
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 19 February 2019
Sec. Gastrointestinal Cancers
Volume 9 - 2019 | https://doi.org/10.3389/fonc.2019.00063
 Jing Lin1,2†
Jing Lin1,2† Yu Chen1,2,3†
Yu Chen1,2,3† Wei-feng Tang4†
Wei-feng Tang4† Chao Liu4
Chao Liu4 Sheng Zhang5
Sheng Zhang5 Zeng-qing Guo1,2,3
Zeng-qing Guo1,2,3 Gang Chen3,6*
Gang Chen3,6* Xiong-wei Zheng3,6*
Xiong-wei Zheng3,6*Purpose: Functional variants in the peroxisome proliferator-activated receptor gamma (PPARG) and PPARG co-activator 1 (PPARGC1) family (e.g., PPARGC1A and PPARGC1B) genes were predicted to confer susceptibility to colorectal cancer (CRC). The aim of the present study was to explore the relationship between PPARG, PPARGC1A, PPARGC1B polymorphism and the risk of CRC.
Patients and methods: We conducted a case-control study with 1,003 CRC cases and 1,303 controls. We selected the PPARG rs3856806 C>T, PPARGC1A rs2970847 C>T, rs8192678 C>T, rs3736265 G>A and PPARGC1B rs7732671 G>C and rs17572019 G>A SNPs to assess the relationship between PPARG, PPARGC1A, PPARGC1B their variants and risk of CRC.
Results: We found that the PPARG rs3856806 C>T polymorphism increased the risk of CRC (TT vs. CC: adjusted OR, 1.59, 95% CI 1.08–2.35, P = 0.020; TT/CT vs. CC: adjusted OR, 1.26; 95% CI 1.06–1.49; P = 0.009 and TT vs. CC/CT: adjusted OR, 1.54; 95% CI 1.05–2.26; P = 0.028), even after a Bonferroni correction test. The stratified analysis revealed that the PPARG rs3856806 C>T polymorphism also increased the risk of CRC, especially in male, ≥61 years old, never smoking, never drinking, BMI ≥ 24 kg/m2, colon cancer and rectum cancer subgroups.
Conclusion: Our findings highlight that the PPARG rs3856806 C>T polymorphism may increase the risk of CRC. In the future larger sample size case-control studies with a detailed functional assessment are needed to further determine the relationship of the PPARG rs3856806 C>T polymorphism with CRC risk.
Colorectal cancer (CRC) is one of the most common type of malignancies, accounting for 1.8 million cases in GLOBOCAN 2018 (1). The incidence of CRC is increasing in China, where it ranks as the fifth most common carcinoma in male and the fourth in female, with a total of 215,700 patients diagnosed in 2015 (2). Epidemiologic investigations have attributed most of CRC to some important environmental factors (3). The increase of the incidence of CRC is proposed to correlate with an unhealthy lifestyle, including drinking, smoking, low intake of dietary fiber, high intake of dietary fat, decreased consumption of vegetables, and fruits and being physically inactive (4–7). Accumulating evidence highlighted that besides these unhealthy lifestyles and environmental factors, some additional inherited susceptibility factors may be associated with the development of CRC. As CRC is associated with obesity and Waist-to-Hip Ratio (WHR) (8–10), the peroxisome proliferator-activated receptor gamma (PPARG), PPARG co-activator 1 (PPARGC1) family (e.g., PPARGC1A and PPARGC1B) may be strong candidate genes predisposing to CRC (11).
PPARG is located in 3p25. PPARG is also known as NR1C3 (nuclear receptor subfamily 1, group C, member 3) which shares some common conservative domains with other steroid receptors (e.g., estrogen, progesterone, retinoid, vitamin D and thyroid receptors). It was reported that PPARG is a regulator of adipocyte differentiation, energy homeostasis and obesity (12–14). PPARG decreases the inflammatory response of cells (15) and increases synthesis and release of paraoxonase 1 (16). Wang et al. reported that PPARG gene might be one of the targets of miRNA-34a and a conceivable therapeutic targets for CRC (17)..PPARGC1A and PPARGC1B, transcriptional co-activators of PPARG, may control transcription in adipogenesis, oxidative metabolism genes (18). Thus, PPARG, PPARGC1A, and PPARGC1B might be implicated in the development of cancer.
Pro12Ala and His449His (rs3856806 C>T) polymorphisms in the PPARG gene are two of the most common variants in the PPARG gene. Recently, a meta-analysis confirmed that the PPARG Pro12Ala polymorphism might decreased the risk of CRC (19). Several case-control studies focused on the potential role of PPARG variants in determining CRC susceptibility. The PPARG rs3856806 C>T is a common single-nucleotide polymorphism (SNP) in the coding region. Recently, a meta-analysis indicated that the PPARG rs3856806 C>T polymorphism may increase the susceptibility of overall cancer (20). In this pooled study, there were seven independent case-control studies with 1,720 cases and 3,458 controls focusing on the association of the PPARG rs3856806 C>T polymorphism with CRC risk (21–24). As well, a tendency to increased CRC susceptibility was noted. Because of the lack of sufficient sample sizes, the evidence may be limited. Additionally, (25) reported that the PPARGC1B rs7732671 G>C polymorphism may decrease the susceptibility of breast cancer. However, the association between PPARGC1A and PPARGC1B SNPs and the risk of CRC was unknown. The aim of this case-control study was to assess the association of PPARG, PPARGC1A, and PPARGC1B polymorphisms with CRC risk. We selected PPARG rs3856806 C>T, PPARGC1A rs2970847 C>T, rs8192678 C>T, rs3736265 G>A, and PPARGC1B rs7732671 G>C and rs17572019 G>A SNPs to determine the relationship between their variants and CRC risk in an Eastern Chinese Han population.
This cohort was in part previously studied (19, 26). The CRC cases were recruited from Fujian Medical University Union Hospital (Fuzhou city, China) and the Affiliated People's Hospital of Jiangsu University (Zhenjiang city, China) between October 2014 and August 2017. The major inclusion criteria of CRC cases were: (1) sporadic CRC cases; (2) newly diagnosed CRC patients via pathology; and (3) Han population who living in Eastern China. And the exclusion criteria were: (1) hereditary non-polyposis CRC; (2) CRC cases who have been treated with chemoradiotherapy and (3) with another malignancy history. During the period, a total of 1,186 CRC patients were diagnosed in those local hospitals. Our study includes 1,003 (84.57%) patients, who agree to attend this study and provided blood samples for SNP analysis. The mean age of CRC patients was 61.10 ± 12.17 years. From 1,521 selected controls, 1,303 (85.67%) agreed to participate and donated a biological sample in this study. The controls included 1,303 healthy volunteers who participated in a routine examination in these hospitals, with a mean age of 61.40 ± 9.61 years. For selecting controls, the inclusion criteria were: (1) without a carcinoma history subjects; (2) similar age matched to CRC group; and (3) Han population who is a resident of Eastern China. Additionally, subjects who had a cancer history were excluded. The controls were matched with CRC patients by age and sex. The information on risk factor was obtained from the CRC cases and controls during a medical interview. And weight and height were measured. The body mass index (BMI) was calculated as weight/height2 (kg/m2) and BMI ≥ 24 kg/m2 was considered as overweight and obesity for Chinese (27, 28). All participants enrolled in the present study signed the informed consent and were of Chinese origin. The study protocol was approved by the Ethics Committee of Fujian Medical University and Jiangsu University.
Two milliliters of Ethylenediamine tetra acetic acid (EDTA)-anticoagulated blood was collected from each participant. Blood samples were stored in a −80° C freezer. Using a Promega DNA Blood Mini Kit (Promega, Madison, USA), genomic DNA was isolated from lymphocytes. We placed the cryopreserved specimen at room temperature for an hour. After red blood cell removal, nuclear releasing and protein precipitation, we obtained genomic DNA. We add 300 μl of DNA solution (pH 8.0) and placed the sample in a refrigerator at 4°C for 1–2 weeks. A NanoDrop ND-1000 micro spectrophotometer was used to determine DNA concentration and purity. As described in previous studies, the genotypes of the PPARG rs3856806 C>T, PPARGC1A rs2970847 C>T, rs8192678 C>T, rs3736265 G>A, and PPARGC1B rs7732671 G>C and rs17572019 G>A SNPs was determined by a custom-by-design 48-Plex SNPscan Kit (Genesky Biotechnologies Inc., Shanghai, China) (29, 30). This genotyping method was designed as a multiplex fluorescence PCR (31). Ninety-two DNA samples (4%) were randomly selected and tested by another technician for quality control. The genotypes of these SNPs were not changed.
We used an online Chi-square software (http://ihg.gsf.de/cgi-bin/hw/hwa1.pl) to test deviation from the Hardy-Weinberg equilibrium (HWE) by using Pearson's goodness-of-fit chi-square. The genotype frequencies of the PPARG rs3856806 C>T, PPARGC1A rs2970847 C>T, rs8192678 C>T, rs3736265 G>A, and PPARGC1B rs7732671 G>C and rs17572019 G>A variants among CRC cases were compared to those of controls using a χ2 test or Fisher's exact test. Multivariate logistic regression analysis was harnessed to obtain crude and adjusted odds ratios (ORs) with their 95% confidence intervals (CIs) to predict the relationship of the PPARG rs3856806 C>T, PPARGC1A rs2970847 C>T, rs8192678 C>T, rs3736265 G>A, and PPARGC1B rs7732671 G>C and rs17572019 G>A polymorphisms with susceptibility to CRC. Dominant, recessive, heterozygote and homozygote models were used to evaluate the association of these SNPs with CRC risk. The χ2 test or Fisher's exact test was first applied to compare the distribution of age, sex, alcohol consumption, smoking status, and BMI between CRC patients and controls. A P < 0.05 (two–tailed) was defined as a significant association. All data were analyzed by SAS software for Windows (9.4 version, SAS Institute, Cary, USA). In this case-control study, a Bonferroni correction test was applied for multiple testing (32, 33). An internal validation the through bootstrap method was applied to PPARG rs3856806 C>T. We used 0.623 bootstrap method to resample 1,003 cases from the CRC patient group and 1,303 cases from the control group to validate our results.
Selected demographic variables and risk factors in the enrolled population and the correlation with CRC are summarized in Table 1. There was no significant difference between CRC patients and controls regarding sex (P = 0.867), age (61.10 ± 12.17 years for cases and 61.40 ± 9.61 years for controls, P = 0.496), suggesting that these variables were well-matched. Alcohol consumption, BMI and smoking status were statistically different (P < 0.001, P < 0.001, and P = 0.002, respectively) between two groups. The primary information of PPARG, PPARGC1A, and PPARGC1B SNPs is displayed in Table 2. The genotype distributions of PPARG rs3856806 C>T, PPARGC1A rs2970847 C>T, rs8192678 C>T, rs3736265 G>A, and PPARGC1B rs7732671 G>C and rs17572019 G>A are in accordance with HWE in controls (P = 0.143, 0.925, 0.800, 0.059, 0.970, and 0.372, respectively).
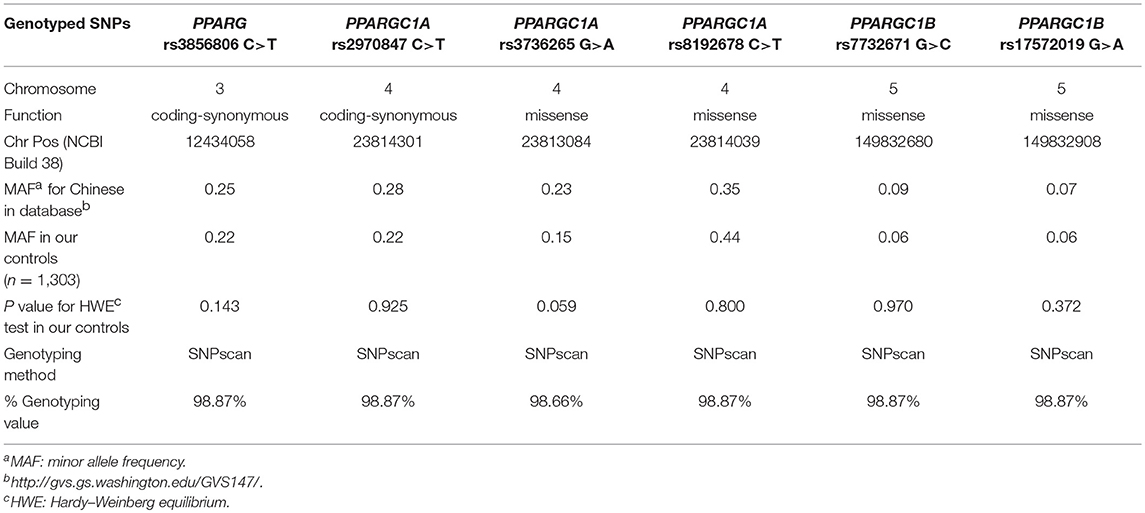
Table 2. Primary information for PPARG rs3856806 C>T, PPARGC1A rs2970847 C>T, rs8192678 C>T, rs3736265 G>A, and PPARGC1B rs7732671 G>C and rs17572019 G>A polymorphisms.
Table 3 summarizes the genotype distributions of PPARG rs3856806 C>T, PPARGC1A rs2970847 C>T, rs8192678 C>T, rs3736265 G>A, and PPARGC1B rs7732671 G>C and rs17572019 G>A SNPs in CRC cases and controls. The genotype frequencies of PPARG rs3856806 C>T were 55.51% (CC), 38.16% (CT), and 6.33% (TT) in CRC cases and 60.69% (CC), 35.31% (CT), and 4.00% (TT) in controls. When the frequency of PPARG rs3856806 CC genotype was used as a reference, individuals carrying the PPARG rs3856806 TT genotype had an increased risk to CRC (crude OR = 1.67, 95% CI 1.13–2.45 for TT vs. CC, P = 0.009). When compared with the frequency of PPARG rs3856806 CC genotype, individuals carrying the PPARG rs3856806 TT/CT genotype also had an increased the risk of CRC (crude OR = 1.24, 95% CI 1.05–1.46 for TT/CT vs. CC, P = 0.013). When the frequency of the PPARG rs3856806 CC/CT genotype was used as a reference, individuals carrying the PPARG rs3856806 TT genotype had a significantly increased susceptibility to CRC (crude OR = 1.62, 95% CI 1.11–2.37 for TT vs. CC/CT, P = 0.012). After adjustments for age, sex, smoking, BMI, and drinking, the observed increased susceptibility of CRC was not essentially altered (TT vs. CC: adjusted OR, 1.59, 95% CI 1.08–2.35, P = 0.020; TT/CT vs. CC: adjusted OR, 1.26; 95% CI 95% CI 1.06–1.49; P = 0.009 and TT vs. CC/CT: adjusted OR, 1.54; 95% CI 95% CI 1.05–2.26; P = 0.028), Table 3.
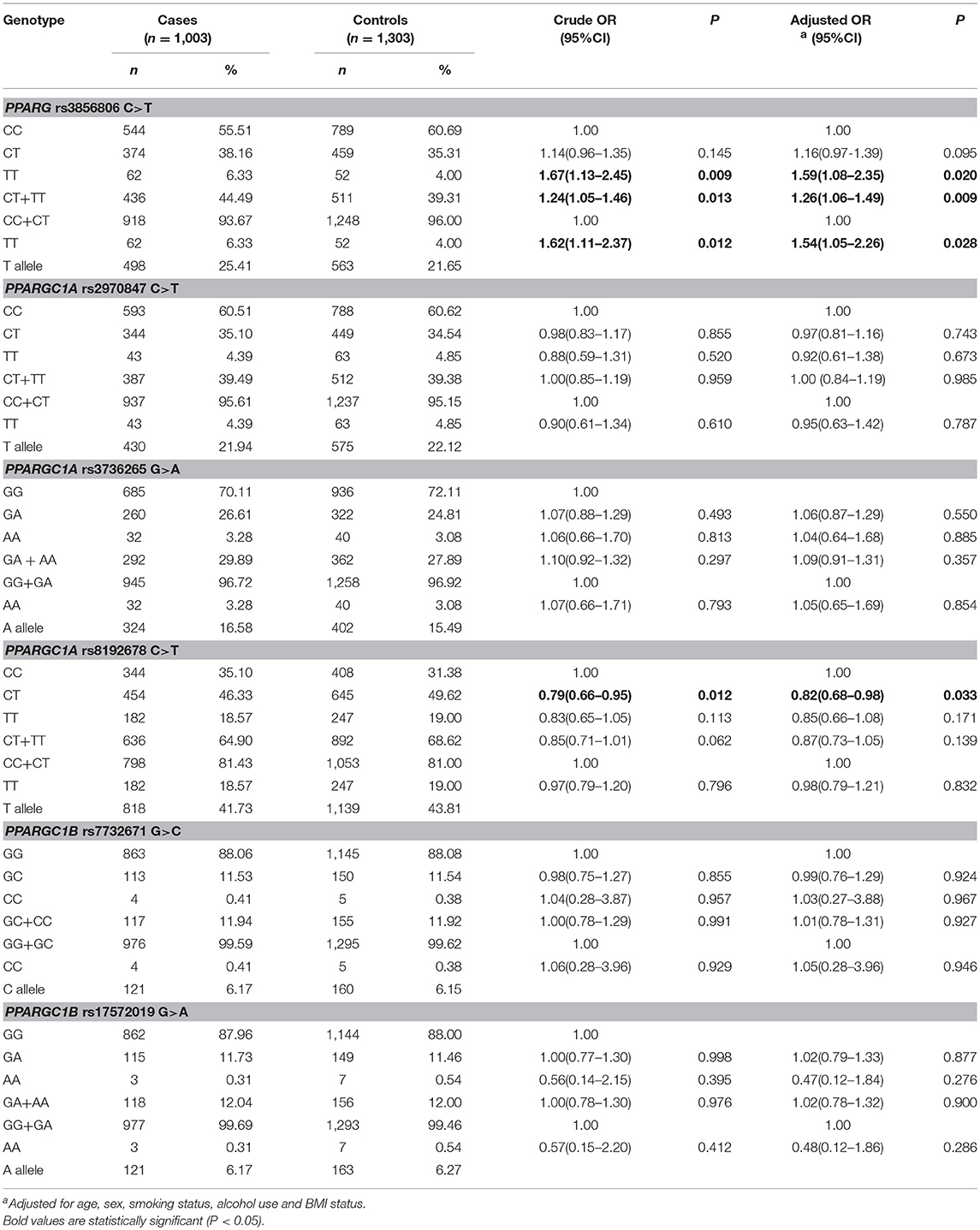
Table 3. Logistic regression analyses of associations between PPARG rs3856806 C>T, PPARGC1A rs2970847 C>T, rs8192678 C>T, rs3736265 G>A, and PPARGC1B rs7732671 G>C and rs17572019 G>A polymorphisms and risk of CRC.
Table S1 shows the internal validation results through the bootstrap method. When compared with the PPARG rs3856806 CC genotype, the PPARG rs3856806 TT, and TT/CT genotypes also indicate an increased CRC risk (crude OR = 1.56, 95% CI 1.09–2.23 for TT vs. CC, P = 0.015; crude OR = 1.20, 95% CI 1.02–1.42 for TT/CT vs. CC, P = 0.033). When compared with the PPARG rs3856806 CC/CT genotype, PPARG rs3856806 TT genotype also suggest an increased CRC risk (crude OR = 1.53, 95% CI 1.08–2.18 for TT vs. CC/CT, P = 0.017). After being adjusted by age, sex, smoking BMI, and drinking, the increased susceptibility of CRC was not essentially altered.
The genotype frequencies of PPARGC1A rs8192678 C>T were 35.10% (CC), 46.33% (CT), and 18.57% (TT) in CRC patients and 31.38% (CC), 49.62% (CT), and 19.00% (TT) in healthy controls. When the frequency of the PPARGC1A rs8192678 CC genotype was used as a reference, individuals carrying the PPARGC1A rs8192678 CT genotype had a decreased susceptibility to CRC (crude OR = 0.79, 95% CI 0.66–0.95 for CT vs. CC, P = 0.012). After adjustments for age, sex, smoking, BMI and drinking, this association was also found (CT vs. CC: adjusted OR, 0.82; 95% CI 95% CI 0.68–0.989; P = 0.033), Table 3.
We found no significant difference in the genotype distribution of the PPARGC1A rs3736265 G>A, rs2970847 C>T and PPARGC1B rs7732671 G>C, rs17572019 G>A polymorphisms among CRC cases and controls, Table 3.
The Bonferroni correction test was applied to determine whether the association of the PPARG rs3856806 C>T and rs8192678 C>T polymorphisms with the risk of CRC was reliable. We defined the statistical significance level at 0.0125 (0.05/4 genetic models). We found the genotype distribution of that the PPARG rs3856806 C>T polymorphism was still significantly different between CRC patients and controls (TT/CT vs. CC: adjusted OR, 1.26; 95% CI 95% CI 1.06–1.49; P = 0.009).
To further assess the association of the PPARG rs3856806 C>T polymorphism with CRC risk, we conducted a stratified analysis by BMI, gender, age, tobacco using and alcohol consumption. Table 4 presents the different genotype frequencies of the PPARG rs3856806 C>T polymorphism in a subgroup analysis. After an adjustment by logistic regression analysis with gender, age, BMI, tobacco using and drinking status, we found that the PPARG rs3856806 C>T polymorphism significantly increased the risk of CRC in several subgroups:1) male subgroup, TT vs. CC, adjusted OR = 1.88, 95% CI 1.14–3.10, P = 0.014 and TT vs. CT/CC, adjusted OR = 1.84, 95% CI 1.12–3.02, P = 0.016; 2) ≥61 years subgroup, CT/TT vs. CC, adjusted OR = 1.36, 95% CI 1.08–1.71, P = 0.010; 3) never smoking subgroup, CT/TT vs. CC, adjusted OR = 1.27, 95% CI 1.05–1.55, P = 0.015; 4) never drinking subgroup, CT/TT vs. CC, adjusted OR = 1.27, 95% CI 1.06–1.53, P = 0.011; 5) BMI ≥ 24 kg/m2 subgroup, TT vs. CC: adjusted OR = 2.65, 95% CI 1.36–5.17, P = 0.004; CT/TT vs. CC, adjusted OR = 1.38, 95% CI 1.05–1.81, P = 0.022, and TT vs. CT/CC, adjusted OR = 2.51, 95% CI 1.03–4.86, P = 0.006 (Table 4).
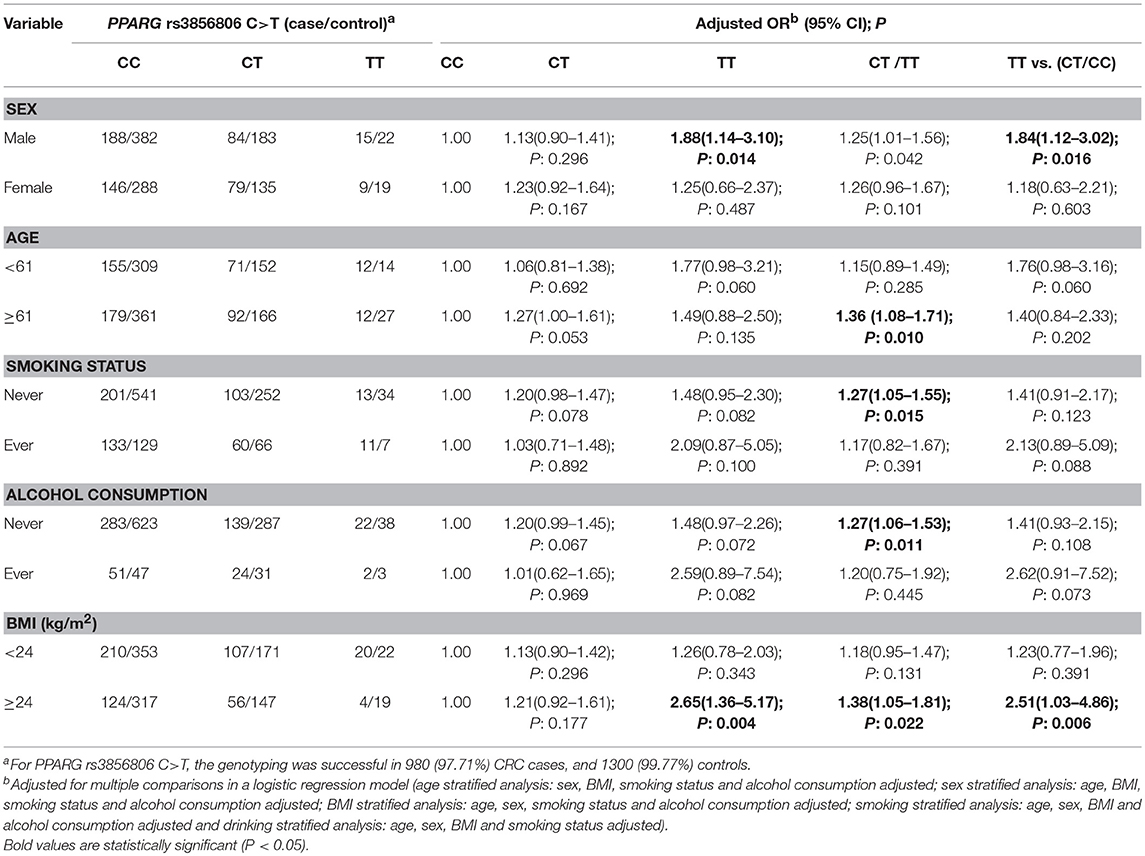
Table 4. Stratified analyses between PPARG rs3856806 C>T polymorphism and CRC risk by sex, age, BMI, smoking status, and alcohol consumption.
To determine whether the association between the PPARG rs3856806 C>T polymorphism and CRC risk was modified by the site of CRC, we conducted stratified analyses. The results of the stratified analyses suggested this SNP increased the risk of colon cancer (CT vs. CC: adjusted OR = 1.27, 95% CI 1.01–1.60, P = 0.044 and TT/CT vs. CC: adjusted OR = 1.34, 95% CI 1.07–1.68, P = 0.011) and rectum cancer (TT vs. CC: adjusted OR = 1.58, 95% CI 1.01–2.49, P = 0.045 and TT vs. CC/CT: adjusted OR = 1.58, 95% CI 1.01–2.46, P = 0.043), Table 5.
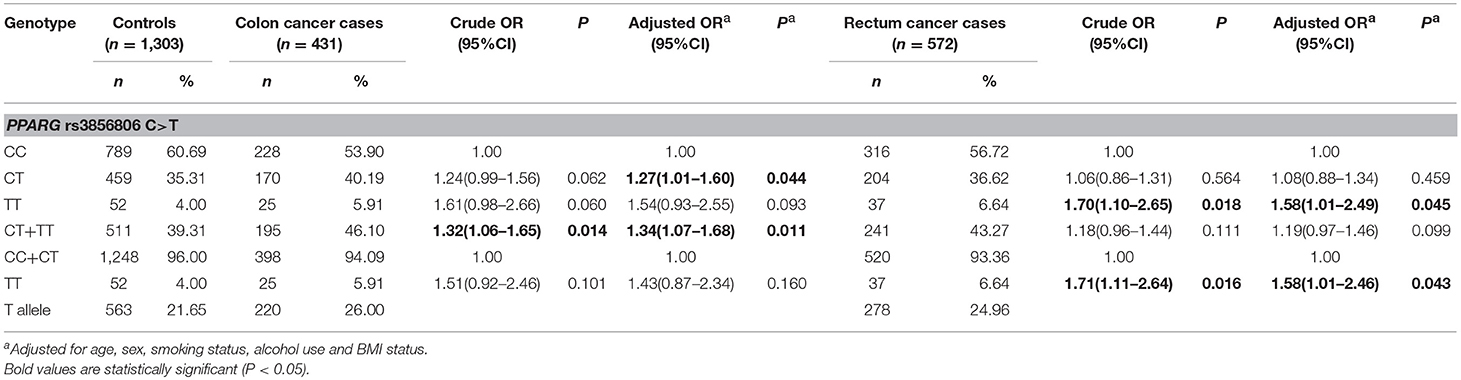
Table 5. Stratified analyses between PPARG rs3856806 C>T polymorphism and CRC risk by site of tumor.
Accumulating evidence has highlighted that CRC is associated with obesity and Waist-to-Hip Ratio (WHR) (8–10). Some important metabolism-related genes may be strong candidates for predisposing to CRC (11). PPARG may be implicated in metabolism, inflammatory response, adipose cell differentiation, and cellular apoptosis (34–37). The PPARGC1 family (e.g., PPARGC1A, PPARGC1B) also regulate fatty acid oxidation, gluconeogenesis and adaptive thermogenesis (38). These proteins may be involved in the development of obesity. Several studies have focused on the association between the PPARG rs3856806 C>T polymorphism and the risk of CRC (21–24). However, the results were inconsistent. In addition, the potential relationships of the PPARGC1A, PPARGC1B SNPs with the development of CRC are unknown. To shed some light on this issue, we carried out a case-control study in Eastern Chinese Han population. Our findings suggested that the PPARG rs3856806 C>T polymorphism is associated with an increased risk of CRC, especially in male, ≥ 61 years old, never smoking, never drinking, BMI ≥24 kg/m2, colon cancer, and rectum cancer subgroups.
PPARG is one of the three subtypes of peroxisome proliferator-activated receptors (PPARs). The PPARG gene encodes a member of the PPAR subfamily of nuclear receptors, which form heterodimers with retinoid X receptors (RXRs) and then influence the transcription of many target genes. A previous study concluded that there was evidence for a relationship between obesity and overweight with a risk of colon and rectum cancer (39). A common functional polymorphism (His449His; rs3856806) in PPARG is a C → T coding-synonymous substitution in codon 449 of exon 6. Grygiel-Górniak and colleagues reported that higher BMI and visceral fat deposition were promoted by the presence of the PPARG rs3856806 T allele (40). Previous studies suggested a potential correlation of this SNP with atherosclerosis, type 2 diabetes and cancer (20, 41–44). Although rs3856806 is a coding-synonymous SNP, it is proposed that a C → T substitution could alter the expression of PPARG protein by altering mRNA processing or translation. Doecke et al. reported that the PPARG rs3856806 CT genotype may increase the susceptibility of adenocarcinoma of the esophagus in an obesity subgroup (BMI ≥ 30 kg/m2) (45). The PPARG rs3856806 C>T polymorphism was also found to be significantly over-represented in sporadic glioblastoma multiforme in American populations (46). Jiang et al. reported that the PPARG rs3856806 C>T polymorphism was associated with an increased risk of CRC in India (21). However, other case-control studies suggested that PPARG rs3856806 C>T might not influence the development of CRC (22–24). Thus, the results were inconsistent and ambiguous. Considering a common SNP having low penetrance susceptibility to cancer, we performed a case-control study with large sample sizes to obtain a more precise assessment. As demonstrated in the results, we found that the PPARG rs3856806 C>T polymorphism was associated with an increased risk of CRC, even after a Bonferroni correction test. Thus, our findings were reliable. Recently, a meta-analysis reported that the PPARG rs3856806 C>T polymorphism increased the risk of overall cancer (20). Our findings were very similar to this pooled-analysis. Additionally, it is worth noting that we found the that the PPARG rs3856806 C>T polymorphism was associated with an increased risk of CRC in the BMI ≥ 24 kg/m2 subgroup. It suggested that this SNP might be implicated in the development of obesity and overweight, and subsequently lead to an increased risk to CRC.
There are, however, several limitations in this case-control study. First, the CRC patients and non-cancer controls were from two local hospitals. The potential selection bias might have occurred. Second, a replicated study focusing on the association of these SNPs with CRC risk was not carried out. Third, although we took some risk factors into consideration such as BMI, gender, age, drinking, and smoking status, many other environmental and lifestyle factors, possibly related to the development of CRC, were not collected in this study. Fourth, due to the moderate sample size in some subgroups, the power might be limited. Fifth, a functional study for the PPARG rs3856806 C>T polymorphism has not been conducted. Finally, in the future, it is necessary to carry out a functional study to identify the mechanism of the PPARG rs3856806 C>T polymorphism.
In conclusion, our findings suggest that the PPARG rs3856806 C>T polymorphism may increase the risk of CRC. In the future, larger sample size case-control studies with a detailed functional assessment are needed to further evaluate the relationship of PPARG rs3856806 C>T polymorphism with CRC risk.
JL, YC, GC, and XZ conceived and designed the experiments. YC, WT, CL, and GC performed the experiments. JL, YC, SZ, and ZG analyzed the data. JL, YC, and XZ contributed reagents, materials, and analysis tools. JL, YC, and WT wrote the paper.
The project was supported by the National Natural Science Foundation of China (Grant No. U1705282), the Ministry of Health P.R. China (Grant No. WKJ2016-2-05), Natural Science Foundation of Fujian Province (Grant No. 2016J01513, 2017J01259, 2018J01267), Fujian provincial health and family planning research talent training program (Grant No. 2015-CX-7, 2018-ZQN-13, 2016-1-11, 2018-1-13), Joint Funds for the innovation of science and Technology, Fujian province (Grant No. 2017Y9077), and the National Clinical Key Specialty Construction Program. Science and Technology Program of Fujian Province, China, No. 2018Y2003.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We appreciate all subjects who participated in this study. We wish to thank Dr. Yan Liu (Genesky Biotechnologies Inc., Shanghai, China) for technical support.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2019.00063/full#supplementary-material
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2018) 68:394–424. doi: 10.3322/caac.21492
2. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin. (2016) 66:115–32. doi: 10.3322/caac.21338
3. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. (2011) 61:69–90. doi: 10.3322/caac.20107
4. Weitz J, Koch M, Debus J, Hohler T, Galle PR, Buchler MW. Colorectal cancer. Lancet (2005) 365:153–65. doi: 10.1016/S0140-6736(05)17706-X
5. Gerber M. Background review paper on total fat, fatty acid intake and cancers. Ann Nutr Metabol. (2009) 55:140–61. doi: 10.1159/000229000
6. Xu M, Chen YM, Huang J, Fang YJ, Huang WQ, Yan B, et al. Flavonoid intake from vegetables and fruits is inversely associated with colorectal cancer risk: a case-control study in China. Br J Nutr. (2016) 116:1275–87. doi: 10.1017/S0007114516003196
7. Nagle CM, Wilson LF, Hughes MC, Ibiebele TI, Miura K, Bain CJ, et al. Cancers in Australia in 2010 attributable to inadequate consumption of fruit, non-starchy vegetables and dietary fibre. Aust N Zealand J Public Health (2015) 39:422–8. doi: 10.1111/1753-6405.12449
8. Dong Y, Zhou J, Zhu Y, Luo L, He T, Hu H, et al. Abdominal obesity and colorectal cancer risk: systematic review and meta-analysis of prospective studies. Biosci Rep. (2017) 37:BSR20170945. doi: 10.1042/BSR20170945
9. Shirakami Y, Ohnishi M, Sakai H, Tanaka T, Shimizu M. Prevention of colorectal cancer by targeting obesity-related disorders and inflammation. Intl J Mol Sci. (2017) 18:E908. doi: 10.3390/ijms18050908
10. Lund EK, Belshaw NJ, Elliott GO, Johnson IT. Recent advances in understanding the role of diet and obesity in the development of colorectal cancer. Proc Nutr Soc. (2011) 70:194–204. doi: 10.1017/S0029665111000073
11. Motawi TK, Shaker OG, Ismail MF, Sayed NH. Peroxisome proliferator-activated receptor gamma in obesity and colorectal cancer: the role of epigenetics. Sci Reports (2017) 7:10714. doi: 10.1038/s41598-017-11180-6
12. Guazzoni G, Montorsi F, Colombo R, Di Girolamo V, Da Pozzo L, Rigatti P. Long term experience with the prostatic spiral for urinary retention due to benign prostatic hyperplasia. Scand J Urol Nephrol. (1991) 25:21–4. doi: 10.3109/00365599109024523
13. AlSaleh A, Sanders TA, O'Dell SD. Effect of interaction between PPARG, PPARA and ADIPOQ gene variants and dietary fatty acids on plasma lipid profile and adiponectin concentration in a large intervention study. Proc Nutr Soc. (2012) 71:141–53. doi: 10.1017/S0029665111003181
14. Barbieri M, Rizzo MR, Papa M, Acampora R, De Angelis L, Olivieri F, et al. Role of interaction between variants in the PPARG and interleukin-6 genes on obesity related metabolic risk factors. Exp Gerontol. (2005) 40:599–604. doi: 10.1016/j.exger.2005.05.004
15. Hamblin M, Chang L, Fan Y, Zhang J, Chen YE. PPARs and the cardiovascular system. Antioxidants Redox Signal. (2009) 11:1415–52. doi: 10.1089/ars.2008.2280
16. Khateeb J, Gantman A, Kreitenberg AJ, Aviram M, Fuhrman B. Paraoxonase 1 (PON1) expression in hepatocytes is upregulated by pomegranate polyphenols: a role for PPAR-gamma pathway. Atherosclerosis (2010) 208:119–25. doi: 10.1016/j.atherosclerosis.2009.08.051
17. Wang T, Xu H, Liu X, Chen S, Zhou Y, Zhang X. Identification of Key Genes in Colorectal Cancer Regulated by miR-34a. Med Sci Monitor (2017) 23:5735–43. doi: 10.12659/MSM.904937
18. Puigserver P, Spiegelman BM. Peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1 alpha): transcriptional coactivator and metabolic regulator. Endocr Rev. (2003) 24:78–90. doi: 10.1210/er.2002-0012
19. Jiang J, Xie Z, Guo J, Wang Y, Liu C, Zhang S, et al. Association of PPARG rs 1801282 C>G polymorphism with risk of colorectal cancer: from a case-control study to a meta-analysis. Oncotarget (2017) 8:100558–69. doi: 10.18632/oncotarget.20138
20. Ding H, Chen Y, Qiu H, Liu C, Wang Y, Kang M, et al. PPARG c.1347C>T polymorphism is associated with cancer susceptibility: from a case-control study to a meta-analysis. Oncotarget (2017) 8:102277–90. doi: 10.18632/oncotarget.20925
21. Jiang J, Gajalakshmi V, Wang J, Kuriki K, Suzuki S, Nakamura S, et al. Influence of the C161T but not Pro12Ala polymorphism in the peroxisome proliferator-activated receptor-gamma on colorectal cancer in an Indian population. Cancer Sci. (2005) 96:507–12. doi: 10.1111/j.1349-7006.2005.00072.x
22. Vogel U, Christensen J, Dybdahl M, Friis S, Hansen RD, Wallin H, et al. Prospective study of interaction between alcohol, NSAID use and polymorphisms in genes involved in the inflammatory response in relation to risk of colorectal cancer. Mutation Res. (2007) 624:88–100. doi: 10.1016/j.mrfmmm.2007.04.006
23. Siezen CL, Bueno-de-Mesquita HB, Peeters PH, Kram NR, van Doeselaar M, van Kranen HJ. Polymorphisms in the genes involved in the arachidonic acid-pathway, fish consumption and the risk of colorectal cancer. Int J Cancer (2006) 119:297–303. doi: 10.1002/ijc.21858
24. Kuriki K, Hirose K, Matsuo K, Wakai K, Ito H, Kanemitsu Y, et al. Meat, milk, saturated fatty acids, the Pro12Ala and C161T polymorphisms of the PPARgamma gene and colorectal cancer risk in Japanese. Cancer Sci. (2006) 97:1226–35. doi: 10.1111/j.1349-7006.2006.00314.x
25. Martinez-Nava GA, Burguete-Garcia AI, Lopez-Carrillo L, Hernandez-Ramirez RU, Madrid-Marina V, Cebrian ME. PPARgamma and PPARGC1B polymorphisms modify the association between phthalate metabolites and breast cancer risk. Biomarkers (2013) 18:493–501. doi: 10.3109/1354750X.2013.816776
26. Zhang S, Chen S, Chen Y, Kang M, Liu C, Qiu H, et al. Investigation of methylenetetrahydrofolate reductase tagging polymorphisms with colorectal cancer in Chinese Han population. Oncotarget (2017) 8:63518–27. doi: 10.18632/oncotarget.18845
27. Zhai Y, Zhao WH, Chen CM. [Verification on the cut-offs of waist circumference for defining central obesity in Chinese elderly and tall adults]. Zhonghua Liu Xing Bing Xue Za Zhi. (2010) 31:621–5.
28. Zhang X, Zhang S, Li Y, Detrano RC, Chen K, Li X, et al. Association of obesity and atrial fibrillation among middle-aged and elderly Chinese. Int J Obes. (2009) 33:1318–25. doi: 10.1038/ijo.2009.157
29. Chen X, Li S, Yang Y, Yang X, Liu Y, Liu Y, et al. Genome-wide association study validation identifies novel loci for atherosclerotic cardiovascular disease. J Thromb Haemost. (2012) 10:1508–14. doi: 10.1111/j.1538-7836.2012.04815.x
30. Chen Y, Tang W, Liu C, Lin J, Wang Y, Zhang S, et al. miRNA-146a rs2910164 C>G polymorphism increased the risk of esophagogastric junction adenocarcinoma: a case-control study involving 2,740 participants. Cancer Manage Res. (2018) 10:1657–64. doi: 10.2147/CMAR.S165921
31. Yin J, Wang X, Wei J, Wang L, Shi Y, Zheng L, et al. Interleukin 12B rs3212227 T > G polymorphism was associated with an increased risk of gastric cardiac adenocarcinoma in a Chinese population. Dis Esophagus (2015) 28:291–8. doi: 10.1111/dote.12189
32. Bland JM, Altman DG. Multiple significance tests: the Bonferroni method. BMJ (1995) 310:170. doi: 10.1136/bmj.310.6973.170
33. Lesack K, Naugler C. An open-source software program for performing Bonferroni and related corrections for multiple comparisons. J Pathol Inform. (2011) 2:52. doi: 10.4103/2153-3539.91130
34. Elrod HA, Sun SY. PPARgamma and Apoptosis in Cancer. PPAR Res. (2008) 2008:704165. doi: 10.1155/2008/704165
35. Girnun GD, Smith WM, Drori S, Sarraf P, Mueller E, Eng C, et al. APC-dependent suppression of colon carcinogenesis by PPARgamma. Proc Natl Acad Sci USA. (2002) 99:13771–6. doi: 10.1073/pnas.162480299
36. Sarraf P, Mueller E, Jones D, King FJ, DeAngelo DJ, Partridge JB, et al. Differentiation and reversal of malignant changes in colon cancer through PPARgamma. Nat Med. (1998) 4:1046–52.
37. Tontonoz P, Spiegelman BM. Fat and beyond: the diverse biology of PPARgamma. Annu Rev Biochem. (2008) 77:289–312. doi: 10.1146/annurev.biochem.77.061307.091829
38. Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell (1998) 92:829–39. doi: 10.1016/S0092-8674(00)81410-5
39. Lauby-Secretan B, Scoccianti C, Loomis D, Grosse Y, Bianchini F, Straif K, et al. Body fatness and cancer–viewpoint of the IARC working group. N Engl J Med. (2016) 375:794–8. doi: 10.1056/NEJMsr1606602
40. Grygiel-Gorniak B, Kaczmarek E, Mosor M, Przyslawski J, Bogacz A. Genetic background, adipocytokines, and metabolic disorders in postmenopausal overweight and obese women. Biochem Genet. (2016) 54:636–52. doi: 10.1007/s10528-016-9743-z
41. Lv X, Zhang L, Sun J, Cai Z, Gu Q, Zhang R, et al. Interaction between peroxisome proliferator-activated receptor gamma polymorphism and obesity on type 2 diabetes in a Chinese Han population. Diabetol Metabol Syndrome (2017) 9:7. doi: 10.1186/s13098-017-0205-5
42. Lu Y, Ye X, Cao Y, Li Q, Yu X, Cheng J, et al. Genetic variants in peroxisome proliferator-activated receptor-gamma and retinoid X receptor-alpha gene and type 2 diabetes risk: a case-control study of a Chinese Han population. Diabetes Technol Therap. (2011) 13:157–64. doi: 10.1089/dia.2010.0122
43. Du J, Shi H, Lu Y, Du W, Cao Y, Li Q, et al. Tagging single nucleotide polymorphisms in the PPAR-gamma and RXR-alpha gene and type 2 diabetes risk: a case-control study of a Chinese Han population. J Biomed Res. (2011) 25:33–41. doi: 10.1016/S1674-8301(11)60004-3
44. Wang P, Wang Q, Yin Y, Yang Z, Li W, Liang D, et al. Association between peroxisome proliferator-activated receptor gamma gene polymorphisms and atherosclerotic diseases: a meta-analysis of case-control studies. J Atheroscler Thromb. (2015) 22:912–25. doi: 10.5551/jat.26138
45. Doecke JD, Zhao ZZ, Stark MS, Green AC, Hayward NK, Montgomery GW, et al. Single nucleotide polymorphisms in obesity-related genes and the risk of esophageal cancers. Cancer Epidemiol Biomarkers Prev. (2008) 17:1007–12. doi: 10.1158/1055-9965.EPI-08-0023
46. Zhou XP, Smith WM, Gimm O, Mueller E, Gao X, Sarraf P, et al. Over-representation of PPARgamma sequence variants in sporadic cases of glioblastoma multiforme: preliminary evidence for common low penetrance modifiers for brain tumour risk in the general population. J Med Genet. (2000) 37:410–4. doi: 10.1136/jmg.37.6.410
Keywords: PPARG, PPARGC1A, PPARGC1B, polymorphism, colorectal cancer, risk
Citation: Lin J, Chen Y, Tang W, Liu C, Zhang S, Guo Z, Chen G and Zheng X (2019) PPARG rs3856806 C>T Polymorphism Increased the Risk of Colorectal Cancer: A Case-Control Study in Eastern Chinese Han Population. Front. Oncol. 9:63. doi: 10.3389/fonc.2019.00063
Received: 22 October 2018; Accepted: 22 January 2019;
Published: 19 February 2019.
Edited by:
Mark De Ridder, Vrije University Brussel, BelgiumReviewed by:
Francesco Caiazza, University of California, San Francisco, United StatesCopyright © 2019 Lin, Chen, Tang, Liu, Zhang, Guo, Chen and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiong-wei Zheng, YWd1MTk2MEBmam11LmVkdS5jbg==
Gang Chen, bmFpY2hlbmdhbmdAZmptdS5lZHUuY24=
† These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.