committee of the Laboratory Animal Research Center of Zhejiang Medical Academy.
CN and JH conceived and designed the study. JY, YS, XH, HY, and QX performed experiment and data collecting. JY, QF, JL, and WX analyzed data and performed the statistical analysis. CN critically revised the manuscript. All authors commented on drafts of the paper and approved the final manuscript.
This work was supported by Natural Science Foundation of Zhejiang Province (grant number LY13H160039, LR19H160001); the Natural Science Foundation of China (grant number 81772575, 81502463); the key Research Project of Science Technology Department of Zhejiang Province (grant number 2015C33097); Key Project of Health Bureau of Zhejiang Province (grant number 20182747347, 2017KY233); and the CSCO Merck Serono Oncology Research Fund, SCORE (grant number Y-MX2015-038).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2019.01251/full#supplementary-material
CCSC, colorectal cancer stem cells; CSC, cancer stem cell; CRC, colorectal cancer; ELISA, enzyme-linked immunosorbent assays; FCM, flow cytometry; FACS, fluorescence activated Cell Sorting; IFNGR, Interferon gamma receptor; NF-κB, nuclear factor kappa-B; MAPK, mitogen-activated protein kinase; PSPC1, paraspeckle component 1; KEGG, Kyoto Encyclopedia of Genes and Genomes; QCCSCs, quiescent cancer stem cells; TCGA, The Cancer Genome Atlas; TNM, tumor, node, metastasis; VEGF-C, Vascular endothelial growth factor C.
1. Lin HH, Lee HW, Lin RJ, Huang CW, Liao YC, Chen YT, et al. Tracking and finding slow-proliferating/quiescent cancer stem cells with fluorescent nanodiamonds. Small. (2015) 11:4394–402. doi: 10.1002/smll.201500878
2. Ni C, Wu P, Zhu X, Ye J, Zhang Z, Chen Z, et al. IFN-gamma selectively exerts pro-apoptotic effects on tumor-initiating label-retaining colon cancer cells. Cancer Lett. (2013) 336:174–84. doi: 10.1016/j.canlet.2013.04.029
3. Iliopoulos D, Hirsch HA, Wang G, Struhl K. Inducible formation of breast cancer stem cells and their dynamic equilibrium with non-stem cancer cells via IL6 secretion. Proc Natl Acad Sci USA. (2011) 108:1397–402. doi: 10.1073/pnas.1018898108
4. Roesch A, Fukunaga-Kalabis M, Schmidt EC, Zabierowski SE, Brafford PA, Vultur A, et al. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell. (2010) 141:583–94. doi: 10.1016/j.cell.2010.04.020
5. Xi XP, Zhuang J, Teng MJ, Xia LJ, Yang MY, Liu QG, et al. MicroRNA-17 induces epithelial-mesenchymal transition consistent with the cancer stem cell phenotype by regulating CYP7B1 expression in colon cancer. Int J Mol Med. (2016) 38:499–506. doi: 10.3892/ijmm.2016.2624
6. Yu X, Mi L, Dong J, Zou J. Long intergenic non-protein-coding RNA 1567 (LINC01567) acts as a “sponge” against microRNA-93 in regulating the proliferation and tumorigenesis of human colon cancer stem cells. BMC Cancer. (2017) 17:716. doi: 10.1186/s12885-017-3731-5
7. Ottaviani S, Stebbing J, Frampton AE, Zagorac S, Krell J, de Giorgio A, et al. TGF-beta induces miR-100 and miR-125b but blocks let-7a through LIN28B controlling PDAC progression. Nat Commun. (2018) 9:1845. doi: 10.1038/s41467-018-03962-x
8. Dokanehiifard S, Soltani BM. TrkC-miR2 regulates TGFbeta signaling pathway through targeting of SMAD3 transcript. J Cell Biochem. (2018) 120:2634–41. doi: 10.1002/jcb.27572
9. Li J, Zou K, Yu L, Zhao W, Lu Y, Mao J, et al. MicroRNA-140 Inhibits the epithelial-mesenchymal transition and metastasis in colorectal cancer. Mol Ther Nucleic Acids. (2018) 10:426–37. doi: 10.1016/j.omtn.2017.12.022
10. Wang K, Liu L, Zhang T, Zhu YL, Qiu F, Wu XG, et al. Oxaliplatin-incorporated micelles eliminate both cancer stem-like and bulk cell populations in colorectal cancer. Int J Nanomed. (2011) 6:3207–18. doi: 10.2147/IJN.S26268
11. Wang YK, Zhu YL, Qiu FM, Zhang T, Chen ZG, Zheng S, et al. Activation of Akt and MAPK pathways enhances the tumorigenicity of CD133+ primary colon cancer cells. Carcinogenesis. (2010) 31:1376–80. doi: 10.1093/carcin/bgq120
12. Li L, Liu CC, Chen X, Xu S, Hernandez Cortes-Manno S, Cheng SH. Mechanistic study of bakuchiol-induced anti-breast cancer stem cell and in vivo anti-metastasis effects. Front Pharmacol. (2017) 8:746. doi: 10.3389/fphar.2017.00746
13. Jin M, Zhang T, Liu C, Badeaux MA, Liu B, Liu R, et al. miRNA-128 suppresses prostate cancer by inhibiting BMI-1 to inhibit tumor-initiating cells. Cancer Res. (2014) 74:4183–95. doi: 10.1158/0008-5472.CAN-14-0404
14. Cammareri P, Vincent DF, Hodder MC, Ridgway RA, Murgia C, Nobis M, et al. TGFbeta pathway limits dedifferentiation following WNT and MAPK pathway activation to suppress intestinal tumourigenesis. Cell Death Differ. (2017) 24:1681–93. doi: 10.1038/cdd.2017.92
15. Arena S, Salati M, Sorgentoni G, Barbisan F, Orciani M. Characterization of tumor-derived mesenchymal stem cells potentially differentiating into cancer-associated fibroblasts in lung cancer. Clin Transl Oncol. (2018) 20:1582–91. doi: 10.1007/s12094-018-1894-4
16. Rahn S, Zimmermann V, Viol F, Knaack H, Stemmer K, Peters L, et al. Diabetes as risk factor for pancreatic cancer: hyperglycemia promotes epithelial-mesenchymal-transition and stem cell properties in pancreatic ductal epithelial cells. Cancer Lett. (2018) 415:129–50. doi: 10.1016/j.canlet.2017.12.004
17. Li B, Huang M, Liu M, Wen S, Sun F. MicroRNA329 serves a tumor suppressive role in colorectal cancer by directly targeting transforming growth factor beta1. Mol Med Rep. (2017) 16:3825–32. doi: 10.3892/mmr.2017.7077
18. Ye J, Wu X, Wu D, Wu P, Ni C, Zhang Z, et al. miRNA-27b targets vascular endothelial growth factor C to inhibit tumor progression and angiogenesis in colorectal cancer. PloS ONE. (2013) 8:e60687. doi: 10.1371/journal.pone.0060687
19. Wang T, Xu Z. miR-27 promotes osteoblast differentiation by modulating Wnt signaling. Biochem Biophys Res Commun. (2010) 402:186–9. doi: 10.1016/j.bbrc.2010.08.031
20. Krausova M, Korinek V. Wnt signaling in adult intestinal stem cells and cancer. Cell Signal. (2014) 26:570–9. doi: 10.1016/j.cellsig.2013.11.032
21. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. (2017) 16:203–22. doi: 10.1038/nrd.2016.246
22. Huang S, Guo W, Tang Y, Ren D, Zou X, Peng X. miR-143 and miR-145 inhibit stem cell characteristics of PC-3 prostate cancer cells. Oncol Rep. (2012) 28:1831–7. doi: 10.3892/or.2012.2015
23. Pan Q, Meng L, Ye J, Wei X, Shang Y, Tian Y, et al. Transcriptional repression of miR-200 family members by Nanog in colon cancer cells induces epithelial-mesenchymal transition (EMT). Cancer Lett. (2017) 392:26–38. doi: 10.1016/j.canlet.2017.01.039
24. Jiang YX, Yang SW, Li PA, Luo X, Li ZY, Hao YX, et al. The promotion of the transformation of quiescent gastric cancer stem cells by IL-17 and the underlying mechanisms. Oncogene. (2017) 36:1256–64. doi: 10.1038/onc.2016.291
25. Zubeldia IG, Bleau AM, Redrado M, Serrano D, Agliano A, Gil-Puig C, et al. Epithelial to mesenchymal transition and cancer stem cell phenotypes leading to liver metastasis are abrogated by the novel TGFbeta1-targeting peptides P17 and P144. Exp Cell Res. (2013) 319:12–22. doi: 10.1016/j.yexcr.2012.11.004
26. Chen W, Zhao K, Miao C, Xu A, Zhang J, Zhu J, et al. Silencing Trim59 inhibits invasion/migration and epithelial-to-mesenchymal transition via TGF-beta/Smad2/3 signaling pathway in bladder cancer cells. Oncotargets Ther. (2017) 10:1503–12. doi: 10.2147/OTT.S130139
27. Yang F, Strand DW, Rowley DR. Fibroblast growth factor-2 mediates transforming growth factor-beta action in prostate cancer reactive stroma. Oncogene. (2008) 27:450–9. doi: 10.1038/sj.onc.1210663
28. Yusra, Semba S, Yokozaki H. Biological significance of tumor budding at the invasive front of human colorectal carcinoma cells. Int J Oncol. (2012) 41:201–10. doi: 10.3892/ijo.2012.1459
29. Rybicka A, Mucha J, Majchrzak K, Taciak B, Hellmen E, Motyl T, et al. Analysis of microRNA expression in canine mammary cancer stem-like cells indicates epigenetic regulation of transforming growth factor-beta signaling. J Physiol Pharmacol. (2015) 66:29–37.
30. Boix L, López-Oliva JM, Rhodes AC, Bruix J. Restoring miR122 in human stem-like hepatocarcinoma cells, prompts tumor dormancy through Smad-independent TGF-beta pathway. Oncotarget. (2016) 7:71309–29. doi: 10.18632/oncotarget.11885
31. Liu Z, Bandyopadhyay A, Nichols RW, Wang L, Hinck AP, Wang S, et al. Blockade of autocrine TGF-beta signaling inhibits stem cell phenotype, survival, and metastasis of murine breast cancer cells. J Stem Cell Res Ther. (2012) 2:1–8. doi: 10.4172/2157-7633.1000116
32. Yeh HW, Hsu EC, Lee SS, Lang YD, Lin YC, Chang CY, et al. PSPC1 mediates TGF-beta1 autocrine signalling and Smad2/3 target switching to promote EMT, stemness and metastasis. Nat Cell Biol. (2018) 20:479–91. doi: 10.1038/s41556-018-0062-y
33. Yin J, Shen X, Li M, Ni F, Xu L, Lu H. miR-329 regulates the sensitivity of 5-FU in chemotherapy of colorectal cancer by targeting E2F1. Oncol Lett. (2018) 16:3587–92. doi: 10.3892/ol.2018.9121
34. Wu L, Pei F, Men X, Wang K, Ma D. miR-329 inhibits papillary thyroid cancer progression via direct targeting WNT1. Oncol Lett. (2018) 16:3561–8. doi: 10.3892/ol.2018.9102
35. Ayyar KK, Reddy KVR. MAPK and NF-kappaB signalling pathways regulate the expression of miRNA, let-7f in human endocervical epithelial cells. J Cell Biochem. (2018) 119:4751–9. doi: 10.1002/jcb.26665
Keywords: colorectal cancer, miR-4666-3p, miR-329, cancer stem cells, TGF-β/smad
Citation: Ye J, Lei J, Fang Q, Shen Y, Xia W, Hu X, Xu Q, Yuan H, Huang J and Ni C (2019) miR-4666-3p and miR-329 Synergistically Suppress the Stemness of Colorectal Cancer Cells via Targeting TGF-β/Smad Pathway. Front. Oncol. 9:1251. doi: 10.3389/fonc.2019.01251
Received: 28 August 2019; Accepted: 30 October 2019;
Published: 19 November 2019.
Edited by:
Kuzhuvelil B. Harikumar, Rajiv Gandhi Centre for Biotechnology, IndiaReviewed by:
Jun Zhao, First Affiliated Hospital of Soochow University, ChinaCopyright © 2019 Ye, Lei, Fang, Shen, Xia, Hu, Xu, Yuan, Huang and Ni. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chao Ni, MTExMTgxMThAemp1LmVkdS5jbg==; Jian Huang, ZHJodWFuZ2ppYW5Aemp1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.