- 1National Laboratory CIB, Trieste, Italy
- 2Department of Life Sciences, University of Trieste, Trieste, Italy
- 3IFOM – Istituto FIRC Oncologia Molecolare, Milan, Italy
Cellular choices are determined by developmental and environmental stimuli through integrated signal transduction pathways. These critically depend on attainment of proper activation levels that in turn rely on post-translational modifications (PTMs) of single pathway members. Among these PTMs, post-phosphorylation prolyl-isomerization mediated by PIN1 represents a unique mechanism of spatial, temporal and quantitative control of signal transduction. Indeed PIN1 was shown to be crucial for determining activation levels of several pathways and biological outcomes downstream to a plethora of stimuli. Of note, studies performed in different model organisms and humans have shown that hormonal, nutrient, and oncogenic stimuli simultaneously affect both PIN1 activity and the pathways that depend on PIN1-mediated prolyl-isomerization, suggesting the existence of evolutionarily conserved molecular circuitries centered on this isomerase. This review focuses on molecular mechanisms and cellular processes like proliferation, metabolism, and stem cell fate, that are regulated by PIN1 in physiological conditions, discussing how these are subverted in and hijacked by cancer cells. Current status and open questions regarding the use of PIN1 as biomarker and target for cancer therapy as well as clinical development of PIN1 inhibitors are also addressed.
Introduction
Cells respond to a continuous flow of intra- and extra-cellular stimuli through integrated signal transduction pathways. Within these signaling cascades proper levels, subcellular localization, complex formation, and activation timing of proteins are fundamental for an appropriate cellular response. Mechanistically, post-translational modifications (PTMs) of proteins underlie the dynamic molecular changes necessary to activate, transmit, and tune signals through the signal transduction network. PTMs impact on protein structure conferring new biochemical and biological properties. In this way, PTMs control several biological processes that require molecular switches, such as cell cycle progression, metabolic changes and cell fate decisions throughout organismal life (1).
Phosphorylation of Serines or Threonines preceding a Proline (S/T-P) is one of the most frequent PTM occurring on a wide range of proteins of eukaryotic cells. This modification is tightly regulated by Proline-directed kinases and phosphatases devoted to their addition and removal, respectively (2). This PTM deserves particular attention, since Prolines adopt either a cis or a trans conformation, a slow but spontaneous structural change, that can elicit profound functional consequences. Peptidyl-prolyl cis-trans isomerases (PPIases) are the enzymes that accelerate the cis-trans rotation to a biologically relevant timescale. Among them, the prolyl-isomerase PIN1 uniquely recognizes S/T-P motifs when they become phosphorylated (pS/T-P). This substrate specificity is based on a conserved two-domain structure, where an N-terminal WW domain specifically recognizes pS/T-P sites (3) and a C-terminal PPIase domain performs the cis-trans isomerization (4). Hence, following Proline-directed phosphorylation, interaction with PIN1, and subsequent isomerization, the shape of target proteins undergoes changes that affect their stability, subcellular localization, protein-protein interaction, occurrence of other PTMs and ultimately activity (5). Accordingly, isomerization by PIN1 adds a new layer of control in signaling pathways that are regulated by phosphorylation, most notably the growth factor/RAS-MAPK, pRB/CDK/CYCLIN D1, p53, NOTCH, c-MYC, WNT/β-CATENIN, NF-kappaB, PI3K/AKT, and several other pathways (6–9). Through these signaling pathways PIN1 has been shown to impact several cellular processes, such as cell cycle progression, regulation of cellular metabolism and stem cell maintenance (Figure 1).
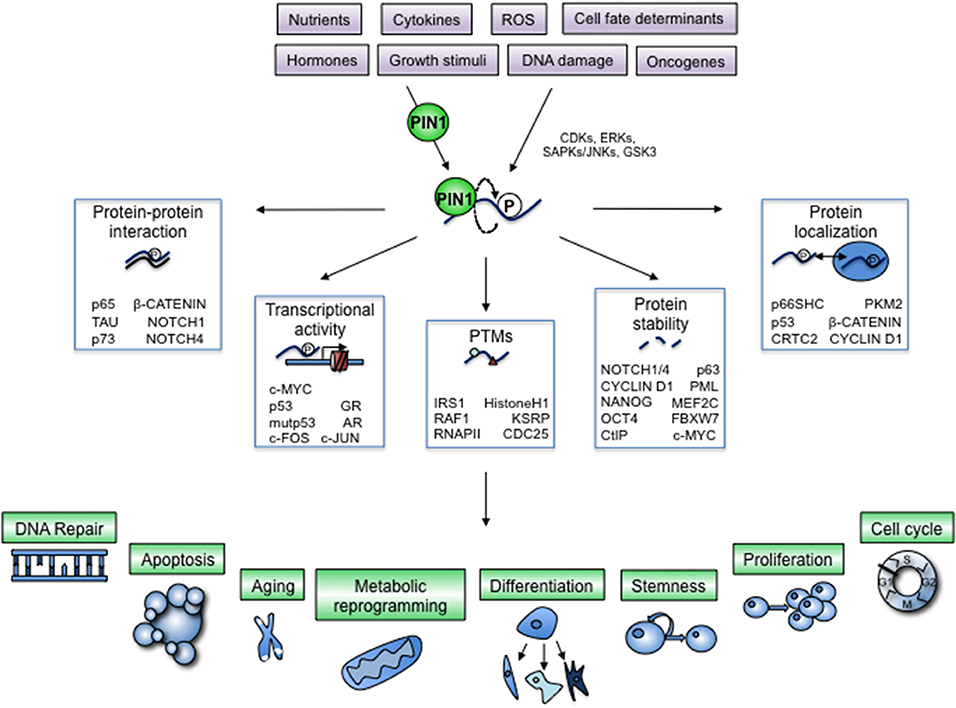
Figure 1. Regulation of cellular processes by PIN1-dependent modification of key cellular proteins. The prolyl-isomerase PIN1 is controlled by physiologic and pathologic cues and helps transducing phosphorylation signaling exerted by prolyl-directed kinases, often activated by the same stimuli (violet boxes). A growing list of cellular proteins are phosphorylated by these kinases and subsequently bound by PIN1, which induces a conformational change (bent arrow) with profound impact on their activation status by means of critical modifications of different biochemical and cellular properties (white boxes). By doing so, PIN1 adds a further layer of control to signaling pathways that are regulated by proline-directed phosphorylation. Proteins modified by PIN1 in fact can elicit cellular responses that involve multiple cellular and organismal processes, depending on the specific protein and on the cellular context (green boxes and blue drawings).
Both cell-autonomous and systemic effects of PIN1, through global regulation of phosphorylation-dependent events, are required for proper embryonic development and maintenance of tissue integrity in adulthood. As a consequence, subtle alterations of PIN1 expression or activity or of the phosphorylation status of its targets have been linked to a number of pathologies, ranging from inflammation to neurodegeneration and cancer. The observed alterations are likely the result of chronically compromised protein folding causing improper tuning or timing of relevant signaling pathways (5, 6, 8, 10–13).
PIN1 function is required for several biological hallmarks of cancer, as has been described in depth (7, 8, 10). PIN1 is overexpressed or hyperactivated in many types of cancers and its loss or inactivation blocks tumor growth. Indeed, Pin1 knockout in NOTCH3-dependent T-ALL (9), Eμ-myc lymphomas (14), or in MMTV-Ras/neu mammary tumors (15) in mice curbs tumorigenesis. Similarly, knockdown of PIN1 in breast cancer xenografts was shown to curb tumor growth and metastasis formation and synergize with chemotherapy by dampening mutant p53 (mutp53) (16) and NOTCH1 (17) signaling, respectively. In addition, pharmacological inhibition of PIN1 dampens mammary tumor growth in a Myc/NeuNT mouse model (18) and metastasis development in a breast cancer xenograft (19). According to published results, PIN1 was shown to boost dozens of oncogenes or factors that promote proliferation, while inactivating several tumor suppressors (20), but which are the deranged underlying cellular processes is still incompletely understood.
In this review we will describe how cellular processes normally regulated by PIN1 could impact the pathogenesis of cancer. We will thus emphasize the relevance of PIN1 for cancer development and progression by orchestrating cellular processes that are emerging to be controlled by PIN1 in normal organismal development and that are hijacked in cancer, like metabolic reprogramming and response to cellular stresses, as well as dichotomous cell choices like stemness or differentiation, cell proliferation or cell death (Figure 2).
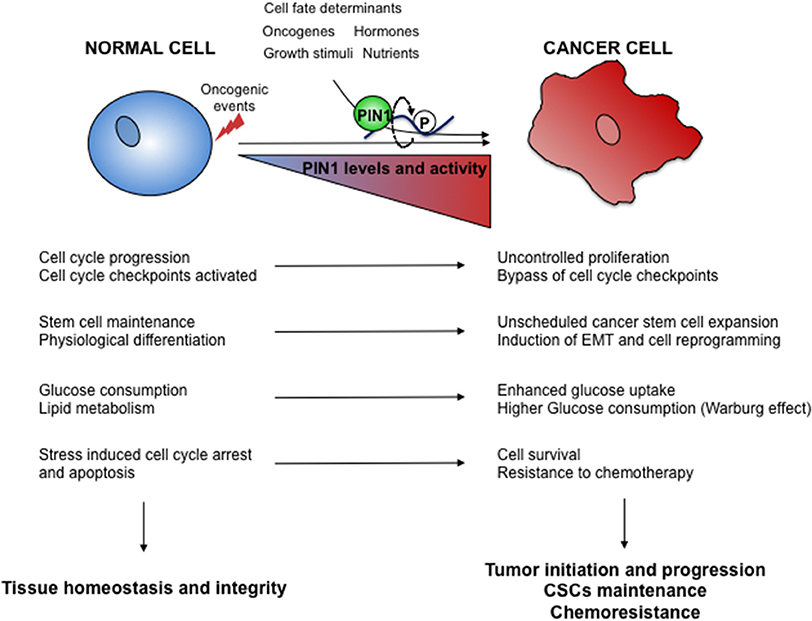
Figure 2. Oncogenic hijacking of the PIN1 signaling network. Oncogene activation and environmental cues cooperate in tumor development and progression by boosting PIN1 levels and activity as well as of its signaling networks. Several cellular processes regulated by PIN1 in normal cells (left) are thus subverted in cancer cells (right) to gain growth advantages.
Biological Roles of PIN1: Insights From Animal Models and Studies in Humans
PPIases are subdivided in three families, namely Cyclophilins, FK506 Binding Proteins (FKBPs) and Parvulins, predominantly defined on structural characteristics. The two former are large PPIases, also called immunophilins that are inhibited by immunosuppressants like Cyclosporin and FK506. The third family of Parvulins, to which PIN1 belongs, comprises smaller PPIases that do not bind to these drugs (21).
In yeast the PIN1 homolog ESS1 is the sole parvulin, while in mice and humans another Parvulin gene, encoding the proteins PAR14 and PAR17 (PAR14/17) (22) has been cloned. Cyclophilins, FKBPs, and PAR14/17 have specific substrate recognition motifs that differ from those of PIN1, nevertheless, these PPIases can share client proteins by targeting different portions on them (23).
More recently, within the human and mouse genomes, Pin1-like transcripts (Pin1L) have been found (24, 25). The human PIN1L encodes a putative protein of 100-amino-acids of which the N-terminal 63 amino acids are almost identical to PIN1 while it is fused to a different N-terminal 37-amino-acid tail. Instead, mouse Pin1L mRNA is widely expressed, showing highest levels in the testis and the protein shares 92% sequence identity with PIN1. Still it is not known whether these genes have a functional role or if they act as pseudogenes (24).
The biological functions of PIN1 have raised interest since its discovery in yeast (ScEss1p/Ptf1) (26, 27), where its deficiency leads to mitotic arrest and nuclear fragmentation (27). Since then, a number of in vivo loss-of-function and gain-of-function genetic models of PIN1 have been established and the biological function of PIN1 orthologs has been characterized in different model organisms, including Saccharomyces pombe (Pin1p) (28), Aspergillus nidulans (PinA) (29), Neurospora crassa (Ssp1) (30), Candida albicans (Ess1) (31), Drosophila melanogaster (Dodo) (32, 33), Danio Rerio (pin1) (34), Xenopus laevis (xPin1) (35), and Mus musculus (mPin1) (36).
Studies in these model organisms indicate functional conservation of PIN1, mainly related to cell proliferation and metabolism. Moreover, while in unicellular organisms like A. nidulans, and S. cervisiae PIN1 is functionally essential, its ablation is associated with developmental defects in C. albicans (31), and metazoans like X. laevis (37), D. melanogaster (38) and M. musculus (36, 39–41). Indeed, lack of Ess1 in C. albicans hampers cell growth and abrogates morphogenetic switching (31), while dodo serves for timely activation of the circadian rhythm (42) and its ablation impairs dorsoventral patterning of the follicular epithelium in the egg chamber in D. melanogaster (38). In X. laevis lack of xPin1 impairs survival of embryos during development (35, 37). Pin1 knock-out (Pin1KO) mice display impaired neonatal motor activity (43), decreased fertility both in males and females (41), alteration of the mammary gland that fails to undergo proliferation during pregnancy, and decreased body weight (39). Moreover, these mice show premature aging phenotypes, such as testicular atrophy (39), alterations of bone homeostasis (44), retinal atrophy (39), and neurodegeneration (45). In addition, in aged mutant APP transgenic mice, knock-out of Pin1 incites deposition of insoluble beta amyloids (46).
Human PIN1 was discovered by a two-hybrid screening aimed at identifying proteins that interact with NIMA-1, the key mitotic kinase of Aspergillus nidulans (47). In humans, deregulation of PIN1 levels or activity has been correlated to inflammation, obesity, diabetes, cancer, and neurodegeration (6, 48, 49). Indeed, neurodegeneration is mainly associated to low levels or activity of PIN1, while on the contrary overexpression of PIN1 is prevalently found in cancers and inflammatory diseases. Of note, the rs2233678 polymorphism in the human PIN1 promoter is associated to decreased gene expression and to a lower risk of breast (50) and lung (51) cancer incidence, while the rs2287839 polymorphism hampers suppression of PIN1 transcription by the AP4 transcription factor and correlates with delayed onset of Alzheimer's disease (52).
Global Roles of PIN1 in Signal Transduction
The dynamicity of reversible phosphorylation of proteins represents a crucial step to transmit cellular inputs through the generation of integrating platforms for signal transduction (53). PIN1 acts on many cellular proteins and pathways in a phosphorylation-dependent manner, displaying a variety of functional consequences. Here we will describe the biochemical consequences of the structural modifications exerted by PIN1 on several proteins and their impact on global functions in signal transduction and cellular processes in different animal and human models.
PIN1 Deciphers the Protein pS/T-P Code
Signal transduction by phosphorylation often involves sets of protein kinases (writers), phospho-binding targets (readers) and protein phosphatases that remove individual phosphorylations (erasers). Typically, the proteins that comprise this system are modular in nature harboring peptide domains with binding and/or catalytic properties (54). In this view, PIN1 might be considered as a “reader” in association with proline-directed kinases as “writers,” which include Cyclin-dependent protein kinases (CDKs), extracellular signal-regulated kinases (ERKs), stress-activated protein kinases/c-Jun N-terminal kinases (SAPKs/JNKs), p38 mitogen-activated protein kinases (MAPK), and glycogen synthase kinase 3 beta (GSK3β). Indeed, several reports have demonstrated that following proline-directed phosphorylation events on many proteins, PIN1 causes further modulation by inducing a conformational change and affecting their pattern of PTMs, for instance by favoring their association with other kinases, phosphatases, acetyl-transferases, etc. (55). In this regard, a biological interdependence has been found between PIN1 and the protein phosphatase 2A (PP2A), since PP2A is conformation-specific and effectively dephosphorylates only trans pS/T-P isomers (56). PIN1 and PP2A share many client proteins and show reciprocal genetic interactions (5). A paradigmatic example for such a PIN1-dependent sequence of PTMs is the priming phosphorylation of c-Myc by RAS/MAPK, followed by GSK3β phosphorylation and PIN1 binding, whose isomerization elicits the PP2A-mediated dephosphorylation and subsequent proteasome dependent degradation by the E3 ubiquitin-ligase FBXW7 (57). Other examples of PIN1-dependent regulation of PTMs at multiple levels are NOTCH1 (17, 58), RAF-1 (59), CYCLIN E (60, 61), KSRP (62, 63), TAU (56), and pRB (64, 65).
PIN1 Optimizes Signal Transduction and Pathway Crosstalk
When a cell receives a signal, a physiological response is initiated by activating a string of biochemical reactions. Indeed signal transduction is accomplished by receptors, adaptors and amplifiers able to modulate a number of effectors like transcription factors that elicit a cellular response (1). Pathway activation can be achieved in many different ways, for instance by changing subcellular localization or enhancing the duration and quantity of the effectors. In this scenario, PIN1 fulfills the role of both fine-tuner and amplifier of signal transduction pathways by regulating single members of a pathway and also their cognate activators and inhibitors, finally determining the level of their activation state. The best described example concerns the growth factor activated RAS/MAPK pathway in human breast epithelial cells, since PIN1 was shown to act as an amplifier of this cascade at all the levels of signal transduction, including stabilization of the receptor (EGFR), enhancing activation of the transducing kinase (RAF-1), increasing the stability of the transcription factor (c-JUN) and of the effector (CYCLIN D1), while its ablation dampened this signal transduction with a consequent impairment of cell cycle progression (39, 59, 65–67).
As mentioned above, PIN1 activity is involved in many other signaling pathways (17, 43, 68–70) and controls the levels and activity of several kinases beyond those of the RAS/MAPK pathway as well as protein phosphatases (55, 56). Therefore, PIN1 could control not only the amplification of a single signaling cascade but also the cross-talk between these pathways, with important consequences for organismal development (Figure 3).
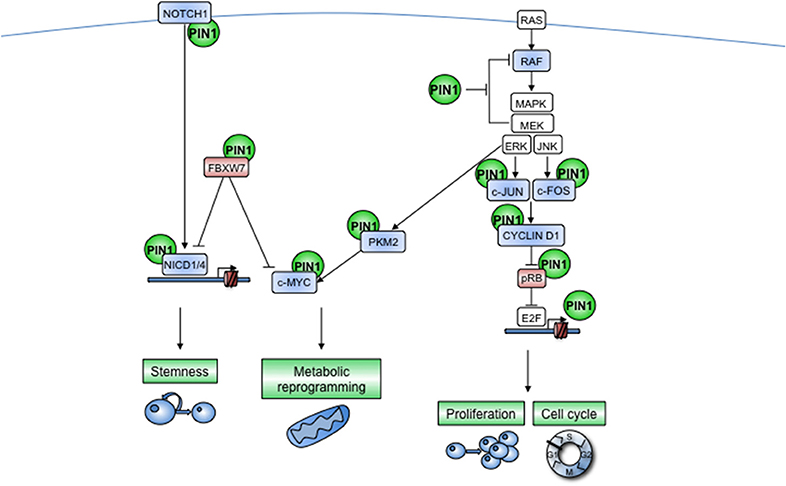
Figure 3. PIN1 acts at multiple levels of cooperating cellular pathways. NOTCH receptor and RAS-dependent pathways are indicated as examples of signal transduction pathways that are regulated by PIN1 at multiple levels. Furthermore, PIN1 enables active cross-talk between different pathways, e.g., by promoting nuclear translocation of PKM2 after EGFR-stimulated phosphorylation by ERKs, that in turn serves to enhance c-MYC transcriptional activity. (Left) The NOTCH pathway is activated by a ligand-dependent cleavage of the membrane-bound receptor that releases the Notch intracellular domain (NICD), which translocates into the nucleus where it promotes the transactivation of several target genes. This signal is turned off by the E3-ubiquitin ligase FBXW7 as a safeguard mechanism against excessive NOTCH signaling. PIN1 is harnessed by the NOTCH pathway to sustain its own signaling, indeed PIN1 boosts the cleavage of the NOTCH1 receptor and prevents the interaction of NICD1 and NICD4 with FBXW7, increasing in this way pathway activation, protein stability and transcriptional activity. In some contexts, PIN1 can also directly block FBXW7 activity. Moreover, some NOTCH transcriptional targets, such as CYCLIN D1 and c-MYC, are also direct PIN1 targets, thus suggesting that PIN1 may amplify also the NOTCH transcriptional program. Notably NICD1 and NICD4 directly promote Pin1 transcription. Blue color indicates PIN1-activated proteins, red color indicates PIN1-inhibited proteins. Arrows and blocked lines show positive and negative effects, respectively. (Right) In response to growth stimuli, RAS activates the Raf kinase, which in turn activates downstream MAPKs that phosphorylate c-FOS and c-JUN causing the formation of the AP-1 transcription factor that transactivates the CYCLIN D1 gene. CYCLIN D1-CDK4/6 complex, in turn, blocks pRb, unleashing the transcription factor E2F, boosting in this way cell cycle progression. PIN1 itself is a direct transcriptional target of E2F and strongly promotes the RAS/MAPKs/CYCLIN/CDK cascade at multiple levels: (i) by blocking negative feed-back regulation of RAF by MAPKs, (ii) by enhancing both transcription and protein stability of CYCLIN D1, and (iii) by blocking the activity of pRB.
In the majority of cases, PIN1-induced conformational changes determine the direction and intensity of cellular responses and hence impact on cell fate by affecting the levels of key client proteins (5). Indeed, PIN1 controls the turnover of many substrates either favoring or blocking their recognition by E3 ubiquitin-ligases, enzymes that often require particular PTMs on their substrates (71). Moreover, PIN1 directly regulates the activity of the ubiquitin-ligase FBXW7 in particular cellular contexts (72), and, since PIN1 and FBXW7 in part share the same consensus on proteins, it can also antagonize FBXW7-mediated degradation of certain substrates. This occurs in the case of the intracellular domains of the NOTCH1 and NOTCH4 receptors, whose stability and transcriptional activity depend on the presence of these two enzymes. Moreover, in breast cancer cells with high levels of PIN1, NOTCH1 signaling is strongly activated despite presence of FBXW7, inciting expansion of breast cancer stem cells (CSC) and tumorigenesis (17) (Figure 3). The protein stability of key drivers of signaling pathways fundamental for organismal development and integrity, i.e., c-MYC (18), NANOG (73), OCT4 (74), p53 (6), p63 (75), β-CATENIN (68), relies on PIN1 levels and activity, thus strengthening the idea that PIN1 can impact cell signaling pathways by calibrating the amount of target proteins.
In addition PIN1 has been shown to affect the subcellular localization of many proteins (5). Nevertheless, the mechanism by which these proteins are shuttled as an effect of PIN1-dependent cis/trans isomerization is varied. While on one hand PIN1 can promote nuclear retention of CYCLIN D1 (39) and NF-kappaB (69) proteins, it induces a direct change in protein subcellular localization of some substrates, as in the case of Pyruvate Kinase 2 (PKM2), whose association with Importin alpha5 and its consequent nuclear translocation, a crucial step in the activation of the Warburg effect in cancer cells, is promoted by PIN1 (76). Conversely, PIN1 binds to and prevents the nuclear translocation of CREB-regulated transcriptional co-activator 2 (CRTC2) thus crippling transcription from cAMP-responsive elements, and ultimately fostering hyperglycemia in diabetic mice (77). Moreover, PIN1 is required for mitochondrial translocation of cytosolic phospho-glycero-kinase 1 (PGK-1) to promote the Warburg effect in tumor cells (78) and of the tumor suppressor p53, following stress-induced phosphorylation, to induce transcription-independent apoptosis (79). Thus, PIN1 activity is essential to determine the correct localization of client proteins within the cell at a given time.
PIN1 Controls Gene Expression: From Transcription Factor-Specific to Global Gene Expression Mechanisms
A number of transcriptome profiles obtained in recent years from PIN1 silenced or knock-out cells demonstrated that this enzyme, although not a transcription factor (TF), has a sizeable impact on gene transcription (16, 18, 19, 62). Notably, PIN1 is able to impinge on gene transcription through the regulation of particular TFs and also by modulation of proteins recruited during mRNA synthesis and processing as well as factors governing chromatin accessibility (21) (Figure 4).
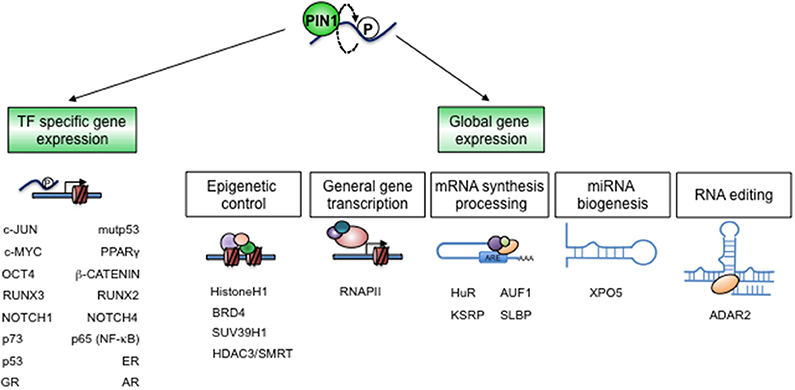
Figure 4. PIN1 controls gene expression. The establishment and dynamic regulation of specific cellular states largely depend on gene expression programs. PIN1 critically impacts these programs by direct structural modifications of specific TFs and co-factors (left), as well as of chromatin and RNA regulators (right), in a cell- and context-specific fashion.
For several TFs, isomerization by PIN1 helps converting phosphorylation events in transcriptional outcomes (10, 80). In particular PIN1 can (i) modulate the association between TFs and co-activators, as described for Estrogen Receptor (ER) (81, 82), p53 (83), p73 (84), c-MYC (18), and several others (10), (ii) increase TFs protein stability thereby prolonging their transcriptional activity as described for NOTCH1 (17), and (iii) regulate the subcellular localization of TFs, allowing for their function, as shown for β-CATENIN (68).
An intriguing connection has been found between PIN1 and c-MYC. PIN1 binds to and controls the extent of c-MYC-dependent gene transcription in a bimodal way: on one hand it facilitates the loading of c-MYC to certain sets of genes, on the other hand it promotes its dephosphorylation and proteasomal-dependent degradation, thus tuning the timing of c-MYC transcriptional activity. In tumors, where c-MYC degradation is impaired, PIN1 promotes c-MYC-dependent gene transcription boosting the expression of genes involved in cell growth and metabolism (18, 57, 76). Interestingly, it has been shown that PIN1 is able to pre-anchor to un-phosphorylated c-MYC and the subsequent phosphorylation event increases the strength of this interaction (85).
Alteration of the epigenetic landscape is a hallmark of cancer. Epigenetic changes including DNA methylation, histone modifications, nucleosome positioning, and non-coding RNA expression, are responsible for cancer cell reprogramming (86). Knowledge about a role of PIN1 in controlling several epigenetic regulators in normal and pathological settings is growing and suggests a role for deregulated PIN1 in fostering cancer development also through epigenetic reprogramming of gene transcription.
In yeast, the HDAC subunits Rpd3 and Sap30 are targets of PIN1 that elicit repression of cell cycle genes: in this case they are negatively regulated by PIN1, and likely underlie the mitotic arrest of PIN1 mutants (87). A role for mammalian PIN1 in the control of HDAC family members has been unveiled in a study performed in human colon cancer cells treated with the HDAC inhibitor sulforaphane. It was proposed that, following treatment with this inhibitor, the nuclear HDAC3/SMRT co-repressor complex is phosphorylated by CK2 and the complex becomes dissociated and exported into the cytoplasm. It is hypothesized that, in this compartment, HDAC3 could interact with the phospho-serine or phospho-threonine binding cofactor 14-3-3 and shuttle back into the nucleus, but there is competition with cytoplasmic PIN1 that sequesters HDAC3, targeting it for degradation, thus hampering its gene silencing function (88).
In metastatic breast cancer cells, PIN1 overexpression controls histone H3K9 trimethylation (H3K9me3) through downregulation of the methyltransferase SUV39H1, fostering tumor growth. This result was validated in human breast cancer samples, showing an inverse correlation between PIN1 levels and those of SUV39H1 and H3K9me3. Importantly, reversal of the repressive chromatin state following PIN1 overexpression could elicit upregulation of CYCLIN D1 (89).
Another PIN1-dependent epigenetic mechanism in cancer cells occurs through stabilization of BRD4, a member of the bromodomain and extraterminal (BET) family of proteins that interact with acetylated histones and TFs. In human gastric cancer cells PIN1 induces a conformational change of phosphorylated BRD4 supporting its interaction with CDK9, thus improving BRD4-mediated gene expression relevant for gastric cancer cell proliferation and tumor development (90).
Phosphorylated histone H1 is found at sites of active transcription and its phosphorylation is required for induction of gene expression. Notably, PIN1 associates with phosphorylated Histone H1 at transcriptionally active sites and controls its turnover and binding to chromatin in order to reduce chromatin decondensation (91).
PIN1 also globally affects mRNA synthesis and processing. First, it has been shown that PIN1 directly impacts on gene transcription by assisting RNA polymerase II (RNAPII) function during the cell cycle where RNAPII preferentially associates with transcribed genes in S phase, while it dissociates from chromatin in a phosphorylation-dependent manner during M phase (92–94). Second, PIN1 affects the stability of mRNAs by acting on AU-rich element (ARE)-binding proteins HuR and AUF1, the KH-type splicing regulatory protein (KSRP), and the Stem–loop–binding protein (SLBP) (62, 63, 95). In particular, the abundance of many ARE-containing mRNAs with short half-lives, such as GM-CSF, TGF-β, and PTH mRNAs depend on PIN1 activity (62, 63, 96–98). Recent data also involve PIN1 in the regulation of microRNA biogenesis since it affects their nuclear-cytoplasmic shuttling in hepatocellular carcinoma (99).
Adenosine-to-inosine (A-to-I) RNA editing accounts for transcript diversification by modifying selected nucleotides in RNA molecules. Major enzymes responsible for this post-transcriptional modification are proteins of the ADAR family. One of these, ADAR2, was shown to be stabilized by PIN1 and to affect both transcript levels and permeability of the glutamate-gated ion channel receptor (GluR) (100). Later studies, investigating the landscape and regulation of editing in different tissues from a cohort of human and mouse samples revealed small effects of Pin1KO on global editing levels in a subset of mouse tissues like CNS, lung, spleen and small intestine (101).
All together these data clearly support a role for PIN1 in both gene-specific and global regulation of gene expression by modulating epigenetic regulators and transcription factors, as well as directly impacting the transcription machinery and post-transcriptional events. These data suggest that PIN1 could act as a determinant of cell identity by controlling the activity and expression programs of master TFs that define differentiation toward specific cellular lineages in normal cells (e.g., MEF2C for muscle cell identity, see section PIN1 Controls Stem Cell Reprogramming and Fate), while accumulating data argue for a potential role of PIN1 in supporting cancers that appear to be addicted to dysregulated transcriptional programs and elevated transcription rates, such as leukeamias and breast cancers (102). These tumors are frequently more sensitive to inhibition of the responsible transcriptional regulators than normal cells and thus these molecular circuitries represent interesting starting points for therapeutic interventions.
Impact of PIN1-Mediated Prolyl-Isomerization on Cellular Processes Connected With Tissue Growth, Development and Cancer
PIN1 Controls the Cell Cycle and Its Checkpoints
Like no other process cell cycle entry and progression, characterized by phosphorylation-dependent events, emphasize the multiple levels of intervention of PIN1. During the various steps of the cell cycle, the role of PIN1 in fine-tuning and regulating several phospho-proteins and thus the duration and intensity of various signals, is crucial to elicit cell cycle progression and cell division in response to pro-proliferative stimuli. Here we will only briefly review the role of PIN1 in key events of cell cycle and proliferation, and refer the reader to other reviews that have focused on this topic (7, 103).
While initial studies in yeast revealed a role for PIN1 in mitosis (27), further studies in mammalian cells unveiled that PIN1 dynamically controls cell cycle regulatory proteins and is required in all phases of the cell cycle (7, 10). PIN1 itself experiences moderate fluctuations of its levels and phosphorylation status in normal cells, which is critical to progress from one phase into the other: in particular CYCLIN D and E, whose levels are subjected to a rapid increase followed by degradation, strongly depend on PIN1 activity at both the transcriptional and post-transcriptional levels after phosphorylation by specific CDKs.
PIN1 controls the G1-S transition by directly repressing pRB. As a consequence pRB no longer inhibits E2F, and expression of CYCLIN D1 is induced (64, 65). Being also PIN1 an E2F target gene, its protein levels increase during the G1-S transition (67). The activation of CYCLIN D by PIN1 in turn further incites G1 phase CDKs to promote the transition from G1 to S-phase. PIN1 promotes CYCLIN E degradation and modulates p27Kip1 activity at a later stage (104–106), thus suggesting that PIN1 is controlling also the duration of the G1-S transition. Not surprisingly, Pin1KO MEFs exhibit defects in cell cycle re-entry after serum starvation (36). In S phase, PIN1 controls DNA and centrosome duplication (107) as well as the dynamic association of RNAPII on the promoters of transcribed genes (92). PIN1 also controls the G2-M transition, acting on the early mitotic inhibitor-1 (Emi1) and on the anaphase promoting complex (APC), thus coordinating the S and M phases (108). Moreover, PIN1 acts on CDC25C and WEE1 phosphatases, regulators of the CyclinB-CDC2 phosphorylation status (56, 109), thereby influencing entry in mitosis, and on separase and centrosomal proteins like CEP55 and Septin 9 during cytokinesis. All this evidence supports the idea that PIN1 is a fine tuner of cell cycle progression coordinating, in concert with cell cycle phase-specific CDKs and phosphatases, every phase of the cell cycle (103).
In addition, as shown in human, mouse, and zebrafish models, PIN1 participates in cell cycle checkpoints and promotes cell cycle arrest and/or apoptosis through activation of the tumor suppressor p53 (34, 79, 83, 110, 111) and its family member p73 (84), or by downregulating Che-1 (112) following genotoxic stress [for a comprehensive review see (6)]. Depending on the cell context and consistent with its role in boosting proliferation, PIN1 counteracts senescence (113, 114), an event that could be explained by the fact that PIN1 is able to regulate telomere length through TRF1 (115), cause inactivation of pRB (65), or target the senescence-inducing promyelocytic protein PML for degradation (116).
In human cancers PIN1 is prevalently overexpressed and essential for cancer cell proliferation and induction of centrosome amplification, two early clinico-pathological hallmarks of cancer (8). This has been observed in cell culture, in human tumors, and in MMTV-PIN1 mice and shown to depend on the activity that PIN1 exerts on a plethora of cell cycle regulators, such as CYCLIN D, CYCLIN E, and c-MYC, just to cite some of the most relevant (8). The question arises as of how this can be reconciled with the function of PIN1 in inducing cell cycle arrest or apoptosis in cooperation with p53? As already mentioned (6–8), PIN1 can impact on the fate of cancer cells depending on the levels and activity of relevant client tumor suppressors and proto-oncogenes (7, 8, 20). Indeed, PIN1 may eliminate incipient cancer cells by cooperating with wild-type p53. Instead, as during tumor development p53 inactivation often occurs either by alteration of its pathway or by deletion of its gene (TP53), high levels of PIN1 might boost the activity of oncogenes. Even worse, TP53 missense mutations that confer neomorphic oncogenic activities (117) are very frequent in human cancers and we have shown that PIN1 supports the oncogenic activities of these mutant p53 proteins in breast (16) and hepatocellular carcinomas (118).
PIN1 Behaves as a Metabolic Adaptor
Proper cell growth, proliferation and survival depend on cellular metabolism (119). Old and recent findings have implicated PIN1 in the context of cell metabolism and metabolic diseases (48) (Figure 5). For instance in yeast, Ess1 deletion causes a lethal phenotype that is rescued by overexpression of the delta-9-fatty-acid desaturase enzyme OLE1, or by supplying cells with unsaturated fatty acids. A key transcription factor of Ole1 is the endoplasmatic reticulum (ER) membrane protein Spt23 that gets activated by regulated proteolysis of the C-terminal membrane anchor through the ER-associated degradation (ERAD) pathway following mono/oligo-ubiquitylation. The N-terminal, cleaved part of Spt23 then translocates into the nucleus to drive transcription of Ole1. Ess1 finely tunes the ubiquitylation of Spt23, counteracting its poly-ubiquitylation and full degradation by ERAD, thus eliciting Ole1 expression and exerting a protective function against lipotoxic stress (120).
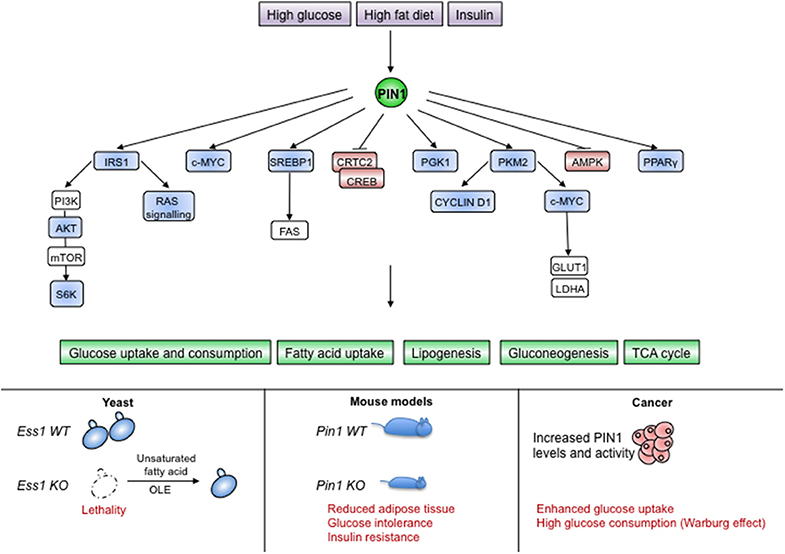
Figure 5. PIN1 is involved in sensing and transducing metabolic cues. (Upper part) Different nutritional and hormonal stimuli impinge on PIN1 levels and activity, which is necessary for the proper activation of metabolic pathways. (Lower part) Absence of PIN1 in unicellular organisms or metazoans causes lethality or metabolic alterations as shown for yeast and mice, respectively. Upregulation of PIN1 in human cancers supports the Warburg effect. Blue color indicates PIN1-activated proteins, red color indicates PIN1-inhibited proteins.
The involvement of PIN1 in fatty acid metabolism seems to be conserved also in mammals, as PIN1 was shown to favor the transcriptional activity of the Serum Response Element Binding Protein 1c (SREBP1c), a master regulator of the fatty acid synthesis pathway and homolog of Spt23. In fact, in response to EGF signaling in breast cancer cells, PIN1 spurs SREBP1-dependent induction of the fatty acid synthase FAS, the rate-limiting enzyme of the fatty acid biosynthesis pathway (121). Another report showed that PIN1 had no effects on SREBP1 mRNA levels in mice (122), suggesting that it could act on SREBP proteins in correspondence of the ER membrane or of the transcriptional activity in the nucleus.
In mice it has been shown that PIN1 becomes upregulated in response to high glucose (123), insulin, and a high-fat diet (124, 125) and that it manages metabolic responses to diet and hormone stimulation (125, 126). Intriguingly, PIN1 augments the uptake of triglycerides and fosters differentiation of MEFs into adipose cells in response to insulin, whereas PIN1KO mice are insulin resistant, have glucose intolerance and display lower adipose tissue weight when fed with high fat diet (125). These reports suggest a role for PIN1 as metabolic adaptor. Accordingly, PIN1 is strongly intertwined with insulin signaling, as witnessed by its requirement in converting signals from insulin receptor (IR), Insulin-like Growth Factor-1 receptor (IGF-1R), and Insulin Receptor Substrate 1 (IRS-1) into intracellular activation of the RAS-MAPKs cascade and the PI3K-AKT-mTOR-S6K axis (127, 128). PIN1 promotes this signaling by fostering phosphorylation and activation of IRS-1 (125) and it is in turn transcriptionally upregulated by this pathway (70). In addition PIN1 has been shown to regulate proteins connected to insulin signaling such as PPARγ (129), AMPK (130, 131), and CRTC1-2 (77).
Most functional changes within a cell come along with an adapted metabolic switch. For instance quiescent and differentiated cells display a metabolism in which glucose is fully degraded to CO2 by glycolysis coupled to the Tri-Carboxylic-Acid Cycle. On the contrary, rapidly proliferating cells switch to aerobic glycolysis in which glucose is fermented to lactate even in the presence of oxygen (Warburg effect) (119). Of note, PIN1 crucially impacts EGFR-promoted Warburg effect in cancer cells by inciting nuclear localization of the Pyruvate Kinase 2 (PKM2), which induces c-MYC expression, resulting in turn in the upregulation of the glucose transport protein 1 (GLUT1) and lactate dehydrogenase A (LDHA) (76). Nuclear PKM2 causes transcription of several other genes, including CYCLIN D1, directly linking glycolytic metabolism with cell cycle progression. In turn, both c-MYC and CYCLIN D1 are central regulators of G1/S progression of the cell cycle, eliciting PIN1 expression via E2F transcription factors (67). Accordingly, PIN1 and c-MYC were found co-over-expressed in breast cancer, driving a gene expression pattern largely composed of genes involved in ribosome biogenesis/ translation and metabolism and enriched in poor-outcome breast cancer subtypes (18). Similarly, activation of ERK by hypoxia and oncogenic mutations can induce PGK1 phosphorylation, PIN1-mediated isomerization and consequent mitochondrial translocation to suppress metabolism of mitochondrial pyruvate further promoting the Warburg effect (78).
It seems that the general action of PIN1 downstream of glucose, insulin, fat or other growth factors that cause a metabolic switch, is to improve glucose uptake, glycolysis and lipogenesis. This is relevant, since metabolic pathways are not only engaged as a biochemical response to diet or energy demand, but also as mechanisms by which cells adapt to different stimuli and control their fate (132). In this view, we might speculate that metabolic reprogramming by PIN1 could be determinant for its pro-oncogenic effects. Indeed, reconsidering older studies where lack of PIN1 was shown to curb tumor growth induced by oncogenes such as RAS, HER2 (15), or NOTCH1 (17, 58), we might speculate that this might result not from a block of proliferation per se, but rather by prior restriction of the glycolytic pathway.
PIN1 Controls Stem Cell Reprogramming and Fate
Organismal development and maintenance of tissue integrity are guaranteed by a correct balance between differentiation and stemness and by a proper determination of cell identity (133, 134).
High levels of PIN1 were shown to be relevant for the maintenance of pluripotency of mouse embryonic stem cells (ESCs) by upregulating the stemness factor NANOG (73). Later on it was shown that PIN1 is indispensable for the induction and maintenance of pluripotency in induced pluripotent stem cells (iPS) by interacting with phosphorylated OCT4, preventing its ubiquitination, boosting its protein levels and activity. Accordingly, PIN1 levels are higher in iPS and ablation of its activity suppresses colony formation (74) (Figure 6).
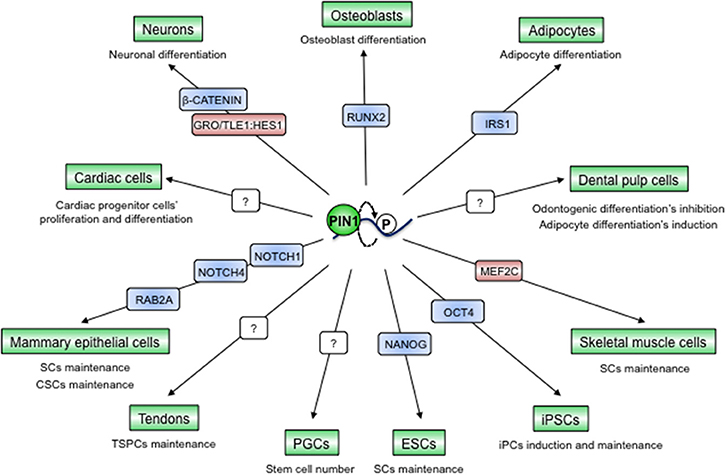
Figure 6. PIN1-dependent prolyl-isomerization serves as a pivotal hub to preserve tissue integrity and to maintain cancers' regenerative potential. PIN1 has been described as a fundamental element for the determination of cell fates in different tissues through interaction with and modulation of different proteins, mainly TFs. Blue color indicates PIN1-activated proteins, red color indicates PIN1-inhibited proteins, question marks indicate unknown protein substrates of PIN1.
The ability of PIN1 to control stemness features seems to be conserved also in stem cell niches of the adult. Indeed, the defects in many tissue compartments observed in Pin1KO mice could be partially explained by a role of PIN1 in regulating the balance between stemness and differentiation. For instance, PIN1 serves to maintain the numbers of primordial germ cells (PGCs) and, as a consequence, Pin1KO mice have impaired fertility (41). In addition, in the developing mouse cortex PIN1 promotes the neurogenic fate through repression of GRO/TLE1:HES1-mediated inhibition of neuronal differentiation on one hand (135) and through upregulation of β-CATENIN in neuronal progenitors on the other (43). On the contrary, PIN1 preserves muscle stem cells by targeting the MEF2C myogenic transcription factor for proteasomal degradation in the nucleus, while the nuclear-cytoplasmic shuttling of PIN1 triggers skeletal muscle cells' differentiation (136). Similar scenarios have been observed also in cardiac progenitor cells (114), adipocytes (126), osteoblasts (137, 138), dental pulp cells (139), and mammary stem cells (17, 140, 141) (Figure 6). All together these data raise the question as of whether the phenotypes observed in adult Pin1KO mice could derive also from impaired stem cell potency and a lower regenerative potential of particular tissues.
Like in the normal mammary gland, PIN1 acts as a fundamental regulator of stem cell features also in cancer stem cells (CSCs) of this tissue compartment. Indeed PIN1 critically controls CSC self-renewal, replicative potential and frequency and these cells are strongly addicted to high PIN1 levels and activity, as demonstrated in both human and mouse tumors (17, 140, 141). PIN1 was shown to regulate breast CSCs' expansion through regulation of the NOTCH1/4 and Rab2A GTPase pathways (17, 141).
A growing body of evidence highlighted that PIN1 can globally affect cellular plasticity and reprogramming by acting upstream and downstream to regulators of epithelial-to-mesenchymal transition (EMT) (17, 140, 142), an embryonic process that is hijacked during cancer progression. Absence of epithelial constraints underlies this phenomenon that preempts the detachment of invasive cancer cells from the primary tumor. High levels of PIN1 were shown to induce EMT and causing E-cadherin downregulation and Vimentin upregulation in both normal and cancerous breast epithelial cells (17, 66, 140) by activating Snail or Stat3 that act via PTEN-PI3K/AKT, NF-kappaB, NOTCH1/4 (17, 143), or oncostatin M (144), respectively.
All these results clearly indicate that PIN1 can be considered as a cell-fate determinant both in normal and in neoplastic contexts and that alterations of its levels, activity or sub-cellular localization could compromise the integrity of many tissues or incite neoplastic proclivity.
PIN1 Contributes to Orchestrate Cellular Responses to Cell-Intrinsic and -Extrinsic Cues
Cells exposed to bioactive stimuli and metabolites from inwards the cell or from the environment, such as growth factors, hormones, and xenobiotics, undergo adaptive responses to properly direct their fate and to safeguard cell and tissue integrity. PIN1 has a central role in tuning the ability of cells to both sense and transduce various stimuli and to elicit integrated cellular responses, thus determining cell fates during dynamic biological processes. While in normal conditions PIN1 activity ensures a homeostatic equilibrium, it appears that during pathological processes, such as inflammation and cancer, deregulated PIN1 exacerbates the diseased state.
For example PIN1 tunes the abundance and activity of both growth factor and hormone nuclear receptors and their molecular networks (10). Most notable examples are EGF and estrogen receptors (EGFR and ER) whose signaling cascades require the activity of PIN1. In fact, PIN1 stabilizes EGFR proteins and activates downstream kinases, such as MAPKs, PI3K-AKT, and CDK2 following growth factor stimulation. These kinases in turn phosphorylate ER, its co-repressor SMRT and its co-activator SRC-3 and elicit their interaction with PIN1. SMRT is subsequently targeted for degradation, while SRC-3 is activated, enhancing ultimately ER-dependent gene transcription (81). This cross-talk appears to be relevant for morphogenetic processes such as expansion of the mammary gland during pregnancy, while in breast cancers with high levels of PIN1, it confers resistance to selective estrogen receptor modulators, such as tamoxifen (81). PIN1 acts as a coactivator of several other hormone or growth factor receptor signaling pathways, ranging from Progesterone receptor to parathyroid hormone synthesis (63, 80, 145–147). Further investigation of the developmental consequences of such interactions is needed.
Integrated responses are required also following genotoxic stress. This is particularly relevant for DNA damage induction following hyperproliferative stimuli in pre-cancerous settings or by means of radio- and chemotherapies in tumor cells. Depending on the stimulus, the entity and the cell type, DNA damage can elicit cell cycle arrest, DNA repair, or apoptosis. As mentioned above, PIN1 coordinates all these processes by orchestrating both upstream and downstream functions of several factors, including ATR (148), p53, and p73 (6). For instance, PIN1 activity helps displacing negative regulators, such as Mdm2 (110, 111) and iASPP (83, 149), while activating upstream kinases, such as HIPK2 (79), to promote p53 nuclear and mitochondrial pro-apoptotic functions, respectively. Interestingly, downstream of chemotherapeutic treatments that induce double strand breaks (DSBs), PIN1 influences the choice of DNA DSB repair mechanisms. In particular high PIN1 levels promote non-homologous end-joining (NHEJ) at the expense of homology-directed recombination (HR), directly impinging on DNA ends resection by interacting with CtIP following its phosphorylation by CDK2 (150). Given that p53 regulates HR, the choice of the DSB repair pathway in cancer cells with high PIN1 levels may thus depend on whether p53 function is still present or lost (150).
Apart from its action in managing the consequences of cell damage induced by chemotherapeutics, in tumors PIN1 strongly correlates with chemoresistance, a major problem for the eradication of tumors and an unresolved cause of death. Indeed, we found that PIN1 inhibition in breast cancer xenografted mice sensitized tumors cells to paclitaxel treatment, not only by reducing the tumor mass but also by preventing the enrichment of the CSC population, a mechanism known to spur tumor recurrence after therapy (17). Such strong effects can be explained by considering the key role of PIN1 in boosting CSCs number, EMT features and the protein levels of factors involved in chemoresistance (NOTCH1, NOTCH4), pro-survival (BIRC-5, MCL-1), and detoxification (ABCG2) (17, 58, 151). In addition, PIN1 confers resistance to taxanes by reducing the activity of the Hippo pathway kinase LATS2, following CDK1 phophorylation (152).
An intriguing interplay between PIN1 and oxidative stress has emerged since the discovery that PIN1 is able to potentiate the mitochondrial accumulation of the lifespan regulator p66shc, thus inducing apoptosis in response to ROS (153). In accord with this finding, in human aortic endothelial cells exposed to high glucose, PIN1 is upregulated and promotes p66shc mitochondrial translocation, reduction of nitric oxide and activation of NF-kappaB dependent inflammation. Consequently, Pin1KO mice are protected against diabetes-induced mitochondrial oxidative stress, endothelial dysfunction, and vascular inflammation (123). In another publication Pin1KO MEFs were shown to resist to H2O2-induced cell death because of upregulation of phosphatidylinositol-5-phosphate (PtdIns5P) (154). The impact of PIN1 on the oxidative stress response is still poorly characterized and seems to be context and tissue dependent, and thus deserves further studies. Nevertheless, given that tumor progression depends on oxidative stress (155), the question arises as to whether PIN1 pro-oxidant activity could foster tumor growth and aggressiveness.
Regulation of Pin1 Expression, Subcellular Localization and Activity
Under physiological conditions, PIN1 levels and function are subjected to tight transcriptional and post-transcriptional regulation, at variance with other PPIases that are mainly constitutively expressed (156). First, scattered information about PIN1 expression in different organisms, such as mouse and zebrafish, show that PIN1 is prevalently expressed in embryonic and proliferating tissues (39), except for the central nervous system, in which PIN1 expression is induced following neuronal differentiation (34, 43, 135, 157). Moreover, in zebrafish embryos it was clearly demonstrated that mRNA and protein levels do not always correspond and, while established cell lines have prevalently nuclear PIN1, primary cell cultures or tissues often display cytoplasmic re-localization of PIN1 (see Ibarra et al. (34) and references therein). These and other studies suggest that in different developmental stages and tissues, PIN1 can be regulated not only at the level of mRNA expression, but also at those of mRNA stability and processing, as well as by critical PTMs and localization to specific cellular microdomains, that ensure proper adaptive cellular processes. Notably, while PIN1 function seems to be dispensable during development and adulthood of higher metazoans, its levels and activity enhance and become crucial for stress-induced cellular responses in vivo, such as during ionizing radiation in zebrafish (158), high glycemia (123), and high fat diet in mice (124).
The above-mentioned observations hold true also in conditions of tumor growth in mice and human models. Indeed, in spite of a very low mutation frequency of the PIN1 gene (8), in human cancers PIN1 is prevalently overexpressed (159). High PIN1 mRNA expression is strongly associated with cell proliferation. Indeed, PIN1 is transcriptionally regulated by E2F in response to growth factors (67, 70) and by NOTCH1/4 activation in cancer (17, 58). On the contrary, the PIN1 rs2233678 (−842G>C) promoter polymorphism is associated to a reduced expression of the gene and a reduced risk of cancer (50, 51). PIN1 is also negatively controlled by tumor-suppressor microRNAs, such as miR-200b (160), a promoter of anoikis, miR-200c (140) that restrains EMT in breast cancer, and by miR-296-5p in prostate cancer (161), and some others only recently identified (20). In addition to PIN1 transcription, the binding and enzymatic activities of PIN1, its cellular localization and turnover are also finely regulated at the post-translational level, mainly through phosphorylation, SUMOylation, and oxidation (162–170).
All these data clearly show that PIN1 levels and activity are regulated at several levels that can be deranged during oncogenesis and in turn have positive repercussions on oncogenic signaling and tumor phenotypes (Figure 2).
PIN1 Targeted Therapies to Treat Cancer
Current challenges for cancer therapy regard the possibility to simultaneously target multiple deregulated pathways that are causative for cancer progression and to effectively hit CSCs, while having none or acceptable side effects. PIN1 seems to be an ideal target that could conciliate all these requirements. Indeed, first of all, PIN1 is a unique enzyme that is overexpressed in many tumors. Second, Pin1KO or pharmacological inhibition drastically impairs tumor growth and dissemination (8, 19). Third, Pin1KO mice are viable and show only marginal defects. Such robust cellular effects on tumor growth and development that arise from loss of PIN1 function, while having milder consequences on normal tissues, are likely due to the modest effect on many members of the same oncogenic signal transduction pathways, rather than from a stark effect on one single client protein. Fourth, PIN1 has been shown to boost the CSC population in breast cancers and it represents a functional node for the cooperation between oncogenic circuitries, reason why tumors, but not normal tissues, are addicted to PIN1. Fifth, PIN1 knockdown or pharmacological inhibition sensitizes cancer cells of different origins to chemo- and target therapy, such as breast cancers to 5-fluorouracil, Taxol (151), and rapamycin (171), hepatocellular carcinoma to sorafenib (172), colon cancer to Taxol (72) and acute myeloid leukemia to retinoic acid (173). All this evidence and the enzymatic nature of PIN1 make it an attractive drug target.
Considerable effort for developing a PIN1 target therapy has been made, yet the provided inhibitors mainly display low potency, specificity, stability, and permeability [for comprehensive reviews about PIN1 inhibitors, see (7, 8, 174)]. Nonetheless, the use of liposomal formulations to vehicle a potent, but not cell-permeable, PIN1 inhibitor in cancer cells has shown effectiveness in curbing tumor growth in vivo (175), implying that such a strategy could be applied to other inhibitors with scarce pharmacokinetic properties.
We have recently discovered a PIN1 inhibitor with a peculiar mechanism of action. This new small molecule, KPT-6566, acts as a specific and covalent inhibitor of PIN1 that in addition causes its degradation. Intriguingly, from the KPT-6566-PIN1 binding reaction, a ROS-producing and DNA damaging molecule is released with exacerbated cytotoxic activity toward cancer cells. The robust tumor-killing activity observed both in vitro and in vivo very likely relies on the exhaustion of CSCs, ROS overload and impairment of anti-apoptotic pathways in bulk tumor cells (19).
All-trans retinoic acid (ATRA) and arsenic trioxide (ATO), recently discovered to inhibit PIN1 (166, 176), are the most attractive compounds to be rapidly repurposed as PIN1 inhibitors in clinical trials, being already FDA approved and showing efficacy against acute promyelocytic leukemia (APL) both alone and in combination. Still their usage poses challenges for an application against solid tumors, mainly because of the poor stability of ATRA (166, 176). Novel formulations of ATRA have now shown efficacy against liver cancers (177), paving the way toward an effective PIN1 therapy in humans.
Drug screenings with both synthetic and natural compound libraries have revealed several interesting hits that deserve further pharmacological development (7, 20, 178). Some of the drugs derived from natural sources have gained interest due to a reported effective PIN1 inhibition in vitro and in pre-clinical models, such as Juglone from Walnut tree or Epigallo-cathechin-gallate (EGCG) from green tea, even though they have toxic or non-specific activities against other cellular targets, respectively (7, 8, 174). Nevertheless, this prompted new efforts for finding PIN1 inhibitors by screening libraries of molecules derived from natural products (179, 180). Of special interest are also intracellular compounds, such as 4-Hydroxynonenal (HNE), a product arising from lipid oxidation, able to inhibit PIN1 by adducting Cysteine 113 within its catalytic pocket (181).
Conclusions and Perspectives
Regulation of the phosphorylation and transcriptional landscapes by PIN1 establishes this enzyme as a central effector of dynamic molecular switches imposed by environmental and cell-intrinsic cues. But, in spite of a huge amount of data reporting a role for PIN1 on specific substrates and pathways, a comprehensive picture of its biological effects is still elusive. In particular, a still puzzling aspect of PIN1 regards its ability in conferring different and often opposite cell fates by operating concomitantly on a multitude of proteins. This is an important question, since deregulation of such molecular programs likely underlies cancer development, where tumor suppressors are inactivated while oncogenes simultaneously upregulated. In addition, not much is known about how PIN1 function is developmentally regulated in vivo under defined circumstances and which are the biological consequences for single cells, tissues, and the whole organism.
Studies performed in animal and human models have begun to shed light on these issues. Indeed, they indicate that PIN1 expression, activity, and subcellular localization are complex and regulated at multiple levels in vivo. Moreover, they demonstrate how particular stress signals impact PIN1 activity, which is in turn crucial in transducing adequate or pathogenic cellular responses through reprogramming of different molecular networks at several levels (Figure 1). It will be important to find out which are the clinical and therapeutic consequences of blocking such signal transduction or the downstream molecular responses.
Along this vein, it has been observed that PIN1 is overexpressed in the majority of human tumors and higher levels of PIN1 are generally associated with poorer prognoses (7). However, PIN1 does not work as an independent prognostic factor in most human cancers, except for prostate cancer (182). We might speculate that PIN1 association with prognosis very likely is still ill defined, since most studies were based on either RNA or protein levels, that do not necessarily correlate with its activity. Indeed, both activating and inactivating PTMs on PIN1 have been observed in different cancers that often also correlated with specific subcellular localizations (7, 8). It is thus conceivable that an appropriate evaluation of the PIN1 activation status in tumor tissues will be more predictive of its activity rather than solely total protein or mRNA levels. In addition, the importance of PIN1 in cancer development may depend on the levels of key oncogenic substrates, such as mutant p53 proteins, that act as prognostic markers in association with PIN1 in breast cancer (16).
Another important level of regulation regards the impact of nutritional cues, such as glucose and lipids, on PIN1 biology. Enhanced glucose levels in fact cause transcriptional de-repression of the mouse Pin1 promoter, and upregulation of PIN1 protein levels that in turn spur ROS production and inflammation, highlighting possible novel mechanisms of PIN1 deregulation in pathogenic conditions such as obesity, diabetes, and cancer (48, 123). These insights obtained from studies in mouse models, coupled with the fact that PIN1 helps enforcing the Warburg effect at multiple levels, support the notion that one crucial developmental function of PIN1 is to integrate metabolic cues with cell proliferation, and that this is deregulated in cancer.
Such information will crucially impact future cancer prevention programs and therapies based on PIN1 inhibition. For instance, it will be important to assess the impact of the diet and life style on PIN1 also in humans, and to validate PIN1 as a potential biomarker and target for metabolic diseases such as diabetes (123) and cancer. In this context, also the nutritional status of neoplastic patients might be an important issue to consider since it is associated with tolerability to conventional therapies (183). Moreover, given the impact of PIN1 in p53-dependent apoptosis, PIN1 based therapies could underlie also radiation- and chemotherapy induced sickness in healthy tissues, especially in those with high PIN1 levels (6). It will thus be relevant to develop more specific and potent PIN1 inhibitors, and elaborate better combination therapies and strategies for drug delivery. In addition more sophisticated animal models are required to improve preclinical investigation, such as stage- and tissue-specific Pin1 conditional knock-out mice (124), or human genetically modified (e.g., gene edited) organoids that could open new horizons for the clinical investigation of PIN1-based therapies with immediate translational potential (184).
Author Contributions
AZ, AR, and GDS: conception and design and writing of the manuscript; EC: assisted in the preparation of the manuscript.
Funding
Work in our lab is supported by the Italian Association for Cancer Research (AIRC) Special Program Molecular Clinical Oncology 5 per mille (Grant n. 10016), (AIRC IG Grant n. 17659 and n. 22174), the Cariplo Foundation (Grant n. 2014-0812), funds from the Italian University and Research Ministry (PRIN-2015-8KZKE3), the European Regional Development Fund Interreg Italia-Österreich (PreCanMed ITAT1009), Regione Autonoma Friuli Venezia Giulia (Contributo ex art.15 L.R. 17/2014), Beneficentia-Stiftung, and Fondazione CRTrieste to GDS; EC is supported by a Fondazione Umberto Veronesi Post-doctoral fellowship.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Fiamma Mantovani, Francesco Napoletano, and Anna Testa for reading and discussion on the manuscript. We apologize to the colleagues whose contributions could not be cited due to space constraints.
References
1. Hynes NE, Ingham PW, Lim WA, Marshall CJ, Massagué J, Pawson T. Signalling change: signal transduction through the decades. Nat Rev Mol Cell Biol. (2013) 14:393–8. doi: 10.1038/nrm3581
2. Khoury GA, Baliban RC, Floudas CA. Proteome-wide post-translational modification statistics: frequency analysis and curation of the swiss-prot database. Sci Rep. (2011) 1:90. doi: 10.1038/srep00090
3. Verdecia MA, Bowman ME, Lu KP, Hunter T, Noel JP. Structural basis for phosphoserine-proline recognition by group IV WW domains. Nat Struct Biol. (2000) 7:639–43. doi: 10.1038/77929
4. Ranganathan R, Lu KP, Hunter T, Noel JP. Structural and functional analysis of the mitotic rotamase Pin1 suggests substrate recognition is phosphorylation dependent. Cell (1997) 89:875–86. doi: 10.1016/S0092-8674(00)80273-1
5. Liou YC, Zhou XZ, Lu KP. Prolyl isomerase Pin1 as a molecular switch to determine the fate of phosphoproteins. Trends Biochem Sci. (2011) 36:501–14. doi: 10.1016/j.tibs.2011.07.001
6. Mantovani F, Zannini A, Rustighi A, Del Sal G. Interaction of p53 with prolyl isomerases: healthy and unhealthy relationships. Biochim Biophys Acta Gen Subj. (2015) 1850:2048–60. doi: 10.1016/j.bbagen.2015.01.013
7. Zhou XZ, Lu KP. The isomerase PIN1 controls numerous cancer-driving pathways and is a unique drug target. Nat Rev Cancer (2016) 16:463–78. doi: 10.1038/nrc.2016.49
8. Rustighi A, Zannini A, Campaner E, Ciani Y, Piazza S, Del Sal G. PIN1 in breast development and cancer: a clinical perspective. Cell Death Differ. (2017) 24:200–11. doi: 10.1038/cdd.2016.122
9. Franciosa G, Diluvio G, Del Gaudio F, Giuli MV, Palermo R, Grazioli P, et al. Prolyl-isomerase Pin1 controls Notch3 protein expression and regulates T-ALL progression. Oncogene (2016) 35:4741–51. doi: 10.1038/onc.2016.5
10. Lu Z, Hunter T. Prolyl isomerase Pin1 in cancer. Cell Res. (2014) 24:1033–49. doi: 10.1038/cr.2014.109
11. La Montagna R, Caligiuri I, Giordano A, Rizzolio F. Pin1 and nuclear receptors: a new language? J Cell Physiol. (2013) 228:1799–801. doi: 10.1002/jcp.24316
12. Polonio-Vallon T, Krüger D, Hofmann TG. ShaPINg cell fate upon DNA damage: role of Pin1 isomerase in DNA damage-induced cell death and repair. Front Oncol. (2014) 4:148. doi: 10.3389/fonc.2014.00148
13. Lu KP, Zhou XZ. The prolyl isomerase PIN1: a pivotal new twist in phosphorylation signalling and disease. Nat Rev Mol Cell Biol. (2007) 8:904–16. doi: 10.1038/nrm2261
14. D'Artista L, Bisso A, Piontini A, Doni M, Verrecchia A. Pin1 is required for sustained B cell proliferation upon oncogenic activation of Myc. Oncotarget (2016) 7:21786–98. doi: 10.18632/oncotarget.7846
15. Wulf G, Garg P, Liou Y-C, Iglehart D, Lu KP. Modeling breast cancer in vivo and ex vivo reveals an essential role of Pin1 in tumorigenesis. EMBO J. (2004) 23:3397–407. doi: 10.1038/sj.emboj.7600323
16. Girardini JE, Napoli M, Piazza S, Rustighi A, Marotta C, Radaelli E, et al. A Pin1/mutant p53 axis promotes aggressiveness in breast cancer. Cancer Cell (2011) 20:79–91. doi: 10.1016/j.ccr.2011.06.004
17. Rustighi A, Zannini A, Tiberi L, Sommaggio R, Piazza S, Sorrentino G, et al. Prolyl-isomerase Pin1 controls normal and cancer stem cells of the breast. EMBO Mol Med. (2014) 6:99–119. doi: 10.1002/emmm.201302909
18. Farrell AS, Pelz C, Wang X, Daniel CJ, Wang Z, Su Y, et al. Pin1 regulates the dynamics of c-Myc DNA binding to facilitate target gene regulation and oncogenesis. Mol Cell Biol. (2013) 33:2930–49. doi: 10.1128/MCB.01455-12
19. Campaner E, Rustighi A, Zannini A, Cristiani A, Piazza S, Ciani Y, et al. A covalent PIN1 inhibitor selectively targets cancer cells by a dual mechanism of action. Nat Commun. (2017) 8:15772. doi: 10.1038/ncomms15772
20. Chen Y, Wu T, Yang H, Li X, Jie M, Hu C, et al. Prolyl-isomerase Pin1: a promoter of cancer and a target for therapy. Cell Death Dis. (2018) 9:883. doi: 10.1038/s41419-018-0844-y
21. Hanes SD. Prolyl isomerases in gene transcription. Biochim Biophys Acta Gen Subj. (2015) 1850:2017–34. doi: 10.1016/j.bbagen.2014.10.028
22. Mueller JW, Bayer P. Small family with key contacts: Par14 and Par17 parvulin proteins, relatives of Pin1, now Emerge in biomedical research. Perspect Medicin Chem. (2008) 2008:11–20. doi: 10.4137/PMC.S496
23. Zhang J, Nakatsu Y, Shinjo T, Guo Y, Sakoda H, Yamamotoya T, et al. Par14 protein associates with insulin receptor substrate 1 (IRS-1), thereby enhancing insulin-induced IRS-1. J Biol Chem. (2013) 288:20692–701. doi: 10.1074/jbc.M113.485730
24. Campbell HD, Webb GC, Fountain S, Young IG. The human PIN1 peptidyl-prolyl cis/trans isomerase gene maps to human chromosome 19p13 and the closely related PIN1L gene to 1p31. Genomics (1997) 44:157–62. doi: 10.1006/geno.1997.4854
25. Zhu JX, Dagostino E, Rejto PA, Mroczkowski B, Murray B. Identification and characterization of a novel and functional murine Pin1 isoform. Biochem Biophys Res Commun. (2007) 359:529–35. doi: 10.1016/j.bbrc.2007.05.124
26. Hani J, Stumpf G, Domdey H. PTF1 encodes an essential protein in Saccharomyces cerevisiae, which shows strong homology with a new putative family of PPIases. FEBS Lett. (1995) 365:198–202. doi: 10.1016/0014-5793(95)00471-K
27. Hanes SD, Shank PR, Bostian KA. Sequence and mutational analysis of ESS1, a gene essential for growth in Saccharomyces cerevisiae. Yeast (1989) 5:55–72. doi: 10.1002/yea.320050108
28. Huang HK, Forsburg SL, John UP, O'Connell MJ, Hunter T. Isolation and characterization of the Pin1/Ess1p homologue in Schizosaccharomyces pombe. J Cell Sci. (2001) 114:3779–88.
29. Joseph JD, Daigle SN, Means AR. PINA is essential for growth and positively influences NIMA function in Aspergillus nidulans. J Biol Chem. (2004) 279:32373–84. doi: 10.1074/jbc.M405415200
30. Kops O, Eckerskorn C, Hottenrott S, Fischer G, Mi H, Tropschug M. Ssp1, a site-specific parvulin homolog from Neurospora crassa active in protein folding. J Biol Chem. (1999) 273:31971–6. doi: 10.1074/jbc.273.48.31971
31. Devasahayam G, Chaturvedi V, Hanes SD. The ESS1 prolyl isomerase is required for growth and morphogenetic switching in Candida albicans. Genetics (2002) 160:37–48. doi: 10.1093/ser/mwx024
32. Maleszka R, Hanes SD, Hackett RL, de Couet HG, Miklos GL. The Drosophila melanogaster dodo (dod) gene, conserved in humans, is functionally interchangeable with the ESS1 cell division gene of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. (1996) 93:447–51. doi: 10.1073/pnas.93.1.447
33. Maleszka R, Lupas A, Hanes SD, Miklos GLG. The dodo gene family encodes a novel protein involved in signal transduction and protein folding. Gene (1997) 203:89–93. doi: 10.1016/S0378-1119(97)00522-2
34. Ibarra MS, Etichetti CB, Di Benedetto C, Rosano GL, Margarit E, Del Sal G, et al. Dynamic regulation of Pin1 expression and function during zebrafish development. PLoS ONE (2017) 12:e0175939. doi: 10.1371/journal.pone.0175939
35. Winkler KE, Swenson KI, Kornbluth S, Means AR. Requirement of the prolyl isomerase Pin1 for the replication checkpoint. Science (2000) 287:1644–7. doi: 10.1126/science.287.5458.1644
36. Fujimori F, Takahashi K, Uchida C, Uchida T. Mice lacking Pin1 develop normally, but are defective in entering cell cycle from G(0) arrest. Biochem Biophys Res Commun. (1999) 265:658–63. doi: 10.1006/bbrc.1999.1736
37. Wildemann D, Hernandez Alvarez B, Stoller G, Zhou XZ, Lu KP, Erdmann F, et al. An essential role for Pin1 in Xenopus laevis embryonic development revealed by specific inhibitors. Biol Chem. (2007) 388:1103–11. doi: 10.1515/BC.2007.127
38. Hsu T, McRackan D, Vincent TS, Gert de Couet H. Drosophila Pin1 prolyl isomerase Dodo is a MAP kinase signal responder during oogenesis. Nat Cell Biol. (2001) 3:538–43. doi: 10.1038/35078508
39. Liou Y-C, Ryo A, Huang H-K, Lu P-J, Bronson R, Fujimori F, et al. Loss of Pin1 function in the mouse causes phenotypes resembling cyclin D1-null phenotypes. Proc Natl Acad Sci USA. (2002) 99:1335–40. doi: 10.1073/pnas.032404099
40. Atchison FW, Means AR. Spermatogonial depletion in adult Pin1-deficient mice. Biol Reprod. (2003) 69:1989–97. doi: 10.1095/biolreprod.103.020859
41. Atchison FW, Capel B, Means AR. Pin1 regulates the timing of mammalian primordial germ cell proliferation. Development (2003) 130:3579–86. doi: 10.1242/dev.00584
42. Kang SW, Lee E, Cho E, Seo JH, Ko HW, Kim EY. Drosophila peptidyl-prolyl isomerase Pin1 modulates circadian rhythms via regulating levels of PERIOD. Biochem Biophys Res Commun. (2015) 463:235–40. doi: 10.1016/j.bbrc.2015.05.033
43. Nakamura K, Kosugi I, Lee DY, Hafner A, Sinclair DA, Ryo A, et al. Prolyl isomerase Pin1 regulates neuronal differentiation via β-catenin. Mol Cell Biol. (2012) 32:2966–78. doi: 10.1128/MCB.05688-11
44. Shen Z-J, Hu J, Ali A, Pastor J, Shiizaki K, Blank RD, et al. Pin1 null mice exhibit low bone mass and attenuation of BMP signaling. PLoS ONE (2013) 8:e63565. doi: 10.1371/journal.pone.0063565
45. Liou Y-C, Sun A, Ryo A, Zhou XZ, Yu Z-X, Huang H-K, et al. Role of the prolyl isomerase Pin1 in protecting against age-dependent neurodegeneration. Nature (2003) 424:556–61. doi: 10.1038/nature01832
46. Pastorino L, Sun A, Lu PJ, Xiao ZZ, Balastik M, Finn G, et al. The prolyl isomerase Pin1 regulates amyloid precursor protein processing and amyloid-β production. Nature (2006) 440:528–34. doi: 10.1038/nature04543
47. Lu KP, Hanes SD, Hunter T. A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature (1996) 380:544–7. doi: 10.1038/380544a0
48. Nakatsu Y, Matsunaga Y, Yamamotoya T, Ueda K, Inoue Y, Mori K, et al. Physiological and pathogenic roles of prolyl isomerase pin1 in metabolic regulations via multiple signal transduction pathway modulations. Int J Mol Sci. (2016) 17:E1495. doi: 10.3390/ijms17091495
49. Driver JA, Zhou XZ, Lu KP. Pin1 dysregulation helps to explain the inverse association between cancer and Alzheimer's disease. Biochim Biophys Acta Gen Subj. (2015) 1850:2069–76. doi: 10.1016/j.bbagen.2014.12.025
50. Han CH, Lu J, Wei Q, Bondy ML, Brewster AM, Yu TK, et al. The functional promoter polymorphism (-842G>C) in the PIN1 gene is associated with decreased risk of breast cancer in non-Hispanic white women 55 years and younger. Breast Cancer Res Treat. (2010) 122:243–249. doi: 10.1007/s10549-009-0682-9
51. Tao LJ, Chen YS, Yao L, Zou B, Tao LS, Kong J, et al. PIN1 promoter polymorphism (-842 G>C) contributes to a decreased risk of cancer: evidence from meta-analysis. Oncol Lett. (2014) 8:1360–6. doi: 10.3892/ol.2014.2280
52. Ma SL, Tang NLS, Tam CWC, Lui VWC, Lam LCW, Chiu HFK, et al. A PIN1 polymorphism that prevents its suppression by AP4 associates with delayed onset of Alzheimer's disease. Neurobiol Aging (2012) 33:804–13. doi: 10.1016/j.neurobiolaging.2010.05.018
53. Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell (2006) 127:635–48. doi: 10.1016/j.cell.2006.09.026
54. Jin J, Pawson T. Modular evolution of phosphorylation-based signalling systems. Philos Trans R Soc B Biol Sci. (2012) 367:2540–55. doi: 10.1098/rstb.2012.0106
55. Litchfield DW, Shilton BH, Brandl CJ, Gyenis L. Pin1: intimate involvement with the regulatory protein kinase networks in the global phosphorylation landscape. Biochim Biophys Acta Gen Subj. (2015) 1850:2077–86. doi: 10.1016/j.bbagen.2015.02.018
56. Zhou XZ, Kops O, Werner A, Lu PJ, Shen M, Stoller G, et al. Pin1-dependent prolyl isomerization regulates dephosphorylation of Cdc25C and Tau proteins. Mol Cell (2000) 6:873–83. doi: 10.1016/S1097-2765(05)00083-3
57. Yeh E, Cunningham M, Arnold H, Chasse D, Monteith T, Ivaldi G, et al. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat Cell Biol. (2004) 6:308–18. doi: 10.1038/ncb1110
58. Rustighi A, Tiberi L, Soldano A, Napoli M, Nuciforo P, Rosato A, et al. The prolyl-isomerase Pin1 is a Notch1 target that enhances Notch1 activation in cancer. Nat Cell Biol. (2009) 11:133–42. doi: 10.1038/ncb1822
59. Dougherty MK, Müller J, Ritt DA, Zhou M, Zhou XZ, Copeland TD, et al. Regulation of Raf-1 by direct feedback phosphorylation. Mol Cell (2005) 17:215–24. doi: 10.1016/j.molcel.2004.11.055
60. Yeh ES, Lew BO, Means AR. The loss of PIN1 deregulates cyclin E and sensitizes mouse embryo fibroblasts to genomic instability. J Biol Chem. (2006) 281:241–51. doi: 10.1074/jbc.M505770200
61. van Drogen F, Sangfelt O, Malyukova A, Matskova L, Yeh E, Means AR, et al. Ubiquitylation of cyclin E requires the sequential function of SCF complexes containing distinct hCdc4 isoforms. Mol Cell (2006) 23:37–48. doi: 10.1016/j.molcel.2006.05.020
62. Krishnan N, Titus MA, Thapar R. The prolyl isomerase pin1 regulates mRNA levels of genes with short half-lives by targeting specific RNA binding proteins. PLoS ONE (2014) 9:e85427. doi: 10.1371/journal.pone.0085427
63. Nechama M, Uchida T, Mor Yosef-Levi I, Silver J, Naveh-Many T. The peptidyl-prolyl isomerase Pin1 determines parathyroid hormone mRNA levels and stability in rat models of secondary hyperparathyroidism. J Clin Invest. (2009) 119:3102–14. doi: 10.1172/JCI39522
64. Rizzolio F, Lucchetti C, Caligiuri I, Marchesi I, Caputo M, Klein-Szanto AJ, et al. Retinoblastoma tumor-suppressor protein phosphorylation and inactivation depend on direct interaction with Pin1. Cell Death Differ. (2012) 19:1152–61. doi: 10.1038/cdd.2011.202
65. Tong Y, Ying H, Liu R, Li L, Bergholz J, Xiao Z-X. Pin1 inhibits PP2A-mediated Rb dephosphorylation in regulation of cell cycle and S-phase DNA damage. Cell Death Dis. (2015) 6:e1640. doi: 10.1038/cddis.2015.3
66. Wulf GM, Ryo A, Wulf GG, Lee SW, Niu T, Petkova V, et al. Pin1 is overexpressed in breast cancer and cooperates with Ras signaling in increasing the transcriptional activity of c-Jun towards cyclin D1. EMBO J. (2001) 20:3459–72. doi: 10.1093/emboj/20.13.3459
67. Ryo A, Liou Y-C, Wulf G, Nakamura M, Lee SW, Lu KP. PIN1 is an E2F target gene essential for Neu/Ras-induced transformation of mammary epithelial cells. Mol Cell Biol. (2002) 22:5281–95. doi: 10.1128/MCB.22.15.5281
68. Ryo A, Nakamura M, Wulf G, Liou YC, Lu KP. Pin1 regulates turnover and subcellular localization of beta-catenin by inhibiting its interaction with APC. Nat Cell Biol. (2001) 3:793–801. doi: 10.1038/ncb0901-793
69. Ryo A, Suizu F, Yoshida Y, Perrem K, Liou YC, Wulf G, et al. Regulation of NF-κB signaling by Pin1-dependent prolyl isomerization and ubiquitin-mediated proteolysis of p65/RelA. Mol Cell (2003) 12:1413–26. doi: 10.1016/S1097-2765(03)00490-8
70. You H, Zheng H, Murray SA, Yu Q, Uchida T, Fan D, et al. IGF-1 induces Pin1 expression in promoting cell cycle S-phase entry. J Cell Biochem. (2001) 84:211–6. doi: 10.1002/jcb.10037
71. Skaar JR, Pagan JK, Pagano M. SCF ubiquitin ligase-targeted therapies. Nat Rev Drug Discov. (2014) 13:889–903. doi: 10.1038/nrd4432
72. Min SH, Lau AW, Lee TH, Inuzuka H, Wei S, Huang P, et al. Negative regulation of the stability and tumor suppressor function of Fbw7 by the Pin1 prolyl isomerase. Mol Cell (2012) 46:771–83. doi: 10.1016/j.molcel.2012.04.012
73. Moretto-Zita M, Jin H, Shen Z, Zhao T, Briggs SP, Xu Y. Phosphorylation stabilizes Nanog by promoting its interaction with Pin1. Proc Natl Acad Sci USA. (2010) 107:13312–7. doi: 10.1073/pnas.1005847107
74. Nishi M, Akutsu H, Masui S, Kondo A, Nagashima Y, Kimura H, et al. A distinct role for Pin1 in the induction and maintenance of pluripotency. J Biol Chem. (2011) 286:11593–603. doi: 10.1074/jbc.M110.187989
75. Restelli M, Lopardo T, Lo Iacono N, Garaffo G, Conte D, Rustighi A, et al. DLX5, FGF8 and the Pin1 isomerase control ΔNp63α protein stability during limb development: a regulatory loop at the basis of the SHFM and EEC congenital malformations. Hum Mol Genet. (2014) 23:3830–42. doi: 10.1093/hmg/ddu096
76. Yang W, Zheng Y, Xia Y, Ji H, Chen X, Guo F, et al. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat Cell Biol. (2012) 14:1295–304. doi: 10.1038/ncb2629
77. Nakatsu Y, Sakoda H, Kushiyama A, Ono H, Fujishiro M, Horike N, et al. Pin1 associates with and induces translocation of CRTC2 to the cytosol, thereby suppressing cAMP-responsive element transcriptional activity. J Biol Chem. (2010) 285:33018–27. doi: 10.1074/jbc.M110.137836
78. Li X, Jiang Y, Meisenhelder J, Yang W, Hawke DH, Zheng Y, et al. Mitochondria-translocated PGK1 functions as a protein kinase to coordinate glycolysis and the TCA cycle in tumorigenesis. Mol Cell (2016) 61:705–19. doi: 10.1016/j.molcel.2016.02.009
79. Sorrentino G, Mioni M, Giorgi C, Ruggeri N, Pinton P, Moll U, et al. The prolyl-isomerase Pin1 activates the mitochondrial death program of p53. Cell Death Differ. (2013) 20:198–208. doi: 10.1038/cdd.2012.112
80. La Montagna R, Caligiuri I, Maranta P, Lucchetti C, Esposito L, Paggi MG, et al. Androgen receptor serine 81 mediates Pin1 interaction and activity. Cell Cycle (2012) 11:3415–20. doi: 10.4161/cc.21730
81. Ryo A, Wulf G, Lee TH, Lu KP. Pinning down HER2-ER crosstalk in SMRT regulation. Trends Biochem Sci. (2009) 34:162–5. doi: 10.1016/j.tibs.2008.12.004
82. Lucchetti C, Caligiuri I, Toffoli G, Giordano A, Rizzolio F. The prolyl isomerase Pin1 acts synergistically with CDK2 to regulate the basal activity of estrogen receptor α in breast cancer. PLoS ONE (2013) 8:e55355. doi: 10.1371/journal.pone.0055355
83. Mantovani F, Tocco F, Girardini J, Smith P, Gasco M, Lu X, et al. The prolyl isomerase Pin1 orchestrates p53 acetylation and dissociation from the apoptosis inhibitor iASPP. Nat Struct Mol Biol. (2007) 14:912–20. doi: 10.1038/nsmb1306
84. Mantovani F, Piazza S, Gostissa M, Strano S, Zacchi P, Mantovani R, et al. Pin1 links the activities of c-Abl and p300 in regulating p73 function. Mol Cell (2004) 14:625–36. doi: 10.1016/j.molcel.2004.05.007
85. Helander S, Montecchio M, Pilstål R, Su Y, Kuruvilla J, Elvén M, et al. Pre-anchoring of Pin1 to unphosphorylated c-Myc in a fuzzy complex regulates c-Myc activity. Structure (2015) 23:2267–79. doi: 10.1016/j.str.2015.10.010
86. Flavahan WA, Gaskell E, Bernstein BE. Epigenetic plasticity and the hallmarks of cancer. Science (2017) 357:eaal2380. doi: 10.1126/science.aal2380
87. Arévalo-Rodríguez M, Cardenas ME, Wu X, Hanes SD, Heitman J. Cyclophilin A and Ess1 interact with and regulate silencing by the Sin3-Rpd3 histone deacetylase. EMBO J. (2000) 19:3739–49. doi: 10.1093/emboj/19.14.3739
88. Rajendran P, Delage B, Dashwood WM, Yu TW, Wuth B, Williams DE, et al. Histone deacetylase turnover and recovery in sulforaphane-treated colon cancer cells: competing actions of 14-3-3 and Pin1 in HDAC3/SMRT corepressor complex dissociation/reassembly. Mol Cancer (2011) 10:e85427. doi: 10.1186/1476-4598-10-68
89. Khanal P, Kim G, Lim SC, Yun HJ, Lee KY, Choi HK, et al. Prolyl isomerase Pin1 negatively regulates the stability of SUV39H1 to promote tumorigenesis in breast cancer. FASEB J. (2013) 27:4606–18. doi: 10.1096/fj.13-236851
90. Hu X, Dong SH, Chen J, Zhou XZ, Chen R, Nair S, et al. Prolyl isomerase PIN1 regulates the stability, transcriptional activity and oncogenic potential of BRD4. Oncogene (2017) 36:5177–88. doi: 10.1038/onc.2017.137
91. Raghuram N, Strickfaden H, McDonald D, Williams K, Fang H, Mizzen C, et al. Pin1 promotes histone H1 dephosphorylation and stabilizes its binding to chromatin. J Cell Biol. (2013) 203:57–71. doi: 10.1083/jcb.201305159
92. Xu Y-X, Manley JL. Pin1 modulates RNA polymerase II activity during the transcription cycle. Genes Dev. (2007) 21:2950–62. doi: 10.1101/gad.1592807
93. Xu Y-X, Hirose Y, Zhou XZ, Lu KP, Manley JL. Pin1 modulates the structure and function of human RNA polymerase II. Genes Dev. (2003) 17:2765–76. doi: 10.1101/gad.1135503
94. Kops O, Zhou XZ, Lu KP. Pin1 modulates the dephosphorylation of the RNA polymerase II C-terminal domain by yeast Fcp1. FEBS Lett. (2002) 513:305–11. doi: 10.1016/S0014-5793(02)02288-3
95. Krishnan N, Lam TT, Fritz A, Rempinski D, O'Loughlin K, Minderman H, et al. The prolyl isomerase Pin1 targets stem-loop binding protein (SLBP) to dissociate the SLBP-histone mRNA complex linking histone mRNA decay with SLBP ubiquitination. Mol Cell Biol. (2012) 32:4306–22. doi: 10.1128/MCB.00382-12
96. Shen Z-J, Esnault S, Malter JS. The peptidyl-prolyl isomerase Pin1 regulates the stability of granulocyte-macrophage colony-stimulating factor mRNA in activated eosinophils. Nat Immunol. (2005) 6:1280–7. doi: 10.1038/ni1266
97. Shen Z-J, Esnault S, Rosenthal LA, Szakaly RJ, Sorkness RL, Westmark PR, et al. Pin1 regulates TGF-beta1 production by activated human and murine eosinophils and contributes to allergic lung fibrosis. J Clin Invest. (2008) 118:479–90. doi: 10.1172/JCI32789
98. Esnault S, Shen Z-J, Whitesel E, Malter JS. The peptidyl-prolyl isomerase Pin1 regulates granulocyte-macrophage colony-stimulating factor mRNA stability in T lymphocytes. J Immunol. (2006) 177:6999–7006. doi: 10.4049/jimmunol.177.10.6999
99. Li J, Pu W, Sun HL, Zhou JK, Fan X, Zheng Y, et al. Pin1 impairs microRNA biogenesis by mediating conformation change of XPO5 in hepatocellular carcinoma. Cell Death Differ. (2018) 25:1612–24. doi: 10.1038/s41418-018-0065-z
100. Marcucci R, Brindle J, Paro S, Casadio A, Hempel S, Morrice N, et al. Pin1 and WWP2 regulate GluR2 Q/R site RNA editing by ADAR2 with opposing effects. EMBO J. (2011) 30:4211–22. doi: 10.1038/emboj.2011.303
101. Tan MH, Li Q, Shanmugam R, Piskol R, Kohler J, Young AN, et al. Dynamic landscape and regulation of RNA editing in mammals. Nature (2017) 550:249–54. doi: 10.1038/nature24041
102. Bradner JE, Hnisz D, Young RA. Transcriptional addiction in cancer. Cell (2017) 168:629–43. doi: 10.1016/j.cell.2016.12.013
103. Lin CH, Li HY, Lee YC, Calkins MJ, Lee KH, Yang CN, et al. Landscape of Pin1 in the cell cycle. Exp Biol Med. (2015) 240:403–8. doi: 10.1177/1535370215570829
104. Zhou W, Yang Q, Low CB, Karthik BC, Wang Y, Ryo A, et al. Pin1 catalyzes conformational changes of Thr-187 in p27Kip1and mediates its stability through a polyubiquitination process. J Biol Chem. (2009) 284:23980–8. doi: 10.1074/jbc.M109.022814
105. Brenkman AB, De Keizer PLJ, Van Den Broek NJF, Der Van Groep P, Van Diest PJ, Van Der Horst A, et al. The peptidyl-isomerase Pin1 regulates p27kip1 expression through inhibition of Forkhead box O tumor suppressors. Cancer Res. (2008) 68:7597–605. doi: 10.1158/0008-5472.CAN-08-1059
106. Cheng CW, Leong KW, Ng YM, Kwong YL, Tse E. The peptidyl-prolyl isomerase PIN1 relieves cyclin-dependent kinase 2 (CDK2) inhibition by the CDK inhibitor p27. J Biol Chem. (2017) 292:21431–41. doi: 10.1074/jbc.M117.801373
107. Suizu F, Ryo A, Wulf G, Lim J, Lu KP. Pin1 regulates centrosome duplication, and its overexpression induces centrosome amplification, chromosome instability, and oncogenesis. Mol Cell Biol. (2006) 26:1463–79. doi: 10.1128/MCB.26.4.1463-1479.2006
108. Bernis C, Vigneron S, Burgess A, Labbé JC, Fesquet D, Castro A, et al. Pin1 stabilizes Emi1 during G2 phase by preventing its association with SCFβtrcp. EMBO Rep. (2007) 8:91–8. doi: 10.1038/sj.embor.7400853
109. Okamoto K, Sagata N. Mechanism for inactivation of the mitotic inhibitory kinase Wee1 at M phase. Proc Natl Acad Sci USA. (2007) 104:3753–8. doi: 10.1073/pnas.0607357104
110. Zacchi P, Gostissa M, Uchida T, Salvagno C, Avolio F, Volinia S, et al. The prolyl isomerase Pin1 reveals a mechanism to control p53 functions after genotoxic insults. Nature (2002) 419:853–7. doi: 10.1038/nature01120
111. Zheng H, You H, Zhou XZ, Murray SA, Uchida T, Wulf G, et al. The prolyl isomerase Pin1 is a regulator of p53 in genotoxic response. Nature (2002) 419:849–53. doi: 10.1038/nature01220
112. De Nicola F, Bruno T, Iezzi S, Di Padova M, Floridi A, Passananti C, et al. The prolyl isomerase Pin1 affects Che-1 stability in response to apoptotic DNA damage. J Biol Chem. (2007) 282:19685–91. doi: 10.1074/jbc.M610282200
113. Wheaton K, Muir J, Ma W, Benchimol S. BTG2 antagonizes Pin1 in response to mitogens and telomere disruption during replicative senescence. Aging Cell (2010) 9:747–60. doi: 10.1111/j.1474-9726.2010.00601.x
114. Toko H, Hariharan N, Konstandin MH, Ormachea L, McGregor M, Gude NA, et al. Differential regulation of cellular senescence and differentiation by prolyl isomerase Pin1 in cardiac progenitor cells. J Biol Chem. (2014) 289:5348–56. doi: 10.1074/jbc.M113.526442
115. Lee TH, Tun-Kyi A, Shi R, Lim J, Soohoo C, Finn G, et al. Essential role of Pin1 in the regulation of TRF1 stability and telomere maintenance. Nat Cell Biol. (2009) 11:97–105. doi: 10.1038/ncb1818
116. Yuan W-C, Lee Y-R, Huang S-F, Lin Y-M, Chen T-Y, Chung H-C, et al. A Cullin3-KLHL20 Ubiquitin ligase-dependent pathway targets PML to potentiate HIF-1 signaling and prostate cancer progression. Cancer Cell (2011) 20:214–28. doi: 10.1016/j.ccr.2011.07.008
117. Mantovani F, Walerych D, Del Sal G. Targeting mutant p53 in cancer: a long road to precision therapy. FEBS J. (2017) 284:837–50. doi: 10.1111/febs.13948
118. Liao P, Zeng SX, Zhou X, Chen T, Zhou F, Cao B, et al. Mutant p53 gains its function via c-Myc activation upon CDK4 phosphorylation at serine 249 and consequent PIN1 binding. Mol Cell (2017) 68:1134–46.e6. doi: 10.1016/j.molcel.2017.11.006
119. Ward PS, Thompson CB. Signaling in control of cell growth and metabolism. Cold Spring Harb Perspect Biol. (2012) 4:1–15. doi: 10.1101/cshperspect.a006783
120. Siepe D, Jentsch S. Prolyl isomerase Pin1 acts as a switch to control the degree of substrate ubiquitylation. Nat Cell Biol. (2009) 11:967–72. doi: 10.1038/ncb1908
121. Yun HJ, Kim JY, Kim G, Choi HS. Prolyl-isomerase pin1 impairs trastuzumab sensitivity by up-regulating fatty acid synthase expression. Anticancer Res. (2014) 34:1409–16.
122. Nakatsu Y, Otani Y, Sakoda H, Zhang J, Guo Y, Okubo H, et al. Role of Pin1 protein in the pathogenesis of nonalcoholic steatohepatitis in a rodent model. J Biol Chem. (2012) 287:44526–35. doi: 10.1074/jbc.M112.397133
123. Paneni F, Costantino S, Castello L, Battista R, Capretti G, Chiandotto S, et al. Targeting prolyl-isomerase Pin1 prevents mitochondrial oxidative stress and vascular dysfunction: insights in patients with diabetes. Eur Heart J. (2015) 36:817–28. doi: 10.1093/eurheartj/ehu179
124. Nakatsu Y, Mori K, Matsunaga Y, Yamamotoya T, Ueda K, Inoue Y, et al. The prolyl isomerase Pin1 increases β-cell proliferation and enhances insulin secretion. J Biol Chem. (2017) 292:11886–95. doi: 10.1074/jbc.M117.780726
125. Nakatsu Y, Sakoda H, Kushiyama A, Zhang J, Ono H, Fujishiro M, et al. Peptidyl-prolyl cis/trans isomerase NIMA-interacting 1 associates with insulin receptor substrate-1 and enhances insulin actions and adipogenesis. J Biol Chem. (2011) 286:20812–22. doi: 10.1074/jbc.M110.206904
126. Uchida T, Furumai K, Fukuda T, Akiyama H, Takezawa M, Asano T, et al. Prolyl isomerase Pin1 regulates mouse embryonic fibroblast differentiation into adipose cells. PLoS ONE (2012) 7:e31823. doi: 10.1371/journal.pone.0031823
127. Liao Y, Wei Y, Zhou X, Yang J-Y, Dai C, Chen Y-J, et al. Peptidyl-prolyl cis/trans isomerase Pin1 is critical for the regulation of PKB/Akt stability and activation phosphorylation. Oncogene (2009) 28:2436–45. doi: 10.1038/onc.2009.98
128. Lee NY, Choi HK, Shim JH, Kang KW, Dong Z, Choi HS. The prolyl isomerase Pin1 interacts with a ribosomal protein S6 kinase to enhance insulin-induced AP-1 activity and cellular transformation. Carcinogenesis (2009) 30:671–81. doi: 10.1093/carcin/bgp027
129. Fujimoto Y, Shiraki T, Horiuchi Y, Waku T, Shigenaga A, Otaka A, et al. Proline cis/trans-isomerase Pin1 regulates peroxisome proliferator- activated receptor γactivity through the direct binding to the activation function-1 domain. J Biol Chem. (2010) 285:3126–32. doi: 10.1074/jbc.M109.055095
130. Khanal P, Kim G, Yun HJ, Cho HG, Choi HS. The prolyl isomerase Pin1 interacts with and downregulates the activity of AMPK leading to induction of tumorigenicity of hepatocarcinoma cells. Mol Carcinog. (2013) 52:813-23. doi: 10.1002/mc.21920
131. Nakatsu Y, Iwashita M, Sakoda H, Ono H, Nagata K, Matsunaga Y, et al. Prolyl isomerase Pin1 negatively regulates AMP-activated protein kinase (AMPK) by associating with the CBS domain in the γ subunit. J Biol Chem. (2015) 290:24255–66. doi: 10.1074/jbc.M115.658559
132. Tatapudy S, Aloisio F, Barber D, Nystul T. Cell fate decisions: emerging roles for metabolic signals and cell morphology. EMBO Rep. (2017) 18:2105–18. doi: 10.15252/embr.201744816
133. Keller G. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes Dev. (2005) 19:1129–55. doi: 10.1101/gad.1303605
134. Chen L, Daley GQ. Molecular basis of pluripotency. Hum Mol Genet. (2008) 17:R23–7. doi: 10.1093/hmg/ddn050
135. Ciarapica R, Methot L, Tang Y, Lo R, Dali R, Buscarlet M, et al. Prolyl isomerase Pin1 and protein kinase HIPK2 cooperate to promote cortical neurogenesis by suppressing groucho/TLE:Hes1-mediated inhibition of neuronal differentiation. Cell Death Differ. (2014) 21:321–32. doi: 10.1038/cdd.2013.160
136. Magli A, Angelelli C, Ganassi M, Baruffaldi F, Matafora V, Battini R, et al. Proline isomerase pin1 represses terminal differentiation and myocyte enhancer factor 2C function in skeletal muscle cells. J Biol Chem. (2010) 285:34518–27. doi: 10.1074/jbc.M110.104133
137. Lee SH, Choi YH, Kim Y-J, Choi HS, Yeo C-Y, Lee KY. Prolyl isomerase Pin1 enhances osteoblast differentiation through Runx2 regulation. FEBS Lett. (2013) 587:3640–7. doi: 10.1016/j.febslet.2013.09.040
138. Lee SH, Jeong HM, Han Y, Cheong H, Kang BY, Lee KY. Prolyl isomerase Pin1 regulates the osteogenic activity of Osterix. Mol Cell Endocrinol. (2015) 400:32–40. doi: 10.1016/j.mce.2014.11.017
139. Lee Y-M, Shin S-Y, Jue S-S, Kwon I-K, Cho E-H, Cho E-S, et al. The role of PIN1 on odontogenic and adipogenic differentiation in human dental pulp stem cells. Stem Cells Dev. (2014) 23:618–30. doi: 10.1089/scd.2013.0339
140. Luo ML, Gong C, Chen CH, Lee DY, Hu H, Huang P, et al. Prolyl isomerase pin1 acts downstream of mir200c to promote cancer stem-like cell traits in breast cancer. Cancer Res. (2014) 74:3603–16. doi: 10.1158/0008-5472.CAN-13-2785
141. Luo ML, Gong C, Chen CH, Hu H, Huang P, Zheng M, et al. The Rab2A GTPase promotes breast cancer stem cells and tumorigenesis via erk signaling activation. Cell Rep. (2015) 11:111–24. doi: 10.1016/j.celrep.2015.03.002
142. Matsuura I, Chiang KN, Lai CY, He D, Wang G, Ramkumar R, et al. Pin1 promotes transforming growth factor-β-induced migration and invasion. J Biol Chem. (2010) 285:1754–64. doi: 10.1074/jbc.M109.063826
143. Kim MR, Choi HK, Cho KB, Kim HS, Kang KW. Involvement of Pin1 induction in epithelial-mesenchymal transition of tamoxifen-resistant breast cancer cells. Cancer Sci. (2009) 100:1834–41. doi: 10.1111/j.1349-7006.2009.01260.x
144. Lufei C, Koh TH, Uchida T, Cao X. Pin1 is required for the Ser727 phosphorylation-dependent Stat3 activity. Oncogene (2007) 26:7656–64. doi: 10.1038/sj.onc.1210567
145. Yi P, Wu R-C, Sandquist J, Wong J, Tsai SY, Tsai M-J, et al. Peptidyl-prolyl isomerase 1 (Pin1) serves as a coactivator of steroid receptor by regulating the activity of phosphorylated steroid receptor coactivator 3 (SRC-3/AIB1). Mol Cell Biol. (2005) 25:9687–99. doi: 10.1128/MCB.25.21.9687-9699.2005
146. Poolman TM, Farrow SN, Matthews L, Loudon AS, Ray DW. Pin1 promotes GR transactivation by enhancing recruitment to target genes. Nucleic Acids Res. (2013) 41:8515–25. doi: 10.1093/nar/gkt624
147. Rajbhandari P, Finn G, Solodin NM, Singarapu KK, Sahu SC, Markley JL, et al. Regulation of estrogen receptor α N-terminus conformation and function by peptidyl prolyl isomerase Pin1. Mol Cell Biol. (2012) 32:445–57. doi: 10.1128/MCB.06073-11
148. Hilton BA, Li Z, Musich PR, Wang H, Cartwright BM, Serrano M, et al. ATR plays a direct antiapoptotic role at mitochondria, which is regulated by prolyl isomerase Pin1. Mol Cell (2015) 60:35–46. doi: 10.1016/j.molcel.2015.08.008
149. Grison A, Mantovani F, Comel A, Agostoni E, Gustincich S, Persichetti F, et al. Ser46 phosphorylation and prolyl-isomerase Pin1-mediated isomerization of p53 are key events in p53-dependent apoptosis induced by mutant huntingtin. Proc Natl Acad Sci USA. (2011) 108:17979–84. doi: 10.1073/pnas.1106198108
150. Steger M, Murina O, Hühn D, Ferretti LP, Walser R, Hänggi K, et al. Prolyl isomerase PIN1 regulates DNA double-strand break repair by counteracting DNA end resection. Mol Cell (2013) 50:333–43. doi: 10.1016/j.molcel.2013.03.023
151. Ding Q, Huo L, Yang JY, Xia W, Wei Y, Liao Y, et al. Down-regulation of myeloid cell leukemia-1 through inhibiting Erk/Pin 1 pathway by sorafenib facilitates chemosensitization in breast cancer. Cancer Res. (2008) 68:6109–17. doi: 10.1158/0008-5472.CAN-08-0579
152. Yeung B, Khanal P, Mehta V, Trinkle-Mulcahy L, Yang X. Identification of Cdk1-LATS-Pin1 as a novel signaling axis in anti-tubulin drug response of cancer cells. Mol Cancer Res. (2018) 16:1035-1045. doi: 10.1158/1541-7786.MCR-17-0684
153. Pinton P, Rimessi A, Marchi S, Orsini F, Migliaccio E, Giorgio M, et al. Protein kinase C beta and prolyl isomerase 1 regulate mitochondrial effects of the life-span determinant p66Shc. Science (2007) 315:659–63. doi: 10.1126/science.1135380
154. Keune W-J, Jones DR, Divecha N. PtdIns5P and Pin1 in oxidative stress signaling. Adv Biol Regul. (2013) 53:179–89. doi: 10.1016/j.jbior.2013.02.001
155. Gill JG, Piskounova E, Morrison SJ. Cancer, oxidative stress, and metastasis. Cold Spring Harb Symp Quant Biol. (2016) 81:163–75. doi: 10.1101/sqb.2016.81.030791
156. Lu KP, Finn G, Lee TH, Nicholson LK. Prolyl cis-trans isomerization as a molecular timer. Nat Chem Biol. (2007) 3:619–29. doi: 10.1038/nchembio.2007.35
157. Balastik M, Zhou XZ, Alberich-Jorda M, Weissova R, Žiak J, Pazyra-Murphy MF, et al. Prolyl isomerase Pin1 regulates axon guidance by stabilizing CRMP2A selectively in distal axons. Cell Rep. (2015) 13:812–28. doi: 10.1016/j.celrep.2015.09.026
158. Bitomsky N, Conrad E, Moritz C, Polonio-Vallon T, Sombroek D, Schultheiss K, et al. Autophosphorylation and Pin1 binding coordinate DNA damage-induced HIPK2 activation and cell death. Proc Natl Acad Sci USA. (2013) 110:E4203–12. doi: 10.1073/pnas.1310001110
159. Bao L, Kimzey A, Sauter G, Sowadski JM, Lu KP, Wang D-G. Prevalent overexpression of prolyl isomerase Pin1 in human cancers. Am J Pathol. (2004) 164:1727–37. doi: 10.1016/S0002-9440(10)63731-5
160. Zhang X, Zhang B, Gao J, Wang X, Liu Z. Regulation of the MicroRNA 200b (miRNA-200b) by transcriptional regulators PEA3 and ELK-1 protein affects expression of Pin1 protein to control anoikis. J Biol Chem. (2013) 288:32742–52. doi: 10.1074/jbc.M113.478016
161. Lee K-H, Lin F-C, Hsu T-I, Lin J-T, Guo J-H, Tsai C-H, et al. MicroRNA-296-5p (miR-296-5p) functions as a tumor suppressor in prostate cancer by directly targeting Pin1. Biochim Biophys Acta (2014) 1843:2055–66. doi: 10.1016/j.bbamcr.2014.06.001
162. Lu P-J, Zhou XZ, Liou Y-C, Noel JP, Lu KP. Critical role of WW domain phosphorylation in regulating phosphoserine binding activity and Pin1 function. J Biol Chem. (2002) 277:2381–4. doi: 10.1074/jbc.C100228200
163. Lee Y-C, Que J, Chen Y-C, Lin J-T, Liou Y-C, Liao P-C, et al. Pin1 acts as a negative regulator of the G2/M transition by interacting with the Aurora-A-Bora complex. J Cell Sci. (2013) 126:4862–72. doi: 10.1242/jcs.121368
164. Cho YS, Park SY, Kim DJ, Lee S-H, Woo K-M, Lee K-A, et al. TPA-induced cell transformation provokes a complex formation between Pin1 and 90 kDa ribosomal protein S6 kinase 2. Mol Cell Biochem. (2012) 367:85–92. doi: 10.1007/s11010-012-1322-y
165. Lee TH, Chen CH, Suizu F, Huang P, Schiene-Fischer C, Daum S, et al. Death-associated protein kinase 1 phosphorylates Pin1 and inhibits its prolyl isomerase activity and cellular function. Mol Cell (2011) 42:147–59. doi: 10.1016/j.molcel.2011.03.005
166. Wei S, Kozono S, Kats L, Nechama M, Li W, Guarnerio J, et al. Active Pin1 is a key target of all-trans retinoic acid in acute promyelocytic leukemia and breast cancer. Nat Med. (2015) 21:457–66. doi: 10.1038/nm.3839
167. Rangasamy V, Mishra R, Sondarva G, Das S, Lee TH, Bakowska JC, et al. Mixed-lineage kinase 3 phosphorylates prolyl-isomerase Pin1 to regulate its nuclear translocation and cellular function. Proc Natl Acad Sci USA. (2012) 109:8149–54. doi: 10.1073/pnas.1200804109
168. Chen C-H, Chang C-C, Lee TH, Luo M, Huang P, Liao P-H, et al. SENP1 deSUMOylates and regulates Pin1 protein activity and cellular function. Cancer Res. (2013) 73:3951–62. doi: 10.1158/0008-5472.CAN-12-4360
169. Innes BT, Sowole MA, Gyenis L, Dubinsky M, Konermann L, Litchfield DW, et al. Peroxide-mediated oxidation and inhibition of the peptidyl-prolyl isomerase Pin1. Biochim Biophys Acta Mol Basis Dis. (2015) 1852:905–12. doi: 10.1016/j.bbadis.2014.12.025
170. Sultana R, Boyd-Kimball D, Poon HF, Cai J, Pierce WM, Klein JB, et al. Oxidative modification and down-regulation of Pin1 in Alzheimer's disease hippocampus: a redox proteomics analysis. Neurobiol Aging (2006) 27:918–25. doi: 10.1016/j.neurobiolaging.2005.05.005
171. Lam PB, Burga LN, Wu BP, Hofstatter EW, Lu KP, Wulf GM. Prolyl isomerase Pin1 is highly expressed in Her2-positive breast cancer and regulates erbB2 protein stability. Mol Cancer (2008) 7:91. doi: 10.1186/1476-4598-7-91
172. Zheng M, Xu H, Liao X-H, Chen CP, Zhang AL, Lu W, et al. Inhibition of the prolyl isomerase Pin1 enhances the ability of sorafenib to induce cell death and inhibit tumor growth in hepatocellular carcinoma. Oncotarget (2017) 8:29771–84. doi: 10.18632/oncotarget.15967
173. Gianni M, Boldetti A, Guarnaccia V, Rambaldi A, Parrella E, Raska I, et al. Inhibition of the peptidyl-prolyl-isomerase Pin1 enhances the responses of acute myeloid leukemia cells to retinoic acid via stabilization of RARα and PML-RARα. Cancer Res. (2009) 69:1016–26. doi: 10.1158/0008-5472.CAN-08-2603
174. Moore JD, Potter A. Pin1 inhibitors: pitfalls, progress and cellular pharmacology. Bioorganic Med Chem Lett. (2013) 23:4283–91. doi: 10.1016/j.bmcl.2013.05.088
175. Russo Spena C, De Stefano L, Palazzolo S, Salis B, Granchi C, Minutolo F, et al. Liposomal delivery of a Pin1 inhibitor complexed with cyclodextrins as new therapy for high-grade serous ovarian cancer. J Control Release (2018) 281:1–10. doi: 10.1016/j.jconrel.2018.04.055
176. Kozono S, Lin YM, Seo HS, Pinch B, Lian X, Qiu C, et al. Arsenic targets Pin1 and cooperates with retinoic acid to inhibit cancer-driving pathways and tumor-initiating cells. Nat Commun. (2018) 9:3069. doi: 10.1038/s41467-018-05402-2
177. Yang D, Luo W, Wang J, Zheng M, Liao XH, Zhang N, et al. A novel controlled release formulation of the Pin1 inhibitor ATRA to improve liver cancer therapy by simultaneously blocking multiple cancer pathways. J Control Release (2018) 269:405–22. doi: 10.1016/j.jconrel.2017.11.031
178. Pu W, Li J, Zheng Y, Shen X, Fan X, Zhou JK, et al. Targeting Pin1 by inhibitor API-1 regulates microRNA biogenesis and suppresses hepatocellular carcinoma development. Hepatology (2018) 68:547–60. doi: 10.1002/hep.29819
179. Mori T, Hidaka M, Ikuji H, Yoshizawa I, Toyohara H, Okuda T, et al. A high-throughput screen for inhibitors of the prolyl isomerase, Pin1, identifies a seaweed polyphenol that reduces adipose cell differentiation. Biosci Biotechnol Biochem. (2014) 78:832–8. doi: 10.1080/09168451.2014.905189
180. Senyuk V, Zhang Y, Liu Y, Ming M, Premanand K, Zhou L, et al. Critical role of miR-9 in myelopoiesis and EVI1-induced leukemogenesis. Proc Natl Acad Sci. (2013) 110:5594–9. doi: 10.1002/asia.201701216
181. Aluise CD, Rose K, Boiani M, Reyzer ML, Manna JD, Tallman K, et al. Peptidyl-prolyl cis/trans-isomerase A1 (Pin1) is a target for modification by lipid electrophiles. Chem Res Toxicol. (2013) 26:270–9. doi: 10.1021/tx300449g
182. Ayala G, Wang D, Wulf G, Frolov A, Li R, Sowadski J, et al. The prolyl isomerase Pin1 is a novel prognostic marker in human prostate cancer. Cancer Res. (2003) 63:6244–51.
183. Sukkar SG. The impact of clinical nutrition on cancer therapy: a frequently underestimated perspective. A complementary approach to cancer patients. Med J Nutrition Metab. (2012) 5:75–9. doi: 10.1007/s12349-012-0105-z
Keywords: signal transduction, tissue integrity, cancer target therapy, post-translational modification, prolyl-isomerase PIN1, organismal development, tumor development and progression
Citation: Zannini A, Rustighi A, Campaner E and Del Sal G (2019) Oncogenic Hijacking of the PIN1 Signaling Network. Front. Oncol. 9:94. doi: 10.3389/fonc.2019.00094
Received: 07 December 2018; Accepted: 01 February 2019;
Published: 25 February 2019.
Edited by:
Flavio Rizzolio, Università Ca' Foscari, ItalyReviewed by:
Maurizio Mongiat, Centro di Riferimento Oncologico di Aviano (IRCCS), ItalyRoberta Moschini, University of Pisa, Italy
Copyright © 2019 Zannini, Rustighi, Campaner and Del Sal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Giannino Del Sal, ZGVsc2FsQGxuY2liLml0
†These authors have contributed equally to this work.
 Alessandro Zannini
Alessandro Zannini Alessandra Rustighi
Alessandra Rustighi Elena Campaner
Elena Campaner Giannino Del Sal
Giannino Del Sal