- 1Institute for Experimental Biomedicine II, University Clinic, Wuerzburg, Germany
- 2Department of Cancer Biology, University of Toledo Health Science Campus, Toledo, OH, United States
In the recent two decades, LIM and SH3 protein 1 (LASP1) has been developed from a simple actin-binding structural protein to a tumor biomarker and subsequently to a complex, nuclear transcriptional regulator. Starting with a brief historical perspective, this review will mainly compare and contrast LASP1 and LASP2 from the angle of the newest data and importantly, examine their role in transcriptional regulation. We will summarize the current knowledge through pictorial models and tables including the roles of different microRNAs in the differential regulation of LASP1 levels and patient outcome rather than specify in detail all tumor entities. Finally, the novel functional roles of LASP1 in secretion of vesicles, expression of matrix metalloproteinases and transcriptional regulation as well as the activation of survival and proliferation pathways in different cancer types are described.
Introduction
LIM and SH3 protein1 (LASP1) was originally identified in human metastatic lymph nodes from breast cancer and was named as MLN50 (1). LASP1 is located on the chromosome 17q21, a region often mutated or amplified in 20–30% of breast cancer patients and in the neighborhood of the proto-oncogene c-ERBB2 or human epidermal growth factor receptor 2 (HER2) (2) and the breast cancer susceptibility gene1 (BRCA1) (3). Though LASP1 was originally identified as a structural cytoskeletal protein, an explosive production of data in recent years provided evidence that it can orchestrate and execute multivarious roles ranging from cell signaling to transcriptional regulation.
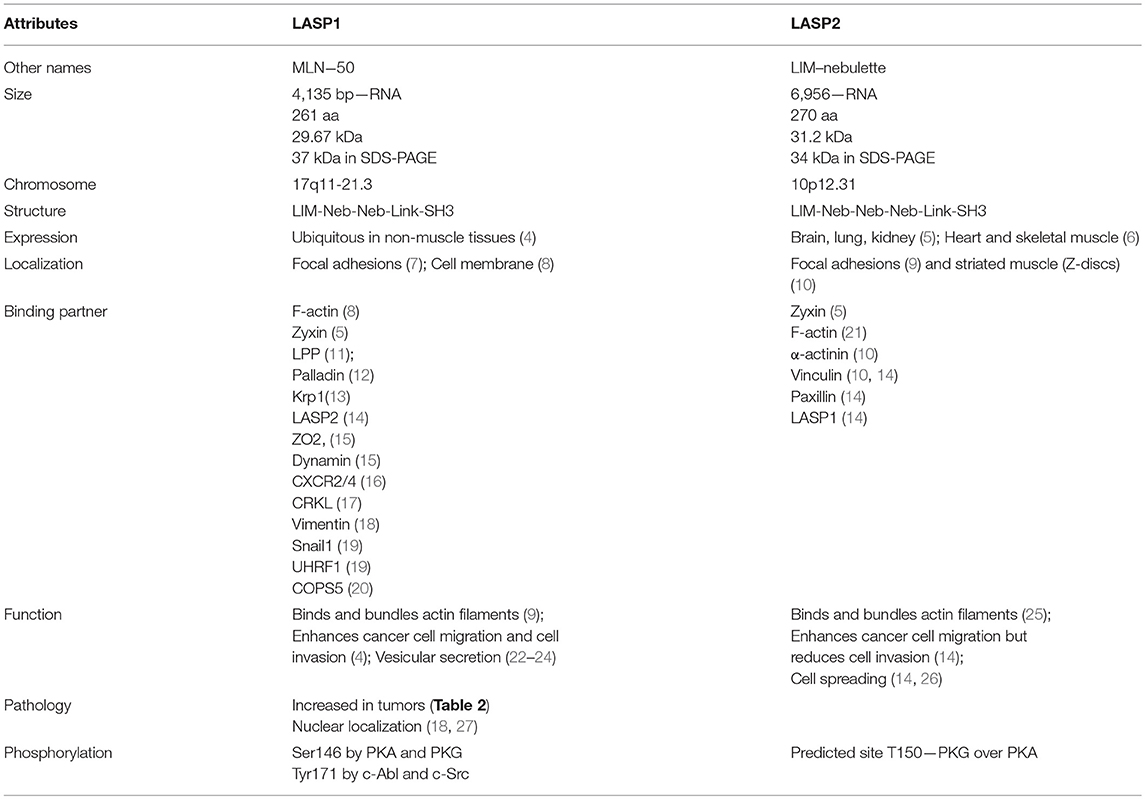
Comparative Structural Organization of LASP1 and LASP2
The mRNA for LASP1 is 4,135 bp long (NM_006148.3) and codes for a protein of 261 amino acid residues with a molecular mass of 29.7 kDa. Table 1 summarizes the similarities and the differences between LASP1 and LASP2. The LASP1 protein runs at 37–38 kDa in Western blot analysis and so far there are no structural explanations provided for this anomalous migration. Proteins with high proline content are notorious for running slower than their actual molecular mass during sodium dodecyl sulfate-polyacrylamide gel electrophoretic (SDS-PAGE) analysis. However, with a proline content of only 6.8% in LASP1, compared to 5.6% median, this does not explain the observed retarded migration in SDS-PAGE. The mRNA for LASP2 is encoded by 6,956 bp (NM_213569.2) that codes for a protein of 270 amino acid residues with a molecular mass of 31.3 kDa. LASP2 is a human ortholog of LASP1 and it was first predicted in silico (28) and was subsequently characterized in detail (9, 10, 14, 21). LASP1 and LASP2 are members of the large actin-binding nebulin family. However, both proteins harbor significantly lower nebulin-like repeats (NR) when compared to nebulin and nebulette and are more widely expressed (29). In addition to the characteristic NRs (two in LASP1and LASP2 and a predicted third one in LASP2) and a variable linker region, both proteins contain a src homology 3 (SH3) domain at the carboxyl-terminus that is absent in nebulin and nebulette (1, 8, 30, 31) (Figure 1). Phylogenetic analysis revealed an early expression of orthologous proteins in insects and invertebrates (Bombyx mori—silk worm, Caenorhabditis elegans—nematode, Drosophila melanogaster—fruit fly) that are ancestral to LASP1 in vertebrates (Gallus gallus—red jungle fowl) and mammals (Oryctolagus cuniculus—rabbit, Mus musculus—mouse, and Homo sapiens—Human) (32).
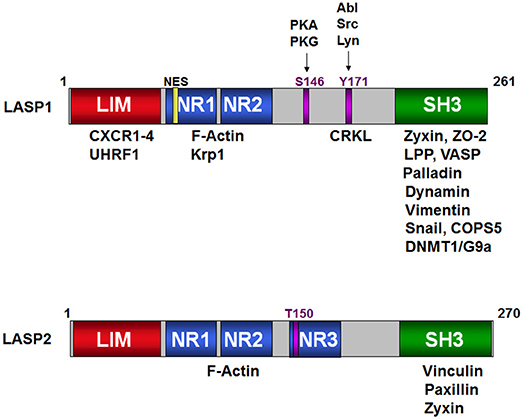
Figure 1. Schematic structure of LASP1 and LASP2. Binding of the different domains of LASP1 to a variety of signaling, structural, epigenetic proteins and transcription factors is shown on the top. The S146 and T171 phosphorylation sites are shown against their respective kinases. NR1, NR2, and NR3 are the nebulin repeats that bind to F-actin. Similarly, the proteins that bind to LASP2 are shown in the cartoon at the bottom. T150 is the predicted phosphorylation site in analogy to S146 in LASP1.
Expression
LASP1 is ubiquitously expressed in normal human tissues (except smooth muscle) at low levels. High expression is observed in the hematopoietic system (blood cells) and the cells of the gastrointestinal tract. Interestingly, enhanced levels of LASP1 are also detected in fetal tissue, such as umbilical vein endothelial cells, fetal brain and liver, suggesting a prominent role in fetal development (4). However, LASP1 knockout mice develop normally (22) assuming a compensation for LASP1 deletion.
LASP1 is mainly located at focal adhesions (8, 7), in podosomes (33), at the leading edges of lamellipodia (9) and tips of filopodia in vitro and in animal models. The “Fluorescence Recovery After Photobleaching (FRAP)” approach demonstrated convincingly that the fluorescence from enhanced green fluorescent protein (EGFP)-LASP1 recovered from the base of actin bundles against the retrograde flow of actin filaments to the tip complex of the cell (9). In a similar experiment, GFP-LASP1, GFP-LASP2, and GFP-nebulette were shown to colocalize with α-actinin and vinculin at sarcomeric Z-lines or Z-disc in the periphery of spreading cardiomyocytes. However, the interaction with the A-band was only observed for LASP1 (10). LASP1 binding to F-actin and accumulation at Z-edges was also seen in Drosophila (10, 34), supporting the importance of this protein in evolutionary development. In contrast, localization of LASP2 at focal adhesion increased the rate of attachment and spreading for cells (26).
In several cancer types, an overexpression of LASP1 has been reported. Table 2 summarizes upregulation of LASP1 in different tumor entities. In this respect, an increased nuclear translocation of LASP1 into the nucleus inversely correlated with patient survival (i.e., poor prognosis) in breast cancer (27), prostate cancer (55), medulloblastoma (59), and hepatocellular carcinoma (18, 48). This will be discussed in detail later.
Structure of LASP1 Domains and Its Interacting Proteins
The N-terminal cysteine rich LIM domain (residues 5–57) is composed of two zinc finger domains and functions as an adaptor for multimeric protein complexes (Figure 1, Schematic structure). LIM motif defines one class of zinc binding domains originally observed in lin-11, Isl-1, and mec-3 proteins (Lin/Isl/Mec). Generally, LIM domain with double finger binds to two zinc ions based on spectroscopic observations (72). Solution structure of the LIM1 domain of cysteine- and glycine-rich protein 2 (CRP2) from Quail revealed that a flexible hydrophobic core of the LIM1 domain provides an optimal binding interface for its physiological targets (73). For this domain, a direct binding to the carboxyl-termini of CXC chemokine receptors 1–4 has been shown (16). While binding of LASP1 to CXCR1-3 is independent of the phosphorylation status of LASP1, the interaction with CXCR4 requires LASP1 phosphorylation at S146. Recently, binding of UHRF1 to LASP1, predominantly to the LIM domain and also in association with DNMT1 and G9a at the SH3 domain, was shown (19) (Table 3). This protein complex regulates chromatin structure and gene expression at late G1 phase and continues during G2 and M phases of the cell cycle, thus nicely explaining the observed G2/M block in LASP1 depleted cells (27, 50, 69).
Two nebulin-like repeats (NR) (residues 62–92 and 98–128) (R1 and R2 in Figure 1) are present in LASP1 following the N-terminal LIM domain. They vary in length (30–35 amino acid residues) and display a conserved motif: SDXXYK (76, 77). The NRs of LASP1 directly interact with filamentous actin (F-actin) (8). Photobleaching experiments with GFP-actin suggested that LASP1 is involved in actin bundling but not in polymerization of actin (9). Interestingly, in human macrophages, LASP1 has been shown to associate with annular F-actin in podosomes and to facilitate proteolysis of extracellular matrix components (33). NR of LASP1 also associates with Kelch-related protein 1 (Krp1) (13, 74) (Table 3). It remains to be elucidated if the association of Krp1 and binding of F-actin to NR of LASP1 is mutually exclusive as the authors employed whole cell lysate for the pulldown experiments and did not control for indirect Krp1 association to tagged LASP1 via F-actin binding.
A linker region (residues 129–202) that follows NR, harbors two phosphorylation sites: S146, which is phosphorylated by protein kinase A (PKA) and protein kinase G (PKG) (78) and is dephosphorylated by protein phosphatase 2B (PP2B) (15), and Y171 that is phosphorylated by c-Src (79) and c-Abl non-receptor tyrosine kinases (27, 80). Phosphorylation at S146 reduces affinity of LASP1 for F-actin, zyxin, and lipoma protein partner (LPP) and allows subcellular relocalization of LASP1 to the cytosol (15, 78). In activated platelets, phosphorylation at Y171 by c-Src kinase leads to relocalization from focal contacts into the leading lamellae of the migrating/spreading cell. In apoptotic cells, activation of c-Abl prevented localization of LASP1 to focal contacts and that possibly disrupted survival signals emanating from these structures (80). Recently, phosphorylation of LASP1 at Y171 by the oncogenic BCR-ABL tyrosine kinase in chronic myeloid leukemia (CML) patients was reported. This study further demonstrated a physiological interaction between pY171-LASP1 and the src homology 2 (SH2) domain of CRK-like protein (CRKL) at amino acid sequence 36–41 that was absent under pathophysiological hyperactivation of BCR-ABL (17). In rabbit, a second PKA site at S99 was described (81); however, the sequence with just one arginine at −2 position of the phosphoserine is not an optimal consensus sequence for PKA and PKG (82). In mouse, T156 compensates for human S146 (11). The tyrosine sequence is conserved (Figure 2, Alignment). For LASP2, no functional phosphorylation sites have been reported, so far. Nevertheless, a putative consensus sequence for PKG over PKA, that resembles S146 in LASP1, is present at T150 (RKNTQ) in the LASP2 sequence while Y184 in LASP2, corresponding to Y171 in human LASP1, is unspecific to any tyrosine kinase when analyzed by NetPhos 3.1 program.

Figure 2. The sequence alignment of human LASP1 with its orthologous proteins. The protein sequences for human LASP1, LASP2, and mouse and rabbit LASP1 were aligned using clustalW program. The phosphorylation sites and different antibody recognition sites are color coded and depicted.
The carboxy-terminal SH3 domain (residues 203–261) of LASP1 binds to proteins with proline-rich motifs. Based on biochemical interaction studies, a differential comparison of proteins associating with LASP1 is shown in Table 3 and reviewed in more detail by Orth et al. (4). A recent bioinformatics approach listed putative LASP1 binding proteins mainly identified by Affinity-Capture-MS and Two-hybrid approach (83).
LASP2 Domain Structure and Binding Partners
Like LASP1, binding of LASP2 to F-actin (9) has been demonstrated and the nebulin repeats are required for this interaction (Figure 1; Table 4). Binding of LASP2 to zyxin via SH3 domain was demonstrated by Li et al. showing an interaction between the SH3 domain of LASP1 and the PXXP-motif at the N-terminus of zyxin, localizing LASP1 to the focal adhesions (5). Recently, binding to vinculin and paxillin at focal contacts was described (14) (Table 4) and mapped to the SH3 domain of LASP2. Interestingly, the authors detected no interaction between LASP1 and vinculin or paxillin, although the SH3 domains of LASP1 and LASP2 are highly homologous (Figure 2). Furthermore, co-immunoprecipitation confirmed a binding between LASP2 and LASP1 (heterodimerization) concomitant with a reduced binding of LASP2 to vinculin and paxillin. A displacement of both proteins by LASP1 was discussed. Like LASP1, overexpression of LASP2 enhanced cell migration and cell spreading when ectopically expressed in fibroblasts (26). However, in contrast to LASP1, LASP2 is associated with reduced cell invasion (14). In this context, it is interesting to note, that high LASP1 expression was accompanied with downregulated levels of LASP2 in colorectal cancer (CRC) cell lines and tissues (84). This is in agreement with a report demonstrating enhanced expression and release of matrix metalloproteinases (MMP) by LASP1 in breast cancer cells with high metastatic potential (24). It should be noted that the rabbit polyclonal antibody against LASP2 (ELQRLKRTQE) used by Wang et al. (84) showed partial homology with the LASP1 sequence (ELQRIKKTQD). Therefore, cross-reactivity cannot be excluded and might explain in part the observed inconsistency at the translational level for LASP1 in this paper.
A similar problem occurred in the recent paper by Zhang et al. when studying the effect of LASP2 on non-small cell lung cancer (NSCLC) and phosphorylation of focal adhesion kinase (FAK) (85). The used antibody, claimed to be specific for LASP2 was actually generated against nebulette (NEBL). The immunogen sequence given by the company does not correspond in any part to LASP2. Therefore, the data should be handled with precaution. Overall, there are clear functional differences between LASP1 and LASP2 (see Table 1) which also explain the fact that LASP2 did not compensate for LASP1 knockout in mice (22).
Regulation of LASP1 Expression
There is strong evidence that LASP1 is upregulated in tumors under hypoxic conditions (86, 87). Hypoxia response elements were identified in the LASP1 promoter and were shown to stimulate LASP1 expression in pancreatic cells in vitro and in mouse tumor xenografts (71). Furthermore, several reports demonstrate an overexpression of LASP1 in response to microRNA (miRNA) downregulation (Table 2). Prediction of microRNA target sequences at the 3‘UTR region of LASP1 by “TargetScanHuman ver 7.1” revealed more than 15 putative miRNAs including miR-145, miR-218, miR-133a, miR-1, miR-29, miR-218, and mainly miR-203 that have been connected to LASP1 overexpression (Table 2). Conversely, in preeclampsia, hypoxia induces miRNA-218 expression, resulting in a downregulation of LASP1 and inhibition of trophoblast invasion (87).
Besides the regulation by HIF1-α and miRNAs, LASP1 is also regulated by the tumor suppressor p53, at least in hepatocellular carcinoma (88), gastric cancer (89), and endometrial cancer (90). As 50% of human tumors show somatic mutations in p53, loss of p53 activity might account for LASP1 overexpression in several cancer types, however, not all tumors with a defect in p53 show increased LASP1 levels (27). A bioinformatics approach identified Foxa1 and Foxa2 binding sites in the LASP1 promoter region, however, whether these two transcription factors are capable of regulating LASP1 expression remains to be elucidated (18). Recently, the presence of a consensus-binding site for SOX9 in the promoter region of LASP1 was detected. Luciferase reporter and ChIP assays verified LASP1 transcriptional regulation by SOX9 (91).
Functional Roles of LASP1 in Cancer
Vesicular Secretion
Since the discovery of LASP1 in 1995, the protein was observed to be overexpressed in a variety of tumor types (Table 2) and the list is still increasing. Interestingly in a melanoma study, LASP1 was described to be distinctly expressed only in the basal epidermal layer of the normal skin and in melanocytes while in primary melanoma and in metastases, a reduced LASP1 expression was noticed and no nuclear presence could be detected (23). The immunofluorescence approach identified LASP1 to be co-localized with dynamin and visualized an overlapping with melanosomes at the tips of melanocyte dendrites. It is assumed, that LASP1 is part of the F-actin-dynamin mediated budding of melanosome-containing vesicle into the extracellular matrix (92). Knockdown of LASP1 led to a distinct reduction in melanosome vesicle shedding in normal human epidermal melanocytes (NHEM) cells.
A similar mechanism is assumed for LASP1 in vesicular secretion of matrix metalloproteinases (MMPs) (24). Again, a complex of LASP1 with dynamin and F-actin, is observed and studies promote a fractionated release of vesicular MMPs into the surrounding tissue, endowing cancer cells with increased invasiveness (Figure 3A). Depletion of LASP1 led to reduced MMP levels that are rescued or restored after LASP1 overexpression (24).

Figure 3. Schematic representation of MMP secretion from invadopodia and podosomes. (A) Schematic representation of the invadopodia: During tumor invasion, invadopodia promote the degradation of the extracellular cell matrix (ECM) by coordinating the secretion of matrix metalloproteases MMP1, MMP3 and MMP9 at the tip of the protruding structure. The detailed mechanism is still unknown but involvement of LASP1 and dynamin is discussed. (B) Schematic representation of the podosome: The top picture depicts the different patterns of distribution of the podosomes (belt, rosettes, and clusters). An expanded view of a single podosome is shown below. LASP1was documented to co-localize with zyxin and vinculin in human macrophages in the outer ring area but not at the inner F-actin core. Vin, Vinculin; Zyx, Zyxin; Pax, Paxillin; LASP, LASP1 or LASP2; WASP, Wiskott-Aldrich syndrome protein; Arp 2/3, Actin-related protein 2/3.
Earlier, LASP1 was observed to have an impact on hydrochloric acid (HCl) secretion in gastric parietal cells. The authors suggested a phosphorylation-dependent alteration of LASP1 binding to F-actin, ezrin, and dynamin as likely mediators linking the vesicular trafficking/activation of H+,K+,-ATPase to the cytoskeleton (22). It remains to be seen whether this would play a role in gastric cancer.
LASP1 and Matrix Metalloproteinases
Matrix metalloproteinase (MMPs) are proteolytic enzymes capable of degrading extracellular matrices (ECM) like collagen, elastin, and fibronectin and therefore are involved in cell proliferation, migration, differentiation, angiogenesis, apoptosis, and defense. During tumor cell metastasis, cells disrupt cadherin-based intercellular junctions and initiate detachment from the primary site. This is enhanced by MMPs which digest the basal lamina components and facilitates cell movement through the ECM (93). In this respect, LASP1 plays a pivotal role in tumor invasion and metastasis by releasing MMPs into the ECM via specialized cell membrane domains called invadopodia (Figure 3A) akin to podosomes in normal cells (94). Stolting et al. showed that LASP1 colocalizes with zyxin and vinculin in the podosome ring structure of human macrophages though LASP1 has not been shown to bind to vinculin biochemically (Figure 3B). Subsequently, knockdown of LASP1 affected podosome dynamics and impaired matrix degradation capacity in these cells (33). Recent studies provided evidence that stable silencing of LASP1 also reduced gene expression levels of MMP9 and 1 in MDA-Bone-Un breast cancer cells (MDA-MB-231 cells that were re-isolated from mouse bone metastatic lesions) (19). This was further supported by reports indicating that transient knockdown of LASP1 led to reduced MMP1, 3 and 9 expression in MDA-MB-231 breast cancer cells by affecting the MMP transcription factor AP1 (24). Reduced MMP1 levels after LASP1 depletion were also observed in LNCaP prostate cancer and T24 bladder cancer cell lines (24) suggesting a general role of LASP1 in favoring distant metastasis by enhanced transcription and secretion of MMPs from invadopodia (Figure 3A).
Nuclear Function
In 2007, Grunewald et al. (35) first reported a nuclear localization of LASP1 in 29% of breast carcinoma patients and later on nuclear localization was correlated with a reduced overall survival rate of invasive breast tumor patients (27). Nuclear LASP1 localization was confirmed in several breast cancer cell lines where the protein increased at S-phase and peaked at a G2/M phase (27). Likewise, a small proportion of medulloblastoma samples showed a nuclear localization of LASP1 (59). Recently, nuclear LASP1 distribution was also described for hepatocellular carcinoma (HCC) (48) and is observed in human bladder cancer tissue (46). As LASP1 only harbors a nuclear export signal (NES) within amino acid sequence 71–77 (Figure 1), the protein is reliant on a nuclear shuttle partner. Phosphorylation of LASP1 at S146 reduces the binding to F-actin and zyxin at the membrane and induces translocation of LASP1 from the cytoplasm into the nucleus through the interaction with nuclear shuttling protein zona occludens 2 (ZO2) (15). There, it was shown to bind to Snail1 and a stabilization of the protein by LASP1 was discussed (19). As Snail forms a transcriptional repressor complex and lowers gene expression of adherens junction proteins like E-cadherin and occludin, stabilization of this complex may enhance mesenchymal cell formation and induce tumor cell migration (95) (Figure 4). Furthermore, the work by Duvall-Noelle et al. showed association of LASP1 with UHRF1 and G9a in a CXCL12-dependent manner, assuming that LASP1, in addition, serves as a nuclear hub for the epigenetic machinery in breast cancer cells (19). Recently, this work was confirmed by data showing an enhanced Snail expression and decreased E-cadherin levels also in NSCLC after LASP1 overexpression (63).

Figure 4. Proposed model for LASP1-Snail1 interaction. A model has been depicted in which a G-protein coupled receptor (GPCR) coupled to Gαs activates protein kinase A (PKA) that in turn phosphorylates LASP1 on S146 and facilitates its binding to CXCR4. Upon activation of CXCR4 by its ligand CXCL12, Gαi is activated and inhibits adenylyl cyclase (AC)/PKA signaling thus preventing accumulation of pS146-LASP1. Upon dephosphorylation of CXCR4, bound phospho-LASP1 is released and co-imported with ZO2 into the nucleus. There, LASP1 stabilizes Snail1 to impart its epigenetic silencing of E-cadherin expression leading to mesenchymal, migratory, and invasive morphology which is typical of CXCR4 activation.
There is growing evidence that LASP1 can also act as a transcriptional co-factor. Microarray analysis of MDAMB-231 breast cancer cells before and after LASP1 depletion revealed 39 regulated genes, thereof 8 (22%) regulated by AP-1 (24). AP-1 is a heterodimer that comprises members of the proto-oncogene c-Jun and c-Fos protein family and may form ternary complexes with transcriptional co-factors (96). AP-1 luciferase reporter assay confirmed LASP1-inducible AP-1 response, however, the authors failed to show a direct binding of LASP1 to either c-Jun or c-Fos or regulation of c-Fos protein levels assuming a more complex regulation.
LASP1, S100A, 14-3-3, PI3K/AKT, and COPS5—Some Critical Remarks
During the last years, the group by Liang Zhao from the Nanfang Hospital in Guangzhou, China, published several papers suggesting a fundamental role for LASP1 in epithelial-mesenchymal transition (EMT) by enhancing or modulating the expression of several proteins involved in EMT, like S100A4 (97), S100A11 (98), 14-3-3σ (99), and COPS5 (20). These studies suggest an interaction between LASP1 and aforementioned proteins, however, for evidence mainly co-localization of LASP1 with S100A11, 14-3-3σ, and COPS5 in colorectal cancer (CRC) cells is shown. The reported associations of LASP1 described for S100A11 or COPS5 lack proof for a direct interaction with LASP1. Direct binding of COP5 was only shown to the truncated SH3 domain of LASP1 but not to the full-length LASP1 protein and the proposed (but not shown) LASP1/14-3-3σ protein complex would require LASP1 phosphorylation as 14-3-3σ binding is primarily serine phosphorylation dependent (100) Most likely, the observed interactions are indirect by association to a ubiquitous protein like F-actin that is often pulled down using Sepharose beads in these experiments, with high interference observed especially with the paramagnetic beads and F-actin or glutathione S-transferase (GST)-tags. The same holds true for the proposed LASP1-FAK interaction shown in NSCLC (63). Here, a weak but detectable binding of FAK to IgG control is observed and therefore, a direct relationship between LASP1 and FAK has to be confirmed.
Concerning AKT, several papers discussed an influence of LASP1 on PI3K/AKT pathway. Depletion of LASP1 resulted in decreased pAKT-S473 phosphorylation while overexpression of LASP1 showed enhanced pAKT accumulation (20, 69, 99, 101). It has been suggested that an inverse correlation of the expression levels between LASP1-COPS5 complex and 14-3-3σ somehow affects the phosphorylation of AKT at S473. However, the exact molecular mechanisms underlying the activation of AKT by LASP1 are unclear. In Figure 5, the LASP1 promoted effects on PI3K/AKT activation are schematically presented (99). Very recently, an association of LASP1 with the phosphatase and tensin homolog (PTEN) was shown by co-immunoprecipitation but co-localization in nasopharyngeal carcinoma cells was not convincing due to big nuclei and a thin rim of cytoplasm (102). The authors claim an inverse correlation of the protein levels between LASP1 and PTEN by immunohistochemistry and that again suggested a LASP1-dependent degradation of PTEN through ubiquitination which may allow an enhanced activation of ligand-activated PI3K pathway (Figure 5) (102).

Figure 5. Proposed model for LASP1-PTEN-COPS5-14-3-3σ interaction. Model illustrate discussed LASP1/AKT signaling pathways. 14-3-3σ somehow inhibits AKT activation. LASP1 and COPS5 (COP9 signalosome subunit 5) synergistically stimulate ubiquitination and degradation of 14-3-3σ, resulting in AKT-S473 phosphorylation. In a second hypothesis, LASP1 enhances ubiquitination and degradation of PTEN (phosphatase and tensin homolog), thus enhancing PIP2 phosphorylation to PIP3 and concomitant AKT activation.
Author Contributions
EB constructed all tables and figures except Figure 3B which is drawn by DR.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This manuscript has been supported in part by National Institute of Health (NIH)/National Cancer Institute (NCI) grant R21CA202176 (to DR), Ohio Cancer Research foundation (OCR) (to DR), University of Toledo startup grants (F110796 to DR), and the German Cancer Foundation (70112717 to EB).
References
1. Tomasetto C, Moog-Lutz C, Regnier CH, Schreiber V, Basset P, Rio MC. Lasp-1 (MLN 50) defines a new LIM protein subfamily characterized by the association of LIM and SH3 domains. FEBS Lett. (1995) 373:245–9. doi: 10.1016/0014-5793(95)01040-L
2. Glynn RW, Miller N, Mahon S, Kerin MJ. Expression levels of HER2/neu and those of collocated genes at 17q12-21, in breast cancer. Oncol Rep. (2012) 28:365–9. doi: 10.3892/or.2012.1780
3. Chamberlain JS, Boehnke M, Frank TS, Kiousis S, Xu J, Guo SW, et al. BRCA1 maps proximal to D17S579 on chromosome 17q21 by genetic analysis. Am J Hum Genet. (1993) 52:792–8.
4. Orth MF, Cazes A, Butt E, Grunewald TG. An update on the LIM and SH3 domain protein 1 (LASP1): a versatile structural, signaling, and biomarker protein. Oncotarget (2015) 6:26–42. doi: 10.18632/oncotarget.3083
5. Li B, Zhuang L, Trueb B. Zyxin interacts with the SH3 domains of the cytoskeletal proteins LIM-nebulette and Lasp-1. J Biol Chem. (2004) 279:20401–10. doi: 10.1074/jbc.M310304200
6. Bang ML, Chen J. Roles of Nebulin family members in the heart. Circ J. (2015) 79:2081–7. doi: 10.1253/circj.CJ-15-0854
7. Chew CS, Chen X, Parente JA Jr, Tarre S, Okamoto C, Qin HY. Lasp-1 binds to non-muscle F-actin in vitro and is localized within multiple sites of dynamic actin assembly in vivo. J Cell Sci. (2002) 115(Pt 24):4787–99. doi: 10.1242/jcs.00174
8. Schreiber V, Moog-Lutz C, Regnier CH, Chenard MP, Boeuf H, Vonesch JL, et al. Lasp-1, a novel type of actin-binding protein accumulating in cell membrane extensions. Mol Med. (1998) 4:675–87.
9. Nakagawa H, Terasaki AG, Suzuki H, Ohashi K, Miyamoto S. Short-term retention of actin filament binding proteins on lamellipodial actin bundles. FEBS Lett. (2006) 580:3223–8. doi: 10.1016/j.febslet.2006.04.082
10. Panaviene Z, Moncman CL. Linker region of nebulin family members plays an important role in targeting these molecules to cellular structures. Cell Tissue Res. (2007) 327:353–69. doi: 10.1007/s00441-006-0305-2
11. Keicher C, Gambaryan S, Schulze E, Marcus K, Meyer HE, Butt E. Phosphorylation of mouse LASP-1 on threonine 156 by cAMP- and cGMP-dependent protein kinase. Biochem Biophys Res Commun. (2004) 324:308–16. doi: 10.1016/j.bbrc.2004.08.235
12. Rachlin AS, Otey CA. Identification of palladin isoforms and characterization of an isoform-specific interaction between Lasp-1 and palladin. J Cell Sci. (2006) 119(Pt 6):995–1004. doi: 10.1242/jcs.02825
13. Spence HJ, McGarry L, Chew CS, Carragher NO, Scott-Carragher LA, Yuan Z, et al. AP-1 differentially expressed proteins Krp1 and fibronectin cooperatively enhance Rho-ROCK-independent mesenchymal invasion by altering the function, localization, and activity of nondifferentially expressed proteins. Mol Cell Biol. (2006) 26:1480–95. doi: 10.1128/MCB.26.4.1480-1495.2006
14. Bliss KT, Chu M, Jones-Weinert CM, Gregorio CC. Investigating lasp-2 in cell adhesion: new binding partners and roles in motility. Mol Biol Cell (2013) 24:995–1006. doi: 10.1091/mbc.e12-10-0723
15. Mihlan S, Reiss C, Thalheimer P, Herterich S, Gaetzner S, Kremerskothen J, et al. Nuclear import of LASP-1 is regulated by phosphorylation and dynamic protein-protein interactions. Oncogene (2013) 32:2107–13. doi: 10.1038/onc.2012.216
16. Raman D, Sai J, Neel NF, Chew CS, Richmond A. LIM and SH3 protein-1 modulates CXCR2-mediated cell migration. PLoS ONE (2010) 5:e10050. doi: 10.1371/journal.pone.0010050
17. Frietsch JJ, Kastner C, Grunewald TG, Schweigel H, Nollau P, Ziermann J, et al. LASP1 is a novel BCR-ABL substrate and a phosphorylation-dependent binding partner of CRKL in chronic myeloid leukemia. Oncotarget (2014) 5:5257–71. doi: 10.18632/oncotarget.2072
18. Salvi A, Bongarzone I, Ferrari L, Abeni E, Arici B, De Bortoli M, et al. Molecular characterization of LASP-1 expression reveals vimentin as its new partner in human hepatocellular carcinoma cells. Int J Oncol. (2015) 46:1901–12. doi: 10.3892/ijo.2015.2923
19. Duvall-Noelle N, Karwandyar A, Richmond A, Raman D. LASP-1: a nuclear hub for the UHRF1-DNMT1-G9a-Snail1 complex. Oncogene (2016) 35:1122–33. doi: 10.1038/onc.2015.166
20. Zhou R, Shao Z, Liu J, Zhan W, Gao Q, Pan Z, et al. COPS5 and LASP1 synergistically interact to downregulate 14-3-3sigma expression and promote colorectal cancer progression via activating PI3K/AKT pathway. Int J Cancer (2017) 142:1853–64. doi: 10.1002/ijc.31206
21. Nakagawa H, Suzuki H, Machida S, Suzuki J, Ohashi K, Jin M, et al. Contribution of the LIM domain and nebulin-repeats to the interaction of Lasp-2 with actin filaments and focal adhesions. PLoS ONE (2009) 4:e7530. doi: 10.1371/journal.pone.0007530
22. Chew CS, Chen X, Bollag RJ, Isales C, Ding KH, Zhang H. Targeted disruption of the Lasp-1 gene is linked to increases in histamine-stimulated gastric HCl secretion. Am J Physiol Gastrointest Liver Physiol. (2008) 295:G37–44. doi: 10.1152/ajpgi.90247.2008
23. Vaman VSA, Poppe H, Houben R, Grunewald TG, Goebeler M, Butt E. LASP1, a newly identified melanocytic protein with a possible role in melanin release, but not in melanoma progression. PLoS ONE (2015) 10:e0129219. doi: 10.1371/journal.pone.0129219
24. Endres M, Kneitz S, Orth MF, Perera RK, Zernecke A, Butt E. Regulation of matrix metalloproteinases (MMPs) expression and secretion in MDA-MB-231 breast cancer cells by LIM and SH3 protein 1 (LASP1). Oncotarget (2016) 7:64244–59. doi: 10.18632/oncotarget.11720
25. Zieseniss A, Terasaki AG, Gregorio CC. Lasp-2 expression, localization, and ligand interactions: a new Z-disc scaffolding protein. Cell Motil Cytoskeleton (2008) 65:59–72. doi: 10.1002/cm.20244
26. Deng XA, Norris A, Panaviene Z, Moncman CL. Ectopic expression of LIM-nebulette (LASP2) reveals roles in cell migration and spreading. Cell Motil Cytoskeleton (2008) 65:827–40. doi: 10.1002/cm.20304
27. Frietsch JJ, Grunewald TG, Jasper S, Kammerer U, Herterich S, Kapp M, et al. Nuclear localisation of LASP-1 correlates with poor long-term survival in female breast cancer. Br J Cancer (2010) 102:1645–53. doi: 10.1038/sj.bjc.6605685
28. Katoh M, Katoh M. Identification and characterization of LASP2 gene in silico. Int J Mol Med. (2003) 12:405–10. doi: 10.3892/ijmm.12.3.405
29. Pappas CT, Bliss KT, Zieseniss A, Gregorio CC. The Nebulin family: an actin support group. Trends Cell Biol. (2011) 21:29–37. doi: 10.1016/j.tcb.2010.09.005
30. Schreiber V, Masson R, Linares JL, Mattei MG, Tomasetto C, Rio MC. Chromosomal assignment and expression pattern of the murine Lasp-1 gene. Gene (1998) 207:171–5. doi: 10.1016/S0378-1119(97)00622-7
31. Terasaki AG, Suzuki H, Ando J, Matsuda Y, Ohashi K. Chromosomal assignment of LASP1 and LASP2 genes and organization of the LASP2 gene in chicken. Cytogenet Genome Res. (2006) 112:141–7. doi: 10.1159/000087526
32. Gao K, Deng XY, Qian HY, Wu P, Qin GX, Liu T, et al. cDNA cloning and characterization of LASP1 from silkworm, Bombyx mori, involved in cytoplasmic polyhedrosis virus infection. Gene (2012) 511:389–97. doi: 10.1016/j.gene.2012.09.062
33. Stolting M, Wiesner C, van Vliet V, Butt E, Pavenstadt H, Linder S, et al. Lasp-1 regulates podosome function. PLoS ONE (2012) 7:e35340. doi: 10.1371/journal.pone.0035340
34. Suyama R, Jenny A, Curado S, Pellis-van Berkel W, Ephrussi A. The actin-binding protein Lasp promotes Oskar accumulation at the posterior pole of the Drosophila embryo. Development (2009) 13:95–105. doi: 10.1242/dev.027698
35. Grunewald TG, Kammerer U, Kapp M, Eck M, Dietl J, Butt E, et al. Nuclear localization and cytosolic overexpression of LASP-1 correlates with tumor size and nodal-positivity of human breast carcinoma. BMC Cancer (2007) 7:198. doi: 10.1186/1471-2407-7-198
36. Moazzeni H, Najafi A, Khani M. Identification of direct target genes of miR-7, miR-9, miR-96, and miR-182 in the human breast cancer cell lines MCF-7 and MDA-MB-231. Mol Cell Probes (2017) 34:45–52. doi: 10.1016/j.mcp.2017.05.005
37. Wang C, Zheng X, Shen C, Shi Y. MicroRNA-203 suppresses cell proliferation and migration by targeting BIRC5 and LASP1 in human triple-negative breast cancer cells. J Exp Clin Cancer Res. (2012) 31:58. doi: 10.1186/1756-9966-31-58
38. Sui Y, Zhang X, Yang H, Wei W, Wang M. MicroRNA-133a acts as a tumour suppressor in breast cancer through targeting LASP1. Oncol Rep. (2018) 39:473–82. doi: 10.3892/or.2017.6114
39. Zhao L, Wang H, Liu C, Liu Y, Wang X, Wang S, et al. Promotion of colorectal cancer growth and metastasis by the LIM and SH3 domain protein 1. Gut (2009) 59:1226–35. doi: 10.1136/gut.2009.202739
40. Wang H, An H, Wang B, Liao Q, Li W, Jin X, et al. miR-133a represses tumour growth and metastasis in colorectal cancer by targeting LIM and SH3 protein 1 and inhibiting the MAPK pathway. Eur J Cancer (2013) 49:3924–35. doi: 10.1016/j.ejca.2013.07.149
41. Xu L, Zhang Y, Wang H, Zhang G, Ding Y, Zhao L. Tumor suppressor miR-1 restrains epithelial-mesenchymal transition and metastasis of colorectal carcinoma via the MAPK and PI3K/AKT pathway. J Transl Med. (2014) 12:244. doi: 10.1186/s12967-014-0244-8
42. Wang W, Ji G, Xiao X, Chen X, Qin WW, Yang F, et al. Epigenetically regulated miR-145 suppresses colon cancer invasion and metastasis by targeting LASP1. Oncotarget (2016) 7:68674–87. doi: 10.18632/oncotarget.11919
43. Grunewald TG, Kammerer U, Winkler C, Schindler D, Sickmann A, Honig A, et al. Overexpression of LASP-1 mediates migration and proliferation of human ovarian cancer cells and influences zyxin localisation. Br J Cancer (2007) 96:296–305. doi: 10.1038/sj.bjc.6603545
44. Dimova I, Orsetti B, Negre V, Rouge C, Ursule L, Lasorsa L, et al. Genomic markers for ovarian cancer at chromosomes 1, 8 and 17 revealed by array CGH analysis. Tumori (2009) 95:357–66. doi: 10.1177/030089160909500315
45. Chiyomaru T, Enokida H, Kawakami K, Tatarano S, Uchida Y, Kawahara K, et al. Functional role of LASP1 in cell viability and its regulation by microRNAs in bladder cancer. Urol Oncol. (2012) 30:434–43. doi: 10.1016/j.urolonc.2010.05.008
46. Ardelt P, Grunemay N, Strehl A, Jilg C, Miernik A, Kneitz B, et al. LASP-1, a novel urinary marker for detection of bladder cancer. Urol Oncol. (2013) 31:1591–8. doi: 10.1016/j.urolonc.2012.02.002
47. Salvi A, Bongarzone I, Micciche F, Arici B, Barlati S, De Petro G. Proteomic identification of LASP-1 down-regulation after RNAi urokinase silencing in human hepatocellular carcinoma cells. Neoplasia (2009) 11:207–19. doi: 10.1593/neo.81076
48. Wang H, Li W, Jin X, Cui S, Zhao L. LIM and SH3 protein 1, a promoter of cell proliferation and migration, is a novel independent prognostic indicator in hepatocellular carcinoma. Eur J Cancer (2013) 49:974–83. doi: 10.1016/j.ejca.2012.09.032
49. Li H, Xiang Z, Liu Y, Xu B, Tang J. MicroRNA-133b inhibits proliferation, cellular migration, and invasion via targeting LASP1 in hepatocarcinoma cells. Oncol Res. (2017) 25:1269–82. doi: 10.3727/096504017X14850151453092
50. Shimizu F, Shiiba M, Ogawara K, Kimura R, Minakawa Y, Baba T, et al. Overexpression of LIM and SH3 Protein 1 leading to accelerated G2/M phase transition contributes to enhanced tumourigenesis in oral cancer. PLoS ONE (2013) 8:e83187. doi: 10.1371/journal.pone.0083187
51. Takeshita N, Mori M, Kano M, Hoshino I, Akutsu Y, Hanari N, et al. miR-203 inhibits the migration and invasion of esophageal squamous cell carcinoma by regulating LASP1. Int J Oncol. (2012) 41:1653–61. doi: 10.3892/ijo.2012.1614
52. Benaich N, Woodhouse S, Goldie SJ, Mishra A, Quist SR, Watt FM. Rewiring of an epithelial differentiation factor, miR-203, to inhibit human squamous cell carcinoma metastasis. Cell Rep. (2014) 9:104–17. doi: 10.1016/j.celrep.2014.08.062
53. Du YY, Zhao LM, Chen L, Sang MX, Li J, Ma M, et al. The tumor-suppressive function of miR-1 by targeting LASP1 and TAGLN2 in esophageal squamous cell carcinoma. J Gastroenterol Hepatol. (2016) 31:384–93. doi: 10.1111/jgh.13180
54. Yang F, Zhou X, Du S, Zhao Y, Ren W, Deng Q, et al. LIM and SH3 domain protein 1 (LASP-1) overexpression was associated with aggressive phenotype and poor prognosis in clear cell renal cell cancer. PLoS ONE (2014) 9:e100557. doi: 10.1371/journal.pone.0100557
55. Hailer A, Grunewald TG, Orth M, Reiss C, Kneitz B, Spahn M, et al. Loss of tumor suppressor mir-203 mediates overexpression of LIM and SH3 Protein 1 (LASP1) in high-risk prostate cancer thereby increasing cell proliferation and migration. Oncotarget (2014) 5:4144–53. doi: 10.18632/oncotarget.1928
56. Viticchie G, Lena AM, Latina A, Formosa A, Gregersen LH, Lund AH, et al. MiR-203 controls proliferation, migration and invasive potential of prostate cancer cell lines. Cell Cycle (2011) 10:1121–31. doi: 10.4161/cc.10.7.15180
57. Hudson RS, Yi M, Esposito D, Watkins SK, Hurwitz AA, Yfantis HG, et al. MicroRNA-1 is a candidate tumor suppressor and prognostic marker in human prostate cancer. Nucleic Acids Res. (2012) 40:3689–703. doi: 10.1093/nar/gkr1222
58. Nishikawa R, Goto Y, Sakamoto S, Chiyomaru T, Enokida H, Kojima S, et al. Tumor-suppressive microRNA-218 inhibits cancer cell migration and invasion via targeting of LASP1 in prostate cancer. Cancer Sci. (2014) 105:802–11. doi: 10.1111/cas.12441
59. Traenka C, Remke M, Korshunov A, Bender S, Hielscher T, Northcott PA, et al. Role of LIM and SH3 protein 1 (LASP1) in the metastatic dissemination of medulloblastoma. Cancer Res. (2010) 70:8003–14. doi: 10.1158/0008-5472.CAN-10-0592
60. Pan X, Wang Z, Wan B, Zheng Z. MicroRNA-206 inhibits the viability and migration of medulloblastoma cells by targeting LIM and SH3 protein 1. Exp Ther Med. (2017) 14:3894–900. doi: 10.3892/etm.2017.5016
61. Jiang N, Jiang X, Chen Z, Song X, Wu L, Zong D, et al. MiR-203a-3p suppresses cell proliferation and metastasis through inhibiting LASP1 in nasopharyngeal carcinoma. J Exp Clin Cancer Res. (2017) 36:138. doi: 10.1186/s13046-017-0604-3
62. Lin X, Liu X, Fang Y, Weng X. LIM and SH3 protein 1 promotes tumor proliferation and metastasis in lung carcinoma. Oncol Lett. (2016) 12:4756–60. doi: 10.3892/ol.2016.5225
63. Zhang X, Liu Y, Fan C, Wang L, Li A, Zhou H, et al. Lasp1 promotes malignant phenotype of non-small-cell lung cancer via inducing phosphorylation of FAK-AKT pathway. Oncotarget (2017) 8:75102–13. doi: 10.18632/oncotarget.20527
64. Hu Z, Cui Y, Zhou Y, Zhou K, Qiao X, Li C, et al. MicroRNA-29a plays a suppressive role in non-small cell lung cancer cells via targeting LASP1. Onco Targets Ther. (2016) 9:6999–7009. doi: 10.2147/OTT.S116509
65. Zheng J, Wang F, Lu S, Wang X. LASP-1, regulated by miR-203, promotes tumor proliferation and aggressiveness in human non-small cell lung cancer. Exp Mol Pathol. (2016) 100:116–24. doi: 10.1016/j.yexmp.2015.11.031
66. Wang LL, Wang L, Wang XY, Shang D, Yin SJ, Sun LL, et al. MicroRNA-218 inhibits the proliferation, migration, and invasion and promotes apoptosis of gastric cancer cells by targeting LASP1. Tumour Biol. (2016) 37:15241–52. doi: 10.1007/s13277-016-5388-0
67. Fahrmann JF, Grapov D, Phinney BS, Stroble C, DeFelice BC, Rom W, et al. Proteomic profiling of lung adenocarcinoma indicates heightened DNA repair, antioxidant mechanisms and identifies LASP1 as a potential negative predictor of survival. Clin Proteomics (2016) 13:31. doi: 10.1186/s12014-016-9132-y
68. Segerer SE, Bartmann C, Kaspar S, Muller N, Kapp M, Butt E, et al. The cytoskeletal protein LASP-1 differentially regulates migratory activities of choriocarcinoma cells. Arch Gynecol Obstet. (2016) 293:407–14. doi: 10.1007/s00404-015-3830-y
69. Li Z, Chen Y, Wang X, Zhang H, Zhang Y, Gao Y, et al. LASP-1 induces proliferation, metastasis and cell cycle arrest at the G2/M phase in gallbladder cancer by down-regulating S100P via the PI3K/AKT pathway. Cancer Lett. (2016) 372:239–50. doi: 10.1016/j.canlet.2016.01.008
70. Gao W, Han J. Silencing of LIM and SH3 protein 1 (LASP-1) inhibits thyroid cancer cell proliferation and invasion. Oncol Res. (2017) 25:879–86. doi: 10.3727/096504016X14785415155643
71. Zhao T, Ren H, Li J, Chen J, Zhang H, Xin W, et al. LASP1 is a HIF1alpha target gene critical for metastasis of pancreatic cancer. Cancer Res. (2015) 75:111–9. doi: 10.1158/0008-5472.CAN-14-2040
72. Konrat R, Weiskirchen R, Krautler B, Bister K. Solution structure of the carboxyl-terminal LIM domain from quail cysteine-rich protein CRP2. J Biol Chem. (1997) 272:12001–7. doi: 10.1074/jbc.272.18.12001
73. Kontaxis G, Konrat R, Krautler B, Weiskirchen R, Bister K. Structure and intramodular dynamics of the amino-terminal LIM domain from quail cysteine- and glycine-rich protein CRP2. Biochemistry (1998) 37:7127–34. doi: 10.1021/bi973055v
74. Gray CH, McGarry LC, Spence HJ, Riboldi-Tunnicliffe A, Ozanne BW. Novel beta-propeller of the BTB-Kelch protein Krp1 provides a binding site for Lasp-1 that is necessary for pseudopodial extension. J Biol Chem. (2009) 284:30498–507. doi: 10.1074/jbc.M109.023259
75. Raman D, Neel NF, Sai J, Mernaugh RL, Ham AJ, Richmond AJ. Characterization of chemokine receptor CXCR2 interacting proteins using a proteomics approach to define the CXCR2 “chemosynapse”. Meth Enzymol. (2009) 460:315–30. doi: 10.1016/S0076-6879(09)05215-X
76. Labeit S, Gibson T, Lakey A, Leonard K, Zeviani M, Knight P, et al. Evidence that nebulin is a protein-ruler in muscle thin filaments. FEBS Lett. (1991) 282:313–6. doi: 10.1016/0014-5793(91)80503-U
77. Labeit S, Kolmerer B. The complete primary structure of human nebulin and its correlation to muscle structure. J Mol Biol. (1995) 248:308–15. doi: 10.1016/S0022-2836(95)80052-2
78. Butt E, Gambaryan S, Gottfert N, Galler A, Marcus K, Meyer HE. Actin binding of human LIM and SH3 protein is regulated by cGMP- and cAMP-dependent protein kinase phosphorylation on serine 146. J Biol Chem. (2003) 278:15601–7. doi: 10.1074/jbc.M209009200
79. Traenka J, Hauck CR, Lewandrowski U, Sickmann A, Gambaryan S, Thalheimer P, et al. Integrin-dependent translocation of LASP-1 to the cytoskeleton of activated platelets correlates with LASP-1 phosphorylation at tyrosine 171 by Src-kinase. Thromb Haemost. (2009) 102:520–8. doi: 10.1160/TH09-03-0143
80. Lin YH, Park ZY, Lin D, Brahmbhatt AA, Rio MC, Yates JR III, et al. Regulation of cell migration and survival by focal adhesion targeting of Lasp-1. J Cell Biol. (2004) 165:421–32. doi: 10.1083/jcb.200311045
81. Chew CS, Parente JA Jr, Zhou C, Baranco E, Chen X. Lasp-1 is a regulated phosphoprotein within the cAMP signaling pathway in the gastric parietal cell. Am J Physiol. (1998) 275(1 Pt 1):C56–67. doi: 10.1152/ajpcell.1998.275.1.C56
82. Kennelly PJ, Krebs EG. Consensus sequences as substrate specificity determinants for protein kinases and protein phosphatases. J Biol Chem. (1991) 266:15555–8.
83. Kong FY, Zhu T, Li N, Cai YF, Zhou K, Wei X, et al. Bioinformatics analysis of the proteins interacting with LASP-1 and their association with HBV-related hepatocellular carcinoma. Sci Rep. (2017) 7:44017. doi: 10.1038/srep44017
84. Wang B, Zhang L, Zhao L, Zhou R, Ding Y, Li G, et al. LASP2 suppresses colorectal cancer progression through JNK/p38 MAPK pathway meditated epithelial-mesenchymal transition. Cell Commun Signal. (2017) 15:21. doi: 10.1186/s12964-017-0179-9
85. Zhang X, Cai L, Zhou H, Liu Y, Fan C, Wang L, et al. Lasp2 enhances tumor invasion via facilitating phosphorylation of FAK and predicts poor overall survival of non-small cell lung cancer patients. Mol Carcinog. (2017) 56:2558–65. doi: 10.1002/mc.22700
86. Fjeldbo CS, Aarnes EK, Malinen E, Kristensen GB, Lyng H. Identification and validation of reference genes for RT-qPCR studies of hypoxia in squamous cervical cancer patients. PLoS ONE (2016) 11:e0156259. doi: 10.1371/journal.pone.0156259
87. Fang M, Du H, Han B, Xia G, Shi X, Zhang F, et al. Hypoxia-inducible microRNA-218 inhibits trophoblast invasion by targeting LASP1: implications for preeclampsia development. Int J Biochem Cell Biol. (2017) 87:95–103. doi: 10.1016/j.biocel.2017.04.005
88. Wang B, Feng P, Xiao Z, Ren EC. LIM and SH3 protein 1 (Lasp1) is a novel p53 transcriptional target involved in hepatocellular carcinoma. J Hepatol. (2009) 50:528–37. doi: 10.1016/j.jhep.2008.10.025
89. Zheng J, Yu S, Qiao Y, Zhang H, Liang S, Wang H, et al. LASP-1 promotes tumor proliferation and metastasis and is an independent unfavorable prognostic factor in gastric cancer. J Cancer Res Clin Oncol. (2014) 140:1891–9. doi: 10.1007/s00432-014-1759-3
90. Li W, Li H, Zhang L, Hu M, Li F, Deng J, et al. Long non-coding RNA LINC00672 contributes to p53 protein-mediated gene suppression and promotes endometrial cancer chemosensitivity. J Biol Chem. (2017) 292:5801–13. doi: 10.1074/jbc.M116.758508
91. Shi J, Guo J, Li X. Role of LASP-1, a novel SOX9 transcriptional target, in the progression of lung cancer. Int J Oncol. (2018) 52:179–88. doi: 10.3892/ijo.2017.4201
92. Morlot S, Roux A. Mechanics of dynamin-mediated membrane fission. Annu Rev Biophys. (2013) 42:629–49. doi: 10.1146/annurev-biophys-050511-102247
93. Davies KJ. The complex interaction of matrix metalloproteinases in the migration of cancer cells through breast tissue stroma. Int J Breast Cancer (2014) 2014:839094. doi: 10.1155/2014/839094
94. Jacob A, Linklater E, Bayless BA, Lyons T, Prekeris R. The role and regulation of Rab40b-Tks5 complex during invadopodia formation and cancer cell invasion. J Cell Sci. (2016) 129:4341–53. doi: 10.1242/jcs.193904
95. Wang Y, Shi J, Chai K, Ying X, Zhou BP. The Role of Snail in EMT and Tumorigenesis. Curr Cancer Drug Targets (2013) 13:963–72. doi: 10.2174/15680096113136660102
96. Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer (2003) 3:859–68. doi: 10.1038/nrc1209
97. Wang H, Shi J, Luo Y, Liao Q, Niu Y, Zhang F, et al. LIM and SH3 protein 1 induces TGFbeta-mediated epithelial-mesenchymal transition in human colorectal cancer by regulating S100A4 expression. Clin Cancer Res. (2014) 20:5835–47. doi: 10.1158/1078-0432.CCR-14-0485
98. Niu Y, Shao Z, Wang H, Yang J, Zhang F, Luo Y, et al. LASP1-S100A11 axis promotes colorectal cancer aggressiveness by modulating TGFbeta/Smad signaling. Sci Rep. (2016) 6:26112. doi: 10.1038/srep26112
99. Shao Z, Cai Y, Xu L, Yao X, Shi J, Zhang F, et al. Loss of the 14-3-3sigma is essential for LASP1-mediated colorectal cancer progression via activating PI3K/AKT signaling pathway. Sci Rep. (2016) 6:25631. doi: 10.1038/srep25631
100. Dougherty MK, Morrison DK. Unlocking the code of 14-3-3. J Cell Sci. (2004) 117:1875–84. doi: 10.1242/jcs.01171
101. Zhong C, Chen Y, Tao B, Peng L, Peng T, Yang X, et al. LIM and SH3 protein 1 regulates cell growth and chemosensitivity of human glioblastoma via the PI3K/AKT pathway. BMC Cancer (2018) 18:722. doi: 10.1186/s12885-018-4649-2
Keywords: LASP1, LASP2, transcriptional regulation, nuclear role, matrix metalloproteinases, AP1
Citation: Butt E and Raman D (2018) New Frontiers for the Cytoskeletal Protein LASP1. Front. Oncol. 8:391. doi: 10.3389/fonc.2018.00391
Received: 28 April 2018; Accepted: 30 August 2018;
Published: 21 September 2018.
Edited by:
Tao Liu, University of New South Wales, AustraliaReviewed by:
Luisa Lanfrancone, Istituto Europeo di Oncologia s.r.l., ItalyAnne George, University of Illinois at Chicago, United States
Copyright © 2018 Butt and Raman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Elke Butt, YnV0dF9lQHVrdy5kZQ==
Dayanidhi Raman, ZGF5YW5pZGhpLnJhbWFuQHV0b2xlZG8uZWR1
 Elke Butt
Elke Butt Dayanidhi Raman
Dayanidhi Raman