
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CASE REPORT article
Front. Oncol. , 30 May 2016
Sec. Cancer Epidemiology and Prevention
Volume 6 - 2016 | https://doi.org/10.3389/fonc.2016.00124
 Muhammad Asif1,2
Muhammad Asif1,2 Mohammad Sarwar Jamal3*
Mohammad Sarwar Jamal3* Abdul Rehman Khan4
Abdul Rehman Khan4 Muhammad Imran Naseer5
Muhammad Imran Naseer5 Abrar Hussain1
Abrar Hussain1 Hani Choudhry6
Hani Choudhry6 Arif Malik7
Arif Malik7 Shahida Aziz Khan3
Shahida Aziz Khan3 Maged Mostafa Mahmoud3,8
Maged Mostafa Mahmoud3,8 Ashraf Ali3
Ashraf Ali3 Saima Iram9
Saima Iram9 Kashif Kamran10
Kashif Kamran10 Asim Iqbal10
Asim Iqbal10 Zainularifeen Abduljaleel11
Zainularifeen Abduljaleel11 Peter Natesan Pushparaj5
Peter Natesan Pushparaj5 Mahmood Rasool5*
Mahmood Rasool5*
Philadelphia (Ph) chromosome (9;22)(q34;q11) is well established in more than 90% of chronic myeloid leukemia (CML) patients, and the remaining 5–8% of CML patients show variant and complex translocations, with the involvement of third, fourth, or fifth chromosome other than 9;22. However, in very rare cases, the fourth chromosome is involved. Here, we found a novel case of four-way Ph+ chromosome translocation involving 46,XY,t(4;9;19;22)(q25:q34;p13.3;q11.2) with CML in the chronic phase. Complete blood cell count of the CML patient was carried out to obtain total leukocytes count, hemoglobin, and platelets. Fluorescence in situ hybridization technique was used for the identification of BCR–ABL fusion gene, and cytogenetic test for the confirmation of Ph (9;22)(q34;q11) and the mechanism of variant translocation in the bone marrow. The patient is successfully treated with a dose of 400 mg/day imatinib mesylate (Gleevec). We observed a significant decrease in white blood cell count of 11.7 × 109/L after 48-month follow-up. Patient started feeling better generally. There was a reduction in the swelling of the body, fatigue, and anxiety.
Chronic myeloid leukemia (CML) is triggered due to the t(9;22)(q34;q11) translocation between the long arms of chromosomes 9 and 22, called as the Philadelphia (Ph) chromosome (1). In these patients, bone marrow myeloid hyperplasia, an elevated myeloid and erythroid cells, and platelets in the peripheral blood were observed (2). This translocation was identified in more than 90% of the CML patients (3, 4), and the variant/complex translocation was observed in 5–8% of cases with an involvement of additional third, fourth, or fifth chromosome (5–7).
The imatinib mesylate is commonly used as the first-line oral treatment of CML patients (6). It blocks the BCR–ABL tyrosine kinase activity and subsequently induces apoptosis followed by the reduction in the proliferation of BCR–ABL-expressing cells in both CML and acute lymphocytic leukemia (ALL). The treatment of CML patients with imatinib significantly increased the survival and improved the quality of life (8).
Potentially imatinib meysylate inhibits the BCR/ABL and platelet-derived growth factor receptor (PDGFR) tyrosine kinase activities. This deactivates downstream signaling by reducing cell proliferation and augmenting apoptosis. Imatinib mesylate (Gleevec) therapy has significantly increased the efficacy in 0–34% of Ph-positive cells with t(9;22) translocation along with other complex translocations causing BCR/ABL gene fusion and subsequent clonal evolution (9, 10). In this study, for the first time, we present a four-way Ph translocation 46,XY,t(4;9;19;22)(q25:q34;p13.3;q11.2) in a CML patient with a new complex rearrangement between chromosomes 4 and 19 as well as 9 and 22.
A 45-year-old male patient was diagnosed with CML on 19 October, 2012. The hematological parameters were hemoglobin (Hb) 10.0 g/dL (normal range, 14–18 mg/dL), MCV 59.6 fL (normal range, 76–95 fL), MCH 19 pg (normal range, 27–32 pg), MCHC 31.9% (normal range, 30–35), hematocrit 31.4% (normal range, 40–54%), red blood cell (RBC) 5.26 × 109/L (normal range, 4.5–6.5 × 109/L), white blood cell (WBC) 160.7 × 109/L (normal range, 4–11 × 103/μL × 109/L), neutrophils 58% (normal range, 40–75%), lymphocytes 2% (normal range, 20–45), eosinophils 3% (normal range, 1–6%), monocytes 4% (normal range, 2–10%), basophils 4% (normal range, 0–1%), metamyelocytes 15% (normal range, 0–0%), myelocytes 15% (normal range, 0–0%), blast cells 2% (normal range, 0–0%), and platelets 223 × 109/L (normal range, 150–400 × 109/L). Peripheral film showed dimorphic picture, anisocytosis, hypochromic, microcytic, polychromasia, polychromasia, tear drop cells, and nucleated RBC. A written informed ethical consent was taken from the patient before the study according to Helsinki declaration. The ethical committee of Balochistan University of Information Technology, Engineering and Management Sciences (BUITEMS), Quetta, Pakistan, had given the approval for this study.
Complete blood count (CBC) was performed to measure the levels of WBCs, RBCs, Hb, and platelets in the CML patients. CBC was performed using an Automatic Hematological Analyzer (Nihon Khoden, Japan) within 2 h of blood sampling. An increase in WBCs and lower levels of RBCs and platelets confirmed the leukemia, and these patients were considered for further examination.
Chromosome analysis using GTG banding was done as described previously (9). Karyotyping was performed in 25 metaphases from unstimulated bone marrow samples according to the nomenclature of the International System for Human Cytogenetics (11).
Fluorescence in situ hybridization was performed to detect BCR/ABL as described previously (12).
The cytogenetic analysis showed 46,XY,t(4;9)(q25:q34)t(9;22)(q34;q11.2). Twenty-five cells were counted, and all were positive for Ph chromosome (Figure 1).
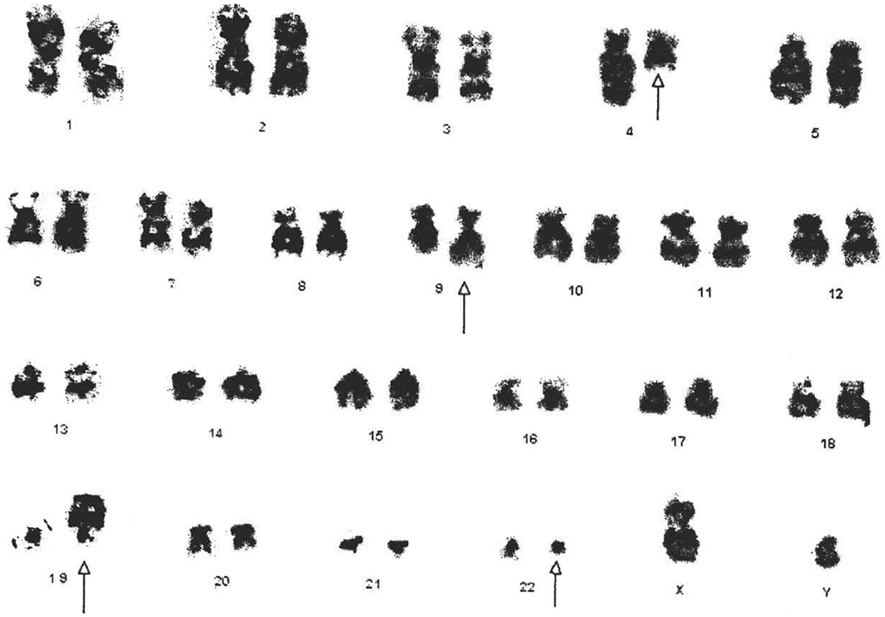
Figure 1. Cytogenetic analysis shows the karyotype of the CML patient with 46,XY,t(4;9;19;22)(q25:q34;p13.3;q11.2). All derivative chromosomes are highlighted by arrow heads.
The BCR–ABL translocation was detected by fluorescence in situ hybridization (FISH) analysis in 91% of the 500 nuclei counted. In this study, FISH analysis of the ABL (9q34) gene was identified by fluorescent red dots and BCR (22q11) gene by green dots. Therefore, a cell exhibiting two separate green and red dots counted as a normal cell shows no translocation. However, the irregular translocation in a cell was identified by one red and one green and fused red, yellow, and green signal. Dual color, dual fusion translocation probes were hybridized to patient’s interphase nuclei (Figure 2).
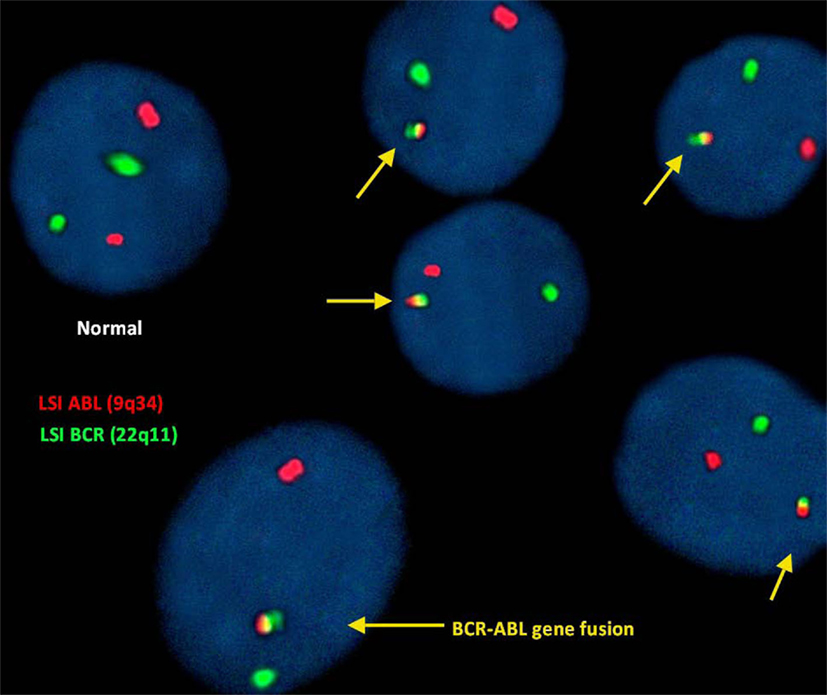
Figure 2. Fluorescence in situ hybridization (FISH) for the detection of (9;22)(q34;q11). BCR–ABL translocation was detected in 91% of the 500 nuclei counted.
Clinical analysis shows the induced level of WBC (160.7 × 109/L) and low level of Hb (10 mg/dL), which indicates anemia in the CML patient.
The X-ray analysis studies showed no active pulmonary or pleural lesion, normal cardiac and aorta, normal hilar and mediastinal shadows, normal costophrenic angles, normal domes of diaphragm, and normal bony thoracic cage with the normal finding in the chest study. The analysis of liver showed a slight enlargement in size, whereas spleen was massively enlarged, with normal gallbladder, pancreas, kidneys, and retroperitoneum with mild hepatomegaly.
HBsAg and anti-HCV tests were performed by an immunochromatographic screening method, which showed negative results. The patient was treated with antibiotics and Gleevec (imatinib mesylate) 400 mg/day. Socioeconomic status of the CML patient was middle class, and he was aware of this disease.
Chronic myeloid leukemia is primarily caused by the balanced translocation between the long arms of 9;22 chromosomes and secondarily by the variant and complex translocation patients. In such cases, the third, fourth, or even fifth chromosome was involved and is termed as four-, five-, or six-way translocation (6). The four-way translocation is rare; only 59 cases are reported in the literature. The four-way translocation is observed more in male than in female. The five-way translocation is very rare in the CML patients, with only few cases are on record (Table 1).

Table 1. Number of complex variant four-way Ph chromosome translocations reported in the literature.
In this study, we reported this new four-way translocation 46,XY,t(4;9;19;22)(q25:q34;p13.3;q11.2) in a CML patient for the first time and was cross-checked in Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer. In four-way Ph chromosome translocation, male patients are 46-XY,t(56%) who are more affected compared to female patients who are 46-XX,t(39%) (Figure 3), whereas in addition to chromosome 9;22, chromosome 4 (2%) is least compared to chromosome 19, 12, 6 (9%) in four-way Ph chromosome translocation (Figure 3). BCR–ABL translocation was detected in 91% of the 500 nuclei counted. Dual color, dual fusion translocation probes were hybridized to patient’s interphase nuclei. Normal nuclei lacking the t(9;22) translocation showed two green and two orange signals. In the nucleus containing a simple balanced t(9;22), one green and one orange signal from the normal 9 and 22 chromosomes and two green/orange (yellow) fusion signals, one each from the chromosomes 9 and 22.
Mkrtchyan et al. have described two sustainable mechanisms for the formation of variant complex translocation, namely, a single incident rearrangement via the simultaneous breakage of few chromosomes followed by mismatched joining and a multistep process of classical Ph translocation followed by additional translocations in chromosomes 9 and 22, as well as other chromosomes (49).
Imatinib mesylate (Gleevec) potentially inhibits BCR–ABL protein tyrosine kinase. Furthermore, it inhibits the tyrosine kinase activities of the platelet-derived growth factor (PDGF) receptor β and c-Kit, but it does not inhibit Flt-3 and Fms belonging to type III tyrosine kinase family (50, 51). Being a first-line oral therapy, imatinib mesylate, an inhibitor of tyrosine kinase activity of BCR–ABL protein, is recommended for Ph-positive chromosome-associated abnormalities. Hence, it would be useful in three-way, four-way, and five-way complex variant translocations, as reported earlier by other research groups (6, 21).
Finally, a unique case of four-way complex variant Ph-positive translocation involving chromosome 46,XY,t(4;9;19;22)(q25:q34;p13.3;q11.2) in a CML patient was reported in this study.
MA, AH, MR, and PP: concept design execution. MA, AM, and AH: experimental lab work, execution. SK, MM, AA, and SI: data collection. MR, MJ, AK, MN, HC, KK, AI, ZA, and PP: data analysis and writing.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This study was partially funded by the Faculty of Life Sciences, BUITEMS, Quetta, Pakistan, registration No: 27934 and by the Kind Abdul Aziz City for Science and Technology (KACST) Strategic Project No: 12-MED3078-03, Saudi Arabia.
1. Aguayo A, Garcia-Alvarez E, Cazares-Ordonez Y, et al. Chronic myeloid leukemia: a clinicoepidemiologic and therapeutic description of a single institution in Mexico City. Clinical Leukemia (2008) 2:261–6. doi:10.3816/CLK.2008.n.036
2. Sawyers CL. Chronic myeloid leukemia. New Eng J of Med (1999) 340:1330–40. doi:10.1056/NEJM199904293401706
3. Faderl S, Talpaz M, Estrov Z, O’Brien S, Kurzrock R, Kantarjian HM. The biology of chronic myeloid leukemia. N Engl J Med (1999) 341:164–72. doi:10.1056/NEJM199907153410306
4. Quintas-Cardama A, Cortes JE. Chronic myeloid leukemia: diagnosis and treatment. Mayo Clin Proc (2006) 81:973–88. doi:10.4065/81.7.973
5. La Starza R, Testoni N, Lafage-Pochitaloff M, Ruggeri D, Ottaviani E, Perla G, et al. Complex variant Philadelphia translocations involving the short arm of chromosome 6 in chronic myeloid leukemia. Haematologica (2002) 87:143–7.
6. Al-Achkar W, Wafa A, Ikhtiar A, Liehr T. Three-way Philadelphia translocation t(9;10;22)(q34;p11.2;q11.2) as a secondary abnormality in an imatinib mesylate-resistant chronic myeloid leukemia patient. Oncol Lett (2013) 5:1656–8. doi:10.3892/ol.2013.1228
7. Sessarego M, Fugazza G, Bruzzone R, Ballestrero A, Miglino M, Bacigalupo A. Complex chromosome rearrangements may locate the BCR/ABL fusion gene sites other than 22q11. Haematologica (2000) 85:35–9.
8. Cowan-Jacob SW, Fendrich G, Floersheimer A, Furet P, Liebetanz J, Rummel G, et al. Structural biology contributions to the discovery of drugs to treat chronic myelogenous leukaemia. Acta Crystallogr D Biol Crystallogr (2007) 63:80–93. doi:10.1107/S0907444906047287
9. Claussen U, Michel S, Mühlig P, Westermann M, Grummt UW, Kromeyer-Hauschild K, et al. Demystifying chromosome preparation and the implications for the concept of chromosome condensation during mitosis. Cytogenet Genome Res (2002) 98:136–46. doi:10.1159/000069817
10. Griffin J. The biology of signal transduction inhibition: basic science to novel therapies. Semin Oncol (2001) 28:3–8. doi:10.1016/S0093-7754(01)90097-1
11. Simons A, Shaffer LG, Hastings RJ. Cytogenetic nomenclature: changes in the ISCN 2013 compared to the 2009 edition. Cytogenet Genome Res (2013) 141(1):1–6. doi:10.1159/000353118
12. Froncillo MC, Maffei L, Cantonetti M, Del Poeta G, Lentini R, Bruno A, et al. FISH analysis for CML monitoring? Ann Hematol (1996) 73:113–9. doi:10.1007/s002770050211
13. Acar H, Stewart J, Boyd E, Connor MJ. Identification of variant translocations in chronic myeloid leukemia by fluorescence in situ hybridization. Cancer Genet Cytogenet (1997) 93:115–8. doi:10.1016/S0165-4608(96)00168-9
14. Bernstein R, Pinto MR, Wallace C, Penfold G, Mendelow B. The incidence, type, and subsequent evolution of 14 variant Ph1 translocations in 180 South African patients with Ph1-positive chronic myeloid leukemia. Cancer Genet Cytogenet (1984) 12:225–38. doi:10.1016/0165-4608(84)90034-7
15. Bennour A, Sennana H, Laatiri MA, Elloumi M, Khelif A, Saad A. Molecular cytogenetic characterization of variant Philadelphia translocations in chronic myeloid leukemia: genesis and deletion of derivative chromosome 9. Cancer Genet Cytogenet (2009) 194:30–7. doi:10.1016/j.cancergencyto.2009.05.010
16. Adriana Z, Al Bahar S. Novel four-way Ph translocation t(9;22;7;1)(q34;q11;q22;p13) in a chronic myeloid leukemia patient receiving tyrosine kinase inhibitor therapy. Int J Hematol (2012) 95:315–9. doi:10.1007/s12185-012-1018-9
17. Borgstrom GH, Vuopio P, de la Chapelle A. Abnormalities of chromosome No. 17 in myeloproliferative disorders. Cancer Genet Cytogenet (1982) 5:123–35. doi:10.1016/0165-4608(82)90003-6
18. Carbonell F, Kratt E, Neuhaus K. Complex translocations between chromosomes #6, #9, #22, and #11 in a patient with chronic myelocytic leukemia: 46, XX, t (6; 9; 22; 11)(p21; q34; q11; q13). Cancer Genet Cytogenet (1980) 2:139–43. doi:10.1016/0165-4608(80)90057-6
19. Costa D, Carrió A, Madrigal I, Arias A, Valera A, Colomer D, et al. Studies of complex Ph translocations in cases with chronic myelogenous leukemia and one with acute lymphoblastic leukemia. Cancer Genet Cytogenet (2006) 166:89–93. doi:10.1016/j.cancergencyto.2005.08.024
20. Dubé I, Dixon J, Beckett T, Grossman A, Weinstein M, Benn P, et al. Location of breakpoints within the major breakpoint cluster region (BCR) in 33 patients with BCR rearrangement-positive chronic myeloid leukemia (CML) with complex or absent Philadelphia chromosomes. Genes Chromosomes Cancer (1989) 1:106–11. doi:10.1002/gcc.2870010116
21. El-Zimaity MM, Kantarjian H, Talpaz M, O’Brien S, Giles F, Garcia-Manero G, et al. Results of imatinib mesylate therapy in chronic myelogenous leukaemia with variant Philadelphia chromosome. Br J Haematol (2004) 125:187–95. doi:10.1111/j.1365-2141.2004.04899.x
22. Endo K, Sato A, Sugawara T, Kameoka J, Fukuhara O, Meguro K, et al. A novel translocation involving chromosomes 2, 9, 14, and 22 in chronic myeloid leukemia. Cancer Genet Cytogenet (1995) 80:155–7. doi:10.1016/0165-4608(94)00163-6
23. Fabarius A, Leitner A, Hochhaus A, Müller MC, Hanfstein B, Haferlach C, et al. Impact of additional cytogenetic aberrations at diagnosis on prognosis of CML: long-term observation of 1151 patients from the randomized CML Study IV. Blood (2011) 118:6760–8. doi:10.1182/blood-2011-08-373902
24. Groupe Francais de Cytogenetique Hematologique. Unusual Ph translocations in the French prospective study on chronic myeloid leukemia. Cancer Genet Cytogenet (1985) 16:305–9. doi:10.1016/0165-4608(85)90238-9
25. Gorusu M, Benn P, Li Z, Fang M. On the genesis and prognosis of variant translocations in chronic myeloid leukemia. Cancer Genet Cytogenet (2007) 173:97–106. doi:10.1016/j.cancergencyto.2006.10.006
26. Gödde-Salz E, Schmitz N, Bruhn H-D. Philadelphia chromosome (Ph) positive chronic myelocytic leukemia (CML): frequency of additional findings. Cancer Genet Cytogenet (1985) 14:313–22. doi:10.1016/0165-4608(85)90197-9
27. Ishihara T, Minamihisamatsu M. The Philadelphia chromosome. Considerations based on studies of variant Ph translocations. Cancer Genet Cytogenet (1988) 32:75–92. doi:10.1016/0165-4608(88)90314-7
28. Jabbour E, Kantarjian H, O’Brien S, Rios MB, Abruzzo L, Verstovsek S, et al. Sudden blastic transformation in patients with chronic myeloid leukemia treated with imatinib mesylate. Blood (2006) 107:480–2. doi:10.1182/blood-2005-05-1816
29. Kerim S, Stul M, Mecucci C, Vandenberghe E, Cuneo A, Dal Cin P, et al. Rearrangement of immunoglobulin and TCR genes in lymphoid blast crisis of Ph+ chronic myeloid leukaemia. Br J Haematol (1990) 74:414–9. doi:10.1111/j.1365-2141.1990.tb06328.x
30. Koshiyama DB, Capra ME, Paskulin GA, Rosa RF, Oliveira CA, Vanelli T, et al. Cytogenetic response to imatinib treatment in Southern Brazilian patients with chronic myelogenous leukemia and variant Philadelphia chromosome. Ann Hematol (2013) 92:185–9. doi:10.1007/s00277-012-1598-8
31. Kubota Y, Waki M. Chronic myeloid leukemia with a novel four-way t(6;13;9;22)(p21;q32;q34;q11.2) successfully treated with imatinib mesylate. Cancer Genet Cytogenet (2010) 201:135–6. doi:10.1016/j.cancergencyto.2010.05.017
32. Lafage-Pochitaloff-Huvalé M, Sainty D, Adriaanssen HJ, Lopez M, Maraninchi D, Simonetti J, et al. Translocation (3;21) in Philadelphia positive chronic myeloid leukemia: high resolution chromosomal analysis and immunological study on five new cases. Leukemia (1989) 3:554–9.
33. Legues ME, Encina A, Valenzuela M, Palma T, Undurraga MS. Cytogenetic and molecular characteristics of 25 Chilean patients with a variant Ph translocation. Cancer Genet (2011) 204:410–2. doi:10.1016/j.cancergen.2011.06.004
34. Markovic VD, Bouman D, Bayani J, Al-Maghrabi J, Kamel-Reid S, Squire JA. Lack of BCR/ABL reciprocal fusion in variant Philadelphia chromosome translocations: a use of double fusion signal FISH and spectral karyotyping. Leukemia (2000) 14:1157–60. doi:10.1038/sj.leu.2401718
35. Marzocchi G, Castagnetti F, Luatti S, Baldazzi C, Stacchini M, Gugliotta G, et al. Variant Philadelphia translocations: molecular-cytogenetic characterization and prognostic influence on frontline imatinib therapy, a GIMEMA Working Party on CML analysis. Blood (2011) 117:6793–800. doi:10.1182/blood-2011-01-328294
36. Morris CM, Fitzgerald PH. Complexity of an apparently simple variant Ph translocation in chronic myeloid leukemia. Leuk Res (1987) 11:163–9. doi:10.1016/0145-2126(87)90022-1
37. Pieńkowska-Grela B, Rygier J, Woroniecka R, Grygalewicz B, Pastwińska A, Krawczyk P, et al. Karyotype changes during long-term targeted therapy of chronic myeloid leukemia with imatinib. Leuk Lymphoma (2009) 50:952–65. doi:10.1080/10428190902838384
38. Prakash O, Yunis JJ. High resolution chromosomes of the t(9;22) positive leukemias. Cancer Genet Cytogenet (1984) 11:361–7. doi:10.1016/0165-4608(84)90015-3
39. Rajcan-Separovic E, Bence-Bruckler I, Wells P, Wang H. Fluorescence in situ hybridization analysis of complex translocations in two newly diagnosed Philadelphia chromosome-positive chronic myelogenous leukemia patients. Cancer Genet Cytogenet (1999) 114:71–4. doi:10.1016/S0165-4608(99)00047-3
40. Ramirez GM, Macera MJ, Verma RS. Two new chromosomal abnormalities in chronic myelogenous leukemia 46,XY,t(9;15;22)(q34;q22;q11) and 46,XY,t(6;9;12;22)(p21;q34;q24;q11). Cancer Genet Cytogenet (1989) 38:115–9. doi:10.1016/0165-4608(89)90171-4
41. Reddy KS, Sulcova V. A FISH study of variant Philadelphia rearrangements. Cancer Genet Cytogenet (2000) 118:121–31. doi:10.1016/S0165-4608(99)00187-9
42. Reid AG, Huntly BJ, Grace C, Green AR, Nacheva EP. Survival implications of molecular heterogeneity in variant Philadelphia-positive chronic myeloid leukaemia. Br J Haematol (2003) 121:419–27. doi:10.1046/j.1365-2141.2003.04291.x
43. Richebourg S, Eclache V, Perot C, Portnoi MF, Van den Akker J, Terré C, et al. Mechanisms of genesis of variant translocation in chronic myeloid leukemia are not correlated with ABL1 or BCR deletion status or response to imatinib therapy. Cancer Genet Cytogenet (2008) 182:95–102. doi:10.1016/j.cancergencyto.2008.01.005
44. Stagno F, Vigneri P, Del Fabro V, Stella S, Cupri A, Massimino M, et al. Influence of complex variant chromosomal translocations in chronic myeloid leukemia patients treated with tyrosine kinase inhibitors. Acta Oncol (2010) 49:506–8. doi:10.3109/02841861003660031
45. Yang CP, Wu JH, Hung IJ, Jaing TH. Cytogenetic pattern of childhood leukemia in Taiwan. J Formos Med Assoc (2000) 99:281–9.
46. Yehuda O, Abeliovich D, Ben-Neriah S, Sverdlin I, Cohen R, Varadi G, et al. Clinical implications of fluorescence in situ hybridization analysis in 13 chronic myeloid leukemia cases: Ph-negative and variant Ph-positive. Cancer Genet Cytogenet (1999) 114:100–7. doi:10.1016/S0165-4608(99)00067-9
47. Yin CC, Medeiros LJ, Glassman AB, Lin P. t(8;21)(q22;q22) in blast phase of chronic myelogenous leukemia. Am J Clin Pathol (2004) 121:836–42. doi:10.1309/H8JH6L094B9U3HGT
48. Zagaria A, Anelli L, Albano F, Vicari L, Schiavone EM, Annunziata M, et al. Molecular cytogenetic characterization of deletions on der(9) in chronic myelocytic leukemia. Cancer Genet Cytogenet (2006) 167:97–102. doi:10.1016/j.cancergencyto.2006.01.011
49. Mkrtchyan H, Ghazaryan S, Avetisyan G, Hovhannisyan A, Muradyan L, Daghbashyan S, et al. Novel complex t(V;9;22) rearrangements in three cases with chronic myeloid leukemia and a rare translocation in a case with classical Philadelphia chromosome. Oncol Rep (2008) 20:99–104. doi:10.3892/or.20.1.99
50. Cohen MS, Hussain HB, Moley JF. Inhibition of medullary thyroid carcinoma cell proliferation and RET phosphorylation by tyrosine kinase inhibitors. Surgery (2002) 132:960–6. doi:10.1067/msy.2002.128562
Keywords: Philadelphia chromosome, complex variant translocation, BCR–ABL gene, chronic myeloid leukemia
Citation: Asif M, Jamal MS, Khan AR, Naseer MI, Hussain A, Choudhry H, Malik A, Khan SA, Mahmoud MM, Ali A, Iram S, Kamran K, Iqbal A, Abduljaleel Z, Pushparaj PN and Rasool M (2016) A Novel Four-Way Complex Variant Translocation Involving Chromosome 46,XY,t(4;9;19;22)(q25:q34;p13.3;q11.2) in a Chronic Myeloid Leukemia Patient. Front. Oncol. 6:124. doi: 10.3389/fonc.2016.00124
Received: 23 March 2016; Accepted: 02 May 2016;
Published: 30 May 2016
Edited by:
Imtiaz Ahmad Siddiqui, University Of Wisconsin-Madison, USAReviewed by:
Mohammad Asim, University of Cambridge, UKCopyright: © 2016 Asif, Jamal, Khan, Naseer, Hussain, Choudhry, Malik, Khan, Mahmoud, Ali, Iram, Kamran, Iqbal, Abduljaleel, Pushparaj and Rasool. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohammad Sarwar Jamal, c2Fyd2FyNHVAZ21haWwuY29t;
Mahmood Rasool, bWFobW9vZHJhc29vbEB5YWhvby5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.