- Department of Surgery, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai Key Laboratory for Gastric Neoplasms, Shanghai, China
The citrullination modification (Cit) of proteins has received increasing attention in recent years. This kind of protein modification was first discovered in autoimmune diseases such as rheumatoid arthritis. The citrullination modification process is catalyzed by the peptidyl arginine deiminases (PADIs) family. A well-known citrullination of histone involves the key mechanism of neutrophil extracellular traps (NETs) of inflammation in the peripheral blood. Further studies revealed that citrullination modification of proteins also involves in carcinogenesis in human being. Citrullinated proteins disturbed the stability of proteins and caused DNA damages. There is increasing evidence that citrullinated proteins can be used as potential targets for cancer diagnosis or treatment. This review introduces the concept of citrullination modification of proteins, substrate proteins, examining methods and biological significances.
Introduction
Proteins are the main executor of life activities. The epigenetics and post-translational modification of proteins, such as phosphorylation, acetylation, glycosylation, methylation, ubiquitination and citrullination have been found to play important roles on pathogenesis and carcinogenesis (1–3). Citrullination of proteins is a new kind of post-translational modification, which has been reported to be involved in large numbers of autoimmune diseases and cancers. This review focuses on the mechanisms, regulation, and the clinical significance of citrullinated proteins in the field of gastrointestinal diseases.
Definition of Citrullinated Proteins
Citrullination of protein refers to the process by which the peptidyl arginine residue is converted to citrulline by a catalytic enzyme (Figure 1). Since this process is accompanied by the removal of an amino group, it is also called a peptidyl arginine deamination reaction. This chemical reaction is accompanied by a change in electrostatic charge, which may affect the folding state and function of protein, especially on histones. To date, it has been confirmed that arginine residues of dozens of proteins can undergo citrullination modifications. The substrates could be enolase, vimentin, keratin, filaggrin, serine protease inhibitors, proteases and metabolic enzymes (4). Moreover, arginine residues of histones such as H3R2/R8/R17/R26, H4R3, H2A, and H1 could be citrullinated by peptidyl arginine deiminases (PADIs) (5–8).
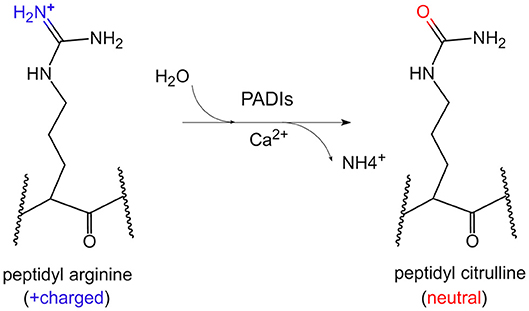
Figure 1. The chemical conversion process of protein citrullination. In the presence of calcium ions, PADIs could catalyze the formation of peptidyl citrulline from peptidyl arginine, which removes an amino group, accompanied by the positive charge becoming electrically neutral.
Citrullination of proteins is catalyzed by PADIs, which include five isoenzymes (PADI1-4 and PADI6) in humans. The genes of these five isozymes are located on chromosome 1p36.13. The coding regions of PADIs are about 2k in size, and consist of three parts: the nitrogen end, middle part and catalytic groups of carbon end. Regarding the subcellular localization, the PADI4 is located in the nucleus with a nuclear localization signal, while others are mainly localized in the cytosol (9) (Figure 2). PADI2 had been shown to be undergoing nuclear translocation in some cells for modifying histones (10). Therefore, citrullinated modification of histones may catalyzed by PADI4 and PADI2. The citrullination of proteins occurs in various life processes, including regulation of gene expression, immune response and protein degradation (10, 11). The citrullination of proteins is also associated with carcinogenesis in the stomach (12, 13), the large intestine (13–15), the pancreas (16), the liver (13), and so on.
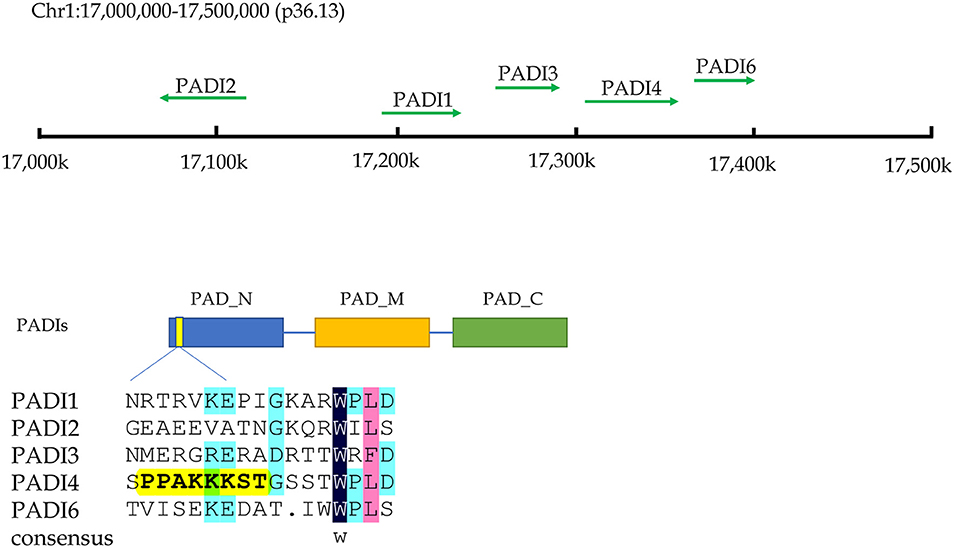
Figure 2. The chromosome location and structural characteristics of PADIs family. Five PADIs genes (PADI 1, 2, 3, 4, and 6) are located in the p36.13 of chromosome 1 across a region of approximately 500 k bases. PADIs members have similar structural regions, which can be divided into three sections: nitrogen zone, middle zone, and carbon end catalytic zone. Only PADI4 contains a nuclear localization signal at the nitrogen end of sequence, implying that PADI4 may play a role in the cell nucleus.
Citrullination of Non-histone Proteins
Citrullination of proteins could be induced by chemical compounds. Qu et al. reported that the antiparasitic drug nitazoxanide could induce citrullination of protein β-catenin in colorectal cancer cells via up-regulation of PADI2 enzyme. Citrullination of β-catenin resulted in the instability of the protein, and then inhibited the Wnt signaling pathway. ING4, a tumor suppressor protein, was identified as a substrate of PADI4 enzyme. Citrullination of ING4 interfered with its interaction with p53, and then decreased the tumor suppressor function in colon cancer cells (17). On the other hand, some research indicated that DNA damage induced PADI4, and then increased the citrullination of NPM1 and lamine C, which inhibited cell growth through the p53 pathway in colon cancer cells (18). Cantarino and colleagues found that down-regulation of PADI2 is an early event in the pathogenesis of colorectal cancer and is associated with poor prognosis (14). Overexpression of PADI2 inhibited cell growth and was accompanied with an increase in citrullinated protein in colon cancer cells. Overexpression of PADI2 did not increase cell apoptosis, but arrested the cell cycle in G1 phase (15). The exact effect of citrullination of proteins on cancer should be studied further.
Citrullination of proteins is not only detected in in vitro experiments, but also in human blood. Ordóñez et al. (19) reported that up-regulation of citrullinated antithrombin in peripheral blood of patients with rheumatoid arthritis and colorectal cancer predicted higher risk of thrombosis. Yuzhalin et al. (20) found that PADI4 could be secreted into the extracellular matrix by colorectal cancer cells, catalyzing the citrullination of proteins, thereby promoting distant metastasis of cancer cells to liver. Increased PADI4 could be found in the peripheral blood of patients with various malignancies such as gastric cancer, lung cancer, hepatocellular carcinoma, esophageal squamous cell carcinoma and breast cancer (13, 21). Until now, multiple proteins have been found as substrates of citrullination, including NF-κB p65 (22), CXCL8 (23), CXCL12 (24), E2F-1 (25), GSK3β (26), MEK1 (27), VEGFR2 (28), and so on. Obviously, citrullination of proteins involve double-sided roles in promoting both inflammation and anti-inflammation, as well as cancer promotion and inhibition.
Citrullination of Histone PROTEINs
Citrullinated modification of histones is an epigenetic event. As introduced above, both PADI2 and PADI4 involve the citrullination process of histones in the nucleus. Recently, increased citrullinated histone H3 (H3Cit) has been considered a novel prognostic blood marker in patients with advanced cancer, due to its higher levels compared to healthy controls (29). PADI2 has been found playing an important role in mediating histone H3Cit modification, and promoting disease progression in some non-digestive cancers (30, 31). McNee et al. (32) found that PADI2 could up-regulate IL-6 expression by catalyzing H3R26Cit of bone marrow mesenchymal stem cells of multiple myeloma, which ultimately lead to chemo-resistance to bortezomib. PADI4 is another important enzyme in catalyzing the citrullination of histones. DNA damage could activate the PADI4-p53 network and catalyze histone chaperone protein, nucleophosmin (NPM1) (18). In addition, DNA damage could catalyze citrullination of the arginine 3 residue of histone H4 (H4R3cit) through the p53-PADI4 pathway in non-small cell lung cancer (33).
Citrullination of Proteins and Immune Response
The immune system is a major weapon against cancer. Citrullination of proteins exist widely in immune-related diseases and cancers. Makrygiannakis and colleagues examined biopsy tissues from rheumatoid arthritis, myositis, tonsillitis and inflammatory bowel disease via immunohistochemistry. They found that there is a significant increase in citrullinated proteins in inflammatory tissues, compared to corresponding normal controls (34). The immune system is composed of innate immunity and acquired immunity. Neutrophils are a member of the cells of innate immunity. In process of clearing bacteria, the neutrophils secrete cell DNA, histones, and intracellular proteins to the extracellular space or circulatory system, forming so-called neutrophil extracellular traps (NETs). The citrullination of histones is involved in the process of NETs. In this process, PADI4 mediates the citrullination of histones, and results in the unwinding of DNA and subsequently excreting into the extracellular space (35–37). NETs are a self-protective mechanism against harmful bacteria. Recently, Thalin et al. found that H3Cit was significantly increased in the peripheral blood of advanced cancer patients (29). The proportion of H3Cit-positive neutrophils was increased in more serious patients. The expression level of H3Cit of serum was strongly correlated with the neutrophil activation markers, such as neutrophil elastase, myeloperoxidase and NETs-induced factors IL-6, as well as IL-8. Therefore, H3Cit is considered a useful blood biomarker for evaluating inflammatory response and prognosis in advanced cancers. Up-regulation of NETs was also identified in pancreatic ductal adenocarcinoma. The histone modification of H3Cit was proposed as a marker of NETs (16). In the pancreas, stimulating factors such as pancreatic juice could induce NETs in pancreatic ducts. Excess in NETs blocks the pancreatic duct and eventually causes pancreatitis (38).
In the cancer immunity area, the new epitopes caused by post-translational modification of proteins may provide a novel target for cancer-specific immune therapy. The condition of the cancer microenvironment including nutrient deficiency, hypoxia, redox stress and DNA damage could irritate active expression of PADIs, and catalyze production of citrullinated peptides. Increased content of citrullinated peptides may be a good target for the immune system. The cancer-specific microenvironment could induce the immune response by citrullinated peptides, and this is non-toxic and safe to the host. Carbohydrate metabolizing enzyme α-enolase is a substrate of citrullinated modification. Cook et al. (39) found that citrullination significantly induced elevation of α-enolase in Th1 immune cells, while unmodified wild-type peptides of α-enolase did not show this efficacy. Citrullinated peptides of α-enolase also induced CD4+ T activation (40, 41). The results suggested that developing tumor vaccines against citrullinated peptides of α-enolase may be a useful strategy (39). The function of citrullinated protein epitope has revealed promising utility in anti-cancer immunity.
Detection and Biological Significance of Citrullination Modification
Citrullination modification of proteins has been reported in several fields of cancer research. Along with the progression of biomedical techniques, detection and identification of citrullinated proteins in complex biological systems becomes more feasible. Clinically, the detection of anti-cyclic citrulline antibody has been used as an assistive method for diagnosis and monitoring clinical rheumatoid arthritis (42, 43). Since the citrullination modification itself leads to 1Da mass change only, detection of the change of low abundance is still a challenging work. Phenylglyoxal (PG) could be covalently bonded with citrullinated residues specifically, and used for specific probes of labeling citrullinated proteins. The reaction could be colored by coupling dyes such as rhodamine (Rh) or biotin, and then identified by ELISA or mass spectrometry (13, 21, 29, 42, 43). By means of this technology, more and more antigens with citrullinated modification could be found, which will provide new targets for diagnosis and treatment of cancers.
In an animal experiment, Mohamed and colleagues found that nanomaterials could induce production of citrullinated protein and auto-antibodies in mice. In their study, after injection of nickel nanowires into mice, the levels of citrullinated protein and PADIs enzymes were elevated in the spleen, kidney and lymph nodes of mice, implying a systemic response to environmental materials (44). Their results suggested that safety of the nanoparticles needs to be evaluated further. Citrullination modification of proteins may be an important event for the host to recognize foreign antigens. Citrullinated proteins may be recognized as new antigens, and are promising for targeted therapy or CAR-T/NK cell-specific recognition targets.
Inhibitors of PADIs demonstrated strong potential of anti-autoimmune and anti-cancer functions in vitro and in vivo. PADI4 is the only member of the PADI family containing a nuclear localization signal, and can citrullinate many substrates including histones. PADI4 functions as a corepressor of p53 and cooperates with a histone deacetylase HDAC2 to repress the expression of tumor suppressor genes. Chlor-amidine (Cl-amidine) is a pan-PADI inhibitor that shows inhibitory effects on several members of PADIs family. However, its higher IC50 (150–200 μm) limit its preclinical exploration in cancer study and treatment (44–47). Recently, Wang and colleagues found a lead compound, YW3-56, which could activate a cohort of p53 target genes, and realize inhibitory efficacy on the mTORC1 signaling pathway, thereby disturbing autophagy and inhibiting cancerous cell growth (45). However, since the feature of a pan-PADIs inhibitor, Cl-amidine, is still be used in experimental study (48), and many new small molecule inhibitors of PADI4 are being developed by pharmacologists (49).
In summary, compared to other modification of proteins, citrullination modification is relatively novel. The exact regulatory mechanisms and biological significance in carcinogenesis are largely unclear. As shown in Figure 3, many substrates of citrullination modification are very important in life processes and development of cancers. The accurate identification of citrullination sites may help researchers to elucidate the underlying molecular mechanisms of citrullination and designing drugs for related human diseases. Several groups made efforts to predict citrullination sites by bioinformatics. Ju and Wang (50) provided a user-friendly web-server for CKSAAP_CitrSite. Zhang et al. (51) published their pioneering work of maximum-relevance-minimum-redundancy to analyze citrullination sites, and constructed classifier by random forest algorithm. We believe that in citrullination research area, bioinformatics will provide some useful insights and assistance.
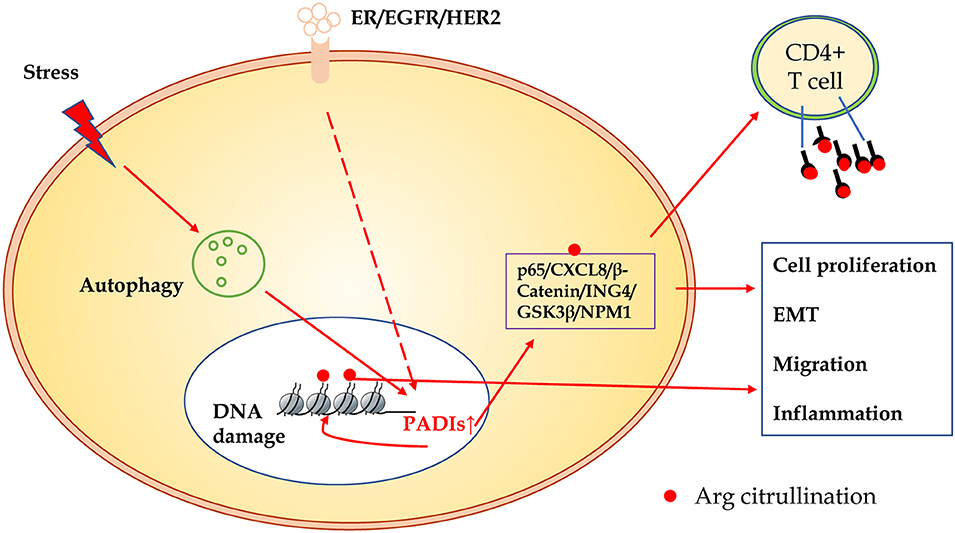
Figure 3. The schematic presentation of citrullinated modification of proteins and its biological significance. PADIs enzymes are activated through receptors of ER/EGFR/HER2, oxidative stress, hypoxia, and other microenvironment factors, which initiate autophagy and DNA damage. Increased PADIs catalyze citrullination modification of histones and non-histone proteins, and result in cell proliferation, epithelial-mesenchymal transition, migration, and inflammation. Citrullinated proteins as a new antigen may activate immune response of T cells or induce specific antibodies.
Author Contributions
SS and YY were involved in concept and design. All authors wrote, reviewed and revised the manuscript.
Funding
This project was supported by National Key R&D Program of China (2017YFC0908300, 2016YFC1303200), the National Natural Science Foundation of China (81772505), Shanghai Science and Technology Committee (18411953100), the Cross-Institute Research Fund of Shanghai Jiao Tong University (YG2017ZD01, YG2015MS62), Innovation Foundation of Translational Medicine of Shanghai Jiao Tong University School of Medicine (15ZH4001, TM201617, and TM201702), and Technology Transfer Project of Science & Technology Department Shanghai Jiao Tong University School of Medicine.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Bettermann K, Benesch M, Weis S, Haybaeck J. SUMOylation in carcinogenesis. Cancer Lett. (2012) 316:113–25. doi: 10.1016/j.canlet.2011.10.036
2. Okudela K, Mitsui H, Suzuki T, Woo T, Tateishi Y, Umeda S, et al. Expression of HDAC9 in lung cancer–potential role in lung carcinogenesis. Int J Clin Exp Pathol. (2014) 7:213–20.
3. Chrun ES, Modolo F, Daniel FI. Histone modifications: a review about the presence of this epigenetic phenomenon in carcinogenesis. Pathol Res Pract. (2017) 213:1329–39. doi: 10.1016/j.prp.2017.06.013
4. Tilvawala R, Nguyen SH, Maurais AJ, Nemmara VV, Nagar M, Salinger AJ, et al. The rheumatoid arthritis-associated citrullinome. Cell Chem Biol. (2018) 25:691–704.e6. doi: 10.1016/j.chembiol.2018.03.002
5. Hagiwara T, Nakashima K, Hirano H, Senshu T, Yamada M. Deimination of arginine residues in nucleophosmin/B23 and histones in HL-60 granulocytes. Biochem Biophys Res Commun. (2002) 290:979–83. doi: 10.1006/bbrc.2001.6303
6. Arita K, Shimizu T, Hashimoto H, Hidaka Y, Yamada M, Sato M. Structural basis for histone N-terminal recognition by human peptidylarginine deiminase 4. Proc Natl Acad Sci USA. (2006) 103:5291–6. doi: 10.1073/pnas.0509639103
7. Saiki M, Watase M, Matsubayashi H, Hidaka Y. Recognition of the N-terminal histone H2A and H3 peptides by peptidylarginine deiminase IV. Protein Pept Lett. (2009) 16:1012–6. doi: 10.2174/092986609789055449
8. Christophorou MA, Castelo-Branco G, Halley-Stott RP, Oliveira CS, Loos R, Radzisheuskaya A, et al. Citrullination regulates pluripotency and histone H1 binding to chromatin. Nature (2014) 507:104–8. doi: 10.1038/nature12942
9. Nakashima K, Hagiwara T, Yamada M. Nuclear localization of peptidylarginine deiminase V and histone deimination in granulocytes. J Biol Chem. (2002) 277:49562–8. doi: 10.1074/jbc.M208795200
10. Amin B, Voelter W. Human deiminases: isoforms, substrate specificities, kinetics, and detection. Prog Chem Org Nat Prod. (2017) 106:203–40. doi: 10.1007/978-3-319-59542-9_2
11. Fert-Bober J, Giles JT, Holewinski RJ, Kirk JA, Uhrigshardt H, Crowgey EL, et al. Citrullination of myofilament proteins in heart failure. Cardiovasc Res. (2015) 108:232–42. doi: 10.1093/cvr/cvv185
12. Zheng Y, Zhao G, Xu B, Liu C, Li C, Zhang X, et al. PADI4 has genetic susceptibility to gastric carcinoma and upregulates CXCR2, KRT14 and TNF-alpha expression levels. Oncotarget (2016) 7:62159–76. doi: 10.18632/oncotarget.11398
13. Guo W, Zheng Y, Xu B, Ma F, Li C, Zhang X, et al. Investigating the expression, effect and tumorigenic pathway of PADI2 in tumors. Onco Targets Ther. (2017) 10:1475–85. doi: 10.2147/OTT.S92389
14. Cantariño N, Musulén E, Valero V, Peinado MA, Perucho M, Moreno V, et al. Downregulation of the deiminase PADI2 is an early event in colorectal carcinogenesis and indicates poor prognosis. Mol Cancer Res. (2016) 14:841–8. doi: 10.1158/1541-7786.MCR-16-0034
15. Funayama R, Taniguchi H, Mizuma M, Fujishima F, Kobayashi M, Ohnuma S, et al. Protein-arginine deiminase 2 suppresses proliferation of colon cancer cells through protein citrullination. Cancer Sci. (2017) 108:713–8. doi: 10.1111/cas.13179
16. Boone BA, Orlichenko L, Schapiro NE, Loughran P, Gianfrate GC, Ellis JT, et al. The receptor for advanced glycation end products (RAGE) enhances autophagy and neutrophil extracellular traps in pancreatic cancer. Cancer Gene Ther. (2015) 22:326–34. doi: 10.1038/cgt.2015.21
17. Guo Q, Fast W. Citrullination of inhibitor of growth 4 (ING4) by peptidylarginine deminase 4 (PAD4) disrupts the interaction between ING4 and p53. J Biol Chem. (2011) 286:17069–78. doi: 10.1074/jbc.M111.230961
18. Tanikawa C, Ueda K, Nakagawa H, Yoshida N, Nakamura Y, Matsuda K. Regulation of protein Citrullination through p53/PADI4 network in DNA damage response. Cancer Res. (2009) 69:8761–9. doi: 10.1158/0008-5472.CAN-09-2280
19. Ordóñez A, Yélamos J, Pedersen S, Miñano A, Conesa-Zamora P, Kristensen SR, et al. Increased levels of citrullinated antithrombin in plasma of patients with rheumatoid arthritis and colorectal adenocarcinoma determined by a newly developed ELISA using a specific monoclonal antibody. Thromb Haemost. (2010) 104:1143–9. doi: 10.1160/TH10-05-0297
20. Yuzhalin AE, Gordon-Weeks AN, Tognoli ML, Jones K, Markelc B, Konietzny R, et al. Colorectal cancer liver metastatic growth depends on PAD4-driven citrullination of the extracellular matrix. Nat Commun. (2018) 9:4783. doi: 10.1038/s41467-018-07306-7
21. Chang X, Han J, Pang L, Zhao Y, Yang Y, Shen Z. Increased PADI4 expression in blood and tissues of patients with malignant tumors. BMC Cancer (2009) 9:40. doi: 10.1186/1471-2407-9-40
22. Sun B, Dwivedi N, Bechtel TJ, Paulsen JL, Muth A, Bawadekar M, et al. Citrullination of NF-kappaB p65 promotes its nuclear localization and TLR-induced expression of IL-1beta and TNFalpha. Sci Immunol. (2017) 2:eaal3062. doi: 10.1126/sciimmunol.aal3062
23. Proost P, Loos T, Mortier A, Schutyser E, Gouwy M, Noppen S, et al. Citrullination of CXCL8 by peptidylarginine deiminase alters receptor usage, prevents proteolysis, and dampens tissue inflammation. J Exp Med. (2008) 205:2085–97. doi: 10.1084/jem.20080305
24. Struyf S, Noppen S, Loos T, Mortier A, Gouwy M, Verbeke H, et al. Citrullination of CXCL12 differentially reduces CXCR4 and CXCR7 binding with loss of inflammatory and anti-HIV-1 activity via CXCR4. J Immunol. (2009) 182:666–74. doi: 10.4049/jimmunol.182.1.666
25. Ghari F, Quirke AM, Munro S, Kawalkowska J, Picaud S, McGouran J, et al. Citrullination-acetylation interplay guides E2F-1 activity during the inflammatory response. Sci Adv. (2016) 2:e1501257. doi: 10.1126/sciadv.1501257
26. Stadler SC, Vincent CT, Fedorov VD, Patsialou A, Cherrington BD, Wakshlag JJ, et al. Dysregulation of PAD4-mediated citrullination of nuclear GSK3beta activates TGF-beta signaling and induces epithelial-to-mesenchymal transition in breast cancer cells. Proc Natl Acad Sci USA. (2013) 110:11851–6. doi: 10.1073/pnas.1308362110
27. Qin H, Liu X, Li F, Miao L, Li T, Xu B, et al. PAD1 promotes epithelial-mesenchymal transition and metastasis in triple-negative breast cancer cells by regulating MEK1-ERK1/2-MMP2 signaling. Cancer Lett. (2017) 409:30–41. doi: 10.1016/j.canlet.2017.08.019
28. Sase T, Arito M, Onodera H, Omoteyama K, Kurokawa MS, Kagami Y, et al. Hypoxia-induced production of peptidylarginine deiminases and citrullinated proteins in malignant glioma cells. Biochem Biophys Res Commun. (2017) 482:50–6. doi: 10.1016/j.bbrc.2016.10.154
29. Thålin C, Lundström S, Seignez C, Daleskog M, Lundström A, Henriksson P, et al. Citrullinated histone H3 as a novel prognostic blood marker in patients with advanced cancer. PLoS ONE (2018) 13:e0191231. doi: 10.1371/journal.pone.0191231
30. Wang L, Song G, Zhang X, Feng T, Pan J, Chen W, et al. PADI2-mediated citrullination promotes prostate cancer progression. Cancer Res. (2017) 77:5755–68. doi: 10.1158/0008-5472.CAN-17-0150
31. DeVore SB, Young CH, Li G, Sundararajan A, Ramaraj T, Mudge J, et al. Histone citrullination represses miRNA expression resulting in increased oncogene mRNAs in somatolactotrope cells. Mol Cell Biol. (2018) 38:e00084–18. doi: 10.1128/MCB.00084-18
32. McNee G, Eales KL, Wei W, Williams DS, Barkhuizen A, Bartlett DB, et al. Citrullination of histone H3 drives IL-6 production by bone marrow mesenchymal stem cells in MGUS and multiple myeloma. Leukemia (2017) 31:373–81. doi: 10.1038/leu.2016.187
33. Tanikawa C, Espinosa M, Suzuki A, Masuda K, Yamamoto K, Tsuchiya E, et al. Regulation of histone modification and chromatin structure by the p53-PADI4 pathway. Nat Commun. (2012) 3:676. doi: 10.1038/ncomms1676
34. Makrygiannakis D, af Klint E, Lundberg IE, Löfberg R, Ulfgren AK, Klareskog L, et al. Citrullination is an inflammation-dependent process. Ann Rheum Dis. (2006) 65:1219–22. doi: 10.1136/ard.2005.049403
35. Neeli I, Khan SN, Radic M. Histone deimination as a response to inflammatory stimuli in neutrophils. J Immunol. (2008) 180:1895–902. doi: 10.4049/jimmunol.180.3.1895
36. Wang Y, Li M, Stadler S, Correll S, Li P, Wang D, et al. Histone hypercitrullination mediates chromatin decondensation and neutrophil extracellular trap formation. J Cell Biol. (2009) 184:205–13. doi: 10.1083/jcb.200806072
37. Honda M, Kubes P. Neutrophils and neutrophil extracellular traps in the liver and gastrointestinal system. Nat Rev Gastroenterol Hepatol. (2018) 15:206–21. doi: 10.1038/nrgastro.2017.183
38. Leppkes M, Maueröder C, Hirth S, Nowecki S, Günther C, Billmeier U, et al. Externalized decondensed neutrophil chromatin occludes pancreatic ducts and drives pancreatitis. Nat Commun. (2016) 7:10973. doi: 10.1038/ncomms10973
39. Cook K, Daniels I, Symonds P, Pitt T, Gijon M, Xue W, et al. Citrullinated alpha-enolase is an effective target for anti-cancer immunity. Oncoimmunology (2018) 7:e1390642. doi: 10.1080/2162402X.2017.1390642
40. Brentville VA, Metheringham RL, Gunn B, Symonds P, Daniels I, Gijon M, et al. Citrullinated vimentin presented on MHC-II in tumor cells is a target for CD4+ T-cell-mediated antitumor immunity. Cancer Res. (2016) 76:548–60. doi: 10.1158/0008-5472.CAN-15-1085
41. Durrant LG, Metheringham RL, Brentville VA. Autophagy, citrullination and cancer. Autophagy (2016) 12:1055–6. doi: 10.1080/15548627.2016.1166326
42. Bicker KL, Subramanian V, Chumanevich AA, Hofseth LJ, Thompson PR. Seeing citrulline: development of a phenylglyoxal-based probe to visualize protein citrullination. J Am Chem Soc. (2012) 134:17015–8. doi: 10.1021/ja308871v
43. Lewallen DM, Bicker KL, Subramanian V, Clancy KW, Slade DJ, Martell J, et al. Chemical proteomic platform to identify citrullinated proteins. ACS Chem Biol. (2015) 10:2520–8. doi: 10.1021/acschembio.5b00438
44. Mohamed BM, Boyle NT, Schinwald A, Murer B, Ward R, Mahfoud OK, et al. Induction of protein citrullination and auto-antibodies production in murine exposed to nickel nanomaterials. Sci Rep. (2018) 8:679. doi: 10.1038/s41598-017-19068-1
45. Wang Y, Li P, Wang S, Hu J, Chen XA, Wu J, et al. Anticancer peptidylarginine deiminase (PAD) inhibitors regulate the autophagy flux and the mammalian target of rapamycin complex 1 activity. J Biol Chem. (2012) 287:25941–53. doi: 10.1074/jbc.M112.375725
46. Mondal S, Parelkar SS, Nagar M, Thompson PR. Photochemical control of protein arginine deiminase (PAD) activity. ACS Chem Biol. (2018) 13:1057–65. doi: 10.1021/acschembio.8b00053
47. Bozdag M, Dreker T, Henry C, Tosco P, Vallaro M, Fruttero R, et al. Novel small molecule protein arginine deiminase 4 (PAD4) inhibitors. Bioorg Med Chem Lett. (2013) 23:715–9. doi: 10.1016/j.bmcl.2012.11.102
48. Witalison EE, Cui X, Hofseth AB, Subramanian V, Causey CP, Thompson PR, et al. Inhibiting protein arginine deiminases has antioxidant consequences. J Pharmacol Exp Ther. (2015) 353:64–70. doi: 10.1124/jpet.115.222745
49. Witalison EE, Cui X, Causey CP, Thompson PR, Hofseth LJ. Molecular targeting of protein arginine deiminases to suppress colitis and prevent colon cancer. Oncotarget (2015) 6:36053–62. doi: 10.18632/oncotarget.5937
50. Ju Z, Wang SY. Prediction of citrullination sites by incorporating k-spaced amino acid pairs into Chou's general pseudo amino acid composition. Gene (2018) 664:78–83. doi: 10.1016/j.gene.2018.04.055
Keywords: citrullination, proteins, histone, PADIs, molecular targets
Citation: Song S and Yu Y (2019) Progression on Citrullination of Proteins in Gastrointestinal Cancers. Front. Oncol. 9:15. doi: 10.3389/fonc.2019.00015
Received: 18 November 2018; Accepted: 04 January 2019;
Published: 23 January 2019.
Edited by:
Zhe-Sheng Chen, St. John's University, United StatesReviewed by:
Kathy Keqin LI, Georgia State University, United StatesQingyuan Zhang, Ningxia Medical University, China
Qi Liu, Tongji University, China
Copyright © 2019 Song and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yingyan Yu, eWluZ3lhbjN5QHNqdHUuZWR1LmNu
 Shuzheng Song
Shuzheng Song Yingyan Yu
Yingyan Yu