
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 08 June 2016
Sec. Cancer Genetics
Volume 6 - 2016 | https://doi.org/10.3389/fonc.2016.00136
 Joy Shi1
Joy Shi1 Kristan J. Aronson1
Kristan J. Aronson1 Anne Grundy2
Anne Grundy2 Lindsay C. Kobayashi3
Lindsay C. Kobayashi3 Igor Burstyn4
Igor Burstyn4 Johanna M. Schuetz5
Johanna M. Schuetz5 Caroline A. Lohrisch6
Caroline A. Lohrisch6 Sandip K. SenGupta7
Sandip K. SenGupta7 Agnes S. Lai8
Agnes S. Lai8 Angela Brooks-Wilson5,9
Angela Brooks-Wilson5,9 John J. Spinelli8,10
John J. Spinelli8,10 Harriet Richardson1*
Harriet Richardson1*
Genetic variants of insulin-like growth factor 1 (IGF1) pathway genes have been shown to be associated with breast density and IGF1 levels and, therefore, may also influence breast cancer risk via pro-survival signaling cascades. The aim of this study was to investigate associations between IGF1 pathway single nucleotide polymorphisms (SNPs) and breast cancer risk among European and East Asian women, and potential interactions with menopausal status and breast tumor subtype. Stratified analyses of 1,037 cases and 1,050 controls from a population-based case–control study were conducted to assess associations with breast cancer for 22 SNPs across 5 IGF1 pathway genes in European and East Asian women. Odds ratios were calculated using logistic regression in additive genetic models. Polytomous logistic regression was used to assess heterogeneity by breast tumor subtype. Two SNPs of the IGF1 gene (rs1019731 and rs12821878) were associated with breast cancer risk among European women. Four highly linked IGF1 SNPs (rs2288378, rs17727841, rs7136446, and rs7956547) were modified by menopausal status among East Asian women only and associated with postmenopausal breast cancers. The association between rs2288378 and breast cancer risk was also modified by breast tumor subtype among East Asian women. Several IGF1 polymorphisms were found to be associated with breast cancer risk and some of these associations were modified by menopausal status or breast tumor subtype. Such interactions should be considered when assessing the role of these variants in breast cancer etiology.
The insulin-like growth factor 1 (IGF1) signaling pathway has been implicated in normal cell growth, development, and differentiation in the mammary gland and other tissues (1, 2). Stimulation by growth hormone (GH) results in the production of IGF1, primarily in the liver. While IGF1 mediates its action through the insulin-like growth factor 1 receptor (IGF1R), its bioavailability is regulated by its binding proteins (IGFBPs). Over 90% of circulating IGF1 is bound by IGFBPs, with most being bound to insulin-like growth factor binding protein 3 (IGFBP3) (3). Downstream of IGF1R, signaling transmission by insulin receptor substrate 1 (IRS1) activates multiple intracellular signaling pathways that mediate the actions of IGF1. The two major pathways are the phosphoinositide 3-kinase (PI3K)/protein kinase B (Akt) pathway and the Ras/mitogen-activated protein kinase (MAPK) pathway (4).
Insulin-like growth factor 1 is suggested to play a role in the progression of many cancers, including prostate (5, 6), colorectal (7, 8), and breast (6, 9), as its pro-survival signaling pathways may encourage the proliferation of cancer cells (10). Epidemiological studies have previously demonstrated that elevated IGF1 concentrations are associated with increased risk of premenopausal breast cancer (11, 12), and current evidence also suggests similar associations between IGF1 concentrations and postmenopausal breast cancers (9). However, these associations may be restricted to estrogen receptor (ER)-positive tumors (9), mediated by synergistic growth effects from interactions between the estrogen and IGF1 signaling pathways when both are stimulated (13). Studies have also implicated IGFBP3 in breast cancer risk (6, 11, 12), although it is unclear whether this is a result of IGF-dependent or -independent functions (1).
Variants of the IGF1 (14–16), IGFBP3 (12, 14–17), IGF1R (18, 19), and PI3KCB (18) genes have been implicated in changes in circulating IGF1 and IGFBP3 levels, and may have systemic effects in the regulation of the IGF1 signaling pathway. However, the evidence for a relationship between variants in IGF-related genes and risk of breast cancer is less compelling. While some studies have identified associations with breast cancer risk for certain variants of IGF-related genes (15, 20–22), the Breast and Prostate Cancer Cohort Consortium (BPC3) genotyped 550 single nucleotide polymorphisms (SNPs) across 24 IGF1 pathway genes in a population of predominantly Caucasian postmenopausal women, but found no association with breast cancer risk (23). However, assessment of these associations in other ethnic groups and in premenopausal women, along with consideration of breast tumor subtypes, remains to be conducted.
We examined the associations of 22 polymorphisms across five IGF1 pathway genes (IGF1, IGFBP3, IGF1R, IRS1, and PI3KCB) with risk of breast cancer among women of European and East Asian descent. Potential interactions between IGF-related genes and menopausal status and breast tumor subtype were also assessed.
To investigate the associations between IGF1 pathway genes and breast cancer risk, data from the Canadian Breast Cancer Study, previously known as the Molecular Epidemiology of Breast Cancer (MEBC) Study as described (24), were used. In brief, a case–control study was conducted in Vancouver, BC, and Kingston, ON, Canada. Incident cases from Vancouver and surrounding communities (n = 1,001) were recruited from the BC Cancer Registry and comprised women aged 40–80 newly diagnosed with in situ or invasive breast cancer with no previous history of cancer (except non-melanoma skin cancer). Age frequency-matched cancer-free controls (n = 1,014) were recruited from the Screening Mammography Program of BC. In Kingston, both cases (n = 131) and age frequency-matched controls (n = 163) were recruited from the Hotel Dieu Breast Assessment Program. All participants completed a questionnaire, either self-administered or by telephone interview, and provided a biological sample, either blood or saliva. Sufficient DNA for genotyping was extracted from blood and saliva samples for 92% of participants (n = 2,127) in the study. Pathologic data concerning tumor ER, progesterone receptor (PgR), and human epidermal growth factor receptor 2 (HER2) expression were obtained for most cases (n = 997, 96%) from both Vancouver and Kingston. Written informed consent was obtained from all participants and ethical approval for this study was obtained from the University of British Columbia – BC Cancer Agency Clinical Research Ethics Board and the Queen’s University Health Sciences Research Ethics Board.
The questionnaire completed by participants provided information regarding geographic ancestry; menopausal status; education; health, medical, and reproductive history; family history of cancer; lifestyle characteristics, including lifetime tobacco and alcohol consumption; and lifetime physical activity.
Genes involved in the synthesis (IGF1), bioavailability (IGFBP3), and downstream signaling (IGF1R, IRS1, PI3KCB) of IGF1 were selected. Based on suspected relationships with breast cancer risk or disease susceptibility from existing literature and using SNP tagging methods, a set of 28 SNPs were selected for genotyping. Selection of tagSNPs, using the CEU population from HapMap release 28 with a minimum minor allele frequency (MAF) of 0.10 and r2 threshold of 0.8, was conducted for three genes (IGF1, IGFBP3, and PI3KCB) using Tagger (25) in Haploview (26). Included in a 768-plex Illumina Golden Gate assay among SNPs related to other hypotheses were 11 SNPs of IGF1: rs6214, rs1549593 (15), rs17727841, rs2288378, rs7136446, rs2195239, rs7956547, rs1019731, rs12821878, rs1520220 (17), and rs2162679; 7 SNPs of IGFBP3: rs6670, rs2453839, rs3110697 (27), rs2471551 (15, 27), rs2132572 (15), rs10255707, and rs2854744 (15, 27, 28); 4 SNPs of IGF1R: rs951715 (29), rs2229765 (18, 29), rs8038415 (30), and rs9672965; 1 SNP of IRS1: rs1801278 (17); and 5 SNPs of PI3KCB: rs12493155, rs524164, rs10513055, rs361072 (17, 18), and rs693293. Genotyping was performed by the Genome Quebec/McGill University Innovation Centre in Montreal, Canada.
Quality control of genotype data was performed in Genome Studio v2011.1 (Illumina, San Diego, CA, USA), PLINK v1.07 (31), GRR (32), and Excel 2007 (Microsoft, Redmond, WA, USA), and has been previously described (24). Briefly, SNPs were excluded if they exhibited Gencall Scores <0.25, had GenTrain scores <0.4, exhibited poor clustering, were mono-allelic, showed genotype discrepancies in 126 pairs of replicate samples, had call rates <0.95, had unexpectedly low MAF among Caucasian controls in comparison to HapMap CEU data, or violated the Hardy–Weinberg equilibrium (p < 0.001) in European-ancestry controls. Relevant to this analysis, six SNPs were excluded following quality control: two by the genotyping center (rs1520220 and rs2854744), two for poor clustering (rs9672965 and rs693293), one for low call rate (rs10255707), and one for discrepancies in replicate samples (rs2162679). Therefore, final analyses included 22 IGF1 pathway SNPs.
Exclusion criteria for samples included call rate <0.95 (n = 14); unrelated samples having identical genotypes (n = 6); sex discrepancy, as identified by Y chromosome markers, indicating that the sample was male (n = 1); excess heterozygosity, specifically >3 SDs from the mean of other samples of the same ethnicity (n = 4); discrepancies between self-reported ethnicity and predicted ethnicity from genotype data (n = 5). If participants in the study were related, one was excluded (n = 9). Questionnaire data were used to verify that nine pairs of samples were from relatives. If the pairs were both cases, the individual with the later diagnosis date was excluded; if the pairs were both controls, the individual with the younger age was excluded. To compare self-reported ethnicity to predicted ethnicity, identity-by-state was calculated and multi-dimensional scaling plots (31) were created with HapMap samples selected from the CEU, CHB, CHD, JPT, YRI, and TSI populations. A total of 1,037 cases and 1,050 controls remained after quality control procedures.
ER, PgR, and HER2 status of all invasive tumors was determined using immunohistochemistry (IHC), supplemented by fluorescence in situ hybridization (FISH) if the IHC results for HER2 were equivocal. Tumors were grouped according to biomarker status, regardless of intensity of expression. Four breast tumor subtypes were identified: ER+ and/or PgR+/HER2−, ER+ and/or PgR+/HER2+, ER−/PgR−/HER2+, and ER−/PgR−/HER2−.
Relationships between SNPs and breast cancer risk were investigated using unconditional logistic regression in an additive genetic model adjusted for age and center. All analyses were stratified by ethnicity and conducted in the two largest subgroups: Europeans and East Asians (Chinese, Japanese, Korean, and Filipino). The Benjamini–Hochberg adjustment (33) was used to control the FDR across all SNPs and provide corrected p-values. SNPs with a q-value (FDR adjusted p-value) <0.05 were considered statistically significant. Modification of these relationships by menopausal status was assessed by including an interaction term. Case–case polytomous logistic regression was used to assess heterogeneity of odds ratios between hormone receptor- and HER2-defined tumor subtypes. Assessment for these interactions was restricted to SNPs with p-value <0.1 for an association with breast cancer risk prior to false discovery rate (FDR) adjustment and MAF among controls >0.1 in each respective ethnic subgroup. All analyses were performed using SAS (Version 9.4, SAS Institute, Cary, NC, USA) and PLINK (31).
Characteristics of the study participants are presented in Table 1. In brief, approximately two-thirds of participants were women of European descent and one-quarter were of East Asian descent, although a greater proportion of cases were East Asian compared to controls. On average, breast cancer cases also had lower household income, were less likely to have completed postsecondary education, were older at their first mammogram, were more likely to have a primary family history of breast cancer and consumed less alcohol over their lifetime than controls.
Relationships between SNPs and breast cancer risk among European and East Asian women are presented in Table 2. In the European group, the minor A-allele of three IGF1 SNPs (rs1549593, rs1019731, and rs12821878) was associated with decreased risk of breast cancer, but only two of these SNPs passed adjustment for the FDR: rs1019731 (OR = 0.67, 95% CI: 0.53–0.84) and rs12821878 (OR = 0.73, 95% CI: 0.61–0.88). Among East Asians, no SNP was associated with breast cancer risk even prior to FDR adjustment.
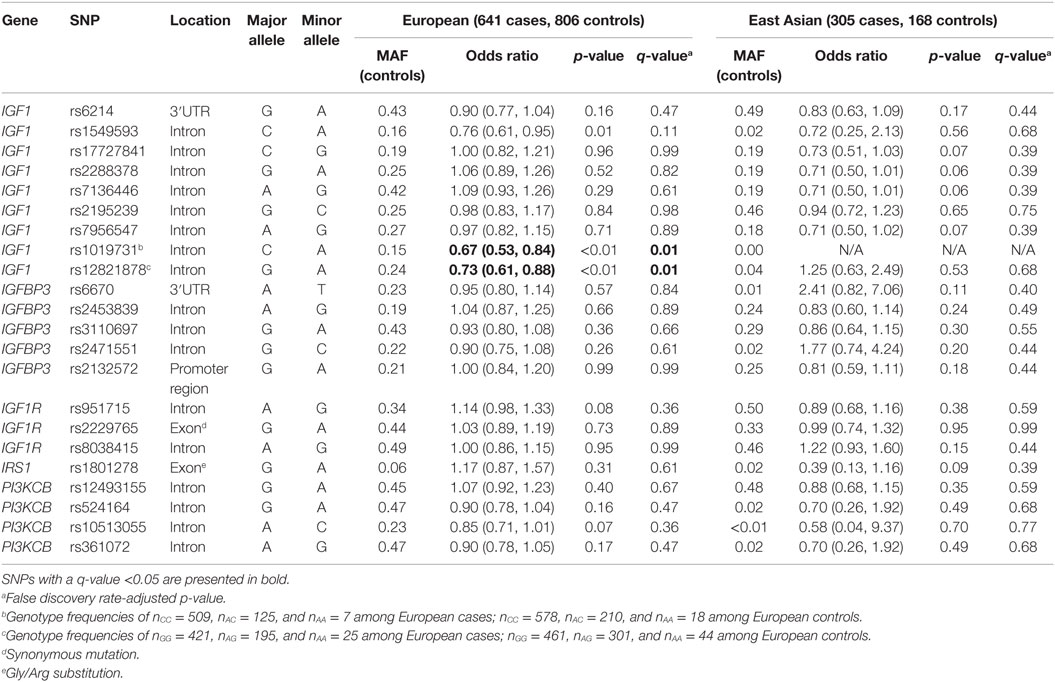
Table 2. Minor allele frequencies (MAF) and age- and center-adjusted odds ratios for the associations between SNPs in insulin-like growth factor signaling genes and breast cancer risk.
Analyses further stratified by menopausal status are presented in Table 3. Among European women, no effect modification was detected. However, there was suggested modification by menopausal status for the rs1801278 polymorphism in the IRS1 gene (interaction p-value, pint = 0.06), which appeared to decrease the risk of premenopausal, while increasing the risk of postmenopausal breast cancers. Among East Asian women, modification of genetic effect by menopausal status was apparent for four IGF1 SNPs: rs17727841, rs2288378, rs7136446, and rs7956547 (pint < 0.01 for all), although these SNPs were in high linkage disequilibrium (LD) (r2 = 0.86–0.98). The minor alleles of these SNPs were associated with decreased risk of breast cancer among postmenopausal women.
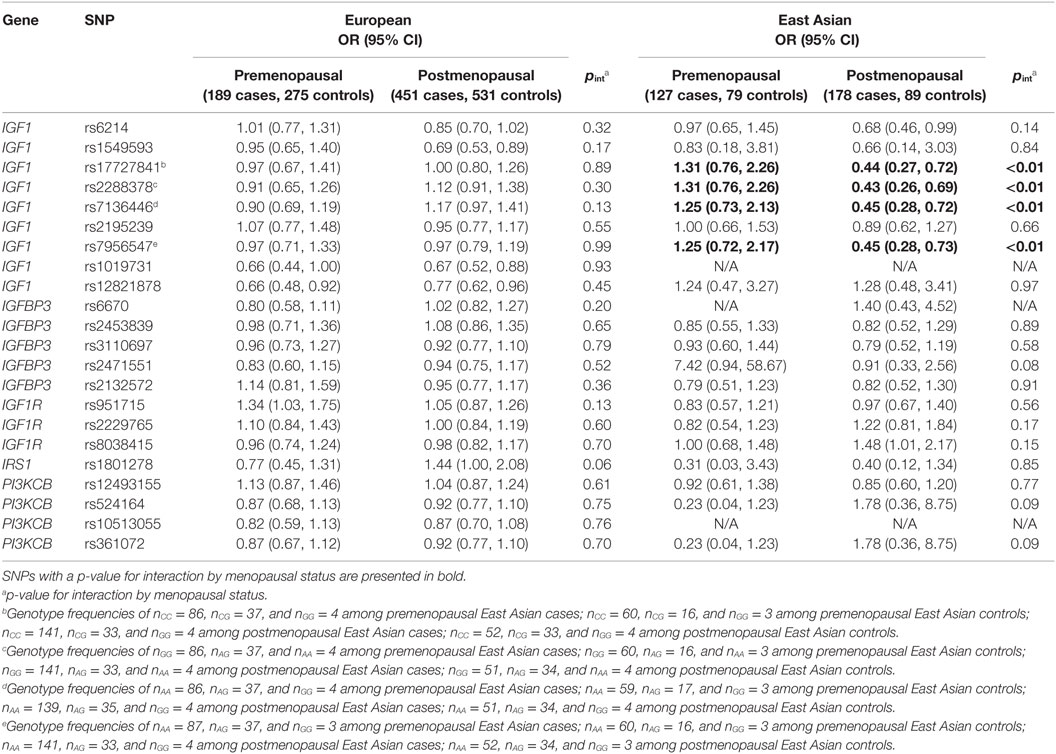
Table 3. Selected interactions of menopausal status and IGF1 pathway SNPs on breast cancer risk among European and East Asian women.
Lastly, heterogeneity in odds ratios between breast tumor subtypes for select IGF1 pathway SNPs among European and East Asian women are presented in Table 4. In European women, heterogeneity between tumor subtypes was not observed for any of the five SNPs that were examined. In East Asian women, tumor heterogeneity was observed for rs2288378 (pTH = 0.04), in which a decreased risk of ER+ and/or PgR+/HER2− tumors, but not other tumor subtypes, was noted.
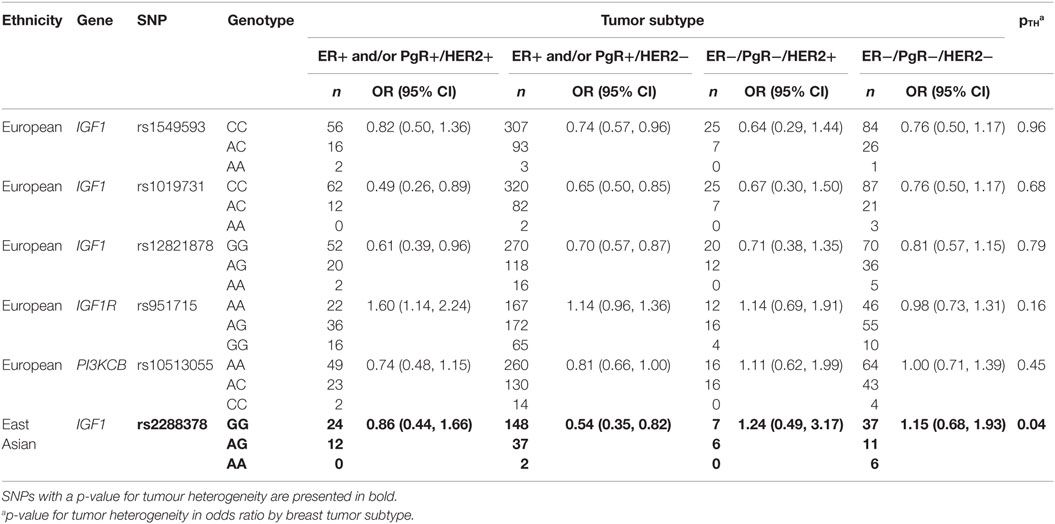
Table 4. Heterogeneity in odds ratios between breast tumour subtypes for select IGF1 pathway SNPs among European and East Asian women.
In our analyses of European and East Asian women, we observed associations between select IGF1 SNPs and breast cancer risk in both European and East Asian women. In Europeans, the minor alleles of two IGF1 SNPs (rs1019731 and rs12821878) were associated with reduced breast cancer risk. In East Asians, the association between four IGF1 SNPs (rs17727841, rs2288378, rs7136446, and rs7956547) in high LD and breast cancer risk was modified by menopausal status and the minor alleles of these SNPs were associated with decreased risk of postmenopausal breast cancer. Lastly, heterogeneity by tumor subtype for rs2288378 was observed in East Asian women.
An association between SNPs of the IGF1 pathway and breast cancer risk has been hypothesized due to the role of circulating IGF1 levels and the IGF1 signaling pathway in risk of breast cancer (3, 9, 23). Variants of the IGF1 gene, which encodes the IGF1 protein, are associated with increased circulating IGF1 levels (14–16) and increased percent breast density (17, 34, 35). However, studies on its association with breast cancer risk have been less conclusive. Our study observed associations between two IGF1 SNPs, rs1019731 and rs12821878, and breast cancer risk among European women; however, a study from the Breast and Prostate Cancer Cohort Consortium reported no association with breast cancer risk for these SNPs (23). A study by Verheus et al. evaluated the relationship between IGF1 variants and mammographic density, and reported that the rs12821878 minor allele may be associated with decreased mammographic density (36), which is congruous with the decrease in risk we observed.
Similarly, the IGFBP3, IGF1R, IRS1, and PI3KCB genes have a purported relationship with breast cancer risk due to either their role in IGF1 signaling regulation (15, 18, 27), or association with strong breast cancer risk factors (17, 28). However, our null results with regard to the relationship between variants of the IGFBP3, IGF1R, IRS1, and PI3KCB genes and, overall, premenopausal and postmenopausal breast cancer risk in European and East Asian women are in concordance with the findings from analyses in the Breast and Prostate Cancer Cohort Consortium (23, 37) and recent GWAS publications (38, 39).
We observed modification of associations with breast cancer risk by menopausal status for four IG1 SNPs (rs17727841, rs2288378, rs7136446, and rs7956547) in high LD among East Asian women. Menopause-associated declines in IGF1 levels have been previously reported (40, 41) and may explain our findings, and is corroborated by reported effect modification by menopausal status of associations between breast cancer risk factors, such as age, recent leisure-time physical activity and alcohol consumption, and IGF1 and IGFBP3 levels (42). However, the decrease in risk as a result of these SNPs among postmenopausal East Asian women is incongruent with the literature. A meta-analysis reported that IGF1 levels were approximately 7 ng/mL higher among individuals carrying at least one minor allele at the rs7136446 polymorphism compared to homozygous carriers of the major allele (43). Genotype imputation of this SNP in another study produced similar findings (16). Increased IGF1 levels would presumably lead to increased risk of breast cancer; however, our study found the opposite direction of effect.
Analyses by breast tumor subtype revealed potential heterogeneity for rs2288378, representing the four high-LD IGF1 SNPs, decreasing risk of only ER+ and/or PgR+/HER2− tumors by approximately 45% among East Asian women. One study suggests that positive associations between IGF1 concentrations and breast cancer risk may be restricted to ER+ tumors (9), which may explain why stronger associations were observed for ER+ and/or PgR+ tumors in our study. In vitro studies have demonstrated that the IGF and ER signaling pathways are positively reinforced by one another, mediating signaling via the other pathway through increasing expression of key signaling pathway proteins, cross-activation, and stimulating transcriptional activation (13, 44, 45). In addition, interactions with HER2 have been suggested, as cross-talk between IGF1R and HER2 has been implicated in resistance against trastuzumab, a monoclonal antibody used in breast cancer treatment to target HER2 receptors (46, 47). This extensive network of interactions between IGF1, ER, and HER2 signaling pathways, and potential heterogeneity of risk by tumor subtype for IGF1 genes observed in our study suggest greater complexities in the relationship between IGF1 pathway genes and breast cancer etiology.
Limitations of our study include limited statistical power in stratified and subgroup analyses. Specifically, with the number of cases and controls in our European subgroup and after accounting for multiple comparisons, we were powered to observe effect sizes as low as OR = 1.34 (or OR = 0.75 if protective) for SNPs with MAFs of 0.50, and OR = 1.80 (or OR = 0.56 if protective) for SNPS with MAFs of 0.05. The small sample sizes in the analyses of East Asian women and of tumor subtypes warrant caution in the interpretations, and further large studies are needed to confirm these findings. Differences in demographic, lifestyle, and reproductive factors between cases and controls of the study may be partly attributed to the recruitment methods used in BC where cases were selected using a population-based cancer registry and controls were recruited from a breast mammography program. If those who elect to participate in screening programs are different than those who do not, there is potential for selection bias to be introduced. However, since the study recruitment strategies are unlikely to be related to individuals’ genetic variants in IGF pathway genes, concerns for selection bias are minimized.
In conclusion, our study observed associations between some IGF1 SNPs and breast cancer risk among European and East Asian women, although evidence of an association for other IGF pathway SNPs was limited. Potential modification of these relationships by menopausal status and breast tumor subtype may warrant further consideration when assessing molecular signaling pathway interactions in the genetic etiology of breast cancer.
JS was responsible for the analysis and drafting the manuscript. Drs. KA and JJS are the co-principal investigators of the Canadian Breast Cancer Study (CBCS). Drs. AG, IB, CL, SS, AB-W, and HR are investigators of the CBCS. All co-PIs and investigators of the CBCS were involved in designing and conducting the original study. AL is the Research Project Coordinator for the CBCS and was involved in study planning. LK assisted in the analysis of the physical activity-related variables, and Dr. JMS was responsible for the quality control for the genotype data. Dr. HR also played a supervisory role in the preparation of this manuscript. All authors reviewed and contributed to the submitted version of the paper.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors thank Dr. Linda Warren, Dr. Philip Switzer, Caroline Speers, the BC Cancer Registry, the Screening Mammography Program of BC, Agnes Bauzon, Alegria Imperial, Betty Hall, Lina Hsu, Maria Andrews, Teresa Pavlin, Dr. Ross Walker, Dr. Ralph George, Celine Morissette, Jane Warner, Hilary Rimmer, Meghan Hamel, and Annie Langley for their assistance in participant recruitment and data collection in Vancouver and Kingston. Finally, we would like to thank McGill University and Genome Quebec Innovation Centre, Montreal, Canada for their high quality genotyping services. The Canadian Breast Cancer Study was supported by the Canadian Institutes of Health Research (Grant #69036).
The Supplementary Material for this article can be found online at http://journal.frontiersin.org/article/10.3389/fonc.2016.00136
1. Christopoulos PFP, Msaouel P, Koutsilieris M. The role of the insulin-like growth factor-1 system in breast cancer. Mol Cancer (2015) 14(1):43. doi:10.1186/s12943-015-0291-7
2. Sachdev D, Yee D. The IGF system and breast cancer. Endocr Relat Cancer (2001) 8(3):197–209. doi:10.1677/erc.0.0080197
3. Pollak M. The insulin and insulin-like growth factor receptor family in neoplasia: an update. Nat Rev Cancer (2012) 12(3):159–69. doi:10.1038/nrc3215
4. Siddle K. Signalling by insulin and IGF receptors: supporting acts and new players. J Mol Endocrinol (2011) 47(1):R1–10. doi:10.1530/JME-11-0022
5. Chan JM, Stampfer MJ, Giovannucci E, Gann PH, Ma J, Wilkinson P, et al. Plasma insulin-like growth factor-I and prostate cancer risk: a prospective study. Science (1998) 279(5350):563–6. doi:10.1126/science.279.5350.563
6. Renehan AG, Zwahlen M, Minder C, O’Dwyer ST, Shalet SM, Egger M. Insulin-like growth factor (IGF)-I, IGF binding protein-3, and cancer risk: systematic review and meta-regression analysis. Lancet (2004) 363(9418):1346–53. doi:10.1016/S0140-6736(04)16044-3
7. Rinaldi S, Cleveland R, Norat T, Biessy C, Rohrmann S, Linseisen J, et al. Serum levels of IGF-I, IGFBP-3 and colorectal cancer risk: results from the EPIC cohort, plus a meta-analysis of prospective studies. Int J Cancer (2010) 126(7):1702–15. doi:10.1002/ijc.24927
8. Ma J, Pollak M, Giovannucci E, Chan JM, Tao Y, Hennekens C, et al. A prospective study of plasma levels of insulin-like growth factor I (IGF-I) and IGF-binding protein-3, and colorectal cancer risk among men. Growth Horm IGF Res (2000) 10(Suppl A):S28–9. doi:10.1016/S1096-6374(00)90013-3
9. Key TJ, Appleby PN, Reeves GK, Roddam AW, Endogenous Hormones and Breast Cancer Collaborative Group. Insulin-like growth factor 1 (IGF1), IGF binding protein 3 (IGFBP3), and breast cancer risk: pooled individual data analysis of 17 prospective studies. Lancet Oncol (2010) 11(6):530–42. doi:10.1016/S1470-2045(10)70095-4
10. Kurmasheva RT, Houghton PJ. IGF-I mediated survival pathways in normal and malignant cells. Biochim Biophys Acta (2006) 1766(1):1–22. doi:10.1016/j.bbcan.2006.05.003
11. Renehan AG, Harvie M, Howell A. Insulin-like growth factor (IGF)-I, IGF binding protein-3, and breast cancer risk: eight years on. Endocr Relat Cancer (2006) 13(2):273–8. doi:10.1677/erc.1.01219
12. Fletcher O, Gibson L, Johnson N, Altmann DR, Holly JMP, Ashworth A, et al. Polymorphisms and circulating levels in the insulin-like growth factor system and risk of breast cancer: a systematic review. Cancer Epidemiol Biomarkers Prev (2005) 14(1):2–19.
13. Yee D, Lee AV. Crosstalk between the insulin-like growth factors and estrogens in breast cancer. J Mammary Gland Biol Neoplasia (2000) 5(1):107–15. doi:10.1023/A:1009575518338
14. Hernandez W, Grenade C, Santos ERE, Bonilla C, Ahaghotu C, Kittles RA. IGF-1 and IGFBP-3 gene variants influence on serum levels and prostate cancer risk in African-Americans. Carcinogenesis (2007) 28(10):2154–9. doi:10.1093/carcin/bgm190
15. Al-Zahrani A, Sandhu MSM, Luben RN, Thompson D, Baynes C, Pooley KA, et al. IGF1 and IGFBP3 tagging polymorphisms are associated with circulating levels of IGF1, IGFBP3 and risk of breast cancer. Hum Mol Genet (2006) 15(1):1–10. doi:10.1093/hmg/ddi398
16. Gu F, Schumacher FR, Canzian F, Allen NE, Albanes D, Berg CD, et al. Eighteen insulin-like growth factor pathway genes, circulating levels of IGF-I and its binding protein, and risk of prostate and breast cancer. Cancer Epidemiol Biomarkers Prev (2010) 19(11):2877–87. doi:10.1158/1055-9965.EPI-10-0507
17. Diorio C, Brisson J, Bérubé S, Pollak M. Genetic polymorphisms involved in insulin-like growth factor (IGF) pathway in relation to mammographic breast density and IGF levels. Cancer Epidemiol Biomarkers Prev (2008) 17(4):880–8. doi:10.1158/1055-9965.EPI-07-2500
18. Bonafè M, Barbieri M, Marchegiani F, Olivieri F, Ragno E, Giampieri C, et al. Polymorphic variants of insulin-like growth factor I (IGF-I) receptor and phosphoinositide 3-kinase genes affect IGF-I plasma levels and human longevity: cues for an evolutionarily conserved mechanism of life span control. J Clin Endocrinol Metab (2003) 88(7):3299–304. doi:10.1210/jc.2002-021810
19. Suh Y, Atzmon G, Cho M-OM, Hwang D, Liu B, Leahy DJ, et al. Functionally significant insulin-like growth factor I receptor mutations in centenarians. Proc Natl Acad Sci U S A (2008) 105(9):3438–42. doi:10.1073/pnas.0705467105
20. Qian B, Zheng H, Yu H, Chen K. Genotypes and phenotypes of IGF-I and IGFBP-3 in breast tumors among Chinese women. Breast Cancer Res Treat (2011) 130(1):217–26. doi:10.1007/s10549-011-1552-9
21. Kang HH-S, Ahn SHS, Mishra SSK, Hong KK-M, Lee ES, Shin KH, et al. Association of polymorphisms and haplotypes in the insulin-like growth factor 1 receptor (IGF1R) gene with the risk of breast cancer in Korean women. PLoS One (2014) 9(1):e84532. doi:10.1371/journal.pone.0084532
22. Slattery ML, Sweeney C, Wolff R, Herrick J, Baumgartner K, Giuliano A, et al. Genetic variation in IGF1, IGFBP3, IRS1, IRS2 and risk of breast cancer in women living in Southwestern United States. Breast Cancer Res Treat (2007) 104(2):197–209. doi:10.1007/s10549-006-9403-9
23. Canzian F, Cox DG, Setiawan VW, Stram DO, Ziegler RG, Dossus L, et al. Comprehensive analysis of common genetic variation in 61 genes related to steroid hormone and insulin-like growth factor-I metabolism and breast cancer risk in the NCI breast and prostate cancer cohort consortium. Hum Mol Genet (2010) 19(19):3873–84. doi:10.1093/hmg/ddq291
24. Grundy A, Schuetz JM, Lai AS, Janoo-Gilani R, Leach S, Burstyn I, et al. Shift work, circadian gene variants and risk of breast cancer. Cancer Epidemiol (2013) 37(5):606–12. doi:10.1016/j.canep.2013.04.006
25. de Bakker PIW, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D, et al. Efficiency and power in genetic association studies. Nat Genet (2005) 37(11):1217–23. doi:10.1038/ng1669
26. Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics (2005) 21(2):263–5. doi:10.1093/bioinformatics/bth457
27. Ren Z, Cai Q, Shu XX-O, Cai H, Li C, Yu H, et al. Genetic polymorphisms in the IGFBP3 gene: association with breast cancer risk and blood IGFBP-3 protein levels among Chinese women. Cancer Epidemiol Biomarkers Prev (2004) 13(8):1290–5.
28. Lai JJH, Vesprini D, Zhang W, Yaffe MMJ, Pollak M, Narod SA. A polymorphic locus in the promoter region of the IGFBP3 gene is related to mammographic breast density. Cancer Epidemiol Biomarkers Prev (2004) 13(4):573–82.
29. Deming SLS, Ren Z, Wen W, Shu XOX, Cai Q, Gao Y-T, et al. Genetic variation in IGF1, IGF-1R, IGFALS, and IGFBP3 in breast cancer survival among Chinese women: a report from the Shanghai Breast Cancer Study. Breast Cancer Res Treat (2007) 104(3):309–19. doi:10.1007/s10549-006-9420-8
30. Neuhausen SLS, Brummel S, Ding YC, Singer CF, Pfeiler G, Lynch HT, et al. Genetic variation in insulin-like growth factor signaling genes and breast cancer risk among BRCA1 and BRCA2 carriers. Breast Cancer Res (2009) 11(5):R76. doi:10.1186/bcr2414
31. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet (2007) 81(3):559–75. doi:10.1086/519795
32. Abecasis GR, Cherny SS, Cookson WO, Cardon LR. GRR: graphical representation of relationship errors. Bioinformatics (2001) 17(8):742–3. doi:10.1093/bioinformatics/17.8.742
33. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B (1995) 57(1):289–300.
34. Tamimi RMR, Cox DGD, Kraft P, Pollak MN, Haiman CA, Cheng I, et al. Common genetic variation in IGF1, IGFBP-1, and IGFBP-3 in relation to mammographic density: a cross-sectional study. Breast Cancer Res (2007) 9(1):R18. doi:10.1186/bcr1655
35. Verheus M, McKay JJD, Kaaks R, Canzian F, Biessy C, Johansson M, et al. Common genetic variation in the IGF-1 gene, serum IGF-I levels and breast density. Breast Cancer Res Treat (2008) 112(1):109–22. doi:10.1007/s10549-007-9827-x
36. Verheus M, Maskarinec G, Woolcott CG, Haiman CA, Le Marchand L, Henderson BE, et al. IGF1, IGFBP1, and IGFBP3 genes and mammographic density: the Multiethnic Cohort. Int J Cancer (2010) 127(5):1115–23. doi:10.1002/ijc.25142
37. Patel AVA, Cheng I, Canzian F, Le Marchand L, Thun MJ, Berg CD, et al. IGF-1, IGFBP-1, and IGFBP-3 polymorphisms predict circulating IGF levels but not breast cancer risk: findings from the Breast and Prostate Cancer Cohort. PLoS One (2008) 3(7):e2578. doi:10.1371/journal.pone.0002578
38. Zheng W, Long J, Gao Y-TY, Li C, Zheng Y, Xiang Y-B, et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat Genet (2009) 41(3):324–8. doi:10.1038/ng.318
39. Thomas G, Jacobs KKB, Kraft P, Yeager M, Wacholder S, Cox DG, et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1). Nat Genet (2009) 41(5):579–84. doi:10.1038/ng.353
40. Nasu M, Sugimoto T, Chihara M, Hiraumi M, Kurimoto F, Chihara K. Effect of natural menopause on serum levels of IGF-I and IGF-binding proteins: relationship with bone mineral density and lipid metabolism in perimenopausal women. Eur J Endocrinol (1997) 136(6):608–16. doi:10.1530/eje.0.1360608
41. Poehlman ET, Toth MJ, Ades PA, Rosen CJ. Menopause-associated changes in plasma lipids, insulin-like growth factor I and blood pressure: a longitudinal study. Eur J Clin Invest (1997) 27(4):322–6. doi:10.1046/j.1365-2362.1997.1160662.x
42. Barnes BBE, Chang-Claude J, Flesch-Janys D, Kinscherf R, Schmidt M, Slanger T, et al. Cancer risk factors associated with insulin-like growth factor (IGF)-I and IGF-binding protein-3 levels in healthy women: effect modification by menopausal status. Cancer Causes Control (2009) 20(10):1985–96. doi:10.1007/s10552-009-9393-0
43. Johansson M, McKay JJD, Wiklund F, Rinaldi S, Verheus M, van Gils CH, et al. Implications for prostate cancer of insulin-like growth factor-I (IGF-I) genetic variation and circulating IGF-I levels. J Clin Endocrinol Metab (2007) 92(12):4820–6. doi:10.1210/jc.2007-0887
44. Lee AV, Weng CN, Jackson JG, Yee D. Activation of estrogen receptor-mediated gene transcription by IGF-I in human breast cancer cells. J Endocrinol (1997) 152(1):39–47. doi:10.1677/joe.0.1520039
45. Stoica A, Saceda M, Fakhro A, Joyner M, Martin MB. Role of insulin-like growth factor-I in regulating estrogen receptor-alpha gene expression. J Cell Biochem (2000) 76(4):605–14. doi:10.1002/(SICI)1097-4644(20000315)76:4<605::AID-JCB9>3.0.CO;2-T
46. Nahta R, Esteva F. HER2 therapy: molecular mechanisms of trastuzumab resistance. Breast Cancer Res (2006) 8(6):215. doi:10.1186/bcr1612
Keywords: IGF, polymorphisms, breast cancer, breast cancer subtypes, interactions, case–control
Citation: Shi J, Aronson KJ, Grundy A, Kobayashi LC, Burstyn I, Schuetz JM, Lohrisch CA, SenGupta SK, Lai AS, Brooks-Wilson A, Spinelli JJ and Richardson H (2016) Polymorphisms of Insulin-Like Growth Factor 1 Pathway Genes and Breast Cancer Risk. Front. Oncol. 6:136. doi: 10.3389/fonc.2016.00136
Received: 02 March 2016; Accepted: 23 May 2016;
Published: 08 June 2016
Edited by:
Sven Bilke, National Institutes of Health, USAReviewed by:
Guanglong Jiang, Indiana University School of Medicine, USACopyright: © 2016 Shi, Aronson, Grundy, Kobayashi, Burstyn, Schuetz, Lohrisch, SenGupta, Lai, Brooks-Wilson, Spinelli and Richardson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Harriet Richardson, aHJpY2hhcmRzb25AY3RnLnF1ZWVuc3UuY2E=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.